Abstract
Proton (H+) and aluminum (Al) rhizotoxicity are two major factors limiting crop production in acid soils. Orthologs of the zinc-finger transcription factor, Sensitive To Proton Rhizotoxicity1 (STOP1), have been found to play an essential role in the tolerance to both stresses by regulating the transcription of multiple H+ and Al tolerant genes. In the present study, color three GmSTOP1 homologs were identified in the soybean genome. All three GmSTOP1 exhibited similar properties as reflected by the harboring of four potential zinc finger domains, localizing in the nucleus, and having transactivation activity. Expression profiling showed that H+ stress slightly modulated transcription of all three GmSTOP1s, while Al significantly up-regulated GmSTOP1-1 and GmSTOP1-3 in root apexes and GmSTOP1-3 in basal root regions. Furthermore, complementation assays in an Arabidopsis Atstop1 mutant line overexpressing these GmSTOP1s demonstrated that all three GmSTOP1s largely reverse the H+ sensitivity of the Atstop1 mutant and restore the expression of genes involved in H+ tolerance. In contrast, only GmSTOP1-1 and GmSTOP1-3 could partially recover Al tolerance in the Atstop1 mutant. These results suggest that the function of three GmSTOP1s is evolutionarily conserved in H+ tolerance, but not in Al tolerance.
Keywords: GmSTOP1, proton rhizotoxicity, Al rhizotoxicity, soybean, transcription factor
Introduction
Agricultural production is limited on acid soils, which comprise approximately 50% of the world’s potentially arable lands (von Uexküll and Mutert, 1995). There are several constraints limiting plant growth on acid soils, including deficiency of mineral nutrients, such as phosphorus (P), calcium (Ca), and magnesium (Mg), as well as, toxicity of excessive ions, including aluminum (Al3+), hydrogen (H+), and manganese (Mn2+) (Ishitani et al., 2004).
Among these stresses, Al toxicity has been widely acknowledged as a major constraint on crop production (Kochian et al., 2004; Ma, 2007; Bojórquez-Quintal et al., 2017). The Al3+ ion can cause rapid and severe impairment of root apical development by damaging cell walls (Horst et al., 1999) and cytoskeletons (Chang et al., 1999; Sivaguru et al., 1999), disturbing DNA and plasma membrane processes (Elstner et al., 1988; Yamaguchi et al., 1999; Meriga et al., 2004), blocking production of callose (Sivaguru et al., 2000), and impeding stress-signaling pathways (Ramos-Díaz et al., 2007). Consequently, plant root growth and nutrient acquisition are inhibited, which leads to significant reductions in crop yields (Ryan et al., 2001; Kochian et al., 2004; Ma, 2007; Bojórquez-Quintal et al., 2017).
Often combined with Al toxicity, H+ rhizotoxicity has also been recognized as a major limiting factor for crop production on acid soils (Kochian et al., 2004). When exposed to strong acid conditions, plant root cells will be structurally and functionally damaged (Foy, 1984). For example, obviously swollen root hairs and cracks between cells in root meristems have been observed in Arabidopsis (Koyama et al., 1995, 2001; Iyer-Pascuzzi et al., 2011) and yorkshire-fog grass (Holcus lanatus) subjected to acid treatments (Kidd and Proctor, 2001). Moreover, a pH drop from 5.5 to 4.0 is associated with significant membrane depolarization, destruction of epidermal and cortical cells, and, ultimately, inhibition of root growth in Lotus corniculatus (Pavlovkin et al., 2009; Palóve-Balang et al., 2012). Similar symptom caused by proton rhizotoxicity have also been observed in many other plant species, such as alfalfa (Medicago sativa) (Yokota and Ojima, 1995), spinach (Spinacia oleracea) (Yang et al., 2005), common bean (Phaseolus vulgaris) (Rangel et al., 2005), and barley (Hordeum vulgare) (Song et al., 2011). Besides direct toxicity, low pH can also increase the solubility of other toxic ions, such as Al3+, in soil, and thus adversely influence plant root growth. In this aspect, Al and H+ toxicities are physiologically linked to one another.
Over the past few decades, mechanisms of plant tolerance to Al and H+ rhizotoxicities have been elucidated in many studies. Among them, identification of the C2H2-type zinc finger transcription factor family, STOP1 (Sensitive to Proton Rhizotoxicity1), contributed considerably to understanding of regulatory mechanisms underlying the integration of Al and H+ tolerance in planta (Iuchi et al., 2007; Yamaji et al., 2009; Ohyama et al., 2013; Sawaki et al., 2014; Fan et al., 2015).
The first STOP1 gene, AtSTOP1, was identified in Arabidopsis (Iuchi et al., 2007). Transcriptome analyses and genetic characterization showed that AtSTOP1 regulates the expression of a set of genes, including three major Al tolerance genes, AtALMT1 (Aluminum activated Malate Transporter1), AtMATE (Multidrug and Toxic Compound Extrusion), and AtALS3 (Aluminum Sensitive3), along with other genes apparently involved in the regulation of cytosolic pH, such as GAD1 (Glutamate Decarboxylase1), ME1/2 (Malic Enzyme1/2), and GDH1/2 (Glutamate Dehydrogenase 1/2) (Liu et al., 2009; Sawaki et al., 2009; Kobayashi et al., 2014). Interestingly, the STOP1 homolog AtSTOP2, which partially accounts for tolerance to Al and H+ rhizotoxicities, is also regulated by AtSTOP1 (Kobayashi et al., 2014). Moreover, a STOP1 ortholog in rice bean (Vigna umbellata), VuSTOP1, was isolated by suppression subtractive hybridization (Fan et al., 2014). In contrast to the constitutive expression exhibited by AtSTOP1, the expression of VuSTOP1 was inducible by both of Al and H+ stresses (Fan et al., 2014). However, the assay of planta complementation in Atstop1 mutant showed that VuSTOP1 could fully restore the transcription of several H+-tolerance related genes, but only partially restores the expression of AtMATE and ALS3, indicating that VuSTOP1 might play a major role in H+ tolerance, but only a minor role in Al tolerance (Fan et al., 2015). Similarly, other STOP1 homologs, including NtSTOP1 in tobacco (Nicotiana tabacum), LjSTOP1 in Lotus japonicas, PnSTOP1 in black poplar (Populus nigra), CsSTOP1 in tea (Camellia sinensis) and EguSTOP1 in Eucalyptus also reportedly possess similar functions in H+ tolerance, and only partial or even no functionality in Al tolerance (Ohyama et al., 2013; Sawaki et al., 2014). On the other hand, the mutation of ART1 (Al Resistance Transcription Factor1), a STOP1 homolog in rice (Oryza sativa), appears to only affect Al hypersensitivity (Yamaji et al., 2009). In short, previous studies suggest that STOP1 transcription factors are ubiquitous in plants and have conserved functions in plant stress (Al and/or H+) tolerance, though specific responses vary among plant species.
Soybean (Glycine max) is one of the most important leguminous crops globally, comprising approximately 68% of crop legume production in the world and 57% of the global oilseed production (Herridge et al., 2008). Though many studies have elucidated the functions of STOP1 orthologs in other plant species, no information is available on whether GmSTOP family members are also involved in H+ and Al tolerance in soybean. In the present study, three GmSTOP1 homologs were isolated and characterized from soybean. The function of each GmSTOP1 gene was analyzed in terms of Al and H+ tolerance in Arabidopsis. The results demonstrate that all three GmSTOP1s play important roles in H+ tolerance, while only GmSTOP1-1 and GmSTOP1-3 could partially recover Al tolerance in Arabidopsis Atstop1 mutant. Taken together, these results strongly suggest that the three GmSTOP1s in soybean share evolutionary conservation of H+ tolerance, but not of Al tolerance.
Materials and Methods
Plant Material and Growth Conditions
The soybean genotype YC03-3 was chosen as the plant material in this study. Soybean seeds were germinated in paper rolls moistened with modified one-half-strength nutrient solution as previously described (Liang et al., 2013). The resultant seedlings were then gown in full strength nutrient solution for 24 h before being used for various treatments. For the low pH treatment, soybean seedlings were subjected to 0.5 mM CaCl2 (pH 4.2) for 0, 2, 4, 6, and 12 h. After low pH treatment, root tips (0–2 cm) were harvested for gene expression assays. For the tissue specific expression experiment, soybean root tips (0–2 cm), which was further divided into two segments (0–1 cm and 1–2 cm), basal roots (>2 cm) and leaves were harvested after 4 h of Al (0 or 50 μM AlCl3 in 0.5 mM CaCl2, pH 4.2) treatment. For the Al dose experiment, soybean seedlings were treated with 0, 10, 50, and 100 μM AlCl3 in 0.5 mM CaCl2 solution (pH 4.2) for 4 h. For the time-course experiment, soybean seedlings were transplanted to Al (50 μM AlCl3 in 0.5 mM CaCl2, pH 4.2) treatments for 0, 2, 4, 6, and 12 h. In both of the concentration response experiment and time-course experiment, root tips (0–2 cm) were separately harvested for gene expression assays. All experiments had four biological replicates.
Phylogenetic Analysis and Characterization of GmSTOP1 Proteins in Soybean
TBLASTN analysis using the AtSTOP1 proteins sequence (accession number: Q9C8N5.1) as the query sequences was conducted at the Phytozome website1. Consequently, three STOP1 homologs with high similarity to AtSTOP1 were identified and designated as GmSTOP1-1 (Glyma10g35940), GmSTOP1-2 (Glyma16g27280), and GmSTOP1-3 (Glyma20g31650). Subsequently, multiple sequence alignment and phylogenetic tree construction were conducted using the deduced protein sequences of all three GmSTOP1s together with other STOP1 homologs, including AtSTOP1 from Arabidopsis, NtSTOP1 from tobacco, LjSTOP1 from Lotus japonicas, PnSTOP1 from black poplar (Populus nigra), CsSTOP1 from tea, EguSTOP1 from Eucalyptus, OsART1 from rice, PpSTOP1 from Physcomitrella patens, and TaSTOP1-A, TaSTOP1-B, TaSTOP1-D from wheat (Triticum aestivum). ClustalX2 and MEGA4.1 were used for the multiple sequence alignment and phylogenetic tree construction, respectively. The phylogenetic tree was constructed using the Neighbor-Joining method with 1,000 bootstrap replicates.
RNA Extraction and Quantitative Real-Time PCR
Total RNA was extracted from plant tissues using RNA-solve reagent (OMEGA Bio-Tek, Norcross, GA, United States). Genomic DNA in the RNA samples was eliminated with RNase-free DNase I (Invitrogen, Carlsbad, CA, United States). The resulting extracts were then used to conduct the reverse transcription via MMLV-reverse transcriptase (Promega, Madison, WI, United States) following the manufacturer’s instructions. Subsequently, SYBR Green monitored qRT-PCR (quantitative real-time PCR) analysis was performed using a ABI Step-one Plus real-time PCR system (Thermo Fisher Scientific, Waltham, MA, United States). The primer pairs used for expression analysis are listed in Supplementary Table S1.
Subcellular Localization of GmSTOP1s
The full length cDNAs of the three identified GmSTOP1 genes were amplified from first strand cDNA derived from soybean roots using gene specific primer pairs as listed in Supplementary Table S1. The PCR products were then cloned into the pMD18-T vector (Takara, Japan) for sequence confirmation.
Full-length cDNA of the three GmSTOP1s was fused with enhanced green fluorescent protein (GFP) to construct 35S::GmSTOP1s-GFP plasmids. Each construct was introduced into tobacco (Nicotiana tabacum) leaf cells according to previously described methods (Liang et al., 2010; Liu et al., 2016). The 35S::GFP construct was used as the control. Fluorescence signals of GFP were detected at 488 nm by confocal scanning microscope (LSM780; Zeiss, Germany).
Transcriptional Activation Activity of GmSTOP1s
Gene specific primers with terminal SfiI and SalI restriction sites were used to amplify full-length cDNA of GmSTOP1-1 and GmSTOP1-3, whereas, primers with BamHI and PstI restriction sites were used to amplify full-length cDNA of GmSTOP1-2. Sequence fragments were digested by the corresponding restriction enzymes and inserted into the pGBKT7 vector (Clontech, Japan), producing pGBKT7-GmSTOP1s plasmids. The resultant plasmids and the pGBKT7 empty vector were then transformed into yeast strain AH109. After verification by PCR, transformed AH109 cells were cultured on either SD-Trp or SD-His medium for 3 days. The yeast cells grown on SD-Trp were then printed onto filter paper moistened with X-gal solution. Subsequently, the filter paper was freeze-thawed with liquid nitrogen and moistened again with X-gal solution. The appearance of blue areas on the filter paper was used to determine β-galactosidase activity.
Complementation of GmSTOP1s in Atstop1 Mutant Plants
The three identified GmSTOP1s were separately introduced into the modified pBEGFP binary vector under the control of a 35S CaMV promoter to produce an over-expression construct that was then transformed into A. tumefaciens strain Gv3101. Subsequently, the constructs were transformed into Arabidopsis Atstop1 mutant plants via the floral dip method (Clough and Bent, 1998). Two independent over-expression lines for each gene were verified by qRT-PCR and used for further analysis as complemented lines.
To investigate the functions of GmSTOP1s in resistance to H+ and Al toxicity, wild type, Atstop1 mutant and the complemented lines overexpressing GmSTOP1 were germinated on solid Murashige and Skoog (MS) medium for 5 days. Uniform seedlings with ∼1.5 cm root lengths were transferred to modified 1/30 strength Hoagland nutrient solution (without NH4H2PO4 and plus 1 mM CaCl2) with different treatments as described (Fan et al., 2015). Control plants were grown in media with pH adjusted to 5.8, while treated plants were grown in low pH media (pH 4.7) or media containing 2 μM AlCl3 (pH 5.0) for 7 days (Fan et al., 2015). Upon harvest, roots of each plant were scanned and analyzed in Image J (National Institutes of Health, United States). All experiments had four biological replicates, each of which contains two plants.
For analysis of H+ and Al genes expression responses, uniform Arabidopsis seedlings were treated with 1/30 strength Hoagland nutrient solution containing 2 μM AlCl3 (pH 5.0) and low pH (pH 4.7) for 24 h (Fan et al., 2015). All experiments were conducted in a growth incubator running a 24°C, 12h/22°C, 12 h day/night cycle. The whole roots were harvested for gene expression assays. The primer pairs of target genes for qRT-PCR analysis are listed in Supplementary Table S1. Arabidopsis UBQ1 was used as housekeeping gene control to normalize the expression of the corresponding genes. All experiments had four biological replicates.
Statistical Analysis
All data were analyzed by Student’s t-tests using SPSS 13.0 (SPSS Institute, Chicago, IL, United States).
Results
Identification of STOP1 Homologs in Soybean
A homolog search resulted in retrieval of three STOP1 homologs in the soybean genome, which were named GmSTOP1-1 (Glyma.10G215200), GmSTOP1-2 (Glyma.16G156400) and GmSTOP1-3 (Glyma.20G176500) based on genome localization. A phylogenetic tree showed that the STOP1 homologs in dicots were differentiated from those in monocots (Figure 1A). Moreover, GmSTOP1-1 and GmSTOP1-3 present as duplicated pair and display high similarity with VuSTOP from rice bean, while GmSTOP1-2 clusters in another sub-clade with LjSTOP1 from Lotus japonicus (Figure 1A). Moreover, the deduced amino acid sequences of all three GmSTOP1s contain four putative C2H2 zinc finger domains that are highly conserved in STOP1 orthologs from other plant species (Figure 1B).
FIGURE 1.
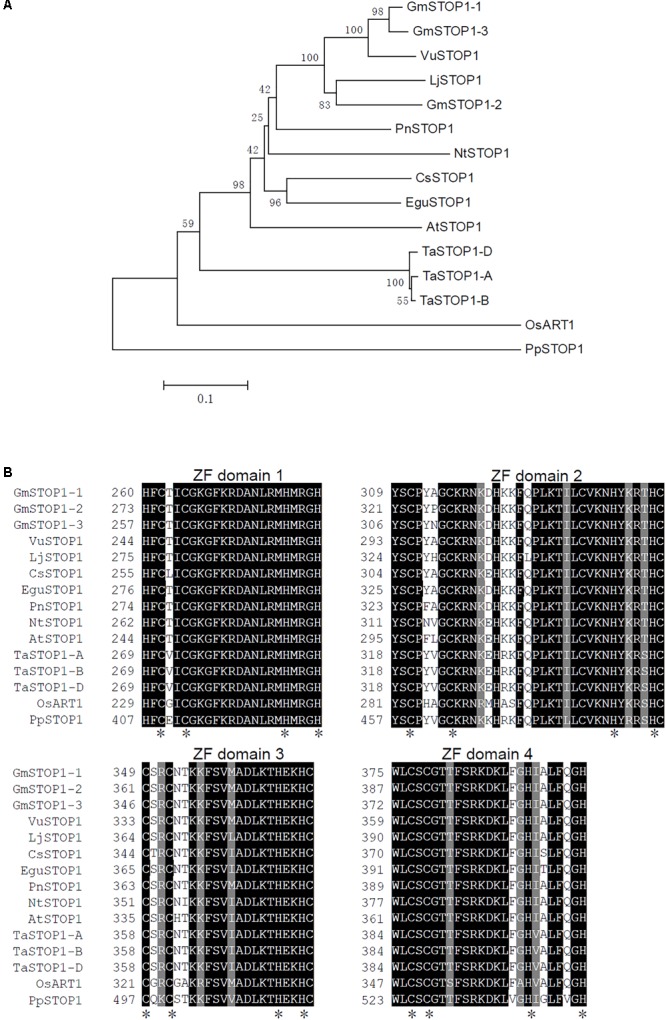
Phylogenetic tree and amino acid alignments of predicted C2H2 zinc finger domains in plant STOP1s. (A) Phylogenetic tree was generated based on an amino-acid alignment with STOP1 orthologs from several plant species. (B) Alignment of the amino acid sequences of predicted C2H2 zinc finger domains in STOP1 proteins. Black background indicates identical residues. Asterisks indicate conserved Cys and His residues of C2H2 motifs. The plant STOP1 proteins aligned include representatives from Glycine max (GmSTOP1-1, XP_006588359.1; GmSTOP1-2, XP_006598713.1; GmSTOP1-3, XP_014628358.1), Arabidopsis thaliana (AtSTOP1, NP_174697.1), Nicotiana tabacum (NtSTOP1, AB811781), Lotus japonicus (LjSTOP1, BAN67817.1), Vigna umbellata (VuSTOP1, KP637172), Camellia sinensis (CsSTOP1, BAN67815.1), Populus nigra (PnSTOP1, BAN67813.1), Eucalyptus (EguSTOP1, BAO56822.1), Triticum aestivum (TaSTOP1-A, AGS15201.1; TaSTOP1-B, AGS15202.1; TaSTOP1-D, AGS15195.1), Oryza sativa (OsART1, AB379846), and Physcomitrella patens (PpSTOP1, BAN67814.1).
Subcellular Localization and Transcription Activation Activity of GmSTOP1s
To determine the subcellular localization of the three identified GmSTOP1s, GmSTOP1-GFP fusion constructs were assayed for transient expression in tobacco leaf cells. The results showed that control GFP fluorescence was detectable in both the nucleus and cytoplasm. In contrast, fluorescence derived from GmSTOP1-GFP constructs was exclusively localized within the nucleus (Figure 2A), strongly suggesting that the three GmSTOP1 members are all nucleus localized proteins.
FIGURE 2.
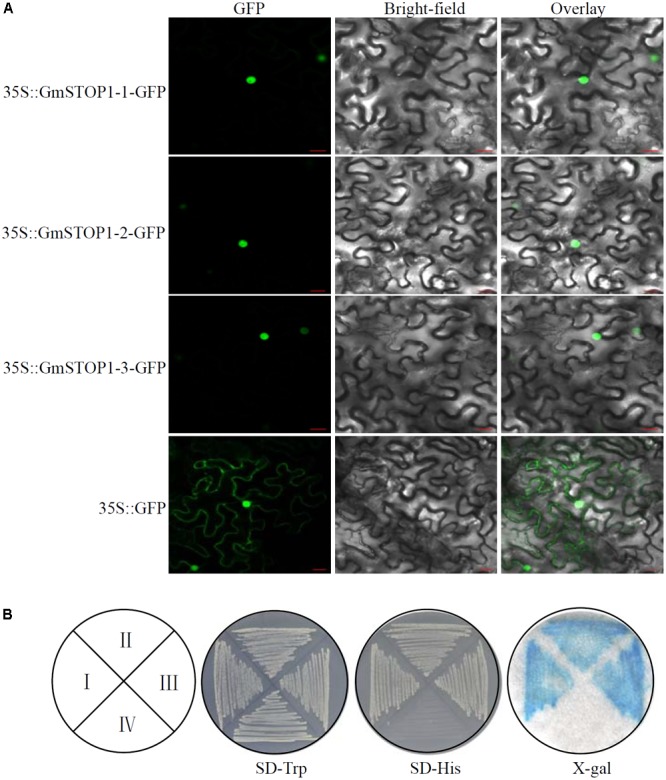
Subcellular localization and transactivation assay of GmSTOP1s. (A) 35S::GmSTOP1s-GFP and 35S::GFP (control) constructs were introduced individually into Nicotiana benthamiana leaves. Scale bars: 20 μm. (B) Transactivation assay of GmSTOP1-1 (I), GmSTOP1-2 (II), GmSTOP1-3 (III), and control (IV). β-galactosidase activity is indicated by blue color on the filter paper using X-gal as the substrate.
Transcription activity of the three GmSTOP1 members was determined in a one-hybridization expression system in yeast. The results showed that the yeast strain AH109 transformed with either a pGBKT7-GmSTOP1 or the pGBKT7 empty vector could grow well on the SD-Trp medium (Figure 2B). However, only the three AH109 strains transformed with pGBKT7-GmSTOP1s grew well on the SD-His medium (Figure 2B). Furthermore, all three of the AH109 strains transformed with a pGBKT7-GmSTOP1 showed high β-galactosidase activity as indicated by the blue color on filter paper using X-gal as a substrate (Figure 2B). Therefore, the ability of all three GmSTOP1 homologs to activate lacZ expression strongly suggests that each one functions as a transcription factor.
Expression Patterns of GmSTOP1s in Response to Al and Low pH Stress
Quantitative real-time PCR was used to analyze GmSTOP1 expression patterns in soybean seedlings. Expression levels of the three GmSTOP1s were hardly affected by low pH stress during the 12 h treatment period (Supplementary Figure S2). On the other hand, transcriptional responses varied among the three GmSTOP1 genes in response to Al stress. As shown in Figure 3A, transcription of GmSTOP1-2 was not significantly affected after 4 h of Al treatment in any tissues, including root tips (0–2 cm), basal regions of roots (>2 cm) and leaves, (Figure 3A). In contrast to the constitutive expression of GmSTOP1-2 in roots, transcript levels of GmSTOP1-1 and GmSTOP1-3 increased by more than 6- and 11-fold, respectively, in root tips after 4 h of Al treatment (Figure 3A). However, in root basal regions, only transcription of GmSTOP1-3 increased by more than 1.7-fold in response to Al stress, while no detectable change was observed for GmSTOP1-1.
FIGURE 3.
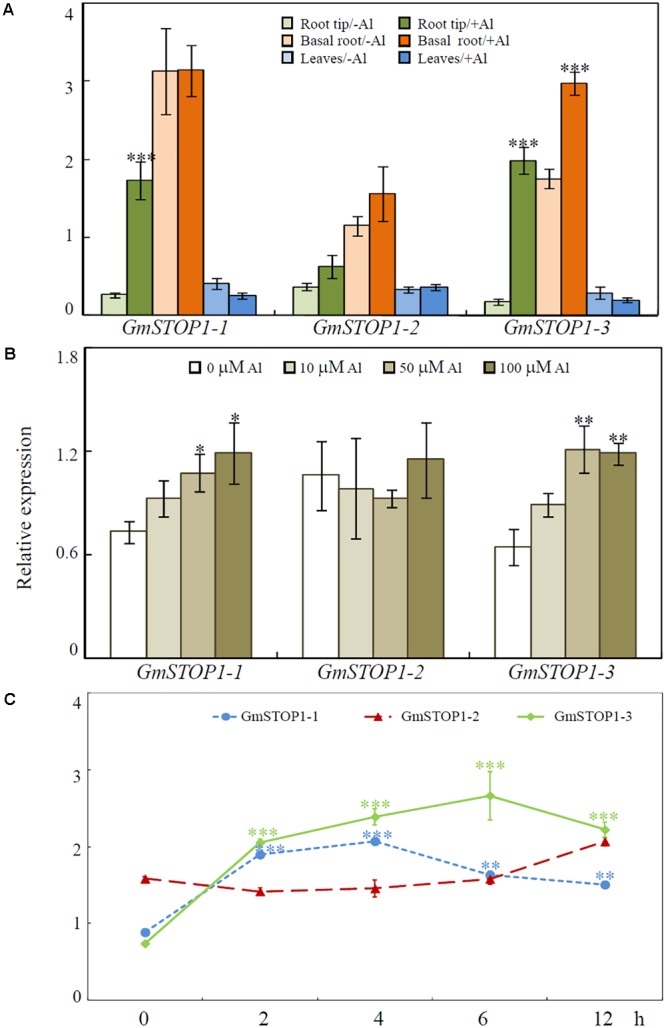
Expression patterns of GmSTOP1s in response to Al toxicity. (A) Relative expression of GmSTOP1s in root tips (0–2 cm), basal roots (>2 cm) and leaves after 4 h of –Al (0 μM) or +Al (50 μM) treatment. (B) Relative expression of GmSTOP1s in soybean root tips (0–2 cm) treated with different concentrations of Al for 4 h. (C) Relative expression of GmSTOP1s in soybean roots tips (0–2 cm) in response to Al (50 μM) for different treatment times. Asterisks indicate significant differences between the +Al treatment and –Al control (∗0.01 < P < 0.05; ∗∗0.001 < P < 0.01; ∗∗∗P < 0.001).
Dose-responses of GmSTOP1s to Al stress were further analyzed in soybean root tips after 4 h of Al treatment. Transcript accumulations of both GmSTOP1-1 and GmSTOP1-3 were strictly dependent on Al concentration in the medium (Figure 3B), with transcript abundances enhanced for both GmSTOP1-1 and GmSTOP1-3 in 50 and 100 μM Al treatments (Figure 3B). The expression of GmSTOP1-2 was constitutively expressed at relatively high levels regardless the external Al concentrations (Figure 3B).
Results from time-course experiments showed that the expression of both GmSTOP1-1 and GmSTOP1-3 were quickly enhanced in response to Al stress by more than twofold after 2 h of Al treatment, and remained high over 12 h (Figure 3C). Meanwhile, the expression of GmSTOP1-2 did not vary during the period of Al treatment (Figure 3C).
Functional Analysis of GmSTOP1s in the Arabidopsis Atstop1 Mutant
In order to examine their functions in plant H+ and Al tolerance, all three GmSTOP1s were overexpressed in the Arabidopsis Atstop1 mutant. The expression of all three of the GmSTOP1 genes in the Atstop1 mutant was verified by qRT-PCR. Under normal growth conditions, no significant differences were observed among wild type, Atstop1 mutant and complemented lines overexpressing any of the GmSTOP1s (Figures 4A,B). However, under low pH condition (pH 4.7), root elongation of wild type and Atstop1 mutant was inhibited by 51% and 80%, respectively (Figures 4A,C). In each of two complemented lines of GmSTOP1-1 (#5 and #6), GmSTOP1-2 (#12 and #15) and GmSTOP1-3 (#47 and #54), root elongation was inhibited much less than that of the Atstop1 mutant (Figures 4A,C). These results suggest that all three GmSTOP1s are able to confer H+ tolerance in Atstop1 mutant plants.
FIGURE 4.
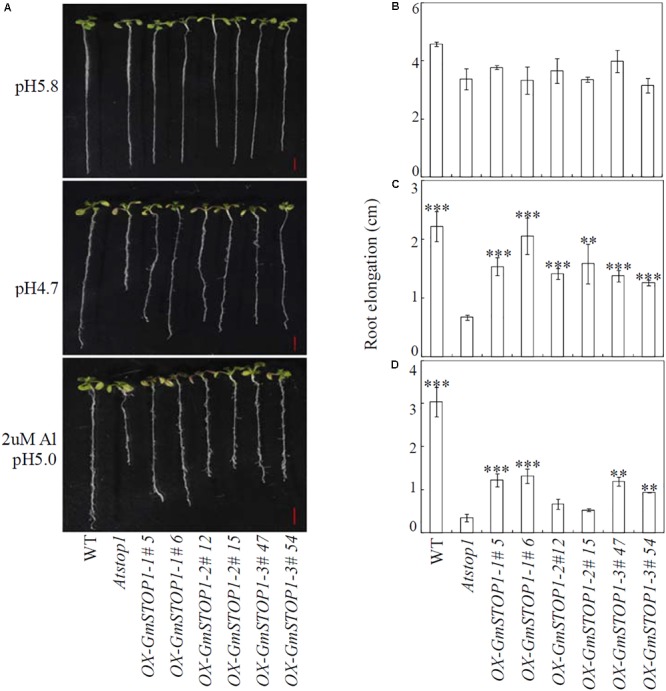
Effects of low pH and Al stresses on the growth of Arabidopsis Atstop1 mutant plants overexpressing GmSTOP1s. Uniform seedlings with ∼1.5 cm root lengths were treated with low pH (pH 4.7) and Al stresses (2 μM Al and pH 5.0) for 7 days. The phenotypes of wild-type (WT), Atstop1 and GmSTOP1 overexpressing Atstop1 lines in response to H+ and Al were photographed (A). Root elongation for each line was measured under control (B), low pH (C), and Al (D) treatments. Each bar represents the mean of four biological replicates with standard error. Asterisks indicate significant differences in comparison to the Atstop1 mutant (∗0.01 < P < 0.05; ∗∗0.001 < P < 0.01; ∗∗∗P < 0.001). Scale bars: 5 mm.
Addition of Al to the low pH culture solution slightly decreased root elongation of wild type plants, but significantly inhibited root elongation of Atstop1 mutants as indicated by a 90% decrease in root elongation compared to root elongation in wild type plants (Figures 4A,D). Unlike the role of GmSTOP1s in H+ tolerance, the functions of GmSTOP1s in Al tolerance varied. Each of the lines complemented with GmSTOP1-1 (#5 and #6) and GmSTOP1-3 (#47 and #54) overexpression recovered elongation to 28% and 32%, and 29% and 19% of that of wild type, respectively (Figures 4A,D). In contrast, lines complemented with GmSTOP1-2 overexpression did not recover root elongation. These results indicate that GmSTOP1-1 and GmSTOP1-3, but not GmSTOP1-2 can partially reverse the Al hypersensitivity of Atstop1 mutant plants.
Transcription of STOP1 Down-Stream Genes in GmSTOP1 Complemented Atstop1 Mutants
The differential contributions of GmSTOP1s to H+ and Al tolerance were further determined by investigating the transcript levels of several related down-stream genes in GmSTOP1 complemented Atstop1 mutants. The results showed that expression of several H+ tolerance genes were significantly restored in all of the complemented Atstop1 mutant lines (Figure 5). Among responsive genes, the expression of GDH2 (At5g07440) was restored the most, with transcription returning to at least 50% of transcript levels in WT plants (Figure 5). Although restored to lesser extents, the expression of three other H+ tolerance genes, GDH1 (At5g18170), GABA-T (At3g22200) and NADP-malate enzyme 2 (NADP-ME2), was restored nonetheless by 37, 25, and 20%, respectively, over expression in the Atstop1 mutant (Figure 5).
FIGURE 5.
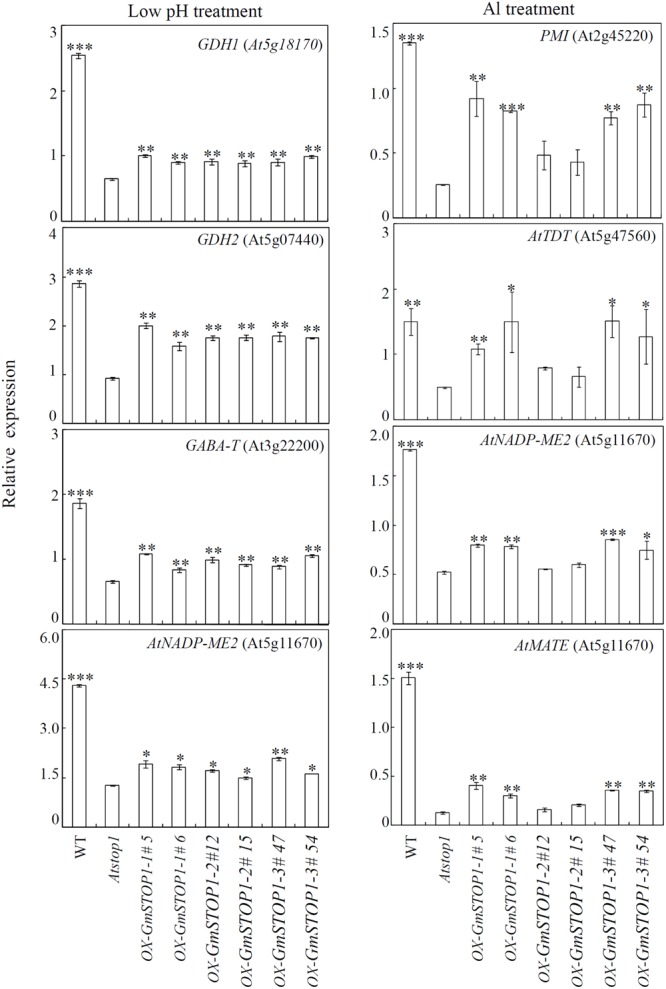
Transcriptional accumulation of genes regulated by GmSTOP1s under low pH and Al stresses. Wild-type (WT), Atstop1 mutant and complemented lines overexpressing GmSTOP1-1 (#5 and #6), GmSTOP1-2 (#12 and #15), and GmSTOP1-3 (#47 and #54) were exposed to low pH (pH 4.7) and Al treatments (AlCl3: 2 μM; pH 5.0) for 24 h. Transcript abundances of GDH1, GDH2, GABA-T and AtNADP-ME2 were quantified from plants grown in the low pH treatment, while expression levels of PMI (At2g45220), AtTDT (At5g47560), AtNADP-ME2 (At5g11670), and AtMATE (At1g51340) were quantified in Al treated samples. UBQ1 transcript levels were used as the internal standard. Data are expressed as means of four replicates. Asterisks indicate significant differences in comparison to the Atstop1 mutant (∗0.01 < P < 0.05; ∗∗0.001 < P < 0.01; ∗∗∗P < 0.001).
Unlike expression patterns of H+ tolerance related genes, expression responses of Al tolerance related genes varied among GmSTOP1 complemented lines. These Al tolerance related genes included pectin methylesterase inhibitor superfamily protein (PMI, At2g45220), AtTDT (At5g47560), NADP-ME2 (At5g11670), and AtMATE (At1g51340). In both GmSTOP1-1 and GmSTOP1-3 complemented lines, the expression of AtPMI, AtTDT, NADP-ME2, and AtMATE were partially restored. Yet, in GmSTOP1-2 complemented lines, the expression of each of these four genes was not affected relative to expression in Atstop1 mutants (Figure 5).
Discussion
Proton and Al rhizotoxicities are two of the major constraints of plant growth and development on acid soil (Kochian et al., 2004). Plants have adapted to these stresses by developing a variety of coping strategies involving a number of genes (Liu et al., 2014; Kochian et al., 2015). Recent studies have revealed that H+ and Al tolerance mechanisms are regulated by STOP1 transcription factors in many plant species (Iuchi et al., 2007; Sawaki et al., 2009, 2014; Yamaji et al., 2009; Garcia-Oliveira et al., 2013; Ohyama et al., 2013; Fan et al., 2015). However, few studies have attempted to systematically dissect the possible roles of all STOP1 members in a single species responding to H+ stress, Al toxicity, or both.
In the present study, a total of three GmSTOP1 genes were identified in the soybean genome. All of these GmSTOP1 homologs are localized to the nucleus, and exhibit transcription activity (Figure 2). Sequence analysis revealed that GmSTOP1-2 is phylogenetically distinct from GmSTOP1-1 and GmSTOP1-3, which appear to be a duplicated pair (Figure 1). As gene duplication provides opportunities for functional divergence (Force et al., 1999; Lynch et al., 2001; Flagel and Wendel, 2009; Libault et al., 2010; Schmutz et al., 2010), we hypothesize that the three GmSTOP1 genes might have divergent functions in regard to H+ and Al tolerance even though they are highly conserved in some features (Figures 1, 2).
To dissect the differential contributions of GmSTOP1s in H+ tolerance, expression analysis was conducted. The results showed that similar to AtSTOP1 in Arabidopsis (Iuchi et al., 2007), all three GmSTOP1 genes were constitutively expressed and hardly affected by H+ treatment (Supplementary Figure S2). Further complementation assays showed that all three GmSTOP1 genes are able to confer H+ tolerance to the Arabidopsis Atstop1 mutant (Figure 4C). These results are consistent with the previous studies reporting that STOP1 orthologs in dicots are able to confer H+ tolerance to the H+ sensitive Atstop1 mutant (Ohyama et al., 2013; Sawaki et al., 2014; Fan et al., 2015). Furthermore, the H+ hypersensitivity of the Atstop1 mutant is the result of down-regulation of genes in several pH regulation pathways caused by the dysfunction of AtSTOP1 (Sawaki et al., 2009).
The expression of several H+ tolerance genes, including STOP2, CIPK23, and PGIP1, has been restored in planta in complementation assays of Atstop1 by STOP1 orthologs from rice bean (VuSTOP1), Eucalyptus (EguSTOP1), tobacco (NtSTOP1), black poplar (PnSTOP1), tea (CsSTOP1), Lotus japonicus (LjSTOP1), or Physcomitrella patens (PpSTOP1) (Ohyama et al., 2013; Sawaki et al., 2014; Fan et al., 2015). However, none of these genes were affected by complementation with any of the GmSTOP1 homologs in the Atstop1 mutant (Supplementary Figure S4). Instead, GmSTOP1 complementation restored the transcription of several other H+ tolerance relative genes, including GDH1, GDH2, GABA-T, and AtNADP-ME2, which are considered to play roles in maintaining pH homeostasis in plants. For example, AtNADP-ME2 has been reported to function in the pH stat pathway through consumption of cytosolic H+ (Roberts et al., 1992; Sakano, 1998). Meanwhile, GDH1, GDH2 and GABA-T are the major isoforms in the “GABA shunt” pathway, which contributes largely to cytosolic pH homeostasis in plant cells (Crawford et al., 1994; Bown and Shelp, 1997). These results strongly suggest that all three GmSTOP1 homologs participate in conserved functions in H+ tolerance mainly through regulation of similar pH stat pathways that are distinct from the pH stat pathways regulated by other plant STOP1 orthologs (Ohyama et al., 2013; Sawaki et al., 2014; Fan et al., 2015).
Interestingly, in the presence of Al, expression of both GmSTOP1-1 and GmSTOP1-3 quickly escalated in root tips, while expression of GmSTOP1-2 was not significantly affected (Figure 3 and Supplementary Figure S3). Similar results have also been reported for bread wheat, in which expression of TaSTOP1-A was found to be responsive to H+ and Al stresses and divergent from the responses of TaSTOP1-B and TaSTOP1-D (Garcia-Oliveira et al., 2013). It has been suggested that this divergence might be mainly due to the presence of a pyrimidine-rich stretch and the absence of a light responsive element in the 5′ UTR of TaSTOP1-A compared to its homologs TaSTOP1-B and TaSTOP1-D (Garcia-Oliveira et al., 2013). Consistent with this, our investigation revealed that the 5′-UTR is more similar between GmSTOP1-1 and GmSTOP1-3 than it is between either of these genes and GmSTOP1-2 (Supplementary Figure S1). Therefore, there is a possibility that the differential expression between GmSTOP1-2 and the other two GmSTOP1s in response to Al stress might be due to divergence in the 5′-UTR. Thereby, divergence between GmSTOP1-2 and the other two GmSTOP1s in both transcriptional regulation and protein sequence further suggests that GmSTOP1-2 functions differently than GmSTOP1-1 and GmSTOP1-3 in Al tolerance responses.
Even more evidence in support of divergence among GmSTOP1s in Al tolerance functionality was gathered in complementation assays (Figure 4). The results strongly indicate that GmSTOP1-1 and GmSTOP1-3 are involved at least partially in AtSTOP1 related Al tolerance responses, whereas GmSTOP1-2 is not. It has been reported that AtSTOP1 regulates transcription of three major Al tolerance genes in Arabidopsis, namely AtALMT1, ALS3, and AtMATE (Kobayashi et al., 2007; Liu et al., 2009; Sawaki et al., 2009; Tokizawa et al., 2015). Among them, AtALMT1 accounts for more than 70% of the Al tolerance phenotype in Arabidopsis (Iuchi et al., 2007; Liu et al., 2012). In the current study, it was interesting to find that none of the GmSTOP1 homologs restores the expression of AtALMT1 or ALS3 in the Atstop1 mutant, while AtMATE expression was recovered slightly in both GmSTOP1-1 and GmSTOP1-3 complemented lines, but not in GmSTOP1-2 complemented lines (Figure 5 and Supplementary Figure S4). Similar results were also reported in other plant species, where most STOP1 orthologs are not able to restore the expression of all three Al tolerance genes in the Atstop1 mutant. For example, LjSTOP1, CsSTOP1, and PnSTOP1 can slightly restore the expression of AtALMT1, but not the expression of ALS3 or AtMATE, while VuSTOP1 can partially restore the expression of ALS3 and AtMATE, but not the expression of AtALMT1 (Ohyama et al., 2013; Fan et al., 2015). Placing the current results in the context of previous reports suggests that the regulatory functions of AtSTOP1 in Al tolerance is not entirely conserved among plant STOP1 orthologs.
Potential phenotypic effects of GmSTOP1-1/GmSTOP1-3 in the Atstop1 mutant in response to Al toxicity are revealed by considering functions of PMI, AtTDT, and NADP-ME2, which are down-regulated in Atstop1 mutants subjected to Al stress (Sawaki et al., 2009), and which had expression restored in the complementation experiments herein. Members of the PMI family have been reported to inhibit pectin methylesterase activity, and thereby increasing Al tolerance (Sénéchal et al., 2015; Geng et al., 2017). The other two genes, AtTDT and NADP-ME2, are involved in malate homeostasis and metabolism in the vacuole and cytosol, respectively (Hurth et al., 2005; Badia et al., 2015). Results in the present study showed that all of these genes were partially restored in both GmSTOP1-1 and GmSTOP1-3 complemented lines, but not in GmSTOP1-2 complemented lines (Figure 5). These expression responses are in accord with the variation in Al tolerance observed among GmSTOP1s complemented lines (Figure 4D). Therefore, it appears that GmSTOP1-1/GmSTOP1-3 might function in Al tolerance through the regulation of cell wall modifications and malate metabolism.
Overall, the present study identifies three GmSTOP1 homologs in the soybean genome, all of which localize in the nucleus and have the transactivation potential. Complementation assays suggest that all three GmSTOP1 homologs play major roles in H+ tolerance through transcriptional regulation of H+ tolerance genes, whereas, only GmSTOP1-1 and GmSTOP1-3 function in Al tolerance. Taken together, the results herein suggest that the functions of the three identified GmSTOP1s are evolutionarily conserved in H+ tolerance responses, but not in Al tolerance responses.
Author Contributions
WW, YL, QC, WP, JP, and CL performed the experiments and collected the data. CL, JT, and HL designed the research, analyzed the data, and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Thomas Walk of Golden Fidelity LLC for critical reading.
Funding. This work was supported by grants from National Natural Science Foundation of China (31672220 and 31422046), the National Key Research and Development Program (2016YFD0100700), Guangdong Natural Science Funds for Distinguished Young Scholars (2015A030306034), Guangdong High-level Personnel of Special Support Program (2015TQ01N078, 2015TX01N042, and YQ201530) and Research Team Project of the Natural Science Foundation of Guangdong Province (2016A030312009).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00570/full#supplementary-material
References
- Badia M. B., Arias C. L., Tronconi M. A., Maurino V. G., Andreo C. S., Drincovich M. F., et al. (2015). Enhanced cytosolic NADP-ME2 activity in A. thaliana affects plant development, stress tolerance and specific diurnal and nocturnal cellular processes. Plant Sci. 240 193–203. 10.1016/j.plantsci.2015.09.015 [DOI] [PubMed] [Google Scholar]
- Bojórquez-Quintal E., Escalante-Magaña C., Echevarría-Machado I., Martínez-Estévez M. (2017). Aluminum, a friend or foe of higher plants in acid soils. Front. Plant Sci. 8:1767. 10.3389/fpls.2017.01767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bown A. W., Shelp B. J. (1997). The metabolism and functions of g-aminobutyric acid. Plant Physiol. 115 1–5. 10.1104/pp.115.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. C., Yamamoto Y., Matsumoto H. (1999). Accumulation of aluminium in the cell wall pectin in cultured tobacco (Nicotiana tabacum L.) cells treated with a combination of aluminium and iron. Plant Cell Environ. 22 1009–1017. 10.1046/j.1365-3040.1999.00467.x [DOI] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Crawford L. A., Bown A. W., Breitkreuz K. E., Guinel F. C. (1994). The synthesis of g-aminobutyric acid in response to treatments reducing cytosolic pH. Plant Physiol. 104 865–871. 10.1104/pp.104.3.865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elstner E. F., Wagner G. A., Schutz W. (1988). Activated oxygen in green plants in relation to stress situations. Curr. Top. Plant Biochem. Physiol. 7 159–187. [Google Scholar]
- Fan W., Lou H. Q., Gong Y. L., Liu M. Y., Cao M. J., Liu Y., et al. (2015). Characterization of an inducible C2H2-type zinc finger transcription factor VuSTOP1 in rice bean (Vigna umbellata) reveals differential regulation between low pH and aluminum tolerance mechanisms. New Phytol. 208 456–468. 10.1111/nph.13456 [DOI] [PubMed] [Google Scholar]
- Fan W., Lou H. Q., Gong Y. L., Liu M. Y., Wang Z. Q., Yang J. L., et al. (2014). Identification of early Al-responsive genes in rice bean (Vigna umbellata) roots provides new clues to molecular mechanisms of Al toxicity and tolerance. Plant Cell Environ. 37 1586–1597. 10.1111/pce.12258 [DOI] [PubMed] [Google Scholar]
- Flagel L. E., Wendel J. F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol. 183 557–564. 10.1111/j.1469-8137.2009.02923.x [DOI] [PubMed] [Google Scholar]
- Force A., Lynch M., Pickett F. B., Amores A., Yan Y. L., Postlethwait J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foy C. D. (1984). “Physiological effects of hydrogen, aluminium and manganese toxicities in acid soil,” in Soil Acidity and Liming 2nd Edn ed. Adams F. (Madison, WI: American Society of Agronomy; ) 57–97. [Google Scholar]
- Garcia-Oliveira A. L., Benito C., Prieto P., de Andrade Menezes R., Rodrigues-Pousada C., Guedes-Pinto H., et al. (2013). Molecular characterization of TaSTOP1 homoeologues and their response to aluminium and proton (H+) toxicity in bread wheat (Triticum aestivum L.). BMC Plant Biol. 13:134. 10.1186/1471-2229-13-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X., Horst W. J., Golz J. F., Lee J. E., Ding Z., Yang Z. B. (2017). LEUNIG_HOMOLOG transcriptional co-repressor mediates aluminium sensitivity through PECTIN METHYLESTERASE46-modulated root cell wall pectin methylesterification in Arabidopsis. Plant J. 90 491–504. 10.1111/tpj.13506 [DOI] [PubMed] [Google Scholar]
- Herridge D. F., Peoples M. B., Boddey R. M. (2008). Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil 311 1–18. 10.1007/s11104-008-9668-3 [DOI] [Google Scholar]
- Horst W. J., Schmoh N., Kollmeier M., Balulška F., Sivaguru M. (1999). Does aluminium affect root growth of maize through interaction with the cell wall-plasma membrane-cytoskeleton continuum? Plant Soil 215 163–174. 10.1023/A:1004439725283 [DOI] [Google Scholar]
- Hurth M. A., Suh S. J., Kretzschmar T., Geis T., Bregante M., Gambale F., et al. (2005). Impaired pH homeostasis in Arabidopsis lacking the vacuolar dicarboxylate transporter and analysis of carboxylic acid transport across the tonoplast. Plant Physiol. 137 901–910. 10.1104/pp.104.058453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M., Rao I., Wenzl P., Beebe S., Tohme J. (2004). Integration of genomics approach with traditional breeding towards improving abiotic stress adaptation: drought and aluminum toxicity as case studies. Field Crop Res. 90 35–45. 10.1016/j.fcr.2004.07.004 [DOI] [Google Scholar]
- Iuchi S., Koyama H., Iuchi A., Kobayashi A., Kitabayashi S., Kobayashi Y., et al. (2007). Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc. Natl. Acad. Sci. U.S.A. 104 9900–9905. 10.1073/pnas.0700117104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer-Pascuzzi A. S., Jackson T., Cui H., Petricka J. J., Busch W., Tsukagoshi H., et al. (2011). Cell identity regulators link development and stress responses in the Arabidopsis root. Dev. Cell 21 770–782. 10.1016/j.devcel.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd P. S., Proctor J. (2001). Why plants grow poorly on very acid soils: are ecologists missing the obvious? J. Exp. Bot. 52 791–799. 10.1093/jexbot/52.357.791 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Hoekenga O. A., Itoh H., Nakashima M., Saito S., Shaff J. E., et al. (2007). Characterization of AtALMT1 expression in aluminum-inducible malate release and its role for rhizotoxic stress tolerance in Arabidopsis. Plant Physiol. 145 843–852. 10.1104/pp.107.102335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Ohyama Y., Kobayashi Y., Ito H., Iuchi S., Fujita M., et al. (2014). STOP2 activates transcription of several genes for Al- and low pH-tolerance that are regulated by STOP1 in Arabidopsis. Mol. Plant 7 311–322. 10.1093/mp/sst116 [DOI] [PubMed] [Google Scholar]
- Kochian L. V., Hoekenga O. A., Piñeros M. A. (2004). How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorus efficiency. Annu. Rev. Plant Biol. 55 459–493. 10.1146/annurev.arplant.55.031903.141655 [DOI] [PubMed] [Google Scholar]
- Kochian L. V., Piñeros M. A., Liu J., Magalhaes J. V. (2015). Plant adaptation to acid soils: the molecular basis for crop aluminum resistance. Annu. Rev. Plant Biol. 66 571–598. 10.1146/annurev-arplant-043014-114822 [DOI] [PubMed] [Google Scholar]
- Koyama H., Toda T., Hara T. (2001). Brief exposure to low-pH stress causes irreversible damage to the growing root in Arabidopsis thaliana: pectin–Ca interaction may play an important role in proton rhizotoxicity. J. Exp. Bot. 52 361–368. [PubMed] [Google Scholar]
- Koyama H., Toda T., Yokota S., Zuraida D., Hara T. (1995). Effects of aluminium and pH on root growth and cell viability in Arabidopsis thaliana strain Landsberg in hydroponic culture. Plant Cell Physiol. 36 201–205. [Google Scholar]
- Liang C., Piñeros M. A., Tian J., Yao Z., Sun L., Liu J., et al. (2013). Low pH, aluminum, and phosphorus coordinately regulate malate exudation through GmALMT1 to improve soybean adaptation to acid soils. Plant Physiol. 161 1347–1361. 10.1104/pp.112.208934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Tian J., Lam H. M., Lim B. L., Yan X., Liao H. (2010). Biochemical and molecular characterization of PvPAP3, a novel purple acid phosphatase isolated from common bean enhancing extracellular ATP utilization. Plant Physiol. 152 854–865. 10.1104/pp.109.147918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M., Farmer A., Joshi T., Takahashi K., Langley R. J., Franklin L. D., et al. (2010). An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 63 86–99. 10.1111/j.1365-313X.2010.04222.x [DOI] [PubMed] [Google Scholar]
- Liu J., Luo X., Shaff J., Liang C., Jia X., Li Z., et al. (2012). A promoter-swap strategy between the AtALMT and AtMATE genes increased Arabidopsis aluminum resistance and improved carbon-use efficiency for aluminum resistance. Plant J. 71 327–337. 10.1111/j.1365-313X.2012.04994.x [DOI] [PubMed] [Google Scholar]
- Liu J., Magalhaes J. V., Shaff J., Kochian L. V. (2009). Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. 57 389–399. 10.1111/j.1365-313X.2008.03696.x [DOI] [PubMed] [Google Scholar]
- Liu J., Piñeros M. A., Kochian L. V. (2014). The role of aluminum sensing and signaling in plant aluminum resistance. J. Integr. Plant Biol. 56 221–230. 10.1111/jipb.12162 [DOI] [PubMed] [Google Scholar]
- Liu P. D., Xue Y. B., Chen Z. J., Liu G. D., Tian J. (2016). Characterization of purple acid phosphatases involved in extracellular dNTP utilization in Stylosanthes. J. Exp. Bot. 67 4141–4154. 10.1093/jxb/erw190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., O’Hely M., Walsh B., Force A. (2001). The probability of preservation of a newly arisen gene duplicate. Genetics 159 1789–1804. 10.1093/jxb/erw190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J. F. (2007). Syndrome of aluminum toxicity and diversity of aluminum resistance in higher plants. Int. Rev. Cytol. 264 225–252. 10.1016/S0074-7696(07)64005-4 [DOI] [PubMed] [Google Scholar]
- Meriga B., Reddy B. K., Rao K. R., Reddy L. A., Kishor P. B. (2004). Aluminium induced production of oxygen radicals, lipid peroxidation and DNA damage in seedlings of rice (Oryza sativa). J. Plant Physiol. 161 63–68. 10.1078/0176-1617-01156 [DOI] [PubMed] [Google Scholar]
- Ohyama Y., Ito H., Kobayashi Y., Ikka T., Morita A., Kobayashi M., et al. (2013). Characterization of AtSTOP1 orthologous genes in tobacco and other plant species. Plant Physiol. 162 1937–1946. 10.1104/pp.113.218958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palóve-Balang P., ćiamporová M., Zelinová V., Pavlovkin J., Gurinová E., Mistrík I. (2012). Cellular responses of two Latin-American cultivars of Lotus corniculatus to low pH and Al stress. Cent. Eur. J. Biol. 7 1046–1054. 10.2478/s11535-012-0098-0 [DOI] [Google Scholar]
- Pavlovkin J., Palóve-Balang P., Kolarović L., Zelinová V. (2009). Growth and functional responses of different cultivars of Lotus corniculatus to aluminum and low pH stress. J. Plant Physiol. 166 1479–1487. 10.1016/j.jplph.2009.03.005 [DOI] [PubMed] [Google Scholar]
- Ramos-Díaz A., Brito-Argáez L., Munnik T., Hernández-Sotomayor S. M. T. (2007). Aluminum inhibits phosphatidic acid formation by blocking the phospholipase C pathway. Planta 225 393–401. 10.1007/s00425-006-0348-3 [DOI] [PubMed] [Google Scholar]
- Rangel A. F., Mobin M., Rao I. M., Horst W. J. (2005). Proton toxicity interferes with the screening of common bean (Phaseolus vulgaris L.) genotypes for aluminium resistance in nutrient solution. J. Plant. Nutr. Soil Sci. 168 607–616. 10.1002/jpln.200520509 [DOI] [Google Scholar]
- Roberts J. K., Hooks M. A., Miaullis A. P., Edwards S., Webster C. (1992). Contribution of malate and amino acid metabolism to cytoplasmic pH regulation in hypoxic maize root tips studied using nuclear magnetic resonance spectroscopy. Plant Physiol. 98 480–487. 10.1104/pp.98.2.480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan P. R., Delhaize E., Jones D. L. (2001). Function and mechanism of organic anion exudation from plant roots. Annu. Rev. Plant Physiol. Mol. Biol. 52 527–560. 10.1146/annurev.arplant.52.1.527 [DOI] [PubMed] [Google Scholar]
- Sakano K. (1998). Revision of biochemical pH-stat: involvement of alternative pathway metabolisms. Plant Cell Physiol. 39 467–473. 10.1093/oxfordjournals.pcp.a029393 [DOI] [Google Scholar]
- Sawaki Y., Iuchi S., Kobayashi Y., Kobayashi Y., Ikka T., Sakurai N., et al. (2009). STOP1 regulates multiple genes that protect Arabidopsis from proton and aluminum toxicities. Plant Physiol. 150 281–294. 10.1104/pp.108.134700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaki Y., Kobayashi Y., Kihara-Doi T., Nishikubo N., Kawazu T., Kobayashi M., et al. (2014). Identification of a STOP1-like protein in Eucalyptus that regulates transcription of Al tolerance genes. Plant Sci. 223 8–15. 10.1016/j.plantsci.2014.02.011 [DOI] [PubMed] [Google Scholar]
- Schmutz J., Cannon S. B., Schlueter J., Ma J., Mitros T., Nelson W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463 178–183. 10.1038/nature08670 [DOI] [PubMed] [Google Scholar]
- Sénéchal F., Mareck A., Marcelo P., Lerouge P., Pelloux J. (2015). Arabidopsis PME17 activity can be controlled by pectin methylesterase inhibitor4. Plant Signal. Behav. 10:e983351. 10.4161/15592324.2014.983351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaguru M., Baluska F., Volkmann D., Felle H. H., Horst W. J. (1999). Impacts of aluminum on the cytoskeleton of the maize root apex. short-term effects on the distal part of the transition zone. Plant Physiol. 119 1073–1082. 10.1104/pp.119.3.1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaguru M., Fujiwara T., Šamaj J., Baluska F., Yang Z., Osawa H., et al. (2000). Aluminum-induced 1-3- b-D-glucan inhibits cell-to-cell trafficking of molecules through plasmodesmata: a new mechanism of aluminum toxicity in plants. Plant Physiol. 124 991–1006. 10.1104/pp.124.3.991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Xu X., Wang H., Tao Y. (2011). Protein carbonylation in barley seedling roots caused by aluminum and proton toxicity is suppressed by salicylic acid. Russ. J. Plant Physiol. 58 653–659. 10.1134/S1021443711040169 [DOI] [Google Scholar]
- Tokizawa M., Kobayashi Y., Saito T., Kobayashi M., Iuchi S., Nomoto M., et al. (2015). SENSITIVE TO PROTON RHIZOTOXICITY1, CALMODULIN BINDING TRANSCRIPTION ACTIVATOR2, and other transcription factors are involved in ALUMINUM-ACTIVATED MALATE TRANSPORTER1 expression. Plant Physiol. 167 991–1003. 10.1104/pp.114.256552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Uexküll H. R., Mutert E. (1995). “Global extent, development and economic impact of acid soils,” in Plant-Soil Interactions at Low pH: Principles and Management eds Date R. A., Grundon N. J., Raymet G. E., Probert M. E. (Dordrecht: Kluwer Academic; ) 5–19. 10.1007/978-94-011-0221-6_1 [DOI] [Google Scholar]
- Yamaguchi Y., Yamamoto Y., Matsumoto H. (1999). Cell death process initiated by a combination of aluminium and iron in suspension cultures of tobacco cells (Nicotiana tabacum): apoptosis-like cell death mediated by calcium and proteinase. Soil Sci. Plant Nutr. 45 647–657. 10.1080/00380768.1999.10415828 [DOI] [Google Scholar]
- Yamaji N., Huang C. F., Nagao S., Yano M., Sato Y., Nagamura Y., et al. (2009). A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 21 3339–3349. 10.1105/tpc.109.070771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. L., Zheng S. J., He Y. F., Matsumoto H. (2005). Aluminium resistance requires resistance to acid stress: a case study with spinach that exudes oxalate rapidly when exposed to Al stress. J. Exp. Bot. 56 1197–1203. 10.1093/jxb/eri113 [DOI] [PubMed] [Google Scholar]
- Yokota S., Ojima K. (1995). Physiological response of root tip of alfalfa to low pH and aluminium stress in water culture. Plant Soil 171 163–165. 10.1007/BF00009581 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


