Abstract
Purpose
The Cancer Genome Atlas data resources represent an opportunity to explore commonalities across cancer types involving multiple molecular levels, but tumor lineage and histology can represent a barrier in moving beyond differences related to cancer type.
Experimental Design
On the basis of gene expression data, we classified 10224 cancers, representing 32 major types, into ten molecular-based “classes.” Molecular patterns representing tissue or histologic dominant effects were first removed computationally, with the resulting classes representing emergent themes across tumor lineages.
Results
Key differences involving mRNAs, miRNAs, proteins, and DNA methylation underscored the pan-cancer classes. One class expressing neuroendocrine and cancer-testis antigen markers represented ~4% of cancers surveyed. Basal-like breast cancers segregated into an exclusive class, distinct from all other cancers. Immune checkpoint pathway markers and molecular signatures of immune infiltrates were most strongly manifested within a class representing ~13% of cancers. Pathway-level differences involving hypoxia, NRF2-ARE, Wnt, and Notch were manifested in two additional classes enriched for mesenchymal markers and miR-200 silencing.
Conclusions
All pan-cancer molecular classes uncovered here, with the important exception of the basal-like breast cancer class, involve a wide range of cancer types and would facilitate understanding the molecular underpinnings of cancers beyond tissue-oriented domains. Numerous biological processes associated with cancer in the laboratory setting were found here to be coordinately manifested across large subsets of human cancers. The number of cancers manifesting features of neuroendocrine tumors may be much higher than previously thought, which disease is known to occur in many different tissues.
Introduction
Cancer is not a single disease, and at the molecular level there is widespread heterogeneity that may be observed from patient to patient. Nevertheless, unsupervised classification of tumors on the basis of molecular profiling data can reveal major subtypes existing within a given cancer type according to tissue of origin(1,2). Such molecular-based subtypes can reflect altered pathways within different cancer subsets, which could have important implications for applying existing therapies or for developing new therapeutic approaches(3). The Cancer Genome Atlas (TCGA), a large-scale effort to comprehensively characterize over 10,000 human cancers at the molecular level, provides a common platform for the study of diverse cancer types, with multiple levels of data including mRNA, miRNA, protein, DNA methylation, copy number, and mutation(2). For most cancer types represented in TCGA, an individual study of the molecular landscape of that cancer type was carried out (2).
With data generation completed, there is opportunity for systematic analyses of the entire TCGA pan-cancer cohort(4), including defining molecular subtypes and associated pathways relevant to multiple cancer types. One challenge, in the identification of cancer subsets transcending the tissue of origin, is that widespread molecular patterns are associated with tumor lineage and histology(5–7). While TCGA datasets have been harmonized to allow for cross-cancer type comparisons—which is useful for addressing a host of questions—an alternative approach to molecular classification on the basis of these data would be to first computationally subtract the molecular differences between cancer types(8,9). This alternative approach would have the effect of consolidating the individual subtypes that might be discoverable in individual cancer types into super-types or pan-cancer “classes” that transcend tissue or histology distinctions.
The aim of our present study was to define pan-cancer molecular-based subtypes or “classes,” which would transcend tumor lineage across the over 10,000 human cancers and 32 cancer types profiled by TCGA, using data from multiple molecular profiling platforms to characterize these classes in terms of associated pathways.
Methods
Results are based upon data generated by TCGA Research Network (http://cancergenome.nih.gov/). Molecular data were aggregated from public repositories. Tumors spanned 32 different TCGA projects, each project representing a specific cancer type, listed as follows: LAML, Acute Myeloid Leukemia; ACC, Adrenocortical carcinoma; BLCA, Bladder Urothelial Carcinoma; LGG, Lower Grade Glioma; BRCA, Breast invasive carcinoma; CESC, Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, Cholangiocarcinoma; CRC, Colorectal adenocarcinoma (combining COAD and READ projects); ESCA, Esophageal carcinoma; GBM, Glioblastoma multiforme; HNSC, Head and Neck squamous cell carcinoma; KICH, Kidney Chromophobe; KIRC, Kidney renal clear cell carcinoma; KIRP, Kidney renal papillary cell carcinoma; LIHC, Liver hepatocellular carcinoma; LUAD, Lung adenocarcinoma; LUSC, Lung squamous cell carcinoma; DLBC, Lymphoid Neoplasm Diffuse Large B-cell Lymphoma; MESO, Mesothelioma; OV, Ovarian serous cystadenocarcinoma; PAAD, Pancreatic adenocarcinoma; PCPG, Pheochromocytoma and Paraganglioma; PRAD, Prostate adenocarcinoma; SARC, Sarcoma; SKCM, Skin Cutaneous Melanoma; STAD, Stomach adenocarcinoma; TGCT, Testicular Germ Cell Tumors; THYM, Thymoma; THCA, Thyroid carcinoma; UCS, Uterine Carcinosarcoma; UCEC, Uterine Corpus Endometrial Carcinoma; UVM, Uveal Melanoma. Cancer molecular profiling data were generated through informed consent as part of previously published studies and analyzed in accordance with each original study’s data use guidelines and restrictions.
For the mRNA platform, log2-transformed expression values within each cancer type (as defined by TCGA project) were normalized to standard deviations from the median. By k-means clustering method (using the “kmeans” R function, with 10 clusters and 1000 maximum iterations and 25 random sets), cancer cases were subtyped, based on the top 2000 features with the most variable expression on average by cancer type (using standard deviation of the log2-transformed expression values as a measure of variability, excluding genes on X or Y chromosomes).
We defined the top differential genes associated with each subtype, or pan-cancer “class”. Taking the top 2000 mRNA features, we first computed the two-sided t-test for each gene and each class, comparing expression levels of each class with that of the rest of the tumors. We then selected the top 100 genes with the lowest p-value for each subtype; however, for the c2 class only three genes were associated, and for the c7 class only 51 genes were associated, resulting in 854 top class-specific genes in all. In a similar manner, top features associated with pan-cancer class for RPPA, miRNA, and DNA methylation platforms were identified.
The Fantom datasets of gene expression by cell type(10) were analyzed using a previously utilized approach(7). Briefly, the top 2000 most variable mRNAs (used above for the clustering analyses) were examined in Fantom. Logged expression values for each gene in the fantom dataset were centered on the median of sample profiles. For each fantom differential expression profile, the inter-profile correlation (Spearman’s) was taken with that of each TCGA pan-cancer differential expression profile (with genes normalized within each TCGA project to standard deviations from the median).
We examined an external gene expression profiling dataset of multiple cancer types from the Expression Project for Oncology (expO) (GSE2109), classifying each external tumor profile by pan-cancer class as defined by TCGA data. Within each cancer type, log2-transformed genes in the expO dataset were normalized to standard deviations from the median. As a classifier, the top set of 854 mRNAs distinguishing between the TCGA pan-cancer classes was used. For each pan-cancer class, the average value for each gene was computed, based on the centered TCGA expression data matrix. The Pearson’s correlation between each expO profile and each TCGA pan-cancer class averaged profile was computed. Each expO case was assigned to TCGA pan-cancer class, based on which class profile showed the highest correlation with the given expO profile.
All p values were two-sided unless otherwise specified. Scoring of expression profiles for pathway-associated gene signatures was carried out as previously described (9,11). Additional Methods details are provided in supplemental. Supplementary Figure 1 provides a schematic of the various analyses performed and their relation to supplementary data files.
Results
Pan-cancer molecular subtypes primarily driven by tumor lineage
Across cancer types, tumor lineage and squamous histology were found to represent major determinates of unsupervised molecular classification. In total, our study involved 11232 human cancer cases representing 32 different major types, for which TCGA generated data on one or more of the following molecular characterization platforms (Supplementary Data 1): whole exome sequencing (WES, n=10224 cases), somatic DNA copy by SNP array (n=10845), RNA-seq (n=10224), miRNA-seq (n=10128), DNA methylation (n=10959), and Reverse Phase Protein Array (RPPA, n=7663). Using established analytical approaches(5–7), 10662 TCGA cases (with data available for at least three platforms) were subtyped according to each of the above data platforms excluding WES, with the various subtype calls for each sample then being consolidated to define multiplatform-based molecular subtypes.
With hierarchical clustering of the platform-level subtype assignments (Supplementary Figures 2A and 3), the cases segregated largely according to cancer type, with higher level branches of the clustering tree representing major tissue-based categories including breast, liver, kidney, brain, gastrointestinal, squamous (head and neck, lung squamous, esophageal, bladder, cervical), lung adenocarcinoma, immune-related, uterus, and skin. In this pan-cancer molecular subtyping, basal-like BRCA (breast cancer) was molecularly distinct from other BRCA. K-means clustering was applied to the platform-level subtype assignment matrix to formally define a discrete number of subtypes (Supplementary Figures 2B, 4A, and 4B, and Supplementary Data 2). With a 25-subtype solution, for example (Supplementary Figure 2B), most subtypes were specific to either a single cancer type or multiple types related by common tissue of origin.
Pan-cancer molecular classes that transcend tumor lineage
Whereas the above pan-cancer molecular subtypes were found to be highly concordant with their tissue-of-origin counterparts, we went on to pursue an alternate analytical strategy to define subtypes that would not be driven by tumor lineage-specific markers or patterns. For expression (mRNA, miRNA, protein) and DNA methylation datasets, values were first normalized or centered within each cancer type, thereby computationally removing any tissue or histology dominant effects. For defining pan-cancer subtypes using the above datasets, we had initially considered multiplatform-based solutions (e.g. analyzing results from the five data platforms together to define subtypes(9)), but instead opted to use RNA-seq platform to define subtypes and then examine the other platforms for correlates of these RNA-seq-based subtypes. RNA-seq data would represent the full range of features for cancer-relevant pathways, while miRNA and protein data platforms both represent expression-based data but with a more limited number of features and represented pathways as compared to RNA-seq data. In addition, DNA copy alterations (while having a normal control) vary by cancer type (Supplementary Figure 3 and ref(9)), and so would serve to segregate samples by cancer type.
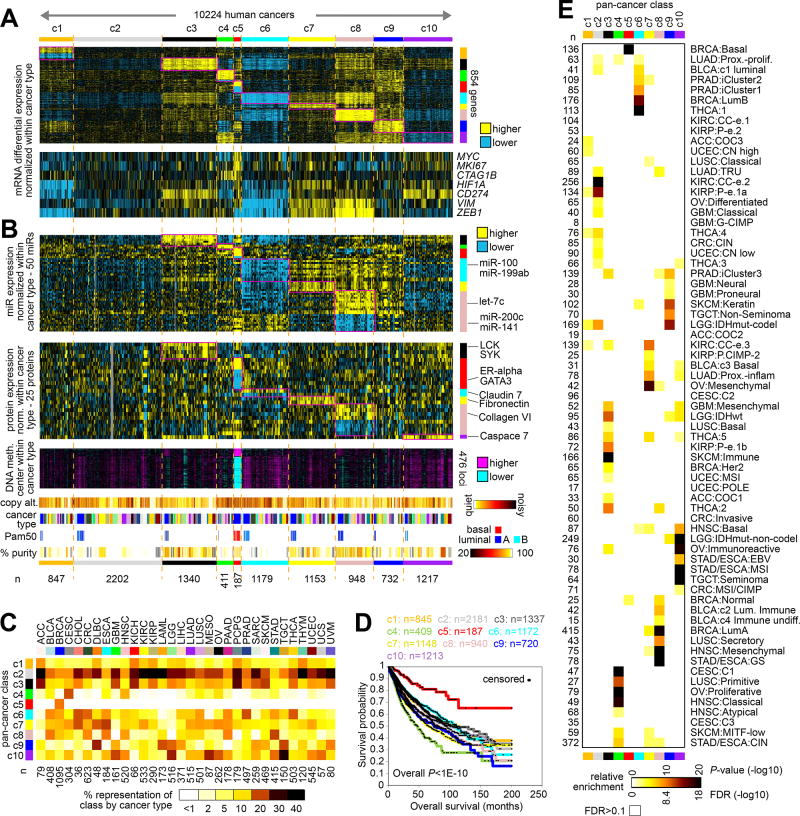
The RNA-seq platform (with values normalized within cancer type) was used to define ten different subtypes or “classes” of cancer (Table 1) across the 10224 cases in TCGA cohort with available data (where with solutions of more than ten subtypes no appreciable increase in clustering consensus was observed, Supplementary Figure 5A), with the other profiling platforms then being used to characterize these classes in terms of associated pathways as described below. These pan-cancer molecular classes, referred to here as “c1” through “c10,” were each characterized by widespread molecular patterns (Figure 1A and Supplementary Figures 5B–5F and 6). For each class, the top genes most differentially expressed in the given class versus the rest of the tumors were identified (Figure 1A and Supplementary Data 2), where the top differential patterns—involving 854 genes in all—could be observed to span cancer type. Notably, very few genes were strongly associated with the c2 class in particular, which represented one distinguishing feature of this class as compared to the other classes, although this c2 class would represent a basis of comparison. Normalized differential expression patterns for specific genes of interest—including MYC, MKI67, CTAG1B, HIF1A, CD274, VIM, and ZEB1—could further distinguish between the classes and suggested associated pathways as further explored below.
Table 1.
Tissue-independent pan-cancer molecular classes in TCGA cohort.
| class | n (%) | description and notable features |
|---|---|---|
| c1 | 847 (8.3) | High differential expression of oxidative phosphorylation genes, glycolysis genes, and pentose phosphate pathway genes. |
| c2 | 2202 (21.5) | Lack of strong associated expression patterns; can serve as a comparison group for the other classes. |
| c3 | 1340 (13.1) | Strong association with immune checkpoint pathway; differential expression profile associated with immune cell infiltration; mesenchymal signature; NRF2/KEAP1 pathway signature; Wnt pathway signature. |
| c4 | 411 (4) | Differential expression profile associated with neuroendocrine tumors and with normal cells and tissues of the central nervous system; CT antigen expression. |
| c5 | 187 (1.8) | Represents basal-like breast cancer; TP53-related alterations, MYC amplification and expression; YAP1 target expression; high expression of pentose phosphate and TCA cycle genes; immune checkpoint pathway; CT antigen expression. |
| c6 | 1179 (11.5) | Epithelial signature; normoxia signature; YAP1 target expression. |
| c7 | 1153 (11.3) | Mesenchymal signature; hypoxia signature; Wnt pathway signature; Notch pathway signature; NRF2/KEAP1 pathway signature; low differential expression of miR-200. |
| c8 | 948 (9.3) | High differential expression of fatty acid metabolism genes; mesenchymal signature; hypoxia signature; Wnt pathway signature; Notch pathway signature; NRF2/KEAP1 pathway signature; high differential DNA methylation and low differential expression of miR-200; differential expression profile associated with normal cells and tissues of the central nervous system; immune checkpoint pathway (observed in TCGA cohort only). |
| c9 | 732 (7.2) | Wnt pathway signature; Notch pathway signature; NRF2/KEAP1 pathway signature. |
| c10 | 1217 (11.9) | Immune checkpoint pathway; differential expression profile associated with immune cell infiltration; YAP1 target expression. |
CT, cancer-testis.
Figure 1. Molecular classes of TCGA cancers that transcend tumor lineage or tissue-of-origin.
(A) Using an alternative molecular classification approach, whereby differences between cancer types were first removed computationally prior to classification on the basis of mRNA expression data, ten major pan-cancer “classes” were identified. The first heat map shows differential mRNA expression patterns (values normalized within each main cancer type) for a set of 854 genes found to best distinguish between the ten subtypes (see Methods). The second shows differential expression patterns for a select set of genes representing pathways of particular interest. Numbers of cases (n=10224) denote representation on RNA-seq data platform. (B) Molecular features from other data platforms associating with pan-cancer molecular class. Top heat map shows differential expression patterns (values normalized within each cancer type), representing a top set of 50 miRNA features that distinguish between the ten molecular classes from part A. The second heat map shows differential protein expression patterns (by RPPA platform, values normalized within each cancer type), representing a top set of 25 features that distinguish between the ten subtypes. The third heat map shows differential DNA methylation patterns (values centered within each cancer type) for a top set of features that distinguish a class associated with basal-like breast cancer. Additional sample-level data tracks denote levels of genome-wide copy number alteration, cancer type (according to TCGA project, color coding in part C), BRCA Pam50 subtype, and estimated tumor sample purity(47) (white, ~100% purity). (C) The percent representations of each pan-cancer class by cancer type (according to TCGA project) are represented using a colorgram. (D) Differences in patient overall survival among the pan-cancer molecular classes. P values by stratified log-rank test incorporating cancer type as a confounder. Overall p value evaluates for significant differences among the groups as defined by pan-cancer class. (E) Significance of overlap between the pan-cancer class assignments made in the present study (columns), with molecular-based subtype assignments (rows) made previously for a subset of cases. P-values by one-sided Fisher’s exact test; only p-values with False Discovery Rate (FDR)<0.1(48) are represented. See Methods for TCGA project abbreviations. See also Supplementary Figures 1 to 7 and Supplementary Data 1 and 2.
Specific miRNAs and proteins (with values normalized within cancer type) could distinguish between the pan-cancer molecular classes (Figure 1B and Supplementary Figure 5E and Supplementary Data 2), with, for example, miR-200 family members showing the lowest differential expression in c8 as well as c7 classes, and with immune cell protein markers LCK and SYK being highest in the c3 class. Differential DNA methylation patterns could also distinguish between the subtypes (Figure 1C, Supplementary Figures 5B and 5F, and Supplementary Data 2), with distinctive patterns associated with c5 class in particular. For a subset of genes, significant anti-correlations between methylation and expression could be identified (Supplementary Figure 5F and Supplementary Data 2). Within the top differentially expressed genes underscoring each pan-cancer molecular class, specific gene categories were over-represented (Supplementary Figure 7), including immune-related genes being most highly expressed in the c3 class, neuron-related genes being highly expressed in the c4 class, and cytoskeleton- and keratin-related genes being highly expressed in the c9 class. The c4, c5, and c6 classes tended to show a higher degree of genome-wide copy alterations (Figure 1B and Supplementary Figure 5B). Each of the pan-cancer classes were found to span cases from multiple cancer types (Figures 1B and 1C), with the notable exception of c5, which consisted entirely of BRCA cases (n=187) and represents the basal-like breast cancer molecular subtype (12)(Figure 1B). As compared to the other pan-cancer classes, the c5 class was associated with better overall patient survival (Figure 1D); however, compared with other breast cancers, c5 has a poorer survival(3). Overall, the pan-cancer molecular classes showed significant concordances with other molecular subtype designations, which had been previously made for a subset of TCGA cases in individual cancer type studies(6,12–24) (Figure 1E).
Associations involving somatic alterations
Across the entire TCGA pan-cancer cohort, assessment of genes within pathways demonstrated a high number of somatic alterations (mutation, copy alteration, or epigenetic silencing) involving p53 (62.7% of 10224 cases with exome data available), PI3K/AKT/mTOR (44.1%), Receptor Tyrosine Kinase signaling (RTK, 43.9%), chromatin modification (40.8%), SWI/SNF complex (32.0%), Wnt/beta-catenin (22.3%), MYC (10.9%), NRF2-ARE (9.3%), and Hippo signaling (4.5%)(Supplementary Figure 8A). The above pathways were found to be altered in different ways involving different genes in different cancer types (Supplementary Figure 9A). A number of pathway-level or individual gene-level alterations surveyed were highly represented within specific cancer types or pan-cancer classes (Supplementary Figures 8B and 9B). In particular, TP53 mutations were highly represented within both c4 and c5 tumors, and MYC amplifications were highly represented in c5 tumors (Supplementary Figures 8B and 9B). Furthermore, c10 tumors were enriched for somatic alterations involving p53 pathway (including TP53 and ATM mutations), RTKs (MET mutations), and numerous genes involving chromatin modification and SWI/SNF complex (Supplementary Figures 8B and 9B). Some of the somatic mutation associations observed might be attributable to cancer type- or mutation rate-specific patterns (Supplementary Figures 10 and 11); for example, TP53 and chromatin modifier mutations being enriched within c4 tumors would reflect in part the types of cancers more highly represented within c4 (BLCA, CESC, HNSC, LUSC, etc.).
We sought to examine the effects on pathway activation—as measured by mRNA or protein signature—of somatic alterations impacting specific pathways noted above. Previously-defined gene expression signatures for p53, k-ras, MTOR, Wnt/beta-catenin, MYC, NRF2, and YAP1 (6,25–27) were applied to the mRNA or protein expression profiles of the TCGA samples, whereby each sample profile was scored for each of the above pathways. For each pathway considered, relative levels of the corresponding signature were significantly different between somatically altered versus unaltered cases for that pathway, and the differences were in the anticipated direction (e.g. p53-related alterations were associated with decreased levels of p53 transcriptional targets, and MTOR-related alterations were associated with increased MTOR proteomic signaling)(Supplementary Figure 8C). These results would demonstrate a widespread level of concordance between observed DNA-level alterations and global expression patterns. At the same time, within the somatically altered and unaltered groups for each pathway, a wide range of genes signature levels were evident (Supplementary Figure 8C), suggesting that other factors besides somatic mutation or copy alteration (e.g. tumor microenvironmental influences) may impact pathways.
Pathway-related gene signatures and mesenchymal cells
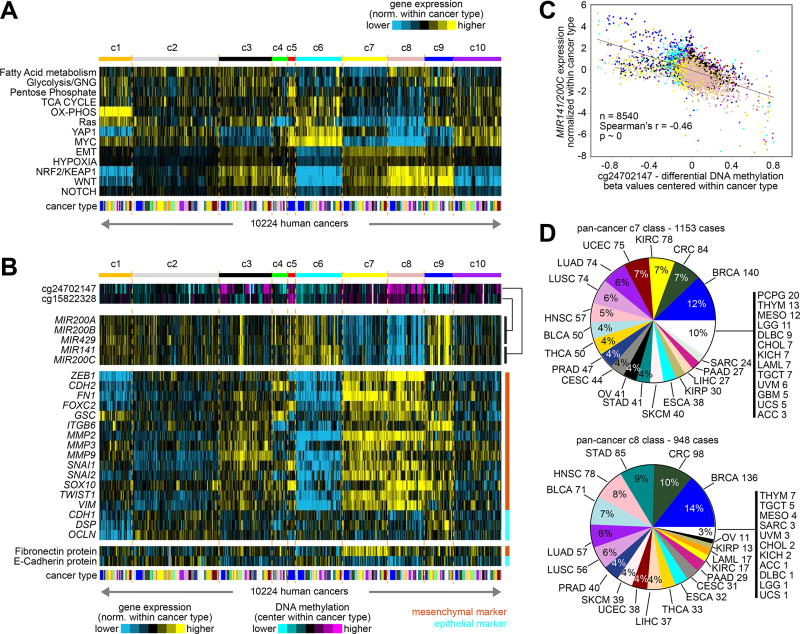
In order to gain insight into pathways that would distinguish between the various pan-cancer molecular classes, we applied a number of gene signatures to TCGA expression profiles with values normalized within each cancer type. A number of pathways appeared more or less active for different pan-cancer classes (Figure 2A), as further explored below. For example, gene signature scores for epithelial-mesenchymal transition (EMT), hypoxia, NRF2/KEAP1, Wnt, and Notch all were higher in c3, c7, and c8 classes, as compared to the rest of the tumors. Focusing here on EMT, representing mesenchymal features, we observed a strong negative association between the expression of miR-200 family members and their promoter methylation levels (Figures 2B and 2C, and Supplementary Figure 12A), which demonstrates epigenetic regulation of these miRNAs across a large subset of human cancers. The miR-200 family suppresses ZEB1, a key transcriptional regulator of EMT. Across the entire TCGA cohort, we examined differential expression values for a set of genes and proteins representing canonical mesenchymal or epithelial markers(11), as well as for miR-200 family and ZEB1 genes (Figure 2B). Manifestation of mesenchymal features was highest in the c7 and c8 tumors and lowest in the c6 tumors (which appeared more epithelial), and miR-200 expression was lowest in c7 and c8 tumors, with associated DNA methylation being highest in c8 tumors; c3 tumors also showed mesenchymal-associated patterns but less strongly than did c7 and c8 tumors, and c9 tumors showed high expression of some mesenchymal and epithelial marker genes, as well as high miR-200. The c7 and c8 pan-cancer classes each involved a wide range of cancer types (Figure 2D).
Figure 2. Pathway-associated gene signatures across pan-cancer molecular classes.
(A) By pan-cancer molecular class, pathway-associated mRNA signatures (using values normalized within each cancer type). See Figure 1C and part D for cancer type color legend. Numbers of cases (n=10224) denote representation on RNA-seq data platform. (B) Corresponding to cases from part A, heat maps showing DNA methylation and expression levels for miR-200 family members (using values normalized or centered within each cancer type). Representative DNA methylation probes(49) that map to the promoter of each miRNA cluster are shown (miR-141/200c = cg24702147, miR-200a/200b/429 = cg15822328). Normalized expression levels for a set of canonical epithelial or mesenchymal markers (11) are also shown. (C) Scatter plot of differential methylation vs differential expression (using values normalized or centered within each cancer type), for cg24702147 versus miR-141/200c (normalized values for the two miRNAs being averaged). Numbers of cases denote representation on all three data platforms for mRNA-seq, miRNA-seq, and 450K DNA methylation. Data points are colored according to pan-cancer class, as represented in parts A and B. (D) For c7 and c8 pan-cancer classes (associated with mesenchymal cells, along with hypoxia, NRF2/KEAP1, Wnt, and Notch signatures), distributions by cancer type. See also Supplementary Figures 8 to 12 and Supplementary Data 3 and 4.
For some tumor subsets, the observed associations with mesenchymal features could conceivably be attributable to surrounding stromal cells within the tumor sample, as well as to changes within the actual cancer cells. For example, invasive lobular carcinoma (ILC), the second most prevalent histologic subtype of invasive breast cancer, is characterized by small discohesive neoplastic cells invading the stroma in a single-file pattern(12). Immunohistochemical analysis in breast ILC has demonstrated that the neoplastic lobular cells tend to retain their epithelial identity, with mesenchymal markers being expressed by the fibroblasts in the prominent stromal component of ILC(28). On the other hand, for other cancer types, changes within cancer cells from epithelial to mesenchymal states may occur at the tumor invasive front, though not within the main tumor bulk(29). Of our ten pan-cancer molecular classes, four were characterized by relatively lower sample purity: c3, c5 (representing basal-like breast cancer), c7, and c8 (Figure 1B and Supplementary Figures 12B and 12C). Interestingly, of these four classes, only the c8 class was strongly associated with the breast ILC cases in TCGA (Supplementary Figure 12B). In contrast, renal cell carcinomas of the previously described “CC-e.3” molecular subtype manifesting EMT(30) associated with the c7 class but not with the c8 class, and the EMT-associated “SQ.1” molecular subtype of lung cancer(7) was distributed among c3, c7, and c8 classes (Supplementary Figure 12B). Three molecular subtypes previously associated with mesenchymal features for specific cancer types, GBM:Mesenchymal, OV:Mesenchymal, and HNSC:Mesenchymal, were each respectively associated with c3, c7, and c8 pan-cancer classes (Figure 1E). In comparison to c7, c8 was further distinguished by high expression of fatty acid metabolism genes (Figure 2A), and associated hypoxia-related changes in c7 and c8, for example, suggest an altered microenvironment. All of this would indicate that the molecular classes associated with lower purity would each represent distinctive biology, apart from the technical aspects involved with sample collection.
Immune checkpoints and neuroendocrine tumors
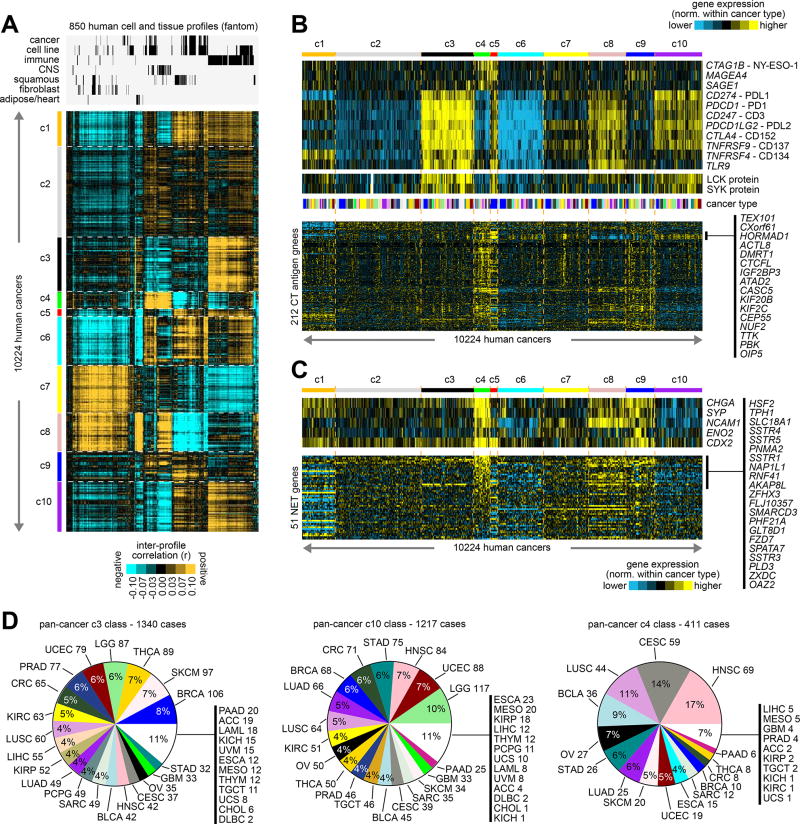
Analysis of gene expression patterns of normal tissues and cells can provide meaningful context to the widespread differential patterns observed in cancer(7,31). We examined the top 2000 differential mRNAs from TCGA cohort (from Supplementary Figure 5B) in a public expression dataset from the Fantom consortium(10) of 850 profiles representing various human cell and tissue specimens. Inter-correlations between Fantom profiles and TCGA profiles revealed each pan-cancer molecular class to manifest distinctive patterns of global similarity or dissimilarity with specific categories of normal cells and tissues (Figure 3A and Supplementary Data 5). In particular, c3, c5, and c10 classes were strongly associated with immune-related cells and tissues; the c4 class was strongly associated with cells and tissues related to the Central Nervous System (CNS), and c8 class was associated with CNS to a somewhat lesser degree; the c7 and c8 classes were associated with fibroblasts and other mesenchymal-related categories (reflecting the mesenchymal marker patterns observed above, Figure 2B); and the c9 class was associated with adipose cells and heart tissues (which may be reflective of the association with fatty acid metabolism observed above, Figure 2A). Analysis of the Fantom mouse expression dataset yielded similar associations (Supplementary Figure 13A). In previous analyses utilizing TCGA expression profiles as normalized across all cancers, brain cancers and blood cancers associated as a group with fantom CNS and immune-related profiles, respectively(7).
Figure 3. Normal tissue and cell type associations with the pan-cancer molecular classes.
(A) Inter-profile correlations were computed between TCGA expression profiles (with values normalized within each cancer type) and profiles from the Fantom consortium expression dataset of various cell types or tissues from human specimens (n=850 profiles)(10). Membership of the Fantom profiles in general categories of “cancer”, “cell line”, “immune” (immune cell types or blood or related tissues), “CNS” (related to central nervous system including brain), “squamous” (including bronchial, trachea, oral regions, throat and esophagus regions, nasal regions, urothelial, cervix, sebocyte, keratin/skin/epidermis), “fibroblast”, or “adipocyte/heart” is indicated. Cancer type color coding in part D. (B) Heat maps of differential expression (values normalized within each cancer type), for genes encoding immunotherapeutic targets (top), for LCK and SYK proteins (middle, representing markers for T-cells and B-cells, respectively), and for genes encoding cancer-testis (CT) antigens (from the CT Gene Database, http://cancerimmunity.org/resources/ct-gene-database/). (C) Heat maps of differential expression (values normalized within each cancer type), for genes encoding canonical markers of neuroendocrine tumors (top), and for a set of 51 genes in a panel of neuroendocrine tumor (NET) markers(34), as uncovered previously using gene expression profiling (bottom). (D) For c3 (immune-associated), c10 (immune-associated), and c4 (CNS- and neuroendocrine-associated) pan-cancer classes, distributions by cancer type. See also Supplementary Figure 13 and Supplementary Data 5.
Focusing here on the immune-related associations found for specific pan-cancer molecular classes, we went on to survey TCGA expression profiles for a set of genes representing potential targets for immunotherapy(6), including cancer-testis (CT) antigen genes and genes involved in immune checkpoint pathway (Figure 3B), such as genes encoding PD1, PDL1, PDL2, and CTLA4. As a group, CT antigen genes were highest in both c4 and c5 classes, with a subset of CT antigen genes being particularly higher in c5 class (see Figure 3B). On the other hand, immune checkpoint genes were most strongly differentially expressed within the c3 class, while the c8 and c10 classes also showed increased though relatively lower expression of these genes. Differential protein expression of T cell marker LCK and B cell marker SYK indicated the presence of T cells in c3, c8, and c10 classes, and the presence of B cells in c3 and c10 classes (Figure 3B). Analysis of gene expression signatures from Bindea et al.(32) suggested that levels of infiltrating immune cell types were highest within the c3 class, followed by the c8 class, while the c10 class showed signatures for some but not all immune cell types (Supplementary Figure 13B).
The above associations of c4 tumors with CNS tissues and cells suggested features of neuroendocrine tumors (NETs), which arise from cells of the endocrine and nervous systems and which occur in many different tissues throughout the human body(33). Genes encoding canonical markers of NETs—including CHGA (chromogranin A), SYP (synaptophysin), NCAM1 (CD56), ENO2 (neuron-specific enolase), and CDX2—were all differentially higher in c4 tumors as compared to the other pan-cancer classes (Figure 3C). In addition, a set of 51 genes in a panel of NET markers previously uncovered using gene expression profiling was examined(34), with the majority of these being differentially higher within the c4 class (Figure 3C). TCGA LUAD cases previously determined to represent neuroendocrine cancer were also significantly enriched within the c4 class (Supplementary Figure 12B, 6/14 cases, p<1E-5, one-sided Fisher’s exact test). The c3, c10, and c4 pan-cancer classes each involved a wide range of cancer types (Figure 3D), with c4 class in particular being comprised of sizable percentages of head and neck, cervical, lung, bladder, ovarian, and gastrointestinal cancers.
Pathway-level differences across pan-cancer molecular classes
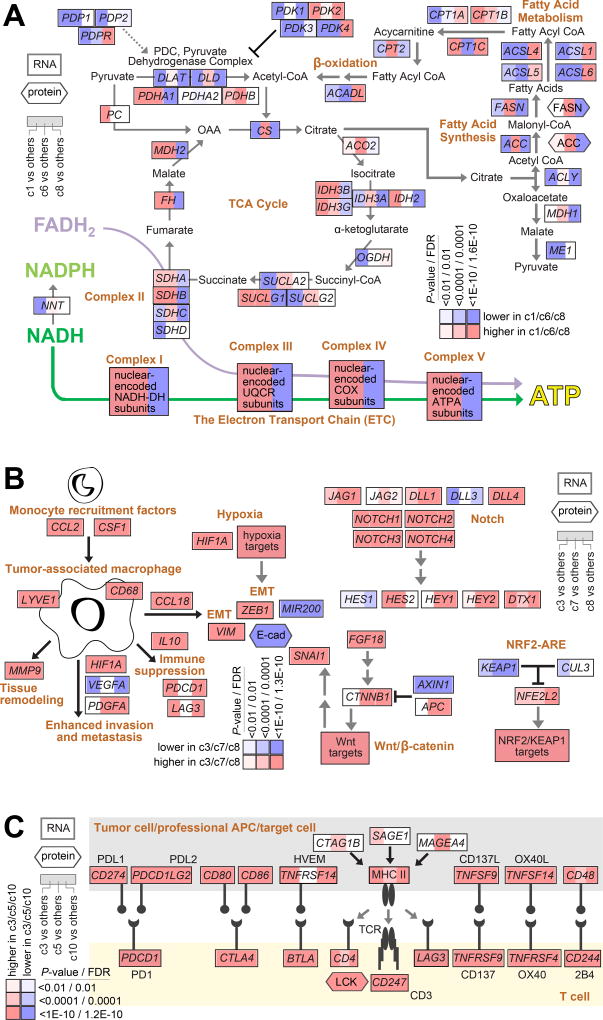
The above pathway-associated gene signatures (Figure 2A) indicated differential activation of specific pathways among the pan-cancer molecular classes. A number of these signatures were related to metabolism, including fatty acid metabolism, glycolysis/gluconeogenesis, pentose phosphate pathway, TCA cycle, and oxidative phosphorylation. The individual genes that comprised these signatures can be represented in a pathway diagram (Figure 4A), whereby the c1, c6, and c8 classes were each observed to show evidence for the altered utilization of specific metabolic pathways, as compared to the rest of the cancer cases. Both c1 and c6 tumors showed increased expression of genes involved with oxidative phosphorylation, while c6 tumors also showed higher expression of genes involved in the TCA cycle and of genes involved in fatty acid synthesis. On the other hand, c8 tumors appeared to down-regulate TCA cycle, oxidative phosphorylation, and fatty acid synthesis pathways, and to up-regulate genes involving in fatty acid metabolism.
Figure 4. Differentially active pathways across pan-cancer molecular classes.
(A) Pathway diagram representing core metabolic pathways, with differential expression patterns represented (using values normalized within cancer type), comparing tumors in pan-cancer classes c1, c6, or c8 with tumors in the other seven classes (red, significantly higher in c1/c6/c8). (B) Diagram of tumor-associated macrophage roles in the tumor microenvironment(35), and of Notch, NRF2-ARE, and Wnt/beta-catenin pathways, with differential expression patterns represented (using values normalized within cancer type), comparing tumors in pan-cancer classes c3, c7, or c8 with tumors in the other seven classes (red, significantly higher in c3/c7/c8). (C) Diagram of immune checkpoint pathway (featuring interactions between T cells and antigen-presenting cells, including tumor cells), with differential expression patterns represented (using values normalized within cancer type), comparing tumors in pan-cancer classes c3, c5, or c10 with tumors in the other seven classes (red, significantly higher in c3/c5/c10). P-values in parts A-C by Mann-Whitney U-test. FDR, false discovery rate.
Deregulated pathways were also reflective of tumor microenvironmental effects at work in distinct subsets of cancers. Differential expression patterns involving the c3, c7, or c8 pan-cancer molecular classes were consistent with a consensus model(35) of tumor-associated macrophage (TAM) roles in the tumor microenvironment (Figure 4B), whereby monocytes are first recruited to the tumor microenvironment (e.g. by CSF1 and CCL2). Tumor-secreted cytokines then have the potential to polarize recruited monocytes into TAMs, which play vital roles in a number of processes including immune suppression and EMT. EMT may also be induced by either hypoxia or Wnt signaling pathway(36–38), both of which appear increased in c3, c7, and c8 tumors (Figures 2A and 4B); NRF2/KEAP1 and Notch pathways also appeared increased in these tumors. Immune suppression may involve multiple redundant immune checkpoints, which were most associated with c3, c5, c8, and c10 classes (Figures 3B and 4C); these tumors showed higher expression of ligands such as PDL1 and PDL2, along with higher expression of the corresponding receptors associated with T cells.
TCGA patterns observable in external cohorts
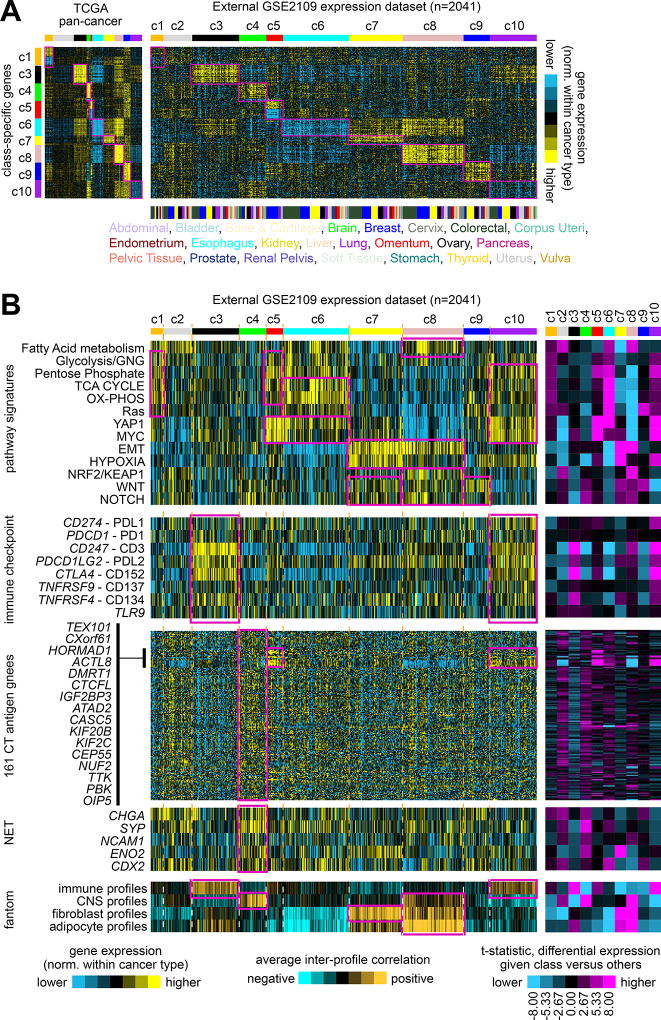
The pan-cancer molecular class associations, as observed in TCGA datasets, were also examined in an external multi-cancer expression dataset. From the Expression Project for Oncology (expO) dataset, mRNA expression profiles of 2041 cancer cases representing 26 different types were obtained. Within each cancer type, expression values for each gene in the expO dataset were first normalized, and then each expO tumor profile was classified by pan-cancer molecular class as defined by TCGA data (Figure 5A and Supplementary Figures 14A and 14B), where the top set of 854 mRNAs distinguishing between our ten classes (Figure 1B) was used as the classifier. In the same manner as carried out above for TCGA datasets, expO expression profiles were also scored for mRNA signatures related to specific pathways or normal cell types (Figure 5B and Supplementary Figure 15), where similar overall trends of pathway-level differences between classes as originally observed in TCGA cohort could also be observed in the expO cohort. In particular, c5 tumors were composed almost entirely of breast cancer cases; anticipated pathway-level differences involving metabolism, YAP1, MYC, EMT, hypoxia, NRF2/KEAP1, WNT, NOTCH, and immune checkpoint were observed in expO cohort; CT antigen genes were higher as a group in c4, with the same subset also being high in c5 class as observed for TCGA; and neuroendocrine-associated global patterns and markers were associated with c4 class.
Figure 5. Observation of patterns associated with TCGA pan-cancer molecular classes in an external multi-cancer expression profiling dataset.
(A) Gene expression profiles of 2041 cancer cases of various pathologically defined cancer types, represented in the Expression Project for Oncology (expO) (GSE2109) dataset (profiles being normalized within their respective cancer type), were classified according to TCGA pan-cancer molecular class. Expression patterns for the top set of 854 mRNAs distinguishing between the ten TCGA molecular classes (from Figure 1A) are shown for both TCGA and GSE2109 datasets. Genes in the GSE2109 sample profiles sharing similarity with TCGA class-specific signature pattern are highlighted. (B) In the same manner as carried out for TCGA datasets, expO expression profiles were scored for pathway-associated gene signatures (from Figure 2A), surveyed for immune checkpoint markers and for CT antigen genes (from Figure 3B, using the same gene ordering), surveyed for canonical neuroendocrine tumor (NET) markers (from Figure 3C), and scored for similarity to normal cell type categories represented in the fantom dataset (from Figure 3A). Pan-cancer class associations of particular interest (which tend to follow the patterns first observed in TCGA cohort) are highlighted. The purple-cyan heat maps off to the right denote t-statistics for comparing the given class versus the rest of the tumors; dark purple or cyan corresponds approximately to p<0.01. Parts A and B have the same ordering of expO expression profiles. See also Supplementary Figures 14 to 16 and Supplementary Data 6.
In a similar manner to that of the expO dataset, cell line profiles from the Cancer Cell Line Encyclopedia (CCLE) expression dataset(39) were each assigned to a pan-cancer class (Supplementary Figure 16). Not all patterns of interest observable in human tumor data were as apparent in the CCLE results, which could be attributed to a number of factors including growth conditions of cell lines lacking tumor microenvironmental effects and differences in the types of cancers represented in CCLE; however, a number of associations were identifiable in CCLE, including c7 and c8 classes with EMT and hypoxia, and c4 class with neuroendocrine-related patterns (e.g. involving small cell lung cancers).
Discussion
While previous studies have greatly served to elucidate the molecular landscape of the individual cancer types represented in TCGA, our pan-cancer molecular classes provide an excellent framework for examining pathways or processes that would cut across these individual types. All but one of these molecular classes each involves a wide range of cancer types and would therefore be relevant to the study of multiple diseases. It is also remarkable that basal-like breast cancer forms its own molecular class—made up only of breast cancers—entirely distinct from all of the other cancer cases examined. While basal-like breast cancer is already understood to represent a fundamentally different disease than other types of breast cancer, questions remain as to the origins of the observed molecular differences(40). The distinct patterns associated with basal-like breast cancer at the epigenetic as well as transcriptional levels, in this present study, could suggest that this disease would have a different cell-of-origin from that of other breast cancers.
The landscape of pan-cancer-associated biological processes and pathways as uncovered here would include many that were well-established as having a functional role in the experimental setting, but for which the full extent of their involvement in human cancers may have been unclear. Processes and pathways such as metabolism, immune checkpoint, hypoxia, NRF2-ARE, HIPPO, Wnt, EMT, and Notch have been well characterized in the experimental setting, using models such as cell lines and mice(41). This present study finds each of the above to involve large subsets of human cancers, as observed based on the coordinate expression patterns involving large numbers of genes. DNA mutation events alone (and tumor evolution by proxy) would not appear to represent the sole driver of these processes in cancer, where tumor microenvironment influences likely play a major role. For example, mutations in VHL or in RTK genes may accelerate processes of hypoxia or RTK signaling, respectively, or microenvironmental conditions such as lack of oxygen or availabilty of growth factors could conceivably achieve the same respective results. Individual molecular markers of our pan-cancer molecular classes that might seem most relevant from a therapeutic standpoint—including markers of processes and pathways noted above—could potentially be evaluated in the setting of patient care in future studies.
Multiple pan-cancer classes manifested patterns that we could ascribe to the non-cancer cellular component of the tumor, including immune infiltrates and cancer-associated fibroblasts. Our finding of different stroma-associated tumor subsets, each with distinctive features that would distinguish it from the other pan-cancer classes, suggests various biological roles for the stromal component in human cancer. Three of our pan-cancer classes (c3, c5, and c10) showed particularly strong patterns of immune cell infiltrates, while one of these (c3) showing stronger patterns of immune checkpoint pathway genes and of genes involved in specific metabolic pathways. Three of our pan-cancer classes (c3, c7, and c8) showed patterns of mesenchymal cells, with two of these (c7 and c8) showing strong patterns of fibroblasts in particular, and with one of these (c8) showing patterns associated with fatty acid metabolism. The tumor microenvironment—which consists of a mixture of fibroblasts, myofibroblasts, endothelial cells, immune cells, other cells, and altered extracellular matrix—is understood to play an important role in the initiation and progression of various cancers(42,43). For example, EMT of the cancer cells can occur in areas of fibrosis and may account for up to 40% of tumor-associated fibroblasts(43). As reflected in TCGA data, the various ways in which the tumor microenvironment can influence cancer may be involved within distinct subsets of human tumors.
Our discovery of a molecular class of cancers manifesting a differential expression profile associated with both neuroendocrine tumors and with normal cells and tissues of the CNS would potentially have implications for a large subset of human cancers, with ~4% of TCGA cases belonging to this “c4” class. Only through our analytical approach of subtracting out tissue-level molecular differences were we able to uncover this neuroendocrine-associated pan-cancer class (where otherwise such CNS-related patterns would be more strongly associated with TCGA brain cancers). Neuroendocrine tumors are understood to occur throughout the body, including in breast, cervical, lung, pancreas, ovarian, and gastrointestinal tissues, though it is believed that the incidence of these tumors is relatively rare, estimated at about 1% of cancers diagnosed in the United States(44,45). If on the order of 4% of human cancers could be considered as neuroendocrine tumors (or at least manifesting a molecular profile associated with these tumors), this would be well outside the range of previous estimates, suggesting that a large proportion of neuroendocrine tumors are not diagnosed as such, but rather are grouped in with other cancers sharing the same tissue of origin. The tumor samples originally contributed to TCGA came from many different treatment facilities, with no uniform practices for determining neuroendocrine features being in place. Current strategies for identifying neuroendocrine tumors would include staging at surgery, pathological grading, blood Chromogranin A (CgA) measurements, and detection of circulating tumor cells, with there being a clear need for additional and better biomarkers(34). The more accurate identification of neuroendocrine-associated cancers in particular would have important implications regarding the treatment of this disease(46).
Supplementary Material
Translational Relevance.
Unsupervised molecular classification of tumors can reveal major subtypes existing within a given cancer type as defined by tissue of origin. Such molecular-based subtypes can reflect different pathways at work within different cancer subsets, which could have important implications for applying existing therapies or for developing new therapeutic approaches. Here we applied an alternative classification approach, in order to consolidate the individual subtypes that might be discoverable in individual cancer types into super-types or pan-cancer “classes” that transcend tissue or histology distinctions. Coordinate pathways and processes were revealed across the ten pan-cancer classes in our cohort. As reflected in these classes, the tumor microenvironment may influence cancer in different ways between distinct subsets of human tumors. Our molecular class manifesting a differential expression profile of neuroendocrine tumors would have therapeutic implications for an appreciable subset of human cancers.
Acknowledgments
This work was supported in part by Cancer Prevention and Research Institute of Texas (CPRIT) grants RP120713 C2 (C. Creighton), RP150405 (D. Gibbons), and RP120713 P2 (D. Gibbons), and by National Institutes of Health (NIH) grant P30CA125123 (C. Creighton).
Abbreviations
- TCGA
The Cancer Genome Atlas
- RNA-seq
RNA sequencing
- RPPA
reverse-phase protein arrays
Footnotes
Disclosure of potential conflicts of interest: The authors have no conflicts of interest.
References
- 1.Perou C, Sørlie T, Eisen M, van de Rijn M, Jeffrey S, Rees C, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Cancer_Genome_Atlas_Research_Network. Weinstein J, Collisson E, Mills G, Shaw K, Ozenberger B, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nature genetics. 2013;45(10):1113–20. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Creighton C. The molecular profile of luminal B breast cancer. Biologics. 2012;6:289–97. doi: 10.2147/BTT.S29923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Kwok-Shing Ng P, Kucherlapati M, Chen F, Liu Y, Tsang Y, et al. A Pan-Cancer Proteogenomic Atlas of PI3K/AKT/mTOR Pathway Alterations. Cancer Cell. 2017 doi: 10.1016/j.ccell.2017.04.013. E-pub May 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoadley K, Yau C, Wolf D, Cherniack A, Tamborero D, Ng S, et al. Multiplatform Analysis of 12 Cancer Types Reveals Molecular Classification within and across Tissues of Origin. Cell. 2014;158(4):929–44. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen F, Zhang Y, Şenbabaoğlu Y, Ciriello G, Yang L, Reznik E, et al. Multilevel Genomics-Based Taxonomy of Renal Cell Carcinoma. Cell Rep. 2016;14(10):2476–89. doi: 10.1016/j.celrep.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen F, Zhang Y, Parra E, Rodriguez J, Behrens C, Akbani R, et al. Multiplatform-based Molecular Subtypes of Non-Small Cell Lung Cancer. Oncogene. 2016 doi: 10.1038/onc.2016.303. E-pub Oct 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akbani R, Ng P, Werner H, Shahmoradgoli M, Zhang F, Ju Z, et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat Commun. 2014 doi: 10.1038/ncomms4887. E-pub May 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen F, Zhang Y, Bossé D, Lalani A, Hakimi A, Hsieh J, et al. Pan-urologic cancer genomic subtypes that transcend tissue of origin. Nat Commun. 2017;8(1):199. doi: 10.1038/s41467-017-00289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.FANTOM_Consortium_and_the_RIKEN_PMI_and_CLST_(DGT) Forrest A, Kawaji H, Rehli M, Baillie J, de Hoon M, et al. A promoter-level mammalian expression atlas. Nature. 2014;507(7493):462–70. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbons D, Creighton C. Pan-cancer survey of epithelial-mesenchymal transition markers across The Cancer Genome Atlas. Dev Dyn. 2017 doi: 10.1002/dvdy.24485. E-pub 2017 Jan 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ciriello G, Gatza M, Beck A, Wilkerson M, Rhie S, Pastore A, et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell Cycle. 2015;163(2):506–19. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer_Genome_Atlas_Research_Network. Kandoth C, Schultz N, Cherniack A, Akbani R, Liu Y, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497(7447):67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cancer_Genome_Atlas_Research_Network. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163(4):1011–25. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cancer_Genome_Atlas_Research_Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315–22. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cancer_Genome_Atlas_Research_Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–50. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cancer_Genome_Atlas_Research_Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cancer_Genome_Atlas_Research_Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cancer_Genome_Atlas_Research_Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cancer_Genome_Atlas_Research_Network. Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543(7645):378–84. doi: 10.1038/nature21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cancer_Genome_Atlas_Research_Network. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541(7636):169–75. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer_Genome_Atlas_Network. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161(7):1681–96. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cancer_Genome_Atlas_Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–82. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cancer_Genome_Atlas_Research_Network. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med. 2015;372(26):2481–98. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gingras M, Covington K, Chang D, Donehower L, Gill A, Ittmann M, et al. Ampullary Cancers Harbor ELF3 Tumor Suppressor Gene Mutations and Exhibit Frequent WNT Dysregulation. Cell Rep. 2016;14(4):907–19. doi: 10.1016/j.celrep.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, et al. A gene expression signature associated with "K-Ras addiction" reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15(6):489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Creighton C. Multiple oncogenic pathway signatures show coordinate expression patterns in human prostate tumors. PloS one. 2008;3(3):e1816. doi: 10.1371/journal.pone.0001816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCart-Reed A, Kutasovic J, Lakhani S, Simpson P. Invasive lobular carcinoma of the breast: morphology, biomarkers and 'omics. Breast Cancer Res. 2015;17:12. doi: 10.1186/s13058-015-0519-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nieto M, Huang R, Jackson R, Thiery J. EMT: 2016. Cell. 2016;166(1):21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 30.Chang J, Wooten E, Tsimelzon A, Hilsenbeck S, Gutierrez M, Tham Y, et al. Patterns of resistance and incomplete response to docetaxel by gene expression profiling in breast cancer patients. J Clin Oncol. 2005;23(6):1169–77. doi: 10.1200/JCO.2005.03.156. [DOI] [PubMed] [Google Scholar]
- 31.Davis C, Ricketts C, Wang M, Yang L, Cherniack A, Shen H, et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell. 2014;26(3):319–30. doi: 10.1016/j.ccr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf A, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–95. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 33.Ramage J, Ahmed A, Ardill J, Bax N, Breen D, Caplin M, et al. Guidelines for the management of gastroenteropancreatic neuroendocrine (including carcinoid) tumours (NETs) Gut. 2012;61(1):6–32. doi: 10.1136/gutjnl-2011-300831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Modlin I, Drozdov I, Kidd M. The identification of gut neuroendocrine tumor disease by multiple synchronous transcript analysis in blood. PloS one. 2013;8(5):e63364. doi: 10.1371/journal.pone.0063364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cook J, Hagemann T. Tumour-associated macrophages and cancer. Curr Opin Pharmacol. 2013;13(4):595–601. doi: 10.1016/j.coph.2013.05.017. [DOI] [PubMed] [Google Scholar]
- 36.Tsai Y, Wu K. Hypoxia-regulated target genes implicated in tumor metastasis. J Biomed Sci. 2012;19:102. doi: 10.1186/1423-0127-19-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kao S, Wu K, Lee W. Hypoxia, Epithelial-Mesenchymal Transition, and TET-Mediated Epigenetic Changes. J Clin Med. 2016;5(2):E24. doi: 10.3390/jcm5020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Micalizzi D, Farabaugh S, Ford H. Epithelial-mesenchymal transition in cancer: parallels between normal development and tumor progression. J Mammary Gland Biol Neoplasia. 2010;15(2):117–34. doi: 10.1007/s10911-010-9178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin A, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skibinski A, Kuperwasser C. The origin of breast tumor heterogeneity. Oncogene. 2015;34(42):5309–16. doi: 10.1038/onc.2014.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanahan D, Weinberg R. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 42.Dakhova O, Ozen M, Creighton C, Li R, Ayala G, Rowley D, et al. Global gene expression analysis of reactive stroma in prostate cancer. Clin Cancer Res. 2009;15(12):3979–89. doi: 10.1158/1078-0432.CCR-08-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franco O, Shaw A, Strand D, Hayward S. Cancer associated fibroblasts in cancer pathogenesis. Semin Cell Dev Biol. 2010;21(1):33–9. doi: 10.1016/j.semcdb.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao J, Hassan M, Phan A, Dagohoy C, Leary C, Mares J, et al. One hundred years after "carcinoid": epidemiology of and prognostic factors for neuroendocrine tumors in 35,825 cases in the United States. J Clin Oncol. 2008;26(18):3063–72. doi: 10.1200/JCO.2007.15.4377. [DOI] [PubMed] [Google Scholar]
- 45.Basuroy R, Srirajaskanthan R, Ramage J. Neuroendocrine Tumors. Gastroenterol Clin North Am. 2016;45(3):487–507. doi: 10.1016/j.gtc.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 46.Strosberg J, El-Haddad G, Wolin E, Hendifar A, Yao J, Chasen B, et al. Phase 3 Trial of 177Lu-Dotatate for Midgut Neuroendocrine Tumors. N Engl J Med. 2017;376(2):125–35. doi: 10.1056/NEJMoa1607427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aran D, Sirota M, Butte A. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6:8971. doi: 10.1038/ncomms9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cherniack A, Shen H, Walter V, Stewart C, Murray B, Bowlby R, et al. Integrated Molecular Characterization of Uterine Carcinosarcoma. Cancer Cell. 2017;31(3):411–23. doi: 10.1016/j.ccell.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.