Abstract
Acinetobacter baumannii is emerging as a challenging nosocomial pathogen due to its rapid evolution of antibiotic resistance. We report characterization of two novel bacteriophages, PBAB08 and PBAB25, infecting clinically isolated, multidrug-resistant (MDR) A. baumannii strains. Both phages belonged to Myoviridae of Caudovirales as their morphology observed under an electron microscope. Their genomes were double stranded linear DNAs of 42,312 base pairs and 40,260 base pairs, respectively. The two phages were distinct from known Acinetobacter phages when whole genome sequences were compared. PBAB08 showed a 99% similarity with 57% sequence coverage to phage AB1 and PBAB25 showed a 97% similarity with 78% sequence coverage to phage IME_AB3. BLASTN significant alignment coverage of all other known phages were <30%. Seventy six and seventy genes encoding putative phage proteins were found in the genomes of PBAB08 and PBAB25, respectively. Their genomic organizations and sequence similarities were consistent with the modular theory of phage evolution. Therapeutic efficacy of a phage cocktail containing the two and other phages were evaluated in a mice model with nasal infection of MDR A. baumannii. Mice treated with the phage cocktail showed a 2.3-fold higher survival rate than those untreated in 7 days post infection. In addition, 1/100 reduction of the number of A. baumannii in the lung of the mice treated with the phage cocktail was observed. Also, inflammatory responses of mice which were injected with the phage cocktail by intraperitoneal, intranasal, or oral route was investigated. Increase in serum cytokine was minimal regardless of the injection route. A 20% increase in IgE production was seen in intraperitoneal injection route, but not in other routes. Thus, the cocktail containing the two newly isolated phages could serve as a potential candidate for therapeutic interventions to treat A. baummannii infections.
Keywords: multidrug-resistance, Acinetobacter baumannii, bacteriophage therapy, mouse model, genome analysis
Introduction
Acinetobacter baumannii is an opportunistic pathogen causing nosocomial infection in hospitals. It is designated as an ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumonia, A. baumannii, Pseudomonas aeruginosa, and Enterobacter species) pathogen by World Health Organization (WHO) (McConnell et al., 2013). It causes mainly pneumonia and additionally burn infections, meningitis, urinary tract infections, and sepsis (Dijkshoorn et al., 2007). Antibiotic- resistant strains are rapidly emerging and even multidrug-resistant (MDR) and pandrug-resistant (PDR) strains are observed (Tuon et al., 2015). Its virulence factors include porins, lipopolysaccharides, capsular polysaccharides, metal acquisition systems, phospholipases, outer membrane vesicles, and protein secretion systems (Weber et al., 2016; Lee et al., 2017). Factors influencing antibiotic resistance includes two-component systems AdeRS, BaeSR, GacSA, and PmrAB (Kröger et al., 2017). Treatment of carbapenem-resistant A. baumannii (CRAB) involves the use of combinations of last resort agents including colistin and tigecycline, but the efficacy and safety issues are not cleared yet (Doi et al., 2015).
Phage therapy is a promising tool for controlling drug-resistant bacteria (Abedon, 2017; Lin et al., 2017). The mechanism of drug-resistance is totally unrelated to that of phage infection. Although bacteriophages have been used as an antibacterial for almost 100 years, mainly in eastern European countries, they are not recognized as drugs due to the lack of a proper documentation needed for drug approval. In addition, we still wait for fulfillment of regulatory affairs to approve phage drugs (Huys et al., 2013).
Novel therapies including bacteriophages against drug-resistant A. baumannii have been reported and reviewed (Mihu and Martinez, 2011; García-Quintanilla et al., 2013; Parasion et al., 2014). Successful phage control of various strains of Acinetobacter was demonstrated not only in vitro but also in vivo. Two newly isolated phages infecting A. baumannii were characterized and suggested as potential candidates for phage cocktail (Merabishvili et al., 2014). Other two newly isolated phages were characterized at genomic DNA level and suggested as potential candidate for phage cocktail against CRAB (Jeon et al., 2016a). Phage BΦ-C62 was used to successfully control CRAB infection via nasal route in mice model (Jeon et al., 2016b). Phage vB-GEC_Ab-M-G7 was used to successfully control wound infection in a rat model (Kusradze et al., 2016). Medes et al. tested phage cocktails against diabetic cutaneous wounds in two animal models. Phage cocktails for Staphylococcus aureus or Pseudomonas aeruginosa improved wound healing, but cocktail for A. baumannii was not as effective. A personalized phage cocktail for treating mouse dorsal wound model was reported (Regeimbal et al., 2016). Phages from multi-institute libraries were used to make a personalized cocktail and proven effective for treating a diabetic human patient with necrotizing pancreatitis complicated by an MDR A. baumannii infection (Schooley et al., 2017). A phage was effective in resolving wound infection caused by multidrug-resistant A. baumannii in an uncontrolled diabetic rat model (Shivaswamy et al., 2015). A bacteriophage-containing aerosol was proven effective for cleaning and decreased the rates of infection caused by CRAB in intensive care units (Wang et al., 2016). A combined lysis spectrum of four lytic phages against clinically isolated CRAB was reported to be 87.5% and phages were proven effective as therapeutic agents for lung infection without deleterious side effects in mice model (Hua et al., 2018).
Here, we report both microbiological characterization of novel phages infecting A. baumannii and the efficacy of a phage cocktail containing the phages in control of nasal infection in a mice model. Further, we provide the basis why the phages can be used as a therapeutic intervention practically.
Materials and methods
Ethics statement
All animal studies were approved by and followed the guidelines and regulations of the Ethical Committee for Animal Experiments of Hankuk University of Foreign Studies (approval number 2017-0001).
Bacterial strains and phages used
Fourteen clinical A. baumannii strains were obtained from patients in Samsung Medical Center, Sungkyukwan University School of Medicine. Antibiotic resistance profile of the strains is shown in Table 1. To determine the genotypes of the A. baumannii isolates, Oxford scheme multilocus sequence typing (MLST) was performed as described previously (Bartual et al., 2005; http://pubmlst.org/abaumannii/info/primers_Oxford.shtml). One of the strains (strain 28) was selected and further used for mice experiment. It was resistant to both kanamycin (3.5 mg/ml, Sigma, USA) and ampicillin (5 mg/ml, Sigma, USA), and this fact was used to enumerate recovered bacteria from mice lung. The 9 bacteriophages tested against the 14 strains were PBAB05, PBAB06, PBAB07, PBAB08, PBAB25, PBAB68, PBAB80, PBAB87, and PBAB93. Bacteriophages used for making the therapeutic cocktail were PBAB08, PBAB25, PBAB68, PBAB80, PBAB93 (Bacteriophage Bank of Korea).
Table 1.
Antibiotic resistance profile of clinically isolated Acinetobacter baumannii strains and their phage susceptibility.
| Antibiotics/ name of isolate | Sequence type (ST) | Imipenem | Meropenem | Ampicillin/sulbactam | Ceftazidime | Cefepime | Gentamicin | Ciprofloxacin | Levofloxacin | Trimethoprim/ Sulfamethoxazole | Piperacillin/tazobac | colistin | Tigecycline | PBAB number* of infecting phages |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain 21 | 191 | >64** | >64 | >64 | >64 | >64 | >64 | >64 | 64 | >64 | >256 | 1 | 8 | none |
| Strain 32-a | 191 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >256 | 1 | 8 | none |
| Strain F-224 | 1240 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >32 | >64 | >256 | 1 | 8 | 5, 6, 7, 87 |
| Strain F-1208 | 357 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | 32 | >64 | >256 | >64 | 1 | 8, 68, 80, 93 |
| Strain F-1510 | 191 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | 16 | >64 | >256 | 2 | 8 | none |
| Strain F-1629 | 357 | >64 | >64 | 32 | >64 | 64 | >64 | >64 | 32 | >64 | >256 | >64 | 8 | 8, 68, 80, 93 |
| Strain 26 | 368 | 16 | 16 | 32 | 64 | 64 | 16 | 4 | 32 | 32 | 128 | 64 | 4 | 8, 68, 80, |
| Strain 28 | 368 | 16 | 16 | 16 | 64 | 64 | 16 | 4 | 32 | 32 | 128 | 1 | 8 | 8, 25, 68, 80, 93 |
| Strain 32-b | 357 | 16 | 16 | 16 | 64 | 64 | 16 | 4 | 32 | 32 | 128 | 4 | 2 | 8, 68, 80, 93 |
| Strain 54 | 208 | 16 | 16 | 32 | 64 | 64 | 16 | 4 | 8 | 32 | 128 | 1 | 2 | none |
| Strain 58 | 208 | 16 | 16 | 32 | 64 | 64 | 16 | 4 | 8 | 32 | 128 | 1 | 2 | none |
| Strain 81 | 191 | 16 | 16 | 32 | 64 | 64 | 16 | 4 | 8 | 32 | 128 | 4 | 4 | 5, 6, 7, 87 |
| Strain K20-B-667 | 191 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >256 | 1 | 8 | none |
| Strain K20-B-890 | 191 | 8 | 64 | >64 | 32 | 64 | >64 | >64 | >64 | >64 | 256 | 1 | 32 | none |
PBAB numbers according to the Bacteriophage Bank of Korea (www.phagebank.or.kr).
The numbers in each box shows the maximum concentration of each antibiotic to which tested bacteria was resistant (mg/L).
Darkly shaded box means “resistant”, lightly shaded box “intermediate”, and white box “susceptible”.
Phage isolation and purification
Bacteriophage isolation and characterization were described thoroughly (Clokie and Kropinski, 2009a,b; Clokie et al., 2018). The isolation and characterization of phages have been described previously (Kim et al., 2013). The phages were purified using a glycerol gradient centrifugation method (Sambrook and Russell, 2006). A single plaque was used to inoculate 5 ml of a mid-exponential-phase culture of the bacterium, followed by incubation at 37°C for 3 h. A phage lysate was obtained by centrifugation at 11,000 xg for 10 min and discarding the supernatant. Five ml of the lysate was used to inoculate 100 ml of a mid-exponential-phase culture of the bacterium, and the mixture was then incubated until lysis was completed. NaCl was added to the lysate at a final concentration of 1 M, and it was incubated at 4°C for 1 h. After centrifugation at 11,000x g for 10 min, 10% (wt/vol) polyethylene glycol 8000 (PEG 8000) was added, and the mixture was then incubated at 4°C for 1 h. The supernatant was discarded after centrifugation at 11,000x g for 10 min. Then the pellet was resuspended in 750 μl of SM buffer [100 mM NaCl, 8 mM MgSO4·7H2O, 50 mM Tris-Cl (pH 7.5)], and chloroform was added at a ratio of 1:1 (vol/vol), followed by vortexing and then centrifugation at 3,000x g for 15 min. The upper phase was isolated and was added to a polycarbonate centrifuge tube containing 3 ml of 40% glycerol in the lower layer and 4 ml of 5% glycerol in the upper layer. After centrifugation at 151,000x g for 1 h, the supernatant was discarded, and the pellet was resuspended in 400 μl of SM buffer. For animal experiments, any contaminating lipopolysaccharide (LPS) was removed before treatment. Triton X-114 (Sigma, USA) was added to phage solution at a final concentration of 1% (vol/vol) and mixed using vortex for 10 s. After incubation on ice for 10 min, the mixture was transferred to 37°C water bath and incubated for 1 min. After a centrifugation at 15,000x g for 5 min, supernatant was collected and used as the final phage solution. Purified phages were stored at 4°C until use. Standard double agar overlay plaque technique was used for phage enumeration (Kropinski et al., 2009).
Transmission electron microscopy (TEM) of phage particles
Purified phage sample was loaded onto a copper grid for 1 min followed by negative staining with 2% (vol/vol) uranyl acetate (pH 6.7) and drying. The phage morphology was observed using a Carl Zeiss LIBRA 120 EF-TEM (Carl Zeiss, Oberkochen, Germany) at an accelerating voltage of 120 kV.
Genome sequencing, annotation, and analysis
Whole genome sequencing of phage DNA was carried out using PacBio RS II system in Macrogen, Korea. Open reading frames (ORFs) were searched using NCBI ORF Finder. Genomic sequence similarity comparison was done using MAUVE. ORF map was drawn using CLC Genomics Workbench 10. Genomic tree was drawn using Mega 7.
One step multiplication
One step growth experiment was described previously (Hyman and Abedon, 2009). Briefly, phage PBAB08 or PBAB25 was added to a fresh culture of A. baumannii at an MOI 0.001 and allowed to adsorb for 5 min. The mixture was then centrifuged at 1,738 xg for 10 min. The supernatant was discarded to remove any free phages and the pellet was resuspended in 3 ml of fresh LB broth incubated at 37°C with shaking. One hundred micro liters of sample was removed from the culture every 5 min and subjected to titration using double-layer agar plate methods (Kropinski et al., 2009). This assay was performed at least in triplicate.
Phage stability
For temperature stability test, phage titer was measured after incubation of phage lysates for 1 h at different temperatures (4, 37, 45, 55, 60, 65, or 80°C) using a double-layer agar plate method (Kropinski et al., 2009). For pH stability test, phage titer was measured after 1 h's incubation of phage lysates at room temperature by mixing with equal volume of pH buffer solutions of different pH values (pH 3, 4, 5, 6, 7, 8, 9, 10, and 11). pH buffer solutions were made by adding HCl drop by drop to 1 M NaOH solution. The resulting pH was measured using S220 seven compact™ pH/Ion (Mettler Toledo, Columbus, USA).
Phage protection studies using a mice model
Six week-old female Balb/c mice were obtained from Young Bio, Korea. Mice were divided into 4 groups; Group 1 was treated with SM buffer only. Group 2 was infected with MDR A. baumannii (strain 28) only. Group 3 was treated with the phage cocktail only. Group 4 was infected with A. baumannii and treated with the phage cocktail. Each group contained 20 mice. Mice were intraperitoneally injected with cyclophosphamide (Sigma, USA) at the concentration of 150 mg/kg at −1 and −3 days of bacterial infection. Before intranasal bacterial or phage injection, mice were anesthetized with 125 mg/kg of avertin (Sigma, USA) by intraperitoneal injection. 1 × 108 CFU of A. baumannii was intranasally injected to each mouse at 0 and +1 days. SM buffer was injected to the control group mice instead. 1 × 109 PFU of phage cocktail was intranasally injected to experimental mice every day from −1 to +7 days. At days 0 and +1, phages were injected 4 h after bacterial injection. Survival of mice was observed until 7 days after bacterial infection. Bacterial load in mice lung was counted as follows; lung was isolated at +3 and +4 days and weight was measured. After homogenization, 500 μl of SM buffer was added and was subjected to centrifugation at 2,000x g. The supernatant was used for counting A. baumannii in a medium containing both kanamycin (3.5 mg/ml) and ampicillin (5 mg/ml).
Cytokine, IgE, and histamine assays
To see immune reactions against phages, the cocktail was introduced every day for 7 days to mice in three different routes; intraperitoneal, intranasal, and oral. Three mice were used for each route. Control group was treated with SM buffer. One day after the last treatment of a phage cocktail, mice were sacrificed and serum was obtained for analysis of cytokine profile using Multi-Analyte ELISArray kits (Qiagen, USA), of IgE profile using RayBio®; Mouse IgE ELISA Kit (RayBiotech, USA), and of histamine using Histamine Enzyme Immunoassay Kit (SPI Bio, France).
Statistical analyses
Student's t-test (Bailey, 1959) was used to calculate the significance of the difference among test groups. For each assay, all determinations were carried out at least in triplicate. Statistically significant values were defined as P < 0.05 or P < 0.01.
Results
Antibiotic resistance of clinically isolated A. baumannii strains
Fourteen bacterial isolates were selected and their antibiotic resistance and phage susceptibility were observed (Table 1). All the isolates were multidrug-resistant (MDR) strains. Many of them were resistant even to colistin or tigecycline. The 14 A. baumannii isolates showed five sequence types (STs) based on MLST (Table 1), and they belonged to the same clonal complex (CC), global complex II. Nine Acinetobacter bacteriophages, PBAB05, PBAB06, PBAB07, PBAB08, PBAB25, PBAB68, PBAB80, PBAB87, and PBAB93, were selected from the Bacteriophage Bank of Korea and tested for infection to the clinical A. baumannii strains. Seven out of 14 isolates could be infected by four or five phages. The phage susceptibility was not related to antibiotic resistance profile. Isolate 28 was susceptible to all five phages and thus selected for mice infection experiments later. We further characterized two of the phages PBAB08 and PBAB25 from which whole genomic DNA sequences were successfully obtained as single contigs after next generation sequencing.
Bacteriophage characterization
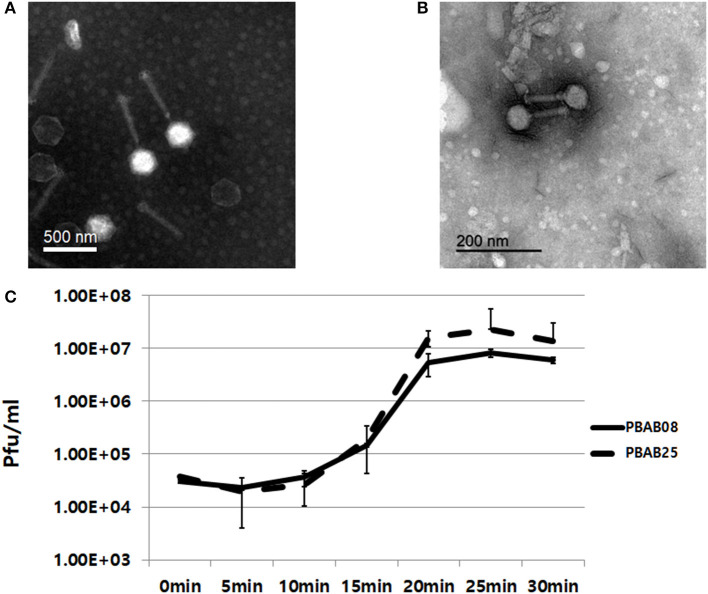
Two phages, PBAB08 and PBAB25, belonged to family Myoviridae of order Caudovirales (Figures 1A,B). Their virions were composed of a spherical head and a long, rigid tail. Phage PBAB08 had a head of 180 nm in diameter and a tail of 360 nm in length. The smaller phage PBAB25 had a head of 80 nm in diameter and a tail of 90 nm in length. A complete lysis of host bacteria infected with PBAB08 or PBAB25 occurred in 25 min post infection (Figure 1C). Burst size for each phage was 215 and 630, respectively.
Figure 1.
Transmission electron micrographs of bacteriophages PBAB08 (A) and PBAB25 (B). Samples were negatively stained with uranyl acetate. Scale bar is shown in each picture. (C) One step multiplication of phages PBAB08 (solid line) and PBAB25 (broken line).
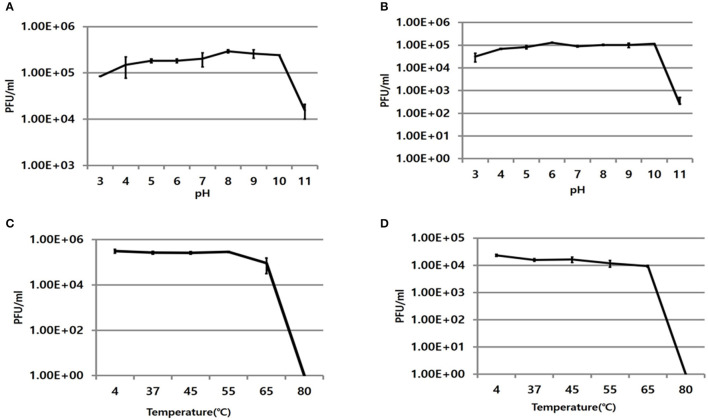
The infectivity of PBAB08 and PBAB25 remained intact when exposed to pHs ranging from 5 to 10 for 1 h (Figures 2A,B). It decreased rapidly at pHs above 10. The infectivity of both phages remained intact when exposed to temperatures ranging from 4 to 55°C for 1 h and dropped rapidly above 65°C (Figures 2C,D).
Figure 2.
Stability of phages at various pHs and temperatures. Remaining infectivity of phages PBAB08 (A) and PBAB25 (B) were measured after exposure of phages to indicated pHs for 1 h. Remaining infectivity of phages PBAB08 (C) and PBAB25 (D) were measured after exposure of phages to indicated temperatures for 1 h. The experiments were carried out in a triplicate.
Genomic sequence analysis
The whole genomes of phages PBAB08 and PBAB25 were sequenced (GenBank accession numbers MG366114 and MG366115, respectively). Their genomes were double stranded linear DNAs of 42,312 base pairs and 40,260 base pairs, respectively. The genome sequences were compared to reported Acinetobacter phages using BLASTN. PBAB08 showed a 99% similarity with 57% sequence coverage to phage AB1 (Yang et al., 2010), and PBAB25 showed a 97% similarity with 78% sequence coverage to phage IME_AB3 (Zhang et al., 2015). BLASTN significant alignment coverage of all other known phages were <30%. A mosaic structure between the genomes of PBAB08 and AB1, and that of PBAB25 and IME_AB3 were observed (Figure 3). An extensive rearrangement occurred between genomes of PBAB08 and AB1, while genomic structure was conserved between PBAB25 and IME_AB3.
Figure 3.
Genomic sequence comparison between phages PBAB08, AB1, PBAB25, and IME_AB3 using MAUVE. Line-connected colored boxes indicate regions of sequence similarity in corresponding phage genomes.
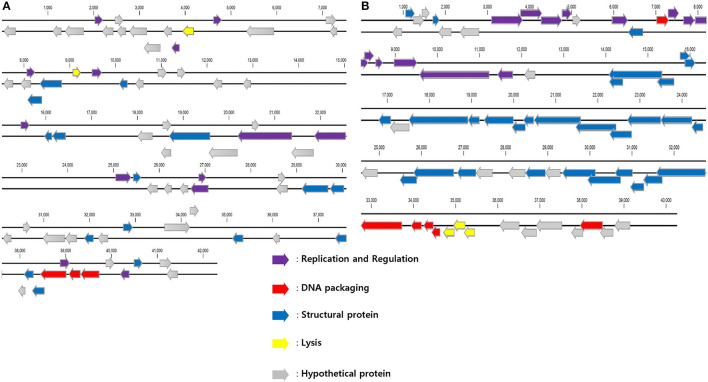
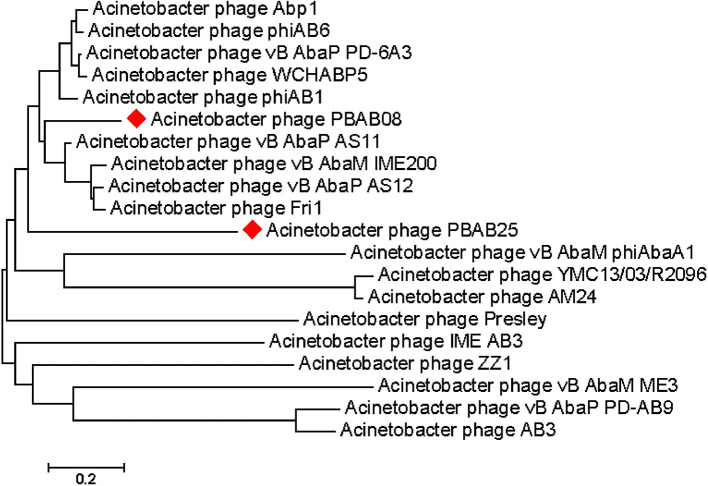
Seventy six and seventy genes encoding phage proteins were found in the genomes of PBAB08 and PBAB25, respectively (Figure 4). Functional annotation of putative ORFs of each phage is shown in Tables 2, 3. They are divided into four categories according to functions; structural proteins, those involved in DNA packaging, those involved in replication and regulation, and those involved in lysis. It was evident that sequence similarities for both pairs were more prominent in virion proteins than in proteins involved in replication and regulation. It is not fair to compare virion proteins of the two pairs in parallel since the extent of annotations were different. Nevertheless, it is notable that PBAB25 had a wide array of tail proteins suggesting a complicated tail structure. Since both phages had their own putative DNA polymerases, a phylogenetic tree of closely related Acinetobacter phages in GenBank was drawn based on the sequence comparison of their DNA polymerase genes (Figure 5). The phages could be grouped as two, in which PBAB08 belonged to one group (upper 10 phages in Figure 5), while PBAB25 belonged to the other group (lower 10 phages). No DNA polymerase was found in functional annotation of ORFs of phage AB1, thus it was not included in the tree.
Figure 4.
Putative open reading frame (ORF) map of phages PBAB08 (A) and PBAB25 (B). ORF map was drawn using CLC Genomics Workbench 10. Each arrow is color-coded according to annotated genomic function.
Table 2.
Functional annotation of putative ORFs found in bacteriophage PBAB08.
| ORF | Function | Related bacteria or phage | Putative encoded phage protein | Similarity (%) |
|---|---|---|---|---|
| 9 | Structural protein | Acinetobacter phage WCHABP5 | Putative internal virion protein B | 100 |
| 11 | Acinetobacter phage vB_AbaP_PD-6A3 | Putative internal virion core protein | 91 | |
| 33 | Acinetobacter phage phiAB6 | Putative internal virion protein | 75 | |
| 46 | Acinetobacter phage AB3 | Tail tubular protein B | 100 | |
| 56 | Acinetobacter phage YMC11/12/R2315 | Putative head protein | 92 | |
| 61 | Acinetobacter phage phiAC-1 | Putative capsid protein | 95 | |
| 66 | Acinetobacter phage phiAC-1 | Putative tail fiber protein | 88 | |
| 68 | Acinetobacter phage AB3 | Putative internal virion protein B | 73 | |
| 69 | Acinetobacter phage AB3 | Putative internal virion protein B | 97 | |
| 73 | Acinetobacter phage IME-AB2 | Putative phage head portal protein | 93 | |
| 79 | Acinetobacter bacteriophage AP22 | Putative portal protein | 91 | |
| 86 | Acinetobacter phage phiAC-1 | Putative capsid protein | 95 | |
| 99 | Acinetobacter phage IME-AB2 | Putative phage head portal protein | 95 | |
| 100 | Acinetobacter phage YMC-13-01-C62 | Putative portal protein | 98 | |
| 111 | Acinetobacter phage YMC-13-01-C62 | Putative portal protein | 90 | |
| 114 | Acinetobacter baumannii 99063 | Putative membrane protein | 94 | |
| 121 | Acinetobacter phage WCHABP12 | Putative tail fiber protein | 75 | |
| 54 | DNA packaging | Acinetobacter phage IME-AB2 | Putative phage terminase large subunit | 98 |
| 78 | Acinetobacter phage IME-AB2 | Putative phage terminase large subunit | 93 | |
| 110 | Acinetobacter phage IME-AB2 | Putative phage terminase large subunit | 90 | |
| 2 | Replication and Regulation | Harpegnathos saltator | Apolipoprotein D | 77 |
| 7 | Acinetobacter phage vB_AbaM_IME200 | DNA polymerase I | 73 | |
| 16 | Stenotrophomonas maltophilia | Adenine phosphoribosyltransferase (fragment) | 63 | |
| 20 | Acinetobacter phage vB_AbaM_IME200 | Carboxypeptidase | 95 | |
| 24 | Massilia sp. JS1662 | Amino acid transporter | 50 | |
| 29 | Staphylococcus aureus subsp. aureus | Nickel ABC transporter, nickel/metallophore periplasmic binding domain protein | 89 | |
| 40 | Rathayibacter sp. VKM Ac-2630 | Transcriptional regulator | 75 | |
| 52 | Burkholderia sp. WSM4176 | TonB-dependent receptor | 60 | |
| 109 | Acinetobacter phage WCHABP12 | Global DNA-binding transcriptional dual regulator | 87 | |
| 116 | Acinetobacter phage YMC-13-01-C62 | Putative RNA polymerase | 92 | |
| 119 | Acinetobacter phage YMC-13-01-C62 | Putative baseplate assembly protein | 98 | |
| 120 | Acinetobacter phage WCHABP1 | Baseplate J-like protein | 98 | |
| 125 | Acinetobacter phage WCHABP1 | Putative RNA polymerase | 89 | |
| 19 | Lysis | Acinetobacter phage WCHABP5 | Putative holin | 51 |
| 74 | Acinetobacter phage WCHABP5 | Putative holin | 83 |
Table 3.
Functional annotation of putative ORFs found in PBAB25.
| ORF | Function | Relative bacteria or phage | Putative encoded phage protein | Similarity |
|---|---|---|---|---|
| 46 | Structural protein | Acinetobacter phage vB_AbaM_ME3 | Baseplate hub protein | 97 |
| 67 | Acinetobacter phage vB_AbaM_ME3 | Head completion protein | 100 | |
| 77 | Acinetobacter phage vB_AbaM_ME3 | Baseplate hub protein | 100 | |
| 106 | Acinetobacter phage IME_AB3 | Putative portal protein | 100 | |
| 107 | Acinetobacter phage IME_AB3 | Putative scaffold protein | 91 | |
| 109 | unclassified Siphoviridae | Putative tail terminator protein | 72 | |
| 110 | Acinetobacter phage IME_AB3 | Putative major tail tube protein | 98 | |
| 112 | Acinetobacter phage IME_AB3 | Putative tail chaperonin protein | 100 | |
| 113 | Acinetobacter phage IME_AB3 | Putative tail tape measure protein | 87 | |
| 114 | unclassified Siphoviridae | Putative distal tail protein | 86 | |
| 116 | Acinetobacter phage IME_AB3 | Putative capsid and scaffold protein | 100 | |
| 135 | Acinetobacter phage IME_AB3 | Putative head protein | 99 | |
| 136 | Acinetobacter phage IME_AB3 | Putative major capsid protein | 100 | |
| 138 | Acinetobacter phage IME_AB3 | Putative structural protein | 95 | |
| 141 | Acinetobacter phage IME_AB3 | Putative tail tape measure protein | 94 | |
| 142 | unclassified Siphoviridae | Putative distal tail protein | 86 | |
| 144 | Acinetobacter phage IME_AB3 | Putative tail protein | 91 | |
| 167 | Acinetobacter phage IME_AB3 | Putative portal protein | 100 | |
| 168 | Acinetobacter phage IME_AB3 | Putative head protein | 98 | |
| 171 | Acinetobacter phage IME_AB3 | Putative major tail tube protein | 98 | |
| 172 | Acinetobacter phage IME_AB3 | Putative tail completion protein | 100 | |
| 173 | Acinetobacter phage IME_AB3 | Putative tail tape measure protein | 67 | |
| 174 | Acinetobacter phage IME_AB3 | Putative tail tape measure protein | 100 | |
| 175 | Acinetobacter phage IME_AB3 | Putative capsid and scaffold protein | 99 | |
| 176 | Acinetobacter phage IME_AB3 | Putative capsid and scaffold protein | 100 | |
| 177 | Acinetobacter phage IME_AB3 | Putative tail protein | 100 | |
| 178 | Acinetobacter phage IME_AB3 | Putative tail protein | 99 | |
| 179 | Acinetobacter phage IME_AB3 | Putative tail protein | 90 | |
| 184 | Acinetobacter phage vB_AbaM_ME3 | Putative membrane protein | 98 | |
| 38 | DNA packaging | Acinetobacter phage IME_AB3 | Putative RecB exonuclease | 100 |
| 101 | Acinetobacter phage IME_AB3 | Putative terminase small subunit | 97 | |
| 103 | Acinetobacter phage IME_AB3 | Putative terminase large subunit | 96 | |
| 128 | Acinetobacter phage IME_AB3 | Putative endonuclease | 81 | |
| 133 | Acinetobacter phage IME_AB3 | Putative terminase large subunit | 97 | |
| 166 | Acinetobacter phage IME_AB3 | Putative terminase small subunit | 94 | |
| 5 | Replication and regulation | Acinetobacter phage IME_AB3 | Putative replicative primase/helicase | 97 |
| 9 | Acinetobacter phage IME_AB3 | Putative single-stranded DNA-binding protein | 100 | |
| 10 | Acinetobacter phage IME_AB3 | Putative DNA/RNA helicase protein | 84 | |
| 11 | Acinetobacter phage IME_AB3 | Putative DNA polymerase subunit | 95 | |
| 36 | Acinetobacter phage IME_AB3 | Putative replicative primase/helicase | 99 | |
| 37 | Acinetobacter phage IME_AB3 | Putative MazG pyrophosphatase | 88 | |
| 39 | Acinetobacter phage IME_AB3 | Putative DNA/RNA helicase protein | 42 | |
| 40 | Acinetobacter phage IME_AB3 | Putative DNA polymerase subunit | 99 | |
| 71 | Acinetobacter phage IME_AB3 | Putative replicative primase/helicase | 100 | |
| 73 | Acinetobacter phage vB_AbaM_ME3 | Putative DNA polymerase subunit | 100 | |
| 74 | Acinetobacter phage IME_AB3 | Putative DNA polymerase subunit | 84 | |
| 121 | Acinetobacter phage vB_AbaM_ME3 | Adenine/guanine phosphoribosyltransferase | 97 | |
| 147 | Acinetobacter phage vB_AbaM_ME3 | Chemical-damaging agent resistance protein C | 94 | |
| 131 | Lysis | unclassified Siphoviridae | Putative holin, class II | 78 |
| 132 | Acinetobacter phage IME_AB3 | Putative endolysin | 99 | |
| 165 | Acinetobacter phage IME_AB3 | putative endolysin | 99 |
Figure 5.
Genomic tree of Acinetobacter phages found in GenBank. The tree was drawn based on each phage's DNA polymerase gene sequence using Mega 7.
In vivo efficacy of phage cocktail
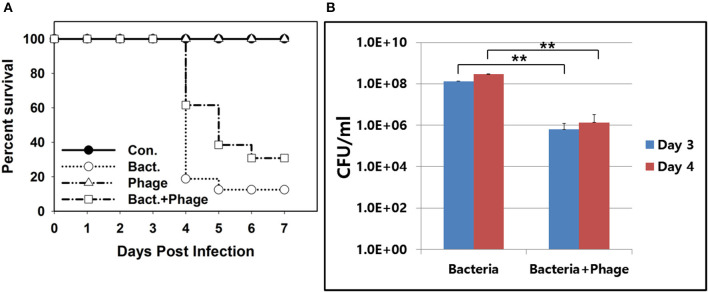
Only 15% of mice infected with A. baumannii intranasally survived at 7 days post infection (Figure 6A). On the contrary, 35% of mice infected with the bacteria followed by treatment with the phage cocktail survived. A better survival of phage-treated mice was observed at day 4 post infection. Sixty percentage of mice survived with phage treatment while 20% of mice survived with only bacterial infection. Mice treated with phage only remained healthy for the entire experimental period.
Figure 6.
In vivo efficacy of the phage cocktail containing PBAB08 and PBAB25 in a mice nasal infection model. (A) Survival of 4 groups of mice observed for 7 days after bacterial infection. Group 1 (closed circle) was inoculated with SM buffer only. Group 2 (open circle) was infected with MDR A. baumannii strain 28 only. Group 3 (triangle) was treated with the phage cocktail only. Group 4 (rectangle) was infected with A. baumannii and treated with the phage cocktail. (B) Bacterial load in lungs of infected mice in 3 and 4 days post-infection with or without phage treatment. **P < 0.01.
Reduction of bacterial load in infected lung of mice was observed (Figure 6B). At 3 and 4 days post bacterial infection, bacteria resistant to both kanamycin and ampicillin were counted. Double-resistant bacterial count was reduced more than 100-fold for both days. Nevertheless, it was clear that a complete elimination of A. baumannii was not achieved with this phage treatment.
Immune reactions against the phage cocktail
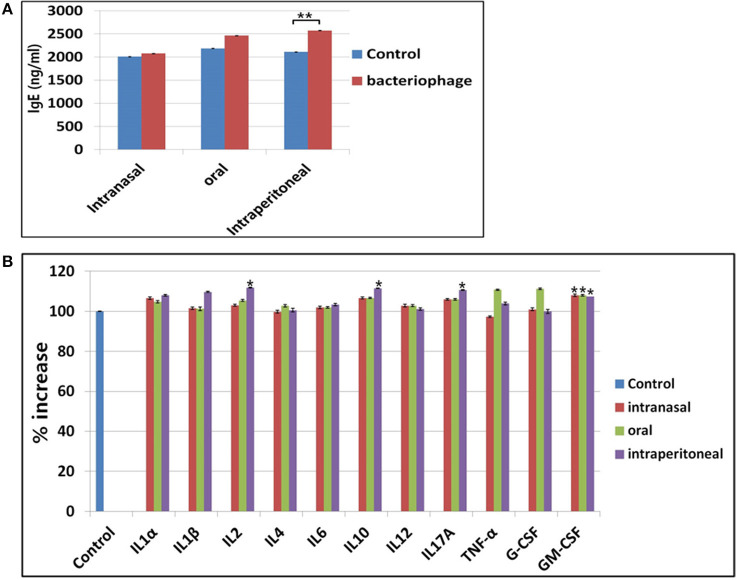
Phages are foreign substances to mice and there are chances they could elicit immune responses. We checked changes in serum IgE level indicating any allergic responses for three different routes of phage injection (Figure 7A). A 20% increase in serum IgE was observed for intraperitoneal injection route, while no significant change was observed for either intranasal or oral administration. For cytokines, a slight increase of serum GM-CSF was observed for all three routes of phage administration (Figure 7B). In intraperitoneal route, slight increases of IL2, IL10, and IL17A were also observed. Thus, inflammatory response against the phage cocktail was minimal. No significant change in histamine level was observed when phage cocktail was administered (Supplementary Figure 1).
Figure 7.
Observation of immune responses against phage treatment in mice. (A) Changes in IgE production after treatment of phages in three different routes. (B) Changes in cytokine production after phage treatment in intranasal, oral, or intraperitoneal route. *P < 0.05, **P < 0.01.
Discussion
Among the clinically isolated MDR A. baumannii strains used in this study, some of them were infected by three or more phages, while the others were not infected by any of the five phages used. The degree of drug-resistance of a host bacterium was not related to the number of phages infecting the same host, indicating that acquirement of drug resistance, probably by horizontal gene transfer (Huang et al., 2015; Krahn et al., 2016; Zhang et al., 2017) did not involve alteration of phage receptor or replication mechanisms. It further suggested that MDR bacteria could be treated with phages as long as infecting phages could be isolated.
Head size of PBAB08 (180 nm) was twice that of PBAB25 (80 nm). Yet, the lengths of genomic DNAs of the two phages were about the same. For example, the genome size of phage T4 is 169 kilobase pair and the head size varies between 85 and 120 nm (Keller et al., 1988). Although a minimal requirement should be enough room for containing entire phage genomic DNA, it seems that there are other determinants of the head size of a phage. In the case of T4, head size determinant included scaffolding core and the shell of the procapsid, mutants of which resulted in varied head sizes (Keller et al., 1988).
From pH and temperature stability data, phage infectivity should remain intact at temperatures and pHs reaching well outside normal human physiological conditions. This should be a good characteristics if the phages are intended for therapeutic applications. A phage preparation in a suitable buffer kept in a refrigerator could remain stable until it encounters target bacteria when applied in vivo.
A mosaic in genomic structure observed from closely related phages in this study is consistent with the modular theory of phage evolution where phage genomes are mosaics of modules that recombines freely in genetic exchanges involving different phages (Botstein, 1980). It was also observed in other viruses including P2-like phages (Davies and Lee, 2006) and even animal viruses (Botstein, 1980). It is no wonder that viruses evolve rapidly by exchanging genetic materials and extend their diversity. The ease of such exchange in a host cell could be one reason why phages are the most divergent biological entities on earth. In the case of phage AB1 which lacks its own DNA polymerase gene, the phage should depend on host DNA polymerase for its own replication. Still its genome retains genomic mosaic to PBAB08, suggesting that modular recombination among phage genomes was independent of their replication strategy. It should be noted that DNA polymerases of PBAB25 and phage Presley was closest in sequence, but overall similarity between their genomic sequences was <30%. It suggests that multiple rounds of recombination events has occurred among various strains of phages during the evolution.
It is noteworthy that only 3 genes encoding putative tail proteins in PBAB08 with a longer tail was annotated while 14 genes annotated in PBAB25 with much shorter tail. It seems that tail length is not related to complexity of its structure. Although electron microscopy of phage IME_AB3, closest to PBAB25 in genomic DNA sequence, was not clear enough to tell the tail structure (Zhang et al., 2015), the two closely related phages of IME_AB3, Acinetobacter phage vB_AbaS_Loki (Turner et al., 2017) and Achromobacter phage phiAxp-1 (Li et al., 2016), have noncontractile tails and are classified as Siphoviridae, while PBAB25 belonged to Myoviridae. Thus it suggests that sequence similarity of tail components does not ensure the similarity in tail morphology.
More than two-fold increase of survival from mice treated with phage cocktail was shown in this study. Although it proved a limited effectiveness, there could be a room for improvements. In a previous report, mice survival after phage treatment in a nasal infection model was higher than this experiment (Jeon et al., 2016b). But it should be noted that in the same report mice were inoculated with phages only after 30 min of bacterial infection, in which bacterial proliferation time was much shorter before confronting phages. In this study, there was a 4 h gap between bacterial infection and phage inoculation. In another report, open wound in rats were treated with phages after A. baumannii infection (Kusradze et al., 2016). Phage treatment was done at 12 h after bacterial infection, but showed a higher reduction of bacterial load than this study. Depending on types of infection and routes of inoculation, the outcome seems to vary. A pretreatment of phages in this study did not help efficiently eliminating incoming bacteria. Most of the phages could be destabilized and lost their infectivity before bacterial infection. In a wound infection model, loss of phages during application would be less than nasal infection model since there would be less chance of immune-mediated clearance. Also, the amount of phages applied seems to be critical. A dose dependency was observed in a previous report (Jeon et al., 2016b).
Increase in serum proinflammaotry cytokines after phage treatment, if any, could reflect a possible inflammatory response elicited by phages. But it was minimal in this study. Mice remained healthy and behaved normally. The observation is coherent with the previous finding that oral inoculation of T7 phages induced a minimal increase in cytokine production of mice (Park et al., 2014). Another phage, SH-Ab 15519, was reported and the therapeutic efficacy was better than the phage cocktail used in this study (Hua et al., 2018). Application of only a single phage showed 90% survival of mice with the M.O.I of 1 or 10. BLAST search revealed 24% coverage with 92% identity when compared to PBAB08, meaning only a low degree of relatedness. As in this study, application of SG-Ab 15519 was generally safe based on histopathological examination of mice lung treated with the phage and cytokine analysis.
Taken together, bacteriophage treatment for mice nasally infected with clinically isolated MDR A. baumannii strain was proven effective and safe. Depending on bacterial strains and phages, an optimal condition for an effective treatment needs to be set. Parameters should include selection of phages from a bank collection based on susceptibility of the causative bacterial strain, composition of a cocktail containing different phage-resistant group members, doses and lengths of phage applications, and routes of phage application. A rapid inoculation of phages after exposure to the pathogen is another critical requirement.
Based on findings that the phage cocktail containing PBAB08 and PBAB25 is effective and safe for treating infections by MDR A. baumannii in vivo, phage therapy would be a viable alternative for antibiotics, especially in cases where antibiotics are not treatment options any more.
Author contributions
HM and KK: designed experiments; KC, YJ, WK, and GH: carried out experiments; HO and JJ: analyzed experimental results; HM: wrote the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by National Research Foundation (NRF) of Korea Funds 2015R1A2A2A01003632 and 2017M3A9B8069292, ATC Program 10076996 from Ministry of Trade, Industry, and Energy of Korea, and HUFS Research Fund of 2017.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00696/full#supplementary-material
Histamine levels in sera from mice treated with phages using three different routes, intraperitoneal (i.p), intranasal (i.n), or oral (p.o). The experiment was performed in triplicate. *P < 0.05, **P < 0.01.
References
- Abedon S. T. (2017). Bacteriophage clinical use as antibacterial “drugs”: utility and precedent. Microbiol. Spectr. 5:BAD-0003-2016. 10.1128/microbiolspec.BAD-0003-2016 [DOI] [PubMed] [Google Scholar]
- Bailey N. T. J. (1959). Statistical Methods in Biology. London: The English Universities Press Ltd. [Google Scholar]
- Bartual S. G., Seifert H., Hippler C., Luzon M. A., Wisplinghoff H., Rodríguez-Valera F. (2005). Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 43, 4382–4390. 10.1128/JCM.43.9.4382-4390.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D. (1980). A theory of modular evolution for bacteriophages. Ann. N.Y. Acad. Sci. 354, 484–490. [DOI] [PubMed] [Google Scholar]
- Clokie M. R. J., Kropinski A. M., Lavigne R. (2018). Bacteriophages methods and protocols, in Methods in Molecular Biology, Vol. 3 (New York, NY: Springer Nature; ). [Google Scholar]
- Clokie M. R. J., Kropinski A. M. (2009a). Bacteriophages methods and protocols, in Methods in Molecular Biology, Vol. 1 (New York, NY: Springer Nature; ). [Google Scholar]
- Clokie M. R. J., Kropinski A. M. (2009b). Bacteriophages methods and protocols, in Methods in Molecular Biology, Vol. 2 (New York, NY: Springer Nature; ). [Google Scholar]
- Davies R. L., Lee I. (2006). Diversity of temperate bacteriophages induced in bovine and ovine Mannheimia haemolytica isolates and identification of a new P2-like phage. FEMS Microbiol. Lett. 260, 162–170. 10.1111/j.1574-6968.2006.00314.x [DOI] [PubMed] [Google Scholar]
- Dijkshoorn L., Nemec A., Seifert H. (2007). An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5, 939–951. 10.1038/nrmicro1789 [DOI] [PubMed] [Google Scholar]
- Doi Y., Murray G. L., Peleg A. Y. (2015). Acinetobacter baumannii: evolution of antimicrobial resistance-treatment options. Semin. Respir. Crit. Care Med. 36, 85–98. 10.1055/s-0034-1398388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Quintanilla M., Pulido M. R., López-Rojas R., Pachón J., McConnell M. J. (2013). Emerging therapies for multidrug resistant Acinetobacter baumannii. Trends Microbiol. 21, 157–163. 10.1016/j.tim.2012.12.002 [DOI] [PubMed] [Google Scholar]
- Hua Y., Luo T., Yang Y., Dong D., Wang R., Wang Y., et al. (2018). Phage therapy as a promising new treatment for lung infection caused by carbapenem-resistant Acinetobacter baumannii in mice. Front. Microbiol. 8:2659. 10.3389/fmicb.2017.02659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T. W., Lauderdale T. L., Liao T. L., Hsu M. C., Chang F. Y., Chang S. C., et al. (2015). Effective transfer of a 47 kb NDM-1-positive plasmid among Acinetobacter species. J. Antimicrob. Chemother. 70, 2734–2738. 10.1093/jac/dkv191 [DOI] [PubMed] [Google Scholar]
- Huys I., Pirnay J.-P., Lavigne R., Jennes S., De Vos D., Casteels M., et al. (2013). Paving a regulatory pathway for phage therapy. EMBO Rep. 14, 951–954. 10.1038/embor.2013.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman P., Abedon S. T. (2009). Practical methods for determining phage growth parameters. Methods Mol. Biol. 501, 175–202. 10.1007/978-1-60327-164-6_18 [DOI] [PubMed] [Google Scholar]
- Jeon J., D'Souza R., Pinto N., Ryu C. M., Park J., Yong D., et al. (2016a). Characterization and complete genome sequence analysis of two myoviral bacteriophages infecting clinical carbapenem-resistant Acinetobacter baumannii isolates. J. Appl. Microbiol. 121, 68–77. 10.1111/jam.13134 [DOI] [PubMed] [Google Scholar]
- Jeon J., Ryu C. M., Lee J. Y., Park J. H., Yong D., Lee K. (2016b). In vivo application of bacteriophage as a potential therapeutic agent to control OXA-66-Like carbapenemase-producing Acinetobacter baumannii strains belonging to sequence ype 357. Appl. Environ. Microbiol. 82, 4200–4208. 10.1128/AEM.00526-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller B., Dubochet J., Adrien M., Maeder M., Wurtz M., Kellenberger E. (1988). Length and shape varients of bacteriophage T4 head: mutations I scaffolding core genes 68 and 22. J. Virol. 62, 2960–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. S., Hong S. S., Park K., Myung H. (2013). Genomic analysis of bacteriophage PBECO4 infecting Escherichia coli O157:H7. Arch. Virol. 158, 2399–2403. 10.1007/s00705-013-1718-3 [DOI] [PubMed] [Google Scholar]
- Krahn T., Wibberg D., Maus I., Winkler A., Bontron S., Sczyrba A., et al. (2016). Intraspecies transfer of the chromosomal Acinetobacter baumannii blaNDM-1 carbapenemase gene. Antimicrob Agents Chemother. 60, 3032–3040. 10.1128/AAC.00124-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröger C., Kary S. C., Schauer K., Cameron A. D. (2017). Genetic regulation of virulence and antibiotic resistance in Acinetobacter baumannii. Genes 8:12. 10.3390/genes8010012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropinski A. M., Mazzocco A., Waddell T. E., Lingohr E., Johnson R. P. (2009). Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol. Biol. 501, 69–76. 10.1007/978-1-60327-164-6_7 [DOI] [PubMed] [Google Scholar]
- Kusradze I., Karumidze N., Rigvava S., Dvalidze T., Katsitadze M., Amiranashvili I., et al. (2016). Characterization and testing the efficiency of Acinetobacter baumannii phage vB-GEC_Ab-M-G7 as an antibacterial agent. Front Microbiol. 7:1590. 10.3389/fmicb.2016.01590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-R., Lee J. H., Park M., Park K. S., Bae I. K., Kim Y. B., et al. (2017). Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front. Cell. Infect. Microbiol. 7:55. 10.3389/fcimb.2017.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E., Zhao J., Ma Y., Wei X., Li H., Lin W., et al. (2016). Characterization of a novel Achromobacter xylosoxidans specific siphoviruse: phiAxp-1. Sci. Rep. 6:21943. 10.1038/srep21943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D. M., Koskella B., Lin H. C. (2017). Phage therapy: an alternative to antibiotics in the age of multi-drug resistance. World J. Gastrointest. Pharmacol. Ther. 8, 162–173. 10.4292/wjgpt.v8.i3.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell M. J., Actis L., Pachon J. (2013). Acinetobacter baumannii: human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol. Rev. 37, 130–155. 10.1111/j.1574-6976.2012.00344.x [DOI] [PubMed] [Google Scholar]
- Merabishvili M., Vandenheuvel D., Kropinski A. M., Mast J., De Vos D., Verbeken G., et al. (2014). Characterization of newly isolated lytic bacteriophages active against Acinetobacter baumannii. PLoS ONE 9:e104853. 10.1371/journal.pone.0104853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihu M. R., Martinez L. R. (2011). Novel therapies for treatment of multi-drug resistant Acinetobacter baumannii skin infections. Virulence 2, 97–102 10.4161/viru.2.2.15061 [DOI] [PubMed] [Google Scholar]
- Parasion S., Kwiatek M., Gryko R., Mizak L., Malm A. (2014). Bacteriophages as an alternative strategy for fighting biofilm development. Pol. J. Microbiol. 63, 137–145. [PubMed] [Google Scholar]
- Park K., Cha K. E., Myung H. (2014). Observation of inflammatory responses in mice orally fed with bacteriophage T7. J. Appl. Microbiol. 117, 627–633. 10.1111/jam.12565 [DOI] [PubMed] [Google Scholar]
- Regeimbal J. M., Jacobs A. C., Corey B. W., Henry M. S., Thompson M. G., Pavlicek R. L., et al. (2016). Personalized therapeutic cocktail of wild environmental phages rescues mice from Acinetobacter baumannii wound infections. Antimicrob. Agents Chemother. 60, 5806–5816. 10.1128/AAC.02877-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W. (2006). Purification of bacteriophage λ particles by centrifugation through a glycerol step gradient. Cold Spring Harb. Protoc. 2006:pdb.prot3969. 10.1101/pdb.prot3969 [DOI] [PubMed] [Google Scholar]
- Schooley R. T., Biswas B., Gill J. J., Hernandez-Morales A., Lancaster J., Lessor L., et al. (2017). Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob. Agents Chemother. 61:e00954-17. 10.1128/AAC.00954-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaswamy V. C., Kalasuramath S. B., Sadanand C. K., Basavaraju A. K., Ginnavaram V., Bille S., et al. (2015). Ability of bacteriophage in resolving wound infection caused by multidrug-resistant Acinetobacter baumannii in uncontrolled diabetic rats. Microb. Drug Resist. 21, 171–177. 10.1089/mdr.2014.0120 [DOI] [PubMed] [Google Scholar]
- Tuon F. F., Rocha J. L., Merlini A. B. (2015). Combined therapy for multi-drug-resistant Acinetobacter baumannii infection – is there evidence outside the laboratory? J. Med. Microbiol. 64, 951–959. 10.1099/jmm.0.000144 [DOI] [PubMed] [Google Scholar]
- Turner D., Wand M. E., Briers Y., Lavigne R., Sutton J. M., Reynolds D. M. (2017). Characterisation and genome sequence of the lytic Acinetobacter baumannii bacteriophage vB_AbaS_Loki. PLoS ONE 12:e0172303. 10.1371/journal.pone.0172303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Mi Z., Niu W., An X., Yuan X., Liu H., et al. (2016). Intranasal treatment with bacteriophage rescues mice from Acinetobacter baumannii-mediated pneumonia. Fut. Microbiol. 11, 631–641. 10.2217/fmb.16.11 [DOI] [PubMed] [Google Scholar]
- Weber B. S., Harding C. M., Feldman M. F. (2016). Pathogenic Acinetobacter: from the cell surface to infinity and beyond. J. Bacteriol. 198, 880–887. 10.1128/JB.00906-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Liang L., Lin S., Jia S. (2010). Isolation and characterization of a virulent bacteriophage AB1 of Acinetobacter baumannii. BMC Microbiol. 10:131. 10.1186/1471-2180-10-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Leclercq S. O., Tian J., Wang C., Yahara K., Ai G., et al. (2017). A new subclass of intrinsic aminoglycoside nucleotidyltransferases, ANT(3")-II, is horizontally transferred among Acinetobacter spp. by homologous recombination. PLoS Genet. 13:e1006602. 10.1371/journal.pgen.1006602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Liu X., Li, Li X. J. (2015). Bioinformatic analysis of phage AB3, a phiKMV-like virus infecting Acinetobacter baumannii. Genet. Mol. Res. 14, 190–198. 10.4238/2015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Histamine levels in sera from mice treated with phages using three different routes, intraperitoneal (i.p), intranasal (i.n), or oral (p.o). The experiment was performed in triplicate. *P < 0.05, **P < 0.01.