Abstract
Background
Recent emergence of long noncoding RNAs in regulating gene expression and thereby modulating physiological functions in brain has manifested their possible role in psychiatric disorders. In this study, the roles of long noncoding RNAs in susceptibility and resiliency to develop stress-induced depression and their response to antidepressant treatment were examined.
Methods
Microarray-based transcriptome-wide changes in long noncoding RNAs were determined in hippocampus of male Holtzman rats who showed susceptibility (learned helplessness) or resiliency (nonlearned helplessness) to develop depression. Changes in long noncoding RNA expression were also ascertained after subchronic administration of fluoxetine to learned helplessness rats. Bioinformatic and target prediction analyses (cis- and trans-acting) and qPCR-based assays were performed to decipher the functional role of altered long noncoding RNAs.
Results
Group-wise comparison showed an overrepresented class of long noncoding RNAs that were uniquely associated with nonlearned helplessness or learned helplessness behavior. Chromosomal mapping within the 5-kbp flank region of the top 20 dysregulated long noncoding RNAs in the learned helplessness group showed several target genes that were regulated through cis- or trans-actions, including Zbtb20 and Zfp385b from zinc finger binding protein family. Genomic context of differentially expressed long noncoding RNAs showed an overall blunted response in the learned helplessness group regardless of the long noncoding RNA classes analyzed. Gene ontology exhibited the functional clustering for anatomical structure development, cellular architecture modulation, protein metabolism, and cellular communications. Fluoxetine treatment reversed learned helplessness-induced changes in many long noncoding RNAs and target genes.
Conclusions
The involvement of specific classes of long noncoding RNAs with distinctive roles in modulating target gene expression could confer the role of long noncoding RNAs in resiliency or susceptibility to develop depression with a reciprocal response to antidepressant treatment.
Keywords: long noncoding RNAs, depression, antidepressant, rat hippocampus, expression
Significance Statement
In the recent past, both the loss and gain of functional activity originating from dysregulated lncRNA expression have been associated with various developmental processes and disease pathogenesis. Although considerable progress has been made in understanding the role of lncRNAs in neuronal functions, their involvement in neurobiology of mental disorders is not known. The present study provides strong evidence of the involvement of lncRNAs in susceptibility or resiliency to develop depression and in the mechanism of action of antidepressants. Moreover, molecular insights from target gene enrichment of dysregulated lncRNAs provide a critical knowledge base to better understand depression resulting from disruptions across cellular ne2rks, leading to aberrant information processing that regulate mood.
Introduction
Earlier findings from both clinical and preclinical studies strongly suggest that stress acts as a major predisposing factor in depression (Gold et al., 1988, 2015; Wong et al., 2017). Negative environmental influences associated with maladaptive stress responsiveness may lead to depressive phenotypes based on an individual’s genetic and epigenetic makeup (Flint and Kendler, 2014; Lopizzo et al., 2015). In an effort to interpret the environmental susceptibility towards depression, various epigenetic modifiers including DNA methylation and microRNAs have been brought into consideration in the recent past (Smalheiser et al., 2011, 2012; Menke and Binder, 2014; Dwivedi et al., 2015; Saavedra et al., 2016; Roy et al., 2017). More recently, lncRNAs have emerged as a critical player in regulating various aspects of coding gene expression (Barry et al., 2014; Spadaro et al., 2015; Zuo et al., 2016). The epigenomic complexity achieved by lncRNAs via dynamically impacting cellular processes and thus brain functioning (Barry, 2014; Briggs et al., 2015) make them an excellent choice to be examined for their role in disease pathophysiology (Iyer et al., 2015), particularly mental disorders.
The majority of the lncRNAs are transcribed by RNA polymerase II. Although they are named noncoding RNAs, infrequently they are annotated for the presence of cryptic open reading frame (Niazi and Valadkhan, 2012; Nelson et al., 2016). lncRNAs differ from protein coding mRNAs despite the common structural features of 5’methyl capping and polyadenylated tail as well as capability of producing splice variants. Most often, lncRNAs have fewer and longer exon with less primary sequence conservation pattern. Their bimodal role in achieving both cis- and trans-regulation comes from their ability to act as scaffold, decoy, or antisense interference (Engreitz et al., 2016a; Quinn and Chang, 2016).
In the recent past, both the loss and gain of functional activity originating from dysregulated lncRNA expression have been associated with various developmental processes and disease pathogenesis (Ng et al., 2013). Although considerable progress has been made in understanding the role of lncRNAs in neuronal functions (Ng et al., 2013; Barry, 2014; Barry et al., 2014; Briggs et al., 2015; Spadaro et al., 2015), their involvement in the neurobiology of mental disorders is not known (Zuo et al., 2016). Except a few recent reports on circulating lncRNAs in blood samples and peripheral blood mononuclear cells of patients with major depression (Liu et al., 2014; Cui et al., 2016), there is no CNS study of lncRNA either in understanding the neuropathology related to depression or their contribution in developing depression-like behavior. In the present study, therefore, we investigated the contribution of lncRNAs and mediated regulatory gene networks in depression using the rodent learned helplessness model. Based on susceptibility (learned helplessness, LH) and resilience (nonlearned helplessness, NLH), this animal model provides a unique opportunity to dissect the molecular mechanisms associated with adaptive and maladaptive responses to stress in the development of depression phenotype. Additionally, we tested whether and how lncRNAs are responsive to antidepressant treatment. We chose hippocampus, as this brain region is closely associated with learning and memory as well emotions. Chronic stress has been shown to decrease hippocampal synaptic plasticity and neuronal atrophy and loss, which consequently influence learning and memory abilities (Howland and Wang, 2008; Kim et al., 2015). Decreased neurotrophic activity in hippocampus of depressed patients as well as in animal models of depression is one of the critical observations, which is reversed by subchronic and chronic administration of antidepressants (Chen et al., 2001; Dwivedi et al., 2003, 2009). In addition, several imaging studies suggest structural abnormalities in hippocampus of depressed patients (Sheline et al., 1996; Frodl et al., 2002, 2006). To our knowledge, this is the first study to shed light on a previously unknown lncRNA-mediated epigenomic regulation underlying depression pathophysiology. Moreover, molecular insights from target gene enrichment of dysregulated lncRNAs provide a critical knowledge base to better understand depression neurobiology resulting from disruptions across cellular networks, leading to aberrant information processing that regulate mood.
Materials and Methods
Detailed methods are provided in the supplementary Methods section.
Animals
Male Holtzman rats (body weight: 350–375 g) were housed 3/cage and maintained under standard laboratory conditions (temperature 21±1°C, humidity 55%±5%, 12-h-light/-dark cycle). All rats received ad libitum food and water and acclimatized for 1 week. All experiments were performed under light cycle (8:00 am and 10:00 am). The study was approved by the Institutional Animal Care and Use Committee of the University of Alabama at Birmingham. Previous reports have shown the usefulness of Holtzman rats in modeling stress-related depression phenotype because of their higher susceptibility to develop depression under stress (Wieland et al., 1986; Padilla et al., 2011). Also, some other strains of rats are either resistant to or less susceptible to this phenotype (Padilla et al., 2009). In addition, in the past, we have successfully used this specific strain of rats in delineating neurobiological changes associated with stress-induced depression (Dwivedi et al., 2005; Smalheiser et al., 2011).
Induction and Assessment of Learned Helpless Behavior and Fluoxetine Treatment
The procedure for the induction of LH behavior in rats was essentially the same as described previously (Dwivedi and Zhang, 2016). Figure 1A provides the paradigms used in induction of inescapable shock (IS) and escape test (ET). The rats were placed in clear acrylic Plexiglas tubes with the tail extending from the rear of the tube (length: 21.6 cm, internal diameter: 6.35 cm). One hundred random ISs were delivered on days 1 and 7 for 5 s at the rate of 1.0 mA to tails. The mean interval between shocks was 60 s. For escape testing (days 2, 8, and 14), foot shocks (0.6 mA) were delivered through the grid floor starting with 5 trials (FR-1) during which a single crossing would terminate the shock. This was followed by 25 trials (FR-2) in which the animal had to cross from one side to the other and come back to terminate the shock. Shocks were terminated automatically after 30 s if there was no response within that time-frame. Rats were divided into 2 groups based on the mean FR-2 escape latencies: (1) LH with mean latency ≥20 s and (2) NLH with mean latency <20 s. Rats confined to Plexiglas tubes but not shocked were termed tested controls (TC). The inclusion of TC rats in the experiment ruled out the nonspecific effects of stress caused by restraint, tail shock, or testing, because TC rats were handled similarly as NLH and LH rats. For lncRNA transcriptomic analyses, hippocampi obtained individually from 6 TCs, 7 NLH, and 7 LH were used. Intraperitoneal injections of fluoxetine (5 mg/kg) were given to 7 randomly selected LH rats (referred to as LH+FLX) once daily for 13 days, which was started right after the first ET on day 2 (Figure 1A). Besides the fluoxetine injected group, all other rats received i.p. injections of an equal volume of normal saline (0.9% w/v) from day 2 to control for injection stress. Twenty-four hours after the final ET, all the rats were decapitated and their hippocampi dissected and flash frozen in liquid nitrogen before storing at -80°C until analysis. As detailed in our earlier study, a 5 mg/kg dose of fluoxetine was based on previous observations showing reversal in depressive behavior in rats and its effect on various neurochemical changes in brain (Dwivedi et al., 2006). We used a 13-d protocol to induce subchronic treatment paradigm, which we have earlier shown to be highly effective in reversing LH behavior (Dwivedi et al., 2002, 2004).
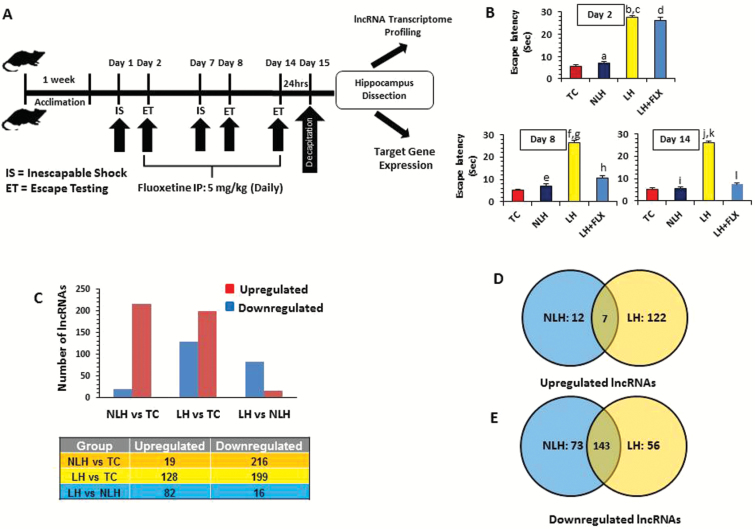
Figure 1.
Stress-induced learned helplessness (LH) model of depression and associated transcriptome-wide changes in long noncoding RNA (lncRNA) expression. (A) Schematic diagram of the timeline followed as part of the stress paradigm to induce the LH behavior in rats. The schematic diagram also represents the timeline followed as part of the fluoxetine treatment paradigm to test the antidepressant effect on LH behavior. (B) The bar diagrams represent escape latencies in tested controls (TC), nonlearned helplessness (NLH), LH, and LH treated with fluoxetine (LH+FLX) rats measured on days 2, 8, and 14. Data are the mean±SEM. On day 2, the NLH rats did not show any significant (aP=.30) difference in escape latency compared with the TC group. A significantly (bP<.001) higher escape latency was observed for LH rats compared with TC on day 2. Similarly LH rats showed significant difference (cP<.001) in mean escape latency compared with the NLH group on day 2. No significant difference was found (dP=.250) when LH+FLX rats were compared with LH rats on day 2. On day 8, NLH rats did not show any significant (eP=.182) difference in escape latency compared with the TC group. A significantly (fP<.001) higher escape latency was noted for LH rats compared with TC rats. Similarly, LH rats showed significant difference (gP<.001) in mean escape latency compared with the NLH group. Comparison on day 8 demonstrated a significant mean escape latency difference in LH+FLX vs LH groups (hP<.001). On day 14, NLH rats did not show any significant (iP=.744) difference in escape latency when compared with the TC group. Individual group comparison identified a significantly (JP<.001) higher escape latency for LH rats compared with TC rats. Similarly, LH rats showed significant difference (kP<.001) in mean escape latency compared with the NLH group. On the other hand, individual comparison on day 14 demonstrated a significant mean escape latency difference in LH+FLX vs LH groups (lP<.001). (C) Bar diagram showing group-wise distribution of differentially regulated lncRNAs, which includes both up- and downregulated sets. The table under the diagram represents the actual number of lncRNAs associated with up- and downregulated sets from each comparison group (TC, NLH, and LH). (D) Venn diagram showing the overlapping sets of upregulated lncRNAs found to be common for both NLH and LH groups. The diagram is also representative of uniquely upregulated lncRNAs in reference to changes associated with the NLH or LH group. (E) Venn diagram shows the overlapping sets of downregulated lncRNAs found to be common for both NLH and LH groups. The diagram is also representative of uniquely downregulated lncRNAs in reference to changes associated with NLH and LH groups.
RNA Isolation and lncRNA Profiling
Total RNA was isolated as described previously (Roy et al., 2017). Based on gel electrophoresis and Nanodrop quantification, only those RNA samples were selected that had high purity (260/280≥1.8) and ribosomal RNA integrity (28S: 18S=2:1).
Transcriptome-wide lncRNA expression was measured using high throughput microarray using Agilent Array platform (Array-Star, Inc). Total RNA from each sample was amplified and transcribed into fluorescent cRNA along the entire length of the transcripts without 3’ bias utilizing a mixture of oligo(dT) and random primers (Arraystar Flash RNA Labeling Kit). The labeled cRNAs were hybridized onto the Rat lncRNA Array (4x44K). After washing the slides, the arrays were scanned by the Agilent Scanner G2505C.
Microarray Data Analysis
Agilent Feature Extraction software (v.11.0.1.1) was used to analyze array images. Quantile normalization and subsequent data processing was performed using the GeneSpring software (GX v12.1; Agilent Technologies). The false discovery rate (FDR) by Benjamini and Hochberg procedure was applied to correct for multiple testing. Analyzed statistical significance was based on P < .05 and a 1.5-fold change between groups. After quantile normalization, lncRNAs were chosen for further analysis. Differentially expressed lncRNAs with statistical significance between the groups were identified through Volcano Plot filtering. Pathway and Gene Ontology (GO) analyses were applied to the predicted targets (cis and trans) of differentially expressed lncRNAs (Bioconductor R). Finally, hierarchical clustering was performed to show the distinguishable lncRNAs expression patterns.
Target Prediction and Functional Analyses
In silico target prediction of cis-acting lncRNA was performed on those protein coding genes that were localized within the 5-kb flanking region of individual lncRNAs. While predicting the target protein coding genes of trans-acting lncRNA, the sequence of each lncRNA was obtained based on the rat genome browser. For the GO analysis, the R package was used to separately prepare biological processes, molecular functions, and cellular components. KEGG pathway-based analysis was done to get the enriched pathways using predicted target genes.
First-Strand cDNA Synthesis and qPCR-Based Transcript Quantification of lncRNAs and mRNAs
Relative quantification of transcripts (both mRNAs and lncRNAs) was determined following the ∆∆Ct method (Livak and Schmittgen, 2001) using the first-strand cDNA synthesized in total RNA. While preparing the first-strand cDNA, the random hexamer-based priming method was followed for lncRNAs, whereas for coding transcripts, conventional oligo dT priming method was used. Primer sequences are provided in supplementaryTable 1. For all qPCR-based expression studies, 4 to 7 animals were used as biological replicates.
Statistical Analyses
SPSS was used for all the data analysis. The data are represented as mean±SEM. TC, LH, and NLH groups were compared using 1-way ANOVA. Posthoc comparisons were calculated based on Tukey’s method of multiple comparisons. Significance level was set at P≤.05.
Results
Escape Latencies
As shown in Figure 1B, 1-way ANOVA followed by a posthoc test showed a significant difference (P<.001) between groups (TC, NLH, LH, and LH+FLX) when compared for mean escape latency on day 2 (df=3; F=147.95), 8 (df=3; F=104.89), and 14 (df=3; F=216.68). Individual group comparison identified significantly (P<.001) higher escape latencies for LH rats compared with the TC or NLH group on days 2, 8, and 14. NLH rats did not show any significant difference in escape latency when compared with the TC group at any time points. Individual comparison on day 2 demonstrated no significant difference (P=.250) when LH+FLX rats were compared with LH rats. On the other hand, both days 8 and 14 show significant effects of fluoxetine on LH behavior, as escape latency was reversed in LH+FLX compared with LH groups (P<.001).
LncRNA Expression Profile in Hippocampus of LH, NLH, and TC Rats
The changes in lncRNA expression in LH rats were compared with rats that did not exhibit LH after receiving IS (NLH) as well as rats that were placed in the apparatus and tested for avoidance but were not given shocks (TC). Based on the detectable florescence signals, 9945 lncRNAs were examined for further analysis. Altered lncRNAs between groups were selected based on ≥1.5-fold change and a statistical significance ≤.05. Group-wise enlisting of differentially regulated lncRNAs is provided in supplementary Table 2. Transcriptome-wide differential expression profiling showed 19 upregulated and 216 downregulated lncRNAs in NLH vs TC groups. On the other hand, LH vs TC group comparison identified 128 upregulated lncRNAs and 199 downregulated lncRNAs. Comparison of LH and NLH groups exhibited 82 upregulated and 16 downregulated lncRNAs (Figure 1C). Since susceptibility or resilience is critical in developing depression, it was examined which lnRNAs were uniquely associated with LH or NLH phenotype individually. For this, NLH and LH groups were first compared separately with the TC group to find out the differentially regulated lncRNAs, and then those lncRNAs were filtered that were exclusively present in the NLH or LH group. As shown in Figure 1D, 12 upregulated lncRNAs were uniquely associated with NLH phenotype and 122 upregulated lncRNAs with LH phenotype. On the other hand, 73 downregulated lncRNAs were exclusively linked to NLH and 56 downregulated lncRNAs with the LH group. Both NLH and LH groups shared an overlapping set of 7 upregulated and 143 downregulated lncRNAs (Figure 1D–E). The IDs of these uniquely associated lncRNAs are provided in supplementary Table 3. Further, the hierarchical clustering of all differentially regulated lncRNAs based on group-wise comparison among TC, NLH, and LH groups is represented as heat maps (Figure 2A).
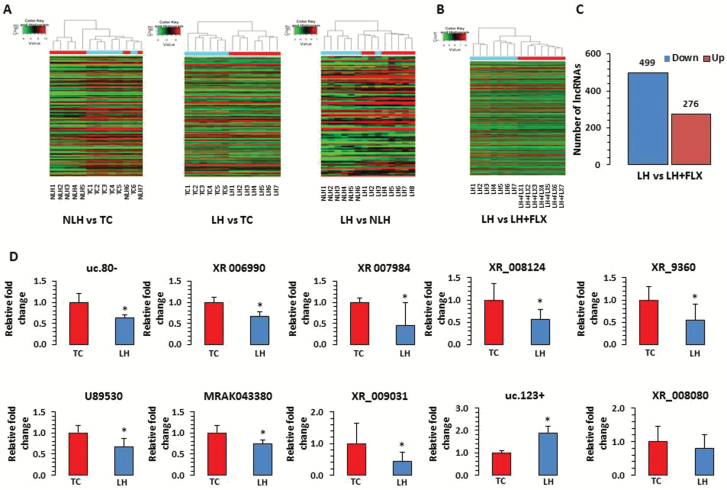
Figure 2.
Expression related changes in long noncoding RNAs (lncRNAs) associated with a rat model of depression. (A) Hierarchical clustering of lncRNAs represents group-wise (nonlearned helplessness [NLH] vs tested controls [TC], learned helplessness [LH] vs TC, and LH vs NLH) expression variability in fold change as plotted with heat map. Individual samples are represented in each column, whereas each row demonstrates lncRNAs. According to the color scheme, red indicates upregulation and green indicates downregulation. (B) Hierarchical clustering of lncRNAs representing the LH vs fluoxetine group is demonstrated with microarray based expression heat map. (C) Bar diagram shows group-wise (LH vs LH rats treated with fluoxetine [LH+FLX]) distribution of differentially regulated lncRNAs including both up- and downregulated sets. The actual number of lncRNAs associated with up- and downregulated sets from the comparison group (LH vs LH-FLX) are indicated on top of each representative bar. (D) qPCR-based expression validation of lncRNA transcripts uc.80-, XR_006990, XR_007984, U89530, MRAK043380, XR_009031, XR_008124, XR_9360, uc.123+, and XR_008080 were analyzed in LH rats using primers mentioned in the Methods section. Gapdh-normalized expression level for each transcript is presented as relative fold change. All data are the mean±SEM. The level of significance was determined using independent-sample t test. *Significant difference between 2 compared groups (uc.80-, P=.01; XR_006990, P=.005; XR_007984, P=.05; U89530, P=.02; MRAK043380, P=.02; XR_009031, P=.05; XR_008124, P=.03; XR_9360, P=.05; uc.123+, P=.01; and XR_008080, P=.30).
Expression Analysis of lncRNAs in LH Rats Treated with Fluoxetine
The hierarchical clustering of lncRNA demonstrating a differential expression profile of lncRNAs based on comparison between the LH and LH+FLX group of rats is represented with a heat map (Figure 2B). Also, a bar diagram depicting the number of differentially expressed lncRNAs in the fluoxetine-treated group are shown in Figure 2C. As can be seen, 13-d treatment in LH+FLX rats impacted many lncRNAs, which included a downregulated group of 499 lncRNAs and an upregulated group of 276 lncRNAs. Further analysis revealed significant reversal in the expression of LH-mediated 52 upregulated and 29 downregulated lncRNAs by fluoxetine (supplementary Table 4).
qPCR-Based Analysis of lncRNAs in Hippocampus of TC and LH Rats
We analyzed the expression of 10 randomly selected lncRNAs (uc.80-, XR_006990, XR_007984, U89530, MRAK043380, XR_009031, XR_008124, XR_9360, uc.123+, and XR_008080) of 20 significantly altered lncRNAs in hippocampus of LH rats by qPCR. It was found that all 10 lncRNAs showed a similar pattern of change in LH rats compared with TC rats as was observed in the microarray analysis. Eight lncRNAs demonstrated significant downregulation (uc.80-, P=.01; XR_006990, P=.005; XR_007984, P=.05; U89530, P=.02; MRAK043380, P=.02; XR_009031, P=.05; XR_008124, P=.03; and XR_9360, P=.05), whereas one (lncR- uc.123+) exhibited significant upregulation (P=.01) in LH rats compared with TC rats. lncR-XR_008080 was also upregulated (~20%), which was in line with the microarray expression data; however, it could not reach statistical significance (P=.30). The expression-arelated changes of these lncRNAs are provided as a bar diagram in Figure 2D.
Genomic Context of Differentially Expressed lncRNAs in LH and NLH Rats
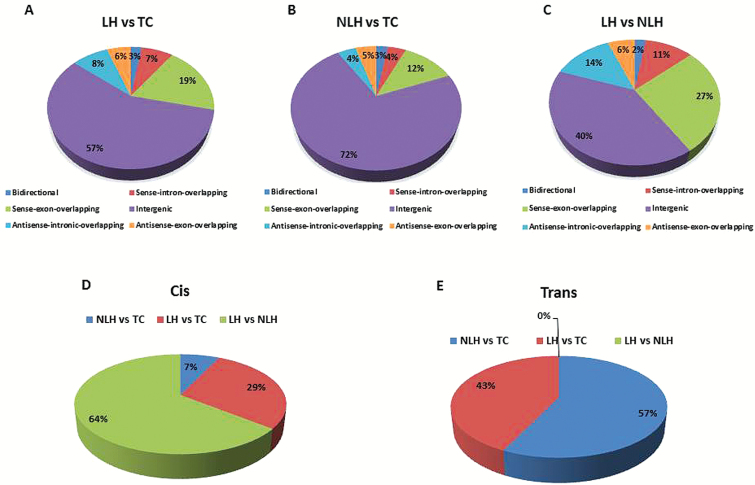
To determine the functional role of lncRNAs, it is essential to dissect their chromosomal localization under stable genomic context. Phenotypic changes are mostly governed by dynamic regulation in protein coding transcripts. Mapping of lncRNAs relative to nearby coding genes classify them into 6 broad categories: bidirectional, sense-intron-overlapping, sense-exon-overlapping, intergenic, antisense-intronic overlapping, and antisense-exon-overlapping. Detailed analysis of microarray data from 3 different group-wise comparisons (TC, LH, and NLH) resulted in identification of major intergenic class of lncRNAs. However, individual group-wise comparison between NLH and TC showed 5 bidirectional, 10 sense-intron-overlapping, 32 sense-exon-overlapping, 135 intergenic, 8 antisense-intronic overlapping, and 10 antisense-exon-overlapping lncRNAs of a total 250 dysregulated lncRNAs (Figure 3A). Interestingly, in the LH vs TC group, more enrichment of altered lncRNAs was noted for all the 6 classes (Figure 3B), whereas an overall blunted response was observed in LH vs NLH groups regardless of the lncRNA classes analyzed (Figure 3C). A comprehensive data table showing the enrichment scheme of 6 different lncRNA subclasses from the 3 group comparisons (NLH vs TC, LH vs TC, and LH vs NLH) is provided in supplementary Table 5 online.
Figure 3.
Class distribution of differentially expressed long noncoding RNAs (lncRNAs) and their predicted target genes across tested control (TC), nonlearned helplessness (NLH), and learned helplessness (LH) groups. (A-C) Distribution of lncRNA classes based on their relative position to protein coding genes on chromosome are represented as pie charts for each pair-wise compared group. Each segment of pie chart is represented with 6 different classes of lncRNAs for their relative genomic context. (D-E) Pie diagrams represent the relative distribution of predicted target genes of differentially regulated lncRNAs based on the cis and trans acting role as identified from pair-wise comparison of 2 groups.
Target Prediction of Differentially Expressed lncRNAs in LH and NLH Rats
Since lncRNAs mediate a bimodal pattern of gene regulation, investigation was extended to both cis and trans mode to predict target genes of significantly dysregulated lncRNAs from 3 separate group-wise comparisons (Figure 3D–E). Among all the differentially expressed lncRNAs, a ranking scheme was followed to short list the top 20 lncRNAs based on their fold change and statistical significance (Table 1). These 20 candidate lncRNAs were further used to conduct a cis- and trans-regulatory analysis to predict potential target genes. As stated in the Methods section, the loci, harboring protein coding genes located within the 5-kbp flank region of a candidate lncRNA were used to screen target genes as part of cis-regulatory analysis. Interestingly, 8 genes were found to be the putative targets of lncRNAs in the LH vs NLH comparison group. LH vs TC group comparison resulted in 3 target genes followed by the identification of one potential target gene from the NLH vs TC group. Furthermore, identification of Zbtb20 and Zfp385b genes from the zinc finger binding protein family indicated the authenticity of this cis target prediction analysis because of their previously discussed role in stress and depression pathology (Liou et al., 2012; Davies et al., 2014). Predicting the trans-regulatory role of differentially regulated lncRNA, 16 protein coding genes were found as targets of 20 dysregulated lncRNAs from NLH vs TC group comparison. A similar trend was observed while predicting the trans action of altered lncRNAs from LH vs TC, which resulted in 12 coding mRNAs across various chromosomal loci in the genome. On the other hand, comparing LH vs NLH group predicted 5 protein coding genes as trans regulated targets of 4 transcriptionally upregulated and one downregulated lncRNAs (supplementary Table 6).
Table 1.
Top 20 Differentially Expressed lncRNAs Based on Fold Change and Statistical Significance
| Differentially Regulated lncRNAs in NLH vs TC | ||||||
|---|---|---|---|---|---|---|
| SeqID | Chromosome | Relationship | Regulation | P Value | FDR | Fold Change |
| XR_008644 | chr17 | Intergenic | Down | .001 | 0.261 | 3.4 |
| XR_007984 | chrX | Intergenic | Down | .004 | 0.275 | 3.33 |
| XR_009360 | chr2 | Others | Down | .005 | 0.285 | 3.27 |
| XR_006015 | chr3 | Intergenic | Down | .019 | 0.363 | 3.26 |
| XR_005855 | chr6 | Intergenic | Down | .002 | 0.261 | 3.02 |
| XR_009031 | chr2 | Intergenic | Down | .006 | 0.285 | 2.95 |
| uc.128- | chr8 | Intergenic | Down | .014 | 0.347 | 2.91 |
| XR_008080 | chr2 | Intergenic | Down | .002 | 0.261 | 2.81 |
| MRAK029283 | chr11 | Intergenic | Down | .002 | 0.261 | 2.75 |
| XR_006363 | chrX | Intergenic | Down | .001 | 0.261 | 2.66 |
| XR_007420 | chr2 | Intergenic | Down | <.001 | 0.22 | 2.64 |
| XR_006210 | chr5 | Intergenic | Down | .003 | 0.27 | 2.59 |
| XR_006429 | chrX | Intergenic | Down | .007 | 0.289 | 2.52 |
| XR_008625 | chr9 | Intergenic | Down | .002 | 0.261 | 2.5 |
| MRAK140228 | chr5 | Sense_intron_overlap | Down | .008 | 0.293 | 2.5 |
| XR_008452 | chrX | Intergenic | Down | .001 | 0.261 | 2.49 |
| XR_006990 | chrUn | Intergenic | Down | .002 | 0.261 | 2.44 |
| U89530 | chr3 | Intergenic | Down | .024 | 0.402 | 2.43 |
| XR_005612 | chr1 | Others | Down | .002 | 0.261 | 2.41 |
| XR_008124 | chr18 | Others | Down | .015 | 0.354 | 2.39 |
| Differentially regulated lncRNAs in LH vs TC | ||||||
| SeqID | Chromosome | Relationship | Regulation | P Value | FDR | Fold Change |
| XR_006990 | chrUn | Intergenic | Down | <.001 | 0.019 | 2.96 |
| XR_008452 | chrX | Intergenic | Down | 4.09E-05 | 0.008 | 2.93 |
| XR_008661 | chr17 | Intergenic | Down | .009 | 0.138 | 2.87 |
| XR_008644 | chr17 | Intergenic | Down | <.001 | 0.037 | 2.8 |
| XR_009360 | chr2 | Others | Down | <.001 | 0.028 | 2.77 |
| XR_006015 | chr3 | Intergenic | Down | .001 | 0.052 | 2.68 |
| uc.123+ | chr8 | Intergenic | Up | 4.79E-06 | 0.002 | 2.65 |
| XR_009031 | chr2 | Intergenic | Down | .001 | 0.047 | 2.63 |
| XR_008124 | chr18 | Others | Down | .001 | 0.055 | 2.57 |
| XR_008080 | chr2 | Intergenic | Down | 1.96E-05 | 0.005 | 2.57 |
| MRAK043380 | chr7 | Sense_intron_overlap | Down | .001 | 0.046 | 2.52 |
| U89530 | chr3 | Intergenic | Down | .001 | 0.055 | 2.49 |
| uc.104- | chr3 | Sense_intron_overlap | Up | <.001 | 0.022 | 2.46 |
| XR_007984 | chrX | Intergenic | Down | .003 | 0.078 | 2.42 |
| MRAK081790 | chrX | Sense_intron_overlap | Down | .003 | 0.074 | 2.41 |
| XR_005855 | chr6 | Intergenic | Down | .001 | 0.043 | 2.38 |
| uc.382+ | chr3 | Intergenic | Up | .003 | 0.077 | 2.36 |
| XR_007260 | chr15 | Intergenic | Down | 1.06E-07 | 0.000476 | 2.36 |
| XR_005612 | chr1 | Others | Down | 8.59E-07 | 0.001 | 2.34 |
| uc.80- | chr3 | Sense_intron_overlap | Down | .003 | 0.08 | 2.34 |
| Differentially regulated lncRNA in LH vs NLH | ||||||
| SeqID | Chromosome | Relationship | Regulation | P Value | FDR | Fold Change |
| uc.396- | chr19 | Intergenic | Up | .003 | 0.371 | 2.17 |
| uc.208+ | chr4 | Antisense_intron_overlap | Up | .002 | 0.371 | 2.14 |
| uc.342+ | chr7 | Others | Up | .002 | 0.351 | 2.14 |
| uc.362+ | chr6 | Intergenic | Up | .003 | 0.371 | 2.13 |
| XR_006950 | chr4 | Intergenic | Up | .005 | 0.4 | 2.11 |
| uc.51+ | chr14 | Intergenic | Up | .001 | 0.351 | 2.09 |
| uc.104- | chr3 | Sense_intron_overlap | Up | .001 | 0.351 | 2.07 |
| uc.224- | chr4 | Intergenic | Up | .007 | 0.425 | 2.02 |
| uc.123+ | chr8 | Intergenic | Up | .007 | 0.422 | 2.02 |
| MRAK012622 | chr2 | Sense_exon_overlap | Up | .009 | 0.435 | 2.01 |
| uc.423+ | chr18 | Others | Down | .008 | 0.435 | 1.98 |
| MRAK015046 | chr5 | Sense_exon_overlap | Up | .013 | 0.435 | 1.96 |
| MRAK015046 | chr5 | Sense_exon_overlap | Up | .013 | 0.435 | 1.96 |
| MRuc009sah | chr8 | Others | Up | .001 | 0.351 | 1.88 |
| MRAK054211 | chr10 | Sense_exon_overlap | Down | .041 | 0.519 | 1.87 |
| AJ535460 | chr20 | Intergenic | Up | .002 | 0.351 | 1.86 |
| MRAK035378 | chr3 | Sense_exon_overlap | Down | .028 | 0.464 | 1.85 |
| uc.120+ | chr11 | Antisense_intron_overlap | Up | .044 | 0.525 | 1.83 |
| MRAK079502 | chrX | Others | Up | .018 | 0.447 | 1.82 |
| uc.312- | chr1 | Sense_intron_overlap | Up | .008 | 0.435 | 1.78 |
Functional Role of Putative Target Genes Based on Gene Ontology and Pathway Analysis
To predict the underlying molecular mechanisms associated with stress-induced depression, a GO analysis using predicted targets of differentially expressed lncRNAs was performed that resulted in a diverse set of molecular functions. While categorizing the 3 functional attributes from GO analysis, namely biological processes, cellular components, and molecular functions, the NLH vs TC group was found to be enriched with GO terms related to ribonucleoprotein complex regulation having functional involvement in gene transcript processing. Targeted protein coding genes from the NLH vs TC group were also found to be enriched with the function of cellular and metabolic processes. On the other hand, the target gene set enrichment of both cis- and trans-acting lncRNAs from other the 2 groups (LH vs TC and LH vs NLH) exhibited the functional clustering towards anatomical structure development, cellular architecture modulation, protein metabolism, and cellular communication. The detailed description of the gene product from the GO analysis for individual group comparison has been provided in supplementary Table 7.
KEGG pathway analysis based on the same predicted target genes across the 3-group comparison showed identification of 3 to 10 different individual pathways (supplementary Table 8). Further screening suggested cell cycle and neurotrophin signaling as the most relevant pathways with functional significance to stress biology in the NLH vs TC group. This observation was further extended to the LH vs TC group, which included 3 pathways related to cellular endocytosis, RNA transport, and mRNA surveillance. On the other hand, in the NLH vs LH group, identification of an exclusive pathway related to spliceosome was noted. This pathway is uniquely involved in producing alternate transcripts through exon splicing mechanism (supplementary Figure 1–3).
qPCR-Based Expression Analysis of Altered lncRNA in LH Rats Treated with Fluoxetine
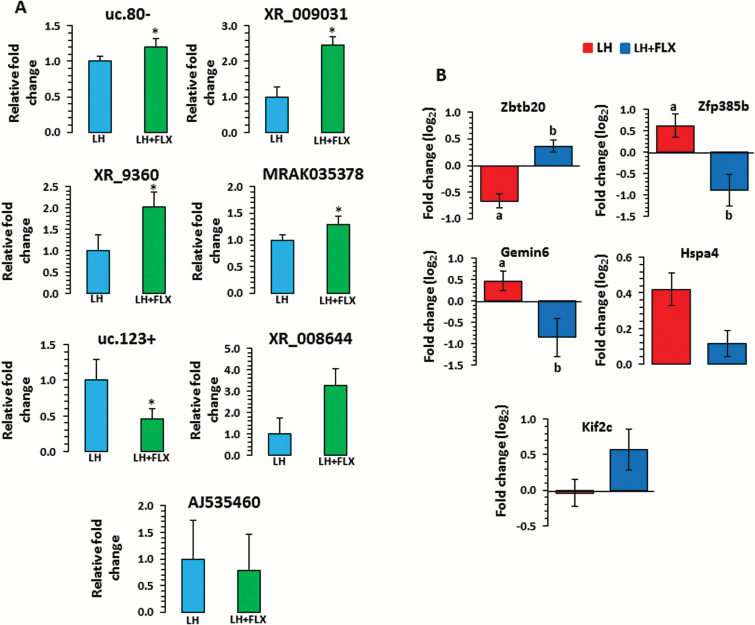
Seven lncRNAs (uc.80-, XR_009031, XR_9360, MRAK035378, uc.123+, XR_008644, AJ535460) were randomly selected to examine their expression status in fluoxetine-treated LH rat hippocampus using qPCR. Similar to the microarray data, a significant (P=.03) upregulation was observed for lnc-uc.80- in LH rats treated with fluoxetine (Figure 4A). Similar expression related changes were noted for XR_009031, XR_9360, and MRAK035378 lncRNAs where a significant increase (XR_009031: P=.003; XR_9360: P=.04; MRAK035378: P=.04) was identified for each lncRNA in fluoxetine-treated LH rats (Figure 4A). On the contrary, a significant (P=.003) downregulation (~55%) was observed for uc.123+ in the fluoxetine-treated LH group (Figure 4A). Two other lncRNAs (XR_008644 and AJ535460), however, did not show any significant changes (XR_008644, P=.07 and AJ535460, P=.35), although their pattern of expression was similar to those observed in microarray analysis (Figure 4A).
Figure 4.
qPCR-based expression-associated changes in long noncoding RNAs (lncRNAs) and target genes in learned helplessness (LH) rats treated with fluoxetine (LH+FLX). (A) Transcript levels of uc.80-, XR_009031, XR_9360, MRAK035378, uc.123+, XR_008644, and AJ535460 were analyzed in LH rats tested under fluoxetine treatment by qPCR using primers mentioned in the Methods section. Gapdh normalized expression level for each transcript is presented as relative fold change. All data demonstrate the mean±SEM (for uc.80, n=5 in LH, n=5 in LH+FLX; for XR_009031, n=5 in LH, n=7 in LH+FLX; for XR_9360, n=4 in LH, n=6 in LH+FLX; for MRAK035378, n=6 in LH, n=7 in LH+FLX; for uc.123+, n=4 in LH, n=5 LH+FLX; for XR_008644, n=7 in LH, n=7 in LH+FLX and for AJ535460, n=6 in LH, n=5 in LH+FLX). The level of significance was determined using independent-sample t test. *Significant difference between 2 compared groups (for uc.80-, P=.03; XR_009031, P=.003; XR_9360, P=.04; MRAK035378, P=.04; uc.123+, P=.003; XR_008644, P=.07 and AJ535460, P=.35). (B) Transcript levels of target genes (log2 fold change) including Zbtb20, Zfp385b, Gemin6, Hspa4, and Kif2c were analyzed in fluoxetine-treated LH (LH+FLX) rats and compared with untreated LH rats. All data are the mean±SEM (for Zbtb20, n=6 in LH and n=7 in LH+Flx; Zfp385b, n=5 in LH and n=4 in LH+Flx; for Gemin6, n=5 in LH and n=5 in LH+Flx; Hspa4, n=7/group and Kif2c, n=5 in LH and n=5 in LH+Flx). The level of significance was determined using independent-sample t test. a denotes significant difference between untreated LH and TC groups (for Zbtb20: P=.0009; Zfp385b: P=.02; and Gemin6: P=.03), whereas b denotes significant difference between LH+FLX and untreated LH groups (for Zbtb20, P=.02; Zfp385b, P=.04; and Gemin6, P=.05). On the contrary, changes associated with Hsp4 and Kif2c were not significant in both LH vs TC and LH+FLX vs LH groups.
Effect of Fluoxetine on the Expression of Cis- and Trans-Regulated Target Genes
Since lncRNA-mediated changes in downstream regulatory functions are primarily transduced via target genes to achieve phenotypic alterations, we anticipated considerable expression alterations of those protein coding genes predicted as cis and trans targets of lncRNAs in LH rats. qPCR-based analyses of 5 such targets (Zbtb20, Zfp385b, Gemin6, Hspa4, and Kif2c) showed marked changes in their expression in LH rats when first compared with TC group of rats. A significant expression upregulation was seen for both Zfp385b (53%) and Gemin6 (38%) genes (Zfp385b: P=.02; Gemin6: P=.03) in LH rats compared with TC rats (Figure 4B). On the other hand, Zbtb20 demonstrated a significant (P=.0009) downregulation (~38%) in the LH vs TC group (Figure 4B). The changes associated with these 3 genes were then found to be significantly reversed by fluoxetine treatment in LH rats compared with untreated LH rats. Individually, the Zfp385b gene was ~66% downregulated (P=.04) and Gemin6 was ~45% downregulated (P=.05) in response to fluoxetine treatment (Figure 4B). Furthermore, in the same group of fluoxetine-treated rats, Zbtb20 was significantly (P=.02) upregulated (~29%; Figure 4B). Although expression changes associated with 2 other target genes (Hsp4 and Kif2c) were not significant in response to fluoxetine treatment, however, a reciprocal relationship was noted for the Kif2c gene, though not statistically significant (Figure 4B).
Discussion
In this study, the lncRNA transcriptome was studied in hippocampus of LH (vulnerable to stress-induced depression), NLH (resilience to depression after receiving similar stress stimuli), and tested control (TC, no shock but tested for escape latency) rats as well as LH rats treated with fluoxetine. The transcriptome analysis showed 103 differentially expressed lncRNAs in hippocampus of LH rats compared with NLH rats. Of them, 87 were upregulated and 16 were downregulated. Interestingly, 33 abundant classes of intergenic lncRNAs, known as lincRNAs, were identified from a total pool of 103 differentially regulated lncRNA transcripts in the LH vs NLH group. These lincRNAs play a critical epigenetic role because of their discrete localization between the protein coding gene loci (Rinn and Chang, 2012). This is evident from studies demonstrating coexistence of lincRNA loci with conserved promoter regions primarily responsible for recruiting key transcription factors for transcription (Engreitz et al., 2016b). There is a possibility that the tight genetic regulation mediated by lincRNAs may be responsible for the underlying causes of susceptibility or resilience to stress, thus inducing or resisting depression-like behavior.
The in-silico cis regulatory target prediction analysis of the top 20 altered lncRNAs from LH rats identified 2 key transcription factors, Zbtb20 and Zfp385b, from the zinc finger binding family of protein. Interestingly, genome-wide association and next generation RNA sequencing studies have found that zing finger proteins may be involved in etiopathogenesis of neuropsychiatric disorders such as schizophrenia, bipolar disorder, and major depression (Davies et al., 2014; Tao et al., 2014; Baek et al., 2017). It has been shown that Zbtb20 plays an essential role in specification of the CA1 field identity in developing hippocampus (Nielsen et al., 2010; Ren et al., 2012). In addition, Zbtb20 is crucial for the regionalization and volume of the archicortex, which plays a role in depression (Rosenthal et al., 2012; Davies et al., 2014). Altered expression of Zbtb20 has also been shown to cause the development of a compact homogenous pyramidal cell layer within the hippocampal region (Nielsen et al., 2010), which is linked to various behavioral abnormalities (Belvindrah et al., 2014). Interestingly, Zbtb20 is hypermethylated in patients with major depressive disorder (Davies et al., 2014). Therefore, the current findings of dysregulated lncRNAs targeting Zbtb20 may provide a crucial mechanism that can be associated with depressive behavior.
Further analysis in the LH vs NLH group predicted Gemin6 gene, which is a target of lncRNA MRAK054211. Gemin6 is well characterized for its role in the cytoplasmic assembly of small nuclear ribonucleoproteins, which serves as a building block of spliceosome complex (Pellizzoni et al., 2002). The overall impact of altered gene expression of Gemin6 mediated through the trans action of lncRNA MRAK054211 may have an important outcome on spliceosome assembly, which in turn may be involved in altering splicing pattern of critical mRNA transcripts frequently observed under compromised synaptic plasticity in depressed brain (Smalheiser, 2014).
The GO and pathway analyses for altered lncRNAs in the LH vs NLH group were in agreement with the functionality of predicted target genes as discussed above. In addition, the predicted biological and molecular attributes from GO analysis were found to be the major governing factors for metabolic processes and cellular architecture (MacQueen and Frodl, 2011). The KEGG analysis based on predicted targets of 20 top-ranking differential lncRNAs from the LH vs NLH group showed the involvement of an antigen processing and presentation pathway. Hspa4 was found to be a putative candidate gene from the family of Hsp70 proteins known for their chromosomal localization within the major histocompability class III cluster between the complement- and tumor necrosis factor locus on the short arm of chromosome 6 (Daugaard et al., 2007). Under stressful conditions, the members from this Hsp70 family are upregulated with the activation of Heat Shock Element on their upstream promoter region (Banerjee Mustafi et al., 2009). In fact, our data showed the presence of lncRNA MRAK054211 upstream to Hspa4 gene, which could possibly be associated with its upregulation under stressful conditions and eventually in modulating the immune reactive functions in depressed brain.
Interesting observations were noted when NLH and LH groups were compared with the TC group. The number of lncRNAs affected in both LH and NLH groups compared with the TC group was much higher than the one with the LH vs NLH group. For example, 235 lncRNAs were dysregulated in the NLH vs TC group, and 327 lncRNAs were altered in the LH vs TC group as opposed to 98 lncRNAs in the LH vs NLH group. More interestingly, many lncRNAs were downregulated in both NLH (216 lncRNA) and LH (199 lncRNAs) groups compared with the TC group. This is in contrast to the observation made when the LH group was compared with the NLH group. Only 16 of 98 lncRNAs were downregulated in this comparison. Thus, there was a blunted response in lncRNAs in both the LH and NLH groups when compared with the TC group. Altogether, it appears that the changes in many lncRNAs (mostly downregulated) may be a consequence of general response to stress given to both the groups in the form of IS. On the other hand, dysregulated lncRNAs in the LH vs NLH group may represent the specific effect of stress-induced susceptibility to develop depression. It is interesting to note that unique sets of lncRNAs were found to be associated with the NLH or LH group. For example, 85 lncRNAs (12 upregulated and 73 downregulated) were associated with NLH group, whereas 178 lncRNAs (122 upregulated and 56 downregulated) were associated with the LH group when independently compared with the TC group. These lncRNAs may be the causative factors in inducing resiliency (NLH) or susceptibility (LH) to develop depression in stressed rats.
It is worth noting that the analysis of predicted target genes and related pathways from the NLH and LH vs TC groups revealed their association with cellular endocytosis, RNA transport, mRNA surveillance, metabolic processes, intercellular communications, and anatomical structure maintenance. Neurotrophin signaling also appeared in the list in the NLH group. It is well known that neurotrophins and their receptors are altered in hippocampus of stressed animals (Jiang and Salton, 2013).
In this study, our microarray data showed strong effects of fluoxetine on restoring the LH-induced altered expression of a large set of lncRNAs. This is further supported by our qPCR-based target gene expression data, which indicate that fluoxetine-mediated phenotypic reversal of rats showing LH behavior could possibly be the direct or indirect result of lncRNA-mediated change in gene regulatory ne2rk as evidenced by the expression modulation of a set of lncRNAs (uc.80-, XR_009031, XR_9360, MRAK035378, uc.123+, XR_008644, and AJ535460) and their cis/trans regulatory target genes (Zbtb20, Zfp385b, Gemin6, Hspa4, and Kif2c). Although the specific mechanism of fluoxetine in causing the transcriptional regulation of lncRNAs is not known, recent reports have shown the epigenetic capability of this pharmacological agent in directly impacting the promoter activity of certain protein coding genes as well as miRNA expression (Bocchio-Chiavetto et al., 2013; Robison et al., 2014).
Altogether, the present study provides strong evidence of the involvement of lncRNAs in susceptibility or resiliency to develop depression and in the mechanism of action of antidepressants. Interestingly, a recent report showed alterations in the expression of lncRNAs in hippocampus of rats who had posttraumatic stress disorder (Qingzhen et al., 2016). The lncRNAs that were affected in this model were different from those observed in our study. It is pertinent to mention that although both the studies were done in hippocampus, the stress paradigms used were completely different. Our study needs to be extended in brain of depressed patients to compare whether the observed changes are similar or dissimilar in a clinical population. So far, there are no studies of lncRNAs in brain of depressed individuals; however, a recent study done in the PBMC of a very small number of depressed patients showed differential regulation of certain lncRNAs, of which 6 lncRNAs (TCONS_00019174, ENST00000566208, NONHSAG045500, ENST00000517573, NONHSAT034045, and NONHSAT142707) were confirmed in many patients who were treated with antidepressants (Cui et al., 2016). Although it is difficult to compare peripheral human studies with those of brain, nevertheless, the peripheral study shows that lncRNAs may serve as a potential biomarker for diagnosis and antidepressant response. One limitation of our study is that it was done in only one brain area. There is a possibility that lncRNAs may be expressed in a brain region-specific manner, and each brain area may respond differently to lncRNAs under stressful conditions. Another limitation is that we used whole hippocampus, and there is a possibility that expression of lncRNAs may show cell-type specific expression. Thus, in the future, it will be interesting to examine not only different brain areas but also specific cell types within the same brain area. Also, it will be interesting to test if the changes in specific lncRNAs can lead to a depression phenotype. One has to be cautious, though, that lncRNAs may function in a coordinated fashion, and therefore, the overall behavioral response may be due to the effect of a cumulative response to overall changes in lncRNAs rather than due to individual lncRNAs. Nevertheless, the present study opens novel avenues to further explore the molecular pathways based on lncRNA functions to dissect the neurobiology associated with vulnerability and resiliency to develop depression.
Supplementary Material
Supplementary data are available at International Journal of Neuropsychopharmacology online.
Statement of Interest
None.
Supplementary Material
Acknowledgments
This research was supported by grants from National Institute of Mental Health (R01MH082802; 1R01MH101890; R01MH100616; 1R01MH107183-01) to Dr. Dwivedi.
References
- Baek JH, Ha K, Kim Y, Yang SY, Cho EY, Choi Y, Ryu S, Lee YS, Park T, Hong KS(2017)Association between the zinc finger protein 804A (ZNF804A) gene and the risk of schizophrenia and bipolar I disorder across diagnostic boundaries. Bipolar Disord 19:305–313. [DOI] [PubMed] [Google Scholar]
- Banerjee Mustafi S, Chakraborty PK, Dey RS, Raha S(2009)Heat stress upregulates chaperone heat shock protein 70 and antioxidant manganese superoxide dismutase through reactive oxygen species (ROS), p38mapk, and akt. Cell Stress Chaperones 14:579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry G.(2014)Integrating the roles of long and small non-coding RNA in brain function and disease. Mol Psychiatry 19:410–416. [DOI] [PubMed] [Google Scholar]
- Barry G, Briggs JA, Vanichkina DP, Poth EM, Beveridge NJ, Ratnu VS, Nayler SP, Nones K, Hu J, Bredy TW, Nakagawa S, Rigo F, Taft RJ, Cairns MJ, Blackshaw S, Wolvetang EJ, Mattick JS(2014)The long non-coding RNA gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol Psychiatry 19:486–494. [DOI] [PubMed] [Google Scholar]
- Belvindrah R, Nosten-Bertrand M, Francis F(2014)Neuronal migration and its disorders affecting the CA3 region. Front Cell Neurosci 8:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocchio-Chiavetto L, Maffioletti E, Bettinsoli P, Giovannini C, Bignotti S, Tardito D, Corrada D, Milanesi L, Gennarelli M(2013)Blood microRNA changes in depressed patients during antidepressant treatment. Eur Neuropsychopharmacol 23:602–611. [DOI] [PubMed] [Google Scholar]
- Briggs JA, Wolvetang EJ, Mattick JS, Rinn JL, Barry G(2015)Mechanisms of long non-coding RNAs in mammalian nervous system development, plasticity, disease and evolution. Neuron 88:861–877. [DOI] [PubMed] [Google Scholar]
- Chen B, Dowlatshahi D, MacQueen GM, Wang JF, Young LT(2001)Increased hippocampal BDNF immunoreactivity in subjects treated with antidepressant medication. Biol Psychiatry 50:260–265. [DOI] [PubMed] [Google Scholar]
- Cui X, Sun X, Niu W, Kong L, He M, Zhong A, Chen S, Jiang K, Zhang L, Cheng Z(2016)Long non-coding RNA: potential diagnostic and therapeutic biomarker for major depressive disorder. Med Sci Monit 22:5240–5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugaard M, Rohde M, Jäättelä M(2007)The heat shock protein 70 family: highly homologous proteins with overlapping and distinct functions. FEBS Lett 581:3702–3710. [DOI] [PubMed] [Google Scholar]
- Davies MN, et al. , UK Brain Expression Consortium (2014)Hypermethylation in the ZBTB20 gene is associated with major depressive disorder. Genome Biol 15:R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi Y.(2009)Brain-derived neurotrophic factor: role in depression and suicide. Neuropsychiatr Dis Treat 5:433–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi Y, Agrawal AK, Rizavi HS, Pandey GN(2002)Antidepressants reduce phosphoinositide-specific phospholipase C (PI-PLC) activity and the mRNA and protein expression of selective PLC beta 1 isozyme in rat brain. Neuropharmacology 43:1269–1279. [DOI] [PubMed] [Google Scholar]
- Dwivedi Y, Mondal AC, Rizavi HS, Shukla PK, Pandey GN(2005)Single and repeated stress-induced modulation of phospholipase C catalytic activity and expression: role in LH behavior. Neuropsychopharmacology 30:473–483. [DOI] [PubMed] [Google Scholar]
- Dwivedi Y, Mondal AC, Shukla PK, Rizavi HS, Lyons J(2004)Altered protein kinase a in brain of learned helpless rats: effects of acute and repeated stress. Biol Psychiatry 56:30–40. [DOI] [PubMed] [Google Scholar]
- Dwivedi Y, Rizavi HS, Conley RR, Roberts RC, Tamminga CA, Pandey GN(2003)Altered gene expression of brain-derived neurotrophic factor and receptor tyrosine kinase B in postmortem brain of suicide subjects. Arch Gen Psychiatry 60:804–815. [DOI] [PubMed] [Google Scholar]
- Dwivedi Y, Rizavi HS, Pandey GN(2006)Antidepressants reverse corticosterone-mediated decrease in brain-derived neurotrophic factor expression: differential regulation of specific exons by antidepressants and corticosterone. Neuroscience 139:1017–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi Y, Roy B, Lugli G, Rizavi H, Zhang H, Smalheiser NR(2015)Chronic corticosterone-mediated dysregulation of microRNA ne2rk in prefrontal cortex of rats: relevance to depression pathophysiology. Transl Psychiatry 5:e682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi Y, Zhang H(2016)Altered ERK1/2 signaling in the brain of learned helpless rats: relevance in vulnerability to developing stress-induced depression. Neural Plast 2016:7383724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Ollikainen N, Guttman M (2016a) Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol 17:756–770. [DOI] [PubMed] [Google Scholar]
- Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES (2016b) Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539:452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J, Kendler KS(2014)The genetics of major depression. Neuron 81:484–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frodl T, Meisenzahl EM, Zetzsche T, Born C, Groll C, Jäger M, Leinsinger G, Bottlender R, Hahn K, Möller HJ(2002)Hippocampal changes in patients with a first episode of major depression. Am J Psychiatry 159:1112–1118. [DOI] [PubMed] [Google Scholar]
- Frodl T, Schaub A, Banac S, Charypar M, Jäger M, Kümmler P, Bottlender R, Zetzsche T, Born C, Leinsinger G, Reiser M, Möller HJ, Meisenzahl EM(2006)Reduced hippocampal volume correlates with executive dysfunctioning in major depression. J Psychiatry Neurosci 31:316–323. [PMC free article] [PubMed] [Google Scholar]
- Gold PW.(2015)The organization of the stress system and its dysregulation in depressive illness. Mol Psychiatry 20:32–47. [DOI] [PubMed] [Google Scholar]
- Gold PW, Goodwin FK, Chrousos GP(1988)Clinical and biochemical manifestations of depression. Relation to the neurobiology of stress (2). N Engl J Med 319:413–420. [DOI] [PubMed] [Google Scholar]
- Howland JG, Wang YT(2008)Synaptic plasticity in learning and memory: stress effects in the hippocampus. Prog Brain Res 169:145–158. [DOI] [PubMed] [Google Scholar]
- Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, Poliakov A, Cao X, Dhanasekaran SM, Wu YM, Robinson DR, Beer DG, Feng FY, Iyer HK, Chinnaiyan AM(2015)The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 47:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Salton SR(2013)The role of neurotrophins in major depressive disorder. Transl Neurosci 4:46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Pellman B, Kim JJ(2015)Stress effects on the hippocampus: a critical review. Learn Mem 22:411–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou YJ, Chen CH, Cheng CY, Chen SY, Chen TJ, Yu YW, Nian FS, Tsai SJ, Hong CJ(2012)Convergent evidence from mouse and human studies suggests the involvement of zinc finger protein 326 gene in antidepressant treatment response. Plos One 7:e32984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Li X, Sun N, Xu Y, Meng Y, Yang C, Wang Y, Zhang K(2014)Microarray profiling and co-expression ne2rk analysis of circulating lncRNAs and mRNAs associated with major depressive disorder. Plos One 9:e93388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD(2001)Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408. [DOI] [PubMed] [Google Scholar]
- Lopizzo N, Bocchio Chiavetto L, Cattane N, Plazzotta G, Tarazi FI, Pariante CM, Riva MA, Cattaneo A(2015)Gene-environment interaction in major depression: focus on experience-dependent biological systems. Front Psychiatry 6:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen G, Frodl T(2011)The hippocampus in major depression: evidence for the convergence of the bench and bedside in psychiatric research?Mol Psychiatry 16:252–264. [DOI] [PubMed] [Google Scholar]
- Menke A, Binder EB(2014)Epigenetic alterations in depression and antidepressant treatment. Dialogues Clin Neurosci 16:395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BR, Makarewich CA, Anderson DM, Winders BR, Troupes CD, Wu F, Reese AL, McAnally JR, Chen X, Kavalali ET, Cannon SC, Houser SR, Bassel-Duby R, Olson EN(2016)A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351:271–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SY, Lin L, Soh BS, Stanton LW(2013)Long noncoding RNAs in development and disease of the central nervous system. Trends Genet 29:461–468. [DOI] [PubMed] [Google Scholar]
- Niazi F, Valadkhan S(2012)Computational analysis of functional long noncoding RNAs reveals lack of peptide-coding capacity and parallels with 3’ utrs. RNA 18:825–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen JV, Blom JB, Noraberg J, Jensen NA(2010)Zbtb20-induced CA1 pyramidal neuron development and area enlargement in the cerebral midline cortex of mice. Cereb Cortex 20:1904–1914. [DOI] [PubMed] [Google Scholar]
- Padilla E, Barrett D, Shumake J, Gonzalez-Lima F(2009)Strain, sex, and open-field behavior: factors underlying the genetic susceptibility to helplessness. Behav Brain Res 201:257–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla E, Shumake J, Barrett DW, Sheridan EC, Gonzalez-Lima F(2011)Mesolimbic effects of the antidepressant fluoxetine in holtzman rats, a genetic strain with increased vulnerability to stress. Brain Res 1387:71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L, Baccon J, Rappsilber J, Mann M, Dreyfuss G(2002)Purification of native survival of motor neurons complexes and identification of gemin6 as a novel component. J Biol Chem 277:7540–7545. [DOI] [PubMed] [Google Scholar]
- Qingzhen L, Jiehua M, Zhiyang Y, Hongjun L, Chunlong C, Weiyan L(2016)Distinct hippocampal expression profiles of lncRNAs in rats exhibiting a PTSD-like syndrome. Mol Neurobiol 53:2161–2168. [DOI] [PubMed] [Google Scholar]
- Quinn JJ, Chang HY(2016)Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 17:47–62. [DOI] [PubMed] [Google Scholar]
- Ren A, Zhang H, Xie Z, Ma X, Ji W, He DZ, Yuan W, Ding YQ, Zhang XH, Zhang WJ(2012)Regulation of hippocampus-dependent memory by the zinc finger protein zbtb20 in mature CA1 neurons. J Physiol 590:4917–4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY(2012)Genome regulation by long noncoding RNAs. Annu Rev Biochem 81:145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison AJ, Vialou V, Sun HS, Labonte B, Golden SA, Dias C, Turecki G, Tamminga C, Russo S, Mazei-Robison M, Nestler EJ(2014)Fluoxetine epigenetically alters the camkiiα promoter in nucleus accumbens to regulate δfosb binding and antidepressant effects. Neuropsychopharmacology 39:1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal EH, Tonchev AB, Stoykova A, Chowdhury K(2012)Regulation of archicortical arealization by the transcription factor zbtb20. Hippocampus 22:2144–2156. [DOI] [PubMed] [Google Scholar]
- Roy B, Dunbar M, Shelton RC, Dwivedi Y(2017)Identification of microRNA-124-3p as a putative epigenetic signature of major depressive disorder. Neuropsychopharmacology 42:864–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra K, Molina-Marquez AM, Saavedra N, Zambrano T, Salazar LA(2016)Epigenetic modifications of major depressive disorder. Int J Mol Sci 17. doi: 10.3390/ijms17081279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheline YI, Wang PW, Gado MH, Csernansky JG, Vannier MW(1996)Hippocampal atrophy in recurrent major depression. Proc Natl Acad Sci U S A 93:3908–3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR.(2014)The RNA-centred view of the synapse: non-coding RNAs and synaptic plasticity. Philos Trans R Soc Lond B Biol Sci 369. doi: 10.1098/rstb.2013.0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR, Lugli G, Rizavi HS, Zhang H, Torvik VI, Pandey GN, Davis JM, Dwivedi Y(2011)MicroRNA expression in rat brain exposed to repeated inescapable shock: differential alterations in learned helplessness vs Non-learned helplessness. Int J Neuropsychopharmacol 14:1315–1325. [DOI] [PubMed] [Google Scholar]
- Spadaro PA, Flavell CR, Widagdo J, Ratnu VS, Troup M, Ragan C, Mattick JS, Bredy TW(2015)Long noncoding RNA-directed epigenetic regulation of gene expression is associated with anxiety-like behavior in mice. Biol Psychiatry 78:848–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, Cousijn H, Jaffe AE, Burnet PW, Edwards F, Eas2od SL, Shin JH, Lane TA, Walker MA, Maher BJ, Weinberger DR, Harrison PJ, Hyde TM, Kleinman JE(2014)Expression of ZNF804A in human brain and alterations in schizophrenia, bipolar disorder, and major depressive disorder: a novel transcript fetally regulated by the psychosis risk variant rs1344706. JAMA Psychiatry 71:1112–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland S, Boren JL, Consroe PF, Martin A(1986)Stock differences in the susceptibility of rats to learned helplessness training. Life Sci 39:937–944. [DOI] [PubMed] [Google Scholar]
- Wong ML, Arcos-Burgos M, Liu S, Vélez JI, Yu C, Baune BT, Jawahar MC, Arolt V, Dannlowski U, Chuah A, Huttley GA, Fogarty R, Lewis MD, Bornstein SR, Licinio J(2017)The PHF21B gene is associated with major depression and modulates the stress response. Mol Psychiatry 22:1015–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Tan Y, Wang Z, Wang KS, Zhang X, Chen X, Li CS, Wang T, Luo X(2016)Long noncoding RNAs in psychiatric disorders. Psychiatr Genet 26:109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.