Abstract
Large expansions of hexanucleotide GGGGCC (G4C2) repeats (hundreds to thousands) in the first intron of the chromosome 9 open reading frame 72 (C9orf72) locus are the strongest known genetic factor associated with amyotrophic lateral sclerosis and frontotemporal lobar degeneration. Different hypotheses exist about the underlying disease mechanism including loss of function by haploinsufficiency, toxicity arising as a result of RNA or dipeptide repeats (DPRs). Five different DPRs are produced by repeat-associated non-ATG-initiated translation of the G4C2 repeats. Though earlier studies have indicated toxicity of the DPRs in worms, flies, primary cultured cells and cell lines, the effect of expressing DPRs of amyotrophic lateral sclerosis-relevant length has not been tested on motor behaviour in vertebrate models. In this study, by expressing constructs with alternate codons encoding different lengths of each DPR (40, 200 and 1000) in the vertebrate zebrafish model, the GR DPR was found to lead to the greatest developmental lethality and morphological defects, and GA, the least. However, expressing 1000 repeats of any DPR, including the ‘non-toxic’ GA DPR led to locomotor defects. Based on these observations, a transgenic line stably expressing 100 GR repeats was generated to allow specific regional and temporal expression of GR repeats in vivo. Expression of GR DPRs ubiquitously resulted in severe morphological defects and reduced swimming. However, when expressed specifically in motor neurons, the developmental defects were significantly reduced, but the swimming phenotype persisted, suggesting that GR DPRs have a toxic effect on motor neuron function. This was validated by the reduction in motor neuron length even in already formed motor neurons when GR was expressed in these. Hence, the expression of C9orf72-associated DPRs can cause significant motor deficits in vertebrates.
Introduction
Intronic GGGGCC (G4C2) expansions in the evolutionarily conserved gene, C9orf72, have been identified in a large proportion of familial and sporadic amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD) patients (1,2). C9orf72 is the most strongly associated gene in ALS/FTD across cohorts and considerable research efforts have been undertaken to elucidate the pathological mechanisms associated with this mutation. Affected individuals typically have large G4C2 expansions in the first intron of C9orf72, ranging from hundreds to several thousands, while in healthy control individuals the G4C2 expansion is far shorter (<30). The disease pathology has been ascribed to three possible mechanisms (3): reduced transcription of C9orf72 leading to haploinsufficiency; toxicity of the transcript from the expanded repeats, which forms RNA foci and can sequester RNA binding proteins; and repeat-associated non-ATG-initiated (RAN) translation of the mutant transcript in the sense and antisense direction resulting in the generation of five different dipeptide repeats (DPRs): Gly-Ala (poly-GA), Gly-Arg (poly-GR), Gly-Pro (poly-GP) and Pro-Ala (poly-PA), Pro-Arg (poly-PR) and Gly-Pro (poly-GP) from the sense and antisense transcripts. Experiments using Drosophila have demonstrated that translation of the G4C2 repeat expansion is important for its toxicity (4), leaving an urgent need to further our understanding of how DPRs may impair motor function in a vertebrate model.
Several studies have demonstrated that DPRs are toxic in cell, fly and zebrafish models (4–6). Among the DPRs, arginine-containing DPRs are especially toxic in cellular models where they are believed to bind to the nucleolus and disrupt ribosome biosynthesis and splicing (4,7). While these studies have focused on understanding the molecular pathways underlying the toxic nature of the arginine-rich DPRs, whether DPRs of ALS-relevant length actually differ in their effect on motor neurons and hence on motor behaviour has not been explored. In this study, we aimed at investigating in vivo and in a vertebrate model: (1) the effects of expressing different chromosome 9 open reading frame 72 (C9orf72)-associated DPRs on survival and morphology, (2) the correlation between DPR length and motor impairment and (3) the effects of stable expression of a toxic DPR on motor neurons and motor function. To do so, DPRs of different lengths (40, 200, 1000) were transiently expressed in zebrafish embryos and morphological defects were assessed to estimate toxicity and motor activity was quantified in these embryos. Though some of the peptides showed differential length-dependent toxicity, expressing 1000 DPRs resulted in impaired swimming in all cases irrespective of the developmental defects and lethality caused by the expression of the DPR. Since GR was found to be the most toxic DPR upon transient expression, a transgenic zebrafish line was generated to stably express 100 GR repeats. While ubiquitous GR dipeptide expression led to severe developmental defects, specific expression in motor neurons resulted in an impairment of swimming specifically, without major defects. Moreover, shortening of motor neurons and increased cell death in the spinal cord were observed upon specific expression of GR repeats in motor neurons.
Hence, this study demonstrates that expression of C9orf72-associated DPRs of physiologically relevant length affects motor neurons and motility in a vertebrate model which could be important for furthering our understanding of the mechanisms underlying DPR toxicity in vivo.
Results
DPRs show length-dependent and peptide-dependent differences in toxicity and motor function
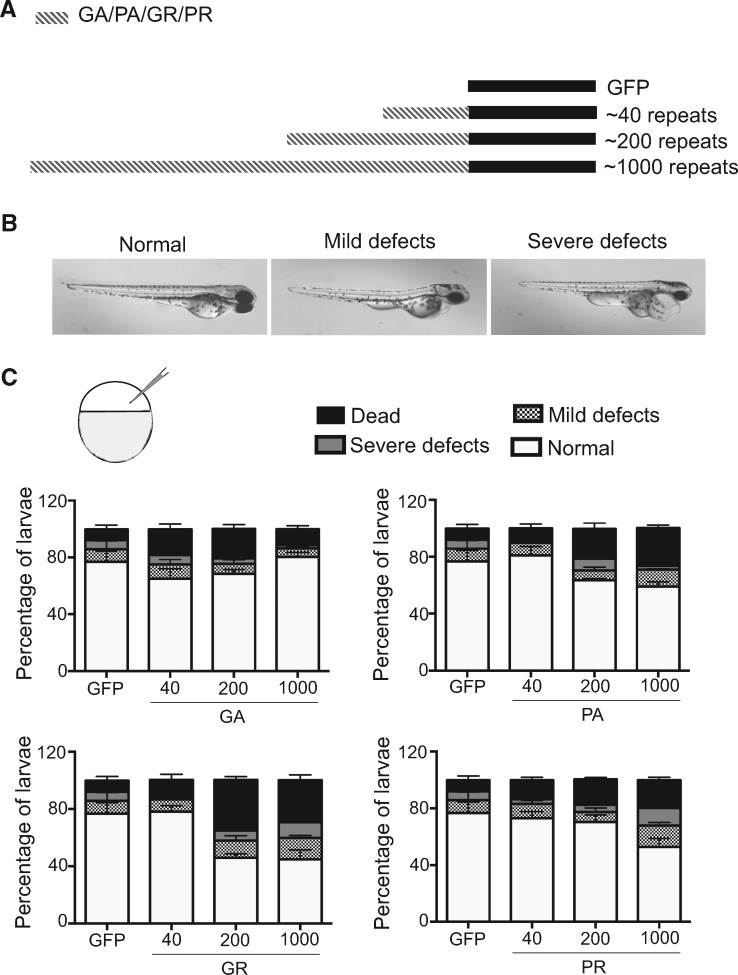
Studies using flies, cultured cells and primary neurons (8,9) showing that DPR expression (especially arginine-containing DPRs) is toxic, but there is no in vivo study comparing the differential toxicity of DPRs of ALS-relevant lengths in vertebrates and its correlation to motor behaviour. External fertilization and development of zebrafish embryos makes this vertebrate model conducive to the study of developmental toxicity and defects. In order to perform comparative toxicity studies, constructs encoding DPRs of different sizes used previously on cultured cells (10) were injected into zebrafish embryos and these were monitored over 3 days (post fertilization and hatching) (Fig. 1A). These constructs utilize alternative and non-repeating codons rather than the G4C2 repeats to encode specific DPRs, without the formation of other DPRs by RAN translation or transcription of G4C2 RNA. Embryos injected with empty vector were considered controls (expressing GFP) and 40 repeats were used to resemble the normal individual condition while 200 and 1000 repeats were used to represent pathological conditions.
Figure 1.
Expression of DPRs of physiologically relevant length has differential effects of survival and morphology. (A) Scheme showing the different constructs used for transient expression. GA, PA, GR and PR DPRs of different lengths (40, 200 and 1000) with C-terminal GFP were used for the toxicity analyses. (B) Different types of defects were observed in the injected larvae at 48 hpf, ranging from slight edema (mild defects) to severe pericardiac edema (severe defects). (C) Quantification of the number of normal, slightly defective, severely defective and dead embryos at 30 hpf after injecting different constructs showed that GA was the least toxic DPR, and GR was the most toxic (N = 3, n = 100 embryos per construct).
Apart from lethality, the phenotypes were classified as mild (cardiac edema) or severe (pericardiac expansion) depending on the intensity of the defects observed at 48 hours post fertilization (hpf) (Fig. 1B). Due to technical difficulty in cloning large GP repeats (10), GP could not be tested in these experiments. When quantified, specific patterns depending on peptide type and length were observed (Fig. 1C). Of the DPRs, GA was observed to be the least toxic, correlating with previous reports (4,8,9), and expression of even 1000 GA repeats did not result in increased lethality or developmental defects. Expression of 1000 repeats of the other DPRs led to increased lethality and developmental defects, but interestingly the intermediate length showed differences between the DPRs and between GR, AP and PR, GR showed the greatest effect on survival and development.
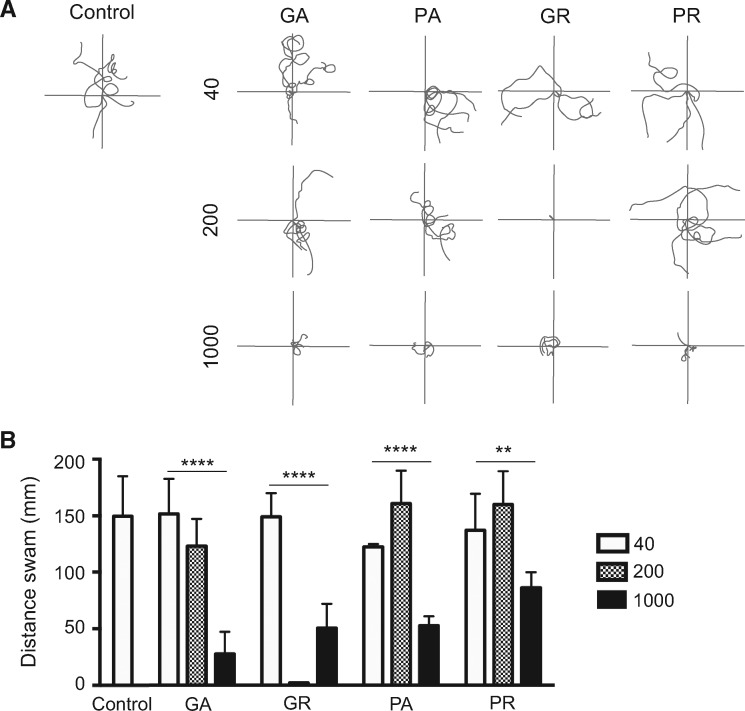
We next sought to determine the effect of DPR expression on the touch-evoked escape response. To specifically assess motor activity without the potentially confounding effects of gross developmental abnormalities, larval zebrafish that did not display any of the abnormal morphological defects described above were selected and assessed. A striking decrease in swimming was observed in embryos expressing the different 1000 DPRs, while no such defects were observed upon expressing 40 DPRs (Fig. 2A and B). Notably, expression of 1000 GA repeats, which did not result in the development of gross abnormalities (Fig. 1C), was observed to impair motor behaviour measured as a reduction in the touch-evoked swim distance. Among the 200 DPRs, GR showed an extreme motor impairment phenotype, but larvae expressing other DPRs appeared to be less affected, including PA200, which showed some effect on survival and overall development. Upon quantifying the distance swam, a clear DPR length-dependent decrease was observed (Fig. 2B).
Figure 2.
DPR expression impairs locomotor activity. (A) Examples of 5 superimposed swimming paths of zebrafish larvae expressing empty vector (control) or 40, 200 or 1000 repeats of different DPRs tested for touch-evoked escape response. While the expression of 1000 DPRs uniformly resulted in impaired touch-evoked escape responses, expression of 200 DPRs showed variability among the dipeptides. (B) Quantification of the mean distance swam upon a light touch on the tail (N = 2, n = 15 larvae per construct). Expression of 1000 DPRs resulted in impaired swimming in all treatment groups including the GA construct. (Statistical significance was estimated by one-way ANOVA was performed for the different groups: GA: P < 0.0001, GR: P < 0.0001, PA: P < 0.0001, PR: P = 0.003.)
Generation and characterization of a transgenic zebrafish model expressing C9orf72-associated GR repeats
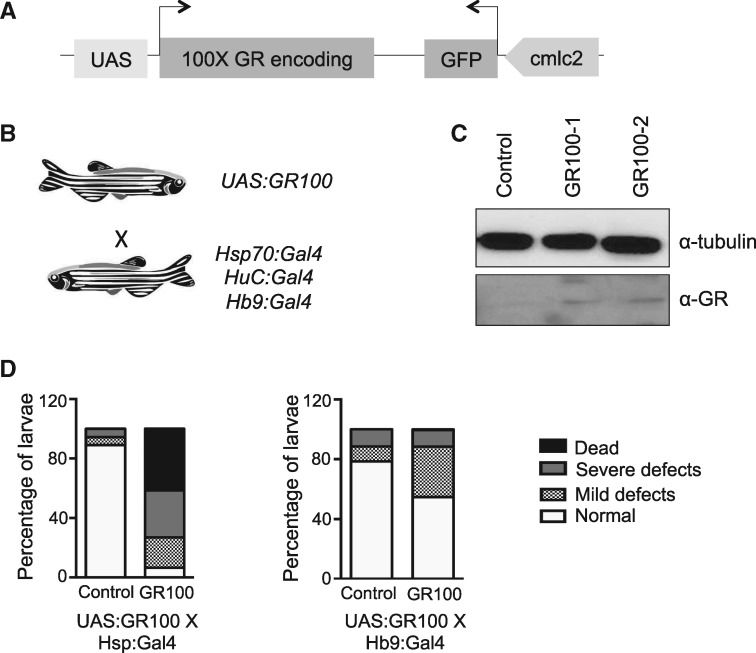
Since GR was the most toxic of the DPRs causing severe developmental defects, and mosaic expression of 200 GR repeats was sufficient to cause a strong effect on touch-evoked escape responses, a transgenic zebrafish line was generated. We overexpressed the less toxic 100 GR repeats using the Tol2 transposon-based system (Fig. 3A) to stably express these under the control of an upstream activation sequence (UAS). In order to facilitate screening, the construct contained the GFP sequence under a cmlc2 promoter for heart-specific expression. Hence, transgenic embryos could be sorted based on the presence of a green heart. In order to drive specific regional or temporal expression of the GR repeats from the UAS, specific Gal4 driver lines were crossed with this transgenic line (Fig. 3B). For this study, three different Gal4 promotor lines were used: heat shock protein 70 (Hsp: Gal4), where expression can be triggered ubiquitously following heat shock; Hb9: Gal4 for motor neuron specific expression; and HuC: Gal4 for pan-neuronal expression.
Figure 3.
Stable expression of GR repeats results in abnormal development. (A) Schematic representation of the construct used for generation of the transgenic zebrafish. (B) Schematic showing the cross between the UAS transgenic line and Gal4 driver line. Gal4 driver lines were used to trigger expression of the construct ubiquitously or in motor neurons specifically (using Gal4 under a ubiquitous Hsp, motor neuron-specific Hb9 or pan-neuronal HuC promoter). (C) Western blotting analysis of lysates from 3 dpf transgenic larvae from two transgenic founder lines (GR100-1 and -2) crossed with a Hsp: Gal4 driver subjected to 1 h heat shock at 48 hpf confirms the expression of the GR DPR. Transgenic embryos not subjected to heat shock were used as control. α-tubulin was used as loading control. (D) Proportion of larvae which were morphologically normal, slightly defective, severely defective or dead upon ubiquitous expression of GR100 (left panel; Hsp: Gal4 driver) or motor neuron specific expression of GR100 (right panel; Hb9-Gal4 driver) at 72 hpf. For the heat shock Gal4 expression, heat shock was performed at 37°C at 48 hpf (N = 2, n = 100 larvae per condition).
In order to confirm the expression in the transgenic line, GR DPRs were expressed ubiquitously (Hsp: Gal4) from 48 to 72 hpf and the expression of the DPR was confirmed by western blotting analysis (Fig. 3C). Many larvae showed cardiac edema and when the proportion of defective larvae was quantified very few larvae were found to be normal after GR expression (Fig. 3D). Non-transgenic siblings also subjected to heat shock did not develop morphological defects. In addition, when GR DPRs were expressed specifically in motor neurons (Hb9: Gal4), the proportion of dead larvae was negligible and few larvae developed minor defects like mild cardiac edema (Fig. 3D), probably due to the restricted expression of the DPRs in motor neurons. In these and in the following experiments, progeny from at least two transgenic founder lines were used for experimentation to account for biological variability of transgenics. These results confirmed that 100 GR DPRs is expressed in our transgenic lines when crossed with a Gal4 line and the extent of defects observed depends on the pattern of expression of the DPR.
Expression of GR DPR results in motor deficits without morphological alterations
We next investigated locomotor behaviour in larvae 7 days post fertilization (dpf) expressing GR. For the swim tests, only normal appearing larvae were chosen so as to avoid the effects of defective circulation or edemas on swimming behaviour. Whether GR was expressed ubiquitously (Hsp: Gal4) or in motor neurons specifically (Hb9: Gal4), it resulted in a reduction in swimming behaviour (Fig. 4A and B). In an attempt to analyse any other possible defects, the heartbeat rate of these larvae was measured, since a large percentage of the larvae expressing GR DPRs ubiquitously showed cardiac edema. However, no differences in heartbeat rate were observed (Fig. 4C), showing the specificity of the swimming phenotype in the GR line. Similar to heartbeat rate, when spontaneous coiling at early stages in development (e.g. the initial stages of Hb9 expression) was analysed, no change was observed (Fig. 4D), showing that the motor defects observed at later stages were specifically due to the expression of GR.
Figure 4.
Expression of GR DPRs results in locomotor defects. (A) Swimming activity assessed at 7 dpf ubiquitously shows a reduction in swimming activity in transgenic larvae expressing GR DPR (with distance swam by the control in mm normalized to 100%). Non-transgenic larvae subjected to heat shock (negative control) and transgenic larvae not subjected to heat shock were used as controls (N = 2, n = 8 larvae per condition, two-tailed P < 0.0001 calculated by unpaired t-test). Larvae subjected to heat shock are represented using black bars. (B) Swimming behaviour assessed at 7 dpf was observed to be reduced in transgenic larvae expressing GR DPRs specifically in motor neurons. Non-transgenic larvae were used as controls (N = 3, n = 8 larvae per condition, two-tailed P < 0.0001 calculated by unpaired t-test). (C) Measurement of heart beats per minute in 72 hpf transgenic zebrafish larvae expressing GR DPR ubiquitously using the Hsp: Gal4 driver, subjected to heat shock at 48 hpf. Non-transgenic larvae subjected to heat shock were used as control. No significant difference was observed (N = 2, n = 5 larvae per condition, two-tailed P = 0.0679 by unpaired t-test). (D) Spontaneous coiling activity in 20 hpf embryos monitored over 20 min represented as percentage burst activity (percentage of total time during which embryos showed activity), which shows no difference in coiling activity between control and transgenic embryos since no DPRs would be expressed at this stage since Hb9 expression has just begun (N = 2, n = 40 larvae/genotype per condition).
Interestingly, the motor phenotype manifested much earlier than 7 dpf, since even at 3 dpf many larvae responded poorly to touch and/or swam short distances in response to touch (movies 1 and 2 show touch response upon ubiquitous expression and motor neuron–specific expression respectively). As with our previous observations in experiments using transient expression of DPRs, assessment of locomotor function in transgenic larvae expressing GR DPRs also revealed impaired motor function.
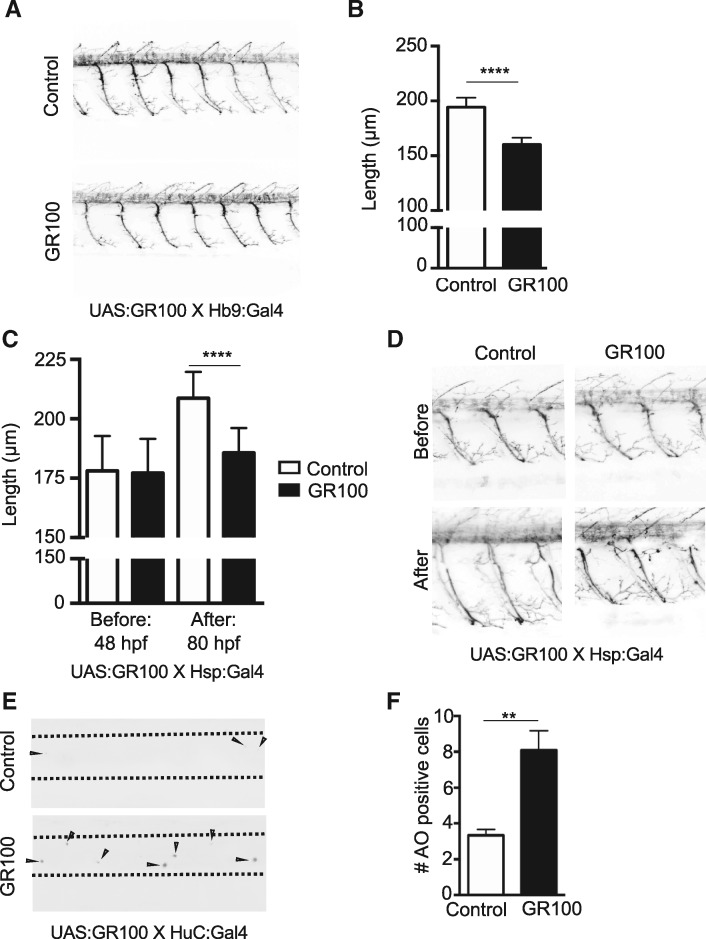
GR DPRs affects motor neuron growth
In an effort to further our understanding of the underlying cause of motor impairment, the morphology of ventral root projections in larvae expressing GR DPRs was examined. When GR was expressed in motor neurons it led to a significant reduction in motor neuron length (Fig. 5A and B). In order to assess if the reduction in length of the motor neurons expressing GR DPR was either because the DPR expression affects early development of motor neuron (thus leading to their shortening), or because of its effect on the growth of an already developed motor neuron, the Hsp: Gal4 driver line was employed to induce GR expression at 48 hpf, i.e. after the formation of motor neurons and elongation of their axons. Motor neuron morphology and length was recorded before heat shock (48 hpf) and after heat shock (80 hpf), i.e. before and after expression of GR DPR. Interestingly, although no difference in motor neuron length was observed before heat shock in transgenic fish, the length was significantly reduced one day after heat shock (Fig. 5C and D), indicating that GR DPR does not necessarily affect motor neuron development, but rather retards the growth of the cell once expressed. Further, analysis of the number of apoptotic cells in the spinal cord also showed a considerable increase (Fig. 5E and F). As a result, expression of GR DPR in motor neurons led to reduction in motor neuron length and increased cell death with reduced swimming but without broad developmental defects.
Figure 5.
GR expression results in reduction of motor neuron length with increased cell death. (A) Example images of GFP-positive motor neurons in 48 hpf zebrafish larvae expressing GR DPR, and a non-transgenic larva as a control. Axons of motor neurons in GR100-expressing larvae appear shorter and less branched. (B) Measurement of the length of motor neurons in the spinal cord showed a significant decrease in motor neuron length when GR DPR is expressed (N = 2, measurement performed from seven embryos per condition, five ventral roots per embryo; two-tailed P < 0.0001 calculated by unpaired t-test). (C). Measurement of the length of motor neurons in the spinal cord before expression of the GR DPR at 48 hpf and at 80 hpf after 24 h of GR100 expression showed a significant decrease in motor neuron length only when GR DPR is expressed (N = 2, measurement performed from seven embryos per condition, five ventral roots per embryo; before heat shock group: two-tailed P = 0.7693 calculated by unpaired t-test; after heat shock group: two-tailed P < 0.0001 calculated by unpaired t-test). (D) Examples of GFP-positive motor neurons in 48 hpf and 80 hpf zebrafish larvae (prior to and after expressing GR DPR), with non-transgenic larvae served as a control. Axons of motor neurons abnormal after GR100 expression. (E) Examples of acridine orange–stained zebrafish larval spinal cords expressing GR DPR in neurons with larvae not expressing the DPR was used as a control. HuC expression was used to set limits while imaging, boundary of the cord is marked with black dotted lines. GR100 DPR expressing larvae show visibly more acridine orange positive cells (black dots, indicated by arrowheads), many of which located in the ventral region of the spinal cord. (F) Quantification of the number of acridine orange positive cells/larva/field in the spinal cord of larvae expressing GR DPR showed significantly greater number of AO positive cells in the spinal cord in comparison to larvae not expressing GR DPR (n = 12 larvae per genotype; two-tailed P = 0.0068 calculated by unpaired t-test).
Discussion
The identification of intronic repeat expansions in a previously uncharacterized gene, C9orf72, as the most common genetic cause of FTD/ALS has led to widespread interest in understanding the disease mechanisms and also the biological role of C9orf72. Among the three possible disease mechanisms, the toxicity of DPRs remains a poorly understood area of disease pathobiology. In this study, we connect DPR toxicity to motor behaviour using transient or stable DPR expression approaches in a vertebrate model. Though earlier studies have compared the toxicity of the DPRs and demonstrated the exceedingly toxic nature of arginine-containing DPRs in flies and cultured cells (4,8,9), this study for the first time shows the effect of DPR expression on motility in a vertebrate model using longer DPRs similar to what has been observed in ALS patients. Furthermore, this study shows that the expression of GR DPRs by itself (i.e. without the expression of RNA from the G4C2 repeats or decreased C9orf72 levels) leads to motor deficits that can be correlated with symptoms observed in ALS patients. These results thus reinforce the notion that DPRs play a role in the pathogenicity of ALS-associated C9orf72 mutations.
With regard to the underlying mechanisms, some studies have suggested a role for C9orf72 loss of function, where a reduction in C9orf72 protein levels were observed in patients (2) and knockdown or mutation of C9orf72 in Caenorhabditis elegans and zebrafish resulted in motor phenotypes (11,12). Though studies in mice have shown no effect of knockdown or knockout of C9orf72 on motor function (13,14), homozygous C9orf72 mutant mice show splenomegaly and defects in immunity (15), which is also observed in patients. A previous transgenic zebrafish model expressing 80 GA DPR (6) reported toxicity but no defects in motor neurons or behaviour, consistent with our observations of weak toxicity and lack of effect on motility of shorter, but not long (1000) repeats. Another very recent report showed that PR and GR DPRs lead to motor axonopathy in zebrafish (16). Here, we show that GR expression on its own is sufficient to induce motor dysfunction and motor neuron length shortening. Thus, although the contribution of reduced expression of C9orf72 and/or of the expression of RNA repeats to the disease pathology cannot be discounted, our results suggest that DPR expression may play a role in the pathogenicity.
Another interesting aspect of the C9orf72 G4C2 repeat expansion is that the spectrum of diseases extends beyond FTD/ALS to a small percentage of Parkinson’s disease, Huntington’s disease and Alzheimer’s disease (17). Further, some animal models also show anxiety and social deficits (18). It would be interesting to also study whether transgenic fish expressing the GR DPR also display other defects in addition to motor defects by performing age-correlated behavioural analysis and analysing other neuronal cell populations.
Owing to the lack of effective drugs to treat ALS and fast progression of this disease, a better understanding to eventually target C9orf72 repeat expansions would help a larger fraction of the patients than before. Though antisense oligonucleotide approaches have proved effective to target the expansion (19), the feasibility of this approach would be dependent on how essential the basic function of C9orf72 is and the challenges associated with efficient delivery of antisense oligonucleotides (20). Hence, the discovery of small molecule drugs to target C9orf72-associated ALS in models such as our GR DPR zebrafish line may aid in the discovery of new effective molecules and potentially lead to the development of novel therapeutic strategies for ALS patients who have intronic expansions in C9orf72. As this vertebrate model facilitates the study of different relevant phenotypes like motor neuron morphology and locomotor activity, it could be an important model to study the disease mechanisms associated with ALS cases with repeat expansions in C9orf72 and especially in chemical screening for new drugs.
Materials and methods
Fish husbandry
Routine zebrafish (Danio rerio) maintenance was performed according to standard protocols (Westerfield) at 28.5°C under 12 h light 12 h dark cycles in the animal facility at the Centre Hospitalier de l’Université de Montréal Research Centre (CRCHUM), Montréal, Québec, Canada. All protocols followed were in compliance with the Canadian Council for Animal Care guidelines.
Transient expression of DPRs of different lengths
DNA sequences containing alternate codons encoding different lengths of the four DPRs: GR, GA, PR and PA were cloned into pEGFP-N1 vector using BamHI and EcoRI restriction enzyme sites. Construction and confirmation of all constructs is described in (10). Each construct was injected into ∼100 single cell embryos at 5 fmol DNA per embryo and expression of GFP in injected embryos was verified at 24 hpf. Counting the dead, severely and slightly defective was performed by an experimenter blind to the construct injected.
Touch-evoked escape response
Zebrafish locomotor activity was assessed by placing 48 hpf larvae in the middle of a circular dish (15 cm diameter) filled with aquarium water at 28.5°C. Swimming was triggered by a slight touch to the tail and the activity was recorded using a Grasshopper 2 camera (Point Grey Research). Path tracing and calculation of swim distance was performed using manual tracking in ImageJ software.
Generation of UAS-GR transgenic line
The multisite Gateway technology-based Tol2kit was used to assemble the construct to generate the transgenic zebrafish line (21). The pDONR221 middle donor vector containing the GR construct was a gift from Prof. Aaron Gitler, Stanford University. This was used along with the 5′ entry clone p5E-UAS and 3′ entry clone p3E-polyA to insert these elements into the pDestTol2CG2 destination vector. For the recombineering reaction, 10 fmol of each of the entry/donor vectors and 20 fmol of the destination vector were mixed with LR Clonase II Plus enzyme mix (Invitrogen), and incubated at 25°C for 16 h. A part of the reaction mix was transformed into chemically competent Escherichia coli following proteinase K treatment. The presence of the GR construct in plasmids extracted from selected clones was confirmed by sequencing.
A mixture containing 40 ng/μl of the destination vector with the GR construct and 40 ng/μl of transposase mRNA was injected into 1 cell stage embryos (∼1 nl per embryo). Screening was done by performing outcrosses and checking for the presence of progeny with green heart.
Western blotting
At 48 hpf larvae sorted as transgenic or non-transgenic were subjected to heat shock at 38.5°C for 60 min with mild agitation. Following this, they were quickly transferred back to 28.5°C for DPR expression and further development. At 80 hpf, larvae were anaesthetized and collected for lysate preparation, deyolked and homogenized in Laemmli lysis buffer. The lysates were centrifuged and protein concentration in the supernatant was estimated using Bradford assay (Biorad). Western blotting was performed using 100 μg total protein per sample which was resolved on a 12% SDS-polyacrylamide gel. After electrophoresis, proteins were electrotransferred from the gel onto a 0.2 micron PVDF membrane. The membrane was blocked with 5% non-fat milk solution in 1× phosphate-buffered saline followed by immunoblotting with α-GR (MABN778; Millipore; rat monoclonal) or α-γ-tubulin (T6557; Millipore Sigma; mouse monoclonal). Detection was performed using anti-rat or anti-mouse antibodies conjugated with horse radish peroxidase. Bands were visualized with ECL and imaged using ChemiDoc (Biorad).
Measurement of activity
Coiling analysis was performed on 20 hpf embryos embedded in low melting agarose and their movements inside the chorion were digitally recorded using a camera for 20 min during which time they were kept hydrated. The Danioscope software (Noldus) was then used to quantify the percentage of time the embryos were active (burst activity).
Swimming activity was measured in 7 dpf larvae transferred individually into a 96-well plate and incubated in the Daniovision recording chamber (Noldus) for 1 h in the dark for habituation. Activity was then recorded over 2 h using Basler GenIcam camera. Analysis was performed using the Ethovision XT 12 software (Noldus) to quantify the distance swam.
Measurement of heartbeat rate
72 hpf larvae which had been subjected to heat shock for 1 h at 48 hpf were mildly anaesthetized so as to not affect heartbeat rate and gently positioned laterally. Videos of the larvae were acquired for 3 min. The boundary of the heart was specified, and heartbeat rate was monitored using the Danioscope software. Non-transgenic larvae after heat shock and transgenic larvae with no heat shock were both used as controls.
Motor neuron morphology
Double transgenic zebrafish with UAS: GR100 and Hb9: GFP were crossed with Hb9: Gal4. At 30 hpf, embryos with GFP in spinal cord with/without green heart were sorted separately. At 48 hpf, larvae were anesthetized, embedded in 1% agarose lateral side facing up and the spinal cord was imaged using a Quorum Technologies spinning disk confocal microscope with CSU10B spinning head (Yokogawa) mounted on an Olympus BX61W1 microscope connected to a Hamamatsu ORCA-ER camera. Further analysis was performed using the Volocity software (Improvision). For temporally regulated expression, double transgenic zebrafish with UAS: GR100 and Hb9: GFP were crossed with Hsp: Gal4. At 48 hpf, larvae were anesthetized, embedded in 1% agarose lateral side facing up and the spinal cord was imaged as described earlier. Following heat shock for 1 hour at 54 hpf at 38.5°C with mild agitation, larvae were incubated at 28.5°C for 24 h and motor neurons were imaged again at 80 hpf.
Acridine orange staining
HuC: Gal4 transgenic zebrafish were crossed with UAS: GR100 line. At 30 hpf, embryos were sorted based on the presence/absence of green heart. At 48 hpf, zebrafish embryos were incubated in 1 μg/ml acridine orange for 15 min followed by washing for about 10 min till no background signal was observed. Larvae were anaesthetized, embedded with lateral view facing up in 1% agarose and imaged using a Quorum Technologies spinning disk confocal microscope with CSU10B spinning head (Yokogawa) mounted on an Olympus BX61W1 microscope connected to a Hamamatsu ORCA-ER camera. The spinal cord was imaged using Volocity (Improvision) setting the limits to the HuC positive neurons. The number of AO positive cells was only counted in the spinal cord (with red signal), and other regions were excluded.
Acknowledgements
We thank Professor Aaron Gitler for kindly sharing the pDONR221 middle donor vector containing the GR construct which was used to generate the stable transgenic line. We thank Marina Drits for assistance with zebrafish maintenance. This work was supported by funding from the Canadian Institutes of Health Research (P.D.) and ALS Canada-Brain Canada (G.A.).
Conflict of Interest statement. None declared.
References
- 1. Renton A.E., Majounie E., Waite A., Simón-Sánchez J., Rollinson S., Gibbs J.R., Schymick J.C., Laaksovirta H., van Swieten J.C., Myllykangas L.. et al. (2011) A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron, 72, 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J.. et al. (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron, 72, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gitler A.D., Tsuiji H. (2016) There has been an awakening: emerging mechanisms of C9orf72 mutations in FTD/ALS. Brain Res., 1647, 19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mizielinska S., Grönke S., Niccoli T., Ridler C.E., Clayton E.L., Devoy A., Moens T., Norona F.E., Woollacott I.O., Pietrzyk J.. et al. (2014) C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science, 345, 1192–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zu T., Liu Y., Bañez-Coronel M., Reid T., Pletnikova O., Lewis J., Miller T.M., Harms M.B., Falchook A.E., Subramony S.H.. et al. (2013) RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl. Acad. Sci. U. S. A., 110, E4968–E4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ohki Y., Wenninger-Weinzierl A., Hruscha A., Asakawa K., Kawakami K., Haass C., Edbauer D., Schmid B. (2017) Glycine-alanine dipeptide repeat protein contributes to toxicity in a zebrafish model of C9orf72 associated neurodegeneration. Mol. Neurodegener., 12, 6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kwon I., Xiang S., Kato M., Wu L., Theodoropoulos P., Wang T., Kim J., Yun J., Xie Y., McKnight S.L. (2014) Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science, 345, 1139–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wen X., Tan W., Westergard T., Krishnamurthy K., Markandaiah S.S., Shi Y., Lin S., Shneider N.A., Monaghan J., Pandey U.B.. et al. (2014) Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron, 84, 1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Freibaum B.D., Lu Y., Lopez-Gonzalez R., Kim N.C., Almeida S., Lee K.H., Badders N., Valentine M., Miller B.L., Wong P.C.. et al. (2015) GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature, 525, 129–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bennion Callister J., Ryan S., Sim J., Rollinson S., Pickering-Brown S.M. (2016) Modelling C9orf72 dipeptide repeat proteins of a physiologically relevant size. Hum. Mol. Genet, 25, 5069–5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Therrien M., Rouleau G.A., Dion P.A., Parker J.A. (2013) Deletion of C9ORF72 results in motor neuron degeneration and stress sensitivity in C. elegans. PLoS One, 8, e83450.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ciura S., Lattante S., Le Ber I., Latouche M., Tostivint H., Brice A., Kabashi E. (2013) Loss of function of C9orf72 causes motor deficits in a zebrafish model of amyotrophic lateral sclerosis. Ann. Neurol., 74, 180–187. [DOI] [PubMed] [Google Scholar]
- 13. Lagier-Tourenne C., Baughn M., Rigo F., Sun S., Liu P., Li H.R., Jiang J., Watt A.T., Chun S., Katz M.. et al. (2013) Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc. Natl. Acad. Sci. U. S. A., 110, E4530–E4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koppers M., Blokhuis A.M., Westeneng H.J., Terpstra M.L., Zundel C.A., Vieira de Sá R., Schellevis R.D., Waite A.J., Blake D.J., Veldink J.H.. et al. (2015) C9orf72 ablation in mice does not cause motor neuron degeneration or motor deficits. Ann. Neurol., 78, 426–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Atanasio A., Decman V., White D., Ramos M., Ikiz B., Lee H.C., Siao C.J., Brydges S., LaRosa E., Bai Y.. et al. (2016) C9orf72 ablation causes immune dysregulation characterized by leukocyte expansion, autoantibody production, and glomerulonephropathy in mice. Sci. Rep., 6, 23204.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Swinnen B., Bento-Abreu A., Gendron T.F., Boeynaems S., Bogaert E., Nuyts R., Timmers M., Scheveneels W., Hersmus N., Wang J.. et al. (2018) A zebrafish model for C9orf72 ALS reveals RNA toxicity as a pathogenic mechanism. Acta Neuropathol., 135, 427–443. [DOI] [PubMed] [Google Scholar]
- 17. Chi S., Jiang T., Tan L., Yu J.T. (2016) Distinct neurological disorders with C9orf72 mutations: genetics, pathogenesis, and therapy. Neurosci. Biobehav. Rev., 66, 127–142. [DOI] [PubMed] [Google Scholar]
- 18. Chew J., Gendron T.F., Prudencio M., Sasaguri H., Zhang Y.J., Castanedes-Casey M., Lee C.W., Jansen-West K., Kurti A., Murray M.E.. et al. (2015) Neurodegeneration. C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science, 348, 1151–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Donnelly C.J., Zhang P.-W., Pham J.T., Haeusler A.R., Mistry N.A., Vidensky S., Daley E.L., Poth E.M., Hoover B., Fines D.M.. et al. (2013) RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron, 80, 415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Juliano R.L. (2016) The delivery of therapeutic oligonucleotides. Nucleic Acids Res., 44, 6518–6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kwan K.M., Fujimoto E., Grabher C., Mangum B.D., Hardy M.E., Campbell D.S., Parant J.M., Yost H.J., Kanki J.P., Chien C.B. (2007) The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn., 236, 3088–3099. [DOI] [PubMed] [Google Scholar]