Abstract
Defects in the MFSD8 gene encoding the lysosomal membrane protein CLN7 lead to CLN7 disease, a neurodegenerative lysosomal storage disorder belonging to the group of neuronal ceroid lipofuscinoses. Here, we have performed a SILAC-based quantitative analysis of the lysosomal proteome using Cln7-deficient mouse embryonic fibroblasts (MEFs) from a Cln7 knockout (ko) mouse model. From 3335 different proteins identified, we detected 56 soluble lysosomal proteins and 29 highly abundant lysosomal membrane proteins. Quantification revealed that the amounts of 12 different soluble lysosomal proteins were significantly reduced in Cln7 ko MEFs compared with wild-type controls. One of the most significantly depleted lysosomal proteins was Cln5 protein that underlies another distinct neuronal ceroid lipofuscinosis disorder. Expression analyses showed that the mRNA expression, biosynthesis, intracellular sorting and proteolytic processing of Cln5 were not affected, whereas the depletion of mature Cln5 protein was due to increased proteolytic degradation by cysteine proteases in Cln7 ko lysosomes. Considering the similar phenotypes of CLN5 and CLN7 patients, our data suggest that depletion of CLN5 may play an important part in the pathogenesis of CLN7 disease. In addition, we found a defect in the ability of Cln7 ko MEFs to adapt to starvation conditions as shown by impaired mammalian target of rapamycin complex 1 reactivation, reduced autolysosome tubulation and increased perinuclear accumulation of autolysosomes compared with controls. In summary, depletion of multiple soluble lysosomal proteins suggest a critical role of CLN7 for lysosomal function, which may contribute to the pathogenesis and progression of CLN7 disease.
Introduction
CLN7 disease represents a severe childhood-onset neurodegenerative disorder caused by mutations in the MFSD8 gene (1). CLN7 disease belongs to the group of neuronal ceroid lipofuscinoses (NCLs) caused by mutations in at least 13 different genes (CLN1–CLN8, CLN10–CLN14) which encode soluble lysosomal proteins (CLN1/palmitoyl protein thioesterase 1 (PPT1), CLN2/tripeptidylpeptidase 1 (TPP1), CLN5, CLN10/cathepsin D and CLN13/cathepsin F), membrane proteins located in lysosomes (CLN3, CLN7 and CLN12) or in the endoplasmic reticulum (CLN6 and CLN8), soluble cytoplasmic proteins (CLN11/progranulin and CLN14) and the synaptic vesicle associated protein CLN4 (2). More than 35 mutations have been identified in MFSD8 leading to CLN7 disease, variant-late infantile phenotype (MIM # 610951), which is characterized by visual impairment, seizures, psychomotor decline and a reduced lifespan (3–11). MFSD8 encodes the lysosomal polytopic CLN7 membrane protein of unknown function which contains sequence similarities with the drug: H+ antiporter family DHA1 of the major facilitator superfamily (MFS) (4,12). The members of the MFS are secondary active, ion-coupled transporters of sugars, amino acids, drugs, nucleosides as well as organic and inorganic cations and anions (13). CLN7 belongs to a group of atypical solute carriers of MFS type which are located at the plasma membrane and/or in intracellular compartments (14). Lysosomal localization of the endogenous CLN7 protein has been demonstrated by proteomic analyses using purified human and rat tritosomes, by immunoblotting of mouse liver tritosomes and by immunohistochemical localization in cultured hippocampal neurons (15–18). We have recently generated a Cln7 knockout (ko) mouse model that recapitulates key features of human CLN7 disease (19,20). In these mice, loss of Cln7 leads to (i) autofluorescence and lysosomal storage of subunit c of mitochondrial ATP synthase and saposin D in the brain and retina, (ii) neurodegeneration in the olfactory bulb, cerebellum, cortex and retina, (iii) neuroinflammation, as well as (iv) reduced lifespan of mutant mice (19). Furthermore, dysregulated expression of several soluble lysosomal proteins and impaired macroautophagy in the Cln7 ko mice suggest that loss of Cln7 results in lysosomal dysfunction in the brain (19). However, the link between deficiency of the putative lysosomal transporter CLN7 and lysosomal dysfunction is unclear.
Autophagy is a catabolic process where cytoplasmic components are delivered to lysosomes for proteolytic degradation by acidic hydrolases (21). Defective autophagy has been shown to be a major pathomechanism contributing to the accumulation of storage material and neurodegeneration in mouse models for CLN2, CLN3, CLN5, CLN6, CLN7 and CLN10 diseases (19,22–26). In this regard, the enzymatic content of lysosomes is an attractive target to study in NCLs. In CLN3 disease, which is caused by defects in the lysosomal membrane protein CLN3, alterations in the amounts of TPP1 (27), lysosomal acid phosphatase (28) and mannose 6-phosphate-containing glycoproteins (29) in the brain have been reported. However, comprehensive profiling of the lysosomal proteome in CLN7 disease and other NCLs has not been performed thus far.
In the present study, we provide quantitative proteomic data that have been obtained by mass spectrometric analysis of isolated lysosomes from Cln7 ko mouse fibroblasts by means of Stable Isotope Labelling by Amino acids in Cell culture (SILAC). We found that the loss of Cln7 results in alterations in lysosomal soluble proteins under steady state conditions. In addition, we observed a defect in the ability of Cln7 ko MEFs to adapt to starvation conditions as shown by impaired mTORC1 reactivation, reduced numbers of cells containing tubules emerging from autolysosomes and increased perinuclear accumulation of autolysosomes compared with controls.
Results
Lysosomal proteome of Cln7 knockout MEFs
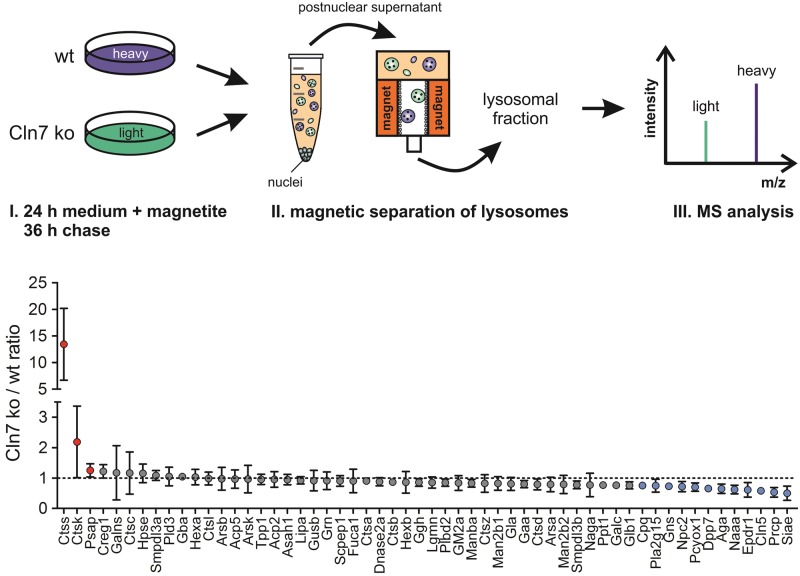
To analyse CLN7 disease-related changes leading to lysosomal dysfunction, we performed a SILAC-based comparative proteomics using mouse embryonic fibroblasts (MEFs) isolated from a Cln7 knockout (Cln7 ko) mouse model that recapitulates major hallmarks of CLN7 disease (19). Using non-radioactive light and heavy isotope amino acids, wild-type and Cln7 ko MEFs were labelled in culture, and magnetite-isolated lysosomal fractions were analysed by mass spectrometry. From 3335 different proteins identified, we detected almost all known soluble lysosomal proteins (number: 56) and 29 highly abundant lysosomal membrane proteins in amounts sufficient for quantification. Mean values of light-to-heavy ratios of >1.25-fold and <0.75-fold determined in four individual SILAC measurements were considered a significant increase and decrease, respectively. Quantification revealed that the amounts of 12 different soluble lysosomal proteins were reduced in Cln7 ko MEFs compared with wild-type controls (Fig. 1). These included soluble lysosomal proteins involved in the degradation of glycans [sialate O-acetylesterase (Siae), aspartylglucosaminidase (Aga), N-acetylglucosamine-6-sulfatase (Gns)], peptides [lysosomal Pro-X carboxypeptidase (Prcp), carboxypeptidase Q (Cpq), dipeptidyl peptidase 2 (Dpp7)], lipids [N-acylethanolamine-hydrolyzing acid amidase (Naaa), group XV phospholipase A2 (Pla2g15)] and fatty acid-modified proteins [prenyl cysteine oxidase (Pcyox1)]. In addition, the levels of lysosomal proteins of unknown function including Niemann Pick type C2 protein (Npc2), neuronal ceroid-lipofuscin protein 5 (Cln5) and mammalian ependymin-related protein 1 (Epdr1) were decreased in Cln7 ko MEFs in comparison to wild-type control cells (Fig. 1). Three soluble lysosomal proteins were detected in significantly elevated amounts in Cln7 ko MEFs compared with wild-type controls: cathepsin S (Ctss, 13.4-fold), cathepsin K (Ctsk, 2.2-fold), and prosaposin (Psap, 1.25-fold). These data showed that the absence of functional Cln7 protein affects the amounts of multiple soluble lysosomal proteins.
Figure 1.
SILAC-based comparative analysis of lysosomal proteomes of wild-type and Cln7 ko MEFs. Wild-type (wt) and Cln7 knockout (ko) MEFs were grown in heavy and light isotope-labelled medium and incubated in medium containing magnetite particles for 24 h, followed by 36 h of chase. Equal amounts of postnuclear supernatants from MEFs of both genotypes were pooled prior to magnetic isolation of lysosomes and MS analysis. Soluble lysosomal proteins identified in the lysosomal fractions by MS analysis were plotted against the Cln7 ko/wt ratio determined in four individual SILAC samples (mean ± SD). Lysosomal proteins with increased and decreased amounts in Cln7 ko lysosomes are indicated with red and blue circles, respectively. A list of all lysosomal proteins identified and the corresponding Cln7 ko/wt ratios are given in the Supplementary Material (Tables S1 and S2).
Alterations of lysosomal proteins in Cln7-deficient cells and tissues
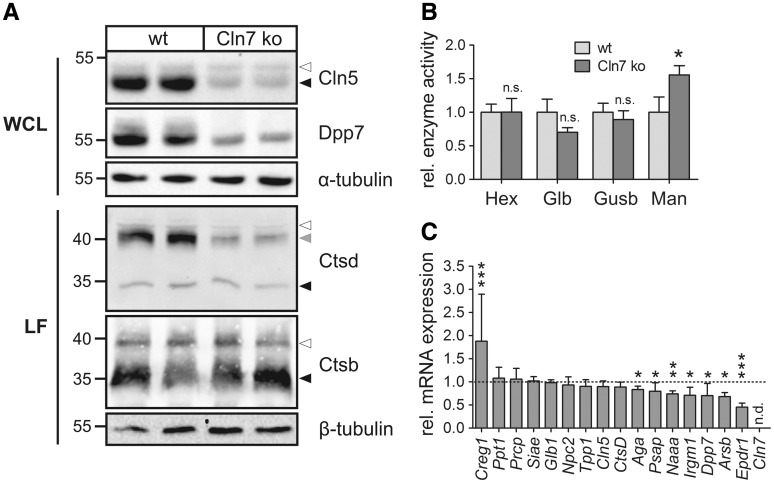
To verify the proteomic data, expression levels of selected lysosomal enzymes were determined by immunoblotting, enzymatic activity measurements and mRNA quantification (Fig. 2). In agreement with the comparative proteomic data, the amounts of Cln5 and Dpp7 were decreased by about 80% and 75%, respectively, in Cln7 ko MEFs compared with wild-type controls as shown by western blotting (Fig. 2A). The enzymatic activities of β-hexosaminidase (Hexb), β-galactosidase (Glb) and β-glucuronidase (Gusb) were not changed in Cln7 ko MEFs compared with controls; however, we observed a 1.5-fold increase in the enzymatic activity of α-mannosidase (Man) in Cln7 ko MEFs (Fig. 2B). To analyse whether changed mRNA amounts contribute to altered levels of dysregulated lysosomal proteins, quantitative RT-PCR analyses were performed. The mRNA amounts of Ppt1, Prcp, Siae, Glb1, Npc2, Tpp1, Cln5 and Ctsd were not changed in Cln7 ko MEFs compared with wild-type controls (Fig. 2C). mRNA expression of Aga, Psap, Naaa, Irgm1, Dpp7, arylsulfatase B (Arsb) and Epdr1 were significantly reduced in Cln7-deficient MEFs.
Figure 2.
Validation of SILAC data using Cln7 ko and wild-type MEFs. (A) Whole cell lysates (WCL) and lysosome-enriched fractions (LF) of Cln7 ko and wild-type MEFs were analysed by Cln5, Dpp7, cathepsin D (Ctsd) and cathepsin B (Ctsb) immunoblotting. α- and β-tubulin western blotting was performed to control equal loading. The positions of the molecular mass markers and the precursor, intermediate and mature forms of Cln5, Ctsb and Ctsd are indicated with white, grey and black arrowheads, respectively. (B) Specific enzymatic activities of β-hexosaminidase (Hex), β-galactosidase (Glb), β-glucuronidase (Gusb) and α-mannosidase (Man) in whole cell lysates of wild-type and Cln7 ko MEFs. The activities relative to wild-type controls are shown (mean ± SD, *P < 0.05, n = 4, two-tailed Student’s t-test). (C) Relative mRNA expression levels of selected lysosomal genes in Cln7 ko MEFs in comparison to wild-type controls (set as 1.0). Data are plotted as mean values ± SD (*P < 0.05, **P < 0.01, ***P < 0.001, n = 3–6, two-tailed Student’s t-test). n.d.: not detected.
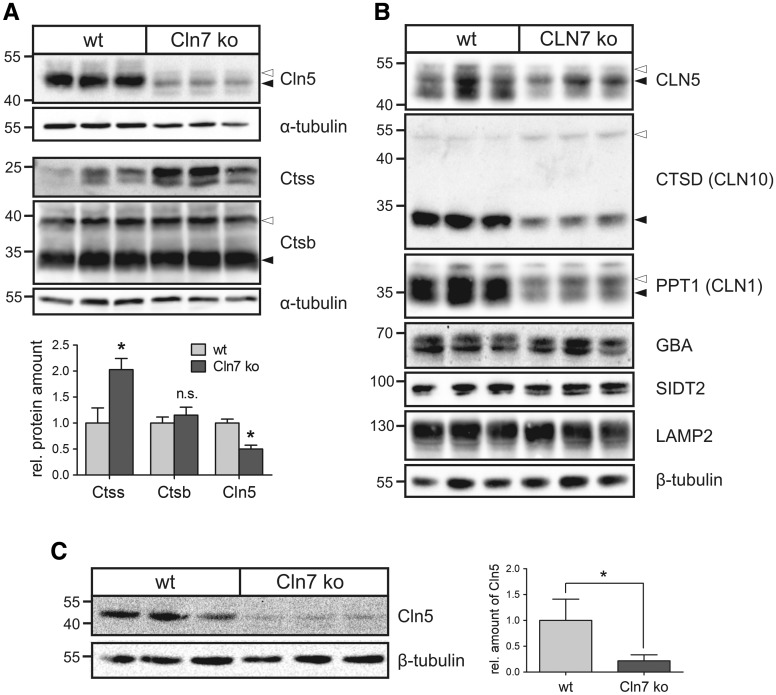
In order to verify dysregulated lysosomal proteins in Cln7 ko cells and tissues, western blot analyses were performed on primary mouse Cln7 ko macrophages, Cln7 ko brain tissue, and in the human HAP1 CLN7 ko cell line (Fig. 3). Immunoblot analyses showed that the Cln5 protein levels were decreased by 50% and the Ctss levels were elevated 2-fold in Cln7 ko macrophages compared with wild-type controls (Fig. 3A). Cathepsin B (Ctsb) levels were not altered in Cln7 ko macrophages. In the human CLN7 ko HAP1 cells, amounts of the NCL-related proteins—CLN5, cathepsin D (CTSD) and PPT1—were significantly reduced in comparison with wild-type cells (Fig. 3B). Both precursor and mature CLN5, CTSD and PPT1 proteins could be detected, indicating their correct targeting and proteolytic processing in CLN7 ko lysosomes. Soluble lysosomal glucocerebrosidase (GBA) and the lysosomal membrane proteins SIDT2 and lysosome-associated membrane protein 2 (LAMP2) remained unaltered in CLN7-deficient HAP1 cells, suggesting that the reduction in the three NCL-related proteins was not due to decreased number of lysosomes or impaired lysosomal integrity (Fig. 3B). Immunoblot analyses showed a reduction of Cln5 protein levels by 75% in Cln7 ko brain tissue compared with wild-type controls (Fig. 3C). These data indicate that depletion of selected soluble lysosomal proteins in Cln7 ko cells and tissues is not cell-type specific.
Figure 3.
Decreased Cln5 protein amounts in Cln7 ko cells and brain tissue. (A) Primary bone macrophages isolated from 5-month-old Cln7 ko and age-matched wild-type mice (N = 3) were cultivated for 14 days. Whole cell lysates were analysed by immunoblotting using antibodies against Cln5, cathepsins S (Ctss) and B (Ctsb). Equal loading was verified by α-tubulin western blotting. The positions of the molecular mass markers and the precursor (open arrowhead) and mature (filled arrowhead) Cln5 and Ctsb proteins, respectively, are indicated. Densitometric quantification of the immunoreactive band intensities has been performed and the relative protein amounts are shown in a bar diagram (mean ± SD, n = 3–5). n.s.: not significant, *P < 0.05 (two-tailed Student’s t-test). (B) Lysosome-enriched fractions of CLN7 ko and wild-type HAP1 cells were analysed by western blotting using antibodies against CLN5, cathepsin D (CTSD), PPT1, glucocerebrosidase (GBA), SIDT2 and LAMP2. Equal loading was confirmed by α-tubulin western blotting. The positions of the molecular mass markers and the precursor (open arrowhead) and mature (filled arrowhead) forms of CLN5, CTSD and PPT1, respectively, are indicated. (C) Whole brain lysates from three 10-month-old Cln7 ko and age-matched wild-type mice were analysed by Cln5 immunoblotting. β-Tubulin western blotting was used as loading control. The positions of the molecular mass markers are indicated. Bar diagram represents densitometric analysis of Cln5 protein levels normalized to the loading control (mean ± SD, n = 5). *P < 0.05 (two-tailed Student’s t-test).
Enhanced degradation of Cln5 by cysteine proteases in Cln7-deficient MEFs
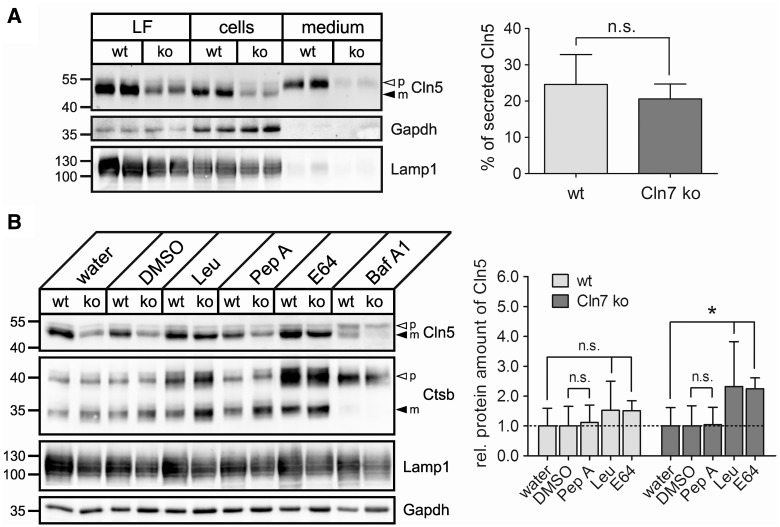
To examine whether mistargeting contributes to the depletion of mature Cln5 protein in Cln7-deficient cells, total cell extracts, lysosome-enriched fractions and conditioned media from Cln7 ko and control MEFs were collected and analysed by immunoblotting (Fig. 4A). In cell extracts and lysosomal fractions of Cln7 ko and wild-type MEFs, a 55 kDa precursor and a 50 kDa mature Cln5 form were detected, indicating correct targeting and proteolytic processing of Cln5 in lysosomes in the absence of Cln7. Similarly to the previous observations, the amounts of mature Cln5 were reduced in both total protein extracts and lysosomal fractions of Cln7 ko MEFs compared with controls. The amounts of secreted Cln5 precursor protein were comparable in MEFs of both genotypes indicating that Cln5 is not missorted into the medium in the absence of Cln7 (Fig. 4A). The levels of the lysosomal-associated membrane protein 1 (Lamp1) were unchanged in Cln7 ko MEFs compared with controls, suggesting no difference in the number of lysosomes between MEFs of both genotypes.
Figure 4.
Increased turnover of Cln5 by cysteine proteases in Cln7 ko lysosomes. (A) Wild-type and Cln7 ko MEFs were incubated in Opti-MEM medium for 24 h. Lysosome-enriched fractions (LF, 50 µg protein), whole cell extracts (100 µg protein) and conditioned media (20%) were analysed by western blotting using anti-Cln5 and anti-Lamp1 antibodies. Equal loading in LF and total cell extracts was confirmed by Lamp1 and Gapdh immunoblotting, respectively. The positions of the molecular mass markers and the precursor (p, open arrowhead) and mature (m, filled arrowhead) Cln5 proteins are indicated. Densitometric quantification of the immunoreactive band intensities from four independent experiments has been performed and the percentage of secreted Cln5 precursor forms related to the total amount of Cln5 is shown in a bar diagram (mean ± SD). n.s.: not significant (two-tailed Student’s t-test). (B) Wild-type and Cln7 ko MEFs were incubated in the presence of leupeptin (Leu, 100 µm), pepstatin A (Pep A, 30 µm), E64 (50 µm) or bafilomycin A1 (Baf A1, 100 nm) for 20 h. Water- and DMSO-treated cells served as negative controls for leupeptin/E64- and pepstatin/bafilomycin-treated cells, respectively. Whole cell extracts (75 µg protein) were analysed by immunoblotting using antibodies against Cln5, cathepsin B (Ctsb) and Lamp1. Equal loading was confirmed by Gapdh western blotting. The positions of the molecular mass markers as well as the precursor (p, open arrowhead) and mature (m, filled arrowhead) Cln5 and Ctsb proteins, respectively, are indicated. Densitometric quantification of the immunoreactive band intensities from at least three independent experiments was performed, and the total Cln5 protein amount was calculated (mean ± SD) relative to water or DMSO controls (set as 1.0). n.s.: not significant, *P < 0.05 compared with a corresponding negative control (two-tailed Student’s t-test).
To analyse whether increased turnover contributes to reduced amounts of mature Cln5 in Cln7 ko lysosomes, we cultured MEFs in the presence of cysteine and aspartyl protease inhibitors and analysed total cell extracts by immunoblotting (Fig. 4B). Treatment of Cln7 ko cells with the cysteine protease inhibitors leupeptin or E64 resulted in significantly increased amounts of mature Cln5 compared with water-treated control MEFs. In contrast, incubation of Cln7 ko MEFs with the aspartyl protease inhibitor pepstatin A did not affect the levels of Cln5 compared with DMSO-treated control MEFs. Inhibition of lysosomal acidification by the vacuolar-type H+-ATPase inhibitor bafilomycin A1 led to a reduced proteolytic processing to the mature Cln5 protein in MEFs of both genotypes and abolished processing of Ctsb. The levels of Ctsb and Lamp1 were not changed between Cln7 ko and wild-type MEFs at steady state (Fig. 4B). These data indicate that the depletion of mature Cln5 protein in Cln7 ko MEFs is due to its increased proteolytic degradation by cysteine proteases in lysosomes. CLN5 has been proposed to be synthesized as a type II membrane precursor protein which undergoes proteolytical cleavage in acidic compartments generating a mature protein that is tightly associated with lysosomal membranes via an amphipathic helix (30). To analyse whether altered membrane association of mature Cln5 might contribute to its increased proteolytic degradation in Cln7 ko lysosomes, we performed subcellular fractionation studies. In cells treated in the presence of E64, the majority of Cln5 was found in the soluble lysosomal fraction with a minor amount in the membrane fraction of both wild-type and Cln7 ko MEFs (Supplementary Material, Fig. S1A). Incubation of membranes with sodium carbonate completely released the membrane-associated fraction of Cln5 in MEFs of both genotypes. The amounts of Cln5 secreted into the media were not altered in Cln7 ko MEFs incubated in the presence of E64 compared with wild-type controls (Supplementary Material, Fig. S1B). Our data suggest that the distribution of soluble and membrane-associated Cln5 in lysosomes is not changed due to the absence of Cln7.
Attenuated mTORC1 reactivation during starvation in Cln7 knockout MEFs
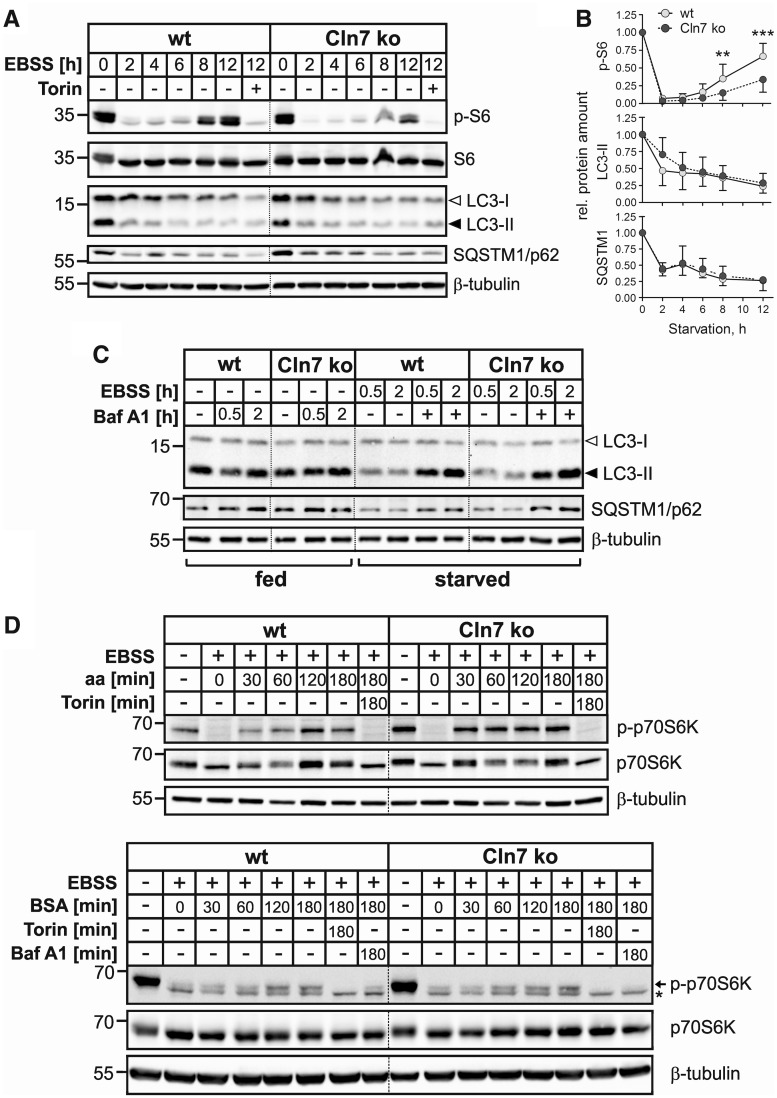
The mTOR complex 1 (mTORC1) regulates cell metabolism in response to environmental stimuli. Under nutrient starvation mTOR signalling is inactive, whereas amino acids provided by the degradation of autophagic substrates reactivate the mTORC1 pathway (31,32). Since dysregulated Mfsd8 mRNA expression levels upon amino acid starvation in a mouse hypothalamic cell line were reported (14), we analysed the effects of amino acid withdrawal in Cln7 ko MEFs. To investigate whether loss of Cln7 affects inactivation and reactivation of mTORC1, we monitored phosphorylation of the S6 ribosomal protein (S6), a downstream target of the mTORC1 pathway, during prolonged starvation in wild-type and Cln7 ko MEFs (Fig. 5A and B). In cells of both genotypes starved for 2 h, phosphorylation of S6 was completely inhibited, whereas after 4 h of starvation we observed recovery of S6 phosphorylation which was gradually increasing over time in MEFs of both genotypes. However, in Cln7 ko MEFs, the levels of phospho-S6 were significantly decreased after 8 and 12 h of starvation in comparison with control cells, indicating impaired mTOR signalling during ongoing starvation. Both in Cln7 ko and wild-type MEFs, phosphorylation of S6 was completely blocked in the presence of torin 1, a selective inhibitor of mTOR (Fig. 5A and B). To identify possible defects in basal and starvation-induced autophagy, Cln7 ko and control MEFs were starved for up to 12 h in amino acid- and serum-free medium and were analysed by immunoblotting using microtubule-associated protein 1 light chain 3B (MAP1LC3B) and the autophagic cargo receptor SQSTM1/p62 as marker proteins for autophagic protein turnover (Fig. 5A and B). Under basal conditions, levels of LC3-II in MEFs of both genotypes were comparable, which indicated similar number of autophagosomes in these cells. Under prolonged starvation, LC3-II and p62 levels in Cln7 ko MEFs decreased similarly to those in control MEFs, reflecting unchanged autophagy-mediated degradation of both proteins in Cln7 ko lysosomes (Fig. 5A and B). To measure autophagic flux, Cln7 ko and wild-type MEFs were incubated in the presence of bafilomycin A1 in complete or in amino acid- and serum-free starvation medium for 0.5 or 2 h (Fig. 5C). By inhibiting lysosomal acidification, bafilomycin A1 blocks the degradation inside lysosomes, resulting in the accumulation of both LC3-II and p62 (33). Western blot analyses of total cell extracts revealed comparable levels of LC3-II and p62 in bafilomycin-treated cells of both genotypes, suggesting that neither basal nor starvation-induced autophagic flux was compromised in Cln7 ko MEFs (Fig. 5C). By performing a similar experiment using primary hepatocytes we confirmed that autophagic flux was not affected in cells lacking Cln7 (Supplementary Material, Fig. S2B).
Figure 5.
mTORC1 reactivation upon starvation is impaired in Cln7 ko MEFs. (A) Wild-type (wt) and Cln7 ko MEFs were either incubated in the presence of serum and amino acids for 12 h or starved for 2, 4, 6, 8 or 12 h in amino acid-deficient EBSS medium lacking serum. MEFs starved in EBSS in the presence of the mTOR inhibitor torin 1 for 12 h were used as controls. Total cell extracts were prepared and analysed by phospho-S6 (p-S6; Ser235/236), S6, LC3 and SQSTM1/p62 immunoblotting. Equal loading was confirmed by β-tubulin western blotting. The positions of the molecular mass markers are indicated. (B) Densitometric analysis of the immunoreactive band intensities shown in (A). Data from at least three independent experiments were analysed. Results are shown as mean ± SD. ***P < 0.001, **P < 0.01 (two-tailed Student’s t-test). (C) Wild-type and Cln7 ko MEFs were incubated either in complete culture medium (fed) containing bafilomycin A1 (Baf A1, 100 nm) or in amino acid- and serum-free EBSS medium in the absence or presence of Baf A1 (starved) for 0.5 or 2 h. Total cell extracts were analysed by LC3 and SQSTM1/p62 immunoblotting. β-Tubulin immunoblotting was used as loading control. The positions of the molecular mass markers are indicated. (D) Wild-type and Cln7 ko MEFs were starved for amino acids and serum in EBSS medium for 1 h. Cells were either harvested or mTORC1 was re-activated by DMEM containing amino acids (aa, upper panel) or EBSS containing 3% (w/v) BSA (lower panel) for 30, 60, 120 and 180 min. MEFs incubated in DMEM in the presence of the mTOR inhibitor torin 1 (upper panel) or incubated in EBSS/3% BSA containing torin 1 or Baf A1 (lower panel) for 180 min were used as controls. Non-starved MEFs incubated in medium containing amino acids and serum were used as additional controls. Total cell extracts were prepared and analysed by phospho-p70S6K (p-p70S6K; Thr389) and p70S6K immunoblotting. The positions of the band corresponding to phospho-p70S6K and an unspecific band are marked with an arrow and an asterisk, respectively (lower panel). Equal loading was confirmed by β-tubulin western blotting. The positions of the molecular mass markers are indicated.
To analyse whether Cln7 deficiency impairs amino acid-dependent mTORC1 reactivation, MEFs were starved for 1 h in serum- and amino acid-free medium followed by the re-addition of amino acids and incubation for 0.5–3 h. mTORC1 inactivation and reactivation was monitored by the phosphorylation status of the mTORC1 downstream target p70 S6 kinase (p70S6K). As demonstrated by immunoblotting, withdrawal of amino acids and serum for 1 h resulted in a complete loss of phosphorylation of p70S6K in both Cln7 ko and wild-type MEFs (Fig. 5D, upper panel). Amino acid replenishment rapidly restored phosphorylation of p70S6K in MEFs of both genotypes, indicating that mTORC1 reactivation by amino acids is independent of Cln7. As negative control, starved cells were incubated with amino acids in the presence of the mTOR inhibitor torin 1 (Fig. 5D, upper panel). In both genotypes, phosphorylation of the mTORC1 substrate p70S6K was completely blocked by torin 1. To analyse whether mTORC1 reactivation by amino acids generated by protein degradation in starved Cln7 ko MEFs is impaired, we incubated Cln7 ko and wild-type MEFs in the presence of BSA after amino acid/serum starvation (Fig. 5D, lower panel). In both Cln7 ko and wild-type MEFs we observed phosphorylation of p70S6K which increased over time and was not altered compared with control cells. In control experiments using Cln7 ko and control MEFs incubated in the presence of torin 1, phosphorylated p70S6K was not detected indicating a complete silencing of BSA-dependent mTOR signalling by the inhibitor (Fig. 5D, lower panel). To rule out proteolytic degradation of added BSA in the medium, we analysed phosphorylation of p70S6K in cells treated with BSA in the presence of bafilomycin A1. We did not observe phosphorylation of p70S6K in bafilomycin-treated cells, indicating that mTORC1 reactivation by BSA in starved wild-type and Cln7 ko cells was due to the amino acids generated by degradation of intact endocytosed BSA in lysosomes (Fig. 5D, lower panel).
At the terminating stages of autophagy, when mTORC1 activity increases, the process of autophagic lysosome reformation (ALR) causes the generation of tubular structures extending from autolysosomes (34). To analyse whether ALR is also affected in the absence of Cln7, we starved wild-type and Cln7 ko MEFs for up to 12 h in amino acid- and serum-free medium and visualized Lamp1-positive lysosomes by immunofluorescence microscopy. After 12 h of starvation we observed formation of Lamp1-positive tubular structures in ∼65% of wild-type cells (Supplementary Material, Fig. S4A and B). Quantification revealed that the fraction of Cln7 ko MEFs containing tubular structures was significantly lower compared with wild-type controls after 12 h of starvation (20% versus 65%, Supplementary Material, Fig. S4B). These Lamp1-positive tubular structures emerging from autolysosomes were associated with microtubules which are required for ALR (Supplementary Material, Fig. S4A) (34). In addition, both under non-starvation and 12-h starvation conditions the perinuclear Lamp1 intensity was significantly higher in Cln7 ko MEFs compared with wild-type controls (Supplementary Material, Fig. S4C).
Discussion
Loss of Cln7 in the brain and retina of mice leads to lysosomal dysfunction, impaired macroautophagy and neurodegeneration (19,20). The molecular mechanisms and the link between the deficiency of the putative lysosomal transporter CLN7 and lysosomal dysfunction are unclear. Our comparative analyses of the lysosomal proteome of Cln7 ko MEFs showed that multiple soluble lysosomal enzymes involved in the degradation of glycans, peptides, lipids and fatty acid-modified proteins were significantly depleted in lysosomes in the absence of functional Cln7 (Fig. 1 and Supplementary Material, Table S1). In contrast, none of the identified membrane proteins was significantly altered in Cln7-deficient lysosomes (Supplementary Material, Table S2). To our knowledge, this report is the first to describe the lysosomal proteome of cells defective in an NCL-related lysosomal membrane protein.
Since Cln5 was one the most significantly depleted soluble lysosomal protein in Cln7 ko lysosomes and mutations in the CLN5 gene lead to CLN5 disease with a phenotype resembling that observed in CLN7 patients (1), we focused our analyses on expression, sorting and processing of Cln5 in Cln7-deficient MEFs (Figs 2–4). The function of the highly N-glycosylated Cln5 protein is unknown, but it has recently been proposed to function as a glycoside hydrolase in Dictyostelium and in humans (35). In the present study, decreased levels of CLN5 protein were observed in human CLN7 ko HAP1 cells, in Cln7 ko MEFs, in primary Cln7 ko mouse macrophages and in mouse brain, indicating cell type-independent depletion of CLN5 in the absence of CLN7. Our studies showed that reduced Cln5 levels in Cln7 ko lysosomes were not due to decreased mRNA transcription, missorting, or altered proteolytic processing. Rather, expression analyses in the presence of lysosomal protease inhibitors demonstrated that enhanced degradation by lysosomal cysteine proteases was primarily responsible for depletion of Cln5 in Cln7 ko lysosomes (Fig. 4). The similar disease onset, progression and phenotypes of CLN5 and CLN7 patients in combination with the observed depletion of Cln5 in Cln7 ko cells and tissues suggest that CLN7 and CLN5 may act in a common pathway which is disturbed in both diseases. Our findings along with the reported interactions of CLN5 with many of the known NCL proteins like PPT1, TPP1, CLN3, CLN6 and CLN8, support an essential role of CLN5 in the NCL network (36,37).
Our quantitative proteome and immunoblot analyses revealed a depletion of PPT1 and CTSD in lysosomes in the absence of CLN7 (Fig. 3). Significant deficiency in CTSD and PPT1 in humans leads to CLN1 and CLN10 disease, respectively (1). Of note, PPT1 has previously been reported to interact both with CTSD in the neuroblastoma cell line SH-SY5Y (38) and with CLN5 (37), whereas the Dictyostelium discoideum Ctsd homologue has been found in the CLN5 interactome (35). The combined depletion of Cln5, Ppt1 and Ctsd, which we observed in Cln7 ko MEFs, may play a specific role in the pathogenesis of CLN7 disease. Since changes in lipid and cholesterol metabolism have been shown in the brains of Cln1 ko and Cln5 ko mouse models, it is tempting to speculate that CLN7 deficiency may affect lipid metabolism in lysosomes (39–41).
The comparative proteome analyses of Cln7 ko lysosomes demonstrated quite modest alterations in soluble lysosomal enzymes (<2-fold for most of the altered proteins). Therefore, we hypothesized that challenging Cln7-deficient cells by nutrient depletion may reveal even more prominent defects. Basal mTORC1 activity and starvation-induced mTORC1 inactivation were comparable in Cln7 ko and wild-type MEFs (Fig. 5). However, under prolonged starvation we observed that reactivation of mTOR signalling was significantly impaired in Cln7 ko MEFs compared with controls. We then raised the question whether altered basal autophagy or autophagic flux may cause impaired mTORC1 reactivation during prolonged starvation in the absence of functional Cln7. We did not observe any defects in the autophagic degradation rate due to the absence of Cln7, as shown by an unaltered autophagic flux (Supplementary Material, Fig. S2) and a normal turnover of the autophagosomal marker protein LC3-II and the autophagy substrate p62 during starvation (Fig. 5A and B). Furthermore, we did not detect alterations in autophagosome-lysosome fusion in the course of autophagy, as shown by partial localization of LC3 puncta inside lysosomes (Supplementary Material, Fig. S2A), which suggested that early steps of autophagy were not affected in Cln7 ko cells. Taken together, our findings indicate that impaired mTORC1 reactivation during starvation in the absence of Cln7 is not caused by defects in basal or starvation-induced macroautophagy.
Amino acids present in the lysosomal lumen are potent activators of mTORC1 (42). To analyse whether impaired mTORC1 reactivation in Cln7 ko MEFs during prolonged starvation was due to defective transport of amino acids from lysosomes, we monitored mTOR activity after replenishment with amino acids or BSA (Fig. 5D). Activation of mTORC1 by amino acids was not altered in Cln7 ko cells either, indicating that the mTOR protein complex 1 is recruited to lysosomes, is functional, and responds to stimuli independently of loss of Cln7. Similarly, adding BSA extracellularly was sufficient to reactivate silenced mTOR signalling, which argued against proteolytic dysfunction of Cln7-deficient lysosomes, at least upon short-term starvation. Normal mTORC1 activities under basal and nutrient-replete conditions in Cln7 ko MEFs suggest that Cln7 is not involved in the transport of amino acids outside of lysosomes. Finally, taken together with unaltered turnover of LC3-II and p62, unchanged processing of soluble lysosomal enzymes (Ctsb, Ctsd, Cln5 and Ppt1), and enhanced cysteine protease-mediated degradation of Cln5 in Cln7 ko cells, these data suggest that impaired mTORC1 reactivation during amino acid starvation is not due to defective proteolytic capacity of Cln7 ko lysosomes at steady state.
In agreement with impaired mTORC1 reactivation during starvation in Cln7-defective cells, we observed an increased localization of enlarged autolysosomes in the perinuclear region of Cln7 ko cells and decreased numbers of Cln7 ko MEFs containing tubular structures extending from autolysosomes compared with controls (Supplementary Material, Fig. S4) (34,43). These findings suggest a defect in autophagic lysosome reformation (ALR) during starvation in Cln7 ko MEFs. Impaired ALR in starved Cln7 ko cells may lead to a depletion of newly generated functional lysosomes, which ultimately could aggravate lysosomal dysfunction in Cln7-deficient cells. Interestingly, after prolonged starvation for 24 h, we observed decreased levels of cathepsins B and D in Cln7 ko MEFs compared with controls (Supplementary Material, Fig. S3). Supporting these data, decreased amounts and reduced enzymatic activities of cathepsins B and D have previously been proposed to cause delayed ALR via impairment of mTOR signalling reactivation (44). Impaired mTORC1 reactivation and defective ALR during prolonged starvation have also been demonstrated in fibroblasts derived from patients with lysosomal storage disorders such as Scheie syndrome, Fabry disease and aspartylglucosaminuria (34), in cystinosin-deficient kidney cells (45) and in normal rat kidney cells depleted in the lysosomal MFS transporter Spinster (46). In spin knockdown cells starved for 12 h, reduced enzymatic activities of Ctsd were found, and the authors suggested that defective mTOR signalling causes impaired degradation in autolysosomes (46). Noteworthy, in the brain of Drosophila spin mutants an NCL-like phenotype, characterized by storage of lipofuscin, accumulation of ceramides, age-dependent synaptic dysfunction and progressive neurodegeneration, was reported (47,48), suggesting that impaired ALR and mTORC1 reactivation may contribute to lysosomal/autolysosomal dysfunction in other NCLs. In agreement, downregulation of mTOR signalling and dysfunction in the mTORC1 pathways have been reported in mouse and yeast models of juvenile CLN3 disease, respectively (23,49).
In summary, our data show that loss of functional Cln7 protein affects the amounts of multiple soluble lysosomal proteins. The combined depletion of several NCL-related lysosomal proteins including Ctsd, Ppt1 and Cln5 may contribute to lysosomal dysfunction in Cln7-deficient fibroblasts. In addition, we found a defect in the ability of Cln7 ko MEFs to adapt to prolonged starvation conditions as shown by impaired mTORC1 reactivation, reduced tubulation of autolysosomes and increased perinuclear accumulation of autolysosomes. The exact role of CLN7 in mTOR signalling during long-term starvation and its relevance in neuronal cells, however, remains to be elucidated and will be addressed in future studies.
Materials and Methods
Antibodies and reagents
Primary antibodies were: goat anti-mouse cathepsin B (Neuromics, GT15047), goat anti-mouse cathepsin D (Santa Cruz, sc6486), monoclonal mouse anti-α-tubulin (Sigma, T9026), polyclonal rabbit anti-Cln5 (Abcam, ab170899), polyclonal goat anti-cathepsin S (Santa-Cruz, sc6503), polyclonal goat anti-mouse Dpp7 (R&D, AF3436), polyclonal rabbit anti-glucocerebrosidase (Sigma, G4171), polyclonal rabbit anti-LC3B/MAP1LC3B (Novusbio, NB100–2220), polyclonal rabbit anti-p62 (Enzo Life Sciences, BML-PW9860), polyclonal rabbit anti-human PPT1 (Millipore, ABS1118), polyclonal rabbit anti-glyceraldehyde-3-phosphate dehydrogenase (Gapdh, Santa Cruz, sc25778), polyclonal rabbit anti-SIDT2 (Thermo Fischer Scientific, PA5–34493). Monoclonal rabbit anti-S6 ribosomal protein (clone 5G10), monoclonal rabbit anti-phospho-S6 ribosomal protein (Ser235/236, clone D57.2.2E), monoclonal rabbit anti-p70 S6 kinase (clone 49D7) and monoclonal rabbit anti-phospho-p70 S6 kinase (Thr389, clone 108D2) were from Cell Signalling. The monoclonal rat anti-mouse Lamp1 antibody (clone 1D4B), rat anti-mouse Lamp2 (clone ABL-93) and mouse anti-human LAMP2 (clone H4B4) developed by J. Thomas August, and the monoclonal mouse anti-β-tubulin antibody (clone E7) developed by M. Klymkowsky were obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at the Department of Biology, University of Iowa, Iowa City, USA. Secondary goat anti-mouse, goat anti-rabbit, rabbit anti-goat and goat anti-rat antibodies coupled to horseradish peroxidase or Alexa Fluor®-fluorophores were from Dianova.
KAPA™ Mouse Genotyping Hot Start Kit and Peqgold Total RNA kit were from VWR. Nitrocellulose and PVDF membranes were purchased from GE Healthcare Life Sciences and Roche, respectively. Albumin standard, restriction enzymes, RT-PCR TaqMan® gene expression assays, High Capacity cDNA Reverse Transcription kit, reagents for SILAC labelling of MEFs and enhanced chemiluminescence (ECL) reagent were from Thermo Fisher Scientific. Protease-free bovine albumin (fraction V) was from Serva. Gel extraction and PCR purification kits were from Qiagen. DMEM, IMDM, RPMI-1640, EBSS, Opti-MEM media and Glutamax were purchased from Gibco. Leupeptin, pepstatin A, E64, bafilomycin A1, β-glucosidase from almonds, 4-methylumbelliferone, enzyme substrates (Supplementary Material, Table S3) and protease inhibitor cocktail were from Sigma. Torin 1 was purchased from Tocris Bioscience and phosphatase inhibitor cocktail (PhosSTOP) was from Roche. Macrophage colony-stimulating factor was from Peprotech. Dextran-stabilized magnetite particles were purchased from Liquids Research Ltd., and isolation of lysosomes was performed using MACS LS Separation columns from Miltenyi Biotec.
Cell culture
Immortalized wild-type and Cln7 ko MEFs were described previously (19) and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% foetal bovine serum (FBS), Glutamax and penicillin/streptomycin. A Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-Cas9 edited human haploid HAP1 MFSD8 knockout cell line was commercially obtained from Horizon Discoveries (Vienna, Austria) and was generated by a 20 bp deletion in exon 4 of the human MFSD8 gene. HAP1 cells were cultured in Iscove’s modified Dulbeccos’s medium (IMDM) supplemented with 10% FBS, Glutamax and penicillin/streptomycin. Primary bone marrow macrophages were isolated from 5-month-old mice and maintained in RPMI-1640 medium containing 10% FBS, Glutamax, penicillin/streptomycin and 50 ng/ml macrophage colony-stimulating factor (M-CSF). All cells were cultured at 37°C, in a humidified atmosphere containing 5% CO2.
Stable isotope labelling by amino acids of cultured MEFs and proteome analysis of lysosomal fractions
Labelling of immortalized MEFs with light and heavy isotopes and subsequent isolation of magnetite-loaded lysosomes was performed as described previously (50). Briefly, immortalized Cln7 ko and wild-type MEFs were cultured for six passages in DMEM for SILAC supplemented with 10% FBS and conventional light- (Arg/Lys) or heavy-labelled (Arg (13N4)/Lys (13N2) isotopes at concentrations of 87.8 mg/l (Arg) and 181.2 mg/l (Lys)). Prior to lysosomal isolation, the cells were incubated in the corresponding light- or heavy-labelled medium supplemented with 10% dextran-stabilized magnetite particles for 24 h. After removal of the magnetic beads, fresh medium was added for a 36 h chase time. Postnuclear supernatants of Cln7 ko and wild-type MEFs were prepared, mixed and magnetite-containing lysosomes were isolated using a Miltenyi LS magnetic column. Preparation of lysosomal fractions for mass spectrometry analyses was performed as described previously (50).
Experimental animals
Cln7 ko mice have been described previously (19). Animals were maintained under standard housing conditions on a 12 h light and 12 h dark schedule in a pathogen-free animal facility at the University Medical Center. Removal of tissues was approved by the local animal welfare officer (ORG769).
Isolation and culture of bone marrow derived macrophages
Experimental mice were sacrificed in a CO2 chamber and femurs and tibias were aseptically removed. The bone marrow was then flushed out with RPMI medium supplemented with 10% FBS, Glutamax and penicillin/streptomycin. The obtained cell suspension was filtered through a 70 µm cell strainer, and the cells were finally plated at a density of 5 × 105 cells/cm2. In order to induce macrophage differentiation, M-CSF was added to the culture medium at a final concentration of 50 ng/ml, and the cells were further cultivated for 2 weeks with the medium exchanged every other day.
Isolation and culture of primary mouse hepatocytes
Primary mouse hepatocytes were isolated from livers of anesthetized wild-type and Cln7 ko mice by perfusion with oxygenated perfusion buffer (pH 7.4) containing 0.14 M NaCl, 5 mm KCl, 0.8 mm MgCl2, 1.6 mm Na2HPO4, 0.4 mm KH2PO4, 25 mm NaHCO3, 2.5 mm EDTA, 16.5 mm glucose, 7.5 mm sodium lactate and 0.2 mm sodium pyruvate. Isolated hepatocytes were suspended in culture medium (DMEM supplemented with Glutamax, 10% FBS and penicillin/streptomycin) and centrifuged at 28g. Stromal cells were removed by gradient centrifugation using 60% Percoll. Hepatocytes were seeded in collagen-coated 12-well plates at 2.5 × 105 cells/well, and the experiment was performed the next day.
Treatment with lysosomal protease inhibitors
Immortalized MEFs were seeded in 60 mm dishes and cultured to confluence. The cells were incubated in fresh medium supplemented with lysosomal protease inhibitors at the indicated concentrations for 20 h. Cells were washed twice with ice-cold PBS, harvested and lysed in ice-cold PBS containing 0.5% (v/v) Triton X-100 (TX-100), 1 mm EDTA and protease inhibitors. Total protein extracts were analysed by immunoblotting.
Preparation of lysosome-enriched fractions
Following indicated treatment, MEFs were washed twice with ice-cold PBS and harvested. The cells were resuspended in at least 5-fold volume of ice-cold homogenization buffer (10 mm phosphate buffer, pH 7.3, 250 mm sucrose, 1 mm EDTA, protease inhibitors) and disrupted on ice by trituration (30 times) through a 27-gauge needle attached to a syringe. The obtained homogenate was further clarified from nuclei and unbroken cells at 1000g for 5 min at 4°C, and the post-nuclear supernatant (PNS) was collected in a new tube. The PNS was centrifuged at 20 000g for 10 min at 4°C and the pellet representing the lysosome-enriched fraction (LF) was resuspended in lysis buffer (50 mm Tris–HCl, pH 7.4, 1% (v/v) TX-100, 150 mm NaCl, 1 mm EDTA, protease inhibitors) and incubated for 30 min on ice. Finally, LFs were clarified by centrifugation at 16 000g for 15 min at 4°C and analysed by immunoblotting.
Membrane extraction assay
Wild-type and Cln7 ko MEFs grown in three 10 cm dishes were incubated in the presence of E64 (10 µM) for 24 h. After removal of the media, cells were washed three times with ice-cold PBS and harvested by scraping. Light mitochondrial fractions (LMF), containing lysosomes, prepared by differential centrifugation were resuspended in ice-cold 10 mm Tris–HCl buffer (pH 7.5) supplemented with protease inhibitors, and lysosomes were disrupted by repeating cycles of freezing in dry ice/ethanol and thawing at 37°C for five times. Following sonication, LMFs were centrifuged for 60 min at 100 000g at 4°C to separate fractions containing soluble lysosomal proteins (supernatant) and membrane proteins (pellet). Membranes (100 µg protein) were further extracted with 1% (v/v) TX-100 (total membranes) in 10 mm Tris–HCl (pH 7.5) or 0.1 M Na2CO3 (pH 11) containing protease inhibitors for 30 min on ice. After a final centrifugation at 100 000g for 30 min, supernatants and pellets were analysed by immunoblotting. For quantification of Cln5 in cells and media, MEFs were incubated in Opti-MEM in the absence or presence of E64 (10 µM) for 24 h. Media were collected and concentrated using Ultra centrifugal filters (Amicon). Total cell extracts and aliquots of the conditioned media were analysed by western blotting.
Immunoblotting
SDS-PAGE and immunoblotting were performed as described previously (19). Immunoreactive band intensities were quantified by densitometry using Image Lab software (Bio-Rad).
Enzyme activity measurements
The enzymatic activities of lysosomal β-hexosaminidase (Hexb), β-galactosidase (Glb), β-glucuronidase (Gusb) and α-mannosidase (Man) in whole cell extracts of MEFs were measured in a colorimetric or fluorometric assay using corresponding 4-nitrophenyl- or 4-methylumbelliferyl- (MU) based substrates (Supplementary Material, Table S3) (51, 52). All enzymatic assays were performed in a microplate format. A specific enzyme activity of β-hexosaminidase was assayed at 37°C in a total volume of 30 µl. The assay mixture contained 100 mm sodium citrate (pH 4.6), 0.1% (v/v) TX-100, 0.2% (w/v) BSA and 4 µg of a whole cell extract. The reaction was initiated with 5 mm substrate and terminated after 45 min by adding 140 µl of 0.4 M glycine/NaOH (pH 10.4). The absorbance of the liberated 4-nitrophenol (ɛ = 18 500 M−1*cm−1) was measured at 405 nm. To measure activities of β-galactosidase, β-glucuronidase and α-mannosidase, respectively 4, 8 and 16 µg of whole cell extracts were added to the reaction mixture which consisted of a corresponding substrate at given concentration, 100 mm sodium citrate (pH 4.6), 0.1% (v/v) TX-100 and 150 mm NaCl, in a final volume of 80 µl. After 5 h of incubation at 37°C, the reaction was terminated with 120 µl stop-buffer (0.4 M glycine/NaOH, pH 10.4) and fluorescence of the released 4-methylumbelliferone (4-MU) was measured using a Fluoroscan AscentTM microplate fluorometer (Thermo Fischer Scientific) with excitation at 355 nm and emission at 460 nm. For all fluorometric assays, a set of standard concentrations of 4-MU was used to generate a calibration curve and to calculate the amount of the released product.
RNA isolation and quantitative RT-PCR
Total RNA was isolated from cultured MEFs using Peqgold Total RNA kit according to the manufacturer’s instructions. The purity of the RNA samples was verified by visualization of 18S and 28S RNA bands after agarose gel electrophoresis. The High Capacity cDNA Reverse Transcription kit was used to convert 1 µg of total RNA to cDNA following the manufacturer's instructions. For the mRNA expression analysis, quantitative RT-PCR was performed as described previously (28), on a Mx3000P® QPCR system (Stratagene) using pre-designed probes and primer sets (Supplementary Material, Table S4). The relative mRNA expression levels were normalized to the expression levels of Gapdh mRNA in the same sample using the comparative 2−ΔΔCT method.
Reactivation of mTORC1 activity during prolonged starvation
Prolonged starvation of cultured MEFs was performed as described previously (35). Confluent MEFs were washed three times with PBS and incubated in either serum-containing DMEM or amino acid- and serum-free Earle’s balanced salt solution (EBSS) for 2, 4, 6, 8 and 12 h. EBSS medium containing torin 1 (250 nm) was used for complete inhibition of mTORC1 reactivation in MEFs starved for 12 h.
Reactivation of mTORC1 activity after amino acid starvation
MEFs were grown in complete growth medium to confluence. After washing three times with PBS, cells were incubated for 60 min in amino acid- and serum-free EBSS. Cells were either harvested or incubated in amino acid-containing DMEM medium lacking serum or EBSS containing 3% (w/v) BSA for 30, 60, 120 and 180 min. Cells incubated in the presence of the mTOR inhibitor torin 1 (250 nm) were used as positive control for complete inhibition of mTORC1 activity. Cell pellets were lysed in 50 mm Tris–HCl buffer (pH 7.5) containing 1% (v/v) TX-100, 150 mm NaCl as well as phosphatase and protease inhibitors.
Statistical analysis of the data
The experiments were performed at least in triplicates, and the data were plotted as mean ± SD. The two-tailed Student’s t-test was used to compare the obtained mean values. The difference between the values was considered statistically significant if P < 0.05 (*), P < 0.01 (**) and P < 0.001 (***). Statistical analyses were performed using GraphPad Prism software.
Supplementary Material
Supplementary material is available at HMG online.
Supplementary Material
Acknowledgements
We thank Kerstin Cornils (Research Institute Children’s Cancer Center Hamburg, UKE) for providing SV40 lentiviral supernatants for immortalization of MEFs, Sandra Ehret (Institute of Biochemistry and Molecular Cell Biology, UKE) and the UKE microscopy imaging facility (UMIF) for expert assistance.
Conflict of Interest Statement. None declared.
Funding
This work was supported by the European Union's Horizon 2020 research and innovation programme under grant agreement No. 666918 (BATCure, to T.D., S.M. and S.S.) and the Deutsche Forschungsgemeinschaft research training group (grant GRK1459 to K.A. and S.S.). Funding to pay the Open Access publication charges for this article was provided by the European Union's Horizon 2020 research and innovation programme under grant agreement No. 666918.
References
- 1. Mole S.E., Williams R.E., Goebel H.H. (2011) The Neuronal Ceroid Lipofuscinoses (Batten Disease) Contemporary Neurology Series, 2nd edn Oxford University Press Inc, New York. [Google Scholar]
- 2. Kollmann K., Uusi-Rauva K., Scifo E., Tyynelä J., Jalanko A., Braulke T. (2013) Cell biology and function of neuronal ceroid lipofuscinosis-related proteins. Biochim. Biophys. Acta, 1832, 1866–1881. [DOI] [PubMed] [Google Scholar]
- 3. Di Fruscio G., Schulz A., De Cegli R., Savarese M., Mutarelli M., Parenti G., Banfi S., Braulke T., Nigro V., Ballabio A. (2015) Lysoplex: an efficient toolkit to detect DNA sequence variations in the autophagy-lysosomal pathway. Autophagy, 11, 928–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Siintola E., Topcu M., Aula N., Lohi H., Minassian B.A., Paterson A.D., Liu X.Q., Wilson C., Lahtinen U., Anttonen A.K.. et al. (2007) The novel neuronal ceroid lipofuscinosis gene MFSD8 encodes a putative lysosomal transporter. Am. J. Hum. Genet., 81, 136–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kousi M., Lehesjoki A.E., Mole S.E. (2012) Update of the mutation spectrum and clinical correlations of over 360 mutations in eight genes that underlie the neuronal ceroid lipofuscinoses. Hum. Mutat., 33, 42–63. [DOI] [PubMed] [Google Scholar]
- 6. Aiello C., Terracciano A., Simonati A., Discepoli G., Cannelli N., Claps D., Crow Y.J., Bianchi M., Kitzmüller C., Longo D.. et al. (2009) Mutations in MFSD8/CLN7 are a frequent cause of variant-late infantile neuronal ceroid lipofuscinosis. Hum. Mutat., 30, E530–E540. [DOI] [PubMed] [Google Scholar]
- 7. Aldahmesh M.A., Al-Hassnan Z.N., Aldosari M., Alkuraya F.S. (2009) Neuronal ceroid lipofuscinosis caused by MFSD8 mutations: a common theme emerging. Neurogenetics, 10, 307–311. [DOI] [PubMed] [Google Scholar]
- 8. Kousi M., Siintola E., Dvorakova L., Vlaskova H., Turnbull J., Topcu M., Yuksel D., Gokben S., Minassian B.A., Elleder M.. et al. (2009) Mutations in CLN7/MFSD8 are a common cause of variant late-infantile neuronal ceroid lipofuscinosis. Brain, 132, 810–819. [DOI] [PubMed] [Google Scholar]
- 9. Mandel H., Cohen Katsanelson K., Khayat M., Chervinsky I., Vladovski E., Iancu T.C., Indelman M., Horovitz Y., Sprecher E., Shalev S.A.. et al. (2014) Clinico-pathological manifestations of variant late infantile neuronal ceroid lipofuscinosis (vLINCL) caused by a novel mutation in MFSD8 gene. Eur. J. Med. Genet., 57, 607–612. [DOI] [PubMed] [Google Scholar]
- 10. Stogmann E., El Tawil S., Wagenstaller J., Gaber A., Edris S., Abdelhady A., Assem-Hilger E., Leutmezer F., Bonelli S., Baumgartner C.. et al. (2009) A novel mutation in the MFSD8 gene in late infantile neuronal ceroid lipofuscinosis. Neurogenetics, 10, 73–77. [DOI] [PubMed] [Google Scholar]
- 11. Patino L.C., Battu R., Ortega-Recalde O., Nallathambi J., Anandula V.R., Renukaradhya U., Laissue P. (2014) Exome sequencing is an efficient tool for variant late-infantile neuronal ceroid lipofuscinosis molecular diagnosis. PLoS One, 9, e109576.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Saier M.H. Jr., Reddy V.S., Tsu B.V., Ahmed M.S., Li C., Moreno-Hagelsieb G. (2016) The Transporter Classification Database (TCDB): recent advances. Nucleic Acids Res., 44, D372–D379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reddy V.S., Shlykov M.A., Castillo R., Sun E.I., Saier M.H. Jr. (2012) The major facilitator superfamily (MFS) revisited. FEBS J., 279, 2022–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Perland E., Bagchi S., Klaesson A., Fredriksson R. (2017) Characteristics of 29 novel atypical solute carriers of major facilitator superfamily type: evolutionary conservation, predicted structure and neuronal co-expression. Open Biol., 7, 170142.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bagshaw R.D., Mahuran D.J., Callahan J.W. (2005) Lysosomal membrane proteomics and biogenesis of lysosomes. Mol. Neurobiol., 32, 27–41. [DOI] [PubMed] [Google Scholar]
- 16. Schröder B., Wrocklage C., Pan C., Jäger R., Kösters B., Schäfer H., Elsässer H.P., Mann M., Hasilik A. (2007) Integral and associated lysosomal membrane proteins. Traffic, 8, 1676–1686. [DOI] [PubMed] [Google Scholar]
- 17. Damme M., Brandenstein L., Fehr S., Jankowiak W., Bartsch U., Schweizer M., Hermans-Borgmeyer I., Storch S. (2014) Gene disruption of Mfsd8 in mice provides the first animal model for CLN7 disease. Neurobiol. Dis., 65, 12–24. [DOI] [PubMed] [Google Scholar]
- 18. Sharifi A., Kousi M., Sagne C., Bellenchi G.C., Morel L., Darmon M., Hulkova H., Ruivo R., Debacker C., El Mestikawy S.. et al. (2010) Expression and lysosomal targeting of CLN7, a major facilitator superfamily transporter associated with variant late-infantile neuronal ceroid lipofuscinosis. Hum. Mol. Genet., 19, 4497–4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brandenstein L., Schweizer M., Sedlacik J., Fiehler J., Storch S. (2016) Lysosomal dysfunction and impaired autophagy in a novel mouse model deficient for the lysosomal membrane protein Cln7. Hum. Mol. Genet., 25, 777–791. [DOI] [PubMed] [Google Scholar]
- 20. Jankowiak W., Brandenstein L., Dulz S., Hagel C., Storch S., Bartsch U. (2016) Retinal degeneration in mice deficient in the lysosomal membrane protein CLN7. Invest. Ophthalmol. Vis. Sci., 57, 4989–4998. [DOI] [PubMed] [Google Scholar]
- 21. Damme M., Suntio T., Saftig P., Eskelinen E.L. (2015) Autophagy in neuronal cells: general principles and physiological and pathological functions. Acta Neuropathol., 129, 337–362. [DOI] [PubMed] [Google Scholar]
- 22. Micsenyi M.C., Sikora J., Stephney G., Dobrenis K., Walkley S.U. (2013) Lysosomal membrane permeability stimulates protein aggregate formation in neurons of a lysosomal disease. J. Neurosci., 33, 10815–10827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cao Y., Espinola J.A., Fossale E., Massey A.C., Cuervo A.M., MacDonald M.E., Cotman S.L. (2006) Autophagy is disrupted in a knock-in mouse model of juvenile neuronal ceroid lipofuscinosis. J. Biol. Chem., 281, 20483–20493. [DOI] [PubMed] [Google Scholar]
- 24. Leinonen H., Keksa-Goldsteine V., Ragauskas S., Kohlmann P., Singh Y., Savchenko E., Puranen J., Malm T., Kalesnykas G., Koistinaho J.. et al. (2017) Retinal degeneration in a mouse model of CLN5 disease is associated with compromised autophagy. Sci. Rep., 7, 1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thelen M., Damme M., Schweizer M., Hagel C., Wong A.M., Cooper J.D., Braulke T., Galliciotti G. (2012) Disruption of the autophagy-lysosome pathway is involved in neuropathology of the nclf mouse model of neuronal ceroid lipofuscinosis. PLoS One, 7, e35493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koike M., Nakanishi H., Saftig P., Ezaki J., Isahara K., Ohsawa Y., Schulz-Schaeffer W., Watanabe T., Waguri S., Kametaka S.. et al. (2000) Cathepsin D deficiency induces lysosomal storage with ceroid lipofuscin in mouse CNS neurons. J. Neurosci., 20, 6898–6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mitchison H.M., Bernard D.J., Greene N.D., Cooper J.D., Junaid M.A., Pullarkat R.K., de Vos N., Breuning M.H., Owens J.W., Mobley W.C.. et al. (1999) Targeted disruption of the Cln3 gene provides a mouse model for Batten disease. The Batten Mouse Model Consortium [corrected]. Neurobiol. Dis., 6, 321–334. [DOI] [PubMed] [Google Scholar]
- 28. Pohl S., Mitchison H.M., Kohlschütter A., van Diggelen O., Braulke T., Storch S. (2007) Increased expression of lysosomal acid phosphatase in CLN3-defective cells and mouse brain tissue. J. Neurochem., 103, 2177–2188. [DOI] [PubMed] [Google Scholar]
- 29. Sleat D.E., Sohar I., Pullarkat P.S., Lobel P., Pullarkat R.K. (1998) Specific alterations in levels of mannose 6-phosphorylated glycoproteins in different neuronal ceroid lipofuscinoses. Biochem. J., 334, 547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Larkin H., Ribeiro M.G., Lavoie C. (2013) Topology and membrane anchoring of the lysosomal storage disease-related protein CLN5. Hum. Mutat., 34, 1688–1697. [DOI] [PubMed] [Google Scholar]
- 31. Saxton R.A., Sabatini D.M. (2017) mTOR signaling in growth, metabolism, and disease. Cell, 168, 960–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zoncu R., Bar-Peled L., Efeyan A., Wang S., Sancak Y., Sabatini D.M. (2011) mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science, 334, 678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mizushima N., Yoshimori T., Levine B. (2010) Methods in mammalian autophagy research. Cell, 140, 313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yu L., McPhee C.K., Zheng L., Mardones G.A., Rong Y., Peng J., Mi N., Zhao Y., Liu Z., Wan F.. et al. (2010) Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature, 465, 942–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huber R.J., Mathavarajah S. (2017) Cln5 is secreted and functions as a glycoside hydrolase in Dictyostelium. Cell Signal., 42, 236–248. [DOI] [PubMed] [Google Scholar]
- 36. Vesa J., Chin M.H., Oelgeschlager K., Isosomppi J., DellAngelica E.C., Jalanko A., Peltonen L. (2002) Neuronal ceroid lipofuscinoses are connected at molecular level: interaction of CLN5 protein with CLN2 and CLN3. Mol. Biol. Cell, 13, 2410–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lyly A., von Schantz C., Heine C., Schmiedt M.L., Sipila T., Jalanko A., Kyttälä A. (2009) Novel interactions of CLN5 support molecular networking between Neuronal Ceroid Lipofuscinosis proteins. BMC Cell Biol., 10, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Scifo E., Szwajda A., Soliymani R., Pezzini F., Bianchi M., Dapkunas A., Dębski J., Uusi-Rauva K., Dadlez M., Gingras A.-C.. et al. (2015) Quantitative analysis of PPT1 interactome in human neuroblastoma cells. Data Brief., 4, 207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schmiedt M.L., Blom T., Blom T., Kopra O., Wong A., von Schantz-Fant C., Ikonen E., Kuronen M., Jauhiainen M., Cooper J.D.. et al. (2012) Cln5-deficiency in mice leads to microglial activation, defective myelination and changes in lipid metabolism. Neurobiol. Dis., 46, 19–29. [DOI] [PubMed] [Google Scholar]
- 40. Lyly A., Marjavaara S.K., Kyttälä A., Uusi-Rauva K., Luiro K., Kopra O., Martinez L.O., Tanhuanpaa K., Kalkkinen N., Suomalainen A.. et al. (2008) Deficiency of the INCL protein Ppt1 results in changes in ectopic F1-ATP synthase and altered cholesterol metabolism. Hum. Mol. Genet., 17, 1406–1417. [DOI] [PubMed] [Google Scholar]
- 41. Ahtiainen L., Kolikova J., Mutka A.L., Luiro K., Gentile M., Ikonen E., Khiroug L., Jalanko A., Kopra O. (2007) Palmitoyl protein thioesterase 1 (Ppt1)-deficient mouse neurons show alterations in cholesterol metabolism and calcium homeostasis prior to synaptic dysfunction. Neurobiol. Dis., 28, 52–64. [DOI] [PubMed] [Google Scholar]
- 42. Efeyan A., Zoncu R., Sabatini D.M. (2012) Amino acids and mTORC1: from lysosomes to disease. Trends Mol. Med., 18, 524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Korolchuk V.I., Saiki S., Lichtenberg M., Siddiqi F.H., Roberts E.A., Imarisio S., Jahreiss L., Sarkar S., Futter M., Menzies F.M.. et al. (2011) Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol., 13, 453–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tatti M., Motta M., Di Bartolomeo S., Scarpa S., Cianfanelli V., Cecconi F., Salvioli R. (2012) Reduced cathepsins B and D cause impaired autophagic degradation that can be almost completely restored by overexpression of these two proteases in Sap C-deficient fibroblasts. Hum. Mol. Genet., 21, 5159–5173. [DOI] [PubMed] [Google Scholar]
- 45. Ivanova E.A., van den Heuvel L.P., Elmonem M.A., De Smedt H., Missiaen L., Pastore A., Mekahli D., Bultynck G., Levtchenko E.N. (2016) Altered mTOR signalling in nephropathic cystinosis. J. Inherit. Metab. Dis., 39, 457–464. [DOI] [PubMed] [Google Scholar]
- 46. Rong Y., McPhee C.K., Deng S., Huang L., Chen L., Liu M., Tracy K., Baehrecke E.H., Yu L., Lenardo M.J. (2011) Spinster is required for autophagic lysosome reformation and mTOR reactivation following starvation. Proc. Natl. Acad. Sci. U S A., 108, 7826–7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dermaut B., Norga K.K., Kania A., Verstreken P., Pan H., Zhou Y., Callaerts P., Bellen H.J. (2005) Aberrant lysosomal carbohydrate storage accompanies endocytic defects and neurodegeneration in Drosophila benchwarmer. J. Cell Biol., 170, 127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hebbar S., Khandelwal A., Jayashree R., Hindle S.J., Chiang Y.N., Yew J.Y., Sweeney S.T., Schwudke D. (2017) Lipid metabolic perturbation is an early-onset phenotype in adult spinster mutants: a Drosophila model for lysosomal storage disorders. Mol. Biol. Cell, 28, 3728–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bond M.E., Brown R., Rallis C., Bahler J., Mole S.E. (2015) A central role for TOR signalling in a yeast model for juvenile CLN3 disease. Microb. Cell, 2, 466–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Markmann S., Thelen M., Cornils K., Schweizer M., Brocke-Ahmadinejad N., Willnow T., Heeren J., Gieselmann V., Braulke T., Kollmann K. (2015) Lrp1/LDL receptor play critical roles in mannose 6-phosphate-independent lysosomal enzyme targeting. Traffic, 16, 743–759. [DOI] [PubMed] [Google Scholar]
- 51. Kollmann K., Damme M., Markmann S., Morelle W., Schweizer M., Hermans-Borgmeyer I., Röchert A.K., Pohl S., Lübke T., Michalski J.C.. et al. (2012) Lysosomal dysfunction causes neurodegeneration in mucolipidosis II ‘knock-in’ mice. Brain, 135, 2661–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. van Diggelen O.P., Zhao H., Kleijer W.J., Janse H.C., Poorthuis B.J., van Pelt J., Kamerling J.P., Galjaard H. (1990) A fluorimetric enzyme assay for the diagnosis of Morquio disease type A (MPS IV A). Clin. Chim. Acta, 187, 131–139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.