Abstract
The TMEM127 tumor suppressor gene encodes a transmembrane protein of unknown function mutated in pheochromocytomas and, rarely, in renal cancers. Tumors with inactivating TMEM127 mutations have increased mTORC1 signaling by undefined mechanisms. Here we report that TMEM127 interacts with the lysosome-anchored complex comprised of Rag GTPases, the LAMTOR pentamer (or ‘ragulator’) and vATPase, which controls amino acid-mediated mTORC1 activation. We found that under nutrient-rich conditions TMEM127 expression reduces mTORC1 recruitment to Rags. In addition, TMEM127 interacts with LAMTOR in an amino acid-dependent manner and decreases the LAMTOR1–vATPase association, while TMEM127–vATPase binding requires intact lysosomal acidification but is amino acid independent. Conversely, both murine and human cells lacking TMEM127 accumulate LAMTOR proteins in the lysosome. Consistent with these findings, pheochromocytomas with TMEM127 mutations have increased levels of LAMTOR proteins. These results suggest that TMEM127 interactions with ragulator and vATPase at the lysosome contribute to restrain mTORC1 signaling in response to amino acids, thus explaining the increased mTORC1 activation seen in TMEM127-deficient tumors.
Introduction
TMEM127 is a susceptibility gene in catecholamine-secreting tumors, pheochromocytomas, and, more rarely, in renal cell carcinomas (1–4). Truncating or missense TMEM127 mutations are detected in the germline of these patients, often accompanied by loss of the wild-type (WT) allele in tumor DNA and consistent with a classic tumor suppressor gene mode of inactivation (2). The TMEM127 gene encodes a transmembrane protein that is ubiquitously expressed, highly conserved and has no ascribed function. We previously showed that recombinant WT TMEM127 exhibits colocalization with multiple intracellular endomembrane domains, including early and late endosomes and lysosomes (1). In agreement with these findings, TMEM127 was recovered in an unbiased screen aimed at identifying novel lysosomal membrane proteins (5). Our earlier studies show that tumor-derived TMEM127 mutant constructs display diffuse, as opposed to punctate endomembrane distribution, suggesting that membrane association is required for TMEM127 tumor suppressor function (2,3). A link between TMEM127 and the lysosome was further supported by mouse embryonic fibroblasts (MEFs) lacking Tmem127, which we found to display increased number and size of lysosomal particles (3). However, the specific function of TMEM127 in the lysosome is unclear.
TMEM127-deficient cells, as seen in pheochromocytomas carrying a TMEM127 mutation, cell lines depleted of TMEM127 by short interfering RNA (siRNA) and Tmem127 knockout (KO) MEFs, display increased mTORC1 signaling, while enforced expression of TMEM127 leads to low phosphorylation levels of mTORC1 targets (1,3), suggesting that TMEM127 may function as a negative regulator of mTORC1 signaling. The mTORC1 pathway integrates different environmental inputs to balance multiple cellular physiological processes, including cell growth, proliferation and homeostasis, and is aberrantly activated in multiple pathological conditions, including cancer and metabolic disorders (6). The lysosome is central to the activation of the mTORC1 pathway by amino acids (7). After amino acid stimulation, mTORC1 is recruited to the lysosomal surface by multiple protein complexes, including Rag GTPases, a pentameric complex known as ragulator or LAMTOR, and the lysosomal vacuolar H+-adenosine triphosphatase ATPase (vATPase) (8–10). Rags are small GTPases including Rag A, B, C and D, which function as heterodimers in which RagA or RagB is paired with either RagC or RagD (8). In response to amino acids, RagA or B are activated to a GTP-bound conformation while RagC or D change from GTP to GDP bound—a configuration that functions to recruit mTORC1 to the lysosome (8). The ragulator/LAMTOR complex, consisting of five proteins: LAMTOR1 through LAMTOR5, serves both as a scaffold for the Rag GTPases and mTORC1 on the lysosomal surface, and has guanine nucleotide exchange factor (GEF) activity toward RagA and RagB in response to amino acid stimulation (9,11). A third component of this signaling cascade is vATPase, comprised of multiple subunits that are organized into a cytosolic domain V1 and a membrane integral domain V0, which together function to transfer protons into the lysosomal lumen, leading to acidification of the lysosome (12). vATPase interacts with ragulator and is thought to serve as a sensor of amino acids from the lysosomal lumen (10). Disruption of the cross-talk between these various complexes results in inhibition of mTORC1 pathway (9,11).
As a result of the association of TMEM127 with the endosome/lysosome and its effects on mTORC1 signaling, in this study we sought to investigate whether TMEM127 influences mTORC1 activation at the level of its scaffold complex on the lysosomal surface.
Results
Given the ubiquitous expression of TMEM127 and the inexistence of established human pheochromocytoma cell lines for in vitro studies, we took advantage of our unique mouse model of Tmem127 loss to derive MEFs with depleted or rescued TMEM127. Furthermore, to expand on these findings and gather molecular insights in the context of human samples, we extended these analyses to human cell models of TMEM127 loss generated by CRISPR-Cas9 technology, explored recombinant-mutant TMEM127, and examined TMEM127-mutant pheochromocytoma samples.
Endogenous TMEM127 resides in lysosomal-enriched fractions and affects mTORC1 activation in response to amino acids
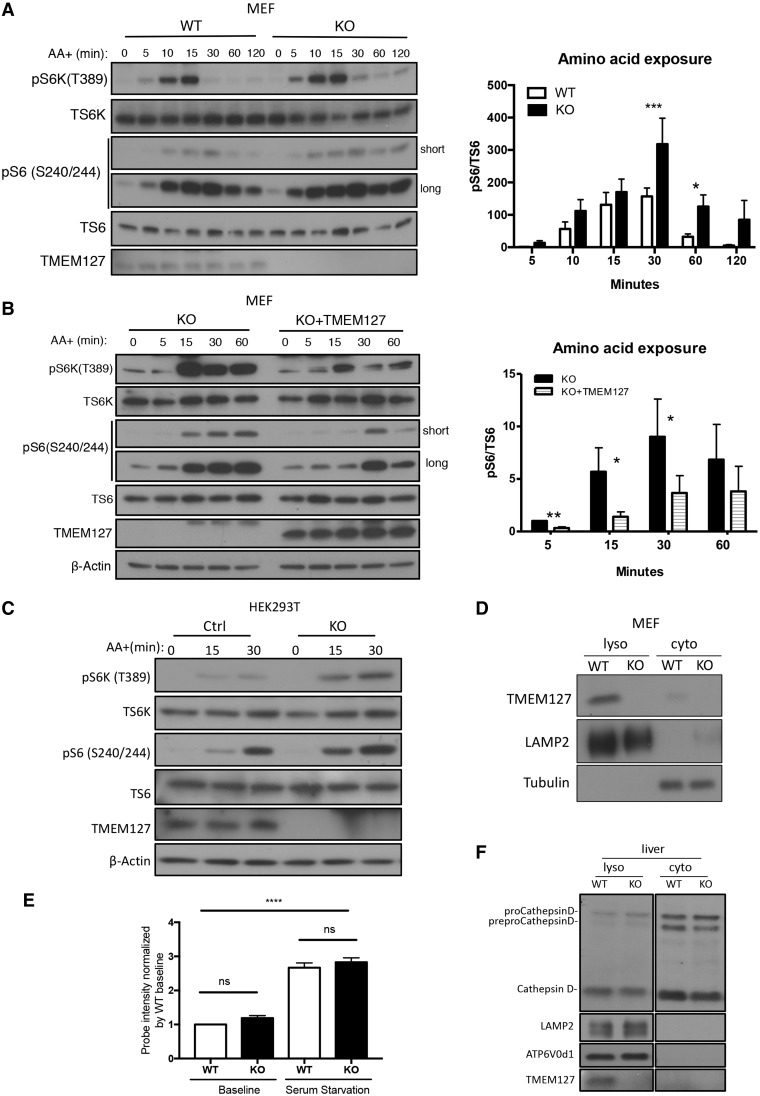
To examine the response of mTORC1 to amino acid stimulation in TMEM127 absence we starved KO or WT primary MEFs of amino acids for 2 h and tracked the phosphorylation levels of mTORC1 downstream targets after amino acid re-exposure. MEFs lacking TMEM127 displayed a modest but sustained increase in S6K and S6 phosphorylation after amino acid stimulation that persisted beyond 1 h, a time at which these targets decrease in WT MEFs (Fig. 1A, Supplementary Material, Fig. S1A). This effect was specific because reconstitution of TMEM127 by stable expression of a retroviral plasmid in Tmem127 KO MEFs led to both lower levels and shorter time of mTORC1 target phosphorylation after amino acid stimulation under the same conditions as above (Fig. 1B, Supplementary Material, Fig. S1B). Furthermore, deprivation of serum in combination to amino acid depletion did not change the dynamic of this response, suggesting that amino acid availability is primarily responsible for the observed effect of TMEM127 on mTORC1 activation (Supplementary Material, Fig. S1C). We validated these findings in human cells by generating TMEM127 KO HEK293T cells by CRISPR-Cas9 editing. Consistent with the murine model, TMEM127 KO HEK293T cells showed increased activation of mTORC1 downstream targets pS6K and pS6 upon amino acid stimulation (Fig. 1C). These results suggest that TMEM127 modifies the response of mTORC1 to amino acid activation, and this effect is not cell type dependent.
Figure 1.
Endogenous TMEM127 resides in lysosomal-enriched fractions and affects mTORC1 activation in response to amino acids. (A) Western blot of WT or Tmem127 KO MEFs cultured under amino acid deprivation for 2 h and re‐exposed to amino acids (AA+) in identical concentration as RPMI media for the designated times; blots were probed with antibodies detecting phosphorylated S6 kinase (p‐S6K T389) and S6 (p‐S6 S240/244), their corresponding total protein (T‐S6K and T‐S6, respectively) and TMEM127. Long and short exp, long and short exposure of blot. Quantification was performed using Image J; bar graphs display mean ± SEM; statistical significance was calculated from five biological replicates using two-way ANOVA with Bonferroni’s post-test, P-values are as follows: *P <0.05; ***P < 0.005 (results from an independent experiment shown in Supplementary Material, Fig. S1A). (B) Western blot of immortal Tmem127 KO MEF stably expressing an empty MSCV retrovirus (KO) or a TMEM127‐expressing MSCV (KO+ TMEM127), treated as in (A) and probed with phosphorylated S6 kinase (p‐S6K T389) and S6 (p‐S6 S240/244), their corresponding total protein (T‐S6K and T‐S6, respectively), TMEM127 and β‐actin antibodies. Quantification was performed using Image J; bar graphs display mean ± SEM; statistical significance was determined from three biological replicates using two-way ANOVA with Bonferroni’s post-test, P-value is as follows: *P <0.05 (results from an independent experiment shown in Supplementary Material, Fig. S1B). (C) Western blot of Control (Ctrl) and TMEM127 KO HEK293FT cells generated by CRISPR-Cas9 editing, starved of amino acid for 2 h and re-exposed to amino acids in identical concentration to those of RPMI media for the designated times. Blots were probed with antibodies detecting phosphorylated S6 kinase (p‐S6K T389) and S6 (p‐S6 S240/244), their corresponding total protein (T‐S6K and T‐S6, respectively), TMEM127 and β‐actin antibodies. Three biological replicates were performed. (D) Western blot of fractionated Tmem127 WT and KO MEF lysates showing detection of endogenous TMEM127 in membrane fraction containing lysosomes (lyso) cell fractions but not in cytosolic (cyto) fractions, LAMP2 and β‐tubulin are lysosomal and cytosolic markers, respectively. More than five biological replicates were performed. (E) Fluorescence levels of the Lysosensor‐DND‐189TM probe in four independent WT and Tmem127 KO MEF pairs, measured by flow cytometry at baseline culture conditions or after overnight serum starvation; each MEF pair was normalized to its respective baseline WT fluorescence, bar graphs display mean ± SEM. Statistical significance was determined from four biological replicates using one-way ANOVA with Tukey’s post-test, P-value is as follows: ****P < 0.001, n.s., nonsignificant. (F) Western blot of lysosomal and cytosolic fractions from WT and KO mouse liver probed with a CathepsinD antibody that detects precursor (preproCathepsinD), intermediate (proCathepsinD) and mature forms of CathepsinD; TMEM127 indicates genotype, LAMP2 and α-tubulin serve as lysosomal and cytosol markers, respectively. Data were obtained from three independent sets of mice.
The lysosome is the site of mTORC1 activation by amino acids (9) and we have previously reported that recombinant TMEM127 localizes to multiple endomembrane domains, including the lysosome, in human (1) and murine (3) cells. To examine the subcellular localization of endogenous TMEM127, we used mouse fibroblasts and confirmed that endogenous TMEM127 is detected in membrane-enriched fractions containing lysosomes (LAMP2 positive) but not in cytosolic fractions of these cells, and was undetectable in samples from Tmem127 null mice, as expected (Fig. 1D).
We considered that TMEM127 might modulate mTOR signaling through disruption of lysosomal function, and evaluated its impact on two key lysosomal properties, vesicle acidification and protein hydrolysis. To measure lysosomal acidity we incubated Tmem127 WT and KO MEFs with LysoSensorTM Green DND-189 probe, which localizes to the lysosome and emits fluorescence upon acidification (13), and quantified the fluorescence intensity of these cells using flow cytometry. We did not observe any significant difference in fluorescent intensity at baseline or after serum starvation, a condition known to alter intravesicular pH (13) (Fig. 1E). These results suggest that acidification is preserved in Tmem127KO MEFs. Next, to evaluate lysosomal enzymatic activity we measured cleavage of the lysosomal aspartyl protease Cathepsin D in Tmem127 KO liver, where expression of this protein is more abundant and more promptly detected than in MEFs (14). We found that the levels of processed Cathepsin D were similar in KO and WT liver (Fig. 1F), in favor of competent proteolytic function of the null cells. These results suggest that TMEM127 loss does not affect acidification or proteolytic activity of the lysosome.
TMEM127 translocates to the lysosomal site of mTORC1 activation upon amino acid stimulation and restricts mTOR recruitment to Rags
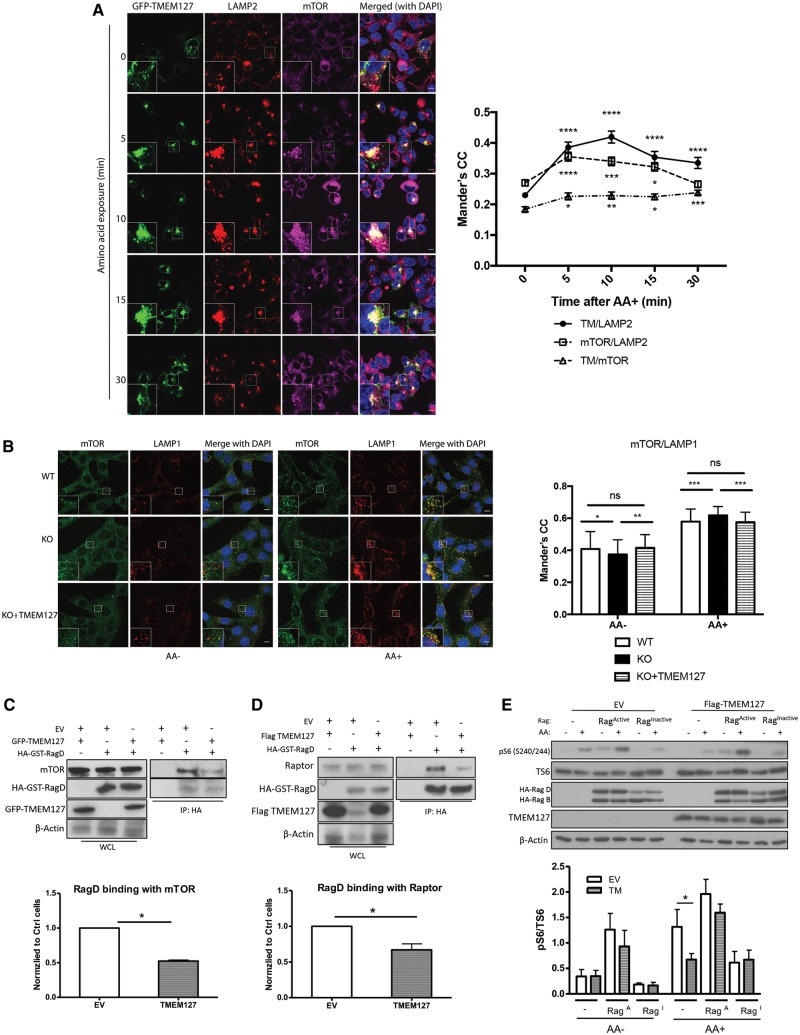
We next asked whether TMEM127 might affect the integrity of the mTORC1 lysosomal scaffold that mediates mTORC1 activation by amino acids. To further explore this hypothesis, we first determined whether TMEM127 lysosomal localization is modified by amino acids with a similar dynamic to that of mTORC1. In the absence of an effective endogenous TMEM127 antibody for immunofluorescence detection, we used confocal microscopy of HEK293T cells stably expressing GFP-TMEM127, which we have previously found to display subcellular distribution similar to other epitope-tagged TMEM127 constructs (1), to measure its colocalization with the lysosome. We found that the LAMP2 lysosomal marker colocalized with TMEM127; this association increased after amino acid exposure and peaked at 10 min, after which it gradually decreased (Fig. 2A). This pattern resembled the dynamic profile that we observed in the amino acid-stimulated mTORC1 association with LAMP2 (Fig. 2A) and is in agreement with the reported data (15). We also noted that, at 30 min, the decrease in TMEM127/LAMP2 colocalization was less marked than the mTOR/LAMP2 colocalization decline (Fig. 2A). An amino acid-dependent increase in the colocalization of TMEM127 and LAMP2 was also seen in another cell line, HeLa, suggesting that these effects are not cell specific (Supplementary Material, Fig. S2A). Importantly, we detected colocalization between GFP-TMEM127 and endogenous mTOR, which was significantly increased after amino acid stimulation in HEK293T (Fig. 2A) and showed similar trend in HeLa cells (Supplementary Material, Fig. S2B). These data suggest that TMEM127 is mobilized to the lysosomal site shared by activated mTORC1 upon amino acid exposure. Next, we used Tmem127 KO MEFs to explore the effect of TMEM127 on mTORC1 lysosomal localization. We observed a modest but significant increase in mTOR/LAMP1 colocalization in amino acid-rich conditions in the null cells, confirming our earlier observations (3). Moreover, we determined that these effects were reversed by TMEM127 re-expression, supporting the specificity of this observation (Fig. 2B).
Figure 2.
TMEM127 translocates to the lysosomal site of mTORC1 activation upon amino acid stimulation, and limits mTOR recruitment to Rags. (A) Confocal microscopy of HEK293FT cells stably expressing GFP‐TMEM127 deprived of amino acids and re‐exposed to amino acids in identical concentration as those of RPMI media for the designated amount of time. Cells were fixed and stained with endogenous LAMP2 (red) or mTOR (magenta); TMEM127 was represented by green fluorescence; DAPI (blue) stained nuclei, scale bar is 10 μm; inset represents 3-fold zoomed image of the indicated square region. Quantification of colocalization between mTOR and LAMP2, GFP‐TMEM127 and LAMP2, and GFP-TMEM127 and mTOR, from the cells shown in (A) was performed using Mander’s correlation coefficient (CC) as detailed in Materials and Methods; at least 70 cells were quantified from 10 independent fields per condition, from two independent experiments. Graph depicts mean ± SEM of each time point. Statistical analysis was performed using one-way ANOVA with Tukey’s post-test, comparing with corresponding 0 time point, P values are as follows: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (B) Confocal microscopy of Tmem127 WT, KO, or KO expressing MSCV‐TMEM127 (KO + TMEM127) cells deprived of both growth factors overnight and amino acids for 50 min (AA−) and re‐exposed to AA for 15 min (AA+), stained with endogenous LAMP1 (red) and endogenous mTOR (green), DAPI (blue) stains nuclei; scale bar is 10 µm; inset represents 3-fold zoomed image of the indicated square region. Quantification of the colocalization between mTOR and LAMP1 was performed using Mander’s coefficient, as detailed in Materials and Methods. At least 60 cells were quantified from 10 independent fields per condition, from three independent experiments. Statistical analysis was performed using one-way ANOVA test with Tukey’s post-test, P values are as follows: *P < 0.05, **P < 0.01, ***P < 0.0001, n.s., not significant. (C) HA IP of HEK293T expressing HA‐GST‐RagD and Flag-TMEM127 or EV as described in Materials and Methods and probed with mTOR, HA, GFP; corresponding WCLs are shown on the left and β-actin controls for loading; quantification from three biological replicates was performed using Image J and is shown below the figure; graph depicts mean ± SEM. Statistical analysis was performed using paired, two-tailed, Student’s t-test, P-value as indicated. (D) HA IP of HEK293T expressing HA‐GST-RagD and GFP‐TMEM127 or EV and probed with Raptor, HA, TMEM127; corresponding WCLs are shown on the left and β-actin controls for loading; quantification from three biological replicate experiments was performed using Image J and is shown below the figure; graph depicts mean ± SEM. Statistical analysis was performed using paired, two-tailed, Student’s t-test, P-value as indicated. (E) HEK293FT cells stably expressing MSCV-EV or Flag-TMEM127 were transfected with constructs expressing HA‐GST‐RagB/RagD mutant heterodimers, either as HA‐GST‐RagBGTP/RagDGDP (RagACTIVE) or HA‐GST‐RagBGDP/RagDGTP (RagINACTIVE) were starved of amino acids (AA−), followed by AA stimulation for 15 min (AA+) as described in Materials and Methods. Blots were probed with phosphorylated S6 (p‐S6 S240/244), total S6 (T‐S6), HA, TMEM127 and β‐actin antibodies; quantification from three biological replicate experiments was performed using Image J and results are shown below the figure; graphs depict mean ± SEM. Statistical analysis was performed using two-way ANOVA with Bonferroni’s post-test, *P <0.05, other comparisons, non-significant.
Rag GTPases have a central role in controlling the recruitment of mTORC1 to the lysosomal surface in response to amino acid stimulation (8,9). To determine whether TMEM127 affected mTORC1 recruitment by Rags we expressed TMEM127 in HEK293T cells carrying HA-GST-RagD and found reduced binding between the RagD construct and both endogenous mTOR (Fig. 2C) and endogenous Raptor, a distinguishing component of mTORC1 (Fig. 2D). The nucleotide conformation of Rag heterodimers affects mTORC1 binding, whereby the RagBGTP/RagDGDP heterodimer is able to activate mTORC1 downstream targets regardless of amino acids (RagACTIVE), while expression of RagBGDP/RagDGTP heterodimer (RagINACTIVE) significantly impairs activation of mTORC1 downstream targets upon amino acid stimulation (8,9). We found that in control cells without ectopic Rag heterodimers, TMEM127 expression led to reduced pS6 levels after amino acids, compared with empty vector (EV) cells, as expected (Fig. 2E). However, TMEM127 expression was not able to inhibit the persistent S6 phosphorylation induced by expression of a constitutively active Rag heterodimer (RagB Q99L+ RagD S77L, RagACTIVE), nor further decrease repressed phosphorylated S6 resulting from inactive Rag (RagB T54L + RagD Q121L, RagINACTIVE) expression (Fig. 2E). Together, these data suggest that TMEM127 restricts mTORC1 recruitment to Rags and functions upstream of these GTPases in the mTORC1 signaling cascade.
TMEM127 interacts with, and regulates the abundance of, the ragulator/LAMTOR complex in cells and human tumors
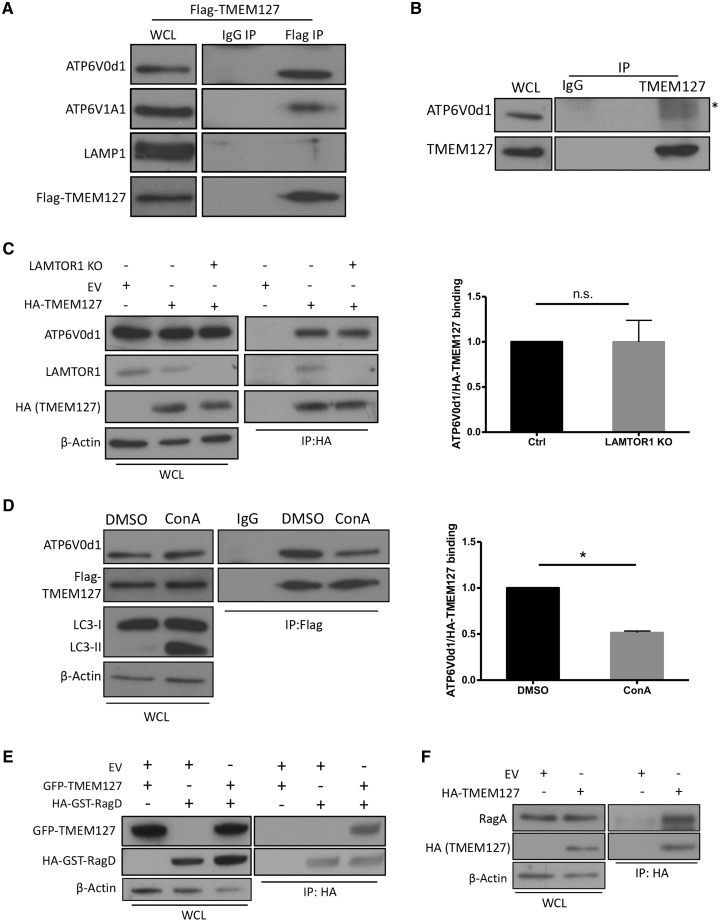
Next, we explored the pathway upstream of Rags, by investigating ragulator, the pentamer complex comprised of LAMTOR proteins, which functions as a GEF to RagA/B (11). Ragulator is an attractive candidate for association with TMEM127 because LAMTOR1 is a lysosomal protein responsible for anchorage of the other LAMTOR components to this organelle’s surface (11,16,17). To explore whether LAMTOR1 is required for the TMEM127 effects on mTOR signaling we knocked down LAMTOR1 by shRNA in TMEM127 KO HEK293T cells. LAMTOR1 deficiency led to reduced mTOR activation both in control and in TMEM127 null cells, confirming its relevance for mTORC1 signaling and suggesting that LAMTOR1 does not require TMEM127 for its actions on this pathway (Fig. 3A, Supplementary Material, Fig. S3A). Given that both TMEM127 and LAMTOR1 reside on the lysosome, we tested whether these two proteins interact. Endogenous TMEM127 was detected by immunoprecipitated recombinant LAMTOR1 expressed in HEK293T cells (Fig. 3B), while HA-TMEM127 immunoprecipitated endogenous LAMTOR1 (Fig. 3C). The interaction was confirmed by co-immunoprecipitation (IP) with distinctly tagged constructs (Supplementary Material, Fig. S3B and C), and, importantly, by IP of endogenous proteins (Fig. 3D). Independent support for the TMEM127 and LAMTOR1 association was seen by confocal microscopy in HeLa cells showing extensive colocalization between these two proteins (Supplementary Material, Fig. S3D, top), which agrees with the colocalization between TMEM127 and mTORC1 at the lysosome described above (Fig. 2A). In contrast, TMEM127 did not colocalize (Supplementary Material, Fig. S3D, bottom) or coimmunoprecipitate (Fig. 3E) with a lipidation-defective LAMTOR1 mutant, G2A, that lacks lysosomal association (11), suggesting that localization of LAMTOR1 to the lysosome is required for its interaction with TMEM127. In further support of a close and selective interaction between TMEM127 and ragulator proteins, we also detected TMEM127 co-IP with LAMTOR2 (Supplementary Material, Fig. S3E), but not with LAMP1 (Fig. 3F), a lysosomal protein which is not a component of ragulator, indicating that the observed interactions with ragulator were specific.
Figure 3.
TMEM127 interacts with and affects ragulator complex abundance. (A) Ctrl and two independent clones of TMEM127 KO HEK293FT cells generated by CRISPR-Cas9 (KO1, KO2) were transduced with lentivirus carrying either pLKO EV or pLKO-shLAMTOR1 (shL1) construct. Cells were stimulated with amino acids for 15 min after a 2-h amino acid starvation, as described in Figure 1, 72 h after transduction. Blots were probed with antibodies for phosphorylated S6 kinase (p‐S6K T389) and S6 (p‐S6 S240/244), their corresponding total protein (T‐S6K and T‐S6, respectively), LAMTOR1, TMEM127 and β‐actin (loading); three biological replicates were performed. (B) Flag IP of HEK293T cells expressing Flag‐LAMTOR1 or Flag-EV and probed for endogenous TMEM127. Corresponding WCLs were probed with Flag, TMEM127 and β-actin; three biological replicates were performed. (C) HA IP of HEK293T cells expressing HA-TMEM127 or HA-EV and probed for endogenous LAMTOR1. Corresponding WCLs were probed with HA, LAMTOR1 and β-actin antibodies; three biological replicates were performed. (D) Endogenous TMEM127 IP in HEK293T cells using a polyclonal TMEM127 antibody and probed for endogenous LAMTOR1; IgG was used as a negative control, corresponding WCLs are shown. (E) Flag IP of HEK293T cells expressing Flag-TMEM127 and HA-LAMTOR1 WT or HA-LAMTOR1 G2A mutant and probed for HA and Flag; corresponding WCLs are shown and β-actin controls for loading; three biological replicates were performed. (F) Flag IP of HEK293T cells expressing Flag-TMEM127 and probed for endogenous LAMP1 and TMEM127 (both Flag-TMEM127 and endogenous TMEM127 bands are seen on WCLs, shown on the left, β-actin controls for loading. (G) Confocal microscopy of GFP‐TMEM127 in HEK293T control (LAMTOR1 WT) and LAMTOR1 KO cells; scar bar: 10 μm. (H) Fractionated lysates from Ctrl, two independent clones of TMEM127 KO (1 and 2) and LAMTOR1 KO HEK293FT cells showing membrane fraction containing lysosomes (Lyso), cytosolic fractions (Cyto) and WCLs, probed for TMEM127, LAMTOR1, LAMTOR2 and LAMTOR4, and RagC. Tubulin and LAMP2 were used as cytosolic and lysosomal markers, respectively; three biological replicates were performed. (I) WCLs of WT and Tmem127 KO MEFs and probed for endogenous LAMTOR1, LAMTOR2, TMEM127, β-actin controls for loading; three biological replicates were performed. (J) Western blot of HEK293T cell lysates expressing increasing amounts of GFP‐TMEM127 (0–500 ng) and probed for endogenous LAMTOR1, LAMTOR4, RagC, LAMP1 and TMEM127, β‐actin is a loading control; three biological replicates were performed (quantification shown in Supplementary Material, Fig. S3G). (K) WCLs of three pheochromocytomas carrying distinct truncating TMEM127 mutations (MUT samples #2, 3, 4) and four pheochromocytomas with WT TMEM127 sequence (WT samples #1, 5, 6, 7) probed with TMEM127, LAMTOR1, LAMTOR2, RagA, RagB, ATP6v0d1 and β-actin, as loading control. Note the absence of detectable WT or truncated TMEM127 protein in mutant lanes; one of two western blots run with the same panel of samples is shown. (L) Flag IP of HEK293T cells expressing Flag-LAMTOR1 and transfected with the indicated amount of HA-TMEM127-WT or HA-TMEM127 532DupT mutant construct, probed for HA and Flag; corresponding WCLs are shown on the left and β-actin controls for loading. Three biological replicates were performed.
Subsequently, we investigated whether TMEM127 and LAMTOR1 influence each other’s subcellular distribution. Depletion of LAMTOR1 by CRISPR-Cas9 editing of HEK293T cells did not affect the characteristic TMEM127 punctate pattern (Fig. 3G) or its association to the lysosomal-enriched cellular fraction (Fig. 3H), although it clearly led to mislocalization of other ragulator proteins, LAMTOR2 and LAMTOR4, as well as RagC, to cytosolic fractions (Fig. 3H—note the presence of these three proteins in the cytosolic fraction of the LAMTOR1 KO cells, unlike the control cells). Conversely, TMEM127 KO cells did not modify the subcellular distribution of ragulator proteins, as they remained enriched in the lysosomal fraction. However, TMEM127 null cells displayed increased levels of LAMTOR1, LAMTOR2 and LAMTOR4 proteins in the lysosome-enriched lysates compared with control cells (Fig. 3H). The enrichment for LAMTOR proteins was also seen in murine Tmem127 null cells (Fig. 3I) and was not due to increased transcription levels of Lamtor1, Lamtor2 and Lamtor3 genes (Supplementary Material, Fig. S3F), suggesting a posttranscriptional effect of TMEM127 on LAMTOR levels. To further verify this phenomenon, we expressed TMEM127 in HEK293T cells and observed a dose-dependent decrease in the abundance of LAMTOR proteins, but not of RagC or LAMP1 levels (Fig. 3J, Supplementary Material, Fig. S3G), suggesting that the effect of TMEM127 expression was selective toward the LAMTOR complex. In turn, ectopic expression of LAMTOR1 did not alter TMEM127 levels (Supplementary Material, Fig. S3H). This finding mirrors the posttranscriptional destabilizing effect toward other LAMTOR proteins that follows the loss of LAMTOR2 (18), and favors a role for TMEM127 on reducing ragulator stability.
To further investigate the significance of the TMEM127 effect on LAMTOR levels in a clinically relevant context, we examined primary pheochromocytomas carrying germline truncating TMEM127 mutations. These tumors have loss of the WT allele and retain no residual TMEM127 expression (Fig. 3K), as we reported (1,2,19). Although few tumors were available for this analysis, lysates from mutant TMEM127 tumors showed consistently higher levels of LAMTOR1 and LAMTOR2 proteins, when compared with pheochromocytomas with WT TMEM127 sequence (Fig. 3K). In the TMEM127 null tumors, we also noted a trend toward increased Rag and vATPase protein levels, potentially suggesting a broader effect of TMEM127 on other lysosomal components of this scaffold complex. These results agree with the cell line data and suggest that accumulation of LAMTOR proteins is a biological consequence of TMEM127 absence.
Finally, to gain additional insight on the interaction between TMEM127 and LAMTOR, we expressed a C-terminally truncated TMEM127 mutant that is diffusely distributed in the cytosol, 532dupT (19), and found that despite not being endomembrane bound, this mutant was still capable of immunoprecipitating LAMTOR1 at levels comparable with or higher than WT-TMEM127 (Fig. 3L). This observation suggests that physical association of TMEM127 with LAMTOR is independent of TMEM127 lysosomal localization and is reminiscent of the membrane independent effect of the LAMTOR2–LAMTOR3 interaction on their reciprocal stability (18). Moreover, in this experiment, lower amounts of WT-TMEM127 input led to a respective decrease in TMEM127 recovery from the LAMTOR1 pulldown, further supporting the specificity of the TMEM127–LAMTOR1 association. Together, these experiments show that TMEM127 both associates with, and reduces the abundance (and likely, the stability), of LAMTOR/ragulator proteins. Conversely, lysosomal localization or protein levels of TMEM127 are not ragulator-dependent.
TMEM127 interacts with vATPase subunits and Rag GTPases
LAMTOR/ragulator interacts with vATPase to promote Rag activation (10). We found that TMEM127 interacts with V0d1 (ATP6v0d1), the vATPase subunit that makes direct contact with LAMTOR1 (10), and to V1A1, as shown by IP of both endogenous subunits with expressed TMEM127 (Fig. 4A). Endogenous ATP6V0d1 was also immunoprecipitated by endogenous TMEM127, confirming the interaction with vATPase (Fig. 4B). Notably, this binding was unchanged in LAMTOR1 KO HEK293T cells, suggesting that ragulator is dispensable for the association between TMEM127 and vATPase (Fig. 4C). We next treated HEK293T cells stably expressing TMEM127 with Concanamycin A (ConA), a selective inhibitor of the vATPase proton pump activity responsible for maintaining lysosomal acidity (20). As predicted, ConA treatment leads to accumulation of LC3, a protein that requires intact lysosomal pH for its degradation, but it does not affect the abundance of vATPase subunits (Fig. 4D). We found that binding between Flag-TMEM127 and ATP6V0d1 is significantly reduced after ConA treatment (Fig. 4D). These observations suggest that TMEM127–vATPase binding is independent of ragulator but requires lysosomal acidification.
Figure 4.
TMEM127 interacts with vATPase and Rag GTPases. (A) Flag IP of HEK293T cells expressing Flag‐TMEM127 and probed for endogenous vATPase subunits (ATP6V0d1 and ATP6V1A1), LAMP1 and Flag; IgG was used as negative control; corresponding WCLs are shown on the left; three biological replicates were performed. (B) IP of endogenous TMEM127 in HEK293T cells using a polyclonal TMEM127 antibody and probed for endogenous ATP6V0d1; IgG was used as a negative control, *indicates a non-specific band, corresponding WCLs are shown on the left. (C) HA IP of Ctrl and LAMTOR1 KO HEK293FT cells expressing HA-TMEM127 or HA-EV and probed for endogenous ATP6V0d1, LAMTOR1 and HA; corresponding WCLs are shown on the left, β-actin controls for loading; quantification was performed with Image J; graph depicts mean ±SEM. Statistics was determined from three biological replicate experiments using paired, two-tailed, Student’s t test, n.s., nonsignificant. (D) Flag IP of HEK293FT cells stably expressing Flag-TMEM127 treated with DMSO or ConA 2 μm for 2 h and probed for ATP6V0d1 and Flag. Corresponding WCLs are shown on the left, LC3 was used as control for ConA treatment and β-actin controls for loading; quantification was performed with Image J; graph depicts mean ±SEM. Statistics was determined from three biological replicate experiments using paired, two-tailed, Student’s t-test, *P < 0.05. (E) HA IP of HEK293T cells expressing GFP‐TMEM127 and HA-GST-RagD probed for GFP, HA; corresponding WCLs are shown on the left; β-actin controls for loading; three biological replicates were performed. (F) HA IP of HEK293T cells expressing HA‐TMEM127 or HA-EV and probed for endogenous RagA or HA; corresponding WCLs are shown on the left and β-actin controls for loading; three biological replicates were performed.
Given our findings of TMEM127 association with ragulator and vATPase, we also asked whether TMEM127 interacts with Rags. We detected binding between TMEM127 and ectopic RagD (Fig. 4E) as well as endogenous RagA (Fig. 4F). Importantly, these interactions were not the result of indiscriminate association to any lysosomal protein because we could not detect TMEM127 IP to the lysosomal protein LAMP1, which is not part of the Rag-ragulator-vATPase complex (Figs 3F and 4A). Together, these results indicate that TMEM127 associates with multiple components of the lysosome-anchored complex related to mTORC1 activation. Noteworthy, all the IPs described in this study were performed under stringent washing conditions that included high salt concentrations, as reported (11), which denote stability of the detected immune complex associations.
TMEM127 interaction with Rag-ragulator-vATPase affects mTORC1 activation at the lysosomal complex
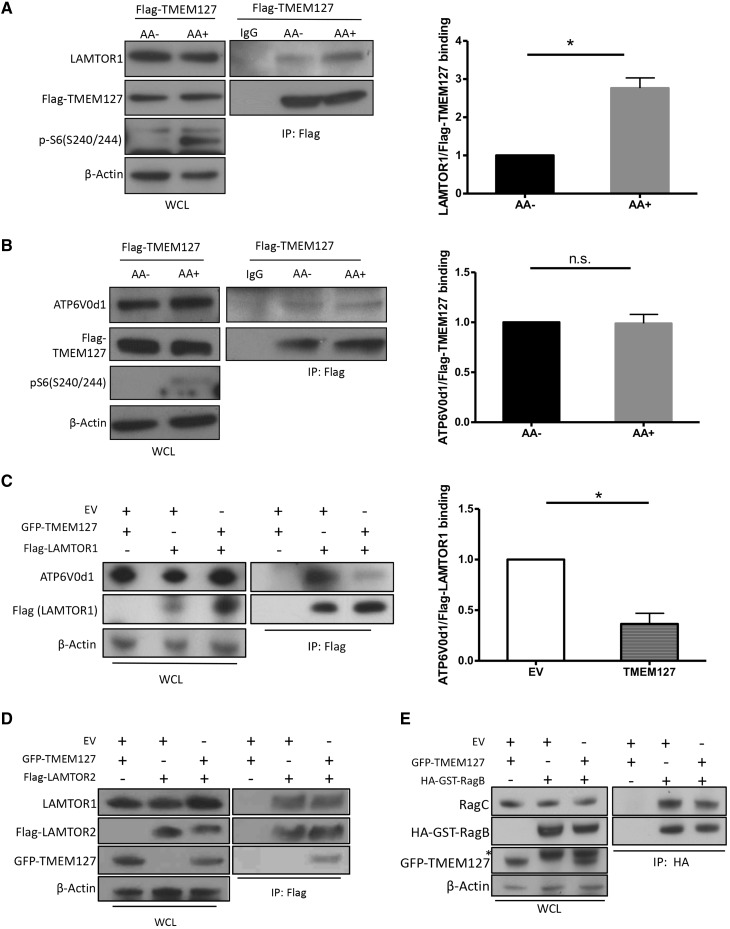
Next, we sought to gain additional insight into the impact of TMEM127 on the assembly of the lysosome-anchored mTORC1 complex. As the full activation of this pathway is dependent on nutrients (11), we examined the effect of amino acids on these TMEM127 interactions. We found that binding of recombinant TMEM127 with LAMTOR1 (Fig. 5A), but not with ATP6V0d1 (Fig. 5B), increases after amino acid stimulation. In addition, TMEM127 expression was able to reduce the binding between ragulator and vATPase in HEK293T cells (Fig. 5C). In contrast, intra-ragulator (LAMTOR1-LAMTOR2, Fig. 5D) or intra-Rag (RagB-RagC heterodimer, Fig. 5E) binding was not affected by TMEM127 expression. Taken together, these data suggest that TMEM127 expression differentially affects Rag-ragulator–vATPase complex interactions.
Figure 5.
TMEM127 association with Rag-ragulator-vATPase modifies interaction between complex components. (A, B) Flag IP of HEK293FT cells stably expressing Flag-TMEM127 (Flag-TM) exposed to amino acids (AA) for 15 min after AA deprivation for 2h, as described in Materials and Methods; blot was probed for LAMTOR1 (A), ATP6V0d1 (B) and Flag (A, B). Corresponding WCLs are shown on the left, phospho-S6 was used as control for AA treatment and β-actin controls for loading; quantification was performed using Image J; graph depicts mean ±SEM. Statistical significance was determined from three biological replicate experiments using paired, two-tailed, Student’s t-test, *P <0.05. (C) Flag IP of HEK293T cells expressing Flag‐LAMTOR1 with GFP-TMEM127 or EV and probed for endogenous ATP6V0d1; corresponding WCLs are shown on the left, β-actin controls for loading; quantification was performed using Image J; graph depicts mean ±SEM. Statistical significance was determined from three biological replicates using paired, two-tailed, Student’s t-test, n.s., nonsignificant. (D) Flag IP of HEK293T cells expressing Flag-LAMTOR2 and GFP-TMEM127 and probed for endogenous LAMTOR1, GFP and Flag; corresponding WCLs are shown on the left and β-actin controls for loading; three biological replicates were performed. (E) HA IP of HEK293T cells expressing HA-GST-RagB and GFP-TMEM127 and probed for endogenous RagC, GFP and HA; Corresponding WCLs are shown on the left, β-actin controls for loading; three biological replicates were performed.
Discussion
The function of TMEM127, a tumor suppressor gene mutated in pheochromocytomas and, more rarely, in renal cancers, has remained poorly defined. We have previously found that tumors carrying TMEM127 mutations display increased mTORC1 signaling (1,19). Our results now reveal that TMEM127 interacts with the multiprotein complex that resides on the lysosomal surface and which functions to regulate mTORC1 activation after amino acid exposure. Dysregulation of the mTORC1 pathway by mutations has been directly implicated in multiple cancers, both sporadic and as part of hereditary tumor syndromes (21). In the context of pheochromocytoma susceptibility genes, tumors carrying loss-of-function mutations of NF1, or activating mutations of HRAS, BRAF, RET or FGFR1 lead to increased signaling via MAPK/ERK and/or PI3K/AKT, upstream of mTOR and result in mTOR activation (22–24). The pathway convergence among these genetic mutations and TMEM127 has been observed previously (1), and was independently supported by recent TCGA findings in pheochromocytomas and paragangliomas (25). Beyond these cancer genes, more direct regulators of mTORC1, including the genes encoding TSC1 and TSC2, involved in mTOR inhibition in response to growth factors, are mutated in the hereditary tumor syndrome tuberous sclerosis, and also somatically disrupted in other cancer types (26,27). Moreover, regulators of the nutrient-sensing branch of mTORC1, including FLCN, mutated in the Bert-Hogg-Dubé cancer syndrome (28), GATOR1 component genes (29), the RRAGC gene, encoding RagC and those encoding for the vATPase isoforms, ATP6V1B2 and ATP6AP1 (47), can also be mutated in cancers. Our current study adds TMEM127, a bona fide tumor suppressor gene in hereditary pheochromocytomas, to this group of cancer genes that code for modulators of the mTORC1 lysosomal machinery.
In the present study we find that endogenous TMEM127 is present in membrane-enriched fractions that contain lysosomes and shares properties of proteins implicated in the mTORC1 nutrient signaling network: it interacts selectively with known components of the pathway (Rag, ragulator and vATPase) and its subcellular distribution is sensitive to variation in nutrient levels. However, unlike classic amino acid sensors of the mTORC1 network, which render the signal either constitutively active (e.g. mutant Rag or their regulatory proteins) or unresponsive to the presence of amino acids (e.g. loss of LAMTOR1) (7,30–32), we find that TMEM127’s inhibitory effects toward mTORC1 and its regulatory lysosomal complex are modest and are enhanced by amino acids, implying a role distinct from other modulators of the pathway. This finding is consistent with the observation that the signals implicated in mTOR activation and its assembly at the lysosome are not always coupled (33), and underscores the complex cross‐talk between mTOR and lysosomal complex components. Interestingly, TMEM127 has only been recognized in vertebrates. Likewise, other members of the mTORC1 nutrient signaling cascade, including CASTORs (31), SLC38A9, SZT2, KPTN, ITHG2 and CI2orf66 (34) also diverge from lower species, suggesting that unique regulatory mechanisms for sensing and regulating nutrient needs evolved differentially in higher species to refine the elaborate mTORC1 response to individual nutrient inputs (34,35), and implying the existence of additional components of this intricate system with specialized roles.
Based on our data, upon amino acid stimulation, TMEM127 is mobilized to the site of mTORC1 scaffolding at the lysosome, where it associates with LAMTOR1 and limits the amount of mTORC1 recruited by active Rags, thus leading to downregulation of mTORC1 signals (Supplementary Material, Fig. S4). In turn, loss of TMEM127 leads to posttranscriptional LAMTOR accumulation. The fact that LAMTOR proteins are also elevated in human pheochromocytomas carrying truncating TMEM127 mutations support the biological relevance of this phenomenon and may explain the mTOR upregulation observed in these tumors (1,19). Our studies indicate that the LAMTOR–TMEM127 interaction is enhanced in the presence of amino acids and it requires that LAMTOR1, but not TMEM127, remain at the lysosome. As the LAMTOR interface with Rags has been recently revealed to involve multiple domains (17), the precise nature of the connection of LAMTOR with other interacting proteins will need to be determined. Our data reveal that the effects of TMEM127 on mTOR signaling are modest, suggesting that TMEM127 associations with LAMTOR may involve functions that are mTOR independent. Recently, LAMTOR1 was found to modulate lysosomal motility through its association with BORC, in a complex that is mutually exclusive with the LAMTOR interaction with mTORC1 and Rags (36,37). Interestingly, BORC also interacts with C17orf59, a protein that binds to ragulator and prevents Rag from recruiting mTORC1 to the lysosome when overexpressed (38), although absence of C17orf59 had no impact on mTORC1 signaling (38). It will be of interest to determine if TMEM127 associates with LAMTOR1 as part of the BORC complex.
In addition to LAMTOR, we found that TMEM127 interacts with vATPase, an enzymatic complex that, similar to TMEM127, localizes to other endomembrane vesicles besides the lysosome, where it contributes to maintaining their intraluminal acidity (39). The TMEM127/vATPase interaction is independent of amino acids or LAMTOR1, but requires vacuolar acidification, suggesting that this association may have other functions, unrelated to the Rag-LAMTOR complex.
In summary, this study introduces the tumor suppressor gene TMEM127 as a component of the multiprotein lysosomal complex involved in mTORC1 activation. Our data reveal that TMEM127 expression limits interactions between Rag, ragulator and vATPase, although the precise mechanisms that dictate this process remain to be defined. Moreover, as the strength of these associations is dictated by different input, it is likely that TMEM127 has mTORC1 independent actions on the lysosome.
Materials and Methods
Reagents
The following antibodies were obtained from Cell Signaling Technologies (CST): mTOR (#2983), LAMTOR1 (#8975), LAMTOR2 (#8145), LAMTOR4 (#12284), RagA (#4357), RagB (#8150), RagD (#4470), RagC (#9480), phospho-S6 ribosomal protein (Ser235/236) (#2211), phospho-S6 ribosomal protein (Ser240/244) (#5364), phospho-p70 S6 kinase (Thr389) (#9206) and LC3A/B (#4108). The following antibodies were obtained from Bethyl Laboratories: TMEM127 (#A303-450A), Raptor (A300-553A), Goat anti-HA antibody (#A190-207A), Rabbit anti-ECS (DDDDK) antibody (#A190-102A). The following antibodies were obtained from Sigma-Aldrich: mouse anti-flag antibody (#F3165), mouse anti-ß-actin antibody (#A5441). Anti-ATP6V0d1 (ab56441) was obtained from Abcam and ATP6V1A1 was from GeneTax (#GTX110815). Rat Anti-Mouse CD107a (LAMP1) antibody (#553792) was obtained from BD Biosciences. GFP antibody (#sc-9996) was obtained from Santa Cruz Biotechnology. Anti-rabbit HRP conjugated secondary antibody (#172-1019), and anti-mouse HRP conjugated secondary antibody (#172-1011) were obtained from Bio-Rad Laboratories. Anti-Mouse Texas Red-Conjugated secondary antibody (#T6390) were obtained from Life Technologies. Anti-Rabbit Cy3-conjugated secondary antibody (#711-166-152) and Anti-Rabbit Cy5-conjugated secondary antibody (#711-606-152) were obtained from Jackson Immuno Research Laboratories.
Clones and Constructs
TMEM127-based constructs and mutants were previously reported (1–3,19). The following constructs were obtained through Addgene from D. Sabatini laboratory (8): PRK5-Flag-LAMTOR1 [#42331], PRK5-HA-LAMTOR1-G2A [#42327], pLKO.1 p18 shRNA [#26631], PRK5-Flag-LAMTOR2 [42330], PRK5-HA-GST-RagB WT [#19301], PRK5-HA-GST-RagB 99L (RagBGTP) [#19303], PRK5-HA-GST-RagB 54L (RagBGDP) [#19315], PRK5-HA-GST-RagD WT [#19307], PRK5-HA-GST-RagD 77L (RagDGDP) [#19308], PRK5-HA-GST-RagD 121L (RagDGTP) [#10309]. pCMV-VSV-G [#8454], pLKO.1puro [#8453] were from R. Weinberg laboratory; pSPAX2 [#12260] and pMD2G [#12259] were from D. Trono laboratory and pLentiCRISPRv2 [#52961] was from F. Zhang laboratory. HA-LAMTOR1 WT was generated by replacing G2A mutations with a WT G2G version with Phusion Polymerase.
Mouse embryonic fibroblast (MEF) generation
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All of the animals were handled according to approved Institutional Animal Care and Use Committee (IACUC) protocols of the University of Texas Health Science Center at San Antonio (protocol #10053x). We previously reported the development of a mouse strain with a targeted deletion of Tmem127 using a broadly expressed promoter, Cre-CMV (3). MEFs from E13.5 embryos of Tmem127+/+ (WT) or Tmem127−/− (KO) genotype, originated from breeding of Cre-negative Tmem127+/− heterozygote pairs, were prepared by digestion with trypsin, followed by mechanical disaggregation, as described previously (40). Genotypes were assigned by three-primer PCR and expression of TMEM127 was verified using quantitative RT-PCR and western blot, as we described (3). In addition, we established immortalized MEF cells by introducing SV40 LargeT antigen (Addgene #14088, from R. Weinberg laboratory) (41) into primary WT and KO MEFs, followed by serial dilution of the cells for 10 consecutive passages (40). TMEM127 expression of established immortal MEFs was verified by western blot. The experiments described here were performed between passages 3 and 8 of primary MEFs, and passages 20 and 50 of immortal MEFs. In addition, some of the MEFs were rescued by transduction of a retroviral vector containing the coding region of human TMEM127 (MSCV-TMEM127), as we reported (1).
Cell culture, transfections and transductions
Cell lines HEK293T, HEK293, HeLa and MEFs were cultured in RPMI1640 or DMEM (Corning) and 10% fetal bovine serum supplemented with 100 U/ml penicillin and 100 mg/ml streptomycin (Corning #30-002-CI). All cells were negative for Mycoplasma infection, using the protocol described previously (42). Plasmid transfections in HEK293T, HeLa or MEFs were performed with Lipofectamine 2000 (Life Technologies #11668019), following the manufacturer’s guidelines. For IP experiments, cells were plated in 10-cm cell culture dishes and 4 ug of total DNA were used for transfection in each plate. Retroviral transduction was used to create HEK239FT cells stably expressing MSCV-Flag-TMEM127. Briefly, packaging plasmids VSV-G (1 ug), pKAT (3 ug) and MSCV-Flag-TMEM127 (6 ug) were transfected into HEK293FT cells using Lipofectamine 2000 as described above. After 48 h culture media containing viral particles were filtered through 0.45 μm filter and added to recipient cells with 1: 1 ratio of fresh media and 8 ng/mL polybrene. Efficiency was determined 72 h after transduction using GFP microscopy. Lentiviral transduction was used to infect HEK239FT cells with PLKO.1 EV or pLKO.1-P18-shRNA. Packaging plasmids pSPAX2 (2.5 ug), pMD2G (3.75 ug) and pLKO.1-EV (5 ug) or pLKO.1-P18-shRNA (5 ug) were transfected into HEK293FT cells using Lipofectamine 2000 as described above. After 48 h culture media containing viral particles were filtered through 0.45 μm filter and added to recipient cells with 1: 1 ratio of fresh media and 8 ng/mL polybrene. Efficiency was determined 72 h after transduction using western blot.
Cell line treatment
Hela, MEFs and HEK293T cells were plated in 10 cm plates for IP, or 24-well plates containing poly-d-lysine-coated coverslips for immunostaining. Two protocols for amino acid starvation and stimulations were used. Protocol 1 (used for MEFs and HEK293T): cells were starved for amino acid using amino acid-free RPMI 1640 medium (US Biological #R8999-04A) for 2 h, and then stimulated with or without 1× amino acid (Sigma #7173) and 1× l-glutamine (Corning #25-005-CI) for 15 min. Protocol 2 (used for MEFs and HeLa): Cells were serum starved overnight, followed by amino acid starvation using DPBS + 4.5 g/l glucose for 50 min. Cells were then stimulated with or without amino acid stimulation buffer [DPBS + 4.5 g/l glucose replenished with amino acid solution (Sigma #M5550, final concentration of 2× to make up to individual amino acid concentration as in DMEM media) and glutamine (Corning #25-005-CI, final concentration of 1×)] for 15 min. ConA treatment was performed with full culture media supplemented with 2 um ConA (Sigma #C9705) for 2 h.
Cell lysis and immunoprecipitation (IP)
Cells were rinsed twice with ice-cold DPBS and lysed in ice-cold CHAPs lysis buffer [0.3% CHAPS (Sigma #C3023), 10 mm beta-glycerol phosphate (Sigma #G9422), 10 mm sodium pyrophosphate (Sigma #221368), 40 mm HEPES (pH 7.4) and 2.5 mm MgCl2] with one tablet of EDTA-free protease inhibitor (Roche #11836170001) per 10 ml as described (11). Lysate were cleared by centrifugation at 16 000g at 4°C for 10 min. For anti-Flag IPs, cleared lysates were normalized for protein content by Bradford assay, and 2–3 mg of protein lysate were incubated with 40 ul of a 50% slurry of Flag-M2 affinity gel (Sigma #A2220) per 1 mg of lysate at 4°C rotating overnight. Immunoprecipitated proteins were washed four times with CHAPs buffer supplemented with 150 mm NaCl, followed by boiling at 100°C in 2× SDS sample buffer for 10 min. In experiments using only cell lysates, without IP, 1% NP40 (Sigma, NP40S) was substituted for CHAPS in the lysis buffer. For experiments with HA, GFP or endogenous RagC IP, 2–4 mg of protein lysate were incubated with primary antibody (at the manufacturer’s recommended dilution) at 4°C rotating overnight, followed by adding 40 ul of 50% slurry protein A/G agarose beads per 1 mg of lysate. Protein A beads (Pierce #22811) were used for IPs performed with rabbit primary antibody; protein G beads (Pierce #22852) were used for IPs containing goat primary antibody; protein A&G mixture were used for IPs with mouse primary antibody. Beads were washed three times with CHAPs lysis buffer, and incubated for 2 h at 4°C. After incubation, samples were washed and denatured as described above.
Primary tumor lysates
Pheochromocytoma samples were obtained after obtaining signed consent according to an UTHSCSA IRB-approved protocol (HSC20060069H, NCT03160274). Whole cell lysates (WCLs) were prepared from three pheochromocytomas carrying distinct truncating TMEM127 mutations (ClinVar rs121908826, rs121908821, SCV000571199.3) and four pheochromocytomas with mutations in other genes (MAX, EPAS1, VHL and SDHB) and WT TMEM127 sequence, as reported previously (19). Lysates were run on SDS-PAGE and probed with the antibodies listed above.
Immunofluorescence microscopy
HEK293T, HeLa cell lines or MEFs were plated on poly-d-lysine-coated glass coverslips, fixed, stained and imaged as described previously (3). Image acquisition of the fluorescence intensity was performed with an Olympus FV1000 confocal and Software 3.2 SP2, SP2 or a Zeiss LSM710 confocal microscope. Microscope images are representative of at least 50 cells examined in two to five independent experiments. Colocalization experiments were quantified using the ImageJ software (NIH). Images were first smoothed and passed by a medium filter to create a local background. Background-subtracted images were generated by subtracting the local background images from the original. Vesicle segmentation was conducted on background subtracted images using the SPOTS program developed by Indiana Center for Biological Microscopy (43). Binary mask images with segmented vesicles were then generated. Pixels from original images of both channels were selected using the mask images (43). The processed images from both channels were used for quantitative colocalization using Intensity Correlation Analysis to Mander’s correlation coefficient, as we applied in our previous work (1), and as reported (44).
Lysosomal acidity measurement
WT and KO MEFs, both primary and immortal, were plated in 60 mm tissue culture plates, treated with or without serum starvation overnight, followed by incubation with 1 uM LysosensorTM Green DND-189 (pKa ≈ 5.2, Life Technologies) at 37°C for 30 min, as reported (13). Fluorescent signals are directly proportional to the acidity of intracellular organelles. Cells were washed with cold Dulbecco-PBS (D-PBS) twice, trypsinized and harvested with D-PBS, followed by two additional cold D-PBS washes. Single cell suspension was stained with propidium iodide to exclude dead cells, and was then subjected to flow cytometry analysis on the BDTM LSR II flow cytometer (45). A total of 30 000 cells were counted for each sample. Four independent KO and WT MEF pairs were used for these experiments.
Cell fractionation
Overnight cultured MEF (6.5 × 106 cells) or approximately 20 mg mouse liver were homogenized in cold fractionation buffer (50 mm KCl, 90 mm potassium gluconate, 1 mm EGTA, 50 mm sucrose, 5 mm glucose, protease inhibitor cocktail tablet and 20 mm HEPES, pH 7.4) and lysed by sonication. Nuclear fraction was removed by centrifugation at 1000g for 10 min at 4°C. The supernatant was further centrifuged at 20 000g for 30 min at 4°C. The precipitated lysosome-enriched fraction representing the heavy membrane fraction was resuspended in fractionation buffer, and the supernatant was aliquoted as cytosolic fraction (14). These samples were also isolated by density-gradient centrifugation using the Lysosome enrichment kit for tissue and cultured cells (Thermofisher), following the manufacturer’s protocol (46).
Quantitative real-time (RT) PCR
RNA was isolated with Trizol (Life Technologies), reverse transcribed and quantitative RT-PCR for Lamtor1 (For-ccagtagcacccctaccaaa, Rev-cggtccatgtactcatgctg), Lamtor2 (For-GGGAACCAAGCGTTTAATGA, Rev-CCTACGGTCTCCTTGGCATA), Lamtor3 (For-GTTTGCCTCTGGTGGTGAGT, Rev-GGTGTGCATCAGAACCACTG), genes was performed in triplicate using the StepOnePlus System (Applied Biosystems/Life Technologies). The TATA box-binding protein (TBP) gene was used as a reference for calculating 2ΔΔ Ct as reported previously (3).
Generation of TMEM127 and LAMTOR1 knockout HEK293T with CRISPR/Cas9
TMEM127-null and LAMTOR1 HEK293 cells were generated by CRISPR-Cas9 technology. Two separate guide RNAs were designed to target the first coding exon of the TMEM127 gene and the LAMTOR1 gene, respectively. Guide RNAs were generated by annealing each following pair of oligonucleotides: TMEM127ex2_g1 For: ATGTACGCCCCCGGAGGCGC and Rev: GCGCCTCCGGGGGCGTACATC; and TMEM127ex2_g2 For: GGCCCCGGGCATGTACGCCCC and Rev: GGGGCGTACATGCCCGGGGCC; LAMTOR1ex1_g2 For: CGAGAACGAGGACTCGGACC and Rev: GGTCCGAGTCCTCGTTCTCGC. All nucleotides contained CACC (For) and AAAC (Rev) sequences attached to the 5′ of the gene-specific target sequence above to allow for ligation into pLentiCRISPRv2 (from F. Zhang laboratory through Addgene) that was previously cut with BsmbI, as reported (47). HEK-293 cells were transfected with LentiCRISPRv2 expressing above guides using Lipofectamine 2000, and single clones were selected with 3 µg/mL puromycin. The KO status of each clone was verified by Sanger sequencing, revealing one to multiple nucleotide deletions that resulted in frameshifts, and by western blot. Cell clones containing EV were sequence verified for intact TMEM127 sequence and used as controls.
Statistical analysis
Statistical analyses were performed by two-way ANOVA with Bonferroni’s post-test in measurements involving two independent variables (e.g. genotype and treatment) (48). One-way ANOVA with Tukey’s post-test was used to determine statistical significance in measurements with one variable in more than two groups (49). Two-tailed Student’s t-test was used in comparing measurement between two groups. The statistical methods used are specified in each corresponding figure legend. Statistical significance was defined by P-values of less than 0.05. Results are representative of two to five individual experiments or as indicated in the figures.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
The authors are grateful to Drs Ricardo C. T. Aguiar, Jim Brugarolas, Jim Lechleiter and LuZhe Sun for their input and many valuable discussions; and to Aaron Rizinger for his advice on imaging data analysis.
Conflict of Interest statement. None declared.
Funding
This work was supported by the Cancer Prevention and Research Institute of Texas (CPRIT) Individual Investigator Grants RP101202 and RP140473 (P.L.M.D), CPRIT Training Grant RP140105 (Y.D.); Department of Defense CDMRP W81XWH-12-1-0508 (P.L.M.D.); NIH/NIGMS GM114102 (P.L.M.D.); NRSA Institutional Predoctoral Training Grant T32CA148724 (S.K.F.); NIA T32AG021890 (S.S.), Max and Minnie Tomerlin Voelcker Fund (P.L.M.D.). The Optical Imaging Core Facility and the Flow Cytometry Core Facility are both supported by NIH-NCI P30-CA54174 (UT Health San Antonio Cancer Center) and NIH-NIA P01-AG19316.
References
- 1. Qin Y., Yao L., King E.E., Buddavarapu K., Lenci R.E., Chocron E.S., Lechleiter J.D., Sass M., Aronin N., Schiavi F.. et al. (2010) Germline mutations in TMEM127 confer susceptibility to pheochromocytoma. Nat. Genet., 42, 229–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yao L., Schiavi F., Cascon A., Qin Y., Inglada-Perez L., King E.E., Toledo R.A., Ercolino T., Rapizzi E., Ricketts C.J.. et al. (2010) Spectrum and prevalence of FP/TMEM127 gene mutations in pheochromocytomas and paragangliomas. J. Am. Med. Assoc., 304, 2611–2619. [DOI] [PubMed] [Google Scholar]
- 3. Qin Y., Deng Y., Ricketts C.J., Srikantan S., Wang E., Maher E.R., Dahia P.L. (2014) The tumor susceptibility gene TMEM127 is mutated in renal cell carcinomas and modulates endolysosomal function. Hum. Mol. Genet., 23, 2428–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Casey R.T., Warren A.Y., Rodrigues J.E., Challis B.G., Rattenberry E., Whitworth J., Andrews K.A., Roberts T., Clark G.R., West H.. et al. (2017) Clinical and molecular features of renal and phaeochromocytoma/paraganglioma tumour association syndrome (RAPTAS): case series and literature review. J. Clin. Endocrinol. Metab., 102, 4013–4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Palmieri M., Impey S., Kang H., di Ronza A., Pelz C., Sardiello M., Ballabio A. (2011) Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet., 20, 3852–3866. [DOI] [PubMed] [Google Scholar]
- 6. Saxton R.A., Sabatini D.M. (2017) mTOR signaling in growth, metabolism, and disease. Cell, 169, 361–371. [DOI] [PubMed] [Google Scholar]
- 7. Bar-Peled L., Sabatini D.M. (2014) Regulation of mTORC1 by amino acids. Trends Cell Biol., 24, 400–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sancak Y., Peterson T.R., Shaul Y.D., Lindquist R.A., Thoreen C.C., Bar-Peled L., Sabatini D.M. (2008) The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science, 320, 1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sancak Y., Bar-Peled L., Zoncu R., Markhard A.L., Nada S., Sabatini D.M. (2010) Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell, 141, 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zoncu R., Bar-Peled L., Efeyan A., Wang S., Sancak Y., Sabatini D.M. (2011) mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science, 334, 678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bar-Peled L., Schweitzer L.D., Zoncu R., Sabatini D.M. (2012) Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell, 150, 1196–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Forgac M. (2007) Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol., 8, 917–929. [DOI] [PubMed] [Google Scholar]
- 13. Rong Y., McPhee C.K., Deng S., Huang L., Chen L., Liu M., Tracy K., Baehrecke E.H., Yu L., Lenardo M.J. (2011) Spinster is required for autophagic lysosome reformation and mTOR reactivation following starvation. Proc. Natl. Acad. Sci. U. S. A., 108, 7826–7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim Y.C., Park H.W., Sciarretta S., Mo J.S., Jewell J.L., Russell R.C., Wu X., Sadoshima J., Guan K.L. (2014) Rag GTPases are cardioprotective by regulating lysosomal function. Nat. Commun., 5, 4241.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Manifava M., Smith M., Rotondo S., Walker S., Niewczas I., Zoncu R., Clark J., Ktistakis N.T. (2016) Dynamics of mTORC1 activation in response to amino acids. eLife, 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang T., Wang R., Wang Z., Wang X., Wang F., Ding J. (2017) Structural basis for ragulator functioning as a scaffold in membrane-anchoring of Rag GTPases and mTORC1. Nat. Commun., 8, 1394.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Araujo M.E.G., Naschberger A., Furnrohr B.G., Stasyk T., Dunzendorfer-Matt T., Lechner S., Welti S., Kremser L., Shivalingaiah G., Offterdinger M.. et al. (2017) Crystal structure of the human lysosomal mTORC1 scaffold complex and its impact on signaling. Science, 358, 377–381. [DOI] [PubMed] [Google Scholar]
- 18. de Araujo M.E., Stasyk T., Taub N., Ebner H.L., Furst B., Filipek P., Weys S.R., Hess M.W., Lindner H., Kremser L.. et al. (2013) Stability of the endosomal scaffold protein LAMTOR3 depends on heterodimer assembly and proteasomal degradation. J. Biol. Chem., 288, 18228–18242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Deng Y., Flores S.K., Cheng Z., Qin Y., Schwartz R.C., Malchoff C., Dahia P.L.M. (2017) Molecular and phenotypic evaluation of a novel germline TMEM127 mutation with an uncommon clinical presentation. Endocr. Relat. Cancer, 24, L79–L82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bowman E.J., Graham L.A., Stevens T.H., Bowman B.J. (2004) The bafilomycin/concanamycin binding site in subunit c of the V-ATPases from Neurospora crassa and Saccharomyces cerevisiae. J. Biol. Chem., 279, 33131–33138. [DOI] [PubMed] [Google Scholar]
- 21. Zoncu R., Efeyan A., Sabatini D.M. (2011) mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol., 12, 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dahia P.L. (2014) Pheochromocytoma and paraganglioma pathogenesis: learning from genetic heterogeneity. Nat. Rev. Cancer, 14, 108–119. [DOI] [PubMed] [Google Scholar]
- 23. Johannessen C.M., Johnson B.W., Williams S.M., Chan A.W., Reczek E.E., Lynch R.C., Rioth M.J., McClatchey A., Ryeom S., Cichowski K. (2008) TORC1 is essential for NF1-associated malignancies. Curr. Biol., 18, 56–62. [DOI] [PubMed] [Google Scholar]
- 24. Polley E., Kunkel M., Evans D., Silvers T., Delosh R., Laudeman J., Ogle C., Reinhart R., Selby M., Connelly J.. et al. (2016) Small cell lung cancer screen of oncology drugs, investigational agents, and gene and microRNA expression. J. Natl. Cancer Inst., 108, djw122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fishbein L., Leshchiner I., Walter V., Danilova L., Robertson A.G., Johnson A.R., Lichtenberg T.M., Murray B.A., Ghayee H.K., Else T.. et al. (2017) Comprehensive molecular characterization of pheochromocytoma and paraganglioma. Cancer Cell, 31, 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kleymenova E., Ibraghimov-Beskrovnaya O., Kugoh H., Everitt J., Xu H., Kiguchi K., Landes G., Harris P., Walker C. (2001) Tuberin-dependent membrane localization of polycystin-1: a functional link between polycystic kidney disease and the TSC2 tumor suppressor gene. Mol. Cell, 7, 823–832. [DOI] [PubMed] [Google Scholar]
- 27. Brugarolas J.B., Vazquez F., Reddy A., Sellers W.R., Kaelin W.G. Jr. (2003) TSC2 regulates VEGF through mTOR-dependent and -independent pathways. Cancer Cell, 4, 147–158. [DOI] [PubMed] [Google Scholar]
- 28. Nickerson M.L., Warren M.B., Toro J.R., Matrosova V., Glenn G., Turner M.L., Duray P., Merino M., Choyke P., Pavlovich C.P.. et al. (2002) Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dube syndrome. Cancer Cell, 2, 157–164. [DOI] [PubMed] [Google Scholar]
- 29. Bar-Peled L., Chantranupong L., Cherniack A.D., Chen W.W., Ottina K.A., Grabiner B.C., Spear E.D., Carter S.L., Meyerson M., Sabatini D.M. (2013) A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science, 340, 1100–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chantranupong L., Wolfson R.L., Orozco J.M., Saxton R.A., Scaria S.M., Bar-Peled L., Spooner E., Isasa M., Gygi S.P., Sabatini D.M. (2014) The Sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell Rep., 9, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chantranupong L., Scaria S.M., Saxton R.A., Gygi M.P., Shen K., Wyant G.A., Wang T., Harper J.W., Gygi S.P., Sabatini D.M. (2016) The CASTOR proteins are arginine sensors for the mTORC1 pathway. Cell, 165, 153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Petit C.S., Roczniak-Ferguson A., Ferguson S.M. (2013) Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J. Cell Biol., 202, 1107–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Carroll B., Maetzel D., Maddocks O.D., Otten G., Ratcliff M., Smith G.R., Dunlop E.A., Passos J.F., Davies O.R., Jaenisch R.. et al. (2016) Control of TSC2-Rheb signaling axis by arginine regulates mTORC1 activity. eLife, 5, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wolfson R.L., Sabatini D.M. (2017) The dawn of the age of amino acid sensors for the mTORC1 pathway. Cell Metab., 26, 301–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hallett J.E., Manning B.D. (2016) CASTORing new light on amino acid sensing. Cell, 165, 15–17. [DOI] [PubMed] [Google Scholar]
- 36. Filipek P.A., de Araujo M.E.G., Vogel G.F., De Smet C.H., Eberharter D., Rebsamen M., Rudashevskaya E.L., Kremser L., Yordanov T., Tschaikner P.. et al. (2017) LAMTOR/ragulator is a negative regulator of Arl8b- and BORC-dependent late endosomal positioning. J. Cell Biol., 216, 4199–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pu J., Schindler C., Jia R., Jarnik M., Backlund P., Bonifacino J.S. (2015) BORC, a multisubunit complex that regulates lysosome positioning. Dev. Cell, 33, 176–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schweitzer L.D., Comb W.C., Bar-Peled L., Sabatini D.M. (2015) Disruption of the Rag-ragulator complex by C17orf59 inhibits mTORC1. Cell Rep., 12, 1445–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hurtado-Lorenzo A., Skinner M., El Annan J., Futai M., Sun-Wada G.H., Bourgoin S., Casanova J., Wildeman A., Bechoua S., Ausiello D.A.. et al. (2006) V-ATPase interacts with ARNO and Arf6 in early endosomes and regulates the protein degradative pathway. Nat. Cell Biol., 8, 124–136. [DOI] [PubMed] [Google Scholar]
- 40. Xu J. (2005) Preparation, culture, and immortalization of mouse embryonic fibroblasts. Curr. Prot. Mol. Biol., 28, 28–21. [DOI] [PubMed] [Google Scholar]
- 41. Hahn W.C., Counter C.M., Lundberg A.S., Beijersbergen R.L., Brooks M.W., Weinberg R.A. (1999) Creation of human tumour cells with defined genetic elements. Nature, 400, 464–468. [DOI] [PubMed] [Google Scholar]
- 42. Uphoff C.C., Denkmann S.A., Drexler H.G. (2012) Treatment of mycoplasma contamination in cell cultures with Plasmocin. J. Biomed. Biotechnol., 2012, 1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Deng L., Jiang C., Chen L., Jin J., Wei J., Zhao L., Chen M., Pan W., Xu Y., Chu H.. et al. (2015) The ubiquitination of rag A GTPase by RNF152 negatively regulates mTORC1 activation. Mol. Cell, 58, 804–818. [DOI] [PubMed] [Google Scholar]
- 44. Li Q., Lau A., Morris T.J., Guo L., Fordyce C.B., Stanley E.F. (2004) A syntaxin 1, Galpha(o), and N-type calcium channel complex at a presynaptic nerve terminal: analysis by quantitative immunocolocalization. J. Neurosci., 24, 4070–4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nofer J.R., Herminghaus G., Brodde M., Morgenstern E., Rust S., Engel T., Seedorf U., Assmann G., Bluethmann H., Kehrel B.E. (2004) Impaired platelet activation in familial high density lipoprotein deficiency (Tangier disease). J. Biol. Chem., 279, 34032–34037. [DOI] [PubMed] [Google Scholar]
- 46. Lukong K.E., Seyrantepe V., Landry K., Trudel S., Ahmad A., Gahl W.A., Lefrancois S., Morales C.R., Pshezhetsky A.V. (2001) Intracellular distribution of lysosomal sialidase is controlled by the internalization signal in its cytoplasmic tail. J. Biol. Chem., 276, 46172–46181. [DOI] [PubMed] [Google Scholar]
- 47. Sanjana N.E., Shalem O., Zhang F. (2014) Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods, 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Talukdar S., Oh D.Y., Bandyopadhyay G., Li D., Xu J., McNelis J., Lu M., Li P., Yan Q., Zhu Y.. et al. (2012) Neutrophils mediate insulin resistance in mice fed a high-fat diet through secreted elastase. Nat. Med., 18, 1407–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bartolomeo R., Cinque L., De Leonibus C., Forrester A., Salzano A.C., Monfregola J., De Gennaro E., Nusco E., Azario I., Lanzara C.. et al. (2017) mTORC1 hyperactivation arrests bone growth in lysosomal storage disorders by suppressing autophagy. J. Clin. Investig., 127, 3717–3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.