Abstract
Amyotrophic lateral sclerosis (ALS) is a devastating, uniformly lethal degenerative disorder of motor neurons that overlaps clinically with frontotemporal dementia (FTD). Investigations of the 10% of ALS cases that are transmitted as dominant traits have revealed numerous gene mutations and variants that either cause these disorders or influence their clinical phenotype. The evolving understanding of the genetic architecture of ALS has illuminated broad themes in the molecular pathophysiology of both familial and sporadic ALS and FTD. These central themes encompass disturbances of protein homeostasis, alterations in the biology of RNA binding proteins, and defects in cytoskeletal dynamics, as well as numerous downstream pathophysiological events. Together, these findings from ALS genetics provide new insight into therapies that target genetically distinct subsets of ALS and FTD.
Amyotrophic lateral sclerosis (ALS or Lou Gehrig's disease) is a devastating neurodegenerative disorder that affects lower motor neurons (LMNs) in the brainstem and spinal cord and upper motor neurons (UMNs) in the motor cortex, which are predominantly large, layer V neurons that comprise the corticospinal tract. The core clinical phenotype is characterized by (1) focal onset of weakness that progresses to involve all of the limbs and bulbar muscles, leading ultimately to total paralysis and death from respiratory failure, and (2) hyperreflexia. At autopsy, degeneration of corticospinal (“upper”) and spinal and bulbar (“lower”) motor neurons is often associated with activation of neuroimmune cells within the central nervous system (CNS) (microglia, astrocytes, and oligodendroglia) (Kang et al. 2013; Philips and Rothstein 2014). In ∼15% of ALS patients, neurons in the prefrontal and temporal cortex are also variably affected (Ringholz et al. 2005), leading to frontal executive dysfunction, speech production disorders, and concurrent frontotemporal dementia (FTD). ALS and FTD can be viewed as divergent ends of the spectrum of a single heterogeneous disease. Moreover, although a combination of UMN and LMN findings characterizes classic ALS, the continuum of disease ranges from exclusively UMN (primary lateral sclerosis or PLS) to exclusively LMN involvement (progressive muscular atrophy or PMA). As discussed below, some cases of ALS with FTD occur with other multisystem manifestations, including pathology in muscle and bone. Rarely, “ALS-plus” syndromes are encountered in which ALS patients also develop autonomic nervous system involvement, extrapyramidal signs, cerebellar findings, and sensory abnormalities in addition to classic ALS signs.
ALS has a worldwide incidence of about two cases per 100,000 per year and a prevalence of about five cases per 100,000. In the United States, ALS affects about 25,000 individuals and thus formally is classified as an “orphan” disease (fewer than 200,000 affected cases) (Rowland and Shneider 2001). However, it is remarkable that ALS accounts for about one in 500 adult deaths, a statistic that predicts that more than 500,000 individuals now in the United States will succumb to this disease. Onset typically occurs between 55 and 60 years of age, with wide-ranging severity. Median survival is 27.5 mo from symptom onset (Robberecht and Philips 2013); the 4-year survival rate is ∼40%. The major cause of death is respiratory failure due to respiratory muscle weakness.
Although most cases of ALS are sporadic (sALS), ∼10% are familial (fALS), with autosomal dominant transmission. Rarely, fALS may be transmitted as either X-linked or recessive traits (Andersen and Al-Chalabi 2011; Deng et al. 2011). fALS and sALS share similar clinical presentations; either can develop concurrently with FTD.
Efforts to identify Mendelian genes whose mutations cause ALS have identified more than 100 gene mutations that increase susceptibility to ALS or modify its phenotype (Fig. 1A,B) (Andersen and Al-Chalabi 2011; Peters et al. 2015b). Multiple genetic variants may interact simultaneously to increase ALS susceptibility; such oligogenic cases of ALS may not appear to be familial in a conventional Mendelian manner yet may underlie the apparently sporadic form of the disease (Luigetti et al. 2011; Chio et al. 2012; van Blitterswijk et al. 2012; Pottier et al. 2015). Analysis of pathways implicated by mutant ALS genes has provided new insights into the pathogenesis of both fALS and sALS, although, regrettably, this has not yet yielded definitive treatments.
Figure 1.
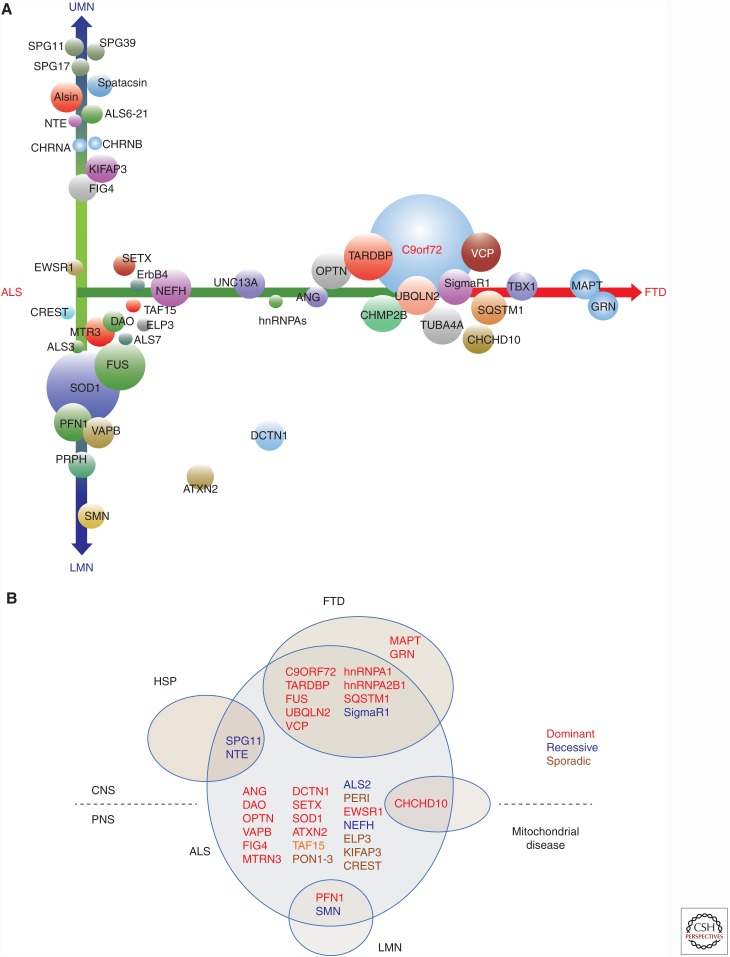
The phenotypic spectrum of amyotrophic lateral sclerosis (ALS) genetics. (A) Forty-six ALS-related genes are arrayed along axes that depict two major phenotypic aspects: the extent to which corticospinal versus lower motor neurons are involved (y-axis) and the overlap with frontotemporal dementia (FTD) (x-axis). The diameters of each gene approximate their relative frequencies. (B) The phenotypic overlap of ALS genes with hereditary spastic paraplegia (HSP), frontotemporal dementia (FTD), mitochondrial disease, and lower motor neuropathies (LMN) is shown.
As summarized in Figure 2, three themes have emerged from the investigations of the molecular biology of diverse mutant ALS genes in fALS and sALS.
Analyses of one set of genes encoding a range of proteins (e.g., cytosolic superoxide dismutase or adaptor proteins that mediate protein quality control such as ubiquilin-2) indicate that motor neuron death in ALS involves misfolding, aggregation, and deposition of proteins that are often ubiquitinated and predominantly cytoplasmic. A growing literature suggests that the mutant proteins may induce misfolding of their wild-type (WT) counterparts and thus have prion-like properties.
Another set of genes (e.g., TAR DNA-binding protein [TDP-43], fused in sarcoma/translocated in liposarcoma [FUS/TLS]) encodes proteins that are implicated in interacting with RNA and DNA and illustrates the principle that perturbations in multiple aspects of RNA function and homeostasis are toxic to motor neurons. Particularly exciting in this gene set is the demonstration of non-nuclear localization and function of protein-bound RNA, with roles in axonal transport and local protein translation in dendrites and neuromuscular junctions.
In most cases, there are disturbances of cytoskeletal architecture and dynamic function of distal axons and dendrites.
Figure 2.
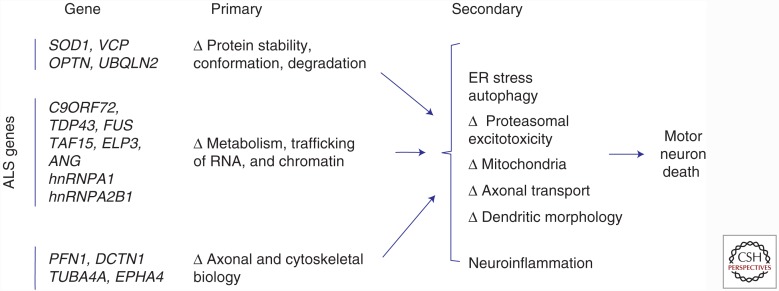
The common ALS genes define three primary themes in pathophysiology: conformational instability and aggregation of proteins, impaired trafficking of RNA, and altered cytoskeletal dynamics. These converge on multiple secondary, downstream pathologic processes that include activation of endoplasmic reticulum (ER) stress and autophagy, proteasomal dysfunction, altered mitochondrial function, disturbed axonal transport, altered dendritic morphology, and excitotoxicity.
Three additional points regarding this formulation deserve mention. First, elegant studies document that the non-cell-autonomous, neuroinflammatory aspect of ALS can accelerate the disease course. Yet, to date, every ALS gene is expressed in motor neurons; none of the known ALS genes are predominantly expressed in non-neural cells. This suggests that expression of a mutant ALS gene within the motor neuron is the sine qua non of motor neuron degeneration in fALS. Second, these themes in pathogenesis are not mutually exclusive; mutations in RNA-binding proteins from category 2 predispose the proteins to self-assembly and to propagating aggregation, as described in category 1. Third, regardless of how the primary gene defects are categorized, they all are associated with common downstream pathologic processes, including induction of endoplasmic reticulum (ER) stress, accelerated autophagy, mitochondrial dysfunction, altered axonal transport, disturbed cytoskeletal dynamics in dendrites and growth cones, and altered membrane excitability (Vucic et al. 2014). A detailed discussion of these diverse but important secondary phenomena, which are also evident in sALS, is beyond the scope of this review. Table 1 provides a detailed summary of the major ALS genes; a full listing can be found at alsod.iop.kcl.ac.uk.
Table 1.
Gene mutations that cause ALS
| Gene | Fraction fALS (%) | Locus | Encoded protein | Functionality | Clinical phenotype | Neuropathology | Reference(s) |
|---|---|---|---|---|---|---|---|
| C9ORF72 | 40–50 | 9p21.3 | C9ORF72 | Transcription and pre-mRNA splicing regulation; membrane traffic via Rab GTPase family | ALS; ALS+ FTLD; FTLD | NCI; DN; GCI; intranuclear RNA foci (sense, antisense); cytoplasmic RNA peptide aggregates | DeJesus-Hernandez et al. 2011; Renton et al. 2011; Cooper-Knock et al. 2012; Diekstra et al. 2014 |
| SOD1 | 20–25 | 21q22 | SOD1 | Major cytosolic antioxidant | ALS; PMA | NCI; NII; DN; GCI; aggregates—p62, C9ORF72, ubiquilin 2, others; impaired axonal transport, mitochondrial function; disturbed dendritic arborization of neurons; oxidative stress-related neuronal toxicity | Rosen et al. 1993; Watanabe et al. 2001; Forsberg et al. 2011 |
| TARDBP | 4–5 | 1p36.2 | TDP-43 | Transcription and pre-mRNA splicing regulation; micRNA biogenesis; RNA transport and stabilization; translational regulation of ApoE-II and CFTR | ALS; ALS+ FTLD; FTLD | NCI; NII; DN; GCI | Rutherford et al. 2008; Sreedharan et al. 2008; Neumann 2009 |
| FUS | 4–5 | 16p11.2 | FUS (or TLS) | Transcription and pre-mRNA splicing regulation; micRNA processing; mRNA transport and stabilization; maintenance of genomic integrity; regulating protein synthesis at synapse | ALS; ALS+ FTLD; FTLD | NCI; DN; GCI | Kwiatkowski et al. 2009; Vance et al. 2009; Huang et al. 2010 |
| OPTN | 2–3 | 10p13 | Optineurin | Golgi maintenance; exocytosis; vesicular trafficking; regulator of NF-kB signaling pathway; autophagy process | ALS; ALS+ FTLD | NCI; ↑ TDP-43, FUS, and SOD1 aggregates | Maruyama et al. 2010 |
| PFN1 | 1–2 | 17p13 | Profilin 1 | Regulates ATP-mediated actin polymerization | ALS | NCI; ↓ axonal distension and growth cone elongation; co-aggregation with TDP-43 | Wu et al. 2012 |
| VCP | 1–2 | 9p13 | VCP or p97 | Protein degradation via UPS, autophagy, and the ER; membrane fusion | ALS; ALS+ FTLD; FTLD | NCI; NII; DN; ↑ TDP-43 aggregates; ↓ stress-granule clearance | Forman et al. 2006; Kimonis et al. 2008; Johnson et al. 2010 |
| ANG | 1–2 | 14q11.2 | Angiogenin | RNA processing and tRNA modification; vascularization; RNAase activity and assembly of stress granules; neurite outgrowth and pathfinding | ALS; ALS+ FTLD | ↓ Stress-granule formation in motor neurons | Greenway et al. 2006; Thiyagarajan et al. 2012 |
| TUBA4A | 1 | 2q35 | Tubulin α4A | Major component of microtubules; neuronal cell skeleton | ALS; ALS+ FTLD | NCI; destabilized microtubule network; ↓ microtubules repolymerization capability | Smith et al. 2014 |
| UBQLN2 | <1 | Xp11 | Ubiquilin 2 | Protein degradation via UPS | ALS; ALS+ FTLD; FTLD (Rare) | NCI; ↑ TDP-43, p62, FUS, and OPTN inclusions | Deng et al. 2011; Ugwu et al. 2014 |
| TAF15 | <1 | 17q11 | TAF15 | Transcription initiation; RNA polymerase II gene component | ALS | NCI | Couthouis et al. 2011 |
| EWSR1 | <1 | 22q12.2 | EWSR1 | Transcriptional repressor | ALS | NCI; DN | Couthouis et al. 2012 |
| hnRNPA1 | <1 | 12q13 | hnRNPA1 | Packing and transport of mRNA; micRNA biogenesis | Rare ALS; ALS+ FTLDa; FTLD (rare) | NCI | Bekenstein and Soreq 2013; Kim et al. 2013; Le Ber et al. 2014 |
| hnRNPA2B1 | <1 | 7p15 | hnRNPA2/B1 | Packing and transport of mRNA; micRNA biogenesis | ALS+ FTLD (rare)a; FTLD (rare) | NCI | Kim et al. 2013; Le Ber et al. 2014 |
| SETX | <1 | 9q34.13 | Senataxin | DNA/RNA helicase activity; DNA/RNA metabolism | ALSb | ↓ neuronal differentiation; ↓ neurite growth | Chen et al. 2004 |
| CREST | <1 | 20q13.3 | SS18L1 | Ca2+-dependent transcriptional activator | ALS | DN; ↓ dendrite outgrowth; ↑ interaction with FUS | Chesi et al. 2013; Teyssou et al. 2014 |
| MATR3 | <1 | 5q31.2 | Matrin 3 | RNA processing; stabilizing mRNAs; gene silencing; chromatin organization | ALS; ALS+ FTLD (Rare) | NCI; NII; ↑ interaction with TDP-43 | Johnson et al. 2014b; Millecamps et al. 2014 |
| ATXN2 | 1-2 | 12q24 | Ataxin 2 | RNA processing; regulation of receptor tyrosine kinase endocytosis | ALS; ALS+ FTLD; PMA | NCI; ↑ interaction with TDP-43 | Elden et al. 2010; Baumer et al. 2014 |
| ELP3 | <1 | 8p21.1 | ELP3 | RNA processing; transcript elongation; histone acetylation; modification of tRNA wobble nucleosides | ALS | NCI; abnormal branching in motor axons; co-localization with TDP-43 and FUS aggregates | Simpson et al. 2009; Fujita et al. 2014 |
| SQSTM1 | <1 | 5q35 | p62 or SQSTM1 | Autophagy and UPS degradation; regulator of NF-kB signaling pathway; immune response | ALS; ALS+ FTLD; FTLD | NCI; NII; GCI; ↓ mutSOD1 autophagic degradation | Gal et al. 2009; Fecto et al. 2011; Rubino et al. 2012 |
| CHMP2B | <1 | 3p11 | CHMP2B | MVBs formation; protein trafficking between plasma membrane, trans-Golgi network, and lysosome | ALSc; PMAd; FTLD | NCI; DN; GCI; disrupted endosomal structure; aggregates of autophagosomes and multilamellar structures; ↑ TDP-43, p62, and ubiquitin inclusions | Cannon et al. 2006; Parkinson et al. 2006 |
| ALS2 | <1 | 2q33.1 | Alsin | Activation of the small GTPase Rac1 macropinocytosis-associated endosome fusion and trafficking; neurite outgrowth | ALSe; PLS | ↓ axonal growth; ↓ lysosome-dependent clearance of p62 and LC3-II | Hadano et al. 2001; Yang et al. 2001; Otomo et al. 2008 |
| VAPB | <1 | 20q13 | VAPB | Regulation of ER–Golgi transport and secretion | ALS; PLS; PMA | NCI; ↑ TDP-43 toxicity and inclusions; aberrant synaptic microtubule cytoskeleton; nuclei mispositioning and aberrant architecture | Nishimura et al. 2004; Sanhueza et al. 2014 |
| SIGMAR1 | <1 | 9p13.3 | SIGMAR1 | Lipid transport through ER; BDNF and EGF signaling | ALS; ALS+ FTLD; FTLD | NCI; ↑ apoptosis induced by ER stress; ↑ interaction with VAPB | Al-Saif et al. 2011; Belzil et al. 2013; Prause et al. 2013 |
| DCTN1 | <1 | 2p13 | Dynactin | ER–Golgi transport; centripetal movement of lysosomes and endosomes; spindle formation, chromosome movement; nuclear positioning; axonogenesis | ALS | NCI; p150glued aggregation; ↑ SOD1 aggregates | Munch et al., 2004 |
| FIG4 | <1 | 6q21 | PI3,5P2 | Phosphoinositide phosphatase activity; endosomal vesicle trafficking to the trans-Golgi network; regulation of autophagy | ALS; PLS | NCI; ↑ swollen intracellular vacuoles; ↑ LC3-II, p62, and LAMP-2 aggregates in neurons and astrocytes | Zhang et al. 2007; Chow et al. 2009 |
| SPG11 | 1 | 15q21.1 | Spatascin | Neuronal cell skeleton; axonal transport; involved in synaptic vesicles | ALS; HSP | NCI; DN; ↓ acetylated stabilized tubulin; ↓ synaptic vesicles in neurites; disrupted anterograde axonal transport | Stevanin et al. 2007; Orlacchio et al. 2010; Manole et al. 2016 |
| NEFH | <1 | 22q12.2 | NEFH | Maintaining axon diameter | ALS | NCI; ↑ neurofilament aggregates | Figlewicz et al. 1994 |
| PRPH | <1 | 12q13 | Peripherin | Regulating neurite elongation during development and axonal regeneration after injury | ALS | NCI; DN; ↓ ability of the neurofilament network to assemble; ↑ ubiquitinated inclusions; coaggregation with mutSOD1 | Beaulieu et al. 1999; Julien and Beaulieu 2000; Gros-Louis et al. 2004 |
| NTE | <1 | 19p13 | Neuropathy target esterase | Regulating the neuronal membrane composition | ALS; HSP | Disruption of ER; ↑ reticular aggregates; ↑ vacuolization of nerve cell bodies | Akassoglou et al. 2004; Rainier et al. 2008; Song et al. 2013 |
| PON1-3 | <1 | 7q21 | Paraoxonase 1-3 | Enzymatic breakdown of nerve toxins | ALS | Oxidative stress-related neuronal toxicity | Saeed et al. 2006; Slowik et al. 2006; Wills et al. 2009 |
| DAO | <1 | 12q22 | DAO | Regulating levels of D-serine, NMDAR function | ALS | NCI; NII; ↑ D-serine levels in motor neurons and glia; ↑ ubiquitinated inclusions | Mitchell et al. 2010 |
| CHRNA3, CHRNA4, CHRNB4 | <1 | 15q24, 20q13, 15q24 | nAChR | Cholinergic neurotransmission | ALS§ | Cationic overloading, Ca+2 toxicity in MNs | Sabatelli et al. 2009, 2012; Moriconi et al. 2011) |
| ERBB4 | <1 | 2q34 | Receptor tyrosine-protein kinase ErbB-4 | Neuronal cell mitogenesis and differentiation | ALS | Takahashi et al. 2013 | |
| CHCHD10 | <1 | 22q11 | Mitochondrial protein | Mitochondrial genome stability; cristae integrity and mitochondrial fusion | ALS+ FTLD | Mitochondrial fragmentation and DNA instability; mitochondrial crystalloid inclusions | Bannwarth et al. 2014; Chaussenot et al. 2014; Johnson et al. 2014a |
| C19orf12 | <1 | 9q12 | Mitochondrial protein | Unknown | ALS | Deschauer et al. 2012 | |
| ALS3 | <1 | 18q21 | Disulfide redox protein | Unknown | ALS | Hand et al. 2002 | |
| ALS7 | <1 | 20p13 | Unknown | Unknown | ALS | Hand et al. 2002 | |
| ALS6-21 | <1 | 6p25, 21q22 | Unknown | Unknown | ALS§ | Butterfield et al. 2009 | |
| ALS-FTD | <1 | 16p12 | Unknown | Unknown | ALS+ FTLD | NCI; DN; Type B TDP pathology; phosphorylated tau pathology | Dobson-Stone et al. 2013 |
| Gene variants that influence ALS phenotype | |||||||
| UNC13A | 19p13 | Unc-13 homolog A | Regulating neurite outgrowth and synaptic neurotransmission | ALS; ALS+ FTLD | ↓ synaptogenesis at neuromuscular junction; possible glutamate excitotoxicity | Varoqueaux et al. 2005; van Es et al. 2009; Diekstra et al. 2014 | |
| EPHA4 | 2q36.1 | Ephrin receptor A4 | Receptor tyrosine kinase activity Modulation of cell morphology and integrin-dependent cell adhesion; regulation of synaptic plasticity and CNS development | ALS | NII; neurite outgrowth deficits in mutant TDP-43 expressed neurons | Van Hoecke et al. 2012; Uyan et al. 2013 | |
| CHGB | 20p12.3 | CHGB | Involved in the ER–Golgi system | ALS | NCI; ↓ density of synaptophysin-like immunoreactivity; ↑ interaction with mutSOD1 | Gros-Louis et al. 2009; Schrott-Fischer et al. 2009 | |
| KIFAP3 | 1q24.2 | Kinesin-associated protein 3 | Tethering chromosomes to spindle pole; chromosome movement; axonal transport of choline acetyl-transferase | ALS§ | NCI; KIFAP3-SOD1 coaggregation in Lewy-body-like hyaline inclusions | Landers et al. 2009; Tateno et al. 2009; Orsetti et al. 2011 | |
| SMN | 5q13 | Germin 1 | Regulating biogenesis of snRNPs | ALS; LMN | NCI; NII; DN; coaggregation with mutFUS, mutSOD1; axonal defects | Jackson et al. 1996; Moulard et al. 1998; Gertz et al. 2012; Groen et al. 2013 | |
ALS, amyotrophic lateral sclerosis; BDNF, brain derived neurotrophic factor; C9ORF72, chromosome 9 open reading frame 72; CHGB, chromogranin B (secretogranin 1); CHMP2B, charged multivesicular body protein 2B; DAO, D-amino acid oxidase; DN, dystrophic neurites; EGF, epidermal growth factor; ELP3, elongator acetyltransferase complex subunit 3; ER, endoplasmic reticulum; EWSR1, Ewing sarcoma breakpoint region 1; FTLD, frontotemporal lobe dementia; FUS, fused in sarcoma; GCIs, glial cell inclusions; hnRNPA1, heterogeneous nuclear ribonucleoprotein A1; hnRNPA2B1, heterogeneous nuclear ribonucleoproteins A2/B1; HSP, hereditary spastic paraplegia; LAMP-2, lysosomal-associated membrane protein 2; LC3-II, microtubule-associated protein 1A/1B-light chain 3-II; LMN, lower motor neuron disease; micRNA, micro RNA; mRNA, messenger RNA; mutSOD1, mutant superoxide dismutase 1; MVBs, multivesicular bodies; nAChR, nicotinic acetylcholine receptor; NCI, neuronal cytoplasmic inclusions; NEFH, neurofilament heavy chain; NII, neuronal intranuclear inclusions; NMDAR, N-methyl-D-aspartate receptor; PI3,5P2, phosphatidylinositol 3,5-bisphosphate 5-phosphatase; PLS, primary lateral sclerosis; PMA, progressive muscular atrophy; SIGMAR1, σ non-opioid intracellular receptor 1; SQSTM1, sequestosome 1; SS18L1, synovial sarcoma translocation gene on chromosome 18-Like 1; TAF15, TATA box binding protein-associated factor 15; TDP-43, TAR DNA-binding protein; TLS, translocated in liposarcoma; UMN, upper motor neuron; UPS, ubiquitin-proteasome system; VAPB, vesicle-associated membrane protein B; VCP, valosin-containing protein.
aAs part of multisystem proteinopathy.
bPhenotype more similar to Silver syndrome than to ALS.
cPredominant LMN phenotype.
dAs part of ALS.
ePredominant UMN phenotype.
OVERVIEW OF ALS GENETICS
Adult-Onset ALS Genes Transmitted as Mendelian Traits
Genes Implicated in Disturbances of Protein Homeostasis
Superoxide Dismutase
The first ALS gene, cytosolic copper-zinc superoxide dismutase (SOD1), was identified in 1993 (Fig. 3) (Rosen et al. 1993). In most families harboring SOD1 gene mutations, disease penetrance is >90% by age 70 (Cudkowicz et al. 1997). More than 170 mutations have now been detected in the SOD1 gene in fALS (Abel et al. 2012); together these account for ∼20% of fALS cases. Almost all of these mutations are missense changes, scattered across the coding sequence without focal mutation hotspots. A few mutations truncate the terminal segment of the SOD1 protein; none of the ALS-related mutations in SOD1 are predicted to eliminate production of the protein. This strongly suggests that the mutant protein must be present to initiate motor neuron death.
Figure 3.
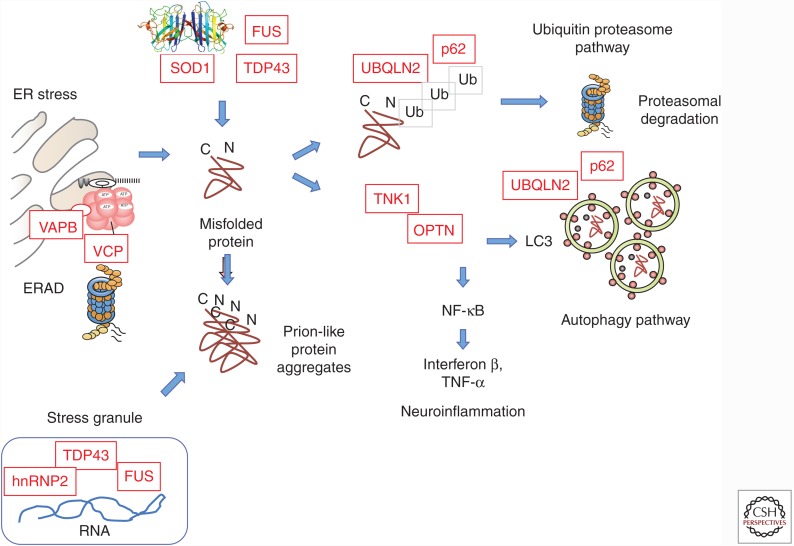
Protein homeostasis is disturbed in ALS. Many ALS genes highlight loss of protein quality control as a central feature of the disease. Misfolding of proteins may reflect many factors including aberrant folding and quality control at the ER (disturbed ER-associated degradation or ERAD), inherent instability due to mutations, aggregation during residence in stress granules, or imperfect clearance via proteasomes or autophagy. Self-assembly of misfolded proteins may lead to propagated instability and prion-like spreading of pathology.
The clinical phenotype associated with SOD1 mutations tends to be only ALS without FTD or other neurological features. In North America, the most common mutation is a substitution of valine for alanine in codon 4 (A4V). This is associated with an extremely aggressive course, with survival in many cases only 11–12 mo; both clinically and pathologically, these individuals usually show relatively little evidence of corticospinal tract involvement (Cudkowicz et al. 1997, 1998).
Although it is distinctly uncommon, there are some instances in which SOD1-related ALS is transmitted as a recessive trait. In northern Scandinavia, the variant D90A is present in ∼1% of the population; in that setting, the mutation only triggers ALS when present homozygously (Andersen et al. 1995). This suggests that there is a dose effect; more of the mutant gene product is required to initiate motor neuron degeneration in this population. The clinical phenotype of ALS associated with the D90A/D90A genotype in northern Sweden is also distinctive, with very slow progression, long survival (12–14 yr), and a predominance of lower limb spasticity at onset. It is intriguing that the D90A mutation can cause ALS as a heterozygous variant in other regions of the world. One hypothesis to explain this is that a closely linked variant co-migrating with the D90A substitution in northern Scandinavia confers neuroprotection; this view is consistent with haplotype analyses showing that the D90A allele in Scandinavia is derived from a single founder. However, extensive analyses have failed to identify the linked beneficial variant (Broom et al. 2008). An alternate hypothesis is that there are several variants across the genome in the northern Scandinavian population that are not linked to the SOD1-D90A locus but are nonetheless neuroprotective. There is at least one other recessive SOD1 mutation (ΔG27/P28). By altering exon splicing, this deletion of two codons may reduce expression of the SOD1 transcript (Zinman et al. 2009); it also is predicted to result in a structurally abnormal, potentially unstable, and cytotoxic protein.
Much excitement resulted shortly after the discovery of mutations in the SOD1 gene from the finding that forced expression of high levels of mutant SOD1 protein from human SOD1 transgenes produced an adult-onset, focal, and then spreading motor neuron disease in transgenic mice (Gurney 1994). This was not seen in mice with WT SOD1 transgenes expressing equivalent levels of WT SOD1. Several alleles of SOD1 transgenic mice have been generated, beginning with SOD1G93A and followed by multiple other mutant alleles. Transgenic rats followed; the first were also SOD1G93A transgenics (Nagai et al. 2001). A general principle that has emerged is that the development of motor neuron disease is dose-dependent, requiring a threshold level of mutant protein.
Microscopically as well as clinically, the pathological features of the transgenic SOD1 mice and rats closely resemble those in human ALS. For this reason, these transgenic models have been used extensively to investigate the molecular pathogenesis of SOD1-mediated ALS as well as a range of therapeutic interventions. The utility of these mice in predicting clinical efficacy in humans has been challenged, in part because minor beneficial effects that are of uncertain significance in underpowered mouse trials have led to unsuccessful ALS treatment trials in humans (Scott et al. 2008).
Valosin-Containing Protein
fALS may sometimes arise in association with three other disorders, including not only FTD but also a myopathy with inclusion bodies and Paget's disease. This constellation of four concurrent disorders has been named multisystem proteinopathy or MSP. Most cases of MSP are caused by dominantly inherited mutations in the gene encoding valosin-containing protein (VCP) (Watts et al. 2007). From a clinical perspective, an important additional observation is that there are some instances of MSP in which the predominant phenotype is ALS (Johnson et al. 2010). It is rare that sALS patients have also been found to have mutations in the VCP gene (Abramzon et al. 2012). There is wide phenotypic variation even within the same family. Findings at autopsy include pathological changes including mislocalization of ubiquitinated TDP-43 (see section below) to the cytoplasm, indicating that the degenerative process initiated by mutant VCP shares common elements with other types of ALS and ALS-FTD.
Ubiquilin-2
In ∼1% of ALS cases, missense mutations are detected in the proline-rich, PXX domain (residues 491–526) of the ubiquilin-2 gene on the X chromosome (Deng et al. 2011). This is important in that the UBQLN2 protein interacts with TDP-43; aggregates that are UBQLN2-positive are detected in many cases of both sporadic and familial ALS and FTD. Because ubiquilin-2 assists in regulating proteasomal degradation of ubiquitinated proteins, it may regulate many protein levels including TDP-43.
Optineurin, TANK-Binding Kinase 1
In patients with fALS, multiple mutations have been identified in the gene encoding optineurin (OPTN). Although some are transmitted as recessive traits (for example, in the initial families from Japan), it is now evident that others are dominantly inherited. According to one estimate, OPTN may account for ∼1% of fALS cases. More recently, a whole-exome mutational screen of 2874 sALS cases and 8475 controls (with a large replication set) concluded that dominant, loss-of-function mutations in the OPTN gene are overrepresented in sALS (Cirulli et al. 2015). OPTN is a protein of ∼58 kDa that serves multiple functions; it is implicated in the activation of inflammatory cascades via regulation of interferon and TNF-α. Coordinately with the protein sequestosome-1/p62 (SQSTM1), OPTN is also involved in membrane trafficking and chaperoning of organelles (e.g., mitochondria) and ubiquitinated, misfolded proteins for degradation via autophagic vacuoles and the proteasome.
The function of OPTN is regulated in part by the protein TANK-binding kinase 1 (TBK1). TBK1 has also been implicated as an ALS gene in the above full exome sequencing scan, which disclosed loss-of-function TBK1 variants in sALS (Cirulli et al. 2015). In a parallel study of 252 fALS pedigrees, 13 families were also found to harbor loss-of-function mutations in the TBK1 gene; in the same report, TBK1 mutations were overrepresented in 1010 sALS cases compared with 650 controls (Freischmidt et al. 2015). Some functions of TBK1 are interlocked with OPTN, which is phosphorylated and activated via the coiled-coil domain of TBK1. TBK1 phosphorylation of OPTN accelerates binding of OPTN to LC3, a protein required in the autophagosome. TBK1 itself is directly involved in activating the inflammatory cascade via NF-kB. Functional studies suggested that a critical aspect of some TBK1 variants is loss of function of the C-terminal coiled-coil CCD2 domain of TBK1, which is the CCD2 domain that interacts with multiple other proteins, including OPTN and SQSTM1. The importance of the OPTN-TBK1 axis was further highlighted by a report that full genome sequencing of 104 cases with pathologically proven FTD (with TDP-43 inclusions) divulged two cases that harbored loss-of-function mutations in both the OPTN and TBK1 genes; three others carried heterozygous missense mutations in the TBK1 gene (Pottier et al. 2015). Combined studies using mRNA and protein expression levels in the cerebellum confirmed marked reductions in OPTN in two cases and reductions in TBK1 levels in two other cases.
CHCHD10 (C22orf2)
Within the last three years, several families have been described in which ALS, FTD, ataxia, and ragged red fibers in muscle are co-inherited with missense mutations in the gene CHCHD10 (Bannwarth et al. 2014; Ajroud-Driss et al. 2015). Several investigators have reproduced this finding; CHCHD10 mutations may account for >2% of fALS cases (Johnson et al. 2014a; Ronchi et al. 2015). This nuclear gene is a member of a family of proteins with coiled-coil helix domains (CHCHD); it encodes a mitochondrial protein within the intermembrane space that is thought to be important in maintaining the architecture of cristae and oxidative phosphorylation. This represents the first gene that directly implicates mitochondrial dysfunction as a cause for ALS.
Genes Implicated in Altered Function and Homeostasis of RNA
Transactive Response DNA Binding Protein
A landmark study in 2006 discovered that TDP-43, a 43-kDa DNA binding protein, is the most abundant ubiquitinated protein in pathological cytosolic inclusions in sALS and sporadic FTD (Fig. 4) (Neumann et al. 2006). Subsequently, the importance of this finding was underscored by reports demonstrating that germline TDP-43 mutations can cause familial forms of both ALS and ALS-FTD (Sreedharan et al. 2008). TDP-43, a heterogeneous nuclear ribonucleoprotein or hnRNP, is predominantly nuclear, but shuttles into the cytoplasm and axons during cellular stress. It is multifunctional, with roles in gene transcription, gene translation, the biogenesis of microRNA, gene silencing and RNA binding and transport, and suppression of aberrantly spliced cryptic RNA transcripts (Ling et al. 2015). The TDP-43 protein has several distinctive domains that underlie RNA binding, nuclear export and import, and protein–protein interactions. The latter are glycine-rich and inherently of low complexity. More than 40 mutations have been identified in the TDP-43 gene; almost all are missense changes (Janssens and Van Broeckhoven 2013; Lattante et al. 2013). Mutations in the TDP-43 gene account for ∼5% of fALS cases. In both ALS and FTD, TDP-43 accumulates in the cytoplasm, where it is cleaved and hyperphosphorylated; C-terminal fragments accumulate in insoluble cytoplasmic aggregates (Neumann et al. 2006). These aggregates are present in both sALS and fALS. Many forms of fALS, regardless of the offending primary gene mutation, show TDP-43 pathology; exceptions are cases associated with SOD1 and FUS gene mutations (see section below). The same types of TDP-43 pathology have also been reported in other CNS disorders including head trauma, stroke, and Parkinson's disease. This observation implicates the biology of TDP-43 broadly across numerous subcategories of neurologic injury (Neumann et al. 2006).
Figure 4.
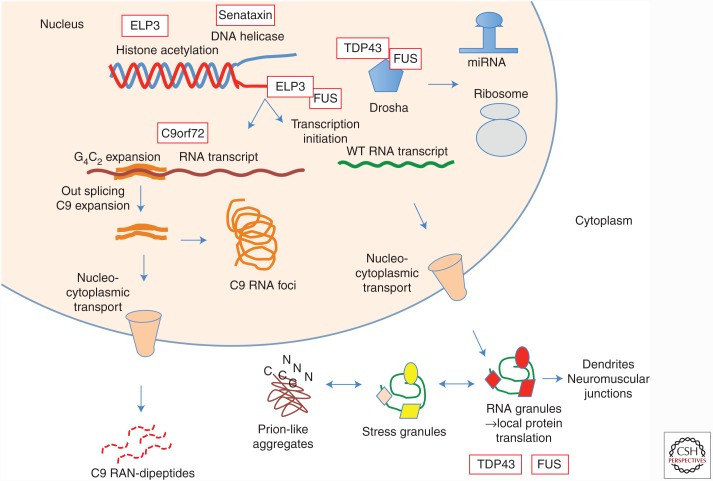
Multiple ALS genes indicate disturbances of RNA-binding proteins and RNA synthesis, trafficking, and function. A diversity of pathologies may arise including altered histone acetylation (elongator acetyltransferase complex subunit 3 [ELP3]), disturbed helicase activity (senataxin), altered splicing and synthesis of microRNA (TAR DNA-binding protein [TDP-43], fused in sarcoma [FUS]), deposition of RNA foci and generation of RNA dipeptides (C9orf72), impaired nucleocytoplasmic transport (C9orf72), and formation of aggregates via stress granules (many of the RNA-binding proteins). Axonal movement of RNA granules may be impaired, with secondary alterations in local protein translation in dendrites and neuromuscular junctions.
Several lines of transgenic TDP-43 mice have been generated. Although some mice develop paralysis, the molecular and clinical correlations are difficult to interpret, in contrast with SOD1 mice, because levels of TDP-43 are tightly autoregulated. Elevated or depressed levels of WT TDP-43 are potentially as adverse as overexpressed mutant TDP-43. These mice have been extensively reviewed (Tsao et al. 2012).
Fused in Sarcoma
In 2009, FUS (fused in sarcoma, also known as TLS, translocated in liposarcoma) was identified as another ALS gene (Kwiatkowski et al. 2009; Vance et al. 2009), a point of considerable interest because FUS shares structural and functional features with TDP–43. FUS is an ∼75-kDa protein that, like TDP-43, is predominantly nuclear and is also found in ALS cases to be mislocalized to aggregates within the cytoplasm. Like TDP-43, FUS serves multiple functions related to the generation and trafficking of diverse types of RNA. It contributes to local protein translation and dynamic morphological changes in dendrites in response to glutamatergic stimulation, a property that bears on synaptic learning. The clinical phenotype of FUS-related ALS is generally not distinctive, except for rare cases in which a missense mutation that alters the nuclear localization signal (P525L) correlates with extremely aggressive, childhood onset ALS. Childhood ALS caused by FUS mutations is associated with basophilic inclusions in the spinal cord (Munoz et al. 2009). Pathologically, it is striking but puzzling that cytoplasmic inclusions with FUS do not also contain TDP-43. This distinction can also be appreciated in FTD cases with frontotemporal lobar degeneration (FTLD), wherein there is yet a third category of inclusions characterized by deposition of tau protein, to the exclusion of TDP43 and FUS. This dichotomy (or trichotomy in the case of FTLD) illustrates that there are patterns of specificity in the molecular events generating these different types of inclusions, although the basis for the specificity remains unclear. Overall, FUS mutations account for about 5% of fALS cases. Although the original FUS mutation was identified as homozygous recessive, most cases are dominantly inherited. FUS shares structural features with two other multifunctional DNA/RNA-binding proteins, Ewing's sarcoma protein (EWS) and TATA-binding associated factor 15 (TAF15) (Morohoshi et al. 1998). This triad has been labeled the TET proteins (translated-in-liposarcoma, Ewing's, and TAF15). Very rare mutations in the EWS and TAF15 genes have been detected in fALS cases (Ticozzi et al. 2011; Couthouis et al. 2012). Two other hnRNPs with domain structures that are broadly homologous to TDP-43, FUS, and the other TET proteins are hnRNPA2B1 and hnRNPA1; rare mutations in the genes encoding each of these proteins have been implicated in cases of ALS associated with MSP (see section below).
There are now two reports of transgenic FUS models of ALS. In one study, rather fulminant motor axonopathy without motor neuron degeneration but with astrogliosis and ubiquitinated aggregates was triggered by transgenic expression of mutant FUS (R521C) under the control of doxycycline. WT FUS induced some late-onset neuron death in frontal regions of the brain without paralysis (Huang et al. 2011). More recently, slowly progressive motor neuron degeneration with some weakness but no paralysis was detected in mice in which conditional WT and mutant FUS genes were introduced in the tau locus (Sharma et al. 2016). The phenotype required expression of the mutant FUS protein in motor neurons; the mechanism was cell autonomous and did not involve loss of FUS function.
hnRNPA2B1 and hnRNPA1
Rare mutations in genes encoding two other RNA binding proteins, hnRNPA2B1 and hnRNPA1, also cause MSP (see above section) (Kim et al. 2013). The mutations in these genes augment the propensity for the low complexity domains to interact with other proteins and thus for the mutant proteins to induce propagated, prion-like misfolding. Both proteins also interact with TDP-43 (Kim et al. 2013).
C9orf72
In 2011, three landmark studies identified the most common cause of ALS and ALS-FTD: an expansion of a hexanucleotide intronic repeat (GGGGCC) within the first intron of a poorly understood gene, termed C9orf72 (chromosome 9 open reading frame 72) (Dejesus-Hernandez et al. 2011; Renton et al. 2011; Gijselinck et al. 2012). Clinical diagnosis presently involves repeat primed polymerase chain reaction (PCR) coupled with Southern blotting to define the size of the expansion. Although the most common repeat size is a triplet of the hexanucleotides, controls may have up to 25 or 30 repeats. By contrast, the disease alleles may have hundreds or even thousands of the hexanucleotide repeats. These C9orf72 intronic expansions underlie 45%–50% of fALS and 20%–25% of familial FTD cases in the United States. They also account for 5%–10% of sporadic ALS and FTLD. There is a tendency for bulbar ALS to be more common in the C9-expanded populations.
The relationship between expansion size and phenotype is not completely delineated. The issue is confounded somewhat by somatic variability in expansion size; in the same individual, the number of repeat expansions in the frontal cortex generally exceeds that in the cerebellum or blood, and there are fewer repeats in samples from fibroblasts. In one analysis (van Blitterswijk et al. 2013), expansion size did not vary with the phenotypes (ALS vs. ALS-FTD vs. FTD). Curiously, there was a positive correlation between the expansion length in the frontal cortex and the age of onset; shorter expansions were associated with earlier onset. In the same study, shorter expansions in the cerebellum conferred a survival advantage.
Little is known about the normal function of the C9orf72 gene. It expresses three transcripts that generate proteins with two isoforms (54 and 24 kDa). These include DENN domains (differentially expressed in normal and neoplastic cells) that are predicted to function as guanine nucleotide exchange factors (GEFs) that interact with Rab GTPases to regulate membrane trafficking events. The scope of this review permits only a brief summary of potential mechanisms of pathogenesis of the hexanucleotide expansion in the C9orf72 gene. Because at autopsy affected brains show a reduction in levels of the full C9 mRNA transcript, it was originally suggested that one mechanism for the pathology of the C9 expansion might be haploinsufficiency. However, there are several arguments against this hypothesis, including the observation that mice that are hemizygous or fully deleted for the C9orf72 gene do not develop significant motor neuron loss. Two other models offer alternate explanations for the neurotoxicity of the C9 intronic expansions. One is based on a major pathological finding in C9orf72 ALS/FTD: the intranuclear deposition of foci of expanded RNA. It is proposed that, as occurs with RNA foci in myotonic dystrophy, the intranuclear RNA deposits may sequester critical intranuclear proteins (like muscleblind proteins in myotonic dystrophy type 1 [DM1]). However, although several candidates for the sequestered protein have been proposed, there is no consensus yet on a single candidate. A second, intriguing model is that the primary pathology is the production, via noncanonical, repeat-associated, non-ATG-dependent translation (RAN-translation), of dipeptide repeat proteins (DPRs) across the expanded G4C2 repeat. The production of such DPRs was first discovered in postmortem tissue from patients with spinocerebellar atrophy (SCA8) (Zu et al. 2013); the expanded domains in C9orf72 undergo RAN-translation across both sense and antisense RNA, in all six possible reading frames; this generates five different DPRs (Ash et al. 2013). Studies indicate that some of these DPRs (such as poly-[proline-arginine]) are highly neurotoxic. Recent studies in flies that examine the respective toxicities of the expanded RNA versus the DPR favor the view that the primary offending agent is the DPR (Mizielinska et al. 2014; Tran et al. 2015). Whether a consequence of RNA foci or the DPR, other potentially downstream pathologies are triggered by the G4C2 expansions, including substantial disruption of the nuclear pore complex and the process of nuclear-cytoplasmic transport (export of RNA, import of proteins) (Freibaum et al. 2015; Jovicic et al. 2015; Zhang et al. 2015).
Four lines of bacterial artificial chromosome (BAC) transgenic C9orf72 mice have been reported. In one, the BAC expresses the short isoform of the C9orf72 protein, whereas the other expresses the full-length isoform. Both lines show the molecular stigmata of the C9orf72 pathology (abundant intranuclear RNA foci, both soluble and aggregated DPR), but curiously neither showed paralysis, even after two years (O'Rourke et al. 2015; Peters et al. 2015a; Jiang et al. 2016). Still another set of BAC C9orf72 transgenics, in a different mouse strain, show paralysis in a subset of female mice (Liu et al. 2016).
Angiogenin and Ribonuclease 4
Angiogenin (ANG, or ribonuclease 5) is a 123-amino-acid protein with multiple functions including stimulation of angiogenesis, hydrolysis of nucleic acids, and context-dependent activation of proteases. In conditions of cellular stress, ANG induces expression of a class of tRNAs (tiRNAs, or transfer RNA-derived, stress-induced, small RNAs) that suppress protein synthesis, presumably thereby conserving cellular energy reserves. Because it has neuroprotective properties, it is of considerable interest that rare variants in ANG are overrepresented in both ALS (sALS and in some small families) and Parkinson's disease (Greenway et al. 2006; van Es et al. 2011). Ribonuclease 4 is a paralog of ANG that also has angiogenic and neuroprotective properties; one study, as yet unconfirmed, has described a hypofunctional missense mutation in this enzyme that is more abundant in ALS than controls (Li et al. 2013).
Matrin-3
In 2014 it was reported that missense mutations in the gene encoding matrin-3 cause motor neuron disease with prominent myopathic features (a scenario not unlike the multisystem proteinopathy caused by VCP gene mutations) (Johnson et al. 2014b; Leblond et al. 2016). Matrin-3 is predominantly a nuclear protein with multiple functions. It binds DNA and RNA and some RNA binding proteins, including TDP-43. Additionally, it is part of the nuclear pore complex and is likely involved in shuttling hnRNP in and out of the nucleus. It probably is also involved in chaperoning edited RNA. How these mutations disrupt these multiple functions is not yet clear. A recent study indicates that, unlike TDP-43 and FUS, in conditions of cellular stress leading to stress-granule formation, matrin-3 remains largely within the nucleus (Gallego-Iradi et al. 2015).
Genes That Implicate Cytoskeletal and Axonal Dynamics
Dynactin (DCTN1)
Genetic analyses have identified several ALS genes that, as a group, identify perturbations of cytoskeletal and axonal dynamics as a central element in ALS pathogenesis (Fig. 5). This concept is not new in the field of motor neuron disease. As early as 1987, studies of median motor nerve twigs isolated from ALS patients revealed altered axonal transport ex vivo (Breuer et al. 1987). Disrupted axonal transport has been documented in numerous studies of transgenic ALS mice (Morfini et al. 2009). It was shown that mutant but not WT SOD1 selectively impairs anterograde axonal transport in the isolated axons from squid (Bosco et al. 2010). For these reasons, it was therefore of considerable interest that mutations have now been documented, albeit infrequently, in the dynactin gene (DCTN1), which encodes an accessory protein, p150glued, that participates in retrograde axonal transport (Puls et al. 2003). At least two clinical phenotypes were reported. In one family, affected individuals developed an early adult onset mainly with lower motor neuropathy with distinctive early laryngeal involvement; in the other, the phenotype was ALS-FTD (Munch et al. 2005).
Figure 5.
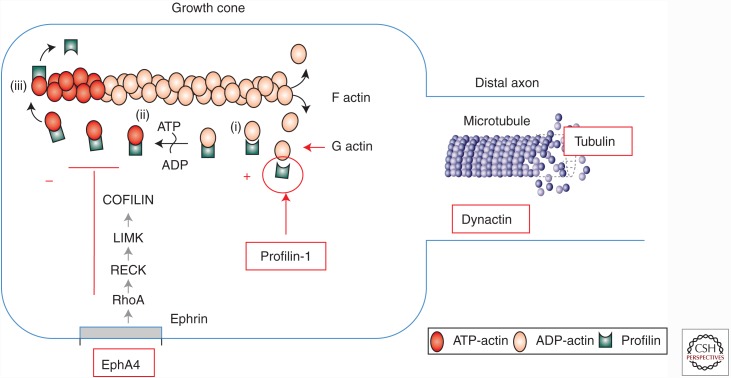
Cytoskeletal dynamics are altered in ALS. Mutations in profilin-1 are likely to impair energy-dependent extension of filamentous actin and elongation of growth cones, a process that is enhanced by reduction in signaling from ephrinA4. Tubulin mutations compromise the structures of microtubules. Mutations in dynactin are predicted to impair retrograde transport along the microtubule backbone.
Profilin-1
PFN1 is a protein that accelerates ATP-initiated polymerization of filamentous actin, a process that is important in many diverse cellular functions including extension of growth cones from the distal motor neuron axon. In 2012, exome sequencing identified mutations in the PFN1 gene in a small set of ALS families, clinically characterized by primarily lower motor neuron disease of adult onset (Wu et al. 2012). Importantly, at least two groups have recently generated transgenic PFN1 mice that, like the transgenic SOD1 mice, develop age- and PFN1-dose-dependent, lethal paralytic disease (Yang et al. 2016; M Kiaei, pers. comm.). This is expected to be an important asset in the field, as comparative studies of the two models should enable definition of the common elements that underlie motor neuron death in both ALS models. The availability of two models will also improve testing of therapeutic compounds.
TubulinA4A
Microtubules, the essential cytoskeletal backbones in axons, are composed of multiple copies of two subunits, α and β tubulin, each a member of a family of tubulin proteins. Four mutations in the gene encoding one form of α tubulin, tubulinA4A (TUBA4A), were identified in an exome scan of 363 cases of fALS and 4300 individuals and confirmed in a replication study of 272 fALS cases and 5510 controls. The same variants were not detected in a large cohort (1355) of sALS cases (Smith et al. 2014). These mutations were predicted to be damaging; all were located within exon 4. In in vitro assays using primary neurons and COS cells, the mutations impaired polymerization of microtubules. Although these mutations were detected in fALS cases, none were tested for co-migration with other affected individuals in the same pedigree, in part because the pedigrees were small. This suggests that penetrance of the TUBA4A variants may be low in contrast with the SOD1 or C9orf72 families.
Juvenile-Onset ALS Genes
Because they are almost always dominantly transmitted, it is thought that most adult-onset neurodegenerative disorders like fALS reflect acquired, adverse properties that are neurotoxic. By contrast, most mutant genes implicated in developmental disorders of juvenile onset often are recessively inherited and reflect the loss of function of genes that are important in development. When the recessive nature of a genetic defect is likely, suggested perhaps by consanguinity, an approach to gene identification is to locate chromosomal regions of preserved homozygosity; fine analysis of the homozygous locus may disclose the mutant disease gene. One example of a recessive, young-onset ALS gene mapped via preserved homozygosity is ALS2, which encodes a rho GEF that is abundant in the brain and spinal cord (Hadano et al. 2001; Yang et al. 2001). As described above for the DENN proteins like C9orf72, this GEF is likely involved in membrane trafficking. It activates membrane-associated Rab5 protein (converting a bound GDP to GTP), permitting its interaction with target proteins and vesicles. In neurons, these activities are important in multiple membrane-processing activities, not only in the Golgi and ER but also in neuronal development and remodeling, synaptic function, and neurotransmitter receptor trafficking. Individuals with loss-of-function mutations in alsin develop early-onset corticospinal tract and lower motor degeneration, which, unlike typical adult-onset ALS, is slowly progressive with survival measured in decades (Flor-de-Lima et al. 2014).
Several other gene defects have now been associated with juvenile ALS. Recessive loss-of-function mutations in the spastic paraplegia gene SPG11 (spatacsin) also commonly cause childhood-onset motor neuron disease; the predominant clinical feature is usually corticospinal tract signs with thinning of the corpus callosum (Del Bo et al. 2007) although a distal axonopathy has also been described (Montecchiani et al. 2016). This protein has an important role in neuronal growth, at least in part because it functions in autophagic pathways (Chang et al. 2014). Several other genes that primarily cause hereditary spastic paraparesis can also include degeneration of spinal motor neurons (Fink 2013). Among these are SPG3 and SPG4, which, respectively, encode atlastin and spastin. Atlastin is a dynamin-like GTPase that mediates ER membrane fusion and interacts with a number of other proteins mutated in hereditary spastic paraplegia (HSP), including spastin, REEP1, and NIPA1 (all of which may be associated with lower motor neuron degeneration, albeit infrequently). Recessive, loss-of-function mutations in the gene-designated neuropathy target esterase also produce early corticospinal tract and more widespread motor neuron degeneration (Rainier et al. 2008). This phospholipase B is localized to ER membranes; it is thought to be neuroprotective following exposure to toxic organophosphorus compounds (Richardson et al. 2013).
Juvenile ALS has also been attributed to recessive mutations in the SIGMAR1 gene, which encodes a σ non-opioid intracellular receptor (Al-Saif et al. 2011). The phenotype is slowly progressive, predominantly with upper motor neuron involvement. The receptor, Sig-1R, is multifunctional, regulating potassium channels, Ca2+ signaling, and chaperone activities in the ER. SigR1 is abundant in motor neurons at excitatory postsynaptic densities; these are reportedly disturbed in sALS, which may reveal cytoplasmic accumulation of SigR1.
Some genes normally associated with adult-onset ALS also are implicated in juvenile ALS. For example, a very specific mutation in the FUS gene, P525L, is now well documented to cause fulminant juvenile-onset motor neuron disease, associated pathologically with distinctive intranuclear basophilic inclusions (Huang et al. 2010). The fulminant toxicity may reflect the fact that this mutation alters a canonical sequence for nuclear import of proteins. Another dominantly inherited form of juvenile ALS (onset in the late teens and early adulthood) is caused by heterozygous mutations in the senataxin gene (Chen et al. 2004), which encodes a helicase that is important in DNA and RNA coiling and repair. This underscores the importance of the DNA repair pathway in neurons, which are terminally differentiated; this is further highlighted by the fact that numerous recessively transmitted mutations in the senataxin gene are associated with a cerebellar and peripheral nerve neurodegenerative syndrome (ataxia with oculomotor apraxia).
Infantile-onset forms of severe pontobulbar palsy that may be associated with hearing loss, oculomotor palsy, and ataxia are also important; historically, these are designated Brown–Vialetto–Van Laere (BVVL) and Fazio–Londe (FL) disease. In some BVVL–FL families, homozygosity mapping and candidate gene sequencing identified homozygous and compound heterozygous mutations in the gene C20orf54 (SLC52A3), which encodes an intestinal riboflavin transporter (Bosch et al. 2011); subsequently, a second riboflavin transporter has also been implicated in BVVL (Johnson et al. 2012). In another family, without the SLC52A3 defect, a missense mutation of UBQLN1 (encoding ubiquilin1) was reported (Gonzalez-Perez et al. 2012). In southern India, many individuals have been identified with elements of the same phenotype: juvenile onset of deafness accompanied by upper motor neuron signs and denervation, sometimes with optic atrophy and behavioral disturbances. Because some cases recur in families, this entity, originally labeled Madras motor neuron disease, is likely to be recessively inherited; however, an offending gene defect has not been identified (Nalini et al. 2013).
Genome-Wide Association Studies
Conventional gene identification strategies (e.g., genetic linkage followed by candidate gene or exome sequencing) have identified many genes related to ALS or ALS-FTD. However, a limitation in this approach is that linkage studies may not have adequate power to detect disease loci when familial traits have low penetrance. If adequately powered with appropriately large sample sizes, genome-wide association studies (GWAS) analyses can define risk alleles with low odds ratios (Purcell et al. 2003). Although early GWAS studies in ALS were only moderately powered, recent collaborative GWAS studies of increasing sizes have identified several potential ALS loci and genes (Table 2).
Table 2.
Genome-wide association studies in ALS
| Single nucleotide polymorphism | Chromosome | Gene | Encoded protein or function | Sample region | Minor allele frequency controls | Patients | Pooled odds ratio (95% confidence interval) | Pooled P value | Reference |
|---|---|---|---|---|---|---|---|---|---|
| rs230667 | 12 | ITPR2 | A Ca2+ channel on the ER | Netherlands/Belgium/Sweden | 0.07 | 0.11 | 1.58 (1.30–1.91) | 3.28 × 10−6 | van Es et al. 2007 |
| Ireland | 0.11 | 0.10 | – | 6.4 × 10−1 | Cronin et al. 2008 | ||||
| USA/UK/France/Netherlands | – | – | – | 4 × 10−2 | Landers et al. 2009 | ||||
| USA/Italy/Germany | 0.11 | 0.11 | 1.01 | 8.7 × 10−1 | Chio et al. 2009 | ||||
| USA/Ireland/Sweden/Belgium/Netherlands | 0.18 | 0.20 | 1.21 | 3.72 × 10−5 | van Es et al. 2009 | ||||
| UK | 0.10 | 0.10 | – | 9.8 × 10−1 | Shatunov et al. 2010 | ||||
| USA | – | – | 1.3 (0.9–1.7) | 1.4 × 10−1 | Kwee et al. 2012 | ||||
| China | 0.20 | 0.21 | 1.02 | 7.4 × 10−1 | Deng et al. 2013 | ||||
| rs6700125 | 1 | FLJ10986 (FGGY) | Unknown; possible role in energy metabolism | USA | 0.32 | 0.41 | 1.35 (1.13–1.62) | 6 × 10−4 | Dunckley et al. 2007 |
| Ireland | 0.33 | 0.34 | – | 7.9 × 10−1 | Cronin et al. 2008 | ||||
| USA/UK/France/Netherlands | – | – | – | 9.9 × 10−2 | Landers et al. 2009 | ||||
| USA/Italy/Germany | – | – | – | 8.0 × 10−2 | Chio et al. 2009 | ||||
| USA/Ireland/Sweden/Belgium/Netherlands | 0.34 | 0.33 | 1.03 | 1.1 × 10−1 | van Es et al. 2009 | ||||
| UK | 0.33 | 0.34 | – | 7.1 × 10−1 | Shatunov et al. 2010 | ||||
| rs10260404 | 7 | DPP6 | A transmembrane protein binding A-type neuronal K+ channel | Ireland | 0.34 | 0.42 | 1.37 (1.2–1.56) | 2.53 × 10−6 | Cronin et al. 2008 |
| USA/UK/France/Netherlands | – | – | – | 1.2 × 10−2 | Landers et al. 2009 | ||||
| USA/Italy/Germany | – | – | – | 6.2 × 10−1 | Chio et al. 2009 | ||||
| Ireland/Poland/USA/Netherland | 0.36 | 0.41 | 1.23 (1.1–1.38) | 2.62 × 10−4 | Cronin et al. 2009 | ||||
| USA/Ireland/Sweden/Belgium/Netherlands | 0.38 | 0.41 | 1.16 | 1.6 × 10−3 | van Es et al. 2009 | ||||
| UK | 0.4 | 0.4 | – | 8.9 × 10−1 | Shatunov et al. 2010 | ||||
| Italy | 0.39 | 0.38 | 0.97 (0.84–1.09) | 6.3 × 10−1 | Fogh et al. 2011 | ||||
| USA | – | – | 1.0 (0.9–1.1) | 9.7 × 10−1 | Kwee et al. 2012 | ||||
| Belgium/Ireland/USA/Sweden/Netherlands | 0.53 | 0.15 | 3.70 | 3.6 × 10−3 | Blauw et al. 2010 | ||||
| China | 0.17 | 0.16 | 0.94 | 3.5 × 10−1 | Deng et al. 2013 | ||||
| rs12608932 | 19 | UNC13A | A presynaptic protein regulating neurotransmitter release in central and neuromuscular synapses | USA/Ireland/Sweden/Belgium/Netherlands | 0.37 | 0.40 | 1.2 | 2.5 × 10−14 | van Es et al. 2009 |
| UK | 0.37 | 0.35 | – | 4.2 × 10−1 | Shatunov et al. 2010 | ||||
| USA | – | – | 1.2 (1.0–1.4) | 4.6 × 10−2 | Kwee et al. 2012 | ||||
| Belgium/Sweden/Netherlands | 0.36 | 0.40 | 1.33 (1.10–1.60) | 1 × 10−3 | Diekstra et al. 2012a | ||||
| Chinaa | 0.26 | 0.39 | – | – | Deng et al. 2013 | ||||
| rs2814707 | 9 | MOBKL2B | May regulate kinase activity | USA/Ireland/Sweden/Belgium/Netherlands | 0.23 | 0.26 | 1.16 | 7.45 × 10−9 | van Es et al. 2009 |
| China | 0.03 | 0.03 | 1.08 | 5.2 × 10−1 | Deng et al. 2013 | ||||
| rs16856202 | 1 | DISC1 | Regulates neurite outgrowth and cortical development | USA/UK/France/Netherlands | 0.04 | 0.02 | 0.499 | 7.98 × 10−6 | Landers et al. 2009 |
| rs3849942 | 9 | C9orf72 | Unknown function | USA/Ireland/Sweden/Belgium/Netherlands | 0.23 | 0.26 | 1.15 | 1.01 × 10−8 | van Es et al. 2009 |
| Chinaa | 0.05 | 0.05 | – | – | Deng et al. 2013 | ||||
| rs2708909 | 7 | SUNC1 (Intronic) | A nuclear envelope protein Sadl and UNC84 domain containing 1 | USA/Italy/Germany | 0.45 | 0.50 | 1.17 (1.11–1.23) | 6.98 × 10−7 | Chio et al. 2009 |
| rs2708851 | 7 | SUNC1 (Intergenic) | 0.45 | 0.50 | 1.17 (1.11–1.23) | 1.16 × 10−6 | |||
| rs2275294 | 20 | ZNF512B | Regulates TGF-β signaling | Japan | 0.41 | 0.48 | 1.32 (1.21–1.44) | 6.70 × 10−10 | Iida et al. 2011 |
| rs6703183 | 1 | CAMK1G | A calmodulin-dependent protein kinase | China | 0.34 | 0.41 | 1.31 | 2.92 × 10−8 | Deng et al. 2013 |
| rs8141797 | 22 | SUSD2 | A novel tumor suppressor in brain, kidney, and lung | China | 0.10 | 0.10 | 1.52 | 2.35 × 10−9 | Deng et al. 2013 |
| rs34517613 | 17 | Unknown | Unknown | China | 0.13 | 0.11 | 0.82 (0.76–0.87) | 1.11 × 10−8 | Fogh et al. 2014 |
| rs1541160 | 1 | KIFAP3 | Regulates Rho and Rap1 family members and Ki-Ras by stimulating their GDP/GTP exchange reactions and inhibiting their interactions with membranes | USA/UK/France/Netherlands | – | – | – | 1.84 × 10−8 | Landers et al. 2009 |
| USA | – | – | 0.95 (0.82–1.2) | 4.9 × 10−1 | Kwee et al. 2012 | ||||
| rs3849942 | 9 | 9p21 locus C9orf72b | – | USA/Ireland/Sweden/Belgium/Netherlands | 0.23 | 0.26 | 1.23 | 1.58 × 10−6 | van Es et al. 2009 |
| UK | 0.24 | 0.30 | 1.39 (1.21–1.59) | 2.22 × 10−6 | Shatunov et al. 2010 | ||||
| Belgium/France/UK/ Netherlands/Ireland/ Italy/Sweden/USA | 0.23 | 0.27 | 1.22 (1.15–1.30) | 4.64 × 10−10 | Shatunov et al. 2010 | ||||
| Finland | 0.16 | 0.29 | 2.16 (1.72–2.70) | 9.11 × 10−11 | Laaksovirta et al. 2010 | ||||
| Belgium/Sweden/USA/Ireland/Netherlands | 0.22 | 0.25 | 1.23 (1.11–1.29) | 1.58 × 10−6 | Deng et al. 2013 | ||||
| rs2814707 | 9 | 9p21 locus | – | USA/Ireland/Sweden/Belgium/Netherlands | 0.23 | 0.26 | 1.22 | 3.33 × 10−6 | van Es et al. 2009 |
| UK | 0.24 | 0.30 | 1.38 (1.20–1.58) | 3.32 × 10−6 | Shatunov et al. 2010 | ||||
| Belgium/France/UK/Netherlands/Ireland/Italy/Sweden/USA | 0.27 | 0.23 | 1.22 (1.15–1.30) | 4.47 × 10−10 | Shatunov et al. 2010 | ||||
| Belgium/Sweden/USA/Ireland/Netherlands | 0.23 | 0.26 | 1.22 (1.10–1.28) | 3.33 × 10−6 | Deng et al. 2013 | ||||
| rs13048019 | 21 | TIAM1 | – | Finland | 0.17 | 0.30 | 4.4 (2.32–7.40) | 2.58 × 10−8 | Laaksovirta et al. 2010 |
| rs2225389 | 9 | MOB3B | May regulate kinase activity | Finland | 0.09 | 0.18 | 2.26 (1.7–3.0) | 2.88 × 10−8 | Laaksovirta et al. 2010 |
| rs75932628 | 6 | TREM2 | Role in glial cell function and inflammation | North America | 0.08 | 0.54 | 2.4 (1.29–4.15) | 4.1 × 10−3 | Cady et al. 2014 |
aThe SNPs failed to be validated in the study for it is significantly deviated from Hardy–Weinberg equilibrium.
bNearby gene.
Prior to the discovery of the role of C9orf72 in ALS, it was of considerable interest that the strongest GWAS peak was directly on top of the ALS-FTD locus on chromosome 9p21, within which the C9orf72 expansions were subsequently found (van Es et al. 2009; Laaksovirta et al. 2010; Shatunov et al. 2010). Another GWAS locus that has been replicated is proximal to the UNC13A gene, which regulates the release of neurotransmitters, including glutamate, and thus is a plausible candidate ALS gene; independent studies have revealed that the minor allele of rs12608932 in UNC13A is not only associated with susceptibility but also with shorter survival of ALS patients (van Es et al. 2009; Diekstra et al. 2012b; Chio et al. 2013).
One of the largest GWAS to date in ALS involved 6100 European cases and 7125 controls (Fogh et al. 2014). In addition to consistent results for chromosome 9p association, this study also showed an intriguing hit at chromosome 17q11.2, near the SARM1 gene. The SARM1 protein participates in active axonal degeneration following axonal injury; it is quite striking that in mice devoid of sarm1, Wallerian degeneration is profoundly attenuated. That said, a focused biological relation of the ALS-associated SARM1 nucleotide polymorphism (SNP) and sarm1 function has not been defined. Another large ALS GWAS has confirmed the SARM1 association and pointed to still other ALS genes (SCFD1, C21orf2, MOBP) (van Rheenen et al. 2016).
Another GWAS-identified gene of interest is ELP3, which encodes an RNA polymerase component. A microsatellite GWAS has identified risk alleles in the ELP3 gene, which were associated with reduced brain expression of ELP3 in postmortem studies (Simpson et al. 2009). A recent GWAS analysis of 533 Han Chinese with ALS failed to define hits related to C9orf72 but did divulge two ALS-associated loci that are not detected in ALS cohorts in Europe or North America (chromosome 1q32 covering CAMK1G [calcium/calmodulin-dependent protein 1] and SUSD2 [sushi domain containing 2]) (Deng et al. 2013). This illustrates the principle that genetic factors that drive susceptibility may differ in populations with differing genetic architectures.
An innovative study by Diekstra and colleagues correlated SNP variants with levels of expression of the corresponding genes, based on mRNA from blood samples, to define a set of SNPs that are expression quantitative trait loci (eQTLs) (Diekstra et al. 2012a). The resulting set of SNPs at eQTLs was tested for association with ALS. After validation, this identified eight SNP pairs in a single gene, CYP27A1, as associated with ALS. The product of this gene, sterol 27-hydroxylase, is necessary for cholesterol breakdown; loss-of-function mutations in the enzyme cause cerebrotendinous xanthomatosis, which is characterized by deposition of cholesterol products in multiple tissues, and progressive multisystem disease with neurodegeneration. The implication of this eQTL association study—that cholesterol metabolism may be relevant in ALS—is further supported by epidemiological studies relating cholesterol levels to ALS (Diekstra et al. 2012a).
Genomic Structural Variations In ALS
The etiologic role of genomic structural variation (e.g., deletions, duplications, copy number variants [CNVs], insertions, inversions, translocations) in late-onset diseases such as ALS has not yet been completely elucidated. Two studies showed chromosomal translocations or inversions in ALS (Kaneko et al. 1995), but in 4 out of 5 families, these aberrations were also found in several related healthy persons (Meyer et al. 2003).
A well-established CNV within the SMN1 gene, whose loss-of-function mutations cause spinal muscular atrophy, has been the subject of several ALS studies. It has been reported that deletions of SMN1 correlate with shortened survival in ALS (Veldink et al. 2005). The most consistent structural variants in the SMN1 gene are duplications, not deletions. The comprehensive investigation by Blauw (2012a) failed to detect changes in copy numbers of SMN2 in ALS; moreover, sequencing of the SMN gene disclosed only one missense variant (G26D) of uncertain significance.
Two studies have failed to find changes in the global genomic burden of CNVs in sALS (Blauw et al. 2008; Blauw et al. 2010). An international study involving more than 19,000 individuals (a discovery cohort of 1875 cases and 8732 controls plus a replication cohort of 2559 cases and 5887 controls) screened ∼ 300,000 SNPs for structural variations across the genome. The overall burden of CNVs in sALS did not differ from controls (Blauw et al. 2010). However, examination of CNVs within genes suggested an association with duplications within the 5′ end of the DPP6 gene on chromosome 7 and deletions in the NIPA1 gene on chromosome 15. The former locus has been implicated but not uniformly confirmed in ALS by prior GWAS studies (see Table 2); variants in the latter gene have been reported to modulate survival in ALS (see below). These findings are consistent with the view that, at least as presently analyzed, common CNVs are not associated with sALS although rare CNVs may contribute to its pathogenesis.
Twin Studies In ALS
One approach to defining the role for genetics in any disease is the analysis of disease concordance in twins. Two studies of twin concordance in ALS are instructive in this regard (Graham et al. 1997; Al-Chalabi et al. 2010). A British motor neuron disease (MND) study of 76 twin pairs found ALS heritability to be between 38% and 85% (Graham et al. 1997). A more recent study examined 171 twin pairs identified by combining data from a British MND twin cohort, the National Swedish Twin Registry, and the Kings College Motor Neuron Clinic Registry. The combined data set encompassed 10,872 MND death certificates over a 10-year period, a country-wide study of 86,441 twin pairs, and a clinic-based study of 4982 individuals with ALS or their family members (Al-Chalabi et al. 2010). Of 49 monozygotic twin pairs, five were concordant and 44 were discordant for ALS. Among dizygotic twin pairs studied, all were discordant for ALS. Analysis of different models concluded that ALS heritability is 76% (95% confidence interval, 60%–86%) when including individuals with known ALS in other, nontwin family members. When familial cases were excluded, the heritability of ALS was 61% (95% confidence interval, 38%–78%). Some studies have not identified concordance for ALS among monozygotic twins (Dellefave et al. 2003).
Genes and Gene Variants That Modify Susceptibility and Phenotype in ALS
Genetic variants that are not primary causes of ALS may nonetheless influence ALS phenotype (e.g., age of onset, duration) or susceptibility (Table 3). As noted above, the minor allele of the ALS-related SNP at the locus UNC13A confers enhanced risk for ALS; it also correlates with shorter survival (van Es et al. 2009; Diekstra et al. 2012b; Chio et al. 2013). More recently, SNP rs3011225 on chromosome 1p has also been shown to correlate with survival (Ahmeti et al. 2013). The biological basis for these associations is poorly understood.
Table 3.
Genes that modify ALS susceptibility or phenotype
| Study | Gene | Susceptibility | Site | Onset | Survival |
|---|---|---|---|---|---|
| GWAS |
UNC13A rs3011225-1p34 |
+++ |
+++ |
+++ | |
| Modifier genes |
EphA4 EphA3 MMP9 Ataxin-2 NIPA1 CX3CR1 DMT1 CHGB P412L TMEM106B TREM2 ARGEF1 |
+++ +++ +++ +++ |
Spinal MNs FTD |
+++ +++ |
+++ +++ +++ +++ +++ +++ |
| Known ALS genes |
C9orf72 C9 repeat length Profilin-1 SOD1-A4V SOD1/SOD1 FUS-P525L |
+++ +++ +++ +++ +++ +++ |
Bulbar Limb |
+++ +++ +++ |
+++ +++ +++ +++ |
Ephrin A4, Ephrin A3
Several genes have now been reported to modify the phenotype of ALS. Perhaps the best example is ephrinA4, a molecule whose normal functions include interacting with tyrosine kinase receptors on the distal terminals of growth cones to attenuate axon extension during neural development. Beginning with a screen to suppress toxicity of mutant SOD1 in zebrafish, Robberecht and colleagues have shown that transgenic SOD1 ALS mice and rats with reduced levels of ephrinA4 have improved survival; analogously, patients with lower circulating ephrinA4 levels have slower disease progression (Van Hoecke et al. 2012). This observation raises the prospect that compounds that selectively inhibit the EphA4 tyrosine kinase receptor or its signaling might therefore be beneficial in ALS. Within the last two years, data from an ALS cohort in Turkey have suggested that genetic variants in the Eph3A gene may also affect neuroprotection and ALS survival (Uyan et al. 2013).
Matrix Metalloprotease 9
Matrix metalloprotease 9 (MMP9) is a zinc metalloenzyme that mediates proteolysis of the extracellular matrix for critical cellular events such as cell migration, development, and angiogenesis. Experiments by Henderson and colleagues using transgenic SOD1G93A mice determined that MMP9 is expressed shortly after birth in mice and is selectively found in fast-fatigable spinal motor neurons but not in oculomotor or sphincteric neurons (Kaplan et al. 2014). This expression pattern mirrors that of motor neuron susceptibility in ALS, implicating MMP9 as a determinant of motor neuron degeneration. This possibility was underscored by the finding that genetic ablation or enzyme inhibition of MMP slowed endplate denervation and prolonged survival in the mice. Moreover, these observations suggest that interventions that block MMP9 activity may be therapeutic in ALS.
Ataxin 2
Expanded polyglutamate domains in the ataxin 2 gene cause spinocerebellar atrophy type 2 (SCA2). Under normal circumstances, there are less than 22 repeats in the gene; in SCA2 there are usually more than 35 repeats. It was therefore of considerable interest that expansions of intermediate length (27–33) enhance susceptibility to ALS (Elden et al. 2010; Lee et al. 2011). This discovery aligned well with a longstanding clinical observation that SCA2 patients sometimes presented with elements of lower motor neuron degeneration. The finding of the association of intermediate length repeats with ALS was robustly confirmed.
Non-Imprinted in Prader–Willi/Angelman Syndrome
Loss-of-function mutations in non-imprinted in Prader–Willi/Angelman Syndrome (NIPA1) cause juvenile-onset hereditary spastic paraparesis. It now appears that loss of NIPA1 function and expansions of a polyalanine repeat within the protein both increase susceptibility to ALS; furthermore, long (>10) repeats also correlate with earlier age of onset and shorter survival in ALS. Moreover, rare ALS cases harbor potentially damaging missense mutations in the NIPA1 gene (Blauw et al. 2012b). These effects may reflect impairment of the activity in NIPA1 in vesicular trafficking necessary for synaptic function.
CX3CR1
In a Spanish study of 187 cases versus 378 controls, the isoleucine allele of a polymorphic variant in the fractalkine gene CX3CR1 (valine-249-isoleucine, rs3732379) was associated with a faster rate of progression and shorter survival in sALS but not in fALS. This variant did not influence age or site of onset (Lopez-Lopez et al. 2014). The V249I variant of the fractalkine protein, which is expressed only on microglial cells, is associated with reduction in some microglial functions; blunted microglial activity is therefore a potential explanation for the more rapid rate of progression. It is an intriguing parallel that deletion of the mouse CX3CR1 gene also accelerates the course of SOD1G93A ALS in mice (Cardona et al. 2006).
Divalent Metal Transporter 1
In a French analysis of 579 cases and 517 controls, it was observed that one allele of a metal transport protein divalent metal transporter 1 (DMT1) was associated with a shorter course in patients with limb-predominant ALS. This association was attributed provisionally to a role for iron transport and metabolism in ALS (Blasco et al. 2011b).
Chromogranin B
It has been reported in a French Canadian cohort that a variant (P413L) of chromogranin B is overrepresented in ALS cases, which also shows earlier onset (Gros-Louis et al. 2009). This was not confirmed in other cohorts (Blasco et al. 2011a).
TMEM106B
This gene, whose protein functions in endosomal trafficking, modifies the phenotype of FTD caused by progranulin mutations. Specifically, the major allele of variant rs1990622 in TMEM106B is associated with both lower levels of serum progranulin and earlier onset of progranulin-mediated FTD. TMEM106B also is a modifier of FTD mediated by C9orf72 expansions but in a manner opposite that in progranulin FTD. In C9orf72 FTD, the major allele of TMEM106B correlated with later ages of both onset and death (but not in C9orf72 ALS) (van Blitterswijk et al. 2014). TMEM106B did not modify the age of onset or death in non-progranulin, non-C9orf72 FTD.
As summarized in Table 3, some alleles of known ALS genes have an influence not only on the risk of developing ALS but also on its phenotype. Thus, in cohorts of C9orf72 ALS, bulbar onset of the disease is more frequent than in other ALS cohorts. Among the many SOD1 mutations, the A4V allele is associated with a particularly devastating time course, with mean survival barely 1 year. Of the FUS mutations, the missense mutation P525L that ablates nuclear localization can be associated with juvenile onset and a remarkably fulminant course.
TREM2
Triggering receptor expressed on myeloid cells 2 (TREM2) is a 25-kDa receptor protein expressed in a complex on the surface of microglial cells. It participates in the immune response by signaling release of inflammatory cytokines. A rare polymorphism in the extracellular domain of TREM2, R47H, confers heightened risk for Parkinson's and Alzheimer's diseases (Rayaprolu et al. 2013), and was recently documented to increase the risk for ALS (Cady et al. 2014). In ALS cases, TREM2 expression in the spinal cord is elevated, and higher levels correlate with shorter survival (Cady et al. 2014).
ARHGEF1
McLaughlin et al. (2015) and colleagues analyzed runs of homozygosity in a single large Irish ALS cohort and identified potential recessive ALS risk loci by mapping specific homozygous haplotypes that are overrepresented in cases compared with controls. In particular, two top results (S3430_7; chr19:42,369,393–43,534,282) and (S4200_8; chr19:42,328,529–43,287,462) mapped to a location that spans the gene ARHGEF (McLaughlin et al. 2015). This gene also encodes a ρ GEF factor and therefore is a plausible ALS candidate gene, given the emerging importance of GEFs in neuronal homeostasis (as in alsin and C9orf72) (Droppelmann et al. 2013, 2014). The importance of GEF biology in this theme in ALS pathogenesis is further underscored by the observations that (1) a heterozygous frameshift mutation in the ARHGEF28 gene was identified as a possible causative factor in one pedigree with fALS (Droppelmann et al. 2013), and (2) dysfunction of multiple GEFs plays a role in the pathogenesis of many neurodegenerative disorders (reviewed in Droppelmann et al. 2014).
CONCLUSION
In the last 10 years, progress in understanding the genetic architecture of ALS can only be described as remarkable. With the wide availability of affordable high-throughput genetic sequencing, the rate of discovery of genetic variables that play a role in ALS pathogenesis will only increase. Full exome sequencing is now the standard in most gene searches. Furthermore, we anticipate that more than 1000 ALS genomes will be sequenced in full in the next 18–24 months. This permits several predictions. First, the remaining, rare causes of Mendelian ALS will likely all be defined. Second, for the first time we will be able to systematically screen noncoding DNA for disease-related genetic variations (e.g., in long noncoding RNAs or microRNA). Third, it will be feasible to study the role in ALS of epistasis—the interactions between multiple genes that each heighten disease susceptibility but cannot initiate pathogenesis alone. There are already several reports of oligogenic ALS cases, in which more than one ALS-related gene variant has been detected in the same individual (Luigetti et al. 2011; Chio et al. 2012; van Blitterswijk et al. 2012; Dekker et al. 2015; Pottier et al. 2015). As a corollary, it seems likely that an increasing fraction of apparently sALS cases will prove to be a consequence of multiple gene variants that are not individually penetrant. It is already apparent that first-degree relatives of ALS patients have an increased incidence compared with unrelated ones (Fallis and Hardiman 2009; Fang et al. 2009). Finally, improved knowledge of genetic variants and their related pathways to neurodegeneration will improve our capacity to define interactions between genetic topology and the environment (e.g., environmental toxins or elements within the microbiome). Perhaps most important, as the understanding of ALS genetics is refined, opportunities for developing meaningful therapies will improve. In part, this may be a direct consequence of improved methods to attenuate expression and toxicity of mutant ALS genes. Indirectly, one anticipates that it will be increasingly possible to stratify and personalize therapeutic approaches and trials based on molecular distinctions between subsets of ALS cases.
ACKNOWLEDGMENTS
The authors gratefully acknowledge support from the Amyotrophic Lateral Sclerosis (ALS) Association, the Angel Fund for ALS Research, Project ALS, ALS Finding a Cure, the Cellucci Fund for ALS Research, the ALS Therapy Alliance, and the Max Rosenfeld Fund. This work was also supported by grants from the National Institute of Neurological Disorders and Stroke (NINDS) (NS088689, FD004127, NS079836, NS065847, and NS073873).
REFERENCES
- Abel O, Powell JF, Andersen PM, Al-Chalabi A. 2012. ALSoD: A user-friendly online bioinformatics tool for amyotrophic lateral sclerosis genetics. Hum Mutat 33: 1345–1351. [DOI] [PubMed] [Google Scholar]
- Abramzon Y, Johnson JO, Scholz SW, Taylor JP, Brunetti M, Calvo A, Mandrioli J, Benatar M, Mora G, Restagno G, et al. 2012. Valosin-containing protein (VCP) mutations in sporadic amyotrophic lateral sclerosis. Neurobiol Aging 33: 2231.e1–2231.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmeti KB, Ajroud-Driss S, Al-Chalabi A, Andersen PM, Armstrong J, Birve A, Blauw HM, Brown RH, Bruijn L, Chen W, et al. 2013. Age of onset of amyotrophic lateral sclerosis is modulated by a locus on 1p34.1. Neurobiol Aging 34: 357.e7–e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajroud-Driss S, Fecto F, Ajroud K, Lalani I, Calvo SE, Mootha VK, Deng HX, Siddique N, Tahmoush AJ, Heiman-Patterson TD, et al. 2015. Mutation in the novel nuclear-encoded mitochondrial protein CHCHD10 in a family with autosomal dominant mitochondrial myopathy. Neurogenetics 16: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akassoglou K, Malester B, Xu J, Tessarollo L, Rosenbluth J, Chao MV. 2004. Brain-specific deletion of neuropathy target esterase/swisscheese results in neurodegeneration. Proc Natl Acad Sci 101: 5075–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Chalabi A, Fang F, Hanby MF, Leigh PN, Shaw CE, Ye W, Rijsdijk F. 2010. An estimate of amyotrophic lateral sclerosis heritability using twin data. J Neurol Neurosurg Psychiatry 81: 1324–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Saif A, Al-Mohanna F, Bohlega S. 2011. A mutation in σ-1 receptor causes juvenile amyotrophic lateral sclerosis. Ann Neurol 70: 913–919. [DOI] [PubMed] [Google Scholar]
- Andersen PM, Nilsson P, Ala-Hurula V, Keranen ML, Tarvainen I, Haltia T, Nilsson L, Binzer M, Forsgren L, Marklund SL. 1995. Amyotrophic lateral sclerosis associated with homozygosity for an Asp90Ala mutation in CuZn-superoxide dismutase. Nat Genet 10: 61–66. [DOI] [PubMed] [Google Scholar]
- Andersen PM, Al-Chalabi A. 2011. Clinical genetics of amyotrophic lateral sclerosis: What do we really know? Nat Rev Neurol 7: 603–615. [DOI] [PubMed] [Google Scholar]
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW III, Rademakers R, et al. 2013. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77: 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannwarth S, Ait-El-Mkadem S, Chaussenot A, Genin EC, Lacas-Gervais S, Fragaki K, Berg-Alonso L, Kageyama Y, Serre V, Moore D, et al. 2014. Reply: Two novel mutations in conserved codons indicate that CHCHD10 is a gene associated with motor neuron disease. Brain 137: e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumer D, East SZ, Tseu B, Zeman A, Hilton D, Talbot K, Ansorge O. 2014. FTLD-ALS of TDP-43 type and SCA2 in a family with a full ataxin-2 polyglutamine expansion. Acta Neuropathol 128: 597–604. [DOI] [PubMed] [Google Scholar]
- Beaulieu JM, Nguyen MD, Julien JP. 1999. Late onset of motor neurons in mice overexpressing wild-type peripherin. J Cell Biol 147: 531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekenstein U, Soreq H. 2013. Heterogeneous nuclear ribonucleoprotein A1 in health and neurodegenerative disease: From structural insights to post-transcriptional regulatory roles. Mol Cell Neurosci 56: 436–446. [DOI] [PubMed] [Google Scholar]
- Belzil VV, Daoud H, Camu W, Strong MJ, Dion PA, Rouleau GA. 2013. Genetic analysis of SIGMAR1 as a cause of familial ALS with dementia. Eur J Hum Genet 21: 237–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco H, Corcia P, Veyrat-Durebex C, Coutadeur C, Fournier C, Camu W, Gordon P, Praline J, Andres CR, Vourc'h P, et al. 2011a. The P413L chromogranin B variation in French patients with sporadic amyotrophic lateral sclerosis. Amyotroph Lateral Scler 12: 210–214. [DOI] [PubMed] [Google Scholar]
- Blasco H, Vourc'h P, Nadjar Y, Ribourtout B, Gordon PH, Guettard YO, Camu W, Praline J, Meininger V, Andres CR, et al. 2011b. Association between divalent metal transport 1 encoding gene (SLC11A2) and disease duration in amyotrophic lateral sclerosis. J Neurol Sci 303: 124–127. [DOI] [PubMed] [Google Scholar]
- Blauw HM, Veldink JH, van Es MA, van Vught PW, Saris CG, van der Zwaag B, Franke L, Burbach JP, Wokke JH, Ophoff RA, et al. 2008. Copy-number variation in sporadic amyotrophic lateral sclerosis: A genome-wide screen. Lancet Neurol 7: 319–326. [DOI] [PubMed] [Google Scholar]
- Blauw HM, Al-Chalabi A, Andersen PM, van Vught PW, Diekstra FP, van Es MA, Saris CG, Groen EJ, van Rheenen W, Koppers M, et al. 2010. A large genome scan for rare CNVs in amyotrophic lateral sclerosis. Hum Mol Genet 19: 4091–4099. [DOI] [PubMed] [Google Scholar]
- Blauw HM, Barnes CP, van Vught PW, van Rheenen W, Verheul M, Cuppen E, Veldink JH, van den Berg LH. 2012a. SMN1 gene duplications are associated with sporadic ALS. Neurology 78: 776–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blauw HM, van Rheenen W, Koppers M, Van Damme P, Waibel S, Lemmens R, van Vught PW, Meyer T, Schulte C, Gasser T, et al. 2012b. NIPA1 polyalanine repeat expansions are associated with amyotrophic lateral sclerosis. Hum Mol Genet 21: 2497–2502. [DOI] [PubMed] [Google Scholar]
- Bosch AM, Abeling NG, Ijlst L, Knoester H, van der Pol WL, Stroomer AE, Wanders RJ, Visser G, Wijburg FA, Duran M, et al. 2011. Brown-Vialetto-Van Laere and Fazio Londe syndrome is associated with a riboflavin transporter defect mimicking mild MADD: A new inborn error of metabolism with potential treatment. J Inherit Metab Dis 34: 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco DA, Morfini G, Karabacak NM, Song Y, Gros-Louis F, Pasinelli P, Goolsby H, Fontaine BA, Lemay N, McKenna-Yasek D, et al. 2010. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat Neurosci 13: 1396–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer AC, Lynn MP, Atkinson MB, Chou SM, Wilbourn AJ, Marks KE, Culver JE, Fleegler EJ. 1987. Fast axonal transport in amyotrophic lateral sclerosis: An intra-axonal organelle traffic analysis. Neurology 37: 738–748. [DOI] [PubMed] [Google Scholar]
- Broom WJ, Johnson DV, Auwarter KE, Iafrate AJ, Russ C, Al-Chalabi A, Sapp PC, McKenna-Yasek D, Andersen PM, Brown RH Jr. 2008. SOD1A4V-mediated ALS: Absence of a closely linked modifier gene and origination in Asia. Neurosci Lett 430: 241–245. [DOI] [PubMed] [Google Scholar]
- Butterfield RJ, Ramachandran D, Hasstedt SJ, Otterud BE, Leppert MF, Swoboda KJ, Flanigan KM. 2009. A novel form of juvenile recessive ALS maps to loci on 6p25 and 21q22. Neuromusc Disord 19: 279–287. [DOI] [PubMed] [Google Scholar]
- Cady J, Koval ED, Benitez BA, Zaidman C, Jockel-Balsarotti J, Allred P, Baloh RH, Ravits J, Simpson E, Appel SH, et al. 2014. TREM2 variant p.R47H as a risk factor for sporadic amyotrophic lateral sclerosis. JAMA Neurol 71: 449–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon A, Baker M, Boeve B, Josephs K, Knopman D, Petersen R, Parisi J, Dickison D, Adamson J, Snowden J, et al. 2006. CHMP2B mutations are not a common cause of frontotemporal lobar degeneration. Neurosci Lett 398: 83–84. [DOI] [PubMed] [Google Scholar]
- Cardona AE, Pioro EP, Sasse ME, Kostenko V, Cardona SM, Dijkstra IM, Huang D, Kidd G, Dombrowski S, Dutta R, et al. 2006. Control of microglial neurotoxicity by the fractalkine receptor. Nat Neurosci 9: 917–924. [DOI] [PubMed] [Google Scholar]
- Chang J, Lee S, Blackstone C. 2014. Spastic paraplegia proteins spastizin and spatacsin mediate autophagic lysosome reformation. J Clin Invest 124: 5249–5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaussenot A, Le Ber I, Ait-El-Mkadem S, Camuzat A, de Septenville A, Bannwarth S, Genin EC, Serre V, Auge G, Brice A, Pouget J, et al. 2014. Screening of CHCHD10 in a French cohort confirms the involvement of this gene in frontotemporal dementia with amyotrophic lateral sclerosis patients. Neurobiol Aging 35: 2884.e1–2884.e4. [DOI] [PubMed] [Google Scholar]
- Chen YZ, Bennett CL, Huynh HM, Blair IP, Puls I, Irobi J, Dierick I, Abel A, Kennerson ML, Rabin BA, et al. 2004. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4). Am J Hum Genet 74: 1128–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesi A, Staahl BT, Jovicic A, Couthouis J, Fasolino M, Raphael AR, Yamazaki T, Elias L, Polak M, Kelly C, et al. 2013. Exome sequencing to identify de novo mutations in sporadic ALS trios. Nat Neurosci 16: 851–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chio A, Schymick JC, Restagno G, Scholz SW, Lombardo F, Lai SL, Mora G, Fung HC, Britton A, Arepalli S, et al. 2009. A two-stage genome-wide association study of sporadic amyotrophic lateral sclerosis. Hum Mol Genet 18: 1524–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chio A, Restagno G, Brunetti M, Ossola I, Calvo A, Canosa A, Moglia C, Floris G, Tacconi P, Marrosu F, et al. 2012. ALS/FTD phenotype in two Sardinian families carrying both C9ORF72 and TARDBP mutations. J Neurol Neurosurg Psychiatry 83: 730–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chio A, Mora G, Restagno G, Brunetti M, Ossola I, Barberis M, Ferrucci L, Canosa A, Manera U, Moglia C, et al. 2013. UNC13A influences survival in Italian amyotrophic lateral sclerosis patients: A population-based study. Neurobiol Aging 34: 357.e1–357.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CY, Landers JE, Bergre SK, Sapp PC, Grant AE, Jones JM, Everett L, Lenk GM, McKenna-Yasek DM, Weisman LS, et al. 2009. Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS. Am J Hum Genet 84: 85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli ET, Lasseigne BN, Petrovski S, Sapp PC, Dion PA, Leblond CS, Couthouis J, Lu YF, Wang Q, Krueger BJ, et al. 2015. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science 347: 1436–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper-Knock J, Hewitt C, Highley JR, Brockington A, Milano A, Man S, Martindale J, Hartley J, Walsh T, Gelsthorpe C, et al. 2012. Clinico-pathological features in amyotrophic lateral sclerosis with expansions in C9ORF72. Brain J Neurol 135: 751–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couthouis J, Hart MP, Shorter J, DeJesus-Hernandez M, Erion R, Oristano R, Liu AX, Ramos D, Jethava N, Hosangadi D, et al. 2011. A yeast functional screen predicts new candidate ALS disease genes. Proc Natl Acad Sci 108: 20881–20890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couthouis J, Hart MP, Erion R, King OD, Diaz Z, Nakaya T, Ibrahim F, Kim HJ, Mojsilovic-Petrovic J, Panossian S, et al. 2012. Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum Mol Genet 21: 2899–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin S, Berger S, Ding J, Schymick JC, Washecka N, Hernandez DG, Greenway MJ, Bradley DG, Traynor BJ, Hardiman O. 2008. A genome-wide association study of sporadic ALS in a homogenous Irish population. Hum Mol Genet 17: 768–774. [DOI] [PubMed] [Google Scholar]
- Cronin S, Tomik B, Bradley DG, Slowik A, Hardiman O. 2009. Screening for replication of genome-wide SNP associations in sporadic ALS. Eur J Hum Genet 17: 213–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cudkowicz M, McKenna-Yasek D, Sapp P, Chin W, Geller B, Hayden D, Horvtiz H, Brown R. 1997. Epidemiology of SOD1 mutations in amyotrophic lateral sclerosis. Ann Neurol 41: 210–212. [DOI] [PubMed] [Google Scholar]
- Cudkowicz ME, McKenna-Yasek D, Chen C, Hedley-Whyte ET, Brown RH Jr. 1998. Limited corticospinal tract involvement in amyotrophic lateral sclerosis subjects with the A4V mutation in the copper/zinc superoxide dismutase gene. Ann Neurol 43: 703–710. [DOI] [PubMed] [Google Scholar]
- Dejesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. 2011. Expanded ggggcc hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72: 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker AM, Seelen M, van Doormaal PT, van Rheenen W, Bothof RJ, van Riessen T, Brands WJ, van der Kooi AJ, de Visser M, Voermans NC, et al. 2015. Large-scale screening in sporadic amyotrophic lateral sclerosis identifies genetic modifiers in C9orf72 repeat carriers. Neurobiol Aging 39: P220.e9–P220.e15. [DOI] [PubMed] [Google Scholar]
- Del Bo R, Di Fonzo A, Ghezzi S, Locatelli F, Stevanin G, Costa A, Corti S, Bresolin N, Comi GP. 2007. SPG11: A consistent clinical phenotype in a family with homozygous spatacsin truncating mutation. Neurogenetics 8: 301–305. [DOI] [PubMed] [Google Scholar]
- Dellefave L, Bangash MA, Siddique T. 2003. Pairwise concordance rates are similar in monozygotic twins and dizygotic twins for amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord 4: 47. [Google Scholar]
- Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H, et al. 2011. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 477: 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M, Wei L, Zuo X, Tian Y, Xie F, Hu P, Zhu C, Yu F, Meng Y, Wang H, et al. 2013. Genome-wide association analyses in Han Chinese identify two new susceptibility loci for amyotrophic lateral sclerosis. Nat Genet 45: 697–700. [DOI] [PubMed] [Google Scholar]
- Deschauer M, Gaul C, Behrmann C, Prokisch H, Zierz S, Haack TB. 2012. C19orf12 mutations in neurodegeneration with brain iron accumulation mimicking juvenile amyotrophic lateral sclerosis. J Neurol 259: 2434–2439. [DOI] [PubMed] [Google Scholar]
- Diekstra FP, Saris CG, van Rheenen W, Franke L, Jansen RC, van Es MA, van Vught PW, Blauw HM, Groen EJ, Horvath S, et al. 2012a. Mapping of gene expression reveals CYP27A1 as a susceptibility gene for sporadic ALS. PLoS ONE 7: e35333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekstra FP, van Vught PW, van Rheenen W, Koppers M, Pasterkamp RJ, van Es MA, Schelhaas HJ, de Visser M, Robberecht W, Van Damme P, et al. 2012b. UNC13A is a modifier of survival in amyotrophic lateral sclerosis. Neurobiol Aging 33: 630.e3–630.e8. [DOI] [PubMed] [Google Scholar]
- Diekstra FP, Van Deerlin VM, van Swieten JC, Al-Chalabi A, Ludolph AC, Weishaupt JH, Hardiman O, Landers JE, Brown RH Jr, van Es MA, et al. 2014. C9orf72 and UNC13A are shared risk loci for amyotrophic lateral sclerosis and frontotemporal dementia: A genome-wide meta-analysis. Ann Neurol 76: 120–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson-Stone C, Luty AA, Thompson EM, Blumbergs P, Brooks WS, Short CL, Field CD, Panegyres PK, Hecker J, Solski JA, et al. 2013. Frontotemporal dementia–amyotrophic lateral sclerosis syndrome locus on chromosome 16p12.1-q12.2: Genetic, clinical and neuropathological analysis. Acta Neuropathol 125: 523–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droppelmann CA, Wang J, Campos-Melo D, Keller B, Volkening K, Hegele RA, Srong MJ. 2013. Detection of a novel frameshift mutation and regions with homozygosis within ARHGEF28 gene in familial amyotrophic lateral sclerosis. Amyotroph Lateral Scler Frontotemporal Degener 14: 444–451. [DOI] [PubMed] [Google Scholar]
- Droppelmann CA, Campos-Melo D, Volkening K, Strong MJ. 2014. The emerging role of guanine nucleotide exchange factors in ALS and other neurodegenerative diseases. Front Cell Neurosci 8: 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley T, Huentelman MJ, Craig DW, Pearson JV, Szelinger S, Joshipura K, Halperin RF, Stamper C, Jensen KR, Letizia D, et al. 2007. Whole-genome analysis of sporadic amyotrophic lateral sclerosis. New Engl J Med 357: 775–788. [DOI] [PubMed] [Google Scholar]
- Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, et al. 2010. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466: 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallis BA, Hardiman O. 2009. Aggregation of neurodegenerative disease in ALS kindreds. Amyotroph Lateral Scler 10: 95–98. [DOI] [PubMed] [Google Scholar]
- Fang F, Kamel F, Lichtenstein P, Bellocco R, Sparen P, Sandler DP, Ye W. 2009. Familial aggregation of amyotrophic lateral sclerosis. Ann Neurology 66: 94–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fecto F, Yan J, Vemula SP, Liu E, Yang Y, Chen W, Zheng JG, Shi Y, Siddique N, Arrat H, et al. 2011. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol 68: 1440–1446. [DOI] [PubMed] [Google Scholar]
- Figlewicz DA, Krizus A, Martinoli MG, Meininger V, Dib M, Rouleau GA, Julien JP. 1994. Variants of the heavy neurofilament subunit are associated with the development of amyotrophic lateral sclerosis. Hum Mol Genet 3: 1757–1761. [DOI] [PubMed] [Google Scholar]
- Fink JK. 2013. Hereditary spastic paraplegia: Clinico-pathologic features and emerging molecular mechanisms. Acta Neuropathol 126: 307–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor-de-Lima F, Sampaio M, Nahavandi N, Fernandes S, Leao M. 2014. Alsin related disorders: Literature review and case study with novel mutations. Case Rep Genet 2014: 691515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogh I, D'Alfonso S, Gellera C, Ratti A, Cereda C, Penco S, Corrado L, Soraru G, Castellotti B, Tiloca C, et al. 2011. No association of DPP6 with amyotrophic lateral sclerosis in an Italian population. Neurobiol Aging 32: 966–967. [DOI] [PubMed] [Google Scholar]
- Fogh I, Ratti A, Gellera C, Lin K, Tiloca C, Moskvina V, Corrado L, Soraru G, Cereda C, Corti S, et al. 2014. A genome-wide association meta-analysis identifies a novel locus at 17q11.2 associated with sporadic amyotrophic lateral sclerosis. Hum Mol Gene 23: 2220–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman MS, Mackenzie IR, Cairns NJ, Swanson E, Boyer PJ, Drachman DA, Jhaveri BS, Karlawish JH, Pestronk A, Smith TW, et al. 2006. Novel ubiquitin neuropathology in frontotemporal dementia with valosin-containing protein gene mutations. J Neuropathol Exp Neurol 65: 571–581. [DOI] [PubMed] [Google Scholar]
- Forsberg K, Andersen PM, Marklund SL, Brannstrom T. 2011. Glial nuclear aggregates of superoxide dismutase-1 are regularly present in patients with amyotrophic lateral sclerosis. Acta Neuropathol 121: 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee KH, Badders N, Valentine M, Miller BL, Wong PC, et al. 2015. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 525: 129–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freischmidt A, Wieland T, Richter B, Ruf W, Schaeffer V, Muller K, Marroquin N, Nordin F, Hubers A, Weydt P, et al. 2015. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat Neurosci 18: 631–636. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Okamoto K, Fujita S, Ikeda Y. 2014. ELP3-positive inclusions in motor neuron diseases. Eur J Neurol 21: 329. [Google Scholar]
- Gal J, Strom AL, Kwinter DM, Kilty R, Zhang J, Shi P, Fu W, Wooten MW, Zhu H. 2009. Sequestosome 1/p62 links familial ALS mutant SOD1 to LC3 via an ubiquitin-independent mechanism. J Neurochem 111: 1062–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Iradi MC, Clare AM, Brown HH, Janus C, Lewis J, Borchelt DR. 2015. Subcellular localization of Matrin 3 containing mutations associated with ALS and distal myopathy. PLoS ONE 10: e0142144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz B, Wong M, Martin LJ. 2012. Nuclear localization of human SOD1 and mutant SOD1-specific disruption of survival motor neuron protein complex in transgenic amyotrophic lateral sclerosis mice. J Neuropathol Exp Neurol 71: 162–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijselinck I, Van Langenhove T, van der Zee J, Sleegers K, Philtjens S, Kleinberger G, Janssens J, Bettens K, Van Cauwenberghe C, Pereson S, et al. 2012. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: A gene identification study. Lancet Neurol 11: 54–65. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Perez P, Lu Y, Chian RJ, Sapp PC, Tanzi RE, Bertram L, McKenna-Yasek D, Gao FB, Brown RH Jr. 2012. Association of UBQLN1 mutation with Brown-Vialetto-Van Laere syndrome but not typical ALS. Neurobiol Dis 48: 391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham AJ, Macdonald AM, Hawkes CH. 1997. British motor neuron disease twin study. J Neurol Neurosurg Psychiatry 62: 562–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenway MJ, Andersen PM, Russ C, Ennis S, Cashman S, Donaghy C, Patterson V, Swingler R, Kieran D, Prehn J, et al. 2006. ANG mutations segregate with familial and “sporadic” amyotrophic lateral sclerosis. Nat Genet 38: 411–413. [DOI] [PubMed] [Google Scholar]
- Groen EJ, Fumoto K, Blokhuis AM, Engelen-Lee J, Zhou Y, van den Heuvel DM, Koppers M, van Diggelen F, van Heest J, Demmers JA, et al. 2013. ALS-associated mutations in FUS disrupt the axonal distribution and function of SMN. Hum Mol Genet 22: 3690–3704. [DOI] [PubMed] [Google Scholar]
- Gros-Louis F, Lariviere R, Gowing G, Laurent S, Camu W, Bouchard JP, Meininger V, Rouleau GA, Julien JP. 2004. A frameshift deletion in peripherin gene associated with amyotrophic lateral sclerosis. J Biol Chem 279: 45951–45956. [DOI] [PubMed] [Google Scholar]
- Gros-Louis F, Andersen PM, Dupre N, Urushitani M, Dion P, Souchon F, D'Amour M, Camu W, Meininger V, Bouchard JP, et al. 2009. Chromogranin B P413L variant as risk factor and modifier of disease onset for amyotrophic lateral sclerosis. Proc Natl Acad Sci 106: 21777–21782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney M. 1994. Mutant mice, Cu,Zn superoxide dismutase, and motor neuron degeneration: Response. Science 266: 1586. [PubMed] [Google Scholar]
- Hadano S, Hand CK, Osuga H, Yanagisawa Y, Otomo A, Devon RS, Miyamoto N, Showguchi-Miyata J, Okada Y, Singaraja R, et al. 2001. A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet 29: 166–173. [DOI] [PubMed] [Google Scholar]
- Hand CK, Khoris J, Salachas F, Gros-Louis F, Lopes AA, Mayeux-Portas V, Brewer CG, Brown RH Jr, Meininger V, Camu W, et al. 2002. A novel locus for familial amyotrophic lateral sclerosis, on chromosome 18q. Am J Hum Genet 70: 251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EJ, Zhang J, Geser F, Trojanowski JQ, Strober JB, Dickson DW, Brown RH Jr, Shapiro BE, Lomen-Hoerth C. 2010. Extensive FUS-immunoreactive pathology in juvenile amyotrophic lateral sclerosis with basophilic inclusions. Brain Pathol 20: 1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Zhou H, Tong J, Chen H, Liu YJ, Wang D, Wei X, Xia XG. 2011. FUS transgenic rats develop the phenotypes of amyotrophic lateral sclerosis and frontotemporal lobar degeneration. PLoS Genet 7: e1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida A, Takahashi A, Kubo M, Saito S, Hosono N, Ohnishi Y, Kiyotani K, Mushiroda T, Nakajima M, Ozaki K, et al. 2011. A functional variant in ZNF512B is associated with susceptibility to amyotrophic lateral sclerosis in Japanese. Hum Mol Genet 20: 3684–3692. [DOI] [PubMed] [Google Scholar]
- Jackson M, Morrison KE, Al-Chalabi A, Bakker M, Leigh PN. 1996. Analysis of chromosome 5q13 genes in amyotrophic lateral sclerosis: Homozygous NAIP deletion in a sporadic case. Ann Neurol 39: 796–800. [DOI] [PubMed] [Google Scholar]
- Janssens J, Van Broeckhoven C. 2013. Pathological mechanisms underlying TDP-43 driven neurodegeneration in FTLD-ALS spectrum disorders. Hum Mol Genet 22: R77–R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Zhu Q, Gendron TF, Saberi S, McAlonis-Downes M, Seelman A, Stauffer JE, Jafar-Nejad P, Drenner K, Schulte D, et al. 2016. Gain of toxicity from ALS/FTD-linked repeat expansions in C9ORF72 is alleviated by antisense oligonucleotides targeting GGGGCC-containing RNAs. Neuron 90: 535–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, et al. 2010. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron 68: 857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Gibbs JR, Megarbane A, Urtizberea JA, Hernandez DG, Foley AR, Arepalli S, Pandraud A, Simon-Sanchez J, Clayton P, et al. 2012. Exome sequencing reveals riboflavin transporter mutations as a cause of motor neuron disease. Brain 135: 2875–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Glynn SM, Gibbs JR, Nalls MA, Sabatelli M, Restagno G, Drory VE, Chio A, Rogaeva E, Traynor BJ. 2014a. Mutations in the CHCHD10 gene are a common cause of familial amyotrophic lateral sclerosis. Brain 137: e311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Pioro EP, Boehringer A, Chia R, Feit H, Renton AE, Pliner HA, Abramzon Y, Marangi G, Winborn BJ, et al. 2014b. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat Neurosci 17: 664–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicic A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, Paul JW 3rd, Sun S, Herdy JR, Bieri G, et al. 2015. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci 18: 1226–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Beaulieu JM. 2000. Cytoskeletal abnormalities in amyotrophic lateral sclerosis: Beneficial or detrimental effects? J Neurol Sci 180: 7–14. [DOI] [PubMed] [Google Scholar]
- Kaneko K, Saito F, Sunohara N, Ikeuchi T. 1995. Cytogenetic analysis of 23 Japanese patients with amyotrophic lateral sclerosis. Clin Genet 47: 158–160. [DOI] [PubMed] [Google Scholar]
- Kang SH, Li Y, Fukaya M, Lorenzini I, Cleveland DW, Ostrow LW, Rothstein JD, Bergles DE. 2013. Degeneration and impaired regeneration of gray matter oligodendrocytes in amyotrophic lateral sclerosis. Nat Neurosci 16: 571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan A, Spiller KJ, Towne C, Kanning KC, Choe GT, Geber A, Akay T, Aebischer P, Henderson CE. 2014. Neuronal matrix metalloproteinase-9 is a determinant of selective neurodegeneration. Neuron 81: 333–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, et al. 2013. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495: 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimonis VE, Mehta SG, Fulchiero EC, Thomasova D, Pasquali M, Boycott K, Neilan EG, Kartashov A, Forman MS, Tucker S, et al. 2008. Clinical studies in familial VCP myopathy associated with Paget disease of bone and frontotemporal dementia. Am J Med Genet Part A 146A: 745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwee LC, Liu Y, Haynes C, Gibson JR, Stone A, Schichman SA, Kamel F, Nelson LM, Topol B, Van den Eeden SK, et al. 2012. A high-density genome-wide association screen of sporadic ALS in US veterans. PloS ONE 7: e32768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski TJ Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, et al. 2009. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323: 1205–1208. [DOI] [PubMed] [Google Scholar]
- Laaksovirta H, Peuralinna T, Schymick JC, Scholz SW, Lai SL, Myllykangas L, Sulkava R, Jansson L, Hernandez DG, Gibbs JR, et al. 2010. Chromosome 9p21 in amyotrophic lateral sclerosis in Finland: A genome-wide association study. Lancet Neurol 9: 978–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landers JE, Melki J, Meininger V, Glass JD, van den Berg LH, van Es MA, Sapp PC, van Vught PW, McKenna-Yasek DM, Blauw HM, et al. 2009. Reduced expression of the Kinesin-Associated Protein 3 (KIFAP3) gene increases survival in sporadic amyotrophic lateral sclerosis. Proc Natl Acad Sci 106: 9004–9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattante S, Rouleau GA, Kabashi E. 2013. TARDBP and FUS mutations associated with amyotrophic lateral sclerosis: Summary and update. Hum Mutat 34: 812–826. [DOI] [PubMed] [Google Scholar]
- Le Ber I, Van Bortel I, Nicolas G, Bouya-Ahmed K, Camuzat A, Wallon D, De Septenville A, Latouche M, Lattante S, Kabashi E, et al. 2014. hnRNPA2B1 and hnRNPA1 mutations are rare in patients with “multisystem proteinopathy” and frontotemporal lobar degeneration phenotypes. Neurobiol Aging 35: 934.e935–934.e936. [DOI] [PubMed] [Google Scholar]
- Leblond CS, Gan-Or Z, Spiegelman D, Laurent SB, Szuto A, Hodgkinson A, Dionne-Laporte A, Provencher P, de Carvalho M, Orru S, et al. 2016. Replication study of MATR3 in familial and sporadic amyotrophic lateral sclerosis. Neurobiol Aging 37: 209.e17–21. [DOI] [PubMed] [Google Scholar]
- Lee T, Li YR, Ingre C, Weber M, Grehl T, Gredal O, de Carvalho M, Meyer T, Tysnes OB, Auburger G, et al. 2011. Ataxin-2 intermediate-length polyglutamine expansions in European ALS patients. Hum Mol Genet 20: 1697–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Sheng J, Hu JK, Yu W, Kishikawa H, Hu MG, Shima K, Wu D, Xu Z, Xin W, et al. 2013. Ribonuclease 4 protects neuron degeneration by promoting angiogenesis, neurogenesis, and neuronal survival under stress. Angiogenesis 16: 387–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling JP, Pletnikova O, Troncoso JC, Wong PC. 2015. TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science 349: 650–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Pattamatta A, Zu T, Reid T, Bardhi O, Borchelt DR, Yachnis AT, Ranum LP. 2016. C9orf72 BAC mouse model with motor deficits and neurodegenerative features of ALS/FTD. Neuron 90: 521–534. [DOI] [PubMed] [Google Scholar]
- Lopez-Lopez A, Gamez J, Syriani E, Morales M, Salvado M, Rodriguez MJ, Mahy N, Vidal-Taboada JM. 2014. CX3CR1 is a modifying gene of survival and progression in amyotrophic lateral sclerosis. PLoS ONE 9: e96528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luigetti M, Lattante S, Zollino M, Conte A, Marangi G, Del Grande A, Sabatelli M. 2011. SOD1 G93D sporadic amyotrophic lateral sclerosis (SALS) patient with rapid progression and concomitant novel ANG variant. Neurobiol Aging 32: 1924.e15–1924.e18. [DOI] [PubMed] [Google Scholar]
- Manole A, Chelban V, Haridy NA, Hamed SA, Berardo A, Reilly MM, Houlden H. 2016. Severe axonal neuropathy is a late manifestation of SPG11. J Neurol 263: 2278–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, Kinoshita Y, Kamada M, Nodera H, Suzuki H, et al. 2010. Mutations of optineurin in amyotrophic lateral sclerosis. Nature 465: 223–226. [DOI] [PubMed] [Google Scholar]
- McLaughlin RL, Kenna KP, Vajda A, Heverin M, Byrne S, Donaghy CG, Cronin S, Bradley DG, Hardiman O. 2015. Homozygosity mapping in an Irish ALS case-control cohort describes local demographic phenomena and points towards potential recessive risk loci. Genomics 105: 237–241. [DOI] [PubMed] [Google Scholar]
- Meyer T, Alber B, Roemer K, Martin T, Kalscheuer VM, Gottert E, Zang KD, Ludolph AC, Ropers HH, Prudlo J. 2003. High rate of constitutional chromosomal rearrangements in apparently sporadic ALS. Neurology 60: 1348–1350. [DOI] [PubMed] [Google Scholar]
- Millecamps S, De Septenville A, Teyssou E, Daniau M, Camuzat A, Albert M, LeGuern E, Galimberti D, Brice A, Marie Y, et al. 2014. Genetic analysis of matrin 3 gene in French amyotrophic lateral sclerosis patients and frontotemporal lobar degeneration with amyotrophic lateral sclerosis patients. Neurobiol Aging 35: 2882.e13–2882.e15. [DOI] [PubMed] [Google Scholar]
- Mitchell J, Paul P, Chen HJ, Morris A, Payling M, Falchi M, Habgood J, Panoutsou S, Winkler S, Tisato V, et al. 2010. Familial amyotrophic lateral sclerosis is associated with a mutation in D-amino acid oxidase. Proc Natl Acad Sci 107: 7556–7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizielinska S, Gronke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IO, Pietrzyk J, et al. 2014. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345: 1192–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecchiani C, Pedace L, Lo Giudice T, Casella A, Mearini M, Gaudiello F, Pedroso JL, Terracciano C, Caltagirone C, Massa R, et al. 2016. ALS5/SPG11/KIAA1840 mutations cause autosomal recessive axonal Charcot-Marie-Tooth disease. Brain 139: 73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfini GA, Burns M, Binder LI, Kanaan NM, LaPointe N, Bosco DA, Brown RH Jr, Brown H, Tiwari A, Hayward L, et al. 2009. Axonal transport defects in neurodegenerative diseases. J Neurosci 29: 12776–12786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriconi C, Di Angelantonio S, Piccioni A, Trettel F, Sabatelli M, Grassi F. 2011. Mutant human β4 subunit identified in amyotrophic lateral sclerosis patients impairs nicotinic receptor function. Pflugers Arch Eur J Physiol 461: 225–233. [DOI] [PubMed] [Google Scholar]
- Morohoshi F, Ootsuka Y, Arai K, Ichikawa H, Mitani S, Munakata N, Ohki M. 1998. Genomic structure of the human RBP56/hTAFIIv68 and FUS/TLS genes. Gene 221: 191–198. [DOI] [PubMed] [Google Scholar]
- Moulard B, Salachas F, Chassande B, Briolotti V, Meininger V, Malafosse A, Camu W. 1998. Association between centromeric deletions of the SMN gene and sporadic adult-onset lower motor neuron disease. Ann Neurol 43: 640–644. [DOI] [PubMed] [Google Scholar]
- Munch C, Sedlmeier R, Meyer T, Homberg V, Sperfeld AD, Kurt A, Prudlo J, Peraus G, Hanemann CO, Stumm G, et al. 2004. Point mutations of the p150 subunit of dynactin (DCTN1) gene in ALS. Neurology 63: 724–726. [DOI] [PubMed] [Google Scholar]
- Munch C, Rosenbohm A, Sperfeld AD, Uttner I, Reske S, Krause BJ, Sedlmeier R, Meyer T, Hanemann CO, Stumm G, et al. 2005. Heterozygous R1101K mutation of the DCTN1 gene in a family with ALS and FTD. Ann Neurol 58: 777–780. [DOI] [PubMed] [Google Scholar]
- Munoz DG, Neumann M, Kusaka H, Yokota O, Ishihara K, Terada S, Kuroda S, Mackenzie IR. 2009. FUS pathology in basophilic inclusion body disease. Acta Neuropathol 118: 617–627. [DOI] [PubMed] [Google Scholar]
- Nagai M, Aoki M, Miyoshi I, Kato M, Pasinelli P, Kasai N, Brown RH Jr, Itoyama Y. 2001. Rats expressing human cytosolic copper-zinc superoxide dismutase transgenes with amyotrophic lateral sclerosis: Associated mutations develop motor neuron disease. J Neurosci 21: 9246–9254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalini A, Pandraud A, Mok K, Houlden H. 2013. Madras motor neuron disease (MMND) is distinct from the riboflavin transporter genetic defects that cause Brown-Vialetto-Van Laere syndrome. J Neurol Sci 334: 119–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M. 2009. Molecular neuropathology of TDP-43 proteinopathies. Intl J Mol Sci 10: 232–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. 2006. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314: 130–133. [DOI] [PubMed] [Google Scholar]
- Nishimura AL, Mitne-Neto M, Silva HC, Richieri-Costa A, Middleton S, Cascio D, Kok F, Oliveira JR, Gillingwater T, Webb J, et al. 2004. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet 75: 822–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlacchio A, Babalini C, Borreca A, Patrono C, Massa R, Basaran S, Munhoz RP, Rogaeva EA, St George-Hyslop PH, Bernardi G, et al. 2010. SPATACSIN mutations cause autosomal recessive juvenile amyotrophic lateral sclerosis. Brain 133: 591–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke JG, Bogdanik L, Muhammad AK, Gendron TF, Kim KJ, Austin A, Cady J, Liu EY, Zarrow J, Grant S, et al. 2015. C9orf72 BAC transgenic mice display typical pathologic features of ALS/FTD. Neuron 88: 892–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsetti V, Pegoraro E, Cima V, D'Ascenzo C, Palmieri A, Querin G, Volpe M, Ermani M, Angelini C, Soraru G. 2011. Genetic variation in KIFAP3 is associated with an upper motor neuron–predominant phenotype in amyotrophic lateral sclerosis. Neuro-degen Dis 8: 491–495. [DOI] [PubMed] [Google Scholar]
- Otomo A, Kunita R, Suzuki-Utsunomiya K, Mizumura H, Onoe K, Osuga H, Hadano S, Ikeda JE. 2008. ALS2/alsin deficiency in neurons leads to mild defects in macropinocytosis and axonal growth. Biochem Biophys Res Commun 370: 87–92. [DOI] [PubMed] [Google Scholar]
- Parkinson N, Ince PG, Smith MO, Highley R, Skibinski G, Andersen PM, Morrison KE, Pall HS, Hardiman O, Collinge J, et al. 2006. ALS phenotypes with mutations in CHMP2B (charged multivesicular body protein 2B). Neurology 67: 1074–1077. [DOI] [PubMed] [Google Scholar]
- Peters OM, Cabrera GT, Tran H, Gendron TF, McKeon JE, Metterville J, Weiss A, Wightman N, Salameh J, Kim J, et al. 2015a. Human C9ORF72 hexanucleotide expansion reproduces RNA foci and dipeptide repeat proteins but not neurodegeneration in BAC transgenic mice. Neuron 88: 902–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters OM, Ghasemi M, Brown RH Jr. 2015b. Emerging mechanisms of molecular pathology in ALS. J Clin Invest 125: 2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips T, Rothstein JD. 2014. Glial cells in amyotrophic lateral sclerosis. Exp Neurol 262: 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pottier C, Bieniek KF, Finch N, van de Vorst M, Baker M, Perkersen R, Brown P, Ravenscroft T, van Blitterswijk M, Nicholson AM, et al. 2015. Whole-genome sequencing reveals important role for TBK1 and OPTN mutations in frontotemporal lobar degeneration without motor neuron disease. Acta Neuropathol 130: 77–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prause J, Goswami A, Katona I, Roos A, Schnizler M, Bushuven E, Dreier A, Buchkremer S, Johann S, Beyer C, et al. 2013. Altered localization, abnormal modification and loss of function of Sigma receptor-1 in amyotrophic lateral sclerosis. Hum Mol Genet 22: 1581–1600. [DOI] [PubMed] [Google Scholar]
- Puls I, Jonnakuty C, LaMonte B, Holzbaur E, Tokito M, Mann E, Floeter M, Bidus K, Drayna D, Oh S, et al. 2003. Mutant dynactin in motor neuron disease. Nat Genet 33: 455–456. [DOI] [PubMed] [Google Scholar]
- Purcell S, Cherny SS, Sham PC. 2003. Genetic Power Calculator: Design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19: 149–150. [DOI] [PubMed] [Google Scholar]
- Rainier S, Bui M, Mark E, Thomas D, Tokarz D, Ming L, Delaney C, Richardson RJ, Albers JW, Matsunami N, et al. 2008. Neuropathy target esterase gene mutations cause motor neuron disease. Am J Hum Genet 82: 780–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayaprolu S, Mullen B, Baker M, Lynch T, Finger E, Seeley WW, Hatanpaa KJ, Lomen-Hoerth C, Kertesz A, Bigio EH, et al. 2013. TREM2 in neurodegeneration: Evidence for association of the p.R47H variant with frontotemporal dementia and Parkinson's disease. Mol Neurodegener 8: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. 2011. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72: 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson RJ, Hein ND, Wijeyesakere SJ, Fink JK, Makhaeva GF. 2013. Neuropathy target esterase (NTE): Overview and future. Chem Biol Interact 203: 238–244. [DOI] [PubMed] [Google Scholar]
- Ringholz GM, Appel SH, Bradshaw M, Cooke NA, Mosnik DM, Schulz PE. 2005. Prevalence and patterns of cognitive impairment in sporadic ALS. Neurology 65: 586–590. [DOI] [PubMed] [Google Scholar]
- Robberecht W, Philips T. 2013. The changing scene of amyotrophic lateral sclerosis. Nat Rev Neurosci 14: 248–264. [DOI] [PubMed] [Google Scholar]
- Ronchi D, Riboldi G, Del Bo R, Ticozzi N, Scarlato M, Galimberti D, Corti S, Silani V, Bresolin N, Comi GP. 2015. CHCHD10 mutations in Italian patients with sporadic amyotrophic lateral sclerosis. Brain 138: e372. [DOI] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O'Regan JP, Deng HX, et al. 1993. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362: 59–62. [DOI] [PubMed] [Google Scholar]
- Rowland LP, Shneider NA. 2001. Amyotrophic lateral sclerosis. N Engl J Med 344: 1688–1700. [DOI] [PubMed] [Google Scholar]
- Rubino E, Rainero I, Chio A, Rogaeva E, Galimberti D, Fenoglio P, Grinberg Y, Isaia G, Calvo A, Gentile S, et al. 2012. SQSTM1 mutations in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Neurology 79: 1556–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford NJ, Zhang YJ, Baker M, Gass JM, Finch NA, Xu YF, Stewart H, Kelley BJ, Kuntz K, Crook RJP, et al. 2008. Novel mutations in TARDBP (TDP-43) in patients with familial amyotrophic lateral sclerosis. PLoS Genet 4: e1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatelli M, Eusebi F, Al-Chalabi A, Conte A, Madia F, Luigetti M, Mancuso I, Limatola C, Trettel F, Sobrero F, et al. 2009. Rare missense variants of neuronal nicotinic acetylcholine receptor altering receptor function are associated with sporadic amyotrophic lateral sclerosis. Hum Mol Genet 18: 3997–4006. [DOI] [PubMed] [Google Scholar]
- Sabatelli M, Lattante S, Conte A, Marangi G, Luigetti M, Del Grande A, Chio A, Corbo M, Giannini F, Mandrioli J, et al. 2012. Replication of association of CHRNA4 rare variants with sporadic amyotrophic lateral sclerosis: The Italian multicentre study. Amyotroph Lateral Scler 13: 580–584. [DOI] [PubMed] [Google Scholar]
- Saeed M, Siddique N, Hung WY, Usacheva E, Liu E, Sufit RL, Heller SL, Haines JL, Pericak-Vance M, Siddique T. 2006. Paraoxonase cluster polymorphisms are associated with sporadic ALS. Neurology 67: 771–776. [DOI] [PubMed] [Google Scholar]
- Sanhueza M, Zechini L, Gillespie T, Pennetta G. 2014. Gain-of-function mutations in the ALS8 causative gene VAPB have detrimental effects on neurons and muscles. Biol Open 3: 59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrott-Fischer A, Bitsche M, Humpel C, Walcher C, Maier H, Jellinger K, Rabl W, Glueckert R, Marksteiner J. 2009. Chromogranin peptides in amyotrophic lateral sclerosis. Regul Pept 152: 13–21. [DOI] [PubMed] [Google Scholar]
- Scott S, Kranz JE, Cole J, Lincecum JM, Thompson K, Kelly N, Bostrom A, Theodoss J, Al-Nakhala BM, Vieira FG, et al. 2008. Design, power, and interpretation of studies in the standard murine model of ALS. Amyotroph Lateral Scler 9: 4–15. [DOI] [PubMed] [Google Scholar]
- Sharma A, Lyashchenko AK, Lu L, Nasrabady SE, Elmaleh M, Mendelsohn M, Nemes A, Tapia JC, Mentis GZ, Shneider NA. 2016. ALS-associated mutant FUS induces selective motor neuron degeneration through toxic gain of function. Nat Commun 7: 10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatunov A, Mok K, Newhouse S, Weale ME, Smith B, Vance C, Johnson L, Veldink JH, van Es MA, van den Berg LH, et al. 2010. Chromosome 9p21 in sporadic amyotrophic lateral sclerosis in the UK and seven other countries: A genome-wide association study. Lancet Neurol 9: 986–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson CL, Lemmens R, Miskiewicz K, Broom WJ, Hansen VK, van Vught PW, Landers JE, Sapp P, Van Den Bosch L, Knight J, et al. 2009. Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Hum Mol Genetics 18: 472–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slowik A, Tomik B, Wolkow PP, Partyka D, Turaj W, Malecki MT, Pera J, Dziedzic T, Szczudlik A, Figlewicz DA. 2006. Paraoxonase gene polymorphisms and sporadic ALS. Neurology 67: 766–770. [DOI] [PubMed] [Google Scholar]
- Smith BN, Ticozzi N, Fallini C, Gkazi AS, Topp S, Kenna KP, Scotter EL, Kost J, Keagle P, Miller JW, et al. 2014. Exome-wide rare variant analysis identifies TUBA4A mutations associated with familial ALS. Neuron 84: 324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Wang M, Mao F, Shao M, Zhao B, Song Z, Shao C, Gong Y. 2013. Knockdown of Pnpla6 protein results in motor neuron defects in zebrafish. Dis Model Mech 6: 404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, et al. 2008. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319: 1668–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevanin G, Santorelli FM, Azzedine H, Coutinho P, Chomilier J, Denora PS, Martin E, Ouvrard-Hernandez AM, Tessa A, Bouslam N, et al. 2007. Mutations in SPG11, encoding spatacsin, are a major cause of spastic paraplegia with thin corpus callosum. Nat Genet 39: 366–372. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Fukuda Y, Yoshimura J, Toyoda A, Kurppa K, Moritoyo H, Belzil VV, Dion PA, Higasa K, Doi K, et al. 2013. ERBB4 mutations that disrupt the neuregulin-ErbB4 pathway cause amyotrophic lateral sclerosis type 19. Am J Hum Genet 93: 900–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateno M, Kato S, Sakurai T, Nukina N, Takahashi R, Araki T. 2009. Mutant SOD1 impairs axonal transport of choline acetyltransferase and acetylcholine release by sequestering KAP3. Hum Mol Genet 18: 942–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teyssou E, Vandenberghe N, Moigneu C, Boillee S, Couratier P, Meininger V, Pradat PF, Salachas F, Leguern E, Millecamps S. 2014. Genetic analysis of SS18L1 in French amyotrophic lateral sclerosis. Neurobiol Aging 35: 1213.e1219–1213.e1212. [DOI] [PubMed] [Google Scholar]
- Thiyagarajan N, Ferguson R, Subramanian V, Acharya KR. 2012. Structural and molecular insights into the mechanism of action of human angiogenin-ALS variants in neurons. Nat Commun 3: 1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticozzi N, Vance C, Leclerc AL, Keagle P, Glass JD, McKenna-Yasek D, Sapp PC, Silani V, Bosco DA, Shaw CE, et al. 2011. Mutational analysis reveals the FUS homolog TAF15 as a candidate gene for familial amyotrophic lateral sclerosis. Am J Med Genet B Neuropsychiatr Genet 156B: 285–290. [DOI] [PubMed] [Google Scholar]
- Tran H, Almeida S, Moore J, Gendron TF, Chalasani U, Lu Y, Du X, Nickerson JA, Petrucelli L, Weng Z, et al. 2015. Differential toxicity of nuclear RNA foci versus dipeptide repeat proteins in a Drosophila model of C9ORF72 FTD/ALS. Neuron 87: 1207–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao W, Jeong YH, Lin S, Ling J, Price DL, Chiang PM, Wong PC. 2012. Rodent models of TDP-43: Recent advances. Brain Res 1462: 26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugwu F, Rollinson S, Harris J, Gerhard A, Richardson A, Jones M, Mann D, Snowden J, Pickering-Brown S. 2014. A UBQLN2 variant of unknown significance in frontotemporal lobar degeneration. Neurobiol Aging 36: 546.e15–546.e16. [DOI] [PubMed] [Google Scholar]
- Uyan O, Omur O, Agim ZS, Ozoguz A, Li H, Parman Y, Deymeer F, Oflazer P, Koc F, Tan E, et al. 2013. Genome-wide copy number variation in sporadic amyotrophic lateral sclerosis in the Turkish population: Deletion of EPHA3 is a possible protective factor. PLoS ONE 8: e72381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Blitterswijk M, van Es MA, Hennekam EA, Dooijes D, van Rheenen W, Medic J, Bourque PR, Schelhaas HJ, van der Kooi AJ, de Visser M, et al. 2012. Evidence for an oligogenic basis of amyotrophic lateral sclerosis. Human Mol Genet 21: 3776–3784. [DOI] [PubMed] [Google Scholar]
- van Blitterswijk M, DeJesus-Hernandez M, Niemantsverdriet E, Murray ME, Heckman MG, Diehl NN, Brown PH, Baker MC, Finch NA, Bauer PO, et al. 2013. Association between repeat sizes and clinical and pathological characteristics in carriers of C9ORF72 repeat expansions (Xpansize-72): A cross-sectional cohort study. Lancet Neurol 12: 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Blitterswijk M, Mullen B, Nicholson AM, Bieniek KF, Heckman MG, Baker MC, DeJesus-Hernandez M, Finch NA, Brown PH, Murray ME, et al. 2014. TMEM106B protects C9ORF72 expansion carriers against frontotemporal dementia. Acta Neuropathol 127: 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es MA, Veldink JH, Saris CG, Blauw HM, van Vught PW, Birve A, Lemmens R, Schelhaas HJ, Groen EJ, Huisman MH, et al. 2009. Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat Genet 41: 1083–1087. [DOI] [PubMed] [Google Scholar]
- van Es MA, Schelhaas HJ, van Vught PW, Ticozzi N, Andersen PM, Groen EJ, Schulte C, Blauw HM, Koppers M, Diekstra FP, et al. 2011. Angiogenin variants in Parkinson disease and amyotrophic lateral sclerosis. Ann Neurol 70: 964–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoecke A, Schoonaert L, Lemmens R, Timmers M, Staats KA, Laird AS, Peeters E, Philips T, Goris A, Dubois B, et al. 2012. EPHA4 is a disease modifier of amyotrophic lateral sclerosis in animal models and in humans. Nat Med 18: 1418–1422. [DOI] [PubMed] [Google Scholar]
- van Rheenen W, Shatunov A, Dekker AM, McLaughlin RL, Diekstra FP, Pulit SL, van der Spek RA, Võsa U, de Jong S, Robinson MR, et al. 2016. Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat Genet 48: 1043–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, et al. 2009. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323: 1208–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoqueaux F, Sons MS, Plomp JJ, Brose N. 2005. Aberrant morphology and residual transmitter release at the Munc13-deficient mouse neuromuscular synapse. Mol Cell Biol 25: 5973–5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldink JH, Kalmijn S, Van der Hout AH, Lemmink HH, Groeneveld GJ, Lummen C, Scheffer H, Wokke JH, Van den Berg LH. 2005. SMN genotypes producing less SMN protein increase susceptibility to and severity of sporadic ALS. Neurology 65: 820–825. [DOI] [PubMed] [Google Scholar]
- Vucic S, Rothstein JD, Kiernan MC. 2014. Advances in treating amyotrophic lateral sclerosis: insights from pathophysiological studies. Trends Neurosci 37: 433–442. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Dykes-Hoberg M, Culotta VC, Price DL, Wong PC, Rothstein JD. 2001. Histological evidence of protein aggregation in mutant SOD1 transgenic mice and in amyotrophic lateral sclerosis neural tissues. Neurobiol Dis 8: 933–941. [DOI] [PubMed] [Google Scholar]
- Watts GD, Thomasova D, Ramdeen SK, Fulchiero EC, Mehta SG, Drachman DA, Weihl CC, Jamrozik Z, Kwiecinski H, Kaminska A, et al. 2007. Novel VCP mutations in inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia. Clin Genet 72: 420–426. [DOI] [PubMed] [Google Scholar]
- Wills AM, Cronin S, Slowik A, Kasperaviciute D, Van Es MA, Morahan JM, Valdmanis PN, Meininger V, Melki J, Shaw CE, et al. 2009. A large-scale international meta-analysis of paraoxonase gene polymorphisms in sporadic ALS. Neurology 73: 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Fallini C, Ticozzi N, Keagle PJ, Sapp PC, Piotrowska K, Lowe P, Koppers M, McKenna-Yasek D, Baron DM, et al. 2012. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature 488: 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Hentati A, Deng HX, Dabbagh O, Sasaki T, Hirano M, Hung WY, Ouahchi K, Yan J, Azim AC, et al. 2001. The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat Genet 29: 160–165. [DOI] [PubMed] [Google Scholar]
- Yang C, Danielson EW, Qiao T, Metterville J, Brown RH, Landers JE, Xu Z. 2016. Mutant PFN1 causes ALS phenotypes and progressive motor neuron degeneration in mice by a gain of toxicity. Proc Natl Acad Sci 113: E6209–E6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zolov SN, Chow CY, Slutsky SG, Richardson SC, Piper RC, Yang B, Nau JJ, Westrick RJ, Morrison SJ, et al. 2007. Loss of Vac14, a regulator of the signaling lipid phosphatidylinositol 3,5-bisphosphate, results in neurodegeneration in mice. Proc Natl Acad Sci 104: 17518–17523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, et al. 2015. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525: 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinman L, Liu HN, Sato C, Wakutani Y, Marvelle AF, Moreno D, Morrison KE, Mohlke KL, Bilbao J, Robertson J, et al. 2009. A mechanism for low penetrance in an ALS family with a novel SOD1 deletion. Neurology 72: 1153–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH, et al. 2013. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci 110: E4968–E4977. [DOI] [PMC free article] [PubMed] [Google Scholar]