Abstract
Mounting evidence in recent years supports the extensive interaction between the circadian and redox systems. The existence of such a relationship is not surprising because most organisms, be they diurnal or nocturnal, display daily oscillations in energy intake, locomotor activity, and exposure to exogenous and internally generated oxidants. The transcriptional clock controls the levels of many antioxidant proteins and redox-active cofactors, and, conversely, the cellular redox poise has been shown to feed back to the transcriptional oscillator via redox-sensitive transcription factors and enzymes. However, the circadian cycles in the S-sulfinylation of the peroxiredoxin (PRDX) proteins constituted the first example of an autonomous circadian redox oscillation, which occurred independently of the transcriptional clock. Importantly, the high phylogenetic conservation of these rhythms suggests that they might predate the evolution of the transcriptional oscillator, and therefore could be a part of a primordial circadian redox/metabolic oscillator. This discovery forced the reappraisal of the dogmatic transcription-centered view of the clockwork and opened a new avenue of research. Indeed, the investigation into the links between the circadian and redox systems is still in its infancy, and many important questions remain to be addressed.
NOTHING NEW UNDER THE SUN: THE HISTORY OF THE RESEARCH IN NONTRANSCRIPTIONAL OSCILLATORS
The concept that the central mechanism that drives the circadian clock might be based on a nontranscriptional process is not a recent one. In fact, it dates all the way back to the year 1960 when the first chronobiology meeting took place, as part of the Cold Spring Harbor Symposia on Quantitative Biology. At the time, most of the evidence for the existence of circadian clocks was based on observations of overt rhythms in a variety of organisms, but the underlying fundamental mechanisms were still largely unknown. It was clear that because unicellular organisms show daily oscillations, the timekeeping mechanism must be essentially a cellular phenomenon. What was also evident was that the clockwork must consist of a feedback system capable of generating 24-h cycles in various cellular parameters. Three main models were built around this concept, providing different interpretations on the same theme. The first envisioned the core oscillator as an interconnected network of coupled biochemical events occurring in fixed succession, such that they generate a closed self-sustainable loop (the network hypothesis) (Pavlidis 1969). The second model used the same basic principle but favored transcription as the central mechanism (the chronon model) (Ehret and Trucco 1967). The third interpretation was somewhat different and considered the primary oscillator to be a feedback system involving ion gradients and ion-transport channels (the Njus–Sulzman–Hastings membrane model) (Njus et al. 1974).
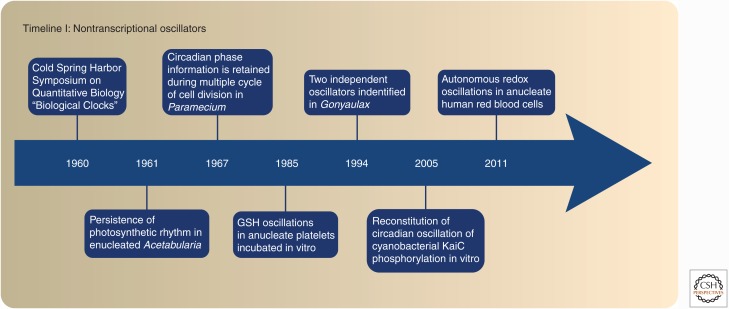
The next 30 years were marked by exciting findings that provided supporting evidence for all three models (Fig. 1). However, because of the discovery of numerous “clock genes” in diverse organisms in the 1990s, the research in nontranscriptional oscillators lost its momentum. The great efforts in finding new “clock genes” culminated in the development of the transcriptional/translation feedback loop (TTFL) model, which seemed to largely account for the generation of circadian rhythmicity in most organisms (Hardin et al. 1990). The overwhelming success of this model led many chronobiologists to adopt an almost dogmatic view of the core clock mechanism, ruling out any other possibilities. Nevertheless, some apparent anomalies to the TTFL (Lakin-Thomas 2006), and the seemingly divergent architecture of the transcriptional clock in different organisms (Young and Kay 2001) preserved the interest in alternative cellular timekeeping models long enough to give rise to the second wave of discoveries in the field of nontranscriptional oscillators (Reddy and O’Neill 2011; van Ooijen and Millar 2012; Reddy and Rey 2014).
Figure 1.
Timeline of the key experiments performed in the field of nontranscriptional oscillators. The timeline begins with the first chronobiology meeting at the Cold Spring Harbor Symposia for Quantitative Biology, where the foundations of the field were laid. The next five decades yielded exciting discoveries in the area of nontranscriptional oscillators, culminating with the finding of the cyanobacterial KaiC oscillator and the autonomous redox oscillations in peroxiredoxins in red blood cells (RBCs). GSH, glutathione.
Indeed, the biggest obstacle that chronobiologists faced in their attempt to investigate nontranscriptional oscillations was the lack of a straightforward procedure for the complete exclusion of the transcriptional contributions from the nucleus. Nevertheless, there are several approaches and model systems that have been successfully used over the years and these are summarized in Table 1.
Table 1.
Model systems and methodological approaches for studying nontranscriptional oscillations
| Method/approach | Model system | Overt rhythm | References |
|---|---|---|---|
| Anucleate cells (rhythms measured in vitro) | Red blood cells | Rhythms in enzymatic activity; rhythms in PRDX overoxidation; rhythms in NAD(P)H and ATP | Cornelius and Rensing 1976; O’Neill and Reddy 2011; Cho et al. 2014; Homma et al. 2015 |
| Platelets | Rhythms in GSH in vitro | Radha et al. 1985 | |
| Removal of the nucleus | Acetabularia | Rhythms in photosynthesis and chloroplast shape | Sweeney and Haxo 1961; Mergenhagen and Schweiger 1975a |
| “Clockless” cells/organisms | Mouse embryonic stem cells | Rhythms in 2-DG uptake precede TTFL expression | Paulose et al. 2012 |
| Caenorhabditis elegans | Rhythms in PRDX overoxidation; olfactory cued behavior; mRNA and protein levels | Olmedo et al. 2012 | |
| Cell-free systems | Kai oscillator | Rhythms in KaiC phosphorylation | Nakajima et al. 2005 |
| Isolated membrane fractions (RBCs) | Rhythms in enzymatic activity | Cornelius and Rensing 1976 | |
| Pharmacological inhibition of transcription and translation | Acetabularia | Rhythm in photosynthesis persists | Mergenhagen and Schweiger 1975b |
| Bulla gouldina | Transcription and translation required only in a phase-dependent manner | Khalsa et al. 1992, 1996 | |
| Pharmacological manipulation of clock activity | REV-ERB agonist; CK1 inhibitor | Could be used to uncouple the transcriptional and metabolic oscillators | Meng et al. 2010; Solt et al. 2012 |
| Environmental perturbations | Transcriptional arrest in Ostreococcus tauri | PRDX overoxidation rhythms; Mg2+ rhythms | O’Neill et al. 2011; Feeney et al. 2016 |
| Treatment with light of different wavelengths in Gonyaulax | Showed the existence of two independent oscillators | Morse et al. 1994 | |
| Genetic manipulation of core clock elements | Constitutive bmal1 expression | Rhythmicity in bmal1 transcription is dispensable for functional TTFL | Liu et al. 2008 |
| Constitutive expression of tim in Drosophila | Restores molecular and behavioral rhythmicity in tim−/− flies | Yang and Sehgal 2001 |
PRDX, Peroxiredoxin; GSH, glutathione; 2-DG, 2-deoxyglucose; TTFL, transcriptional/translation feedback loop; mRNA, messenger RNA; RBCs, red blood cells.
In the 1960s, experiments performed in the unicellular alga Acetabularia for the first time showed the existence of nontranscriptional oscillations. This organism consists of a giant cell (≈1–10 cm at maturity) that contains a single nucleus residing in a basal root-like structure, called rhizoid (Mandoli 1998). Interestingly, it was shown that Acetabularia remains viable for several weeks after amputation of the nucleus-bearing rhizoid, which made the alga an important model system for studying the nuclear–cytoplasmic interactions (Mandoli 1998). Taking advantage of this peculiarity, it was shown that enucleated Acetabularia major plants show endogenous rhythms in photosynthesis and chloroplast shape in constant conditions (Sweeney and Haxo 1961; Driessche 1966). These results were later confirmed in another two Acetabularia species—A. medeternea (Mergenhagen and Schweiger 1975a) and A. crenulata (Terborgh and McLeod 1967). Although early grafting experiments (whereby the nucleus-containing rhizoid of one plant is transplanted onto another) indicated that the nucleus is capable of imposing control over the phase of the overt rhythms, these studies lacked sufficient sampling resolution to make firm conclusions (Schweiger et al. 1964; Vanden Driessche 1966). Indeed, later it was shown that when a nucleus entrained in the opposite phase is reintroduced into the cell, the cytoplasmic oscillator dominates over the nuclear to maintain the phase the photosynthetic rhythm (Woolum 1991). Consistently, experiments using pharmacological inhibitors of transcription showed that the plants remain rhythmic for the initial 14 days of treatment, thus suggesting that transcription is not required to generate rhythmicity, but is rather involved in maintaining the levels of key components of the oscillator (Mergenhagen and Schweiger 1975b).
A few years after the initial experiments in Acetabularia, studies in the unicellular eukaryote Paramecium multimicronucleatum showed that information about the circadian phase is transferred during cell division over many generations (Barnett 1966). These results were later validated in two additional model systems: in the cyanobacteria, Synechococcus elongatus, which have a division time much shorter than 24 h (Kondo et al. 1997), and in mouse fibroblasts cells (Nagoshi et al. 2004). Indeed, this spurred one of the biggest controversies in the field of chronobiology. If transcription is the sole timekeeping mechanisms in the cell, then how do actively dividing cells keep track of time? A possible explanation is that the cell harbors a second oscillator, which runs in parallel to the transcriptional clock and becomes the dominant timekeeper during transcriptional arrest. Evidence for this model was shown in the unicellular photosynthetic alga, Ostreococcus tauri, in which transcription is completely abolished under conditions of constant darkness (O’Neill et al. 2011). Even after complete termination of transcription, O. tauri cultures could restore transcriptional rhythms once transferred to constant light conditions. Perhaps, more importantly, the phase of the oscillations is not dictated by the transfer to light, which suggests that a nontranscriptional mechanism runs in parallel to preserve the phase of the clock.
In 1974, Njus, Sulzman, and Hastings proposed that the timekeeping mechanism is a property of the cell membrane (Njus et al. 1974). This model postulated that the ion gradients across biological membranes and the ion transporters engage in a self-sustained feedback loop. The most convincing results in support of this model came from studies in the fruit fly Drosophila melanogaster, where electrical silencing of the pacemaker neurones causes complete loss of behavioral and transcriptional rhythmicity in constant darkness (Nitabach et al. 2002). However, data from other organisms did not prove sufficiently compelling to attribute a central role for the membrane model in the clock mechanism (Nitabach et al. 2005). It is interesting to note that a recent study showed the presence of circadian oscillations in magnesium (Mg2+) and potassium (K+) levels in cultured human cells and O. tauri under constant conditions. The latter was shown to show the rhythms even under conditions of transcriptional arrest (Feeney et al. 2016). Although the authors did not relate their findings to the membrane model, it is plausible that a similar mechanism operates to generate these rhythms, at least in O. tauri.
A series of experiments performed in red blood cells (RBCs) in the late 1970s yielded rather controversial results, mostly because of the lack of an appropriate experimental readout. In 1976, Cornelius and Rensing showed rhythms in Mg2+-dependent ATPase activity in the membranes of RBCs kept in vitro (Cornelius and Rensing 1976). These results were later challenged by several studies failing to replicate the observed oscillations in enzymatic activities (Mabood et al. 1978; Schrader and Holzapfel 1980). Nevertheless, in 1985, it was shown that human platelets incubated in vitro display oscillations in the levels of the low-molecular weight thiol, glutathione (GSH) (Radha et al. 1985). In the same year, work in the single-cell flagellate, Euglena, proposed an explanation for the overt rhythms in the nicotine adenine dinucleotides, NAD+ and NADP+, based on a purely nontranscriptional feedback loop. In this model, an increase in the levels of NAD+ drives Ca2+ efflux from the mitochondria, which activates calmodulin in the cytoplasm. The activated Ca2+-calmodulin complex, in turn, activates NAD+ kinase and inhibits NADP+ phosphatase, thus generating a decrease in NAD+ via its conversion to the phosphorylated form (Goto et al. 1985). Indeed, it is well established that the daily oscillations in Ca2+ levels are not a mere readout of clock functions but constitute an integral part of the timekeeping fundamental mechanism (O’Neill and Reddy 2012).
In the late 1980s and early 1990s, experiments in the unicellular dinoflagellate, Gonyaulax polyedra (Lingulodinium polyedrum), showed that the bioluminescence rhythms shown by this organism are transcriptionally independent and are controlled at the translational level (Morse et al. 1989). Later, it was also shown that, in Gonyaulax, the rhythms in bioluminescence and phototaxis are powered by two independent mechanisms as the phasing of the two oscillations can be segregated using different wavelengths of light (Roenneberg and Morse 1993; Morse et al. 1994). These studies provided the first example of the coexistence of multiple independent oscillators in the same cell, which was later found to be the case for other unicellular organisms such as Neurospora crassa and S. elongatus (Bell-Pedersen et al. 2005).
In the 1990s, experiments with the marine mollusc, Bulla gouldiana, were used to test the contribution of transcription and translation to circadian rhythmicity. The optic nerves in this organism show circadian oscillations in action potential firing, which can be recorded both in vivo and in explanted eye cultures. By using pharmacological inhibition of transcription and translation, it was shown that rhythmicity in this model system is dependent on the two processes but only during a certain time of the day (in a phase-dependent manner).
A significant drawback of all these studies is that they lacked conclusive mechanistic proof. If the nucleus is not essential for maintaining cellular rhythmicity, then what constitutes the oscillator? The first compelling evidence for an autonomous nontranscriptional oscillator came in the early 2000s from studies in the cyanobacteria S. elongatus, the simplest organism known to show daily oscillations. In cyanobacteria, three genes (kaiA, kaiB, kaiC) and their protein products were shown to constitute the core oscillator that drives the rhythmic transcription of the whole genome (Nishiwaki et al. 2000; Nakajima et al. 2005; Tomita et al. 2005). In this three-component system, KaiC displays autokinase and autophosphatase activities; KaiA stimulates the kinase and inhibits the phosphatase activities of KaiC, whereas KaiB opposes the action of KaiA. The complex dynamic interactions between these three proteins result in rhythms in the phosphorylation state of KaiC, which, in turn, drives rhythmic genome-wide transcriptional changes, including that of the kaiBC operon. Later, it was shown that the rhythmic transcription of the kaiBC operon was not required for the oscillations in the phosphorylation of KaiC, suggesting that the cyanobacterial timekeeping mechanism does not use a transcription–translation feedback loop, but rather a purely nontranscriptional mechanism (Tomita et al. 2005). In a series of elegant biochemical experiments, Nakajima and colleagues showed that the 24-h cycles of alternating phosphorylation and dephosphorylation of the KaiC protein could be reconstituted in vitro using a simple four-component system consisting of the three Kai proteins and ATP (Nakajima et al. 2005). Although the Kai proteins are not conserved across taxa, these results provided a novel paradigm in circadian timekeeping; a purely nontranscriptional process acts as the primary oscillator, and this biochemical cycle drives rhythms in gene expression.
Despite the general skepticism in the field that nontranscriptional oscillators are compatible only with “simple” organisms, such as cyanobacteria, the search for an evolutionary conserved nontranscriptional oscillator continued. In 2011, this search came to an end with the discovery of the circadian redox cycles in the antioxidant peroxiredoxin (PRDX) proteins (O’Neill and Reddy 2011; O’Neill et al. 2011). These rhythms were found to occur in the anucleate human RBCs and in the unicellular alga O. tauri, even during transcriptional arrest. Moreover, these oscillations showed all the key characteristics of circadian clocks. But perhaps the most exciting finding came a year later when it was shown that, in contrast to the divergent evolution of the TTFL in different organisms, PRDX redox oscillations are conserved across all domains of life (Edgar et al. 2012). The high phylogenetic conservation of the PRDX oscillations, together with the fact that they likely evolved to counteract oxidative stress, suggested that this nontranscriptional timekeeping mechanism might be as ancient as the first aerobic organisms (O’Neill and Reddy 2011; O’Neill et al. 2011; Edgar et al. 2012). These findings revived the interest in metabolic oscillators as cellular timekeeping mechanisms and has forced the reappraisal of the gene-centric model of the clockwork.
Shortly after the discovery of the redox cycle in PRDXs, it was shown that, in undifferentiated mouse embryonic stem cells (mESCs), rhythms in the uptake of the glucose analog 2-deoxyglucose (2-DG) precede the development of a functional TTFL (Paulose et al. 2012). Even more interesting is the fact that these apparently “clockless” cells showed rhythms in the transcript of the glucose transporter, mGlut8. This observation implied that a putative metabolic oscillator might drive rhythmic expression of certain genes, analogous to the Kai oscillator.
PEROXIREDOXINS: GUARDIANS AGAINST OXIDATIVE STRESS, REDOX SIGNALING HUBS, AND CELLULAR TIMEKEEPERS
Peroxiredoxins Are the Major Target of H2O2 in the Cell
PRDXs are a large and evolutionarily conserved family of thiol peroxidases that serve to maintain the cellular redox tone by keeping the levels of reactive oxygen and nitrogen species (ROS/RNS) in check (Rhee 2016). PRDXs are also increasingly being recognized for their role in integrating redox signaling into cellular physiology by acting as peroxide sensors (Netto and Antunes 2016). The hallmark feature of this family of antioxidant proteins is a conserved cysteine residue in the active site (also termed peroxidatic cysteine, CysP), which displays unprecedentedly high reactivity toward biological peroxides. The PRDXs owe their incredible specificity and reactivity toward H2O2 to the highly conserved active site. This not only activates the catalytic CysP in the reactive thiolate form but also stabilizes the transition state during catalysis (Perkins et al. 2015). This specificity dictates remarkably high reaction rates on the order of 107 to 109 m−1sec−1, which make PRDXs the preferential targets for H2O2 in the cell (Randall et al. 2013). Interestingly, the specificity of PRDXs to H2O2 is also reflected in their relatively poor affinity toward other electrophiles, such as iodoacetamide (Peskin et al. 2007). Although the main substrate of PRDXs is H2O2, some members of the family react preferentially with peroxynitrite and lipid peroxide species (Fisher 2011; Knoops et al. 2011).
During catalysis, the reactive cysteine performs a nucleophilic attack on one of the oxygens in the H2O2 molecule, which leads to the oxidation of the CysP sulfhydryl (-SH) to a sulfenic acid (-SOH) intermediate. Although there are different classification systems that subdivide this diverse family of peroxidases (Poole and Nelson 2016), the most well-known criterion is the mechanism by which the catalytic intermediate is resolved, which groups the PRDXs into typical 2-Cys, atypical 2-Cys, and 1-Cys (Rhee et al. 2001). In the typical 2-Cys PRDXs, the CysP reacts with another conserved cysteine residue (termed the resolving cysteine, CysR) residing on a second PRDX monomer to form an intermolecular disulfide bond. The PRDXs from this subgroup exist as noncovalent homodimers in a head-to-tail configuration, and thus the CysP of one PRDX molecule is ideally aligned with the CysR of the other monomer. In the atypical 2-Cys PRDX class, the resolving cysteine is positioned on the same PRDX molecule and thus the reaction between CysP and CysR results in an intramolecular disulfide bond. In the third group, the 1-Cys PRDXs, the CysP does not rely on a partnering cysteine, and the resolving power originates from low-molecular-weight reductants such as ascorbate or GSH (Kang et al. 1998; Manevich et al. 2004). Mammalian cells express four typical 2-Cys PRDXs (PRDX1-4), one atypical (PRDX5), and a single 1-Cys PRDX (PRDX6) (Rhee et al. 2001). The different isoforms display broad cellular distribution with PRDX 1, 2, and 6 being exclusively nucleocytoplasmic, PRDX3 located in the mitochondrion, PRDX4 residing in the endoplasmic reticulum (ER), and PRDX5 being present in the mitochondrion, peroxisomes, and cytosol (Park et al. 2014).
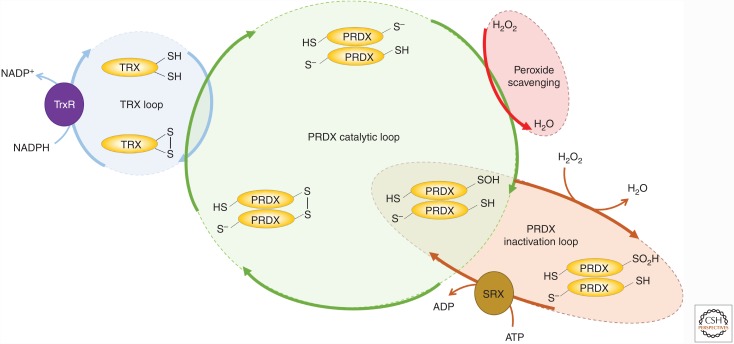
When the reactive cysteines of the 2-Cys PRDXs are in the disulfide form, the proteins are inactive and are recycled by the thioredoxin (TRX) system. The redox-active cysteine couple in TRX extracts the disulfide bond from the PRDXs via a thiol/disulfide exchange mechanism and transfers it to the flavoprotein TRX reductase (TrxR) (Chae et al. 1994). The oxidized TrxR utilizes a charge relay system to transfer electrons from nicotinamide adenine dinucleotide phosphate (NADPH) to the redox-active cofactor flavin adenine dinucleotide (FADH) and finally to the reactive cysteine couple in the active site of the reductase. Thus, the TRX system (including the PRDXs) acts to channel electrons from NADPH to H2O2, reducing the latter to water (Fig. 2).
Figure 2.
Peroxiredoxins (PRDXs) are the primary H2O2 scavengers. The catalytic loop of 2-Cys PRDXs begins with the reaction between the reactive peroxidatic cysteine residues (CysP) and an incoming peroxide molecule (H2O2). This reaction results in the oxidation of CysP to a sulfenic acid (-SOH) intermediate and the detoxification of H2O2 to water. The sulfenic acid intermediate is relatively unstable and can continue its reaction route either via the catalytic or the inactivation loop. In the first case, the sulfenic acid intermediate reacts with another conserved residue, termed the resolving cysteine (CysR), originating from a second PRDX monomer to form an intramolecular disulfide bond. The thioredoxin (TRX) system acts to recycle the disulfide form of the enzyme via the action of TRX and TrxR, with electrons originating from nicotinamide adenine dinucleotide phosphate (NADPH). Occasionally, the sulfenic acid intermediate reacts with a second peroxide molecule, which results in the hyperoxidation to sulfinic acid (SO2H). This form of the enzyme is slowly recycled back to the active thiolate form by the enzyme sulfiredoxin (SRX) in an ATP-consuming reaction. The inactivation loop is considerably slower than the catalytic loop, which allows the accumulation of overoxidized PRDXs. The proportion of PRDX molecules entering the two pathways is dictated by intrinsic factors (e.g., structural characteristics) and dynamic parameters (the local H2O2 concentration).
In 99.3% of the time under unstressed conditions, the catalytic cycle of the PRDXs proceeds via the above-described fast catalytic loop (Perkins et al. 2015). However, a rather unusual characteristic of the PRDXs is that the CysP sulfenic acid intermediate can undergo further oxidation, termed over/hyperoxidation, to form a sulfinic acid (-SO2H) (Rhee 2016). This form of the enzyme is catalytically inactive and is slowly regenerated back to the active sulfhydryl form via an ATP-consuming reaction catalyzed by the only known cellular sulfinic acid reductase, sulfiredoxin (SRX) (Biteau et al. 2003; Woo et al. 2003b; Rhee et al. 2007). The slow reaction rates of the recycling reaction allow the hyperoxidized species to persist in the cell on a timescale of hours, thus suggesting a functional role for this form of the enzyme. In response to increased ROS levels or exogenous application of H2O2, the relative proportion of the hyperoxidized species increases and is frequently used as an indirect measure of oxidative stress (Poynton and Hampton 2014). Initially, the concept that an enzyme is inactivated by an increase in the levels of its substrate seemed counterintuitive. However, soon it became apparent that this paradox is, in fact, an evolutionary adaptation that allows the PRDX proteins to fulfill a broad range of cellular functions in signal transduction networks (Park et al. 2014) as molecular chaperones (Jang et al. 2004; Barranco-Medina et al. 2009) and as intermediates between redox metabolism and the circadian clock. The reason for the hyperoxidation lies in the structural rearrangement that is required for disulfide bond formation (Wood et al. 2003). This reconfiguration allows the solvent exposure of the otherwise shielded sulfenic acid intermediate and therefore the occasional further oxidation. Therefore, the resolution reaction of the sulfenic acid intermediate is in equilibrium between disulfide bond formation and hyperoxidation, and the factors that dictate the direction include intrinsic biochemical characteristics, such as the rate of the unfolding and dynamic parameters, such as the local H2O2 concentration.
Circadian Oscillations in the S-Sulfinylation of the PRDXs
The story of the identification of the circadian hyperoxidation cycles in PRDXs began in 2006 when a global proteomics study identified that up to 20% of the mouse liver proteins show circadian oscillations (Reddy et al. 2006). This result was interesting from a global perspective but what was even more fascinating was that many of the oscillating proteins did not display rhythms in the corresponding transcripts. The study utilized two-dimensional difference gel electrophoresis (2D-DiGE), which allows the segregation of proteins carrying posttranslational modifications (PTMs) from their unmodified counterparts. On examination of the gels, the authors identified two spots corresponding to different forms of PRDX6. The two forms of the protein displayed antiphasic oscillations, which indicated that PRDX6 is rhythmically modified. Initially it was suggested that the forms could represent different phosphorylation states of PRDX6, mostly because of the prevalence of this form of posttranslational regulation in the clockwork. However, around the same time, a series of studies from the Rhee laboratory identified the mechanism of reversible inactivation of PRDXs via hyperoxidation (Yang et al. 2002). More importantly, taking advantage of the fact that the S-sulfinylation form of the enzyme is relatively stable and persists for several hours in vivo, the authors developed an antibody directed specifically against this form of the enzymes (Woo et al. 2003a). The subsequent turn of events can only be described with the words “applying the right tool in the right model systems, asking the right questions.”
In 2011, it was shown that two of the most powerful models for investigating nontranscriptional oscillations—the anucleate RBCs and the photosynthetic alga, O. tauri—show robust circadian oscillations in the S-sulfinylated form of PRDXs. Importantly, the two model systems allow the addressing of two critical issues: (1) do nontranscriptional rhythms occur in the absence of a nucleus, and (2) do these rhythms persist when transcription is stalled in nucleated systems?
The first question was answered by assaying PRDXs hyperoxidation in RBCs under constant environmental conditions (O’Neill and Reddy 2011). Indeed, the results from these experiments showed that the PRDX redox oscillations persisted for three full cycles under constant conditions and were unaffected by incubation at a lower free-running temperature. Even more strikingly, these oscillations could be synchronized by temperature cycles, whereby two groups of RBCs entrained in the opposite temperature cycles displayed antiphasic PRDX oscillations when subsequently released into free running conditions. Because RBCs lack mitochondria, the auto-oxidation of hemoglobin, which occurs naturally in these cells, was proposed as the primary source of H2O2. Consistently, the hemoglobin tetramer-to-dimer ratio was also seen to oscillate in RBCs incubated in vitro. However, one of the most important findings was that the oscillations of PRDX hyperoxidation and hemoglobin oligomerization were paralleled by rhythms in the ratio of the reduced redox cofactors NADH/NADPH as well as ATP. These results indicated that the redox/energy metabolism in RBCs shows self-sustained circadian oscillations without any guidance from the transcription.
Although these initial studies strongly suggested the role of hemoglobin in the generation of the rhythmic oxidation of the PRDXs, this was unequivocally shown a few years later by work from the Rhee laboratory (Cho et al. 2014). Also, contrary to the expectations that SRX plays an important role in the generation of the oscillations, srx-null mice displayed essential normal PRDX rhythms, and the inactivated form was shown to be subjected to degradation by the 20S proteasome. This might appear counterintuitive considering that RBCs have a limited amount of PRDXs owing to the lack of transcription and translation. However, less than 1% of total PRDX2 (the major PRDX isoform in RBCs) undergoes daily oscillation, resulting in ∼30% loss of PRDX2 during the life span (30–40 days) of mouse RBCs (Cho et al. 2014). Therefore, it is possible that the proteasome-mediated oscillation mechanism has evolved for energy conservation because the SRX catalyzed reaction requires ATP.
The exact mechanism that generates the 24-h period of the oscillations in RBCs is still an open question. For instance, are the proteasomal degradation and hemoglobin auto-oxidation required in a phase-dependent manner? Also, because the reduction of the disulfide and hyperoxidized forms of PRDXs are dependent on NADPH and ATP, respectively, is it possible that the rhythmic accumulation of the inactivated enzyme reflects the availability of the two metabolites and therefore of primary metabolism? A recent study in RBCs of superoxide dismutase-1 (Sod1)-deficient mice indicated that, under conditions of elevated ROS, the cycle in PRDXs hyperoxidation is diminished (Homma et al. 2015). This clearly indicates that ROS homeostasis is important for the generation of circadian cycles in PRDX hyperoxidation, but further investigations are required to unravel the oscillation-generating mechanism under normal conditions.
The second question, concerning the possible role of transcription in the hyperoxidation rhythms of PRDXs in nucleated systems was answered by incubating O. tauri under conditions of transcriptional arrest (O’Neill et al. 2011). It was unequivocally shown that the PRDX oscillations persist under such conditions, which indicated that, in this model system, normally expressing a functional TTFL, the nontranscriptional rhythms in PRDX hyperoxidation are independent of the transcriptional clock. As already mentioned, the phase of the transcriptional clock in O. tauri does not reset after alleviation of the transcriptional arrest. This observation indicates that the PRDX hyperoxidation cycles, or a putative metabolic oscillator that they report on, not only act as the primary timekeeping mechanism during periods of attenuated transcription but that can also relay timing information back to the transcriptional clock (O’Neill et al. 2011).
The icing on the cake of these series of discoveries was the demonstration that the PRDX hyperoxidation cycles occur in organisms from all three domains of life, including archaea, eubacteria, plants, worms, flies, mice, and man (Edgar et al. 2012; Olmedo et al. 2012). Importantly, these rhythms were found to be independent of the TTFL because clock mutant organisms display essentially normal rhythms (Edgar et al. 2012). This not only cemented the role of the PRDX oscillations in the clockwork but also challenged the ideas about the evolutionary origins of circadian clocks.
If the genetic clock has been invented several times in the evolutionary history, then it follows that the primary timekeeping mechanism might be encoded by the PRDX oscillations or the process they report on. In this scenario, the transcriptional clock might have evolved to meet the specific requirements of a given organism or system and rather fulfills species-specific functions in the adaptation response to the distinct environmental niche, whereas PRDX/metabolic oscillations might represent a primary timekeeping mechanism. This is supported by studies showing tissue-specific effects of clock function. For instance, disruption of the molecular clockwork in the liver results in fasting-induced hypoglycemia, whereas the same intervention in the pancreas impairs glucose tolerance (Marcheva et al. 2010; Peek et al. 2013). In this respect, analogies such as “time-keeping” versus “time-telling” mechanisms would be appropriate (Rey and Reddy 2015). In simple terms, in the absence of a metabolic/redox oscillator, the cells/organisms would not be able to keep track of time. On the other hand, in the absence of a transcriptional oscillator, the cells/organisms would not be able to make use of the timing information. In a “basic” system such as RBCs, the metabolic oscillator is both the “time-keeping” and “time-telling” mechanism with the only task of organizing the antioxidant defense around the clock. In more “complex” systems that coordinate a variety of processes, another layer of complexity needs to be added, and this is enabled by the diverse cyclic transcriptional programs fulfilled by the TTFL.
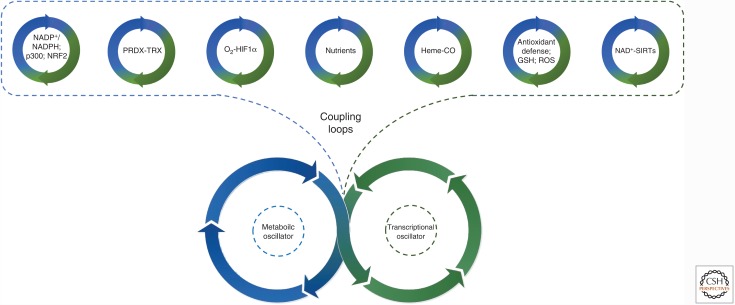
There are many questions that remain to be answered concerning both the mechanistic and functional aspects of the oscillations in the S-sulfinylation of PRDXs. Although it seems unlikely that the oscillations in PRDX hyperoxidation serve as the timekeeping mechanism per se, it is highly possible that they are an important cog in the clockwork, potentially acting as a coupling agent between the metabolic and transcriptional oscillators. How can PRDXs fulfill this role? The last decade has delivered a plethora of examples supporting the important role of this family of peroxidases in integrating ROS into cellular signaling. The subsequent section will outline the different signaling mechanisms and experimental evidence that argue in favor of the functional role of PRDXs in the clockwork.
PRDXs as Mediators of Peroxide Signaling
Although H2O2 is now regarded as a bona fide physiological second messenger (Rhee 2006), the exact details of the signal-transduction mechanism by which the oxidant exerts its effects are still largely unknown. There are three principal issues associated with peroxide signaling that has complicated the identification of novel regulatory targets: specificity, reactivity, and accessibility. The specificity in H2O2 signaling cannot be achieved via molecular recognition because the oxidant has a very simple chemical structure. Also, the unstable nature of H2O2 prevents its reversible interaction with protein targets. It follows that the selectivity must be based on another physical property, such as hierarchy in the reaction rates (Winterbourn and Hampton 2008). This logic unveils yet another problem—although the reaction between H2O2 and cellular thiols is highly thermodynamically favorable, the extremely effective cellular peroxidases display unprecedented kinetic superiority over other proteins. Kinetic measurements indicate that most cellular thiols show moderate reaction rates with H2O2 on the order of 1–10 m−1sec−1, which are several orders of magnitude lower than the ones displayed by peroxidases such as PRDXs and glutathione peroxidase (GPx) (∼107–109 m−1sec−1) (Randall et al. 2013). Moreover, the cellular peroxidases are among the most abundant proteins in the cell, with PRDXs making up to 1%–2% of the total mass in most cells (Chae et al. 1999), which indicates that the reaction between H2O2 and other cellular thiols is a highly unlikely event. Even though PRDXs act as the primary target of H2O2 in the cell, numerous examples in the literature have shown that other proteins undergo reversible oxidation in response to the oxidant. How does this occur in the cell? It appears that there is no clear-cut answer, but the direct oxidation of target proteins by H2O2 seems the least possible explanation (Randall et al. 2013). However, there are several different, but not mutually exclusive, theories that offer plausible and, more importantly, experimentally validated solutions to this apparent paradox. Unsurprisingly, all of these mechanistic models revolve around the PRDX system (Fig. 3).
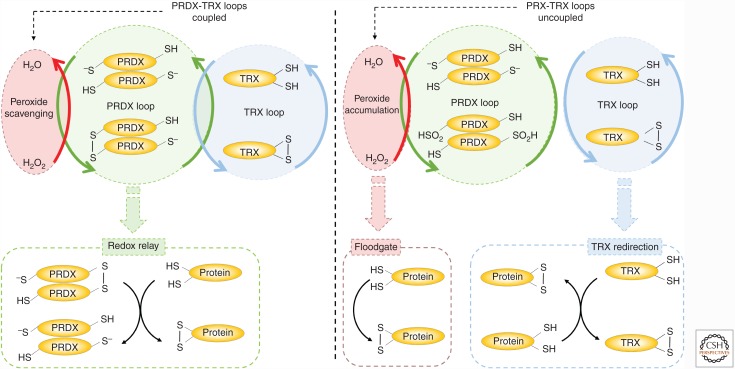
Figure 3.
Peroxiredoxins (PRDXs) are mediators of reactive oxygen species (ROS) signaling. Schematic representation of the three main models for PRDXs signaling. The redox relay model: In this model, the oxidized PRDXs transfer oxidative equivalents to protein targets. This transfer reaction proceeds via the formation of a mixed disulfide intermediate between PRDXs and the client protein. The floodgate model: This model postulates that the inactivation of PRDXs via overoxidation allows the local accumulation of H2O2, which can then react with other cellular targets. The thioredoxin (TRX) redirection model: This model proposes that the inactivation of PRDXs allows the accumulation of active TRX, which can then reduce other cellular targets. Note, that the redox relay model could occur under normal operational conditions when the PRDX and TRX loops are coupled. The floodgate and TRX redirection models are dependent on the uncoupling of the PRDX-TRX loop via the overoxidation of the peroxidases.
Signaling via Hyperoxidation
The structural peculiarity of the carboxy-terminal tail of 2-Cys PRDXs, which enables the S-sulfinylation of the active site cysteine, together with the slow rates of regeneration of the active enzymatic form by SRX, led some authors to suggest that the controlled inactivation of the peroxidases allows H2O2 to act as a second messenger. This theory was coined the floodgate hypothesis, and was later supported by the finding that the hyperoxidation of mitochondrial PRDX3 is essential for the circadian rhythms in adrenal steroidogenesis (Wood et al. 2003; Kil et al. 2012). Adrenocorticotropic hormone (ACTH)-induced corticosterone (CS) synthesis occurs with a concomitant increase in H2O2, driven by cytochrome P450 in the mitochondria. The increase in the levels of the oxidant results in the inactivation of PRDX3 by S-sulfinylation and leads to the accumulation of H2O2 in mitochondria and its overflow into the cytosol, where it triggers the activation of p38 mitogen-activated protein kinase and consequently the down-regulation of both CS synthesis and H2O2 production. Also, ACTH stimulates the expression of SRX, which translocates to the mitochondrial matrix and mediates the regeneration of PRDX3, thus generating a regulatory feedback loop.
A daily oscillation of hyperoxidized PRDX3 was also detected in brown adipose and heart tissues, where a high level of fat metabolism is accompanied by H2O2 production (Kil et al. 2015). In addition, although only a small proportion of SRX was detected in mitochondria, the amount of mitochondrial SRX also undergoes daily oscillation in adrenal gland, as well as in brown adipose and heart tissues, with this oscillation being antiphasic relative to that of hyperoxidized PRDX3. These observations suggest that the mitochondrial abundance of SRX is likely the key determinant of PRDX3 hyperoxidation and, consequently, also that of the timing and extent of mitochondrial H2O2 release. The H2O2 released from mitochondria promotes the formation of a disulfide-linked complex between SRX and heat-shock protein 90 (HSP90), and the resulting SRX–S–S–HSP90 complex is imported into mitochondria by the translocase of the outer membrane (TOM) complex (Kil et al. 2015). A cochaperone of HSP90, FK506-binding protein (FKBP), is also required for the import process. In this context, it is interesting to note that analysis of gene regulatory networks associated with mammalian circadian rhythms revealed that HSP90-FKBP complexes are central to the control of circadian gene expression by diverse environmental signals (Yan et al. 2008). The imported SRX binds tightly to sulfinylated PRDX3 and reduces it. As the amount of sulfinylated PRDX3 then declines, SRX becomes vulnerable to degradation by the protease Lon, resulting in its down-regulation to basal levels and the consequent onset of the next round of sufinylated PRDX3 accumulation and H2O2 release.
Although this is the only existing example in support of the floodgate hypothesis, hyperoxidation has been found to enable other modes of signaling. Analogous to the floodgate hypothesis, some investigators have proposed that when H2O2 levels exceed the antioxidant capacity of the cell, S-sulfinylation acts as a molecular switch that activates the chaperone-like properties of PRDXs at the expense of their peroxidase function (Jang et al. 2004; Barranco-Medina et al. 2009). The important physiological role of the chaperone functions of 2-Cys PRDXs was recently shown in yeast. Tsa1, the major 2-Cys PRDX in Saccharomyces cerevisiae, was shown to be essential for the recruitment of the heat-shock chaperones Hsp70/Hsp104 to protein aggregates, thus preventing the accumulation of aberrantly folded protein species in this organism (Hanzén et al. 2016). Intriguingly, the recruitment of Hsp70/Hsp104 to protein aggregates resulting from aging, but not heat stress, was shown to depend on the hyperoxidation of Tsa1, whereas the disaggregation of the misfolded proteins only occurred after the reduction of the S-sulfinylated form by SRX. Moreover, the overexpression of Tsa1 led to an increase in life span in an Hsp70-dependent manner. Overall, these findings show the complex nature of the PRDX system and underline the importance of this class of enzyme for the maintenance of cellular integrity.
The third mechanism by which hyperoxidation might lead to signaling was recently shown in yeast. The inactivation of PRDXs allows the accumulation of reduced (active) TRX, which could, in turn, target other cellular proteins that have undergone reversible oxidation. This mechanism was found to be essential for the inactivation of the yeast AP-1-like transcription factor, Pap1, and the activation of the enzyme methionine sulfoxide reductase (Mxr1) (Bozonet et al. 2005; Vivancos et al. 2005; Day et al. 2012). Also, expression of a hyperoxidation-resistant form of the yeast 2-Cys PRDX, Tpx1, led to a significant decrease in cell survival after exposure to H2O2 (Day et al. 2012). In support of the role of S-sulfinylation as a switch to allow repair of reversibly oxidized proteins, the deletion of PRDX2 in Caenorhabditis elegans confers increased resistance to oxidative stress (Oláhová et al. 2008). However, the nematodes also displayed shortened life span, which underlines the important role of the PRDXs for longevity.
Interestingly, in vitro studies have revealed that PRDX2, which is located in the cytosol, is more sensitive to hyperoxidation than mitochondrial-located PRDX3 owing to the 10-fold slower formation of a disulfide bond with its resolving cysteine (Haynes et al. 2013; Poynton et al. 2016). In addition to the intrinsic biochemical properties of PRDXs, the hyperoxidation is, in fact, dependent on a functional reducing system, which allows the enzymes to spend more time in the sulfenic acid form. Consistently, although the ER-localized PRDX4 displays similar sensitivity to hyperoxidation to the cytosolic forms in vitro, an insignificant fraction of the enzyme becomes S-sulfinylated in vivo. This is attributed to the low abundance of disulfide recycling systems in this organelle (Cao et al. 2014). The combination of different sensitivities toward hyperoxidation and the modulation of the activity of the disulfide reductase systems allows the PRDXs to be dynamically regulated to meet the specific demands of the different cellular compartments (Cox et al. 2009; Tavender et al. 2010) as well as different cell types (Low et al. 2007) and tissues (Oláhová et al. 2008; Kil et al. 2012). In erythrocytes, for instance, where PRDX2 is one of the most abundant proteins, it was shown that the peroxidase acts mostly to detoxify low-grade H2O2 levels; treatment with 5 µm H2O2 of 5 × 109 erythrocytes dm−3 for 1 min is sufficient to convert the whole PRDX pool into the disulfide-linked form (Low et al. 2007). Also, the reduction by TRX was seen to be considerably slow, occurring gradually over a period of 20 min. This mechanism probably evolved to protect PRDX2 from hyperoxidation because it has been shown recently that, in erythrocytes, this form of the enzyme is targeted for proteasomal degradation rather than recycled by SRX (Cho et al. 2014).
Finally, it should be noted that the regulation of PRDXs could be achieved via other oxidative modifications, such as S-nitrosylation (Fang et al. 2007; Engelman et al. 2013; Chung et al. 2017) and S-glutathionylation (Peskin et al. 2016). It also seems that, in addition to being regulators of kinases, the activity of the PRDXs is modulated by phosphorylation. For example, controlled inactivation of PRDX1 by CDK1 allows the pericentrosomal buildup of H2O2 with potential consequences for protein phosphatases such as Cdc14B (Lim et al. 2015).
Beyond Hyperoxidation—Signaling via Redox Relay Mechanism
An alternative to the floodgate hypothesis is that the fast reaction kinetics of PRDXs with H2O2 allow them to act as sensors and transducers of peroxide signaling via a thiol/disulfide exchange mechanism, also termed redox relay. In this model, PRDXs are first oxidized by H2O2, then form mixed disulfide intermediates with a client protein, and finally transfer the oxidative equivalents, thereby oxidizing their binding partner. This could either occur straight from the sulfenic acid form or from the disulfide form of 2-Cys PRDXs (Sobotta et al. 2014). This model solves the problems related to H2O2 signaling outlined at the beginning of this section. Namely, specificity is provided by the direct protein–protein interactions, the relatively nonreactive H2O2 molecule is activated by its conversion to the much more reactive sulfenic acid, and, finally, the competition is eliminated because now PRDXs themselves act as second messengers. The pioneering work supporting this model was performed in the yeast S. cerevisiae, where the thiol peroxidase Orp1 was shown to form mixed disulfides with the AP-1-like transcription factor Yap1, which ultimately leads to the activation of Yap1 via formation of an intramolecular disulfide (Delaunay et al. 2002). Another example from yeast is the H2O2-induced activation of the p38/JNK homolog, Sty1, by the Schizosaccharomyces pombe 2-Cys PRDX, Tpx1 (Veal et al. 2004). Since these initial discoveries, participation in such signaling events has been described for all four mammalian typical 2-Cys PRDXs. For instance, PRDX1 was shown to transfer oxidative equivalents to the kinase ASK1, which results in the formation of active homodimers and the subsequent phosphorylation of p38 (Jarvis et al. 2012). Interestingly, this result adds an important detail to the mechanism of steroidogenesis in the mouse adrenal cortex described by Kil et al. (2012). It seems likely that the inactivation of PRDX3 in the mitochondria allows the oxidation of PRDX1 in the cytosol, which then acts to activate ASK1 and in turn p38, which then feeds back to induce the SRX-mediated recycling of PRDX3 (Jarvis et al. 2012; Kil et al. 2012). This also strongly suggests that the movement of H2O2 within the cell is mediated by the cellular peroxidases rather than by simple diffusion. More recently, a redox relay between PRDX2 and the oxidative stress-related transcription factor STAT3 was described (Sobotta et al. 2014). In this work, it was shown that PRDX2 transfers oxidative equivalents to STAT3, whereby STAT3 forms transcriptionally inactive oligomers. In a similar fashion, PRDX2 also determines the oxidation status of the human protein deglycase DJ-1 (Fernandez-Caggiano et al. 2016). Of note, PRDX2 nuclear levels oscillate in HaCaT keratinocytes and overexpression of the protein in the nucleus leads to damping of the oscillations in BMAL1 (Avitabile et al. 2014; Ranieri et al. 2015). This clearly indicates that the redox environment in the nucleus affects the transcriptional oscillator and suggests that PRDX2 might be involved in this process. Finally, the ER-resident PRDX4 was shown to be involved in the oxidative folding of proteins by transferring oxidative equivalents to several members of the protein disulfide isomerase (PDI) family (Tavender et al. 2010). The interaction between members of the PDI and PRDX family was also observed in the green alga Chlamydomonas reinhardtii, and, more interestingly, the complex between the two proteins was detected only during the night phase (Filonova et al. 2013).
The PRDXs have often been described as “the major sinks for H2O2.” Considering the data presented above, this analogy is perhaps not the most accurate one. Our notion of these conserved thiol-dependent antioxidant proteins has evolved over the past decade, and now they can be rather classified as major cell-signaling players that integrate redox signals into cellular physiology. Whether PRDXs are actively participating in the cellular timekeeping mechanism remains to be proven but, considering the important role of the peroxidases in cell signaling, this idea seems highly plausible.
REDOX AROUND THE CLOCK
Overt Rhythms in Cellular Redox Couples
Oscillations in the Cellular Nicotinamide Adenine Dinucleotide Redox Couples
Nicotinamide adenine dinucleotide (NAD+) and its phosphorylated form NAD phosphate (NADP+) are the two major hydride ion cofactors, which enable approximately 2000 cellular reactions (Opitz and Heiland 2015). NAD+ could either be synthesized de novo from tryptophan or salvaged using preformed compounds such as niacin. NADP+ is synthesized directly via the phosphorylation of NAD+, catalyzed by NAD+ kinase, and can be converted back to NAD+ via the action of NADP+ phosphatase. Considering that the NADP+/NADPH couple is maintained in the reduced state and serves mainly to shuffle hydride ions, this redox couple could be regarded as the major reducing power of the cell. On the other hand, the metabolism of NAD+ is much more complex. This is because, in addition to redox reactions, NAD+ also participates as an enzyme cofactor in mono- and poly(ADP-ribosylation) as well as NAD+-dependent protein deacetylation. These so-called NAD+-consuming reactions convert NAD+ into nicotinamide mononucleotide, which can be recycled back to NAD+ via the salvage pathway.
In the fungus N. crassa, the NAD+/NADH ratio was shown to resonate with the rhythms in conidiation (Brody and Harris 1973). Similar oscillations were observed for NADP+/NADPH in plant seedlings kept under constant conditions (Wagner and Frosch 1974). Diurnal variation in both redox couples was also observed in rodent liver (Robinson et al. 1981; Kaminsky et al. 1984), although these were assayed under normal feeding regimens and thus might be partially driven by food intake. It is interesting to note that rats adapted to a high-protein diet displayed nearly unchanged oscillations in the cytoplasmic redox couples compared with controls, suggesting that food intake might not be the sole driver of these rhythms. Indeed, oscillations in the levels of NAD(P)H were also observed in RBCs under constant conditions (O’Neill and Reddy 2011) and more recently in human osteosarcoma U2OS cells (Rey et al. 2016). The redox state of the central brain pacemaker, the suprachiasmatic nuclei (SCN) as represented by the ratio between NAD(P)H/FADH, was seen to oscillate (Wang et al. 2012). More recently, with the use of noninvasive methods, it was shown that the NAD/NADH ratio oscillates in vivo in stem cells and HEK293T cells (Stringari et al. 2015; Huang et al. 2016).
Although the rhythms in pyridine dinucleotide redox couples might in part be driven by the transcriptional clock in some of these model systems, the fact that they have also been described to persist in the absence of a nucleus strongly suggests that they might be driven by nontranscriptional mechanisms.
Oscillations in Glutathione
GSH is a tripeptide consisting of l-γ-glutamyl-l-cysteinyl-glycine and constitutes the most abundant low-molecular-weight thiol in eukaryotic cells. As such, GSH is the main thiol buffering system and has often been described as a rheostat of the cellular redox state. In cells, GSH exists in a redox couple with its oxidized form, glutathione disulfide (GSSG). Under unstressed conditions, the GSH/GSSG ratio is kept largely in the reduced state by the flavoprotein, glutathione reductase (GR), which consumes reducing power in the form of NADPH. Owing to its important role in antioxidant defense, GSH has attracted much attention, and consistently its diurnal profile has been assayed in most major organ systems. Oscillations in GSH have been observed in rodent liver (Isaacs and Binkley 1977a,b; Robinson et al. 1981; Farooqui and Ahmed 1984; Bélanger et al. 1991; Neuschwander-Tetri and Rozin 1996; Li et al. 1997; Baydas et al. 2002; Ponce et al. 2012), brain (Farooqui and Ahmed 1984; Baydas et al. 2002; Wang et al. 2012; Kinoshita et al. 2014), lung (Farooqui and Ahmed 1984; Pekovic-Vaughan et al. 2014), spleen (Farooqui and Ahmed 1984), kidney (Farooqui and Ahmed 1984), gut (Li et al. 1997), pancreas (Neuschwander-Tetri and Rozin 1996), stomach (Farooqui and Ahmed 1984), blood plasma (Blanco et al. 2007), and platelets (Radha et al. 1985). Despite the numerous reports on overt rhythms in the levels of GSH, the exact molecular mechanism has not yet been elucidated, and it seems that the regulation might occur in a tissue-specific manner. For example, fasting diminishes GSH oscillations in the liver but not in the pancreas (Neuschwander-Tetri and Rozin 1996) and in the mesencephalon region of the brain (Kinoshita et al. 2014). Interestingly, in the case of the latter, the oscillations in GSH were attributed to a posttranslational mechanism via a microRNA (miRNA), which controls the levels of a neuron-specific membrane transporter for cysteine, the rate-limiting substrate for GSH synthesis. The situation becomes even more complex considering that platelets, which are naturally anucleate, show robust GSH oscillations even when kept in vitro. In stark contrast, Pekovic-Vaughan et al. (2014) found that, in the lung, the transcriptional clock exerts control over GSH metabolism via the transcription factor NRF2. One of the transcriptional targets of NRF2 is glutamate cysteine ligase (GCL), which catalyses the rate-limiting step in GSH synthesis, and thus rhythmic transcription of NRF2 was shown to result in rhythmic GCL transcription. Although nuclear import of NRF2 has been shown to be a redox-regulated process, in the lung, NRF2 displays rhythmic nuclear accumulation under unstressed conditions. A similar mechanism seems to be operating in fruit flies where the circadian rhythmicity in GCL expression and activity, as well as GSH levels, were shown to be directly controlled by the transcriptional clock (Beaver et al. 2012).
It is important to note that the rhythms in GSH translate to daily oscillations in the susceptibility to oxidant insult. Considering that time-dependent protection against oxidative stress has been observed in both liver and the brain (Li et al. 1997; Kinoshita et al. 2014), and that supplementation with N-acetyl-l-cysteine ameliorates the effects of oxidative stress (Kondratov et al. 2009; Berman et al. 2011), the interaction between GSH metabolism and the clock might be more important than previously thought.
Redox Process as Outputs of the Transcriptional Clock
Clock-Mediated Control over the NAMPT-NAD-SIRT Axis
As already described, in addition to its important function in redox metabolism, NAD+ serves as a cofactor for several enzymes, such as the protein deacetylases sirtuins (SIRTs) and poly(ADP-ribose) polymerase 1 (PAPR1). The participation of NAD+ in these reactions results in the hydrolysis of the cofactor to the precursor nicotinamide (NAM). NAD+ can then be regenerated from NAM via a two-step enzymatic pathway termed the NAD+ salvage pathway. The first and rate-limiting step is catalyzed by nicotinamide phosphoribosyltransferase (NAMPT), which converts NAM into NAM mononucleotide (MNM) using ATP and 5-phosphoribosyl 1-pyrophosphate (PRPP) as cofactors. Although NAD+ can also be synthesized de novo from the amino acid tryptophan or directly from diet-derived nicotinic acid (NA), the salvage pathway appears to be the major source of NAD+ in cells (Cantó et al. 2015). Therefore, the discovery that NAMPT is under the direct control of the transcriptional clock and, more importantly, that oscillations in NAMPT generate rhythms in the cellular NAD+ levels not only solidified the connection between the redox system and circadian rhythms, but also indicated that cellular NAD+ metabolism is much more dynamic than previously anticipated (Opitz and Heiland 2015).
The first proof of a mechanistic link between NAD+ metabolism and the circadian clock was the demonstration that the NAD-dependent deacetylase SIRT1 binds to the CLOCK:BMAL1 complex in a circadian fashion and regulates circadian transcriptional programs via the deacetylation of the core clock transcription factors BMAL1 and PER2 and chromatin-associated proteins (Asher et al. 2008; Nakahata et al. 2008). Shortly after, two independent studies established the important role of the cellular clock in controlling the NAMPT-NAD-SIRT axis by showing that the transcriptional oscillator drives the cycling of the NAMPT transcript and protein, which, in turn, generates rhythms in the cellular NAD+ levels (Nakahata et al. 2009; Ramsey et al. 2009). Of note, although fasted animals display NAD+ oscillations in the liver, feeding induces 12-h harmonics in this organ (Ramsey et al. 2009; Peek et al. 2013). The relationship between NAD+ and the circadian clock appears to be reciprocal as deletion of CD38, one of the major NAD+-consuming enzymes in the cell, results in altered behavioral and metabolic rhythms (Sahar et al. 2011). Another NAD+-dependent enzyme, which has been shown to feed back to affect clock function is PARP1. PARP1 was shown to bind to CLOCK and transfer poly (ADP-ribose) moieties, thereby affecting the DNA-binding affinity of the transferrin (TF) and ultimately the phase of the clock (Asher et al. 2010). Of note, the circadian activity of PARP-1 was shown to be independent of a functional clock and rather regulated by feeding. This is in line with the fact that the Km value of PARP1 for NAD+ is relatively low (≈50–59 µM), and thus NAD+ levels are rarely limiting for PARP1 activity (Mendoza-Alvarez and Alvarez-Gonzalez 1993; Cantó et al. 2015). Another way by which PARP-1 can indirectly affect the clock is by limiting the availability of NAD+ for the SIRT enzymes (Cantó et al. 2015). This is supported by the fact that deletion or pharmacological inhibition of PARP-1 results in significantly elevated NAD+ levels and increased SIRT1 activity, indicating that the enzyme is a major NAD+ consumer and in direct competition with the SIRTs for NAD+ availability (Bai et al. 2011). In fact, a recent study used computational simulations to show that increased PARP-1 activity in response to DNA damage might be able to induce SIRT1-dependent clock phase shifts because of competition for limited NAD+ supplies (Luna et al. 2015).
In mammals, there are seven SIRT enzymes (SIRT1-7) that display diverse cellular distribution with SIRT1, SIRT6, and SIRT7 primarily in the nucleus, SIRT3-5 in the mitochondria, and SIRT2 found in the cytoplasm (Cantó et al. 2015). Perhaps, more importantly, the SIRTs display widely different Km values to NAD+, ranging from ∼100 µM (SIRT1/2/4/6) to 1000 µM (SIRT3/5) (Cantó et al. 2015). Indeed, this sensitive range enables the family of deacetylases to perform specific functions in different tissues and cellular compartments, depending on the local daily oscillations in NAD+ levels. Consistently, the intracellular rhythms in NAD+ were shown to modulate the activity of SIRT3, thereby inducing rhythmic cellular respiration through modulation of mitochondrial protein acetylation (Peek et al. 2013). Similarly, SIRT6 was shown to aid the rhythmic recruitment of CLOCK:BMAL1 complexes and another transcription factor SREBP-1 to circadian gene promoters, thereby regulating fatty acid metabolism in the liver (Masri et al. 2014).
Rhythms in Antioxidant Defense Systems
The important roles of the clock for maintaining redox homeostasis have come from three major lines of evidence. First, a high number of studies have shown that various oxidative stress markers, such as DNA damage, lipid peroxidation, and protein carbonylation, display diurnal oscillations (Hardeland et al. 2003; Wilking et al. 2013). This leads to the second line of evidence that comes from studies showing that the rhythms in oxidative stress markers are parallel to oscillations in the levels and activity of a high proportion of antioxidant enzymes, including SOD1, catalase, GPx, and the PRDXs. Finally, studies in clock mutant organisms have shown that the circadian regulation of the antioxidant defense systems is under the control of the transcriptional clock. For example, ROS levels and protein carbonylation display daily oscillations in wide-type flies but not in per01 mutants (Krishnan et al. 2008). Bmal1−/− mice display reduced life span and symptoms of premature aging as well as increased ROS levels in several tissues (Kondratov et al. 2006). Consistently, supplementation with the antioxidant N-acetyl-l-cysteine ameliorates some of the overt effects in these mutant animals (Kondratov et al. 2009). The tau mutant in the Syrian hamster also displays increased levels of oxidative damage, at least in the Harderian gland (Coto-Montes et al. 2001). Mutations in the plant core clock regulator CCA1 affects the transcriptional regulation of many ROS-scavenging enzymes, ROS homeostasis, and the tolerance to oxidative stress (Lai et al. 2012).
It should be noted that in most cases the deletion of the clock gene leads to a general increase in intracellular ROS and oxidative damage. This indicates that the effects might be attributable to noncircadian roles of these transcription factors. For example, Bmal1−/− mice display significantly lower NAD+ and NADH levels compared with wild-type (WT) animals (Peek et al. 2013), and rhythmic expression of Bmal1 was shown to be dispensable for molecular rhythmicity (Liu et al. 2008). Also, in addition to the loss of rhythmicity in response to oxidant treatment, both the arrhythmic per01 and short-period perS mutant flies show higher average levels of lipid peroxidation and carbonylated proteins with aging (Krishnan et al. 2009; Coto-Montes and Hardeland 2010).
Antioxidant Functions of Melatonin
Secreted primarily from the pineal gland, melatonin is involved in the temporal coordination of a plethora of processes, most notably in the control of seasonal physiology (Challet 2015). The antioxidant properties of melatonin were first shown by studies reporting on the in vitro ROS-scavenging properties of the hormone (Ianăş et al. 1991). Since then, the role of melatonin in the antioxidant defense system has been unequivocally shown (Hardeland et al. 2003; Hardeland and Pandi-Perumal 2005; Hardeland 2013). However, the circulating levels of melatonin are too low to account for the observed antioxidant effects of the hormone (Hardeland et al. 2003). In addition, one would expect that if melatonin was to engage in direct ROS scavenging, then the secretion of the hormone would peak during the light phase in diurnal animals to coincide with the maximal locomotor activity. In contrast, melatonin levels peak at the start of the dark phase in both nocturnal and diurnal animals, which suggests that the hormone offers antioxidant support in another form. Several different modes of action have been described for the antioxidant role of melatonin, including up-regulation of the levels of many ROS-scavenging enzymes (e.g., GPx, SOD1), down-regulation of pro-oxidant enzymes (e.g., lipoxygenase, NO synthase), and the activation of antioxidant transcriptional programs via stimulation of nuclear factor (NF)-κB, AP-1, and NRF2 (Luchetti et al. 2010).
Indeed, it is possible that the antioxidant properties of melatonin are a pleiotropic effect and unrelated to its role in circadian and seasonal physiology. However, the fact that melatonin supplementation has been shown to exert positive effects beyond its role in the clock suggests that further investigation is required (Hardeland and Pandi-Perumal 2005).
Redox Control over the Transcriptional Clock
As already alluded to earlier, the connection between the circadian clock and the redox poise is reciprocal and several lines of evidence have shown that the redox state of the cell can influence the function of the molecular clockwork. For example, Sod-1-deficient Neurospora show more robust rhythms in the molecular clockwork (Yoshida et al. 2011). In addition, ROS can modulate the circadian phase and period in the fungi (Gyöngyösi et al. 2013). Consistent with the results in Neurospora, treatment with exogenous oxidants has been shown to reset the phase of the transcriptional oscillator in mammalian cells (Tamaru et al. 2013). In zebrafish, light induces the production of H2O2, which, in turn, acts as a second messenger to transduce the photic signals to the transcriptional oscillator (Hirayama et al. 2007). In cyanobacteria, the light-sensitive protein Ldp-1 functions as a cellular redox sensor, which is capable of transducing redox signals to the core oscillator (Ivleva et al. 2005). In addition, treatment with oxidized quinones resets the cyanobacterial clock, and this effect was also showed using electrochemically controlled extracellular electron transfer, which enables the external modulation of the redox state (Lu et al. 2014). All these studies indicate that the redox state can influence circadian physiology. What are the molecular mechanisms that enable these interactions? Below, we summarize the most well-known examples of the putative mechanistic links between cellular redox and the transcriptional oscillator.
Redox Control over the Positive Elements of the Transcriptional Clock
One of the first indications that the transcriptional oscillator might be able to sense changes in the cellular redox state came from biochemical studies showing that the DNA-binding affinity of CLOCK:BMAL1 and NPAS2:BMAL1 transcriptional complexes can be modulated by varying the NAD(P)+/NAD(P)H ratio in vitro (Rutter et al. 2001). The reduced cofactors were shown to increase binding, whereas their oxidized counterparts exerted an inhibitory effect. Importantly, even subtle changes in the NAD(P)+/NAD(P)H ratio are capable of inducing changes in the binding and dissociation, indicating that these clock transcription factors have the properties of bona fide redox sensors. These findings were later extended by showing that the 1-61 amino-terminal amino acids of the bHLH domain of NPAS2 were sufficient to sense NAD(P)H and that the NPAS2/BMAL1 transcriptional complexes can also sense changes in the cellular pH (Yoshii et al. 2013, 2015). The concentrations of NADH and NADPH used in these studies were considerably higher than the one measured in vivo and whether NPAS2/CLOCK:BMAL1 are capable of sensing redox changes in cells remains an open question (Zhang et al. 2002). However, the direct sensing of changes in the NAD+/NADH ratio has been shown for carboxyl-terminus-binding protein (CtBP), indicating that such a mechanism operates in vivo (Zhang et al. 2002; Fjeld et al. 2003). Indeed, this would also allow the oscillations in NAD+ levels to feed back to the transcriptional clock by directly modulating the transcriptional activity of NPAS2/CLOCK:BMAL1. Indirect evidence for the importance of the nuclear redox state for the functioning of the molecular clockwork came from a recent study showing that the nuclear overexpression of PRDX2 in HaCaT keratinocyte induces perturbations to the transcriptional oscillations of Bmal1 (Ranieri et al. 2015).
As described earlier, another potential way in which oxidative stress might impinge on the molecular clockwork is through the activation of PARP-1 by oxidation-induced DNA damage (Luna et al. 2015). Under such conditions, PARP-1 is expected to consume relatively higher amounts of NAD+ and thus limit the availability of the cofactor for the SIRT deacetylase, which would, in turn, affect clock function.
Redox Control over the Negative Arm of the TTFL
Recently, two independent studies reported on the crystal structure of the mouse PER2:CRY1 complex (Nangle et al. 2014; Schmalen et al. 2014). The structure of the complex between the circadian repressor proteins shows that a zinc ion (Zn2+) binds in the interface between the two proteins and leads to the stabilization of the dimer. Interestingly, Schmalen et al. reported that Zn2+ coordination is governed by the redox state of a regulatory disulfide bond in CRY1. However, these results were questioned by Nangle et al., who showed that the mutation of one of the cysteines from the regulatory couple did not display a significantly different phenotype than the WT counterpart. Of note, the mutation of a second cysteine residue in CRY1 (Cys414) was shown to perturb both behavioral and molecular rhythms (Okano et al. 2009). This residue is essential for Zn2+ coordination, and, although it does not participate in disulfide bond formation, it is plausible that the oxidation of this cysteine in the apo-CRY1 might interfere with its interaction with PER2. To complicate matters even further, it appears that Cys412 in CRY1 might be required for the interaction of the protein with FBXL3 (Xing et al. 2013; Schmalen et al. 2014). The latter promotes the degradation of the CRY proteins via its association with the ubiquitin E3 ligase complex SCF (Skp1-Cul1-F-box protein), and thus this interaction might have a functional consequence. Of note, mutation of another cysteine residue in FBXL3, termed after hours (Afh), results in long free-running rhythms with the loss of circadian transcriptional oscillations (Busino et al. 2007; Godinho et al. 2007), suggesting that the cellular redox state can impinge on the molecular clockwork via reversible oxidation of cysteine residues. Following the same line of thought, several proteins involved in the control of CRY1 stability have been shown to harbor reactive cysteines. For instance, the metabolic sensor AMP-activated protein kinase (AMPK), which promotes the phosphorylation-dependent degradation of CRY1, has recently been shown to be regulated via a redox-active disulfide switch (Lamia et al. 2009). Similarly, CRY1 is stabilized by the deubiquitinase herpes-virus-associated ubiquitin-specific protease (HAUSP), which harbors an active site cysteine residue controlled via reversible oxidation (Lee et al. 2013).
The involvement of redox regulation in the clockwork has also been implied in the control of the accessory loops of the transcriptional oscillator. It was shown that REV-ERBβ harbors a regulatory disulfide bond in the heme-binding pocket, which dictates the binding of the cofactor and thus the activation of the TF (Gupta and Ragsdale 2011). Although these cysteine residues are not conserved in the closely related REV-ERBα, a similar mechanism might still be in place to control this TF because redox regulation appears to be a common theme in the biology of the nuclear receptor superfamily (Carter and Ragsdale 2014).
The Heme and Carbon Monoxide (CO) Loop
Another reciprocal link between the circadian and redox systems centers on the metabolism of the iron-containing porphyrin, heme. Heme forms the nonprotein part of hemoglobin but also acts as a cofactor for many cellular enzymes and transcription factors. The rate-limiting enzyme in heme biosynthesis, amino-levulinate synthase 1 (Alas1), is under transcriptional regulation by NASP2:BMAL1 and the levels of the porphyrin oscillate in a circadian manner. Heme was shown to feed back to the core clock by affecting both the positive and negative limbs of the transcriptional oscillator. For instance, binding of heme to the NPAS2:BMAL1 inhibits the interaction of the complex with DNA in response to CO (Dioum et al. 2002; Ishida et al. 2008). In contrast, an association of the cofactor with the nuclear hormone receptors REV-ERBα and REV-ERBβ induces the recruitment of the corepressor NcoR, which causes repression of target genes, including Bmal1 (Raghuram et al. 2007; Yin et al. 2007). Thus, heme exerts direct control over its levels by inhibiting both NPAS2:BMAL1 activity and Bmal1 transcription, thereby generating a regulatory loop. Finally, heme has been shown to bind to a novel regulatory motif in PER2 (different from the PAS domain), which leads to increased stability of the protein and altered association with CRY1. Of note, the motif binds exclusively to ferric heme, and thus this interaction is governed by the redox state of the iron core of the cofactor (Yang et al. 2008).
A recent study added yet more detail to the mechanism of this feedback loop by showing that heme is rhythmically degraded to CO by the action of the enzyme heme oxygenase 1 and 2 (HO-1 and HO-2) (Klemz et al. 2016). More importantly, rhythmic CO generation was shown to be essential for normal circadian rhythmicity on both the molecular and the behavioral level. The primary action of CO was shown to occur through inhibition of the transcriptional activity of CLOCK/NPAS2:BMAL1 complexes, whereas the depletion of CO was shown to lead to global up-regulation of CLOCK:BMAL1-dependent gene expression. Interestingly, heme degradation via the HOs consumes NADPH, thereby indicating a potential interaction between the cellular redox poise and the heme-CO-clock loop.
The Oxygen—HIF1α Axis
One of the first indications that hypoxia and the circadian system are linked derived from the discovery that both clock and the hypoxia-inducible factors (HIFs) are members of the basic helix–loop–helix PER-ARNT-SIM (bHLH-PAS) transcription factor superfamily. In addition, it was shown that BMAL1 dimerizes with both HIF1α and HIF2α in vitro (Hogenesch et al. 1998). The significance of this interaction in physiological settings remained unexplored up until recently when three independent studies showed the reciprocal interaction between the transcriptional oscillator and oxygen sensing via HIF1α, each one adding novel insights into the underlying mechanism of the coupling and the physiological relevance of this interaction in mammals (Adamovich et al. 2016; Peek et al. 2016; Wu et al. 2016).
Peek et al. (2016) investigated the interaction between hypoxia and circadian clocks in muscle tissue owing to the significant implications of oxygen metabolism in this system. Exercise causes oxygen depletion in skeletal muscle, which forces a metabolic switch to primarily anaerobic glycolysis. This effect is mediated by the HIF1α/β heterodimer, which senses the decrease in oxygen levels and induces the transcription of genes important in glycolytic metabolism. The activity of HIF1α is directly proportional to the stability of the protein, which is under the control of the oxygen-dependent prolyl-hydroxylases and the von Hippel–Lindau (VHL) E3 ubiquitin ligase. Under normoxic conditions, HIF1α is continuously marked for degradation, whereas the decrease in oxygen levels during hypoxia leads to the accumulation of the protein (Weidemann and Johnson 2008). The authors showed that deletion of Bmal1 in myotubes leads to the abrogation of the HIF1α-mediated responses to hypoxia, whereas the absence of the clock repressors CRY1/2 led to the stabilization of the TF. Conversely, pharmacological or genetic stabilization of HIF1α altered the period of PER2 oscillations and led to an increase in the transcript levels of several core clock genes. In addition, it was shown that HIF1α binds to the promoter regions of Per2 and Cry1 and that the TF acts synergistically with BMAL1 to induce expression of Per2. Finally, the authors showed the clear physiological implication of this interaction by showing that the circadian clock gates the hypoxic response to exercise in mice. Considering the major role of HIF1α in the metabolic switch during hypoxia, these findings strongly suggest that the TF might act as a coupling factor between the transcriptional clock and the redox system of the cell.
By performing a series of ChIP-Seq experiments, Wu et al. (2016) elaborated on the mechanism of the interaction between BMAL1 and HIF1α. Interestingly, they found BMAL1/HIF1α co-occupancy at 20%–30% at BMAL1 binding sites and that this interaction is synergistically leading to an increase in the transcriptional activation of these loci. Interestingly, the circadian pathway was found to be significantly overrepresented among the genes co-occupied by both TFs but not in genes occupied only by BMAL1. This implies that HIF1α is a dominant regulator of the circadian clock during hypoxic conditions. Reciprocally, the authors showed that the transcription of HIF1α displays circadian oscillations in mouse liver, supporting the role of the clock in priming the response of HIF1α to hypoxia. To solidify the physiological relevance of the findings, the authors investigated the possible involvement of the circadian clock in the severity of the damage induced by myocardial infarction (MI), one of the most well-known hypoxia-related pathologies. Consistent with previous reports showing higher mortality rates associated with morning heart attacks (Portaluppi and Lemmer 2007), the authors showed that Per1−/−/Per2−/− mutant mice show more severe MI damage compared with WT controls.
Adamovich et al. (2016) focused primarily on the physiological aspects of the interaction between oxygen metabolism and the circadian clock. The authors showed that oxygen consumption is rhythmic in mice and, perhaps more importantly, that this translated to rhythmic oxygenation of the blood and the kidney (Adamovich et al. 2016). One particularly interesting finding is that alternating 12-h low- and high-oxygen cycles can entrain the molecular clockwork in cultured cells in an HIF1α-dependent manner. To examine the relevance of these findings in the context of the whole animal, the authors showed that the application of a low oxygen pulse accelerates the adaptation of mice subjected to a jet lag protocol. This observation unequivocally shows that hypoxia is an important resetting cue and, together with the findings of Peek et al. (2016), might help to explain the positive effect of exercising in accelerating the adaption to jet lag (Mrosovsky and Salmon 1987).
The Cellular Redox Poise and Circadian Timekeeping: A Systems Affair
Although some of the mechanistic details of the PRDX hyperoxidation cycles have now been described, two important issues remain unresolved. First, what is the significance of these oscillations? Is the hyperoxidized form of the enzyme involved in signaling events or does the controlled inactivation of PRDXs allow signaling via rhythmic accumulation of H2O2? Second, what is the driving force behind these oxidation cycles? Are they intrinsic to the biochemical characteristics of PRDXs or are they a by-product of oscillations in a global cellular parameter such as the cellular redox poise?
In RBCs, the PRDXs hyperoxidation cycles are paralleled by robust oscillations in NADH, NADPH, and ATP levels (O’Neill and Reddy 2011). Because the reduction of the disulfide and hyperoxidized forms of PRDXs are dependent on NADPH and ATP, respectively, it seems plausible that the rhythmic accumulation of the inactivated enzyme reflects the availability of the two metabolites. In support of this hypothesis, recent work from our group identified the highly conserved pentose phosphate pathway (PPP), the primary source of NADPH in the cell, as a novel regulator of the cellular redox and transcriptional oscillations (Rey et al. 2016). Using pharmacological and genetic approaches, we found that inhibition of the PPP prolongs the period circadian rhythms in human cells, mouse tissues, and fruit flies.
Inhibition of the PPP leads to significant changes in the period length of the oscillations in NADPH and the hyperoxidized form of PRDX in nucleated cells. This suggests that the fluctuations in the reducing power of the cell are linked to the oscillations in PRDX. Because the catalytic cycle of the TRX system relies on NADPH as the ultimate electron donor, it is possible that the activity of the system is reflecting the levels of the cofactor. This also implies that the hyperoxidation of PRDXs might depend on the activity of the TRX systems, whereby the maximal activity of TRX coincides with the peak in PRDXs hyperoxidation. Although this might appear counterintuitive, active recycling of PRDXs via the TRX system has been shown to be essential for the hyperoxidation of the peroxidases because the disulfide-linked form is protected from further oxidation (Cao et al. 2014).
In addition to the alterations in the cellular redox oscillations, blocking of the PPP leads to profound period changes in the circadian gene-expression programs in cells. By performing a series of ChIP experiments, we found that this global effect is partially mediated by increased activity of the circadian transcription factors BMAL1 and CLOCK, the redox-sensitive transcription factor NRF2, and the chromatin remodeling histone acetyltransferase, P300. In addition, we found that inhibition of the PPP in primary mouse tissues recapitulated the results seen with cells, thus clearly showing the physiological significance of this perturbation.
Finally, inhibition of the PPP in fruit flies leads to behavioral misalignment, which indicates that the perturbation of this highly conserved pathway affects rhythms at the molecular, tissue, and behavioral level. Collectively, these data suggest that the putative redox oscillator might be represented by the rhythmic activity of the highly conserved PPP pathway or the interplay between the PPP and other primary metabolic pathways such as glycolysis.
In fact, the existence of a redox oscillator has been previously described in the literature. In 2005, Tu and colleagues showed that a continuous-culture system in S. cerevisiae shows robust ∼4–5-h cycles of oxygen consumption under nutrient-limited growth conditions (Tu et al. 2005; Tu and McKnight 2007). These cycles were found to consist of three distinct phases: oxidative, reductive/building (R/B), and reductive/charging (R/C). The authors found that over half of the S. cerevisiae genes are expressed periodically as a function of these metabolic oscillations, and that specific clusters of genes are expressed in each phase. For example, cell division occurs only during the R/B phase (minimal oxygen consumption), and never during the oxidative respiratory phase. The R/C phase is defined by up-regulation of genes involved in nonrespiratory modes of metabolism, which ensures the buildup of reducing power before the respiratory burst in the subsequent oxidative phase. In an excellent review on the topic, the authors evolved the hypothesis that such metabolic oscillations might be at the heart of most biological cycles (Tu and McKnight 2006). Based on the data summarized above, it is tempting to speculate that such a model could be applied in higher eukaryotes. Indeed, the rhythms in NADPH shown in RBCs and U2OS cells under constant conditions indicate rhythmicity in the flux through the oxidative branch of the PPP pathway. If we accept such a model, then this would require that the term “redox homoeostasis,” which infers that the system is static, be substituted by a term that factors in the diurnal aspects of the system, such as redox “homeodynamism.”
Importantly, the question of what drives these oxidation-reduction phases remains open. Diurnal flux through the oxidative and reductive branches of metabolic pathways might in part explain this if a regulatory mechanism that dictates the diurnal rate of activity of these pathways is in place. These possibilities remain to be tested.
Intersections with Tissue-Specific Clocks: The Redox Factor
Countless evidence has supported the important role of peripheral clocks for circadian rhythmicity. One of the emerging concepts in the past couple of decades is that the circadian system shows pronounced tissue-specific roles. Indeed, the importance of the cellular redox state in the generation of tissue-specific oscillations has been shown for most major organ systems (Milev and Reddy 2015).
In the liver, the rhythmic activation of SIRT6 by NAD+ was shown to play an important role in lipid homeostasis (Masri et al. 2014). Deletion of Bmal1 in the liver leads to increased anaerobic metabolism and lactic acid production, whereas deletion of the TF in the muscle leads to a decrease in anaerobic metabolism, suggesting that BMAL1 controls distinct transcriptional programs in these tissues (Peek et al. 2016). Oscillations in the hyperoxidation of the mitochondrial PRDX3 were shown to be essential for rhythmic steroidogenesis in the adrenal gland (Kil et al. 2012). Several redox parameters such as the NADPH/FAD ratio and protein glutathionylation display rhythms in the SCN and these oscillations were shown to be essential for neuronal firing (Wang et al. 2012). Numerous studies have shown the prevalence of overt oscillations of the redox state in the hematologic system. RBCs display oscillations in PRDX hyperoxidation, in the pyridine nucleotide levels, and hemoglobin auto-oxidation (O’Neill and Reddy 2011). Platelets display rhythms in GSH, which might be involved in the rhythmic activation of these cells (Radha et al. 1985; Scheer et al. 2011). The levels of GSH and sulfhydryl to disulfide ratio in blood plasma have also been shown to oscillate (Blanco et al. 2007). These findings imply that the redox capacity of the blood resonates with the day–night cycles and implies functional consequences for a variety of processes, such as cell–cell interactions and wound healing (Sen and Roy 2008; Eble and de Rezende 2014). Recently, oscillations in the NAD/NADH ratio were also shown in epidermal stem cells, implying circadian gating of the protective mechanisms against genotoxicity (Stringari et al. 2015).
CONCLUDING REMARKS
Retrospective
In the early days of the research into the molecular mechanism of the circadian clock, the view of most chronobiologists appeared more balanced. The following sentence taken from Goto et al. (1985) embodies the view of the larger part of the field at the time:
Finally, we have proposed a model—not for “the” clock—but for one oscillator in what is most probably a cellular “clockshop” ….
Unfortunately, as summarized in Lakin-Thomas (2006), not long after such statements, this metamorphosed into:
All circadian oscillators are based on transcriptional feedback loops involving the canonical clock genes.
Unfortunately, even nowadays, after the discovery of numerous examples of TTFL anomalies and transcriptionally independent oscillations, the prevalent view remains largely unchanged (Fig. 4). It is of paramount importance that the chronobiologists of the future adopt a more objective view and assume that the circadian system is most probably composed of several coupled oscillators that work in unison in the cell to generate a robust and highly adaptive cellular timekeeping mechanism.
Figure 4.
The unifying model of the transcriptional and metabolic oscillators—coupled oscillators. In the cell, the metabolic and transcriptional oscillators are coupled to generate a robust timekeeper, which is shielded from both metabolic and transcriptional perturbations. The coupling of the two oscillators occurs via the various known and potentially many unknown loops, which are individually dispensable but confer a high degree of flexibility to the clock. The high degree of interplay between the two oscillators in nucleated systems makes it extremely challenging to tease apart the two systems. However, manipulations of the coupling loops have provided ample evidence that the two oscillators are tightly connected. NADPH, Nicotinamide adenine dinucleotide phosphate; PRDX, peroxiredoxin; HIF, hypoxia-inducible factor; CO, carbon monoxide; GSH, glutathione; ROS, reactive oxygen species; SIRT, sirtuin.
Perspective
We believe that the future efforts in the field should focus on unraveling the coupling mechanisms that exist between the different cogs in the clockwork, most notably between the putative metabolic oscillator and the TTFL. Considering the ample evidence for the involvement of the cellular redox system in circadian timekeeping, we expect that this relationship will take an even more central position in the future. However, we do not exclude the possibility that other futures of primary metabolism play an important role in the cellular timekeeping mechanism and hope that no stones will be left unturned.
REFERENCES
- Adamovich Y, Ladeuix B, Golik M, Koeners MP, Asher G. 2016. Rhythmic oxygen levels reset circadian clocks through HIF1α. Cell Metab 25: 93–101. [DOI] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. 2008. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134: 317–328. [DOI] [PubMed] [Google Scholar]
- Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, Hottiger MO, Schibler U. 2010. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell 142: 943–953. [DOI] [PubMed] [Google Scholar]
- Avitabile D, Nicolussi A, Capriotti AL, Cucina A, Samperi R, Bizzarri M, Torrisi MR. 2014. Peroxiredoxin 2 nuclear levels are regulated by circadian clock synchronization in human keratinocytes. Int J Biochem Cell Biol 53: 24–34. [DOI] [PubMed] [Google Scholar]
- Bai P, Cantó C, Oudart H, Brunyánszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, et al. 2011. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab 13: 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett A. 1966. A circadian rhythm of mating type reversals in Paramecium multimicronucleatum, syngen 2, and its genetic control. J Cell Physiol 67: 239–270. [DOI] [PubMed] [Google Scholar]
- Barranco-Medina S, Lázaro JJ, Dietz KJ. 2009. The oligomeric conformation of peroxiredoxins links redox state to function. FEBS Lett 583: 1809–1816. [DOI] [PubMed] [Google Scholar]
- Baydas G, Gursu MF, Yilmaz S, Canpolat S, Yasar A, Cikim G, Canatan H. 2002. Daily rhythm of glutathione peroxidase activity, lipid peroxidation and glutathione levels in tissues of pinealectomized rats. Neurosci Lett 323: 195–198. [DOI] [PubMed] [Google Scholar]
- Beaver LM, Klichko VI, Chow ES, Kotwica-Rolinska J, Williamson M, Orr WC, Radyuk SN, Giebultowicz JM. 2012. Circadian regulation of glutathione levels and biosynthesis in Drosophila melanogaster. PLoS ONE 7: e50454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bélanger PM, Desgagné M, Bruguerolle B. 1991. Temporal variations in microsomal lipid peroxidation and in glutathione concentration of rat liver. Drug Metab Dispos 19: 241–244. [PubMed] [Google Scholar]
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. 2005. Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat Rev Genet 6: 544–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman AE, Chan WY, Brennan AM, Reyes RC, Adler BL, Suh SW, Kauppinen TM, Edling Y, Swanson RA. 2011. N-acetylcysteine prevents loss of dopaminergic neurons in the EAAC1−/− mouse. Ann Neurol 69: 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Labarre J, Toledano MB. 2003. ATP-dependent reduction of cysteine–sulphinic acid by S. cerevisiae sulphiredoxin. Nature 425: 980–984. [DOI] [PubMed] [Google Scholar]
- Blanco RA, Ziegler TR, Carlson BA, Cheng PY, Park Y, Cotsonis GA, Accardi CJ, Jones DP. 2007. Diurnal variation in glutathione and cysteine redox states in human plasma. Am J Clin Nutr 86: 1016–1023. [DOI] [PubMed] [Google Scholar]
- Bozonet SM, Findlay VJ, Day AM, Cameron J, Veal EA, Morgan BA. 2005. Oxidation of a eukaryotic 2-Cys peroxiredoxin is a molecular switch controlling the transcriptional response to increasing levels of hydrogen peroxide. J Biol Chem 280: 23319–23327. [DOI] [PubMed] [Google Scholar]
- Brody S, Harris S. 1973. Circadian rhythms in Neurospora: Spatial differences in pyridine nucleotide levels. Science 180: 498–500. [DOI] [PubMed] [Google Scholar]
- Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SIH, Draetta GF, Pagano M. 2007. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science 316: 900–904. [DOI] [PubMed] [Google Scholar]
- Cantó C, Menzies KJ, Auwerx J. 2015. NAD+ metabolism and the control of energy homeostasis: A balancing act between mitochondria and the nucleus. Cell Metab 22: 31–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Subramaniam S, Bulleid NJ. 2014. Lack of an efficient endoplasmic reticulum-localized recycling system protects peroxiredoxin IV from hyperoxidation. J Biol Chem 289: 5490–5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter EL, Ragsdale SW. 2014. Modulation of nuclear receptor function by cellular redox poise. J Inorg Biochem 133: 92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae HZ, Chung SJ, Rhee SG. 1994. Thioredoxin-dependent peroxide reductase from yeast. J Biol Chem 269: 27670–27678. [PubMed] [Google Scholar]
- Chae HZ, Kim HJ, Kang SW, Rhee SG. 1999. Characterization of three isoforms of mammalian peroxiredoxin that reduce peroxides in the presence of thioredoxin. Diabetes Res Clin Pract 45: 101–112. [DOI] [PubMed] [Google Scholar]
- Challet E. 2015. Keeping circadian time with hormones. Diabetes, Obesity Metab 17: 76–83. [DOI] [PubMed] [Google Scholar]
- Cho CS, Yoon HJ, Kim JY, Woo HA, Rhee SG. 2014. Circadian rhythm of hyperoxidized peroxiredoxin II is determined by hemoglobin autoxidation and the 20S proteasome in red blood cells. Proc Natl Acad Sci 111: 12043–12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MC, Alem F, Hamer SG, Narayanan A, Shatalin K, Bailey C, Nudler E, Hakami RM. 2017. S-nitrosylation of peroxiredoxin 1 contributes to viability of lung epithelial cells during Bacillus anthracis infection. Biochim Biophys Acta 1861: 3019–3029. [DOI] [PubMed] [Google Scholar]
- Cornelius G, Rensing L. 1976. Daily rhythmic changes in Mg2+-dependent ATPase activity in human red blood cell membranes in vitro. Biochem Biophys Res Commun 71: 1269–1272. [DOI] [PubMed] [Google Scholar]
- Coto-Montes A, Hardeland R. 2010. Diurnal rhythm of protein carbonyl as an indicator of oxidative damage in Drosophila melanogaster: Influence of clock gene alleles and deficiencies in the formation of free-radical scavengers. Biol Rhythm Res 30: 383–391. [Google Scholar]
- Coto-Montes A, Tomás-Zapico C, Rodríguez-Colunga MJ, Tolivia-Cadrecha D, Martínez-Fraga J, Hardeland R, Tolivia D. 2001. Effects of the circadian mutation “tau” on the Harderian glands of Syrian hamsters. J Cell Biochem 83: 426–434. [DOI] [PubMed] [Google Scholar]
- Cox AG, Pearson AG, Pullar JM, Jönsson TJ, Lowther WT, Winterbourn CC, Hampton MB. 2009. Mitochondrial peroxiredoxin 3 is more resilient to hyperoxidation than cytoplasmic peroxiredoxins. Biochem J 421: 51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day AM, Brown JD, Taylor SR, Rand JD, Morgan BA, Veal EA. 2012. Inactivation of a peroxiredoxin by hydrogen peroxide is critical for thioredoxin-mediated repair of oxidized proteins and cell survival. Mol Cell 45: 398–408. [DOI] [PubMed] [Google Scholar]
- Delaunay A, Pflieger D, Barrault MB, Vinh J, Toledano MB. 2002. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111: 471–481. [DOI] [PubMed] [Google Scholar]
- Dioum EM, Rutter J, Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA, McKnight SL. 2002. NPAS2: A gas-responsive transcription factor. Science 298: 2385–2387. [DOI] [PubMed] [Google Scholar]
- Driessche TV. 1966. Circadian rhythms in Acetabularia: Photosynthetic capacity and chloroplast shape. Exp Cell Res 42: 18–30. [DOI] [PubMed] [Google Scholar]
- Eble JA, de Rezende FF. 2014. Redox-relevant aspects of the extracellular matrix and its cellular contacts via integrins. Antioxid Redox Signal 20: 1977–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, et al. 2012. Peroxiredoxins are conserved markers of circadian rhythms. Nature 485: 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehret CF, Trucco E. 1967. Molecular models for the circadian clock I. The chronon concept. J Theoret Biol 15: 240–262. [DOI] [PubMed] [Google Scholar]
- Engelman R, Weisman-Shomer P, Ziv T, Xu J, Arnér ESJ, Benhar M. 2013. Multilevel regulation of 2-Cys peroxiredoxin reaction cycle by S-nitrosylation. J Biol Chem 288: 11312–11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Nakamura T, Cho DH, Gu Z, Lipton SA. 2007. S-nitrosylation of peroxiredoxin 2 promotes oxidative stress-induced neuronal cell death in Parkinson’s disease. Proc Natl Acad Sci 104: 18742–18747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooqui MY, Ahmed AE. 1984. Circadian periodicity of tissue glutathione and its relationship with lipid peroxidation in rats. Life Sci 34: 2413–2418. [DOI] [PubMed] [Google Scholar]
- Feeney KA, Hansen LL, Putker M, Olivares-Yañez C, Day J, Eades LJ, Larrondo LF, Hoyle NP, O’Neill JS, van Ooijen G. 2016. Daily magnesium fluxes regulate cellular timekeeping and energy balance. Nature 532: 375–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Caggiano M, Schröder E, Cho HJ, Burgoyne J, Barallobre-Barreiro J, Mayr M, Eaton P. 2016. Oxidant-induced interprotein disulfide formation in cardiac DJ-1 occurs via an interaction with peroxiredoxin 2. J Biol Chem 291: 10399–10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filonova A, Haemsch P, Gebauer C, Weisheit W, Wagner V. 2013. Protein disulfide isomerase 2 of Chlamydomonas reinhardtii is involved in circadian rhythm regulation. Mol Plant 6: 1503–1517. [DOI] [PubMed] [Google Scholar]
- Fisher AB. 2011. Peroxiredoxin 6: A bifunctional enzyme with glutathione peroxidase and phospholipase A2 activities. Antioxid Redox Signal 15: 831–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjeld CC, Birdsong WT, Goodman RH. 2003. Differential binding of NAD+ and NADH allows the transcriptional corepressor carboxyl-terminal binding protein to serve as a metabolic sensor. Proc Natl Acad Sci 100: 9202–9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho SIH, Maywood ES, Shaw L, Tucci V, Barnard AR, Busino L, Pagano M, Kendall R, Quwaillid MM, Romero MR, et al. 2007. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science 316: 897–900. [DOI] [PubMed] [Google Scholar]
- Goto K, Laval-Martin DL, Edmunds LN. 1985. Biochemical modeling of an autonomously oscillatory circadian clock in Euglena. Science 228: 1284–1288. [DOI] [PubMed] [Google Scholar]
- Gupta N, Ragsdale SW. 2011. Thiol-disulfide redox dependence of heme binding and heme ligand switching in nuclear hormone receptor Rev-erbβ. J Biol Chem 286: 4392–4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyöngyösi N, Nagy D, Makara K, Ella K, Káldi K. 2013. Reactive oxygen species can modulate circadian phase and period in Neurospora crassa. Free Radic Biol Med 58: 134–143. [DOI] [PubMed] [Google Scholar]
- Hanzén S, Vielfort K, Yang J, Roger F, Andersson V, Zamarbide-Forés S, Andersson R, Malm L, Palais G, Biteau B, et al. 2016. Lifespan control by redox-dependent recruitment of chaperones to misfolded proteins. Cell 166: 1–13. [DOI] [PubMed] [Google Scholar]
- Hardeland R. 2013. Chronobiology of melatonin beyond the feedback to the suprachiasmatic nucleus—Consequences to melatonin dysfunction. Int J Mol Sci 14: 5817–5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardeland R, Pandi-Perumal SR. 2005. Melatonin, a potent agent in antioxidative defense: Actions as a natural food constituent, gastrointestinal factor, drug and prodrug. Nutr Metab 2: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardeland R, Coto-Montes A, Poeggeler B. 2003. Circadian rhythms, oxidative stress, and antioxidative defense mechanisms. Chronobiol Int 20: 921–962. [DOI] [PubMed] [Google Scholar]
- Hardin PE, Hall JC, Rosbash M. 1990. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343: 536–540. [DOI] [PubMed] [Google Scholar]
- Haynes AC, Qian J, Reisz JA, Furdui CM, Lowther WT. 2013. Molecular basis for the resistance of human mitochondrial 2-Cys peroxiredoxin 3 to hyperoxidation. J Biol Chem 288: 29714–29723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama J, Cho S, Sassone-Corsi P. 2007. Circadian control by the reduction/oxidation pathway: Catalase represses light-dependent clock gene expression in the zebrafish. Proc Natl Acad Sci 104: 15747–15752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenesch JB, Gu YZ, Jain S, Bradfield CA. 1998. The basic-helix–loop–helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Acad Sci 95: 5474–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma T, Okano S, Lee J, Ito J, Otsuki N, Kurahashi T, Kang ES, Nakajima O, Fujii J. 2015. SOD1 deficiency induces the systemic hyperoxidation of peroxiredoxin in the mouse. Biochem Biophys Res Commun 463: 1040–1046. [DOI] [PubMed] [Google Scholar]
- Huang G, Zhang Y, Shan Y, Cheliah Y, Wang H, Takahashi JS. 2016. Circadian oscillations of NADH redox state using heterologous metabolic sensor in mammalian cells. J Biol Chem 291: 23906–23914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianăş O, Olinescu R, Bădescu I. 1991. Melatonin involvement in oxidative processes. Endocrinologie 29:147–153. [PubMed] [Google Scholar]
- Isaacs JT, Binkley F. 1977a. Cyclic AMP-dependent control of the rat hepatic glutathione disulfide–sulfhydryl ratio. Biochim Biophys Acta 498: 29–38. [DOI] [PubMed] [Google Scholar]
- Isaacs J, Binkley F. 1977b. Glutathione dependent control of protein disulfide-sulfhydryl content by subcellular fractions of hepatic tissue. Biochim Biophys Acta 497: 192–204. [DOI] [PubMed] [Google Scholar]
- Ishida M, Ueha T, Sagami I. 2008. Effects of mutations in the heme domain on the transcriptional activity and DNA-binding activity of NPAS2. Biochem Biophys Res Commun 368: 292–297. [DOI] [PubMed] [Google Scholar]
- Ivleva NB, Bramlett MR, Lindahl PA, Golden SS. 2005. LdpA: A component of the circadian clock senses redox state of the cell. EMBO J 24: 1202–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang HH, Lee KO, Chi YH, Jung BG, Park SK, Park JH, Lee JR, Lee SS, Moon JC, Yun JW, et al. 2004. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell 117: 625–635. [DOI] [PubMed] [Google Scholar]
- Jarvis RM, Hughes SM, Ledgerwood EC. 2012. Peroxiredoxin 1 functions as a signal peroxidase to receive, transduce, and transmit peroxide signals in mammalian cells. Free Radic Biol Med 53: 1522–1530. [DOI] [PubMed] [Google Scholar]
- Kaminsky YG, Kosenko EA, Kondrashova MN. 1984. Analysis of the circadian rhythm in energy metabolism of rat liver. Int J Biochem 16: 629–639. [DOI] [PubMed] [Google Scholar]
- Kang SW, Baines IC, Rhee SG. 1998. Characterization of a mammalian peroxiredoxin that contains one conserved cysteine. J Biol Chem 273: 6303–6311. [DOI] [PubMed] [Google Scholar]
- Khalsa SB, Block GD. 1992. Phase-shifting of a neuronal circadian pacemaker in Bulla gouldiana by pentylenetetrazol. Comp Biochem Physiol C 101: 557–560. [DOI] [PubMed] [Google Scholar]
- Khalsa SB, Whitmore D, Bogart B, Block GD. 1996. Evidence for a central role of transcription in the timing mechanism of a circadian clock. Am J Physiol 271: C1646–C1651. [DOI] [PubMed] [Google Scholar]
- Kil IS, Lee SK, Ryu KW, Woo HA, Hu MC, Bae SH, Rhee SG. 2012. Feedback control of adrenal steroidogenesis via H2O2-dependent, reversible inactivation of peroxiredoxin III in mitochondria. Mol Cell 46: 584–594. [DOI] [PubMed] [Google Scholar]
- Kil IS, Ryu KW, Lee SY, Kim YY, Chu SY, Kim JH, Park S, Rhee SG. 2015. Circadian oscillation of sulfiredoxin in the mitochondria. Mol Cell 59: 651–613. [DOI] [PubMed] [Google Scholar]
- Kinoshita C, Aoyama K, Matsumura N, Kikuchi-Utsumi K, Watabe M, Nakaki T. 2014. Rhythmic oscillations of the microRNA miR-96-5p play a neuroprotective role by indirectly regulating glutathione levels. Nat Commun 5: 3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemz R, Reischl S, Wallach T, Witte N, Jürchott K, Klemz S, Lang V, Lorenzen S, Knauer M, Heidenreich S, et al. 2016. Reciprocal regulation of carbon monoxide metabolism and the circadian clock. Nat Struct Mol Biol 24: 15–22. [DOI] [PubMed] [Google Scholar]
- Knoops B, Goemaere J, Van der Eecken V, Declercq JP. 2011. Peroxiredoxin 5: Structure, mechanism, and function of the mammalian atypical 2-Cys peroxiredoxin. Antiox Redox Signal 15: 817–829. [DOI] [PubMed] [Google Scholar]
- Kondo T, Mori T, Lebedeva NV, Aoki S, Ishiura M, Golden SS. 1997. Circadian rhythms in rapidly dividing cyanobacteria. Science 275: 224–227. [DOI] [PubMed] [Google Scholar]
- Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. 2006. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev 20: 1868–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV, Vykhovanets O, Kondratova AA, Antoch MP. 2009. Antioxidant N-acetyl-l-cysteine ameliorates symptoms of premature aging associated with the deficiency of the circadian protein BMAL1. Aging 1: 979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan N, Davis AJ, Giebultowicz JM. 2008. Circadian regulation of response to oxidative stress in Drosophila melanogaster. Biochem Biophys Res Commun 374: 299–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan N, Kretzschmar D, Rakshit K, Chow E, Giebultowicz JM. 2009. The circadian clock gene period extends healthspan in aging Drosophila melanogaster. Aging 1: 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai AG, Doherty CJ, Mueller-Roeber B, Kay SA, Schippers JHM, Dijkwel PP. 2012. Circadian clock-associated 1 regulates ROS homeostasis and oxidative stress responses. Proc Natl Acad Sci 109: 17129–17134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakin-Thomas PL. 2006. Transcriptional feedback oscillators: Maybe, maybe not…. J Biol Rhythms 21: 83–92. [DOI] [PubMed] [Google Scholar]
- Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, et al. 2009. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 326: 437–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JG, Baek K, Soetandyo N, Ye Y. 2013. Reversible inactivation of deubiquitinases by reactive oxygen species in vitro and in cells. Nat Commun 4: 1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XM, Metzger G, Filipski E, Boughattas N, Lemaigre G, Hecquet B, Filipski J, Levi F. 1997. Pharmacologic modulation of reduced glutathione circadian rhythms with buthionine sulfoximine: Relationship with cisplatin toxicity in mice. Toxicol Appl Pharmacol 143: 281–290. [DOI] [PubMed] [Google Scholar]
- Lim JM, Lee KS, Woo HA, Kang D, Rhee SG. 2015. Control of the pericentrosomal H2O2 level by peroxiredoxin I is critical for mitotic progression. J Cell Biol 210: 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA. 2008. Redundant function of REV-ERBα and β and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet 4: e1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low FM, Hampton MB, Peskin AV, Winterbourn CC. 2007. Peroxiredoxin 2 functions as a noncatalytic scavenger of low-level hydrogen peroxide in the erythrocyte. Blood 109: 2611–2617. [DOI] [PubMed] [Google Scholar]
- Lu Y, Nishio K, Matsuda S, Toshima Y, Ito H, Konno T, Ishihara K, Hashimoto K, Nakanishi S. 2014. Regulation of the cyanobacterial circadian clock by electrochemically controlled extracellular electron transfer. Angew Chem Int Ed Engl 53: 2208–2211. [DOI] [PubMed] [Google Scholar]
- Luchetti F, Canonico B, Betti M, Arcangeletti M, Pilolli F, Piroddi M, Canesi L, Papa S, Galli F. 2010. Melatonin signaling and cell protection function. FASEB J 24: 3603–3624. [DOI] [PubMed] [Google Scholar]
- Luna A, McFadden GB, Aladjem MI, Kohn KW. 2015. Predicted role of NAD+ utilization in the control of circadian rhythms during DNA damage response. PLoS Comput Biol 11: e1004144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabood SF, Newman PF, Nimmo IA. 1978. Circadian rhythms in the activity of acetylcholinesterase of human erythrocytes incubated in vitro. Biochem Soc Trans 6: 305–308. [DOI] [PubMed] [Google Scholar]
- Mandoli DF. 1998. Elaboration of body plan and phase change during development of Acetabularia: How is the complex architecture of a giant unicell built? Annu Rev Plant Physiol Plant Mol Biol 49: 173–198. [DOI] [PubMed] [Google Scholar]
- Manevich Y, Feinstein SI, Fisher AB. 2004. Activation of the antioxidant enzyme 1-CYS peroxiredoxin requires glutathionylation mediated by heterodimerization with πGST. Proc Natl Acad Sci 101: 3780–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, et al. 2010. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 466: 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S, Rigor P, Cervantes M, Ceglia N, Sebastian C, Xiao C, Roqueta-Rivera M, Deng C, Osborne TF, Mostoslavsky R, et al. 2014. Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell 158: 659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Alvarez H, Alvarez-Gonzalez R. 1993. Poly(ADP-ribose) polymerase is a catalytic dimer and the automodification reaction is intermolecular. J Biol Chem 268: 22575–22580. [PubMed] [Google Scholar]
- Meng QJ, Maywood ES, Bechtold DA, Lu WQ, Li J, Gibbs JE, Dupre SM, Chesham JE, Rajamohan F, Knafels J, et al. 2010. Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. Proc Natl Acad Sci 107: 15240–15245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergenhagen D, Schweiger HG. 1975a. Circadian rhythm of oxygen evolution in cell fragments of Acetabularia mediterranea. Exp Cell Res 92: 127–130. [DOI] [PubMed] [Google Scholar]
- Mergenhagen D, Schweiger HG. 1975b. The effect of different inhibitors of transcription and translation on the expression and control of circadian rhythm in individual cells of Acetabularia. Exp Cell Res 94: 321–326. [DOI] [PubMed] [Google Scholar]
- Milev NB, Reddy AB. 2015. Circadian redox oscillations and metabolism. Trends Endocrinol Metab 26: 430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse D, Milos PM, Roux E, Hastings JW. 1989. Circadian regulation of bioluminescence in Gonyaulax involves translational control. Proc Natl Acad Sci 86: 172–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse D, Hastings JW, Roenneberg T. 1994. Different phase responses of the two circadian oscillators in Gonyaulax. J Biol Rhythms 9: 263–274. [DOI] [PubMed] [Google Scholar]
- Mrosovsky N, Salmon PA. 1987. A behavioural method for accelerating re-entrainment of rhythms to new light–dark cycles. Nature 330: 372–373. [DOI] [PubMed] [Google Scholar]
- Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. 2004. Circadian gene expression in individual fibroblasts: Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119: 693–705. [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. 2008. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134: 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. 2009. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324: 654–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. 2005. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308: 414–415. [DOI] [PubMed] [Google Scholar]
- Nangle SN, Rosensweig C, Koike N, Tei H, Takahashi JS, Green CB, Zheng N. 2014. Molecular assembly of the period-cryptochrome circadian transcriptional repressor complex. eLife 3: e03674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netto LES, Antunes F. 2016. The roles of peroxiredoxin and thioredoxin in hydrogen peroxide sensing and in signal transduction. Mol Cells 39: 65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuschwander-Tetri BA, Rozin T. 1996. Diurnal variability of cysteine and glutathione content in the pancreas and liver of the mouse. Comp Biochem Physiol B Biochem Mol Biol 114: 91–95. [DOI] [PubMed] [Google Scholar]
- Nishiwaki T, Iwasaki H, Ishiura M, Kondo T. 2000. Nucleotide binding and autophosphorylation of the clock protein KaiC as a circadian timing process of cyanobacteria. Proc Natl Acad Sci 97: 495–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitabach MN, Blau J, Holmes TC. 2002. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell 109: 485–495. [DOI] [PubMed] [Google Scholar]
- Nitabach MN, Holmes TC, Blau J. 2005. Membranes, ions, and clocks: Testing the Njus–Sulzman–Hastings model of the circadian oscillator. Methods Enzymol 393: 682–693. [DOI] [PubMed] [Google Scholar]
- Njus D, Sulzman FM, Hastings JW. 1974. Membrane model for the circadian clock. Nature 248: 116–120. [DOI] [PubMed] [Google Scholar]
- Okano S, Akashi M, Hayasaka K, Nakajima O. 2009. Unusual circadian locomotor activity and pathophysiology in mutant CRY1 transgenic mice. Neurosci Lett 451: 246–251. [DOI] [PubMed] [Google Scholar]
- Oláhová M, Taylor SR, Khazaipoul S, Wang J, Morgan BA, Matsumoto K, Blackwell TK, Veal EA. 2008. A redox-sensitive peroxiredoxin that is important for longevity has tissue- and stress-specific roles in stress resistance. Proc Natl Acad Sci 105: 19839–19844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo M, O’Neill JS, Edgar RS, Valekunja UK, Reddy AB, Merrow M. 2012. Circadian regulation of olfaction and an evolutionarily conserved, nontranscriptional marker in Caenorhabditis elegans. Proc Natl Acad Sci 109: 20479–20484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill JS, Reddy AB. 2011. Circadian clocks in human red blood cells. Nature 469: 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill JS, Reddy AB. 2012. The essential role of cAMP/Ca2+ signaling in mammalian circadian timekeeping. Biochem Soc Trans 40: 44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill JS, van Ooijen G, Dixon LE, Troein C, Corellou F, Bouget FY, Reddy AB, Millar AJ. 2011. Circadian rhythms persist without transcription in a eukaryote. Nature 469: 554–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz CA, Heiland I. 2015. Dynamics of NAD+-metabolism: Everything but constant. Biochem Soc Trans 43: 1127–1132. [DOI] [PubMed] [Google Scholar]
- Park J, Lee S, Lee S, Kang SW. 2014. 2-Cys peroxiredoxins: Emerging hubs determining redox dependency of mammalian signaling networks. Int J Cell Biol 2014: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulose JK, Rucker EB, Cassone VM. 2012. Toward the beginning of time: Circadian rhythms in metabolism precede rhythms in clock gene expression in mouse embryonic stem cells. PLoS ONE 7: e49555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis T. 1969. Populations of interacting oscillators and circadian rhythms. J Theor Biol 22: 418–436. [DOI] [PubMed] [Google Scholar]
- Peek CB, Affinati AH, Ramsey KM, Kuo HY, Yu W, Sena LA, Ilkayeva O, Marcheva B, Kobayashi Y, Omura C, et al. 2013. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science 342: 1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peek CB, Levine DC, Cedernaes J, Taguchi A, Kobayashi Y, Tsai SJ, Bonar NA, McNulty MR, Ramsey KM, Bass J. 2016. Circadian clock interaction with HIF1α mediates oxygenic metabolism and anaerobic glycolysis in skeletal muscle. Cell Metab 25: 86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekovic-Vaughan V, Gibbs J, Yoshitane H, Yang N, Pathiranage D, Guo B, Sagami A, Taguchi K, Bechtold D, Loudon A, et al. 2014. The circadian clock regulates rhythmic activation of the NRF2/glutathione-mediated antioxidant defense pathway to modulate pulmonary fibrosis. Genes Dev 28: 548–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins A, Nelson KJ, Parsonage D, Poole LB, Karplus PA. 2015. Peroxiredoxins: Guardians against oxidative stress and modulators of peroxide signaling. Trends Biochem Sci 40: 435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peskin AV, Low FM, Paton LN, Maghzal GJ, Hampton MB, Winterbourn CC. 2007. The high reactivity of peroxiredoxin 2 with H2O2 is not reflected in its reaction with other oxidants and thiol reagents. J Biol Chem 282: 11885–11892. [DOI] [PubMed] [Google Scholar]
- Peskin AV, Pace PE, Behring JB, Paton LN, Soethoudt M, Bachschmid MM, Winterbourn CC. 2016. Glutathionylation of the active site cysteines of peroxiredoxin 2 and recycling by glutaredoxin. J Biol Chem 291: 3053–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponce IT, Rezza IG, Delgado SM, Navigatore LS, Bonomi MR, Golini RL, Gimenez MS, Anzulovich AC. 2012. Daily oscillation of glutathione redox cycle is dampened in the nutritional vitamin A deficiency. Biol Rhythm Res 43: 351–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole LB, Nelson KJ. 2016. Distribution and features of the six classes of peroxiredoxins. Mol Cells 39: 53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portaluppi F, Lemmer B. 2007. Chronobiology and chronotherapy of ischemic heart disease. Adv Drug Deliv Rev 59: 952–965. [DOI] [PubMed] [Google Scholar]
- Poynton RA, Hampton MB. 2014. Peroxiredoxins as biomarkers of oxidative stress. Biochim Biophys Acta 1840: 906–912. [DOI] [PubMed] [Google Scholar]
- Poynton RA, Peskin AV, Haynes AC, Lowther WT, Hampton MB, Winterbourn CC. 2016. Kinetic analysis of structural influences on the susceptibility of peroxiredoxins 2 and 3 to hyperoxidation. Biochem J 473: 411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radha E, Hill TD, Rao GH, White JG. 1985. Glutathione levels in human platelets display a circadian rhythm in vitro. Thrombosis Res 40: 823–831. [DOI] [PubMed] [Google Scholar]
- Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F. 2007. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBα and REV-ERBβ. Nat Struct Mol Biol 14: 1207–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, et al. 2009. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324: 651- 654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall LM, Ferrer-Sueta G, Denicola A. 2013. Peroxiredoxins as preferential targets in H2O2-induced signaling. Methods Enzymol 527: 41–63. [DOI] [PubMed] [Google Scholar]
- Ranieri D, Avitabile D, Shiota M, Yokomizo A, Naito S, Bizzarri M, Torrisi MR. 2015. Nuclear redox imbalance affects circadian oscillation in HaCaT keratinocytes. Int J Biochem Cell Biol 65: 113–124. [DOI] [PubMed] [Google Scholar]
- Reddy AB, O’Neill JS. 2011. Metaclocks. Nature 12: 612–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AB, Rey G. 2014. Metabolic and nontranscriptional circadian clocks: Eukaryotes. Annu Rev Biochem 83: 165–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AB, Karp NA, Maywood ES, Sage EA, Deery M, O’Neill JS, Wong GK, Chesham J, Odell M, Lilley KS, et al. 2006. Circadian orchestration of the hepatic proteome. Curr Biol 16: 1107–1115. [DOI] [PubMed] [Google Scholar]
- Rey G, Reddy AB. 2015. Interplay between cellular redox oscillations and circadian clocks. Diabetes Obes Metab 17: 55–64. [DOI] [PubMed] [Google Scholar]
- Rey G, Valekunja UK, Feeney KA, Wulund L, Milev NB, Stangherlin A, Ansel-Bollepalli L, Velagapudi V, O’Neill JS, Reddy AB. 2016. The pentose phosphate pathway regulates the circadian clock. Cell Metab 24: 462–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SG. 2006. Cell signaling. H2O2, a necessary evil for cell signaling. Science 312: 1882–1883. [DOI] [PubMed] [Google Scholar]
- Rhee SG. 2016. Overview on peroxiredoxin. Mol Cells 39: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SG, Kang SW, Chang TS, Jeong W, Kim K. 2001. Peroxiredoxin, a novel family of peroxidases. IUBMB Life 52: 35–41. [DOI] [PubMed] [Google Scholar]
- Rhee SG, Jeong W, Chang TS, Woo HA. 2007. Sulfiredoxin, the cysteine sulfinic acid reductase specific to 2-Cys peroxiredoxin: Its discovery, mechanism of action, and biological significance. Kidney Int Suppl 72: S3–S8. [DOI] [PubMed] [Google Scholar]
- Robinson JL, Foustock S, Chanez M, Bois-Joyeux B, Peret J. 1981. Circadian variation of liver metabolites and amino acids in rats adapted to a high protein, carbohydrate-free diet. J Nutr 111: 1711–1720. [DOI] [PubMed] [Google Scholar]
- Roenneberg T, Morse D. 1993. Two circadian oscillators in one cell. Nature 362: 362–364. [DOI] [PubMed] [Google Scholar]
- Rutter J, Reick M, Wu LC, McKnight SL. 2001. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science 293: 510–514. [DOI] [PubMed] [Google Scholar]
- Sahar S, Nin V, Barbosa MT, Chini EN, Sassone-Corsi P. 2011. Altered behavioral and metabolic circadian rhythms in mice with disrupted NAD+ oscillation. Aging 3: 794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer FAJL, Michelson AD, Frelinger AL, Evoniuk H, Kelly EE, McCarthy M, Doamekpor LA, Barnard MR, Steven AS. 2011. The human endogenous circadian system causes greatest platelet activation during the biological morning independent of behaviors. PLoS ONE 6: e24549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalen I, Reischl S, Wallach T, Klemz R, Grudziecki A, Prabu JR, Benda C, Kramer A, Wolf E. 2014. Interaction of circadian clock proteins CRY1 and PER2 is modulated by zinc binding and disulfide bond formation. Cell 157: 1203–1215. [DOI] [PubMed] [Google Scholar]
- Schrader LO, Holzapfel G. 1980. Circadian rhythms in human erythrocytes in vitro not confirmed. J Interdisciplinary Cycle Res 11: 199–207. [Google Scholar]
- Schweiger E, Wallraff HG, Schweiger HG. 1964. Endogenous circadian rhythm in cytoplasm of Acetabularia: Influence of the nucleus. Science 146: 658–659. [DOI] [PubMed] [Google Scholar]
- Sen CK, Roy S. 2008. Redox signals in wound healing. Biochim Biophys Acta 1780: 1348–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobotta MC, Liou W, Stöcker S, Talwar D, Oehler M, Ruppert T, Scharf AN, Dick TP. 2014. Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling. Nat Chem Biol 11: 64–70. [DOI] [PubMed] [Google Scholar]
- Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, Lundasen T, Shin Y, Liu J, Cameron MD, Noel R, et al. 2012. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature 485: 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringari C, Wang H, Geyfman M, Crosignani V, Kumar V, Takahashi JS, Andersen B, Gratton E. 2015. In vivo single-cell detection of metabolic oscillations in stem cells. Cell Rep 10: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney BM, Haxo FT. 1961. Persistence of a photosynthetic rhythm in enucleated Acetabularia. Science 134: 1361–1363. [DOI] [PubMed] [Google Scholar]
- Tamaru T, Hattori M, Ninomiya Y, Kawamura G, Varès G, Honda K, Mishra DP, Wang B, Benjamin I, Sassone-Corsi P, et al. 2013. ROS stress resets circadian clocks to coordinate pro-survival signals. PLoS ONE 8: e82006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavender TJ, Springate JJ, Bulleid NJ. 2010. Recycling of peroxiredoxin IV provides a novel pathway for disulfide formation in the endoplasmic reticulum. EMBO J 29: 4185–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terborgh J, McLeod G. 1967. The photosynthetic rhythm of Acetabularia crenulata. I. Continuous measurements of oxygen exchange in alternating light–dark regimes and in constant light of different intensities. Biol Bull 133: 659–669. [Google Scholar]
- Tomita J, Nakajima M, Kondo T, Iwasaki H. 2005. No transcription–translation feedback in circadian rhythm of KaiC phosphorylation. Science 307: 251–254. [DOI] [PubMed] [Google Scholar]
- Tu BP, McKnight SL. 2006. Metabolic cycles as an underlying basis of biological oscillations. Nat Rev Mol Cell Biol 7: 696–701. [DOI] [PubMed] [Google Scholar]
- Tu BP, McKnight SL. 2007. The yeast metabolic cycle: Insights into the life of a eukaryotic cell. Cold Spring Harb Symp Quant Biol 72: 339–343. [DOI] [PubMed] [Google Scholar]
- Tu BP, Kudlicki A, Rowicka M, McKnight SL. 2005. Logic of the yeast metabolic cycle: Temporal compartmentalization of cellular processes. Science 310: 1152–1158. [DOI] [PubMed] [Google Scholar]
- Vanden Driessche T. 1966. The role of the nucleus in the circadian rhythms of Acetabularia mediterranea. Biochim Biophys Acta 126: 456–470. [DOI] [PubMed] [Google Scholar]
- Van Ooijen G, Millar AJ. 2012. Non-transcriptional oscillators in circadian timekeeping. Trends Biochem Sci 37: 484–492. [DOI] [PubMed] [Google Scholar]
- Veal EA, Findlay VJ, Day AM, Bozonet SM, Evans JM, Quinn J, Morgan BA. 2004. A 2-Cys peroxiredoxin regulates peroxide-induced oxidation and activation of a stress-activated MAP kinase. Mol Cell 15: 129–139. [DOI] [PubMed] [Google Scholar]
- Vivancos AP, Castillo EA, Biteau B, Nicot C, Ayté J, Toledano MB, Hidalgo E. 2005. A cysteine-sulfinic acid in peroxiredoxin regulates H2O2-sensing by the antioxidant Pap1 pathway. Proc Natl Acad Sci 102: 8875–8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E, Frosch S. 1974. Cycles in plants: Endogenous rhythmicity and energy transduction VI. Rhythmicity in reduced and oxidized pyridine nucleotide levels in seedlings of Chenopodium rubrum. J Interdisciplinary Cycle Res 5: 231–239. [Google Scholar]
- Wang TA, Yu YV, Govindaiah G, Ye X, Artinian L, Coleman TP, Sweedler JV, Cox CL, Gillette MU. 2012. Circadian rhythm of redox state regulates excitability in suprachiasmatic nucleus neurons. Science 337: 839–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidemann A, Johnson RS. 2008. Biology of HIF-1α. Cell Death Differ 15: 621–627. [DOI] [PubMed] [Google Scholar]
- Wilking M, Ndiaye M, Mukhtar H, Ahmad N. 2013. Circadian rhythm connections to oxidative stress: Implications for human health. Antioxid Redox Signal 19: 192–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterbourn CC, Hampton MB. 2008. Thiol chemistry and specificity in redox signaling. Free Radic Biol Med 45: 549–561. [DOI] [PubMed] [Google Scholar]
- Woo HA, Kang SW, Kim HK, Yang KS, Chae HZ, Rhee SG. 2003a. Reversible oxidation of the active site cysteine of peroxiredoxins to cysteine sulfinic acid. Immunoblot detection with antibodies specific for the hyperoxidized cysteine-containing sequence. J Biol Chem 278: 47361–47364. [DOI] [PubMed] [Google Scholar]
- Woo HA, Chae HZ, Hwang SC, Yang KS, Kang SW, Kim K, Rhee SG. 2003b. Reversing the inactivation of peroxiredoxins caused by cysteine sulfinic acid formation. Science 300: 653–656. [DOI] [PubMed] [Google Scholar]
- Wood ZA, Poole LB, Karplus PA. 2003. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science 300: 650–653. [DOI] [PubMed] [Google Scholar]
- Woolum JC. 1991. A re-examination of the role of the nucleus in generating the circadian rhythm in Acetabularia. J Biol Rhythms 6: 129–136. [DOI] [PubMed] [Google Scholar]
- Wu Y, Tang D, Liu N, Xiong W, Huang H, Li Y, Ma Z, Zhao H, Chen P, Qi X, et al. 2016. Reciprocal regulation between the circadian clock and hypoxia signaling at the genome level in mammals. Cell Metab 25: 73–85. [DOI] [PubMed] [Google Scholar]
- Xing W, Busino L, Hinds TR, Marionni ST, Saifee NH, Bush MF, Pagano M, Zheng N. 2013. SCFFBXL3 ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature 496: 64–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Wang H, Liu Y, Shao C. 2008. Analysis of gene regulatory networks in the mammalian circadian rhythm. PLoS Comput Biol 4: e1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Sehgal A. 2001. Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron 29: 453–467. [DOI] [PubMed] [Google Scholar]
- Yang KS, Kang SW, Woo HA, Hwang SC, Chae HZ, Kim K, Rhee SG. 2002. Inactivation of human peroxiredoxin I during catalysis as the result of the oxidation of the catalytic site cysteine to cysteine-sulfinic acid. J Biol Chem 277: 38029–38036. [DOI] [PubMed] [Google Scholar]
- Yang J, Kim KD, Lucas A, Drahos KE, Santos CS, Mury SP, Capelluto GS, Finkielstein CV. 2008. A novel heme-regulatory motif mediates heme-dependent degradation of the circadian factor period 2. Mol Cell Biol 28: 4697–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, et al. 2007. Rev-erbα, a heme sensor that coordinates metabolic and circadian pathways. Science 318: 1786–1789. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Iigusa H, Wang N, Hasunuma K. 2011. Cross-talk between the cellular redox state and the circadian system in Neurospora. PLoS ONE 6: e28227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii K, Ishijima S, Sagami I. 2013. Effects of NAD(P)H and its derivatives on the DNA-binding activity of NPAS2, a mammalian circadian transcription factor. Biochem Biophys Res Commun 437: 386–391. [DOI] [PubMed] [Google Scholar]
- Yoshii K, Tajima F, Ishijima S, Sagami I. 2015. Changes in pH and NADPH regulate the DNA binding activity of neuronal PAS domain protein 2, a mammalian circadian transcription factor. Biochemistry 54: 250–259. [DOI] [PubMed] [Google Scholar]
- Young MW, Kay SA. 2001. Time zones: A comparative genetics of circadian clocks. Nat Rev Genet 2: 702–715. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Piston DW, Goodman RH. 2002. Regulation of corepressor function by nuclear NADH. Science 295: 1895–1897. [DOI] [PubMed] [Google Scholar]