Abstract
The Hippo signal transduction pathway is an important regulator of organ growth and cell differentiation, and its deregulation contributes to the development of cancer. The activity of the Hippo pathway is strongly dependent on cell junctions, cellular architecture, and the mechanical properties of the microenvironment. In this review, we discuss recent advances in our understanding of how cell junctions transduce signals from the microenvironment and control the activity of the Hippo pathway. We also discuss how these mechanisms may control organ growth during development and regeneration, and how defects in them deregulate Hippo signaling in cancer cells.
Animals and their organs normally grow to a specific size, which depends on poorly understood signaling mechanisms that coordinate and orchestrate the pattern and timing of cell proliferation during development. Efforts to identify these growth control mechanisms through genetic screens in Drosophila led to the discovery of the Hippo signaling pathway (Harvey and Tapon 2007; Pan 2007; Halder and Johnson 2011). These screens identified several genes that are required for the formation of normal-sized adult structures, among the first ones were warts (wts) (Xu et al. 1995; Justice et al. 1995), salvador (sav) (Kango-Singh et al. 2002; Tapon et al. 2002), and hippo (hpo) (Harvey et al. 2003; Jia et al. 2003; Pantalacci et al. 2003; Udan et al. 2003; Wu et al. 2003). Mutations in hpo, wts, or sav cause similar phenotypes and lead to dramatic overgrowth of imaginal discs and the corresponding adult structures. This is because mutant cells hyperproliferate and are resistant to apoptosis that would normally eliminate extra cells. Genetic and biochemical studies then revealed that Hpo, Wts, and Sav form the core of a conserved signaling pathway, now known as the “Hippo pathway.” Remarkably, mutations in the homologous Hippo pathway genes also cause dramatic tissue overgrowth in mice. For example, conditional deletion of the hpo homologs Mst1/2 in the developing liver causes sustained hepatocyte cell proliferation resulting in severely overgrown livers (Zhou et al. 2009; Lu et al. 2010; Song et al. 2010). Therefore, the Hippo pathway is a conserved signal transduction pathway that is required to restrict excessive organ growth in mice and flies.
The Hippo pathway acts by regulating the localization and activity of its downstream effectors Yorkie (Yki) in flies and Yes-associated-protein (YAP)/WW Domain Containing Transcription Regulator 1 (TAZ) in mammals (Huang et al. 2005; Dong et al. 2007). In brief, the core of the pathway comprises the Hpo kinase (MST1/2 in mammals), which together with the Sav adaptor protein (SAV1 in mammals), phosphorylates and activates a complex of the Wts kinase (LATS1/2 in mammals) and its cofactor Mats (MOBKL1A/B in mammals) (Kango-Singh et al. 2002; Tapon et al. 2002; Harvey et al. 2003; Jia et al. 2003; Pantalacci et al. 2003; Udan et al. 2003; Wu et al. 2003; Lai et al. 2005). When active, Wts and LATS1/2 bind and phosphorylate the transcriptional coactivators Yki and YAP/TAZ, causing their inactivation by nuclear exclusion and subsequent proteasomal degradation (Dong et al. 2007; Hao et al. 2008; Oh and Irvine 2008, 2009; Zhang et al. 2008a). On the other hand, when the Hippo core kinases are not active, Yki/YAP/TAZ accumulate in the nucleus where they bind to TEAD family and other transcription factors and drive the expression of target genes that promote cell proliferation and survival such as myc, diap1, the bantam miRNA, and cyclinE (Nolo et al. 2006; Thompson and Cohen 2006; Goulev et al. 2008; Ota and Sasaki 2008; Zhang et al. 2008b, 2009; Zhao et al. 2008; Wu et al. 2008; Chan et al. 2009; Neto-Silva et al. 2010; Oh and Irvine 2011; Galli et al. 2015; Zanconato et al. 2015). Therefore, reduced Hippo pathway activity causes upregulation of Yki/YAP/TAZ activity, which leads to hyperproliferation and resistance to apoptosis resulting in tissue overgrowth.
Given the striking overgrowth phenotypes caused by loss of Hippo pathway activity in flies and mice, a critical question is how is the activity of the Hippo pathway controlled? Since the discovery of the core components, several upstream regulators have been identified including classical signaling molecules such as G-protein coupled receptors (GPCRs) (Yu et al. 2012) and Ras-mitogen-activated protein kinase (MAPK) signaling (Reddy and Irvine 2013). However, the strongest effects on Hippo pathway activity are exerted by changes in the cytoskeleton, by the action of cell–cell and cell–matrix junction components, and by the physical properties of the extracellular matrix (Halder et al. 2012; Schroeder and Halder 2012; Gaspar and Tapon 2014; Gumbiner and Kim 2014; Yu et al. 2015; Dupont 2016; Sun and Irvine 2016).
The first indications that cell–cell junctions play important roles in the Hippo pathway came from the discovery that the neurofibromatosis type 2 (NF2)/Merlin (Mer) and Expanded (Ex) FERM-domain adaptor proteins are required for Hippo signaling (Hamaratoglu et al. 2006). NF2/Mer and Ex localize to tight and adherens junctions in epithelial cells (Boedigheimer and Laughon 1993; McCartney and Fehon 1996) and Mer is required for the formation of adherens junctions (Lallemand et al. 2003). In addition, it was found that the activity of the Hippo pathway is regulated by contact-dependent inhibition of cell proliferation (a.k.a., contact inhibition) such that YAP is excluded from the nucleus and its activity is inhibited when cultured cells stop proliferating as they become confluent (Zhao et al. 2007; Ota and Sasaki 2008). Since then, many additional molecular links between cell junctions and the Hippo pathway have been discovered and it is now clear that cell junctions are major hubs where complexes of Hippo pathway components are organized and processed (Yu et al. 2015).
The sheer number of molecular mechanisms that are currently known to affect the activity of the Hippo pathway, which often includes multiple independent links between particular junctional complexes and the core of the Hippo pathway, reveals that the Hippo pathway is not a conventional signal transduction pathway with a dedicated set of receptor proteins and signaling molecules. Rather, Hippo pathway components are often deeply embedded into the molecular networks that assemble the different cellular junctions and are often regulated through multiple mechanisms emanating from the junctions including recruitment, phosphorylation, ubiquitination, competitive binding to cofactors, etc. (Fig. 1). This deep connection between the Hippo pathway and cell junctions implies that the Hippo pathway is sensitive to changes in cellular morphology and architecture. This then further implies that signals that may modulate the activity of the Hippo pathway in an organism during development, homeostasis, or regeneration, may not primarily be the type of extracellular peptide signals that command embryonic patterning and differentiation, but may include mechanical forces and signals that affect cell shape and morphogenesis. Measuring, manipulating, and understanding these signals will be an exciting challenge for the Hippo field and for growth control in general for the coming years. In this review, we provide an overview of our current understanding of the molecular mechanisms by which cell junctions affect the activity of the Hippo pathway, and discuss their role during development and tissue homeostasis.
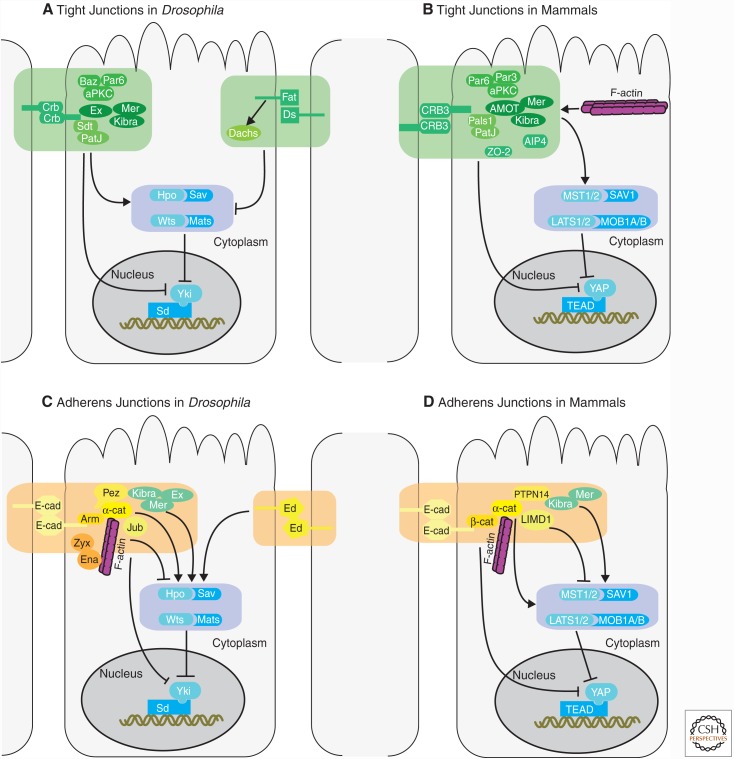
Figure 1.
Schematic representation of molecular mechanisms that connect cellular junctions to the Hippo pathway. (A) Effect of tight junctions in Drosophila. Crumbs (Crb) and atypical protein kinase C (aPKC) polarity complexes promote Hippo signaling through Mer, Ex, and kidney and brain expressed protein (KIBRA), while the Fat-Dachs branch blocks the Warts (Wts) kinase. (B) Effect of tight junctions in mammals. The Crb complex acts through Angiomotin (AMOT), which suppresses Yes-associated protein (YAP) activity by direct binding and activates LATS2 by recruitment. AIP4 inhibits YAP by ubiquitination and stabilizes AMOT. F-actin competes for AMOT binding therefore inducing YAP activity. Zonula Occludens (ZO)-2 binds to YAP and sequesters it in the cytoplasm. (C) Effect of adherens junctions in Drosophila. E-cad-α-cat-β-cat complex, Echinoid (Ed), and Pez promotes Hippo signaling activity while Ajuba (Jub) and Zyxin (Zyx) inhibit it. (D) Effect of adherens junctions in mammals. The E-cad-α-cat-β-cat complex and PTPN14 promote Hippo signaling activity while the Ajuba protein LIMD1 inhibits the activity of LATS1/2 kinases. Also, α-cat and PTPN14 inhibit YAP by direct binding and sequestering it in the cytoplasm.
TIGHT JUNCTIONS AND THE SUBAPICAL REGION IN HIPPO SIGNALING
Among the different cell–cell junctions, the tight junctions are perhaps the best-characterized junctions in terms of their effect on the Hippo pathway. Tight junctions typically localize just apical to adherens junctions in vertebrate epithelial cells, where they form a permeability barrier between adjacent cells (Anderson et al. 2004; Matter et al. 2005). Proteins associated with tight junctions also play instrumental roles in the establishment and maintenance of apical-basal cell polarity, most notably the Crumbs (Crb) and atypical protein kinase C (aPKC) complexes (Bulgakova and Knust 2009; St Johnston and Ahringer 2010; Tepass 2012; Thompson 2013). Several upstream regulators of the Hippo pathway localize to tight junctions, or are themselves integral parts of tight junctions, such as the Crb and aPKC polarity complexes, the Angiomotin (Amot) family proteins, and the Zonula occludens (ZO) factors. Most of these factors and the apical-basal polarity complexes in general, are conserved in Drosophila, where they also localize apical to adherens junctions to a domain called the subapical region or marginal zone (Bulgakova and Knust 2009; Tepass 2012). However, epithelial cells in Drosophila do not have apical tight junctions, but form functionally equivalent septate junctions basal to adherens junctions (Woods and Bryant 1993). Nevertheless, the organization and function of apical-basal cell polarity complexes and apically localized Hippo pathway components is conserved between Drosophila and vertebrates, although some differences exist in the composition of upstream components (Bossuyt et al. 2014).
Not only upstream regulators, but also Hippo pathway core components themselves localize to apical junctions. For example, Wts can localize to the subapical region and to adherens junctions (Sun et al. 2015). In the subapical region, Wts colocalizes with Hpo and Sav, which activates Wts in response to upstream components such as the apically localized Crumbs transmembrane receptor and the Mer and Ex adaptor proteins. On the other hand, Wts at adherens junctions interacts with the LIM-domain protein Ajuba, which inhibits Wts activity (Sun et al. 2015). Thus, the different junctional complexes are major sites of Hippo pathway assembly and regulation. Interestingly, activated Yki promotes the expansion of the apical membrane domain and upregulates the expression of Ex, Kib, Wts, and other upstream regulators of the Hippo pathway in a negative feedback loop (Genevet et al. 2009; Hamaratoglu et al. 2009). In this section of the review, we will summarize what is known about the mechanisms that link tight junctions and the subapical region to the Hippo pathway.
The Crumbs Complex
In Drosophila, the Crumbs complex is composed of the transmembrane protein Crumbs (Crb) and the adaptor proteins Pals1-associated tight junction protein (Patj), Stardust (Sdt) and Lin7, which complex with PDZ (postsynaptic density protein 95 (PSD-95)/discs large (dlg)/Zona Occludens-1 (ZO-1))-binding motif in the intracellular domain of Crb (Bulgakova and Knust 2009; Tepass 2012). The Crb intracellular domain also contains a FERM-domain binding motif, which binds and recruits Ex to the subapical region (Ling et al. 2010; Chen et al. 2010; Robinson et al. 2010). crb mutant cell clones in imaginal discs thus lose membrane localization of Ex, which is essential for recruitment of Hippo core components and their activation (Chen et al. 2010; Ling et al. 2010; Robinson et al. 2010; Yin et al. 2013). Thus, loss of Crb in imaginal discs causes activation of Yki and overgrowth. Similarly, overexpression of Crb, especially a mutant version of Crb that lacks the extracellular domain, also disrupts Ex localization and causes induction of Yki target genes (Chen et al. 2010; Grzeschik et al. 2010; Robinson et al. 2010). This is at least in part caused by induced Ex phosphorylation, which causes its ubiquitin-dependent proteasomal degradation (Ribeiro et al. 2014). Notably, Crb homotypically interacts with Crb molecules on neighboring cells and loss of Crb in a cell results in loss of Crb recruitment to the plasma membrane of adjacent cells. In that manner, Crb non-cell-autonomously affects the localization of Ex in neighboring cells (Chen et al. 2010; Hafezi et al. 2012).
The components of the Crb complex are conserved in mammals where they are also required for apical-basal cell polarity of epithelial cells (Roh et al. 2003; Bulgakova and Knust 2009). Mammals have three Crb homologs of which CRB3 is the best studied regarding effects on the Hippo pathway. CRB3 is widely expressed in epithelial tissues (Lemmers et al. 2004) and distinguished by a short extracellular domain and the presence of a PxxP SH3-domain binding motif (Li et al. 2015b). Loss of CRB3 in different organs or in cultured cells triggers YAP nuclear localization and increased activity (Varelas et al. 2010; Szymaniak et al. 2015; Fernando et al. 2016). CRB3 is required for the differentiation of airway epithelial cells in developing mouse lungs (Szymaniak et al. 2015) and for the control of optimal myelin elongation during axonal growth in Schwann cells (Fernando et al. 2016) through regulation of Hippo signaling. However, these effects may not be transduced by the mammalian Ex homolog Willin/FRMD6, because manipulating Willin function only modestly affects the Hippo pathway in vitro (Angus et al. 2012) and because Willin lacks the domain that signals to the Hippo core (Hamaratoglu et al. 2006; Bossuyt et al. 2014). Rather, CRB3 signals through the Amot family adaptor proteins (Wells et al. 2006) that are not present in flies (Bossuyt et al. 2014). Thus, Crb polarity complexes regulate the Hippo pathway in Drosophila and in mammals, however apparently through different mechanisms that involve Ex in Drosophila and Amot in mammals.
Angiomotin Family Proteins
The Amot proteins are scaffold proteins that complex with several components of the Hippo pathway. The Amot protein family comprises AMOT, AMOTL1, and AMOTL2 (Ernkvist et al. 2006), which all share PPxY motifs, conserved coiled-coil domains, and a carboxy-terminal PDZ-binding motif (Moleirinho et al. 2014). Amot proteins are integral components of the Crumbs complex and essential for its stability (Wells et al. 2006; Heller et al. 2010). The Angiomotins were identified as regulators of the Hippo pathway through proteomics assays because they are major binding partners of YAP and TAZ through the interaction between PPxY motifs and WW-domains of YAP and TAZ (Varelas et al. 2010; Chan et al. 2011; Wang et al. 2011; Zhao et al. 2011). Amot proteins are required to keep YAP and TAZ out of the nucleus in confluent cultured cells (Chan et al. 2011; Wang et al. 2011; Zhao et al. 2011). For this, they interact with YAP/TAZ (Chan et al. 2011; Wang et al. 2011; Zhao et al. 2011), the LATS2 kinase (Paramasivam et al. 2011), and the ubiquitin ligase AIP4 (Adler et al. 2013a), which triggers multiple mechanisms of cross-regulation. In confluent cells, Amot proteins localize to tight junctions and the interaction with YAP/TAZ sequesters these at the plasma membrane (Wang et al. 2011; Zhao et al. 2011). They also bind to LATS2 and recruit it to the plasma membrane, which activates LATS2 thereby also causing inactivation of YAP/TAZ by phosphorylation (Paramasivam et al. 2011). Moreover, recruitment of AIP4 causes ubiquitination and destabilization of YAP (Adler et al. 2013a). The sequestration and inhibition of YAP is further reinforced by feedback loops between AIP4, the Angiomotins, and LATS2 (Adler et al. 2013a,b; Chan et al. 2013; Dai et al. 2013). In subconfluent cells however, Endotubin (EDTB) competes with YAP for AMOT binding and sequesters Amot proteins on endosomal membranes, thereby promoting nuclear localization of YAP (Cox et al. 2015). Thus, Amot proteins act as scaffolds at tight junctions that induce activation of LATS2 and inactivation and sequestration of YAP/TAZ.
The inhibition of YAP by Amots is important during early mouse development when Amots acts downstream of cell polarity (Hirate et al. 2013). In blastocysts, YAP is nuclear in presumptive trophectoderm cells but excluded from the nuclei of inner cell mass (ICM) cells. Amot mutant embryos fail to exclude YAP from the nuclei of ICM cells, which causes failure to specify the ICM (Hirate et al. 2013). AMOT localizes to adherens junctions of the nonpolar inner cells where it is required for activation of Hippo signaling through the recruitment and activation of the LATS1/2 kinases, which directly inhibit YAP by phosphorylation and indirectly by phosphorylation of AMOT, which shifts AMOT binding from F-actin to YAP (Hirate et al. 2013; Hirate and Sasaki 2014). However, in polarized outer cells, AMOT is restricted to apical domains by polarity complexes, which enables AMOT-F-actin interaction, abrogation of LATS1/2 activation, and causes YAP nuclear accumulation (Hirate et al. 2013; Hirate and Sasaki 2014).
Opposite to this inhibitory effect, however, AMOT has a positive effect on YAP function in the mouse liver. Here, conditional deletion of Amot suppresses the liver overgrowth caused by loss of Nf2 and reduced the proliferation of oval cells, a type of liver progenitor cells that are induced upon toxic liver injury and in Nf2 knockout livers (Yi et al. 2013). Experiments in HEK293 cells further showed that interaction of YAP with Amot is required for YAP nuclear localization and proliferation of Nf2-deficient cells. This positive function of AMOT for YAP activity may be explained by AMOT action in the nucleus. Here, Amot-YAP interaction protects YAP from LATS phosphorylation and AMOT also acts as a cofactor in Amot-YAP-TEAD complexes. In summary, AMOT is a molecular hub that forms complexes with several components of the Hippo pathway and other factors that affect the localization and activity of AMOT and thus the activity of YAP. Further experiments are required to untangle the positive and negative effects of AMOT on Hippo signaling.
The aPKC Polarity Module
The aPKC complex comprises the aPKC serine-threonine kinase, and the Par3 and Par6 adaptor proteins. The aPKC complex is highly conserved, localizes to tight junctions in mammalian cells and to the subapical region in Drosophila cells, where it cooperates with the Crb complex to control apical-basal cell polarity (St Johnston and Ahringer 2010; Tepass 2012). Overexpression of hyperactive forms of aPKC in Drosophila imaginal discs lead to excessive cell proliferation and overgrowth by disrupting the Hippo pathway through mislocalization of the Hpo inhibitor RASSF and the Hpo kinase (Rolls et al. 2003, Polesello et al. 2006; Grzeschik et al. 2010). Similar effects were also observed in cultured mammalian cells (Archibald et al. 2015). In addition, aPKC phosphorylates and regulates kidney and brain expressed protein (KIBRA), a WW- and C2-domain containing adaptor protein (Kremerskothen et al. 2003) that functions together with Mer and Ex (Baumgartner et al. 2010; Genevet et al. 2010; Yu et al. 2010). aPKC also phosphorylates and stabilizes Crb in Drosophila follicle cells (Sotillos et al. 2004), which induces a positive feedback loop by suppressing the endocytic removal of the Crb complex from the plasma membrane (Fletcher et al. 2012). Thus, aPKC modulates the activity of multiple Hippo pathway components.
Zonula Occludens Proteins
The ZO proteins ZO-1, ZO-2, and ZO-3 are membrane-associated guanylate kinases (MAGUK) family scaffold proteins that contain PDZ, SH3, and guanylate kinase (GuK) domains and localize to tight junctions where they give structural support. They also link adhesion molecules of the tight junctions to intracellular signaling molecules (González-Mariscal et al. 2011) and ZO-2 directly binds to YAP via one of its PDZ-domains (Oka et al. 2010). Knockdown of ZO-2 in sparse breast and MDCK kidney cells in vitro abrogates nuclear localization of YAP (Oka et al. 2010; Spadaro et al. 2014), but others found that knockdown of ZO-2 in MDCK kidney cells induces nuclear localization and transcriptional activity of YAP (Domínguez-Calderón et al. 2016). How these different results can be reconciled is not resolved.
Signaling by the Fat and Dachsous Protocadherins
The Fat protocadherin is important for the establishment of planar cell polarity (PCP) and for growth control of imaginal discs in Drosophila (Bryant et al. 1988; Mahoney et al. 1991; Yang et al. 2002). Loss-of-function mutations in fat thus cause swirls of bristles instead of regular arrays, and severe overgrowth of imaginal discs and the corresponding adult structures (Bryant et al. 1988; Mahoney et al. 1991). The effects on organ growth, but not those on PCP, are mediated by the Hippo pathway (Bennett and Harvey 2006; Silva et al. 2006; Willecke et al. 2006). Fat acts as a cell-surface receptor that signals through the atypical myosin Dachs (Cho et al. 2006; Mao et al. 2006) and requires several other factors such as the SH3-domain containing protein Vamana (Vam) (Misra and Irvine 2016; Zhang et al. 2016), the palmitoyltransferase Approximated (App) (Matakatsu and Blair 2008; Matakatsu et al. 2017) and the ubiquitin ligase Fbxl7 (Bosch et al. 2014; Rodrigues-Campos and Thompson 2014) to regulate the activity of Wts. Dachs affects the protein levels of Wts (Cho et al. 2006) and its activity through inducing a conformational change that opposes activation by Hpo (Vrabioiu and Struhl 2015).
The activity of Fat for PCP and growth control is regulated by the protocadherin Dachsous (Ds) that acts as a ligand for Fat, and the extracellular kinase Four-jointed (Fj), which modulates the interaction between Fat and Ds (Yang et al. 2002; Sopko et al. 2009; Matakatsu and Blair 2012). Ds and Fj present another case of regulation of Hippo signaling through cell–cell contact mediated signals. Cells upregulate Yki activity when neighboring cells express different amounts of Ds and/or Fj and induction of artificial Ds/Fj boundaries leads to extra proliferation (Rogulja et al. 2008; Willecke et al. 2008). Notably, Ds not only acts as a ligand for Fat but also has cell autonomous effects on the Hippo pathway by acting as a receptor. Here, Ds signals through the WD40 repeat protein Riquiqui (Riq) and the dual-specificity protein kinases (DYRK) family kinase Minibrain (Mnb), which directly phosphorylates and inactivates Wts (Degoutin et al. 2013). Thus, Ds and Fat act as a pair of back-and-forth signaling receptors/ligands that organize PCP and growth. Fat and Ds are conserved in vertebrates where they also command PCP (Fanto and McNeill 2004; Hale and Strutt 2015). However, mammalian Fat/Ds signaling does not directly regulate the Hippo pathway through a conserved mechanism (Bossuyt et al. 2014; Bagherie-Lachidan et al. 2015; Kuta et al. 2016), but rather YAP localization is affected by FAT4 during heart development through sequestration of AMOTL1-YAP complexes to the plasma membrane thereby reducing YAP activity (Ragni et al. 2017).
EFFECTS OF ADHERENS JUNCTIONS ON THE ACTIVITY OF THE HIPPO PATHWAY
Adherens junctions (AJs) are the main sites that mediate adhesion between epithelial cells. They are composed of cadherin-catenin-based molecular complexes that form an adhesion belt around the apical circumference of cells (Baum and Georgiou 2011). Adherens junctions are essential for stable cell–cell adhesion and thus epithelial tissue architecture and also for normal activity of the Hippo pathway (Gumbiner and Kim 2014). Identified molecular links between adherens junctions and the Hippo pathway include the E-cadherin (E-cad) linked adaptor protein α-catenin, Merlin (NF2) and, the LIM-domain containing proteins Ajuba and Zyxin (Schroeder and Halder 2012; Gumbiner and Kim 2014; Sun and Irvine 2016). Moreover, Hippo pathway core components can localize to adherens junctions similar to tight junctions (Sun et al. 2015). However, adherens junctions do not simply provide inert sites for Hippo pathway assembly, but they can transduce the effects of mechanical stress in an epithelial tissue into the Hippo pathway (Kim et al. 2011; Benham-Pyle et al. 2015). Thus, the localization and activity of Hippo pathway components at adherens junctions is dynamically regulated. Wts for example, is active when localized to tight junctions, but inactivated when shuttled to adherens junctions via its interaction with Ajuba (Sun et al. 2015). In this section, we discuss mechanisms of how adherens junctions impact on the activity of the Hippo pathway.
E-Cadherin, α-Catenin, and β-Catenin in Mammals
Loss of the adherens junction components E-cad, α-catenin (α-cat), or β-catenin (β-cat) induces nuclear accumulation and activation of YAP in cultured endothelial cells and breast cancer cells (Kim et al. 2011; Choi et al. 2015). Similarly, expression of mutant forms of E-cad that lack the extracellular domain or the β-cat binding site induces YAP nuclear localization and activity in breast cancer cells in vitro (Kim et al. 2011). In vivo conditional deletion of α-cat in basal skin cells likewise leads to nuclear accumulation of YAP and causes hyperproliferation of progenitor cells and thickening of the epidermis (Schlegelmilch et al. 2011; Silvis et al. 2011). Similarly, loss of α-cat causes nuclear YAP accumulation, hyperproliferation of progenitor cells, and defects in organogenesis in the liver (Herr et al. 2014), heart (Li et al. 2015a), and tooth primordia (Li et al. 2016a). Thus, adherens junctions are required for proper Hippo pathway activity in multiple epithelial tissues.
Several molecular mechanisms connect the E-cad-β-cat-α-cat complex to the Hippo pathway including direct interactions whereby α-cat forms a tripartite complex with phosphorylated YAP and 14-3-3 in keratinocytes that inhibits YAP by keeping it out of the nucleus and preventing its dephosphorylation by PP2A (Schlegelmilch et al. 2011; Silvis et al. 2011). Similarly, VE-cadherin affects YAP localization by recruiting 14-3-3 and YAP to adherens junctions in endothelial cells (Giampietro et al. 2015). In addition to these direct effects, several other mechanisms link adherens junctions to the Hippo pathway. These include MST1/2 independent stimulation of LATS kinases by the Kibra, Merlin, and Na+/H+ exchanger regulatory factor (NHERF) adaptor proteins (Kim et al. 2011); activation of Akt, which can phosphorylate and inhibit YAP in endothelial cells (Choi et al. 2015; Giampietro et al. 2015); inhibition of Src, which otherwise directly activates YAP by phosphorylation (Li et al. 2016b); and other mechanisms that are discussed in the following sections.
Ajuba
Ajuba (Jub) is a Drosophila LIM-domain adaptor protein that associates with α-cat at adherens junctions (Das Thakur et al. 2010; Rauskolb et al. 2014). Loss of Ajuba in imaginal discs causes a decrease in tissue size and Yki activity (Das Thakur et al. 2010). Ajuba can bind to Wts and Sav, which causes sequestration and inactivation of Wts at adherens junctions and thus promotes Yki activity (Das Thakur et al. 2010; Rauskolb et al. 2014). Moreover, Ajuba is required for JNK-mediated activation of Yki during regeneration and tumor growth (Sun and Irvine 2013). Ajuba is also required for regulation of Yki by cytoskeletal tension in which it accumulates at adherens junctions under high cellular tension and inhibits Wts thereby promoting Yki nuclear localization and activity (Rauskolb et al. 2014; Pan et al. 2016). Thus, loss of adherens junctions causes loss of Ajuba function, activation of Wts, and ensuing inactivation of Yki.
Ajuba and its function are conserved in mammals, where its three homologs AJUBA, LIMD1, and WTIP also localize to adherens junctions and suppress the activity of the Hippo pathway by binding to LATS1/2 and SAV1 (Das Thakur et al. 2010; Schimizzi and Longmore 2015; Jagannathan et al. 2016). Moreover, JNK activity induces LATS1/2-WTIP and LATS1/2-LIMD1 interactions downstream of cytoskeletal tension thereby inducing nuclear localization and transcriptional activity of YAP (Sun and Irvine 2013; Codelia et al. 2014). Interestingly however, Ajuba proteins are required for inhibition of LATS1/2 and the Hippo pathway only in proliferating cells while it does not interact or inhibit LATS1/2 in growth-arrested cells in vitro (Jagannathan et al. 2016). Therefore, Ajuba is an adherens junction component that inhibits Hippo pathway activity and its activity is regulated by cell–cell interactions.
Role of NF2/Merlin
Another molecular link between adherens junctions and the Hippo pathways is the FERM-domain adaptor protein Mer. Mer is encoded by the NF2 tumor suppressor gene that is frequently mutant in peripheral nerve sheath tumors (Neurofibromatosis type 2) (McClatchey 2003). Mer colocalizes and interacts with adherens junction components in confluent cells and loss of Mer abrogates the development and stabilization of adherens junctions (Lallemand et al. 2003). In Drosophila, loss of Mer together with the related FERM-domain protein Ex phenocopies the hyperproliferation and overgrowth phenotypes of hpo mutants (Hamaratoglu et al. 2006). The effect of Mer on the activity of the Hippo pathway is conserved in mammals where conditional deletion of NF2 in genetically engineered mice causes YAP-dependent phenotypes in multiple tissues (Zhang et al. 2010). Thus, Mer suppresses the activity of YAP in the mouse liver (Zhang et al. 2010), in cardiomyocytes on oxidative stress (Matsuda et al. 2016), in expansion of neural progenitor cells in the brain during development of the hippocampus (Lavado et al. 2013) and in the developing mouse kidney during branching morphogenesis (Reginensi et al. 2016). Mer activates the Hippo pathway by binding to and recruiting the LATS1/2 kinases to the plasma membrane (Yin et al. 2013). Mer can also activate MST1 directly in response to oxidative stress (Matsuda et al. 2016) and promote LATS1/2 stability by inhibiting the CRL4DCAF1 E3 ubiquitin ligase in the nucleus (Li et al. 2014).
PTPN14/Pez
Pez family phosphatases are FERM-domain containing nonreceptor protein tyrosine phosphatases including PTPD1 and PTPD2 (PTPN14) in mammals and the Drosophila homolog Pez (Møller et al. 1994; Smith et al. 1995; Edwards et al. 2001). They localize to adherens junctions and affect the phosphorylation status of β-cat, which is important for E-cad/β-cat linkage and cell adhesion (Wadham et al. 2003). Pez family phosphatases were recently discovered as modulators of Yki/YAP activity (Poernbacher et al. 2012; Wang et al. 2012; Huang et al. 2013; Liu et al. 2013; Wilson et al. 2014). In confluent mammalian tissue cultures, knockdown of PTPN14 induces YAP nuclear localization, while PTPN14 overexpression in subconfluent cultures induces YAP cytoplasmic translocation (Wang et al. 2012; Liu et al. 2013; Wilson et al. 2014). This effect depends on a direct interaction between the PPxY motifs of PTPN14 and the WW domains of YAP to sequester YAP in the cytoplasm while the phosphatase activity of PTPN14 is dispensable (Wang et al. 2012; Huang et al. 2013; Liu et al. 2013). PTPN14 also binds to KIBRA, which activates LATS1/2 independently of MST1/2 (Wilson et al. 2014). In Drosophila imaginal discs, overexpression of Pez causes a decrease in tissue size but Pez mutant clones had no growth defects (Poernbacher et al. 2012). However, loss of Pez in the fly midgut epithelium resulted in ectopic Yki activation and hyperplasia (Poernbacher et al. 2012). For this function, Pez interacts with Kibra to activate Wts similar to PTPN14, (Poernbacher et al. 2012).
Different Effect of Loss of Adherens Junctions in Flies and Mammals
The stability of adherens junctions affects Hippo signaling in flies. However, the effect of loss of adherens junctions in flies is opposite to that in mammalian cells. In flies, knockdown of E-cad, α-cat, or β-cat in wing imaginal discs causes a cell autonomous decrease in Yki activity (Yang et al. 2015) while, loss of E-cad or α-cat in vertebrates in vitro or in vivo in liver, skin, and heart leads to nuclear localization and hyperactivation of YAP (Schlegelmilch et al. 2011; Silvis et al. 2011; Herr et al. 2014; Li et al. 2015a). This difference indicates different wiring of the Hippo pathway in Drosophila versus mammalian adherens junctions. One possible explanation for this difference is that Drosophila adherens junctions contain the cell adhesion molecule Echinoid (Ed) that is not present in mammals (Wei et al. 2005). Ed physically interacts with Sav and causes activation of the Hippo pathway (Yue et al. 2012). Thus, Ed may still activate the Hippo kinases in the absence of E-cad, which may explain why loss of E-cad suppresses Yki in flies but activates YAP in mammalian cells. The different effects of loss of adherens junctions on the Hippo pathway in flies and mammals, although most of the components of the Hippo pathway are conserved, indicate that the balance of the activities between different branches of the Hippo pathway determines the final Yki/YAP/TAZ activity. In summary, as for tight junctions, multiple molecular interactions connect adherens junctions with the Hippo pathway.
EFFECTS OF FOCAL ADHESIONS ON THE ACTIVITY OF THE HIPPO PATHWAY
In addition to cell–cell interactions, interaction with the extracellular matrix is also an important modulator of Hippo signaling. Focal adhesions are main contact points between cells and the extracellular matrix and harbor integrins as adhesion proteins (Burridge and Chrzanowska-Wodnicka 1996). Integrins signal through a complex of proteins including the actin binding protein Vinculin, and the integrin-linked kinase (ILK), Src, and FAK kinases to anchor and organize stress fibers (Hynes 2002) and to signal to several downstream pathways (Moreno-Layseca and Streuli 2014) including the Hippo pathway (Dupont 2016; Kuroda et al. 2017). The activity of integrins promotes the nuclear accumulation and activity of YAP and TAZ (Tang et al. 2013; Kaneko et al. 2014; Chang et al. 2015; Elbediwy et al. 2016). Thus, β1-integrin is required for YAP/TAZ activity in the interfollicular epidermis (Elbediwy et al. 2016), in hair follicles (Elbediwy et al. 2016), and in skeletal stem cells (Tang et al. 2013) in vivo, while β3-integrin is required for YAP/TAZ activation in response to unidirectional shear stress in endothelium (Wang et al. 2016). Similarly, integrin-αv is required for nuclear accumulation of YAP/TAZ in primary osteoblastic cells (Kaneko et al. 2014), and α6β1 integrin is required for both activity and nuclear accumulation of TAZ in breast cancer stem cells (Chang et al. 2015). Moreover, the extracellular matrix (ECM) protein Agrin signals through integrins and causes activation of YAP (Chakraborty et al. 2017). Several mechanisms mediate the effects of integrins on YAP/TAZ. ILK is an important transducer of integrin signaling (McDonald et al. 2008). ILK modulates Hippo pathway activity in different types of epithelial cells in vitro, including cells from the colon, prostate, and breast (Serrano et al. 2013). ILK phosphorylates the MYPT1 phosphatase, which dephosphorylates and activates Mer (Jin et al. 2006), which in turn activates Hippo signaling and inhibits YAP/TAZ (Serrano et al. 2013).
The integrin-associated FAK and Src kinases also regulate Hippo signaling. Activation of Src and FAK on attachment of cultured cells to fibronectin-coated substrates activates YAP (Kim and Gumbiner 2015). Src and FAK activate PI3K and PDK1, which then inhibit LATS1/2 and enhance nuclear localization of YAP/TAZ (Kim and Gumbiner 2015). Similar effects may activate YAP in epidermal cells in vitro where β1-integrin signaling promotes nuclear localization of YAP through a process that is sensitive to pharmacological inhibition of Src and involves suppression of YAP phosphorylation by LATS1/2 (Elbediwy et al. 2016). Src is also implicated in directly activating YAP in response to inflammation. Here, activation of the IL6 coreceptor gp130 recruits and activates the Src and Yes kinases, which then phosphorylate YAP at tyrosine 357 to increase the stability and activity of YAP (Taniguchi et al. 2015). Yes kinase specific phosphorylation of YAP induces localization of a complex involving YAP, TBX5, and β-cat to the promoters of antiapoptotic BCL2L1 and BIRC5 genes in β-cat active cancer cell lines (Rosenbluh et al. 2012). Although removal of YAP or Yes kinase inhibits proliferation of β-cat active cancer cells, Src kinase has no effect on the growth of these cells (Rosenbluh et al. 2012). YAP nuclear localization also depends on the activity of Src kinase and its substrate p130Cas downstream of integrin-αv in primary osteoblastic cells (Kaneko et al. 2014). All in all, focal adhesions can regulate Hippo signaling and the activity of its effectors YAP and TAZ by transmitting information about the physical environment of the cell and this regulation involves integrins and downstream kinases such as Src, FAK, and ILK.
REGULATION OF HIPPO SIGNALING BY THE CYTOSKELETON
Cell junctions typically connect to and organize the actin cytoskeleton (Collins and Nelson 2015). Several cell junction components that affect the Hippo pathway can also bind to F-actin and induce cytoskeletal remodeling. These include the adherens junction proteins α-cat and Mer, the AMOTs at tight junctions, and integrins in focal adhesions (Gaspar and Tapon 2014; Sun and Irvine 2016). The actin cytoskeleton may thus transduce junctional signals to the Hippo pathway. Indeed, experimentally induced F-actin remodeling has a strong effect on the activity of the Hippo pathway (Dupont et al. 2011; Fernández et al. 2011; Sansores-Garcia et al. 2011; Wada et al. 2011; Zhao et al. 2012). In this section, we discuss recent insights on how cytoskeletal elements affect Hippo signaling.
Structural Changes in the Actin Cytoskeleton
In 2D cultured cells, nuclear accumulation of YAP generally correlates with the presence of F-actin stress fibers (Dupont et al. 2011; Wada et al. 2011; Aragona et al. 2013). Thus, cells grown on stiff substrates, such as in a plastic dish, produce stress fibers and have nuclear active YAP, while cells grown on soft substrates round up, lack stress fibers, and have cytoplasmic YAP. The status of the F-actin cytoskeleton acts upstream of YAP localization because experimental manipulations that induce or disrupt stress fibers affect YAP localization and activity. Thus, pharmacologic disruption of F-actin by administration of latrunculin, cytochalasin D, the Rho inhibitor botulinum toxin C3 or the ROCK inhibitor Y27632 cause nuclear exclusion of YAP in cultured cells (Dupont et al. 2011; Sansores-Garcia et al. 2011; Wada et al. 2011; Zhao et al. 2012). Conversely, inducing the formation of ectopic stress fibers by knockdown of Capping proteins, the actin severing factors Cofilin and Gelsolin, or by expression of a constitutively active form of the Diaphanous formin, induces nuclear accumulation and increased activity of YAP/TAZ (Sansores-Garcia et al. 2011; Aragona et al. 2013). Moreover, Amots have the ability to bind F-actin structures, and expression of Amots can induce the formation of extra F-actin bundles (Ernkvist et al. 2008; Gagné et al. 2009). Notably, F-actin competes with YAP for AMOT binding and ectopic F-actin may thus promote nuclear localization of YAP by freeing it from AMOT (Mana-Capelli et al. 2014). LATS2 also regulates YAP by phosphorylating AMOT which disrupts AMOT-F-actin binding thereby enabling YAP-AMOT interaction (Dai et al. 2013; Mana-Capelli et al. 2014).
The status of the cytoskeleton also affects the Hippo pathway in vivo. In Drosophila, ectopic F-actin accumulation caused by loss of capping proteins or by expression of constitutively active Diaphanous induces Yki activity and causes overgrowth of imaginal discs by affecting the Hippo pathway upstream of Wts (Fernández et al. 2011; Sansores-Garcia et al. 2011). In addition, activation of the LIM-domain protein Zyxin by membrane targeting promotes Yki activity and tissue growth while loss of Zyxin suppresses the growth in imaginal discs (Gaspar et al. 2015). Zyxin antagonizes the function of Ex and it promotes growth by interacting with the actin polymerization factor Enabled (Ena) (Gaspar et al. 2015). Similar to flies, Cdc42-mediated actin polymerization is an important regulator of YAP in mice where loss of Cdc42 induces nuclear exclusion of YAP in both kidney and lung cells (Reginensi et al. 2013; Liu et al. 2016).
Spectrins
The spectrin proteins form a subcortical network to support the mechanical properties of the cell (Machnicka et al. 2012). There are three spectrin proteins α-, β-, and βH-spectrin present in Drosophila and Caenorhabditis elegans while humans have two α, four β, and one βH spectrin (Bennett and Baines 2001). Spectrins exist as α-β heterodimers and these heterodimers form tetramers that assemble into a fibrillar meshwork beneath the plasma membrane by binding to short actin filaments (Machnicka et al. 2012). In Drosophila, α-spectrin localizes apically and basally while βH-spectrin localizes only apically and β-spectrin only basally. The apical spectrins are important for normal activity of the Hippo pathway in imaginal discs. Therefore, loss-of-function mutations in α and βH-spectrins cause a slight hyperactivation of Yki and imaginal disc overgrowth (Deng et al. 2015; Fletcher et al. 2015; Wong et al. 2015). On the other hand, basal spectrins are essential for tissue homeostasis in follicle cells and intestinal stem cells. Thus mutations in α and β-spectrins cause Yki hyperactivity in these cells (Deng et al. 2015; Fletcher et al. 2015; Wong et al. 2015). Mechanistically, apical spectrins bind to Ex and induce clustering of upstream Hippo pathway components including Ex, Crb, Mer, and Kib, which promotes Hippo pathway activity and inhibits Yki (Fletcher et al. 2015). Spectrins may also inhibit Yki activity by modulating the tension in the actomyosin cytoskeleton (Deng et al. 2015). Knockdown of α-spectrin and β-spectrin also causes nuclear localization of YAP in vitro in mammalian cells (Wong et al. 2015; Deng et al. 2015).
Regulation of the Hippo Pathway by Cellular Tension
Cellular tension promotes nuclear accumulation of YAP in mammalian cells (Aragona et al. 2013; Codelia et al. 2014; Benham-Pyle et al. 2015). Although cytoskeletal tension and the architecture of the F-actin cytoskeleton are interconnected and dependent on each other, it appears that their effects on the Hippo pathway are mediated by separable molecular mechanisms (Das et al. 2016). In sparsely cultured MCF10A cells, F-actin stress fibers promote nuclear localization of YAP through a mechanism that is independent of the YAP phosphorylation status (Dupont et al. 2011; Das et al. 2016), whereas inhibition of actomyosin contractility causes YAP phosphorylation at LATS1/2 phosphorylation sites and its nuclear export in sparse mouse embryonic fibroblasts (MEFs) (Das et al. 2016). Therefore, the structure of the F-actin cytoskeleton and the tension that it generates affect Hippo pathway activity through distinct mechanisms.
While the mechanisms at work are poorly understood, there are some glimpses of potential mechanisms. Cyclic stretch of cells activates JNK, which phosphorylates the Ajuba family protein LIMD1 enabling its interaction with LATS1/2 (Codelia et al. 2014). This then inhibits LATS1/2 and promotes nuclear accumulation of YAP (Codelia et al. 2014). Moreover, transfer of cytoskeletal tension to the nucleus regulates the activity of YAP in mesenchymal stem cells through a process that requires Nesprin 1 giant and Lamin A/C on the nuclear envelope (Bertrand et al. 2014; Driscoll et al. 2015).
However, although many different molecular links between the cytoskeletal changes and YAP have been discovered, none of them alone can explain the dramatic effect of modulating the cytoskeleton on YAP. Therefore, how the actomyosin cytoskeleton regulates YAP/TAZ is still not understood.
CELL-JUNCTION-DEPENDENT REGULATION OF THE HIPPO PATHWAY IN DEVELOPMENT AND DISEASE
Modulation of Hippo pathway activity by cell–cell and cell–ECM interactions is important in development and for homeostasis. A prominent example is the regulation of YAP activity in developing mouse embryos (Nishioka et al. 2009). After blastocyst formation, the inner cell mass (ICM) gives rise to embryonic tissues, while the outer cell layer gives rise to the extraembryonic trophectoderm. Differences in Hippo signaling activity determine these two cell types: YAP is nuclear in trophectoderm cells and cytoplasmic in ICM cells (Nishioka et al. 2009). Deletion of Nf2 in all cells causes loss of LATS1/2-dependent YAP phosphorylation in the inner cell mass leading to nuclear YAP accumulation and the formation of extra trophectoderm at the expense of ICM (Cockburn et al. 2013). Mechanistically, AMOT and AMOTL2 localize apically in polarized outer cells where they are sequestered away from LATS1/2 and YAP, which reduces the activity of Hippo signaling resulting in nuclear and active YAP (Hirate et al. 2013; Hirate and Sasaki 2014). In contrast, in ICM cells, AMOT and AMOTL2 localize to adherens junctions where they are phosphorylated and in complex with LATS1/2. This interaction activates LATS1/2, which phosphorylates YAP, causing its nuclear exclusion, and thereby prevents the differentiation of ICM cells into trophectoderm (Hirate et al. 2013; Leung and Zernicka-Goetz 2013).
Another important example is the effect of focal adhesions on YAP activity during stem-cell-fate decisions. Integrin-mediated regulation of YAP activity can affect the differentiation process. For example, decreased β1-integrin activity on loss of membrane-anchored metalloproteinase MT1-MMP (Mmp14) in skeletal stem cells causes decreased YAP activity, which leads to change in the fate of skeletal stem cells from osteogenesis to adipogenesis and chondrogenesis in vivo (Tang et al. 2013).
One of the most prominent consequences of the regulation the Hippo pathway by cell junctions is in cancer cells. Cancer cells often lose junctional integrity and apico-basal cell polarity (Gandalovičová et al. 2016), which may cause or at least contribute to activation of YAP, which is seen with high frequency in many different types of cancer (Harvey et al. 2013; Halaoui and McCaffrey 2014; Zanconato et al. 2016). In addition, regulation by F-actin may be important in cancer cells. For example in uveal melanoma, activating mutations in the catalytic domain of Gq/11, induce Trio/Rho-Rac signaling which causes F-actin polymerization and leads to YAP nuclear localization and activation (Feng et al. 2014; Yu et al. 2014).
CONCLUDING REMARKS: ARE SIGNALS FROM CELL JUNCTIONS PERMISSIVE OR INSTRUCTIVE FOR HIPPO SIGNALING?
Many connections between cell–cell and cell–matrix junctions and the Hippo pathway have been identified. It is clear that defects in cellular junctions can have profound effects on the activity of the Hippo pathway. Often these effects have been interpreted as evidence that these junctions and the proteins involved regulate the activity of the Hippo pathway in the sense that they transduce information and modulate the activity of the pathway in response to changes in junctional strength, extracellular signals, etc. However, while it is clear that differences in junctional strength cause differences in the amount of Hippo pathway activity, it is mostly not obvious under which natural circumstances such changes are actually occurring and impacting the biology of an organism. Thus, it will be important in future experiments to test whether a particular junction acts instructive in that it transduces information into the Hippo pathway and thus leads to a differential Hippo pathway activity between cells, or whether a junction is simply permissive for the assembly of the Hippo pathway complex that is nevertheless essential for normal cellular functioning.
ACKNOWLEDGMENTS
We would like to thank members of the Halder laboratory for helpful discussions in preparation of this manuscript. This work was supported by The Research Foundation - Flanders (FWO, www.fwo.be) Odysseus Type I, and grants G0306.16, G0665.14, and G.0954.16 to G.H.
REFERENCES
- Adler JJ, Heller BL, Bringman LR, Ranahan WP, Cocklin RR, Goebl MG, Oh M, Lim HS, Ingham RJ, Wells CD. 2013a. Amot130 adapts atrophin-1 interacting protein 4 to inhibit yes-associated protein signaling and cell growth. J Biol Chem 288: 15181–15193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler JJ, Johnson DE, Heller BL, Bringman LR, Ranahan WP, Conwell MD, Sun Y, Hudmon A, Wells CD. 2013b. Serum deprivation inhibits the transcriptional co-activator YAP and cell growth via phosphorylation of the 130-kDa isoform of Angiomotin by the LATS1/2 protein kinases. Proc Natl Acad Sci 110: 17368–17373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JM, Van Itallie CM, Fanning AS. 2004. Setting up a selective barrier at the apical junction complex. Curr Opin Cell Biol 16: 140–145. [DOI] [PubMed] [Google Scholar]
- Angus L, Moleirinho S, Herron L, Sinha A, Zhang X, Niestrata M, Dholakia K, Prystowsky MB, Harvey KF, Reynolds PA, et al. 2012. Willin/FRMD6 expression activates the Hippo signaling pathway kinases in mammals and antagonizes oncogenic YAP. Oncogene 31: 238–250. [DOI] [PubMed] [Google Scholar]
- Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, Piccolo S. 2013. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 154: 1047–1059. [DOI] [PubMed] [Google Scholar]
- Archibald A, Al-Masri M, Liew-Spilger A, McCaffrey L. 2015. Atypical protein kinase C induces cell transformation by disrupting Hippo/Yap signaling. Mol Biol Cell 26: 3578–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagherie-Lachidan M, Reginensi A, Zaveri HP, Scott DA, Helmbacher F, McNeill H. 2015. Stromal Fat4 acts non-autonomously with Dachsous1/2 to restrict the nephron progenitor pool. Development 142: 2564–2573. [DOI] [PubMed] [Google Scholar]
- Baum B, Georgiou M. 2011. Dynamics of adherens junctions in epithelial establishment, maintenance, and remodeling. J Cell Biol 192: 907–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner R, Poernbacher I, Buser N, Hafen E, Stocker H. 2010. The WW Domain Protein Kibra Acts Upstream of Hippo in Drosophila. Dev Cell 18: 309–316. [DOI] [PubMed] [Google Scholar]
- Benham-Pyle BW, Pruitt BL, Nelson WJ. 2015. Mechanical strain induces E-cadherin-dependent Yap1 and β-catenin activation to drive cell cycle entry. Science 348: 1024–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett FC, Harvey KF. 2006. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol 16: 2101–2110. [DOI] [PubMed] [Google Scholar]
- Bennett V, Baines AJ. 2001. Spectrin and Ankyrin-Based Pathways: Metazoan Inventions for Integrating Cells Into Tissues. Physiol Rev 81: 1353–1392. [DOI] [PubMed] [Google Scholar]
- Bertrand AT, Ziaei S, Ehret C, Duchemin H, Mamchaoui K, Bigot A, Mayer M, Quijano-Roy S, Desguerre I, Lainé J, et al. 2014. Cellular micro-environments reveal defective mechanosensing responses and elevated YAP signaling in LMNA-mutated muscle precursors. J Cell Sci 127: 2873–2884. [DOI] [PubMed] [Google Scholar]
- Boedigheimer M, Laughon A. 1993. Expanded: A gene involved in the control of cell proliferation in imaginal discs. Development 118: 1291–1301. [DOI] [PubMed] [Google Scholar]
- Bosch JA, Sumabat TM, Hafezi Y, Pellock BJ, Gandhi KD, Hariharan IK. 2014. The Drosophila F-box protein Fbxl7 binds to the protocadherin fat and regulates Dachs localization and Hippo signaling. eLife 3: e03383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossuyt W, Chen CL, Chen Q, Sudol M, McNeill H, Pan D, Kopp A, Halder G. 2014. An evolutionary shift in the regulation of the Hippo pathway between mice and flies. Oncogene 33: 1218–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant PJ, Huettner B, Held LI, Ryerse J, Szidonya J. 1988. Mutations at the fat locus interfere with cell proliferation control and epithelial morphogenesis in Drosophila. Dev Biol 129: 541–554. [DOI] [PubMed] [Google Scholar]
- Bulgakova NA, Knust E. 2009. The Crumbs complex: From epithelial-cell polarity to retinal degeneration. J Cell Sci 122: 2587–2596. [DOI] [PubMed] [Google Scholar]
- Burridge K, Chrzanowska-Wodnicka M. 1996. Focal adhesions, contractility, and signaling. Annu Rev Cell Dev Biol 12: 463–519. [DOI] [PubMed] [Google Scholar]
- Chakraborty S, Njah K, Pobbati AV, Lim YB, Raju A, Lakshmanan M, Tergaonkar V, Lim CT, Hong W. 2017. Agrin as a Mechanotransduction Signal Regulating YAP through the Hippo Pathway. Cell Rep 18: 2464–2479. [DOI] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Chong YF, Pobbati AV, Huang C, Hong W. 2011. Hippo pathway-independent restriction of TAZ and YAP by Angiomotin. J Biol Chem 286: 7018–7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Guo F, Tan I, Leung T, Hong W. 2013. Actin-binding and cell proliferation activities of angiomotin family members are regulated by Hippo pathway-mediated phosphorylation. J Biol Chem 288: 37296–37307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Loo LS, Chong YF, Huang C, Hong W. 2009. TEADs mediate nuclear retention of TAZ to promote oncogenic transformation. J Biol Chem 284: 14347–14358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Goel HL, Gao H, Pursell B, Shultz LD, Greiner DL, Ingerpuu S, Patarroyo M, Cao S, Lim E, et al. 2015. A laminin 511 matrix is regulated by Taz and functions as the ligand for the α6β1 integrin to sustain breast cancer stem cells. Genes Dev 29: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Gajewski KM, Hamaratoglu F, Bossuyt W, Sansores-Garcia L, Tao C, Halder G. 2010. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc Natl Acad Sci 107: 15810–15815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. 2006. Delineation of a Fat tumor suppressor pathway. Nat Genet 38: 1142–1150. [DOI] [PubMed] [Google Scholar]
- Choi HJ, Zhang H, Park H, Choi KS, Lee HW, Agrawal V, Kim YM, Kwon YG. 2015. Yes-associated protein regulates endothelial cell contact-mediated expression of angiopoietin-2. Nat Commun 6: 6943. [DOI] [PubMed] [Google Scholar]
- Cockburn K, Biechele S, Garner J, Rossant J. 2013. The hippo pathway member Nf2 is required for inner cell mass specification. Curr Biol 23: 1195–1201. [DOI] [PubMed] [Google Scholar]
- Codelia VA, Sun G, Irvine KD. 2014. Regulation of YAP by mechanical strain through Jnk and Hippo signaling. Curr Biol 24: 2012–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C, Nelson WJ. 2015. Running with neighbors: Coordinating cell migration and cell-cell adhesion. Curr Opin Cell Biol 36: 62–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox CM, Mandell EK, Stewart L, Lu R, Johnson DL, McCarter SD, Tavares A, Runyan R, Ghosh S, Wilson JM. 2015. Endosomal regulation of contact inhibition through the AMOT:YAP pathway. Mol Biol Cell 26: 2673–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, She P, Chi F, Feng Y, Liu H, Jin D, Zhao Y, Guo X, Jiang D, Guan KL, et al. 2013. Phosphorylation of angiomotin by Lats1/2 kinases inhibits F-actin binding, cell migration, and angiogenesis. J Biol Chem 288: 34041–34051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Fischer RS, Pan D, Waterman CM. 2016. YAP nuclear localization in the absence of cell-cell contact is mediated by a filamentous actin-dependent, Myosin II and Phospho-YAP-independent pathway during extracellular matrix mechanosensing. J Biol Chem 291: 6096–6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Thakur M, Feng Y, Jagannathan R, Seppa MJ, Skeath JB, Longmore GD. 2010. Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Curr Biol 20: 657–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degoutin JL, Milton CC, Yu E, Tipping M, Bosveld F, Yang L, Bellaiche Y, Veraksa A, Harvey KF. 2013. Riquiqui and Minibrain are regulators of the Hippo pathway downstream of Dachsous. Nat Cell Biol 15: 1176–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Wang W, Yu J, Zheng Y, Qing Y, Pan D. 2015. Spectrin regulates Hippo signaling by modulating cortical actomyosin activity. eLife 4: e06567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Calderón A, Ávila-Flores A, Ponce A, López-Bayghen E, Calderón-Salinas JV, Reyes JL, Chávez-Munguía B, Segovia J, Angulo C, Ramírez L, et al. 2016. ZO-2 silencing induces renal hypertrophy through a cell cycle mechanism and the activation of YAP and the mTOR pathway. Mol Biol Cell 27: 1581–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. 2007. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130: 1120–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll TP, Cosgrove BD, Heo SJ, Shurden ZE, Mauck RL. 2015. Cytoskeletal to Nuclear Strain Transfer Regulates YAP Signaling in Mesenchymal Stem Cells. Biophys J 108: 2783–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S. 2016. Role of YAP/TAZ in cell-matrix adhesion-mediated signalling and mechanotransduction. Exp Cell Res 343: 42–53. [DOI] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. 2011. Role of YAP/TAZ in mechanotransduction. Nature 474: 179–183. [DOI] [PubMed] [Google Scholar]
- Edwards K, Davis T, Marcey D, Kurihara J, Yamamoto D. 2001. Comparative analysis of the Band 4.1/ezrin-related protein tyrosine phosphatase Pez from two Drosophila species: Implications for structure and function. Gene 275: 195–205. [DOI] [PubMed] [Google Scholar]
- Elbediwy A, Vincent-Mistiaen ZI, Spencer-Dene B, Stone RK, Boeing S, Wculek SK, Cordero J, Tan EH, Ridgway R, Brunton VG, et al. 2016. Integrin signalling regulates YAP/TAZ to control skin homeostasis. Development 143: 1674–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernkvist M, Aase K, Ukomadu C, Wohlschlegel J, Blackman R, Veitonmäki N, Bratt A, Dutta A, Holmgren L. 2006. Angiomotin associates to actin and controls endothelial cell shape. FEBS J 273: 2000–2011. [DOI] [PubMed] [Google Scholar]
- Ernkvist M, Birot O, Sinha I, Veitonmaki N, Nyström S, Aase K, Holmgren L. 2008. Differential roles of p80- and p130-angiomotin in the switch between migration and stabilization of endothelial cells. Biochim Biophys Acta Mol Cell Res 1783: 429–437. [DOI] [PubMed] [Google Scholar]
- Fanto M, McNeill H. 2004. Planar polarity from flies to vertebrates. J Cell Sci 117: 527–533. [DOI] [PubMed] [Google Scholar]
- Feng X, Degese M, Iglesias-Bartolome R, Vaque JP, Molinolo AA, Rodrigues M, Zaidi MR, Ksander BR, Merlino G, Sodhi A, et al. 2014. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a Trio-regulated Rho GTPase Signaling Circuitry. Cancer Cell 25: 831–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández BG, Gaspar P, Brás-Pereira C, Jezowska B, Rebelo SR, Janody F. 2011. Actin-Capping Protein and the Hippo pathway regulate F-actin and tissue growth in Drosophila. Development 138: 2337–2346. [DOI] [PubMed] [Google Scholar]
- Fernando RN, Cotter L, Perrin-Tricaud C, Berthelot J, Bartolami S, Pereira JA, Gonzalez S, Suter U, Tricaud N. 2016. Optimal myelin elongation relies on YAP activation by axonal growth and inhibition by Crb3/Hippo pathway. Nat Commun 7: 12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher GC, Elbediwy A, Khanal I, Ribeiro PS, Tapon N, Thompson BJ. 2015. The Spectrin cytoskeleton regulates the Hippo signalling pathway. EMBO J 34: 940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher GC, Lucas EP, Brain R, Tournier A, Thompson BJ. 2012. Positive feedback and mutual antagonism combine to polarize crumbs in the Drosophila follicle cell epithelium. Curr Biol 22: 1116–1122. [DOI] [PubMed] [Google Scholar]
- Gagné V, Moreau J, Plourde M, Lapointe M, Lord M, Gagnon É, Fernandes MJG. 2009. Human angiomotin-like 1 associates with an angiomotin protein complex through its coiled-coil domain and induces the remodeling of the actin cytoskeleton. Cell Motil Cytoskeleton 66: 754–768. [DOI] [PubMed] [Google Scholar]
- Galli GG, Carrara M, Yuan WC, Valdes-Quezada C, Gurung B, Pepe-Mooney B, Zhang T, Geeven G, Gray NS, de Laat W, et al. 2015. YAP Drives Growth by Controlling Transcriptional Pause Release from Dynamic Enhancers. Mol Cell 60: 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandalovičová A, Vomastek T, Rosel D, Brábek J. 2016. Cell polarity signaling in the plasticity of cancer cell invasiveness. Oncotarget 7: 25022–25049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar P, Tapon N. 2014. Sensing the local environment: Actin architecture and Hippo signalling. Curr Opin Cell Biol 31: 74–83. [DOI] [PubMed] [Google Scholar]
- Gaspar P, Holder MV, Aerne BL, Janody F, Tapon N. 2015. Zyxin antagonizes the FERM protein expanded to couple F-actin and yorkie-dependent organ growth. Curr Biol 25: 679–689. [DOI] [PubMed] [Google Scholar]
- Genevet A, Polesello C, Blight K, Robertson F, Collinson LM, Pichaud F, Tapon N. 2009. The Hippo pathway regulates apical-domain size independently of its growth-control function. J Cell Sci 122: 2360–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N. 2010. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev Cell 18: 300–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giampietro C, Disanza A, Bravi L, Barrios-Rodiles M, Corada M, Frittoli E, Savorani C, Lampugnani MG, Boggetti B, Niessen C, et al. 2015. The actin-binding protein EPS8 binds VE-cadherin and modulates YAP localization and signaling. J Cell Biol 211: 1177–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Mariscal L, Quirós M, Díaz-Coránguez M. 2011. ZO proteins and redox-dependent processes. Antioxid Redox Signal 15: 1235–1253. [DOI] [PubMed] [Google Scholar]
- Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber J, Zider A. 2008. SCALLOPED interacts with YORKIE, the nuclear effector of the Hippo tumor-suppressor pathway in Drosophila. Curr Biol 18: 435–441. [DOI] [PubMed] [Google Scholar]
- Grzeschik NA, Parsons LM, Allott ML, Harvey KF, Richardson HE. 2010. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo Pathway through two distinct mechanisms. Curr Biol 20: 573–581. [DOI] [PubMed] [Google Scholar]
- Gumbiner BM, Kim NG. 2014. The Hippo-YAP signaling pathway and contact inhibition of growth. J Cell Sci 127: 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafezi Y, Bosch JA, Hariharan IK. 2012. Differences in levels of the transmembrane protein Crumbs can influence cell survival at clonal boundaries. Dev Biol 368: 358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaoui R, McCaffrey L. 2014. Rewiring cell polarity signaling in cancer. Oncogene 34: 1–12. [DOI] [PubMed] [Google Scholar]
- Halder G, Johnson RL. 2011. Hippo signaling: Growth control and beyond. Development 138: 9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Dupont S, Piccolo S. 2012. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat Rev Mol Cell Bio 13: 591–600. [DOI] [PubMed] [Google Scholar]
- Hale R, Strutt D. 2015. Conservation of planar polarity pathway function across the animal kingdom. Annu Rev Genet 49: 529–551. [DOI] [PubMed] [Google Scholar]
- Hamaratoglu F, Gajewski K, Sansores-Garcia L, Morrison C, Tao C, Halder G. 2009. The Hippo tumor-suppressor pathway regulates apical-domain size in parallel to tissue growth. J Cell Sci 122: 2351–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. 2006. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol 8: 27–36. [DOI] [PubMed] [Google Scholar]
- Hao Y, Chun A, Cheung K, Rashidi B, Yang X. 2008. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem 283: 5496–5509. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK. 2003. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114: 457–467. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Tapon N. 2007. The Salvador-Warts-Hippo pathway—an emerging tumour-suppressor network. Nat Rev Cancer 7: 182–191. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Zhang X, Thomas DM. 2013. The Hippo pathway and human cancer. Nat Rev Cancer 13: 246–257. [DOI] [PubMed] [Google Scholar]
- Heller B, Adu-Gyamfi E, Smith-Kinnaman W, Babbey C, Vora M, Xue Y, Bittman R, Stahelin RV, Wells CD. 2010. Amot recognizes a juxtanuclear endocytic recycling compartment via a novel lipid binding domain. J Biol Chem 285: 12308–12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr KJ, Tsang YHN, Ong JWE, Li Q, Yap LL, Yu W, Yin H, Bogorad RL, Dahlman JE, Chan YG, et al. 2014. Loss of α-catenin elicits a cholestatic response and impairs liver regeneration. Sci Rep 4: 6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirate Y, Hirahara S, Inoue KI, Suzuki A, Alarcon VB, Akimoto K, Hirai T, Hara T, Adachi M, Chida K, et al. 2013. Polarity-dependent distribution of angiomotin localizes hippo signaling in preimplantation embryos. Curr Biol 23: 1181–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirate Y, Sasaki H. 2014. The role of angiomotin phosphorylation in the Hippo pathway during preimplantation mouse development. Tissue Barriers 2: e28127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. 2005. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell 122: 421–434. [DOI] [PubMed] [Google Scholar]
- Huang JM, Nagatomo I, Suzuki E, Mizuno T, Kumagai T, Berezov A, Zhang H, Karlan B, Greene MI, Wang Q. 2013. YAP modifies cancer cell sensitivity to EGFR and survivin inhibitors and is negatively regulated by the nonreceptor type protein tyrosine phosphatase 14. Oncogene 32: 2220–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO. 2002. Integrins: Bidirectional, allosteric signaling machines. Cell 110: 673–687. [DOI] [PubMed] [Google Scholar]
- Jagannathan R, Schimizzi GV, Zhang K, Loza AJ, Yabuta N, Nojima H, Longmore GD. 2016. AJUBA LIM proteins limit Hippo activity in proliferating cells by sequestering the Hippo core kinase complex in the cytosol. Mol Cell Biol 36: 2526–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Zhang W, Wang B, Trinko R, Jiang J. 2003. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev 17: 2514–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Sperka T, Herrlich P, Morrison H. 2006. Tumorigenic transformation by CPI-17 through inhibition of a merlin phosphatase. Nature 442: 576–579. [DOI] [PubMed] [Google Scholar]
- Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. 1995. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev 9: 534–546. [DOI] [PubMed] [Google Scholar]
- Kaneko K, Ito M, Naoe Y, Lacy-Hulbert A, Ikeda K. 2014. Integrin αv in the mechanical response of osteoblast lineage cells. Biochem Biophys Res Commun 447: 352–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kango-Singh M, Nolo R, Tao C, Verstreken P, Hiesinger PR, Bellen HJ, Halder G. 2002. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development 129: 5719–5730. [DOI] [PubMed] [Google Scholar]
- Kim NG, Koh E, Chen X, Gumbiner BM. 2011. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc Natl Acad Sci 108: 11930–11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NG, Gumbiner BM. 2015. Adhesion to fibronectin regulates Hippo signaling via the FAK-Src-PI3K pathway. J Cell Biol 210: 503–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremerskothen J, Plaas C, Büther K, Finger I, Veltel S, Matanis T, Liedtke T, Barnekow A. 2003. Characterization of KIBRA, a novel WW domain-containing protein. Biochem Biophys Res Commun 300: 862–867. [DOI] [PubMed] [Google Scholar]
- Kuroda M, Wada H, Kimura Y, Ueda K, Kioka N. 2017. Vinculin promotes nuclear localization of TAZ to inhibit ECM stiffness-dependent differentiation into adipocytes. J Cell Sci 130: 989–1002. [DOI] [PubMed] [Google Scholar]
- Kuta A, Mao Y, Martin T, Ferreira de Sousa C, Whiting D, Zakaria S, Crespo-Enriquez I, Evans P, Balczerski B, Mankoo B, et al. 2016. Fat4-Dchs1 signalling controls cell proliferation in developing vertebrae. Development 143: 2367–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, Li Y. 2005. Control of cell proliferation and apoptosis by Mob as tumor suppressor, Mats. Cell 120: 675–685. [DOI] [PubMed] [Google Scholar]
- Lallemand D, Curto M, Saotome I, Giovannini M, McClatchey AI. 2003. NF2 deficiency promotes tumorigenesis and metastasis by destabilizing adherens junctions. Genes Dev 17: 1090–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavado A, He Y, Paré J, Neale G, Olson EN, Giovannini M, Cao X. 2013. Tumor suppressor Nf2 limits expansion of the neural progenitor pool by inhibiting Yap/Taz transcriptional coactivators. Development 140: 3323–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers C, Michel D, Lane-Guermonprez L, Delgrossi M-H, Médina E, Arsanto JP, Le Bivic A. 2004. CRB3 binds directly to Par6 and regulates the morphogenesis of the tight junctions in mammalian epithelial cells. Mol Biol Cell 15: 1324–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung CY, Zernicka-Goetz M. 2013. Angiomotin prevents pluripotent lineage differentiation in mouse embryos via Hippo pathway-dependent and independent mechanisms. Nat Commun 4: 2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Cooper J, Zhou L, Yang C, Erdjument-Bromage H, Zagzag D, Snuderl M, Ladanyi M, Hanemann CO, Zhou P, et al. 2014. Merlin/NF2 loss-driven tumorigenesis linked to CRL4DCAF1-mediated Inhibition of the Hippo pathway kinases Lats1 and 2 in the nucleus. Cancer Cell 26: 48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Gao E, Vite A, Yi R, Gomez L, Goossens S, Van Roy F, Radice GL. 2015a. Alpha-catenins control cardiomyocyte proliferation by regulating yap activity. Circ Res 116: 70–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Mao X, Ren Y, Liu P. 2015b. Epithelial cell polarity determinant CRB3 in cancer development. Int J Biol Sci 11: 31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Silvis MR, Honaker Y, Lien WH, Arron ST, Vasioukhin V. 2016b. αE-catenin inhibits a Src-YAP1 oncogenic module that couples tyrosine kinases and the effector of hippo signaling pathway. Genes Dev 30: 798–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CY, Hu J, Lu H, Lan J, Du W, Galicia N, Klein OD. 2016a. αE-catenin inhibits YAP/TAZ activity to regulate signalling centre formation during tooth development. Nat Commun 7: 12133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling C, Zheng Y, Yin F, Yu J, Huang J, Hong Y, Wu S, Pan D. 2010. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc Natl Acad Sci 107: 10532–10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yang N, Figel SA, Wilson KE, Morrison CD, Gelman IH, Zhang J. 2013. PTPN14 interacts with and negatively regulates the oncogenic function of YAP. Oncogene 32: 1266–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wu H, Jiang K, Wang Y, Zhang W, Chu Q, Li J, Huang H, Cai T, Ji H, et al. 2016. MAPK-mediated YAP activation controls mechanical-tension-induced pulmonary alveolar regeneration. Cell Rep 16: 1810–1819. [DOI] [PubMed] [Google Scholar]
- Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, Wang Y, Halder G, Finegold MJ, Lee JS, et al. 2010. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci 107: 1437–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnicka B, Grochowalska R, Bogusławska DM, Sikorski AF, Lecomte MC. 2012. Spectrin-based skeleton as an actor in cell signaling. Cell Mol Life Sci 69: 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney PA, Weber U, Onofrechuk P, Biessmann H, Bryant PJ, Goodman CS. 1991. The fat tumor suppressor gene in Drosophila encodes a novel member of the cadherin gene superfamily. Cell 67: 853–868. [DOI] [PubMed] [Google Scholar]
- Mana-Capelli S, Paramasivam M, Dutta S, McCollum D. 2014. Angiomotins link F-actin architecture to Hippo pathway signaling. Mol Biol Cell 25: 1676–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Rauskolb C, Cho E, Hu WL, Hayter H, Minihan G, Katz FN, Irvine KD. 2006. Dachs: An unconventional myosin that functions downstream of Fat to regulate growth, affinity and gene expression in Drosophila. Development 133: 2539–2551. [DOI] [PubMed] [Google Scholar]
- Matakatsu H, Blair SS. 2008. The DHHC palmitoyltransferase approximated regulates Fat signaling and Dachs localization and activity. Curr Biol 18: 1390–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matakatsu H, Blair SS. 2012. Separating planar cell polarity and Hippo pathway activities of the protocadherins Fat and Dachsous. Development 139: 1498–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matakatsu H, Blair SS, Fehon RG. 2017. The palmitoyltransferase Approximated promotes growth via the Hippo pathway by palmitoylation of Fat. J Cell Biol 216: 265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Zhai P, Sciarretta S, Zhang Y, Jeong JI, Ikeda S, Park J, Hsu CP, Tian B, Pan D, et al. 2016. NF2 activates Hippo signaling and promotes ischemia/reperfusion injury in the heart. Circ Res 119: 596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter K, Aijaz S, Tsapara A, Balda MS. 2005. Mammalian tight junctions in the regulation of epithelial differentiation and proliferation. Curr Opin Cell Biol 17: 453–458. [DOI] [PubMed] [Google Scholar]
- McCartney BM, Fehon RG. 1996. Distinct cellular and subcellular patterns of expression imply distinct functions for the Drosophila homologues of Moesin and the Neurofibromatosis 2 tumor suppressor, Merlin. J Cell Biol 133: 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClatchey AI. 2003. Merlin and ERM proteins: Unappreciated roles in cancer development? Nat Rev Cancer 3: 877–883. [DOI] [PubMed] [Google Scholar]
- McDonald PC, Fielding AB, Dedhar S. 2008. Integrin-linked kinase--essential roles in physiology and cancer biology. J Cell Sci 121: 3121–3132. [DOI] [PubMed] [Google Scholar]
- Misra JR, Irvine KD. 2016. Vamana couples Fat signaling to the Hippo pathway. Dev Cell 39: 254–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moleirinho S, Guerrant W, Kissil JL. 2014. The angiomotins—from discovery to function. FEBS Lett 588: 2693–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller NP, Møller KB, Lammers R, Kharitonenkov A, Sures I, Ullrich A. 1994. Src kinase associates with a member of a distinct subfamily of protein-tyrosine phosphatases containing an ezrin-like domain. Proc Nat Acad Sci 91: 7477–7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Layseca P, Streuli CH. 2014. Signalling pathways linking integrins with cell cycle progression. Matrix Biol 34: 144–153. [DOI] [PubMed] [Google Scholar]
- Neto-Silva RM, de Beco S, Johnston LA. 2010. Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev Cell 19: 507–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N, et al. 2009. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell 16: 398–410. [DOI] [PubMed] [Google Scholar]
- Nolo R, Morrison CM, Tao C, Zhang X, Halder G. 2006. The bantam MicroRNA Is a Target of the Hippo Tumor-Suppressor Pathway. Curr Biol 16: 1895–1904. [DOI] [PubMed] [Google Scholar]
- Oh H, Irvine KD. 2008. In vivo regulation of Yorkie phosphorylation and localization. Development 135: 1081–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Irvine KD. 2009. In vivo analysis of Yorkie phosphorylation sites. Oncogene 28: 1916–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Irvine KD. 2011. Cooperative regulation of growth by Yorkie and Mad through bantam. Dev Cell 20: 109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T, Remue E, Meerschaert K, Vanloo B, Boucherie C, Gfeller D, Bader GD, Sidhu SS, Vandekerckhove J, Gettemans J, et al. 2010. Functional complexes between YAP2 and ZO-2 are PDZ domain-dependent, and regulate YAP2 nuclear localization and signalling. Biochem J 432: 461–472. [DOI] [PubMed] [Google Scholar]
- Ota M, Sasaki H. 2008. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development 135: 4059–4069. [DOI] [PubMed] [Google Scholar]
- Pan D. 2007. Hippo signaling in organ size control. Genes Dev 21: 886–897. [DOI] [PubMed] [Google Scholar]
- Pan Y, Heemskerk I, Ibar C, Shraiman BI, Irvine KD. 2016. Differential growth triggers mechanical feedback that elevates Hippo signaling. Proc Natl Acad Sci 113: E6974–E6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantalacci S, Tapon N, Léopold P. 2003. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol 5: 921–927. [DOI] [PubMed] [Google Scholar]
- Paramasivam M, Sarkeshik A, Yates JR, Fernandes MJG, McCollum D. 2011. Angiomotin family proteins are novel activators of the LATS2 kinase tumor suppressor. Mol Biol Cell 22: 3725–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poernbacher I, Baumgartner R, Marada SK, Edwards K, Stocker H. 2012. Drosophila Pez acts in Hippo signaling to restrict intestinal stem cell proliferation. Curr Biol 22: 389–390. [DOI] [PubMed] [Google Scholar]
- Polesello C, Huelsmann S, Brown NH, Tapon N. 2006. The Drosophila RASSF homolog antagonizes the Hippo pathway. Curr Biol 16: 2459–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragni CV, Diguet N, Le Garrec JF, Novotova M, Resende TP, Pop S, Charon N, Guillemot L, Kitasato L, Badouel C, et al. 2017. Amotl1 mediates sequestration of the Hippo effector Yap1 downstream of Fat4 to restrict heart growth. Nat Commun 8: 14582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauskolb C, Sun S, Sun G, Pan Y, Irvine KD. 2014. Cytoskeletal tension inhibits Hippo signaling through an Ajuba-Warts complex. Cell 158: 143–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy BVVG, Irvine KD. 2013. Regulation of Hippo signaling by EGFR-MAPK signaling through Ajuba family proteins. Dev Cell 24: 451–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reginensi A, Scott RP, Gregorieff A, Bagherie-Lachidan M, Chung C, Lim DS, Pawson T, Wrana J, McNeill H. 2013. Yap- and Cdc42-dependent nephrogenesis and morphogenesis during mouse kidney development. PLoS Genet 9: e1003380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reginensi A, Enderle L, Gregorieff A, Johnson RL, Wrana JL, McNeill H. 2016. A critical role for NF2 and the Hippo pathway in branching morphogenesis. Nat Commun 7: 12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro P, Holder M, Frith D, Snijders AP, Tapon N. 2014. Crumbs promotes expanded recognition and degradation by the SCF (Slimb/β-TrCP) ubiquitin ligase. Proc Natl Acad Sci 111: E1980–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson BS, Huang J, Hong Y, Moberg KH. 2010. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein expanded. Curr Biol 20: 582–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Campos M, Thompson BJ. 2014. The ubiquitin ligase FbxL7 regulates the Dachsous-Fat-Dachs system in Drosophila. Development 4098–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogulja D, Rauskolb C, Irvine KD. 2008. Morphogen control of Wing growth through the Fat signaling pathway. Dev Cell 15: 309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh MH, Fan S, Liu CJ, Margolis B. 2003. The Crumbs3-Pals1 complex participates in the establishment of polarity in mammalian epithelial cells. J Cell Sci 116: 2895–2906. [DOI] [PubMed] [Google Scholar]
- Rolls MM, Albertson R, Shih HP, Lee CY, Doe CQ. 2003. Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J Cell Biol 163: 1089–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluh J, Nijhawan D, Cox AG, Li X, Neal JT, Schafer EJ, Zack TI, Wang X, Tsherniak A, Schinzel AC, et al. 2012. β-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell 151: 1457–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansores-Garcia L, Bossuyt W, Wada KI, Yonemura S, Tao C, Sasaki H, Halder G. 2011. Modulating F-actin organization induces organ growth by affecting the Hippo pathway. EMBO J 30: 2325–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimizzi GV, Longmore GD. 2015. Ajuba proteins. Curr Biol 25: R445–R446. [DOI] [PubMed] [Google Scholar]
- Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, Kreger BT, Vasioukhin V, Avruch J, Brummelkamp TR, et al. 2011. Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell 144: 782–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder MC, Halder G. 2012. Regulation of the Hippo pathway by cell architecture and mechanical signals. Semin Cell Dev Biol 23: 803–811. [DOI] [PubMed] [Google Scholar]
- Serrano I, McDonald PC, Lock F, Muller WJ, Dedhar S. 2013. Inactivation of the Hippo tumour suppressor pathway by integrin-linked kinase. Nat Commun 4: 2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. 2006. The Tumor-suppressor gene fat controls tissue growth upstream of expanded in the Hippo signaling pathway. Curr Biol 16: 2081–2089. [DOI] [PubMed] [Google Scholar]
- Silvis MR, Kreger BT, Lien WH, Klezovitch O, Rudakova GM, Camargo FD, Lantz DM, Seykora JT, Vasioukhin V. 2011. α-catenin is a tumor suppressor that controls cell accumulation by regulating the localization and activity of the transcriptional coactivator Yap1. Sci Signal 4: ra33–ra33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AL, Mitchell PJ, Shipley J, Gusterson BA, Rogers MV, Crompton MR. 1995. Pez: A novel human cDNA encoding protein tyrosine phosphatase- and ezrin-like domains. Biochem Biophys Res Commun 209: 959–965. [DOI] [PubMed] [Google Scholar]
- Song H, Mak KK, Topol L, Yun K, Hu J, Garrett L, Chen Y, Park O, Chang J, Simpson RM, et al. 2010. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc Natl Acad Sci 107: 1431–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R, Silva E, Clayton L, Gardano L, Barrios-Rodiles M, Wrana J, Varelas X, Arbouzova NI, Shaw S, Saburi S, et al. 2009. Phosphorylation of the tumor suppressor Fat is regulated by its ligand Dachsous and the kinase Discs Overgrown. Curr Biol 19: 1112–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillos S, Díaz-Meco MT, Caminero E, Moscat J, Campuzano S. 2004. DaPKC-dependent phosphorylation of Crumbs is required for epithelial cell polarity in Drosophila. J Cell Biol 166: 549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadaro D, Tapia R, Jond L, Sudol M, Fanning AS, Citi S. 2014. ZO proteins redundantly regulate the transcription factor DbpA/ZONAB. J Biol Chem 289: 22500–22511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D, Ahringer J. 2010. Cell polarity in eggs and epithelia: Parallels and diversity. Cell 141: 757–774. [DOI] [PubMed] [Google Scholar]
- Sun G, Irvine KD. 2013. Ajuba family proteins link JNK to Hippo signaling. Sci Signal 6: ra81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Irvine KD. 2016. Cellular organization and cytoskeletal regulation of the Hippo signaling network. Trends Cell Biol 26: 694–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Reddy BVVG, Irvine KD. 2015. Localization of Hippo signalling complexes and Warts activation in vivo. Nat Commun 6: 8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymaniak AD, Mahoney JE, Cardoso WV, Varelas X. 2015. Crumbs3-mediated polarity directs airway epithelial cell fate through the Hippo pathway effector Yap. Dev Cell 34: 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Rowe RG, Botvinick E, Kurup A, Putnam A, Seiki M, Weaver V, Keller E, Goldstein S, Dai J, et al. 2013. MT1-MMP-dependent control of skeletal stem cell commitment via a β1-integrin/YAP/TAZ signaling axis. Dev Cell 25: 402–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K, Wu LW, Grivennikov SI, de Jong PR, Lian I, Yu FX, Wang K, Ho SB, Boland BS, Chang JT, et al. 2015. A gp130-Src-YAP module links inflammation to epithelial regeneration. Nature 519: 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapon N, Harvey KF, Bell DW, Wahrer DCR, Schiripo TA, Haber DA, Hariharan IK. 2002. Salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110: 467–478. [DOI] [PubMed] [Google Scholar]
- Tepass U. 2012. The apical polarity protein network in Drosophila epithelial cells: Regulation of polarity, junctions, morphogenesis, cell growth, and survival. Annu Rev Cell Dev Biol 28: 655–685. [DOI] [PubMed] [Google Scholar]
- Thompson BJ. 2013. Cell polarity: models and mechanisms from yeast, worms and flies. Development 140: 13–21. [DOI] [PubMed] [Google Scholar]
- Thompson BJ, Cohen SM. 2006. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell 126: 767–774. [DOI] [PubMed] [Google Scholar]
- Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. 2003. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol 5: 914–920. [DOI] [PubMed] [Google Scholar]
- Varelas X, Samavarchi-Tehrani P, Narimatsu M, Weiss A, Cockburn K, Larsen BG, Rossant J, Wrana JL. 2010. The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-β-SMAD pathway. Dev Cell 19: 831–844. [DOI] [PubMed] [Google Scholar]
- Vrabioiu AM, Struhl G. 2015. Fat/Dachsous signaling promotes Drosophila wing growth by regulating the conformational state of the NDR kinase warts. Dev Cell 35: 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada KI, Itoga K, Okano T, Yonemura S, Sasaki H. 2011. Hippo pathway regulation by cell morphology and stress fibers. Development 138: 3907–3914. [DOI] [PubMed] [Google Scholar]
- Wadham C, Gamble JR, Vadas MA, Khew-Goodall Y. 2003. The protein tyrosine phosphatase Pez is a major phosphatase of adherens junctions and dephosphorylates β-catenin. Mol Biol Cell 14: 2520–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Huang J, Chen J. 2011. Angiomotin-like proteins associate with and negatively regulate YAP1. J Biol Chem 286: 4364–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Huang J, Wang X, Yuan J, Li X, Feng L, Park J Il, Chen J. 2012. PTPN14 is required for the density-dependent control of YAP1. Genes Dev 26: 1959–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Luo JY, Li B, Tian XY, Chen LJ, Huang Y, Liu J, Deng D, Lau CW, Wan S, et al. 2016. Integrin-YAP/TAZ-JNK cascade mediates atheroprotective effect of unidirectional shear flow. Nature 540: 579–582. [DOI] [PubMed] [Google Scholar]
- Wei SY, Escudero LM, Yu F, Chang LH, Chen LY, Ho YH, Lin CM, Chou CS, Chia W, Modolell J, et al. 2005. Echinoid is a component of adherens junctions that cooperates with DE-cadherin to mediate cell adhesion. Dev Cell 8: 493–504. [DOI] [PubMed] [Google Scholar]
- Wells CD, Fawcett JP, Traweger A, Yamanaka Y, Goudreault M, Elder K, Kulkarni S, Gish G, Virag C, Lim C, et al. 2006. A Rich1/Amot complex regulates the Cdc42 GTPase and apical-polarity proteins in epithelial cells. Cell 125: 535–548. [DOI] [PubMed] [Google Scholar]
- Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, Zhang X, Halder G. 2006. The Fat cadherin acts through the Hippo tumor-suppressor pathway to regulate tissue size. Curr Biol 16: 2090–2100. [DOI] [PubMed] [Google Scholar]
- Willecke M, Hamaratoglu F, Sansores-Garcia L, Tao C, Halder G. 2008. Boundaries of Dachsous cadherin activity modulate the Hippo signaling pathway to induce cell proliferation. Proc Natl Acad Sci 105: 14897–14902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KE, Li YW, Yang N, Shen H, Orillion AR, Zhang J. 2014. PTPN14 forms a complex with Kibra and LATS1 proteins and negatively regulates the YAP oncogenic function. J Biol Chem 289: 23693–23700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KKL, Li W, An Y, Duan Y, Li Z, Kang Y, Yan Y. 2015. β-spectrin regulates the Hippo signaling pathway and modulates the basal actin network. J Biol Chem 290: 6397–6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods DF, Bryant PJ. 1993. Apical junctions and cell signalling in epithelia. J Cell Sci Suppl 17: 171–181. [DOI] [PubMed] [Google Scholar]
- Wu S, Huang J, Dong J, Pan D. 2003. Hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114: 445–456. [DOI] [PubMed] [Google Scholar]
- Wu S, Liu Y, Zheng Y, Dong J, Pan D. 2008. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell 14: 388–398. [DOI] [PubMed] [Google Scholar]
- Xu T, Wang W, Zhang S, Stewart RA, Yu W. 1995. Identifying tumor suppressors in genetic mosaics: The Drosophila lats gene encodes a putative protein kinase. Development 121: 1053–1063. [DOI] [PubMed] [Google Scholar]
- Yang CC, Graves HK, Moya IM, Tao C, Hamaratoglu F, Gladden AB, Halder G. 2015. Differential regulation of the Hippo pathway by adherens junctions and apical-basal cell polarity modules. Proc Natl Acad Sci 112: 1785–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH, Axelrod JD, Simon MA. 2002. Regulation of Frizzled by Fat-like cadherins during planar polarity signaling in the Drosophila compound eye. Cell 108: 675–688. [DOI] [PubMed] [Google Scholar]
- Yi C, Shen Z, Stemmer-Rachamimov A, Dawany N, Troutman S, Showe LC, Liu Q, Shimono A, Sudol M, Holmgren L, et al. 2013. The p130 isoform of angiomotin is required for Yap-mediated hepatic epithelial cell proliferation and tumorigenesis. Sci Signal 6: ra77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin F, Yu J, Zheng Y, Chen Q, Zhang N, Pan D. 2013. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell 154: 1342–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. 2010. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell 18: 288–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, et al. 2012. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 150: 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Luo J, Mo JS, Liu G, Kim YC, Meng Z, Zhao L, Peyman G, Ouyang H, Jiang W, et al. 2014. Mutant Gq/11 promote uveal melanoma tumorigenesis by activating YAP. Cancer Cell 25: 822–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Guan KL. 2015. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell 163: 811–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue T, Tian A, Jiang J. 2012. The cell adhesion molecule Echinoid functions as a tumor suppressor and upstream regulator of the Hippo signaling pathway. Dev Cell 22: 255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, Rosato A, Bicciato S, Cordenonsi M, Piccolo S. 2015. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol 17: 1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F, Cordenonsi M, Piccolo S. 2016. YAP/TAZ at the roots of cancer. Cancer Cell 29: 783–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Smolen GA, Haber DA. 2008a. Negative regulation of YAP by LATS1 underscores evolutionary conservation of the Drosophila Hippo pathway. Cancer Res 68: 2789–2794. [DOI] [PubMed] [Google Scholar]
- Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. 2008b. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell 14: 377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu CY, Zha ZY, Zhao B, Yao J, Zhao S, Xiong Y, Lei QY, Guan KL. 2009. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J Biol Chem 284: 13355–13362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Bai H, David KK, Dong J, Zheng Y, Cai J, Giovannini M, Liu P, Anders RA, Pan D. 2010. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev Cell 19: 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang X, Matakatsu H, Fehon R, Blair SS. 2016. The novel SH3 domain protein dlish/CG10933 mediates fat signaling in Drosophila by binding and regulating dachs. eLife 5: e16624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. 2007. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev 21: 2747–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, Guan KL. 2011. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev 25: 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Wang L, Wang CY, Yu J, Guan KL. 2012. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev 26: 54–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, et al. 2008. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 22: 1962–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, Lauwers GY, Thasler W, Lee JT, Avruch J, et al. 2009. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell 16: 425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]