Abstract
The retrotransposon Long Interspersed Element 1 (LINE-1 or L1) is a continuing source of germline and somatic mutagenesis in mammals. Deregulated L1 activity is a hallmark of cancer, and L1 mutagenesis has been described in numerous human malignancies. We previously employed retrotransposon capture sequencing (RC-seq) to analyze hepatocellular carcinoma (HCC) samples from patients infected with hepatitis B or hepatitis C virus and identified L1 variants responsible for activating oncogenic pathways. Here, we have applied RC-seq and whole-genome sequencing (WGS) to an Abcb4 (Mdr2)−/− mouse model of hepatic carcinogenesis and demonstrated for the first time that L1 mobilization occurs in murine tumors. In 12 HCC nodules obtained from 10 animals, we validated four somatic L1 insertions by PCR and capillary sequencing, including TF subfamily elements, and one GF subfamily example. One of the TF insertions carried a 3′ transduction, allowing us to identify its donor L1 and to demonstrate that this full-length TF element retained retrotransposition capacity in cultured cancer cells. Using RC-seq, we also identified eight tumor-specific L1 insertions from 25 HCC patients with a history of alcohol abuse. Finally, we used RC-seq and WGS to identify three tumor-specific L1 insertions among 10 intra-hepatic cholangiocarcinoma (ICC) patients, including one insertion traced to a donor L1 on Chromosome 22 known to be highly active in other cancers. This study reveals L1 mobilization as a common feature of hepatocarcinogenesis in mammals, demonstrating that the phenomenon is not restricted to human viral HCC etiologies and is encountered in murine liver tumors.
Transposable element (TE) sequences make up at least half of the human and mouse genomes (Lander et al. 2001; Waterston et al. 2002; de Koning et al. 2011). While most TE sequences are ancient molecular fossils, a subset of evolutionarily young TEs retain the ability to mobilize (Furano 2000). Both the human and murine genomes are characterized by the activity of the retrotransposon Long Interspersed Element-1 (LINE-1 or L1), which accounts for 19.9% and 17.5% of mouse and human genomic DNA, respectively (Smit et al. 2013). Although the vast majority of L1 copies are no longer mobile, 80–100 human L1s and 2000–3000 mouse L1s, per individual, are full-length and retain retrotransposition potential (Goodier et al. 2001; Brouha et al. 2003). Aside from L1, several retrotransposon families are active in mice, including B1 and B2 SINEs, as well as LTR retrotransposons, such as IAP and ETn elements (Nellaker et al. 2012; Richardson et al. 2017). In humans, there is one presently active L1 subfamily, L1-Ta, while three mouse L1 subfamilies are known to be recently active: TF, GF, and A. These mouse L1 subfamilies are primarily distinguished by the sequence of the ∼200-bp repetitive units, or monomers, comprising their 5′ UTR (DeBerardinis et al. 1998; Naas et al. 1998; Boissinot et al. 2000; Goodier et al. 2001). Full-length L1s (∼6 kb in humans and ∼7 kb in mice) encode proteins required for their own mobilization (i.e., retrotransposition) through reverse transcription of an RNA intermediate (Mathias et al. 1991; Feng et al. 1996). L1 integration into the genome occurs by a process termed target primed reverse transcription (TPRT) (Luan et al. 1993), which produces distinctive structural hallmarks, including variable-length target site duplications (TSDs), a 3′ poly(A) tract, frequent 5′ truncation of the L1 sequence, and insertion at a genomic sequence motif resembling the consensus 5′-TT/AAAA-3′ (Singer et al. 1983; Scott et al. 1987; Feng et al. 1996; Jurka 1997). Critically, characterization of both 5′ and 3′ termini of L1 insertions to discern these structural hallmarks is essential to distinguish bona fide L1 insertions from other types of genomic rearrangements involving L1 sequences (Richardson et al. 2014).
L1 is a potent endogenous insertional mutagen. Exonic L1 insertions are highly detrimental to gene function (Kazazian et al. 1988; Miki et al. 1992; Scott et al. 2016), and intronic insertions can compromise transcript integrity by introducing cryptic splice sites and polyadenylation signals and can interfere with RNA polymerase processivity, particularly when oriented in sense relative to the interrupted gene (Perepelitsa-Belancio and Deininger 2003; Han et al. 2004; Belancio et al. 2008). Furthermore, L1 insertions are occasionally associated with deletions and rearrangements to target site DNA, including extreme examples such as chromosomal translocations (Gilbert et al. 2002, 2005). Indeed, more than 100 cases of human genetic disease have been attributed to L1-mediated retrotransposition events (Hancks and Kazazian 2016).
Due to its mutagenic potential, L1 expression and mobilization is tightly regulated in most cell types and developmental contexts, with CpG methylation of the L1 internal promoter representing a key mechanism for control of L1 activity (Goodier and Kazazian 2008). In the mammalian germline and early embryo, L1 escapes repression to mobilize and create heritable insertions to ensure its evolutionary success (Kano et al. 2009; Faulkner and Garcia-Perez 2017; Richardson et al. 2017). More recently, somatic L1 mobilization has been revealed as a characteristic of normal neuronal cells in rodents and humans, and L1 dysregulation has been associated with neurological diseases (Muotri et al. 2005, 2010; Baillie et al. 2011; Coufal et al. 2011; Evrony et al. 2012; Upton et al. 2015; Erwin et al. 2016; Hazen et al. 2016).
Unchecked somatic L1 mobilization can lead to human disease in the context of oncogenesis and cancer progression. In 1992, Miki et al. identified an exonic L1 insertion into the APC gene that very likely led to tumor initiation in a case of colorectal cancer, providing the first direct evidence for L1 involvement in oncogenesis (Miki et al. 1992). Since then, spurred by the advent of high-throughput sequencing approaches for identifying endogenous somatic L1 insertions, L1 mobilization events have been uncovered in a plethora of tumor types, including lung, ovarian, breast, colorectal, prostate, liver, pancreatic, gastric, endometrial, and esophageal cancers (Iskow et al. 2010; Lee et al. 2012; Solyom et al. 2012; Shukla et al. 2013; Helman et al. 2014; Tubio et al. 2014; Doucet-O'Hare et al. 2015, 2016; Ewing et al. 2015; Paterson et al. 2015; Rodic et al. 2015; Scott et al. 2016). Although these studies have elucidated driver L1 mutations exonic to APC and PTEN, most of the cataloged tumor-specific L1 insertions represent likely passenger events (Helman et al. 2014; Scott et al. 2016). Evidence for L1 mobilization in cancers is largely restricted to tumors of epithelial origin, with somatic L1 insertions conspicuously infrequent in brain and blood cancers (Iskow et al. 2010; Lee et al. 2012; Achanta et al. 2016; Carreira et al. 2016). Thus, some cancer cell types may be intrinsically susceptible to L1 retrotransposition (Carreira et al. 2014; Scott and Devine 2017).
Liver cancer is the sixth most prevalent type of cancer in the world, with 782,000 new cases and 745,000 deaths in 2012 (Ferlay et al. 2015), and is the second most common cause of death from cancer worldwide (Petrick et al. 2016). In previous work (Shukla et al. 2013), we investigated the frequency and impact of L1 mobilization in hepatocellular carcinoma (HCC), which accounts for ∼80% of liver cancer cases (Petrick et al. 2016). Hepatitis B virus (HBV) and hepatitis C virus (HCV) infection are leading causes of chronic liver disease and cirrhosis, which are, in turn, the most important risk factors for the development of HCC (Balogh et al. 2016). We applied retrotransposon-capture sequencing (RC-seq) (Baillie et al. 2011; Shukla et al. 2013) to paired tumor/nontumor samples from 19 donors positive for HBV or HCV and uncovered and PCR-validated 12 tumor-specific somatic L1 insertions. One intronic insertion resulted in the activation of the transcriptional repressor and putative oncogene ST18 by disrupting a binding site required for its own repression, demonstrating the capacity for intronic tumor-specific L1 insertions to contribute to cancer progression. Notably, a recent study confirmed ST18 as a liver oncogene, highlighting the value of mapping L1 insertions in cancer genomes as an endogenous mutagenesis screen (Rava et al. 2017).
In addition to HBV and HCV infection, alcoholism is also a prominent risk factor for HCC (Balogh et al. 2016). However, the involvement of somatic L1 mutagenesis in HCC in patients with a history of alcoholism has not been assessed, leaving the potential restriction of L1 mobilization to viral HCC etiologies unresolved. Indeed, increased accumulation of L1 DNA has been observed in HIV-1 infected cultured cells, suggesting that in some cases viral infection may render cells susceptible to increased L1 activity (Jones et al. 2013). Other liver malignancies, such as intra-hepatic cholangiocarcinoma (ICC), which is the second most common type of liver cancer and arises from the epithelial cells lining the bile ducts (Serafini and Radvinsky 2016), have not been investigated for tumor-specific L1 activity. Finally, it is unknown whether tumor-specific L1 mutagenesis also occurs in rodents, leaving open the question of whether L1 mobilization is peculiar to human epithelial cancers and, if not, whether its characteristics vary among mammalian species, including animal models of hepatocarcinogenesis.
Results
Liver tumor-specific L1 retrotransposition in Abcb4 (Mdr2)−/− mice
To investigate whether L1 mobilization is a feature of hepatocarcinogenesis in rodents, we employed mice with homozygous disruption of the Abcb4 (also known as Mdr2) gene. This Abcb4 (Mdr2)−/− mouse (hereafter Mdr2−/−) is an established model of human HCC in which pathogenesis is attributed to the inability to secrete phospholipid into bile, leading to hepatocellular damage, inflammatory cholangitis, pre-neoplastic lesions and, ultimately, metastatic liver cancer (Smit et al. 1993; Mauad et al. 1994). We isolated 12 HCC tumor nodules and matching tail tissue from 10 Mdr2−/− mice at age 15 mo (Supplemental Table S1) and performed mouse retrotransposon capture sequencing (mRC-seq) (Richardson et al. 2017) on tumor and tail genomic DNA and ∼45× whole-genome sequencing (WGS) on each of five nodules from three animals (Supplemental Table S1). Sequence analysis to identify putative retroelement insertions was performed using the TEBreak bioinformatic pipeline (https://github.com/adamewing/tebreak) and intersected with known nonreference genome TE insertions found in a wide array of mouse strains (Nellaker et al. 2012). Among the 10 animals examined, we bioinformatically identified 748 polymorphic nonreference insertions, including 154 events comprising 42 SINE (11 B1 and 31 B2), three LTR (all ETn), and 109 L1 (48 A, 19 GF, and 42 TF) insertions not found in the parental FVB or 129 mouse strains, or other sequenced strains (Supplemental Table S2). Of the 154 insertions, 141 (91.6%) were found in multiple animals, demonstrating that the vast majority of polymorphic insertions we found were vertically inherited, rather than occurring de novo in the animals analyzed here. Polymorphic insertions were found within known cancer genes. For example, we identified an intact ETn retrotransposon inserted in sense and intronic to Fgf1 (Supplemental Fig. S1A), a ligand for the oncogenic Fgfr1 receptor (Zhang et al. 2006). Further analysis showed that the ETn element was present in the parental FVB genome (Keane et al. 2011) and was homozygous in all of the animals studied here (Supplemental Fig. S1A; Supplemental Table S2). A meta-analysis of published RNA-seq data for HCC nodules obtained from FVB mice (Hashimoto et al. 2015; Kress et al. 2016) did not, however, reveal an alternative Fgf1 promoter located in either of the ETn element LTRs, as has been shown to occur previously for LTRs located proximal to genes in Mdr2−/− liver nodules (Hashimoto et al. 2015). More broadly, polymorphic L1 and SINE insertions typically utilized an L1 EN motif, generated TSDs, and incorporated a poly(A) tail (Supplemental Fig. S1B–D), while ETn LTR insertions carried TSDs with an average length of 6 nt (Supplemental Fig. S1C), in agreement with previous reports (Kaghad et al. 1985). These data highlight ongoing retrotransposon activity in the Mdr2−/− mouse FVB.129 background strain.
We identified by mRC-seq four somatic nodule-specific L1 insertions, which were confirmed by PCR validation using gDNA extracted from nodule and tail samples, and validation products were capillary sequenced to characterize the TPRT-derived structural hallmarks of each insertion (Fig. 1; Supplemental Fig. S2). In one case (Fig. 1B), two nodules from the animal were analyzed, but the insertion was specific to only one of these nodules. All four L1 insertions were full-length (∼1.5–4.5 5′ UTR monomer units), occurred at sites resembling the L1 endonuclease cleavage consensus, bore 15- to 19-bp TSDs, and terminated in 3′ poly(A) tracts ranging in length from 26 to 64 bp (Fig. 1; Supplemental Fig. S2; Supplemental Table S2). Notably, we subsequently performed 45× WGS on five tumor nodes from the three mice in which these full-length tumor-specific L1 insertions arose, to identify potential 5′ truncated L1 insertions that were not detected due to previously described technical difficulties in sequencing the mouse L1 3′ end (Richardson et al. 2017). As in our previous study, analysis of WGS data did not reveal any additional 5′ truncated insertions.
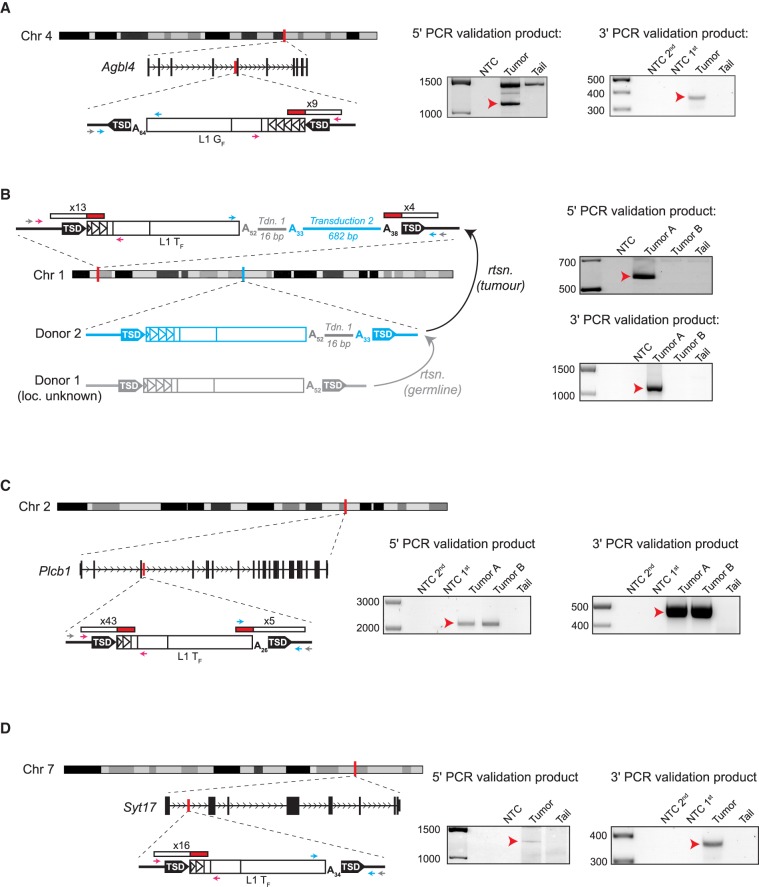
Figure 1.
Tumor-specific L1 insertions in Mdr2−/− animals. For each insertion, the chromosomal location and orientation relative to the interrupted gene, if applicable, are shown. L1 insertions are depicted as white rectangles, with monomer units represented as triangles within the 5′ UTR. Target-site duplications (TSDs) are shown as black arrows. Poly(A) tail length is indicated. The location and count of junction-spanning mRC-seq and WGS reads supporting each insertion are depicted as red and white rectangles. The positions of PCR validation primers are shown as small arrows; where hemi-nested PCR was used, the outer primer is depicted in grey and the inner primers are depicted in pink (5′ junction) and blue (3′ junction). At right, agarose gels containing the 5′ and 3′ junction validation products are shown. Templates are indicated above; for hemi-nested PCR, NTC 1st and NTC 2nd depict the no-template control reactions for the first and second rounds of PCR. Red arrows indicate the validating band. (A) A full-length (4.5 monomer units) L1 GF insertion in antisense orientation within the fourth intron of the gene Agbl4 on Chromosome 4. (B) A full-length (2.5 monomer units) L1 TF insertion in an intergenic region on Chromosome 1. This insertion bore two sequential 3′ transductions of 16 bp (gray line) and 682 bp (blue line). The 682-bp transduction indicated a source L1 for the tumor-specific insertion (Donor 2) on Chromosome 1 (shown in blue). The original source L1 (Donor 1, shown in gray) responsible for the 16-bp transduction could not be identified. (C) A full-length (1.5 monomer units) L1 TF insertion in sense orientation within the third intron of the gene Plcb1 on Chromosome 2. (D) A full-length (1.5 monomer units) L1 TF insertion in sense orientation within the first intron of the gene Syt17 on Chromosome 7.
Three of the four L1 insertions (Fig. 1A,C,D) occurred within the introns of genes (Agbl4, Plcb1, Syt17), and the fourth was inter-genic (Fig. 1B; Supplemental Table S2). Analysis of their 5′ UTR monomers indicated that three of the tumor-specific L1 insertions were L1 TF elements, consistent with previous reports of spontaneous disease-causing L1 insertions in mice (Kingsmore et al. 1994; Mulhardt et al. 1994; Kohrman et al. 1996; Takahara et al. 1996; Perou et al. 1997; Naas et al. 1998; Yajima et al. 1999; Cunliffe et al. 2001). Unexpectedly, the fourth L1 insertion belonged to the L1 GF subfamily, representing the first instance in which an endogenous de novo L1 insertion has been definitively identified as an L1 GF subfamily element (Fig. 1A; Supplemental Fig. S2A).
For one L1 TF insertion, analysis of the 3′ capillary sequencing PCR validation product revealed serial 3′ transductions (Fig. 1B; Supplemental Table S2; Supplemental Fig. S2B). In this case, the L1 was followed by a 52-bp poly(A) tract, a 16-bp non-homopolymer sequence, a 33-bp poly(A) tract, a 682-bp unique sequence, and terminated in a 38-bp poly(A) tract. Such 3′ transductions occur when transcription of a donor L1 bypasses the native L1 polyadenylation signal and terminates at a downstream genomic polyadenylation signal, so that the nascent L1 mRNA incorporates a unique genomic sequence tag that, upon retrotransposition, can potentially be used to identify the donor L1 responsible for the insertion (Holmes et al. 1994; Moran et al. 1996, 1999; Goodier et al. 2000; Pickeral et al. 2000; Macfarlane et al. 2013). Indeed, while the first (16-bp) transduction could not be definitively mapped to a donor L1, the second (682-bp) transduction identified a full-length L1 TF element on Chromosome 1 absent from the C57BL/6J reference genome but annotated as a structural variant in the draft genome of the FVB mouse strain (Yalcin et al. 2012). This donor L1 bore the 16-bp transduced sequence also noted in the tumor-specific L1 insertion. Thus, we identified a specific donor L1 active in the context of liver cancer in mice, as identified similarly in humans (Tubio et al. 2014).
We PCR-amplified this full-length donor L1 from tail DNA from the mouse in which the nascent insertion was found and determined that it contained two intact and functional open reading frames (Fig. 2A). We also cloned the L1 into a retrotransposition indicator plasmid containing a CMV promoter (Moran et al. 1996; Naas et al. 1998; Wei et al. 2000) and tested its retrotransposition efficiency in cultured HeLa cells. This L1 retrotransposed approximately eightfold more efficiently than L1spa, a previously-identified disease causing L1 TF insertion (Fig. 2B; Naas et al. 1998). Thus, the L1 donor represents one of the most highly active natural L1 TF elements tested in cultured cells to date (Naas et al. 1998; Martin et al. 2008; Richardson et al. 2017).
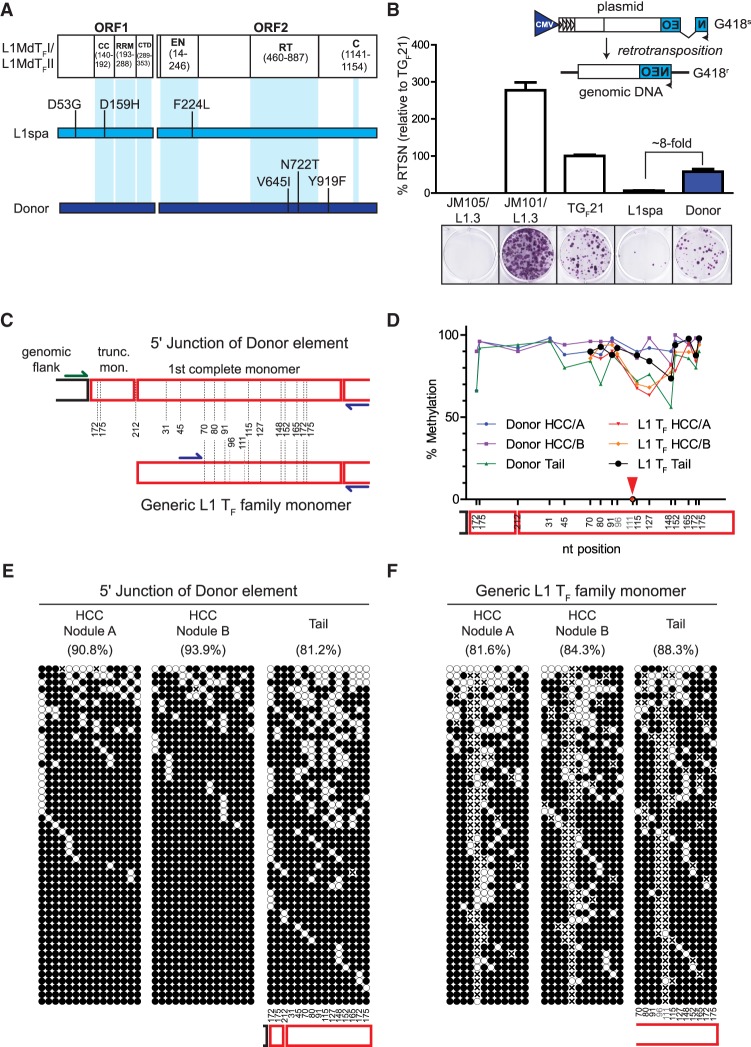
Figure 2.
Retrotransposition competence and epigenetic characterization of an active donor L1. (A) Amino acid sequence comparison between the L1MdTF_I/L1MdTF_II consensus sequence (Sookdeo et al. 2013), the L1 TF element L1spa (Naas et al. 1998), and the donor L1 responsible for the tumor-specific 682-bp transduction-bearing L1 TF insertion (Fig. 1B). The locations of functional domains within ORF1p and ORF2p are depicted along the L1MdTF_I/L1MdTF_II consensus. Amino acid changes within L1spa and the donor L1 relative to the L1MdTF_I/L1MdTF_II consensus are shown. (B) (Top) Rationale of the cultured-cell retrotransposition assay (Moran et al. 1996; Wei et al. 2000). An L1 is tagged in its 3′ UTR with a reporter cassette (in this case, a neomycin phosphotransferase gene) in antisense orientation relative to the L1, which is interrupted by an intron with splice donor and splice acceptor sites in sense orientation relative to the L1. This arrangement ensures that, upon transfection of the construct into cultured cells, G418-resistant foci arise only from cells which have undergone a successful round of retrotransposition. (Bottom) Results of the cultured cell retrotransposition assay. Representative wells for each construct are depicted. JM105/L1.3 is a reverse transcriptase-deficient human L1, JM101/L1.3 is a wild-type human L1, TGF21 is a previously-identified active mouse GF subfamily L1, L1spa is a previously identified disease-causing mouse TF subfamily L1, and Donor is the active L1 TF subfamily element identified in this study. The histogram depicts retrotransposition efficiency relative to TGF21. Data represent the mean of three technical replicates ± SD. This assay was repeated three times (biological replicates) with similar results, and a representative assay is shown. (C) Experimental design for analyzing the methylation status of both the donor L1 and the overall population of L1 TF members. The red boxes represent monomers and the dotted lines represent the different CpG dinucleotides. CpG 96 and 111 are variable in the L1 TF population and are not present in the donor L1. Blue primers are specific for the L1 TF family, while the green primer is specific to the junction of the donor L1. (D) Methylation level of the CpG dinucleotides along the L1 sequence. Note that position 111 was present in <10% of the sequences and was completely unmethylated, and has not been taken into consideration for the line drawing (red arrow). (E) The methylation pattern of 50 random products of locus-specific bisulfite sequencing reactions of the donor L1 5′ junction. (F) The overall L1 TF element population is depicted as in E. In E and F, methylated and unmethylated CpGs are represented by solid and empty circles, respectively, while an “x” means a CpG was not present. CpG dinucleotide positions are indicated below the panels for “Tail” but can be extrapolated to the “HCC” panels. The overall percentage of methylated CpG dinucleotides is indicated above each cartoon.
The in vivo activity of a particular L1 copy depends upon its epigenetic regulation in the relevant biological context, in addition to the enzymatic efficiency of its encoded proteins (Scott and Devine 2017). Therefore, we used bisulfite sequencing to analyze the locus-specific DNA methylation status of the donor L1, as well as the overall methylation status of L1 TF promoter monomeric sequences genome-wide (Fig. 2C,D), in all tumor and tail samples used in this study. Notably, in the locus-specific assay, each sequenced clone represents a discrete donor L1 allele in a particular cell. For the mouse in which the tumor-specific insertion was identified (animal #96109), the donor L1 locus-specific assay unexpectedly revealed significantly higher methylation levels in both tumor nodules relative to the tail (Fig. 2E; Supplemental Fig. S3). This pattern was evident for the methylation status of the donor L1 across all tumor-tail pairs (P = 0.0004, paired t-test, two tailed) (Supplemental Fig. S3). The methylation status of L1 TF monomer sequences was, in contrast, indistinct in most of the tumor nodules relative to tail, with statistical significance (P = 0.0236, paired t-test, two tailed) achieved due to exceptions to this trend (Fig. 2F; Supplemental Fig. S3). However, examination of the locus-specific methylation status of the donor L1 in the remaining tumor-tail pairs revealed multiple clones with complete or near-complete CpG demethylation in two nodules (animals #103699 and #88218) (Supplemental Fig. S3), highlighting the propensity of this locus to become demethylated during tumor development. Thus, we hypothesize that the methylation status of the donor L1 in animal #96109 may have been transiently permissive for L1 expression in a subset of cells during the temporal window when the insertion was generated but may have subsequently undergone remethylation during clonal expansion of daughter cells prior to tumor harvesting. Indeed, the presence of demethylated donor L1 sequences in tumors from other animals (Supplemental Fig. S3) suggests that highly active L1s have the potential to become epigenetically derepressed during murine tumor development.
Liver tumor-specific L1 retrotransposition in human hepatocellular carcinoma
In a previous study (Shukla et al. 2013), we analyzed 19 HCC patients infected with HCV or HBV and determined that L1 retrotransposition is a feature of liver cancers of viral etiologies (Supplemental Table S1). Here, to ask whether L1 retrotransposition is ubiquitous in liver cancers or is a particular feature of viral-driven tumorigenesis, we assessed L1 activity in human HCC patients with a history of alcohol abuse. We performed RC-seq on matched HCC and nontumor liver tissue samples from 25 additional patients with a history of alcohol abuse, as well as five patients with chronic HCV infection and two patients with chronic HBV infection, to bring the total of viral HCC cases investigated by RC-seq to 26 (Supplemental Table S1). In the full data set of 51 HCC patients, and the 10 ICC patients discussed below, we identified 3055 polymorphic retrotransposon insertions, including 414 events not annotated previously (97 L1, 312 Alu, and five SVA) that, collectively, carried the expected hallmarks of TPRT (Supplemental Fig. S1E–G).
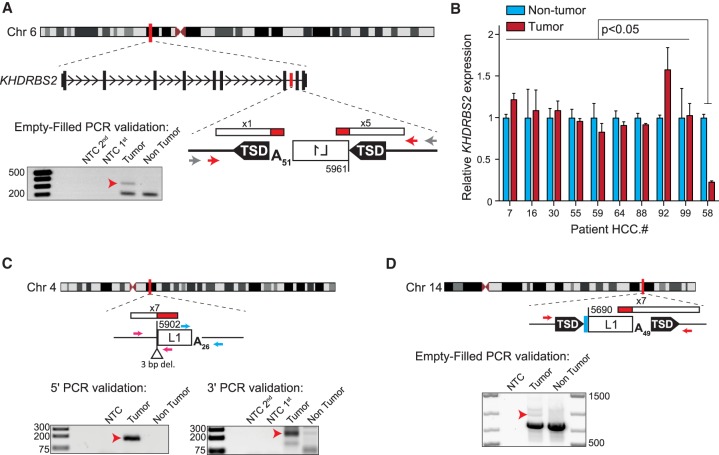
In the 32 new HCC samples, we also identified eight highly 5′ truncated tumor-specific L1 insertions, which were concentrated in three patients with a history of alcoholism (HCC.58, five insertions; HCC.16, two insertions; HCC.99, one insertion) (Fig. 3; Supplemental Figs. S4A–E, S5; Supplemental Table S2). No tumor-specific insertions were identified in the seven additional HBV/HCV cases. Six of the eight insertions bore TSDs ranging in size from 3 to 16 bp, while two lacked TSDs and featured small genomic deletions; all eight insertions occurred at sites resembling the consensus L1 endonuclease motif and ended in poly(A) tracts of 14–121 bp. One insertion was located in the seventh intron of the gene KHDRBS2, in antisense orientation (Fig. 3A), while another occurred in the ninth intron of the SLC10A7 gene, also in antisense orientation (Supplemental Fig. S4A). The remaining six insertions were intergenic (Fig. 3C,D; Supplemental Fig. S4B–E).
Figure 3.
Tumor-specific L1 insertions in HCC patients with a history of alcoholism. For each insertion, the chromosomal location and orientation relative to the interrupted gene, if applicable, are shown. L1 insertions are depicted as white rectangles; the truncation point of each insertion is indicated. Target-site duplications (TSDs) are shown as black arrows; deletions of genomic DNA are shown as white triangles. Poly(A) tail length is indicated. Where applicable, untemplated nucleotides at the 5′ L1-genome junction are indicated as a blue rectangle. The location and count of junction-spanning RC-seq reads supporting each insertion are depicted as red and white rectangles. The positions of PCR validation primers are shown as small arrows. Where empty-filled validation was successful, primers are indicated in red. Where junction PCRs were employed, primers are depicted in pink (5′ junction) and blue (3′ junction). Where nested or hemi-nested PCR was necessary, the outer primer(s) are depicted in gray and the inner primers are depicted in color. Agarose gels containing the empty/filled or 5′ and 3′ junction validation products are shown. Templates are indicated above; for nested and hemi-nested PCR reactions, NTC 1st and NTC 2nd depict the no-template control reactions for the first and second rounds of PCR. Red arrows indicate the validating band. (A) A 5′ truncated L1 insertion in antisense orientation within the seventh intron of the gene KHDRBS2, detected in patient HCC.58 tumor. (B) KHDRBS2 mRNA abundance relative to TBP measured by qRT-PCR using RNA extracted from 10 HCC patient tumor (red) and nontumor (light blue) sample pairs, including patient HCC.58. Data represent the mean of three technical replicates ± SD. For each patient, values were normalized to the nontumor mean. Significance values were obtained via a two-way ANOVA followed by Tukey's post hoc analysis. (C,D) Two additional tumor-specific 5′ truncated L1 insertions, their genomic locations and structural hallmarks, and validating PCR products are depicted as described above.
KHDRBS2 is an RNA-binding protein thought to regulate alternative splicing (Iijima et al. 2014). To investigate the ability of intronic tumor-specific L1 insertions to influence gene expression, we performed qRT-PCR using RNA extracted from tumor and nontumor liver tissue from the patient bearing the KHDRBS2 L1 insertion (HCC.58) and nine other HCC tumor/nontumor tissue pairs. We found that the only individual to present a significant difference in KHDRBS2 mRNA levels between tumor and nontumor liver, when compared to each of the remaining individuals (P < 0.05, two-way ANOVA with Tukey's post hoc analysis) was patient HCC.58, with a 77% reduction in KHDRBS2 expression in tumor (Fig. 3B). Notably, this qRT-PCR assay captured the expression of both major KHDRBS2 protein-coding splice isoforms at an exon–exon junction upstream of the L1 insertion. Repeated attempts to robustly measure KHDRBS2 expression at an exon–exon junction downstream from the L1 insertion were unsuccessful, and the gene was not detected by a previously published genome-wide survey of promoter usage (Hashimoto et al. 2015). Thus, consistent with previous reports, we find that a tumor-specific intronic L1 insertion can coincide with down-regulation of host gene expression (Lee et al. 2012; Shukla et al. 2013; Helman et al. 2014; Tubio et al. 2014; Carreira et al. 2016), but the mechanism for this potential dysregulation remains unresolved.
Liver tumor-specific L1 retrotransposition and comprehensive genomic analysis of intra-hepatic cholangiocarcinoma
We next asked whether tumor-specific L1 insertions arise in intra-hepatic cholangiocarcinoma, a liver malignancy originating from the epithelial cells lining the bile ducts (Serafini and Radvinsky 2016). Incidence of ICC is rapidly increasing worldwide and accounts for 10%–20% of primary hepatic cancers (Gupta and Dixon 2017), and ICC is associated with a poorer 5-yr survival rate than HCC (Lang et al. 2007; Rizvi and Gores 2013). We analyzed tumor/nontumor sample pairs from 10 ICC patients by RC-seq and ∼40× WGS. The WGS data also allowed us to perform a comprehensive genomic analysis (Fig. 4A) to assess the frequency of tumor-specific mutations and the prevalence of tumor and nontumor cells within each sample. We identified between 1.39 and 6.28 mutations per megabase in eight of the 10 ICC tumor/normal pairs analyzed (Supplemental Table S3) and observed a range of different variant allele fraction distributions (Supplemental Figs. S6, S7). The remaining two ICC tumors (ICC.55 and ICC.63) either had very high levels of nontumor sample sequenced or the samples provided were both from the same tissue sample, given the relative lack of deleterious somatic mutations detected (Supplemental Fig. S7).
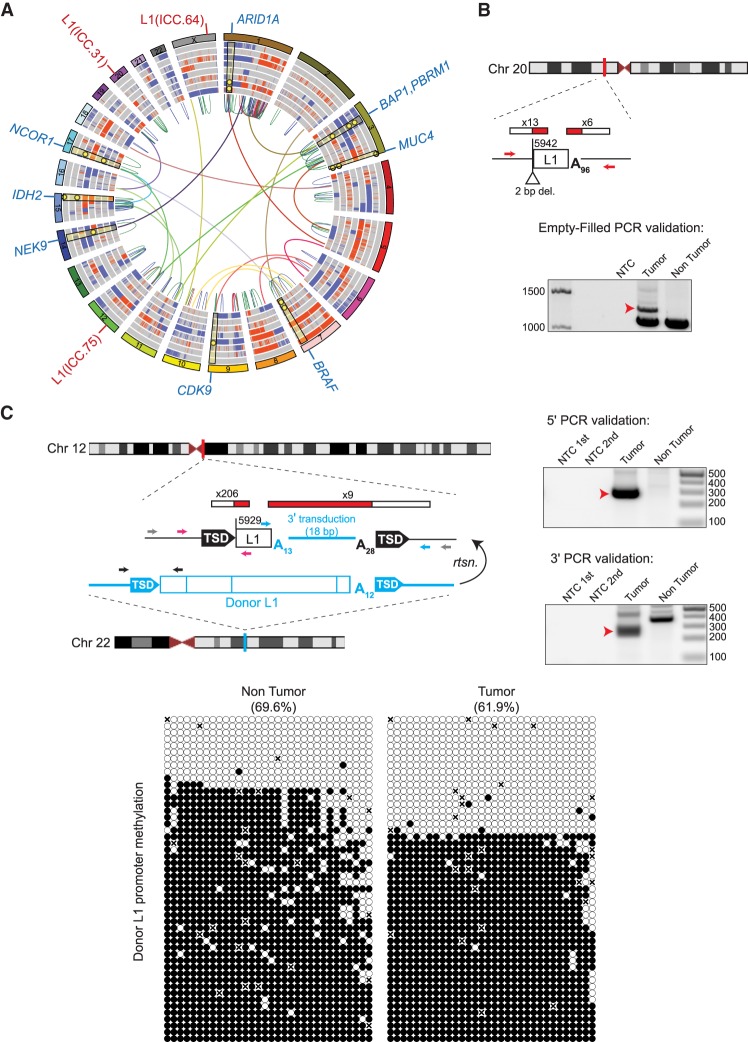
Figure 4.
ICC patient tumor mutational landscape, including tumor-specific L1 insertions. (A) Circos diagram (Krzywinski et al. 2009) summarizing ICC somatic mutations including copy number gains (red) and losses (blue), intra-chromosomal (colored by chromosome pair) and inter-chromosomal (red = duplication, blue = deletion, green = inversion) rearrangements. Rearrangements shown are those that intersect genes identified in prior ICC studies or present in the COSMIC Cancer Gene Census. L1 insertions are highlighted in red text labels on the outside of the circle, and selected genes with somatic mutations are highlighted with blue labels. Selected somatic mutations (SNVs and INDELs) are shown as yellow dots with highlights superimposed on the copy number variation rings. (B,C) Detailed characterization of two intergenic and 5′ truncated tumor-specific L1 insertions. For each insertion, the chromosomal location is shown. L1 insertions are depicted as white rectangles; the truncation point of each insertion is indicated. The location and count of junction-spanning RC-seq and WGS reads supporting each insertion are depicted as red and white rectangles. Target-site duplications are shown as black arrows; deletions of genomic DNA are shown as white triangles. Poly(A) tail length is indicated. The positions of PCR validation primers are shown as small arrows. Where empty-filled validation was successful, primers are indicated in red. Where junction PCRs were employed, primers are depicted in pink (5′ junction) and blue (3′ junction). Where hemi-nested PCR was necessary, the outer primer(s) are depicted in gray and the inner primers are depicted in color. Agarose gels containing the empty/filled or 5′ and 3′ junction validation products are shown. Templates are indicated above; for hemi-nested PCR reactions, NTC 1st and NTC 2nd depict the no-template control reactions for the first and second rounds of PCR. Red arrows indicate the validating band. The L1 insertion shown in C carried an 18-bp 3′ transduced sequence implicating a donor L1 (shown in blue) located in the TTC28 gene on Chromosome 22. The methylation state of this specific donor L1 (below) was determined via targeted bisulfite sequencing of its promoter CpG island (primers are depicted in C as black arrows). Each cartoon represents 50 random, nonidentical sequences matching the donor L1 (black circle, methylated CpG; white circle, unmethylated CpG; “x,” absent) obtained from patient ICC.75 nontumor liver and matched tumor.
Bioinformatic analysis revealed three tumor-specific L1 insertions for patients ICC.33, ICC.64, and ICC.75, which were PCR-validated and fully characterized (Supplemental Fig. S8; Supplemental Table S2). All three insertions were highly 5′ truncated and occurred in intergenic regions. Two insertions (Fig. 4B; Supplemental Figs. S4F, S8A,B) did not bear TSDs and instead exhibited small deletions of target-site DNA (1 and 2 bp) consistent with second-strand cleavage upstream of rather than downstream from the first-strand L1 endonucleolytic nick (Gilbert et al. 2002, 2005). Indeed, as these insertions occurred at perfect (5′-TT/AAAA-3′) and near-perfect (5′-AT/AAAA-3′) L1 endonuclease cleavage motifs and incorporated ∼61- and ∼96-bp poly(A) tracts, they likely represent bona fide TPRT events, despite lacking TSDs.
The third ICC tumor-specific L1 insertion (Fig. 4C; Supplemental Fig. S8C), found in patient ICC.75, bore 16-bp TSDs, occurred at a near-perfect L1 endonuclease cleavage motif (5′-TT/AGAA-3′), and incorporated a short (18-bp) 3′ transduction flanked by 13- and 28-bp poly(A) tracts. Analysis of the 3′ transduced sequence identified a donor L1 on Chromosome 22 and intronic to the gene TTC28. Recent studies of 3′ transduction-bearing L1 insertions in tumor samples pointed to this element as a highly active donor L1 in human tumor cells (Pitkanen et al. 2014; Tubio et al. 2014; Paterson et al. 2015; Philippe et al. 2016; Gardner et al. 2017). We therefore performed targeted bisulfite sequencing of the complete donor L1 promoter region, which was significantly demethylated in patient ICC.75 tumor compared to matched nontumor liver (P < 0.0001, paired t-test, two tailed) (Fig. 4C). Despite this difference, we observed numerous fully demethylated clones in both the tumor and nontumor liver, suggesting this specific donor L1 may have been de-repressed prior to, or as part of, liver dysplasia.
We next analyzed the ICC WGS data to identify deleterious, non-retrotransposition-mediated mutations driving oncogenesis (Supplemental Tables S4, S5). Most of the ICC genomes analyzed had high levels of rearrangement reflected by copy number variation, and inter- and intra-chromosomal variation. In particular, high levels of loss and rearrangement of the p arm of Chromosome 1 involving the telomere were seen in seven out of eight ICC tumors with high tumor cellularity. In patients ICC.75 and ICC.30, high numbers of SV predictions were clustered in Chromosome 1q, across Chromosome 7 in ICC.64, and Chromosome 9p encompassing the tumor suppressor CDKN2A in ICC.17 (Supplemental Fig. S7). Inter-chromosomal rearrangements clustering on Chromosome 4 of ICC.17 and throughout the genome of ICC.64 (notably on Chromosome 17) added to a picture of high plasticity in ICC genomes (Supplemental Fig. S9).
Among the spectrum of tumor-specific genomic aberrations, we identified multiple samples with likely loss-of-function mutations or deletions in tumor suppressor genes found to be frequently mutated in previous genomic studies of ICC and other cholangiocarcinomas (Chan-On et al. 2013; Jiao et al. 2013; Zou et al. 2014; Nakamura et al. 2015; Farshidfar et al. 2017; Jusakul et al. 2017). Examples include the tumor suppressor genes ARID1A, BAP1, and PBRM1, which encode chromatin remodeling factors (Thompson 2009; Scheuermann et al. 2010; Wilson and Roberts 2011; Wu and Roberts 2013). In addition, multiple samples contained inactivating mutations in the tumor suppressor NF2, which encodes a scaffold protein that links extracellular stimuli and intra-cellular signaling pathways involved in cell proliferation and survival (Petrilli and Fernandez-Valle 2016).
The resolution provided via WGS allowed us to identify several likely biallelic losses of known ICC driver genes: For example, we observed possible loss of both ARID1A alleles in the tumors of patients ICC.1, ICC.64, ICC.75, and ICC.81 through a combination of Chromosome 1p loss, localized SVs, frameshift, and stop-gain mutations. When the Chromosome 1p CNV profile was taken into account, all eight tumors were likely to have lost at least one allele of ARID1A (Supplemental Fig. S7), underscoring its importance as a tumor suppressor in ICC. Additionally, in accordance with previous observations (Jiao et al. 2013), we observe frequent loss of at least one allele on Chromosome 3p surrounding the BAP1 and PBRM1 genes (6/8 tumors), further highlighting the importance of chromatin remodeling factors in this tumor type.
In addition to loss-of-function mutations in genes previously implicated in cholangiocarcinomas, we also uncovered inactivating mutations in classical tumor suppressor and proto-oncogenes. We identified two patients (ICC.75, ICC.81) with tumor-specific deletions on Chromosome 13q involving the BRCA2 and RB1 genes. Notably, ICC.81 likely has a biallelic loss of BRCA2 due to an additional frameshift indel mutation in BRCA2 (I1924fs) detectable in the nontumor sample. In addition, two patients (ICC.75 and ICC.81) have oncogenic mutations in BRAF (G466A and G466R), two have IDH2 mutations (R172W and R172M), ICC.64 has a KRAS G12D mutation, and ICC.33 has an amplification involving KRAS. Finally, we identified recurrent mutations in possible, but less well-characterized, cancer drivers in multiple patients, including CDK6, CDK9, NEK9, and EPHA2. This genome-wide view of the ICC mutation landscape highlights mutational processes underpinning ICC, where de novo L1 insertions occur alongside other forms of genomic aberration in these highly mutated tumors.
Discussion
Here, we have conducted an extended analysis of L1 insertions arising in both mouse and human hepatocarcinogenesis of multiple etiologies. We validated each tumor-specific insertion thoroughly by PCR, clearly establishing the hallmarks of bona fide L1 retrotransposition by TPRT. We find that L1 retrotransposition is a feature of hepatocarcinogenesis in mice and humans, suggesting similar L1 dysregulation in cancer genomes across species. To our knowledge, this is the first report of endogenous L1 mobilization being observed directly in a nonhuman cancer, with the possible exception of an L1 mutation found proximal to MYC in canine transmissible venereal tumors and arising either in the dog germline or the somatic evolution of the original tumor (Murgia et al. 2006). Combined with a previous report of L1 and IAP protein expression in tumors obtained from a Myc murine liver cancer model (Wylie et al. 2016), our results highlight mouse as a suitable system to study L1 activity in oncogenesis and cancer progression, which may be useful for future studies that are not feasible using human patient samples. Indeed, the finding that Mdr2−/− mice accommodate tumor-specific L1 retrotransposition is particularly interesting, as these genomic alterations occur alongside massive gene amplifications (Iannelli et al. 2014). All four tumor-specific mouse L1 insertions were full-length and therefore likely to retain retrotransposition competence, allowing them to serve as potential donor L1s for subsequent tumor-specific retrotransposition events, as previously observed in human malignancies (Tubio et al. 2014; Gardner et al. 2017).
Of the four tumor-specific mouse L1 insertions, three occurred in introns and the remaining example was intergenic. None of the three affected genes (Agbl4, Plcb1, Syt17) is well established to play a role in HCC tumorigenesis, although Plcb1, a regulator of signal transduction, has been reported as both a cancer marker and suppressor (Guerrero-Preston et al. 2014; Tan et al. 2015; Li et al. 2016). No exonic tumor-specific mouse L1 insertions were detected. In this regard, the genomic distribution of mouse tumor-specific L1 insertions, although involving at this stage a handful of events, resembles patterns reported for human cancers, where L1 insertions found in exons of known cancer genes (Miki et al. 1992; Helman et al. 2014; Scott et al. 2016) are far rarer than such events located in introns (Lee et al. 2012), including the example of the ST18 oncogene (Shukla et al. 2013; Rava et al. 2017) and a more recent instance in the tumor suppressor gene BRCA1 (Tang et al. 2017). Although the oncogenic impact of intronic tumor-specific L1 insertions is more difficult to mechanistically assess than exonic events, they are also far more numerous and are therefore a potentially important and underexplored class of mutation encountered in mammalian cancers (Scott and Devine 2017).
The prevalence of full-length L1 insertions detected in mouse tumors, as well as the lack of 5′ truncated and inverted/deleted events in this context, is also noteworthy. Used in isolation, the mRC-seq approach can overlook 5′ truncated or inverted/deleted L1 insertions due to (1) the position of the mRC-seq probes at the L1 5′ and 3′ termini, and (2) low efficiency Illumina sequencing across the mouse L1 3′ end (Richardson et al. 2017). However, whole-genome sequencing, which efficiently detects the 5′ end of both full-length and rearranged L1 insertions, even if the corresponding 3′ end is not detected, was applied to five Mdr2−/− nodules and did not reveal any insertions not found by mRC-seq, as we found was also the case in our previous study of heritable L1 insertions in extended mouse pedigrees (Richardson et al. 2017). Hence, at least some mouse L1 families may be highly predisposed to generate full-length offspring L1 insertions in the germline (Hardies et al. 2000; Richardson et al. 2017) as well as in cancer cells. In contrast, L1 insertions detected in human tumors are predominantly 5′ truncated (Iskow et al. 2010; Lee et al. 2012; Solyom et al. 2012; Shukla et al. 2013; Helman et al. 2014; Tubio et al. 2014; Doucet-O'Hare et al. 2015, 2016; Ewing et al. 2015; Paterson et al. 2015; Rodic et al. 2015; Scott et al. 2016) and involve structural rearrangements at a higher frequency than for L1 insertions arising in the human germline (Gardner et al. 2017). These differences are apparent even among hominids. For example, the rate of germline L1 5′ inversion appears to be lower in chimpanzee than in human (Gardner et al. 2017). Whether these observations reflect species-specific differences in the enzymatic properties of L1s or the cellular environment in which retrotransposition events occur, including host factors such as the APOBEC (also known as AID) family proteins that are more diverse in humans than in rodents (Goodier 2016), remains to be determined.
Furthermore, we report the identification of a specific donor mouse L1 TF element responsible for a tumor-specific L1 insertion and demonstrate its retrotransposition competence in a cultured cell assay, implicating this L1 locus as active in cancer. Indeed, promoter methylation analysis of the donor L1 across all Mdr2−/− nodules revealed completely unmethylated copies in two nodules other than the one in which the insertion was found but not in any of the matched tail samples, indicating that this L1 may recurrently become derepressed in the context of oncogenesis and tumor progression. Future identification of 3′ transduction events arising from the same active donor L1 in additional mouse malignancies may further implicate this locus as particularly “hot” in the context of murine tumorigenesis, as has been found in human cancers (Tubio et al. 2014; Scott et al. 2016). We also report the first example of a de novo L1 GF retrotransposition event in vivo. While some L1 GF elements are polymorphic among mouse strains, indicating recent germline mobilization, and L1 GF elements retain retrotransposition capability in cultured cell assays (Goodier et al. 2001), all previously described disease-causing mouse L1 insertions where the subfamily could be determined were identified as TF elements (Kingsmore et al. 1994; Mulhardt et al. 1994; Kohrman et al. 1996; Takahara et al. 1996; Perou et al. 1997; Naas et al. 1998; Yajima et al. 1999; Cunliffe et al. 2001). Indeed, recent work from our laboratory identified 11 de novo L1 insertions occurring in the germline or early embryo in C57BL/6J mice, and all of these insertions belonged to the L1 TF subfamily (Richardson et al. 2017). Whether different mouse L1 subfamilies are prone to mobilization in particular cell types or developmental contexts remains to be determined. Alternatively, the activity of different L1 subfamily elements may be influenced by the genetic background of the inbred mouse strain analyzed (Maksakova et al. 2006; Akagi et al. 2008; Nellaker et al. 2012).
Along similar lines, we identified polymorphic ETn (but not IAP) LTR retrotransposon insertions present in our mice but absent from the C57BL/6J reference genome, and some of these were not detected in the mouse strains contributing the genetic background of the animals in our study (FVB and 129) or any other inbred mouse strain analyzed. This result is in contrast to previous work from our lab, wherein we did not detect any nonreference polymorphic ETn or IAP LTR retrotransposon insertions in wild-type C57BL/6J mice (Richardson et al. 2017). All animals in the present study also harbored an intact ETn element inserted in Fgf1 and found in the parental FVB mouse genome. Although we did not find evidence linking transcription of the ETn LTRs with Fgf1 expression, this result shows that different mouse strains can harbor polymorphic TE variants in oncogenic pathways, as we suggested previously for human individuals (Shukla et al. 2013).
In addition to demonstrating L1 activity in mouse hepatocarcinogenesis, we report tumor-specific human L1 mobilization in HCC from patients with a history of alcoholism. We previously identified tumor-specific L1 mobilization in HCC of viral etiology (HBV or HCV), raising the possibility that viral infection influenced susceptibility to L1 mobilization (Shukla et al. 2013). However, the analysis of these new results in combination with those from our previous study (Shukla et al. 2013) indicates that liver cancer etiology does not appear to be a predictor of L1 activity (12 insertions among 26 HCC tumors of viral etiology, eight insertions among 25 HCC tumors of alcoholic etiology). We also describe an intronic L1 insertion into the gene KHDRBS2 which correlates with tumor-specific KHDRBS2 down-regulation, reinforcing the observation that L1 mutagenesis may result in dysregulation of cellular genes in a tumor-specific manner (Lee et al. 2012; Shukla et al. 2013).
Finally, we report that L1 activity in human liver malignancies is not restricted to HCC, as we identified three tumor-specific L1 insertions among 10 ICC patients. Among these, one insertion originated from a donor L1 on Chromosome 22 previously found to be responsible for tumor-specific L1 insertions in other human cancer types (Pitkanen et al. 2014; Tubio et al. 2014; Paterson et al. 2015). An increasing body of evidence suggests that a particular subset of retrotransposition-competent L1 loci tend to be recurrent contributors to genomic instability in human malignancies and cancer cell lines (Tubio et al. 2014; Philippe et al. 2016; Scott et al. 2016; Gardner et al. 2017). Indeed, here we found that the promoter region of the TTC28 donor L1 was largely de-methylated in both nontumor liver and matched ICC tumor cells, as a prior study found for a Chromosome 17 donor L1 responsible for APC exon mutagenesis in a colorectal cancer patient (Scott et al. 2016). These findings together raise the prospect of multiple donor L1 loci being active in the pre-neoplastic tissues of cancer patients.
Although all three tumor-specific insertions in the ICC patients studied here were intergenic and therefore unlikely to exert a significant functional impact, we also analyzed WGS data from the ICC samples to characterize the spectrum of deleterious tumor-specific mutations occurring in these patients. Our results revealed frequent mutation of several genes, particularly ARID1A, known to be recurrently mutated in ICC and other cholangiocarcinomas, as well as aberrations in classical tumor suppressor and proto-oncogenes. Furthermore, we identified multiple mutations in a handful of genes associated with tumorigenic processes (AMER1, NEK9, NAP1L1, and GAB2) but not previously described as recurrently mutated in ICC. Thus, analysis of additional patients and functional experiments may reveal a recurrent role for these factors in ICC and other cholangiocarcinomas. Together, our results indicate that L1 mobilization is a distinguishing characteristic of epithelial cancers of diverse etiologies and across species, demonstrating that this phenomenon is an intrinsic component of epithelial tumorigenesis.
Methods
Animals
Founders of the FVB.129P2-Abcb4tm1Bor (Mdr2−/−) strain were purchased from The Jackson Laboratory and maintained by mating homozygous siblings. Colonies were maintained under specific pathogen-free conditions. A portion of each nodule and control sample (tail) was snap-frozen for DNA extraction. Furthermore, a portion of each HCC specimen was histologically assessed after overnight fixation in 4% formaldehyde and paraffin inclusion. Slides were finally counterstained with hematoxylin and mounted with Eukitt. Frozen tissue samples were homogenized with a gentleMACS Dissociator (Miltenyi Biotec) before column extraction. Genomic DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen) according to the manufacturer's protocol for all samples.
All analyzed samples were inspected by a mouse pathologist. Tumor growth in Mdr2−/− livers is multicentric; gross detectable masses are often very heterogeneous resulting from the collision of multiple contiguous hepatocellular proliferations with different histologic features and grades. In addition, there is a tendency for hepatocellular carcinoma to develop within an adenoma (as foci of tumor progression). Given these peculiar characteristics of tumor growth in Mdr2−/− mice, the histological composition of gross detectable hepatic nodules was semiquantitatively determined based on reported classification criteria (Thoolen et al. 2010).
Human patient samples
Snap-frozen tissue samples were obtained from 32 patients with HCC induced by a history of alcoholism (25), chronic HBV infection (2), and chronic HCV infection (5), and 10 patients with intrahepatic cholangiocarcinoma (ICC) (Supplemental Table S1). These samples are in addition to those HCC samples previously analyzed by Shukla et al. (2013), comprising HBV (10) and HCV (9) HCC etiologies (Supplemental Table S1). All patients gave written informed consent. Specimens were obtained after surgical resection or from liver explants at transplantation. For each patient, we analyzed tumor and distal nontumor liver tissue (T/NT pairs).
Library preparation and sequencing
For Mdr2−/− mouse samples, genomic DNA was extracted from tumor nodules and tails using standard phenol-chloroform extraction. Illumina libraries were prepared as described in Richardson et al. (2017) using Illumina TruSeq DNA LT library preparation kits incorporating an insert size of ∼450–550 bp. Twenty-two total barcoded libraries were pooled, subjected to the mRC-seq enrichment protocol as described (Richardson et al. 2017), and sequenced on an Illumina MiSeq platform using 600 cycle kits generating 114,638,158 2 × 300-bp paired-end reads. A subset of five pre-enrichment libraries prepared from tumor DNA (Supplemental Table S1) was subjected to ∼45× whole-genome sequencing (Macrogen) on an Illumina HiSeq X Ten platform, generating 2,272,895,769 2 × 150-bp paired-end reads (Supplemental Table S1).
For human HCC patient samples, genomic DNA was extracted from tumor and nontumor liver tissue using standard phenol-chloroform extraction. Illumina libraries were prepared as described (Shukla et al. 2013) using Illumina TruSeq DNA LT library preparation kits incorporating an average insert size of 220 bp. Seventy total barcoded libraries were pooled, subjected to RC-seq enrichment as described in Shukla et al. (2013), and sequenced on an Illumina HiSeq 2500 platform (Ambry Genetics), generating 1,635,587,606 2 × 150-bp paired-end reads.
For human ICC patient samples, total DNA was isolated using the DNeasy Blood and Tissue Kit (Qiagen). Illumina libraries were prepared as described (Shukla et al. 2013) using Illumina TruSeq DNA LT library preparation kits incorporating an average insert size of 250 bp. Barcoded libraries were pooled, subjected to RC-seq enrichment as described (Upton et al. 2015), and sequenced on an Illumina HiSeq 2500 Platform (Ambry Genetics) generating 255,239,605 2 × 150-bp paired-end reads.
Human ICC genomic DNA samples were independently subjected to library preparation by Macrogen, using Illumina TruSeq DNA Nano LT library preparation kits incorporating an average insert size of 350 bp. Libraries were subjected to ∼40× WGS on an Illumina HiSeq X 10 platform, generating 8,482,986,041 2 × 150-bp paired-end reads.
Human ICC WGS analysis
Reads were aligned to the human genome reference sequence (hg19 build) with BWA-MEM (Li 2013), duplicates were marked with Picard MarkDuplicates (http://broadinstitute.github.io/picard), and indel realignment was carried out with GATK (DePristo et al. 2011). Quality assessment via FastQC identified no issues with sequence quality or content, duplication, overrepresented sequences or k-mers. Point mutations (SNVs and indels) were detected with MuTect 2 (Cibulskis et al. 2013), and structural rearrangements were detected using Manta (Chen et al. 2016). CNV profiles were derived from normalized tumor vs. normal read depth in 150-kbp windows using a Python script deposited on GitHub (https://gist.github.com/adamewing/70c8819f6f1eec 0ffbdafa6210f0f677) and also available in Supplemental Materials.
TE insertion detection
WGS- and RC-seq-aligned BAMs (mouse: mm10 build, human: hg19 build) were processed with TEBreak (https://github.com/adamewing/tebreak and Supplemental Materials) to detect nonreference transposable element insertions. Further post-filtering on detected insertions was performed as follows: Reads supporting TE insertions must align to TE consensus sequences at ≥90% identity and to the reference genome at ≥99% identity. TE insertion calls must be supported by at least eight split reads and eight discordant read pairs for mouse and at least four split reads and four discordant read pairs for human. At least 50% of all split reads and discordant reads surrounding an insertion call must be attributable (i.e., remappable) to that insertion. For SINEs, insertions detected with 5′ truncations of more than 1 bp, or with 3′ truncations, or presenting low complexity TSDs (e.g., all “A” or “T” bases), or lacking TSDs, were excluded. Nonreference insertion calls were annotated with known nonreference insertion polymorphisms. For human, this annotation is derived from 15 previous studies included with TEBreak (Ewing 2015). For mouse insertions, the nonreference annotations are derived from running TEBreak on 40 inbred mouse strains available through the Sanger Mouse Genomes Project (http://www.sanger.ac.uk/science/data/mouse-genomes-project) (Keane et al. 2011), which is also included with TEBreak. To ensure that use of the hg19 human reference genome assembly, rather than hg38, did not impact our conclusions, we remapped the genomic consensus sequences for tumor-specific insertions (Supplemental Table S2) to the hg38 assembly. We found that the number of results did not change, the relative chromosome positions were consistent with results obtained via the UCSC liftOver tool, the gene annotations did not change, and all sites mapped uniquely. Therefore, remapping to hg38 would be highly unlikely to alter the results or conclusions that we have highlighted.
Validation and structural characterization of TE insertions
RC-seq, mRC-seq, and WGS reads, and TEBreak breakpoint consensus sequences, for putative tumor-specific insertions were manually inspected using SerialCloner (http://serialbasics.free.fr/Serial_Cloner.html) and the UCSC genome browser BLAT tool (Kent 2002). Primers to test de novo L1 insertions were designed in the 5′ and 3′ flanking genomic DNA regions (Supplemental Table S2). The complete ETn insertion found in Fgf1 was amplified with the primers TCCAGCCTCCTCCTCATCTA (forward) and GGAAGAGGGGGCACTAAACT (reverse). Oligonucleotides were obtained from Integrated DNA Technologies).
Empty-filled validation PCRs were carried out using the Roche Expand Long-Range PCR system (Roche) with primers specific to the 5′ and 3′ genomic sequence flanking putative insertions. 5′ and 3′ junction validation PCRs were carried out using the MyTaq PCR system (Bioline) using the appropriate flanking genomic primer paired with a primer internal to the L1 sequence; where necessary, hemi-nested and fully nested PCR reactions were carried out using appropriately designed genomic and L1-specific primers. Products were visualized on 1%–2.5% agarose gels stained with SYBR Safe stain (Life Technologies). Bands matching the expected size or present in tumor samples and absent from the corresponding control tissue were excised, purified using Qiagen MinElute gel extraction kits (Qiagen), and either capillary sequenced directly or cloned into and capillary sequenced from pGEM-T Easy (Promega) or TOPO-XL (Life Technologies) vectors. Validation product sequences were manually analyzed and inspected for the structural hallmarks of L1 retrotransposition.
Plasmid constructs
pTN201 (Naas et al. 1998) is a neomycin-based L1 retrotransposition indicator plasmid containing L1spa, a wild-type mouse L1 TF element in the pCEP4 plasmid backbone. TGF21 (Goodier et al. 2001) is a neomycin-based L1 retrotransposition indicator plasmid containing TGF21, a wild-type mouse L1 GF element in the pCEP4 plasmid backbone. pJM101/L1.3 (Dombroski et al. 1991; Sassaman et al. 1997) is a neomycin-based L1 retrotransposition indicator plasmid containing L1.3, a wild-type human L1 in the pCEP4 plasmid backbone, and pJM105/L1.3 (Wei et al. 2000) is a neomycin-based L1 retrotransposition indicator plasmid containing L1.3 with an inactivating mutation in the L1 reverse transcriptase active site in the pCEP4 plasmid backbone.
Generation of a mouse donor L1 reporter construct
The mouse donor L1 TF element was PCR-amplified from mouse #96109 tail genomic DNA using the Roche Expand Long-Range PCR system with primers located in the 5′ and 3′ flanking genomic DNA; the 5′ forward primer incorporated a NotI restriction site (fwd: aaaaaaGCGGCCGCcaagaatgcccaGTTCAGCC; rev: GAATTGGGTTGGGTATTCTTCC). Ten replicate PCR reactions were performed in parallel; to determine the sequence of the donor L1, the products were pooled and capillary sequenced using primers located at ∼500-bp intervals along the L1 TF consensus sequence. The donor L1 PCR product was also cloned into a neomycin-based L1 retrotransposition indicator plasmid as described in Richardson et al. (2017). Ten independent clones were sequenced; the L1 shown in Figure 2B contained three PCR-induced silent mutations and no nonsynonymous mutations relative to the sequence derived from the pooled PCR products.
Cultured cell retrotransposition assay
Cultured cell retrotransposition assay: HeLa-JVM cells were seeded at 5 × 103 cells/well in six-well plates and transfected using FuGENE HD Transfection Reagent (Promega) at a ratio of 4 µL to 1 µg plasmid DNA. G418 selection (400 µg/mL) was initiated at 72 h post-transfection and carried out for 12 d (Wei et al. 2000).
Assays for transfection efficiency were performed in parallel by cotransfection of pCAG-EGFP with L1 reporter plasmids. At 48 h post-transfection, cells were subjected to flow cytometry on a CytoFLEX flow cytometer (Beckman-Coulter) at the Translational Research Institute Flow Cytometry Core. The percentage of EGFP positive cells for each L1 reporter construct was used to normalize the G418-resistant colony counts obtained in the retrotransposition assay (Wei et al. 2000; Kopera et al. 2016).
Mouse L1 methylation analysis
Genomic DNA from mouse tumor and tail samples was subjected to bisulfite conversion using the EZ DNA Methylation-Lightning Kit (Zymo Research), following the manufacturer's instructions. To amplify L1 TF monomers genome-wide, PCR amplification was performed using the MyTaq DNA Polymerase PCR kit (Bioline) with primers targeting the internal sequence of the L1 TF monomer (fwd: GTTGAGGTAGTATTTTGTGTGGGT; rev: TTCCAAAAACTATCAAATTCTCTAAC). To amplify the 5′ junction and first monomer of the L1 TF donor element of interest, locus-specific primers were designed targeting the 5′ genomic region (fwd: GAAATTTTGTTTTTAAAAATTAAAAA) and the internal sequence of the L1 promoter (rev: TTCCAAAAACTATCAAATTCTCTAAC). Fifty PCR cycles were carried out using the following conditions: 95°C for 2 min, 50 cycles of 95°C for 30 sec, 54°C for 30 sec, and 72°C for 30 sec, followed by 72°C for 5 min. DMSO at a final concentration of 0.1% was added to the PCR reactions. PCR products were run on a 2% agarose gel, excised, purified using phenol-chloroform extraction, and subjected to library preparation using the NEBNext Ultra II DNA Library Prep Kit (Illumina) following the manufacturer's instructions. Barcoded libraries were pooled and sequenced on an Illumina MiSeq platform using a 2 × 300-bp paired-end reagent kit. Paired-end sequencing reads were assembled into contigs via FLASH (Magoč and Salzberg 2011) using default parameters. Contigs matching each target sequence were identified based on the primers carried at their termini. Per sample, 50 reads were randomly selected and analyzed using QUMA (QUantification tool for Methylation Analysis) (Kumaki et al. 2008) with default parameters, plus requiring strict CpG recognition and, in the case of the overall L1 methylation, excluding identical bisulfite sequences.
Human TTC28 donor L1 methylation analysis
The methylation state of the CpG island present in the TTC28 donor L1 promoter region was analyzed with genomic DNA extracted from patient ICC.75 tumor and nontumor liver samples. Genomic DNA was bisulfite-converted using the EZ DNA Methylation-Gold Kit (Zymo Research), following the manufacturer's instructions with the following specifications: 500 ng gDNA was treated per reaction, the desulfonation reaction was conducted for 20 min at room temperature, and DNA was eluted in 25 µL final volume. PCR amplification was conducted using MyTaq polymerase, with a locus-specific primer placed in the 5′ genomic flank (ATTTTAGTTTGGGAGATAGAGYGA) and a reverse primer located in the donor L1 5′ UTR (ACTATAATAAACTCCACCCAAT). PCR reactions were performed in 20-µL final volume reactions, using 1× reaction buffer, 20 pmol of each primer, and 1 U of enzyme. The following cycling conditions were used: 95°C for 2 min, then 40 cycles of 95°C for 30 sec, 54°C for 30 sec, 72°C for 30 sec, followed by a single extension step at 72°C for 5 min. PCR amplicons were resolved, processed for library preparation, Illumina sequenced, and analyzed via QUMA as detailed above for the mouse methylation analysis, except here a TruSeq DNA PCR-Free Library Preparation Kit (Illumina) was used for library preparation.
qRT-PCR analysis
Snap-frozen tumor and matched nontumor liver samples from 10 patients, including HCC.58, were shaved with a scalpel on dry ice and resuspended in TRIzol Reagent (#15596-026, Invitrogen, Life Technologies) following the manufacturer's instructions for total RNA isolation. Quantification was performed using NanoDrop 1000 (Thermo Fisher Scientific). Two micrograms total RNA were treated with DNase I (#AM1906, Ambion, Life Technologies) and used as template for cDNA synthesis with SuperScript III Reverse Transcriptase (#18080-093, Invitrogen, Life Technologies) following the manufacturer's instructions. Two micrograms total RNA were processed as described with no reverse transcriptase added to the cDNA synthesis reaction for use as negative control (RT−). cDNA was diluted to final concentrations of 1:2, 1:8, 1:32, 1:128, 1:256, 1:512, and 1:1024, and used to generate a standard curve for each primer set. Samples were diluted to 1:20 final concentration for qRT-PCR. Real time PCRs were performed using KiCqStart SYBR Green qPCR ReadyMix Low ROX (Sigma-Aldrich) and run on a ViiA 7 Real-Time PCR System (Life Technologies) with standard curve experiment analysis settings with the following PCR conditions: 20 sec at 95°C, then 40 cycles of 5 sec at 95°C and 20 sec at 60°C, followed by a Melt Curve stage of 15 sec at 95°C, 1 min at 60°C, and 15 sec at 95°C. Negative control qRT-PCRs were performed using water as template (no template control, NTC) and 2 µL of RT− reaction; no amplification was detected. Both KHDRBS2 isoforms (NM_152688) were assessed using 5′-TCTGGTCGTGGCAGAGGTAT and 5′- TCCACGGGTTACAGTGCTTC as forward primers and reverse primers. TATA-box binding protein mRNA (TBP, NM_003194) was amplified using 5′-GCAAGGGTTTCTGGTTTGCC and 5′-GGGTCAGTCCAGTGCCATAA as forward primers and reverse primers. Relative expression levels of KHDRBS2 were calculated using three technical replicates and normalized to TBP. Statistical analysis was performed with Prism5 (GraphPad Software), applying a two-way ANOVA followed by Tukey's post hoc analysis.
Ethics approval and consent to participate
Experiments involving animals were performed in accordance with Italian laws (D.lgs. 116/92), which enforces the EU 86/609 directive. Human samples were analyzed with approval from the French Institute of Medical Research and Health (Ref: 11-047), the East of Scotland Research Ethics Service (Ref: LR/11/ES/0022), and the Mater Health Services Human Research Ethics Committee (Ref: 1915A).
Data access
RC-seq, mRC-seq, and WGS data from this study have been submitted to the European Nucleotide Archive (ENA; http://www.ebi.ac.uk/ena/) under project accession number PRJEB18756. Sanger trace files from this study have been submitted to the NCBI Trace Archive (http://www.ncbi.nlm.nih.gov/Traces/home/index.cgi) with TI numbers 2344290281–2344290314.
Supplementary Material
Acknowledgments
We thank Pr. John V. Moran for the plasmids pJM101/L1.3 and pJM105/L1.3, and Pr. Haig H. Kazazian for the plasmids pTN201 and TGF21. We thank Pr. René Adam, Pr. Daniel Cherqui, Pr. Denis Castaing, Pr. Antonio Sa Cunha, and Pr. Eric Vibert, who are surgeons in the Centre Hépatobiliaire, Villejuif, and the Tissue Biobank Group (AP-HP and Université Paris-Sud) for providing human specimens. We thank members of the Faulkner laboratory for helpful advice and discussion. This study was supported by CSL Centenary Fellowship and National Health and Medical Research Council (NHMRC) Project Grant (GNT1042449, GNT1045991, GNT1067983, GNT1068789, and GNT1106206) funding to G.J.F., the EU FP7 under grant agreement No. 259743 underpinning the MODHEP consortium to J.F., D.S., and G.J.F., and an Australian Research Council Discovery Early Career Researcher Award (DE150101117) to A.D.E.
Author contributions: S.N.S., P.E.C., and R.S. are equal co-authors. S.N.S., R.S., and D.J.G. prepared RC-seq libraries. S.N.S., P.E.C., S.R.R., and M.K. performed PCR validation and capillary sequencing of tumor-specific L1 insertions and associated sequence analyses. S.R.R. performed retrotransposition assays. P.G. and F.J.S.-L. performed methylation analyses. A.D.E. and G.J.F. performed bioinformatics analyses. J.F., A.D.S., D.R., and D.S. recruited patients and collected samples and clinical information. P.N. and S.G. performed animal experiments. R.S., J.F., and G.J.F. initiated the study. G.J.F. funded and coordinated the study. P.E.C., A.D.E., S.R.R., and G.J.F. prepared figures. A.D.E., S.R.R., and G.J.F. wrote the manuscript. All authors read and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.226993.117.
Freely available online through the Genome Research Open Access option.
References
- Achanta P, Steranka JP, Tang Z, Rodic N, Sharma R, Yang WR, Ma S, Grivainis M, Huang CRL, Schneider AM, et al. 2016. Somatic retrotransposition is infrequent in glioblastomas. Mob DNA 7: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi K, Li J, Stephens RM, Volfovsky N, Symer DE. 2008. Extensive variation between inbred mouse strains due to endogenous L1 retrotransposition. Genome Res 18: 869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, Brennan PM, Rizzu P, Smith S, Fell M, et al. 2011. Somatic retrotransposition alters the genetic landscape of the human brain. Nature 479: 534–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balogh J, Victor D III, Asham EH, Burroughs SG, Boktour M, Saharia A, Li X, Ghobrial RM, Monsour HP Jr. 2016. Hepatocellular carcinoma: a review. J Hepatocell Carcinoma 3: 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio VP, Roy-Engel AM, Deininger P. 2008. The impact of multiple splice sites in human L1 elements. Gene 411: 38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot S, Chevret P, Furano AV. 2000. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol 17: 915–928. [DOI] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH Jr. 2003. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci 100: 5280–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira PE, Richardson SR, Faulkner GJ. 2014. L1 retrotransposons, cancer stem cells and oncogenesis. FEBS J 281: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira PE, Ewing AD, Li G, Schauer SN, Upton KR, Fagg AC, Morell S, Kindlova M, Gerdes P, Richardson SR, et al. 2016. Evidence for L1-associated DNA rearrangements and negligible L1 retrotransposition in glioblastoma multiforme. Mob DNA 7: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan-On W, Nairismagi ML, Ong CK, Lim WK, Dima S, Pairojkul C, Lim KH, McPherson JR, Cutcutache I, Heng HL, et al. 2013. Exome sequencing identifies distinct mutational patterns in liver fluke-related and non-infection-related bile duct cancers. Nat Genet 45: 1474–1478. [DOI] [PubMed] [Google Scholar]
- Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Kallberg M, Cox AJ, Kruglyak S, Saunders CT. 2016. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32: 1220–1222. [DOI] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. 2013. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31: 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coufal NG, Garcia-Perez JL, Peng GE, Marchetto MC, Muotri AR, Mu Y, Carson CT, Macia A, Moran JV, Gage FH. 2011. Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proc Natl Acad Sci 108: 20382–20387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunliffe P, Reed V, Boyd Y. 2001. Intragenic deletions at Atp7a in mouse models for Menkes disease. Genomics 74: 155–162. [DOI] [PubMed] [Google Scholar]
- de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. 2011. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet 7: e1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Goodier JL, Ostertag EM, Kazazian HH Jr. 1998. Rapid amplification of a retrotransposon subfamily is evolving the mouse genome. Nat Genet 20: 288–290. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH Jr. 1991. Isolation of an active human transposable element. Science 254: 1805–1808. [DOI] [PubMed] [Google Scholar]
- Doucet-O'Hare TT, Rodic N, Sharma R, Darbari I, Abril G, Choi JA, Young Ahn J, Cheng Y, Anders RA, Burns KH, et al. 2015. LINE-1 expression and retrotransposition in Barrett's esophagus and esophageal carcinoma. Proc Natl Acad Sci 112: E4894–E4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet-O'Hare TT, Sharma R, Rodic N, Anders RA, Burns KH, Kazazian HH Jr. 2016. Somatically acquired LINE-1 insertions in normal esophagus undergo clonal expansion in esophageal squamous cell carcinoma. Hum Mutat 37: 942–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin JA, Paquola AC, Singer T, Gallina I, Novotny M, Quayle C, Bedrosian TA, Alves FI, Butcher CR, Herdy JR, et al. 2016. L1-associated genomic regions are deleted in somatic cells of the healthy human brain. Nat Neurosci 19: 1583–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrony GD, Cai X, Lee E, Hills LB, Elhosary PC, Lehmann HS, Parker JJ, Atabay KD, Gilmore EC, Poduri A, et al. 2012. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell 151: 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing AD. 2015. Transposable element detection from whole genome sequence data. Mob DNA 6: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing AD, Gacita A, Wood LD, Ma F, Xing D, Kim MS, Manda SS, Abril G, Pereira G, Makohon-Moore A, et al. 2015. Widespread somatic L1 retrotransposition occurs early during gastrointestinal cancer evolution. Genome Res 25: 1536–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farshidfar F, Zheng S, Gingras MC, Newton Y, Shih J, Robertson AG, Hinoue T, Hoadley KA, Gibb EA, Roszik J, et al. 2017. Integrative genomic analysis of cholangiocarcinoma identifies distinct IDH-mutant molecular profiles. Cell Rep 18: 2780–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner GJ, Garcia-Perez JL. 2017. L1 mosaicism in mammals: extent, effects, and evolution. Trends Genet 33: 802–816. [DOI] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH Jr, Boeke JD. 1996. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87: 905–916. [DOI] [PubMed] [Google Scholar]
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. 2015. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136: E359–E386. [DOI] [PubMed] [Google Scholar]
- Furano AV. 2000. The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons. Prog Nucleic Acid Res Mol Biol 64: 255–294. [DOI] [PubMed] [Google Scholar]
- Gardner EJ, Lam VK, Harris DN, Chuang NT, Scott EC, Pittard WS, Mills RE, Genomes Project C, Devine SE. 2017. The Mobile Element Locator Tool (MELT): population-scale mobile element discovery and biology. Genome Res 27: 1916–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N, Lutz-Prigge S, Moran JV. 2002. Genomic deletions created upon LINE-1 retrotransposition. Cell 110: 315–325. [DOI] [PubMed] [Google Scholar]
- Gilbert N, Lutz S, Morrish TA, Moran JV. 2005. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol Cell Biol 25: 7780–7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL. 2016. Restricting retrotransposons: a review. Mob DNA 7: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH Jr. 2008. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 135: 23–35. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Kazazian HH Jr. 2000. Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum Mol Genet 9: 653–657. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Du K, Kazazian HH Jr. 2001. A novel active L1 retrotransposon subfamily in the mouse. Genome Res 11: 1677–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Preston R, Michailidi C, Marchionni L, Pickering CR, Frederick MJ, Myers JN, Yegnasubramanian S, Hadar T, Noordhuis MG, Zizkova V, et al. 2014. Key tumor suppressor genes inactivated by “greater promoter” methylation and somatic mutations in head and neck cancer. Epigenetics 9: 1031–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Dixon E. 2017. Epidemiology and risk factors: intrahepatic cholangiocarcinoma. Hepatobiliary Surg Nutr 6: 101–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JS, Szak ST, Boeke JD. 2004. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature 429: 268–274. [DOI] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH Jr. 2016. Roles for retrotransposon insertions in human disease. Mob DNA 7: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardies SC, Wang L, Zhou L, Zhao Y, Casavant NC, Huang S. 2000. LINE-1 (L1) lineages in the mouse. Mol Biol Evol 17: 616–628. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Suzuki AM, Dos Santos A, Desterke C, Collino A, Ghisletti S, Braun E, Bonetti A, Fort A, Qin XY, et al. 2015. CAGE profiling of ncRNAs in hepatocellular carcinoma reveals widespread activation of retroviral LTR promoters in virus-induced tumors. Genome Res 25: 1812–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen JL, Faust GG, Rodriguez AR, Ferguson WC, Shumilina S, Clark RA, Boland MJ, Martin G, Chubukov P, Tsunemoto RK, et al. 2016. The complete genome sequences, unique mutational spectra, and developmental potency of adult neurons revealed by cloning. Neuron 89: 1223–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helman E, Lawrence MS, Stewart C, Sougnez C, Getz G, Meyerson M. 2014. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res 24: 1053–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes SE, Dombroski BA, Krebs CM, Boehm CD, Kazazian HH Jr. 1994. A new retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric insertion. Nat Genet 7: 143–148. [DOI] [PubMed] [Google Scholar]
- Iannelli F, Collino A, Sinha S, Radaelli E, Nicoli P, D'Antiga L, Sonzogni A, Faivre J, Buendia MA, Sturm E, et al. 2014. Massive gene amplification drives paediatric hepatocellular carcinoma caused by bile salt export pump deficiency. Nat Commun 5: 3850. [DOI] [PubMed] [Google Scholar]
- Iijima T, Iijima Y, Witte H, Scheiffele P. 2014. Neuronal cell type-specific alternative splicing is regulated by the KH domain protein SLM1. J Cell Biol 204: 331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, Van Meir EG, Vertino PM, Devine SE. 2010. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 141: 1253–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Pawlik TM, Anders RA, Selaru FM, Streppel MM, Lucas DJ, Niknafs N, Guthrie VB, Maitra A, Argani P, et al. 2013. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet 45: 1470–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RB, Song H, Xu Y, Garrison KE, Buzdin AA, Anwar N, Hunter DV, Mujib S, Mihajlovic V, Martin E, et al. 2013. LINE-1 retrotransposable element DNA accumulates in HIV-1-infected cells. J Virol 87: 13307–13320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. 1997. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci 94: 1872–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jusakul A, Cutcutache I, Yong CH, Lim JQ, Huang MN, Padmanabhan N, Nellore V, Kongpetch S, Ng AWT, Ng LM, et al. 2017. Whole-genome and epigenomic landscapes of etiologically distinct subtypes of cholangiocarcinoma. Cancer Discov 7: 1116–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaghad M, Maillet L, Brulet P. 1985. Retroviral characteristics of the long terminal repeat of murine E.Tn sequences. EMBO J 4: 2911–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano H, Godoy I, Courtney C, Vetter MR, Gerton GL, Ostertag EM, Kazazian HH Jr. 2009. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev 23: 1303–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH Jr, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE. 1988. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 332: 164–166. [DOI] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, et al. 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477: 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsmore SF, Giros B, Suh D, Bieniarz M, Caron MG, Seldin MF. 1994. Glycine receptor β-subunit gene mutation in spastic mouse associated with LINE-1 element insertion. Nat Genet 7: 136–141. [DOI] [PubMed] [Google Scholar]
- Kohrman DC, Harris JB, Meisler MH. 1996. Mutation detection in the med and medJ alleles of the sodium channel Scn8a. Unusual splicing due to a minor class AT-AC intron. J Biol Chem 271: 17576–17581. [DOI] [PubMed] [Google Scholar]
- Kopera HC, Larson PA, Moldovan JB, Richardson SR, Liu Y, Moran JV. 2016. LINE-1 cultured cell retrotransposition assay. Methods Mol Biol 1400: 139–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress TR, Pellanda P, Pellegrinet L, Bianchi V, Nicoli P, Doni M, Recordati C, Bianchi S, Rotta L, Capra T, et al. 2016. Identification of MYC-dependent transcriptional programs in oncogene-addicted liver tumors. Cancer Res 76: 3463–3472. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaki Y, Oda M, Okano M. 2008. QUMA: quantification tool for methylation analysis. Nucleic Acids Res 36: W170–W175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Lang H, Sotiropoulos GC, Brokalaki EI, Schmitz KJ, Bertona C, Meyer G, Frilling A, Paul A, Malago M, Broelsch CE. 2007. Survival and recurrence rates after resection for hepatocellular carcinoma in noncirrhotic livers. J Am Coll Surg 205: 27–36. [DOI] [PubMed] [Google Scholar]
- Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ III, Lohr JG, Harris CC, Ding L, Wilson RK, et al. 2012. Landscape of somatic retrotransposition in human cancers. Science 337: 967–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 1303.3997.
- Li J, Zhao X, Wang D, He W, Zhang S, Cao W, Huang Y, Wang L, Zhou S, Luo K. 2016. Up-regulated expression of phospholipase C, β1 is associated with tumor cell proliferation and poor prognosis in hepatocellular carcinoma. Onco Targets Ther 9: 1697–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH. 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72: 595–605. [DOI] [PubMed] [Google Scholar]
- Macfarlane CM, Collier P, Rahbari R, Beck CR, Wagstaff JF, Igoe S, Moran JV, Badge RM. 2013. Transduction-specific ATLAS reveals a cohort of highly active L1 retrotransposons in human populations. Hum Mutat 34: 974–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoč T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27: 2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksakova IA, Romanish MT, Gagnier L, Dunn CA, van de Lagemaat LN, Mager DL. 2006. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet 2: e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SL, Bushman D, Wang F, Li PW, Walker A, Cummiskey J, Branciforte D, Williams MC. 2008. A single amino acid substitution in ORF1 dramatically decreases L1 retrotransposition and provides insight into nucleic acid chaperone activity. Nucleic Acids Res 36: 5845–5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias SL, Scott AF, Kazazian HH Jr, Boeke JD, Gabriel A. 1991. Reverse transcriptase encoded by a human transposable element. Science 254: 1808–1810. [DOI] [PubMed] [Google Scholar]
- Mauad TH, van Nieuwkerk CM, Dingemans KP, Smit JJ, Schinkel AH, Notenboom RG, van den Bergh Weerman MA, Verkruisen RP, Groen AK, Oude Elferink RP, et al. 1994. Mice with homozygous disruption of the mdr2 P-glycoprotein gene. A novel animal model for studies of nonsuppurative inflammatory cholangitis and hepatocarcinogenesis. Am J Pathol 145: 1237–1245. [PMC free article] [PubMed] [Google Scholar]
- Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler KW, Vogelstein B, Nakamura Y. 1992. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res 52: 643–645. [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH Jr. 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87: 917–927. [DOI] [PubMed] [Google Scholar]
- Moran JV, DeBerardinis RJ, Kazazian HH Jr. 1999. Exon shuffling by L1 retrotransposition. Science 283: 1530–1534. [DOI] [PubMed] [Google Scholar]
- Mulhardt C, Fischer M, Gass P, Simon-Chazottes D, Guenet JL, Kuhse J, Betz H, Becker CM. 1994. The spastic mouse: aberrant splicing of glycine receptor beta subunit mRNA caused by intronic insertion of L1 element. Neuron 13: 1003–1015. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. 2005. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435: 903–910. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH. 2010. L1 retrotransposition in neurons is modulated by MeCP2. Nature 468: 443–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgia C, Pritchard JK, Kim SY, Fassati A, Weiss RA. 2006. Clonal origin and evolution of a transmissible cancer. Cell 126: 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naas TP, DeBerardinis RJ, Moran JV, Ostertag EM, Kingsmore SF, Seldin MF, Hayashizaki Y, Martin SL, Kazazian HH. 1998. An actively retrotransposing, novel subfamily of mouse L1 elements. EMBO J 17: 590–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Arai Y, Totoki Y, Shirota T, Elzawahry A, Kato M, Hama N, Hosoda F, Urushidate T, Ohashi S, et al. 2015. Genomic spectra of biliary tract cancer. Nat Genet 47: 1003–1010. [DOI] [PubMed] [Google Scholar]
- Nellaker C, Keane TM, Yalcin B, Wong K, Agam A, Belgard TG, Flint J, Adams DJ, Frankel WN, Ponting CP. 2012. The genomic landscape shaped by selection on transposable elements across 18 mouse strains. Genome Biol 13: R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AL, Weaver JM, Eldridge MD, Tavare S, Fitzgerald RC, Edwards PA, Consortium OC. 2015. Mobile element insertions are frequent in oesophageal adenocarcinomas and can mislead paired-end sequencing analysis. BMC Genomics 16: 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perepelitsa-Belancio V, Deininger P. 2003. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat Genet 35: 363–366. [DOI] [PubMed] [Google Scholar]
- Perou CM, Pryor RJ, Naas TP, Kaplan J. 1997. The bg allele mutation is due to a LINE1 element retrotransposition. Genomics 42: 366–368. [DOI] [PubMed] [Google Scholar]
- Petrick JL, Braunlin M, Laversanne M, Valery PC, Bray F, McGlynn KA. 2016. International trends in liver cancer incidence, overall and by histologic subtype, 1978–2007. Int J Cancer 139: 1534–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrilli AM, Fernandez-Valle C. 2016. Role of Merlin/NF2 inactivation in tumor biology. Oncogene 35: 537–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe C, Vargas-Landin DB, Doucet AJ, van Essen D, Vera-Otarola J, Kuciak M, Corbin A, Nigumann P, Cristofari G. 2016. Activation of individual L1 retrotransposon instances is restricted to cell-type dependent permissive loci. eLife 5: e13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickeral OK, Makalowski W, Boguski MS, Boeke JD. 2000. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res 10: 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitkanen E, Cajuso T, Katainen R, Kaasinen E, Valimaki N, Palin K, Taipale J, Aaltonen LA, Kilpivaara O. 2014. Frequent L1 retrotranspositions originating from TTC28 in colorectal cancer. Oncotarget 5: 853–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rava M, D'Andrea A, Doni M, Kress TR, Ostuni R, Bianchi V, Morelli MJ, Collino A, Ghisletti S, Nicoli P, et al. 2017. Mutual epithelium-macrophage dependency in liver carcinogenesis mediated by ST18. Hepatology 65: 1708–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson SR, Morell S, Faulkner GJ. 2014. L1 retrotransposons and somatic mosaicism in the brain. Annu Rev Genet 48: 1–27. [DOI] [PubMed] [Google Scholar]
- Richardson SR, Gerdes P, Gerhardt DJ, Sanchez-Luque FJ, Bodea GO, Munoz-Lopez M, Jesuadian JS, Kempen MHC, Carreira PE, Jeddeloh JA, et al. 2017. Heritable L1 retrotransposition in the mouse primordial germline and early embryo. Genome Res 27: 1395–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi S, Gores GJ. 2013. Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology 145: 1215–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodic N, Steranka JP, Makohon-Moore A, Moyer A, Shen P, Sharma R, Kohutek ZA, Huang CR, Ahn D, Mita P, et al. 2015. Retrotransposon insertions in the clonal evolution of pancreatic ductal adenocarcinoma. Nat Med 21: 1060–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, DeBerardinis RJ, Gabriel A, Swergold GD, Kazazian HH Jr. 1997. Many human L1 elements are capable of retrotransposition. Nat Genet 16: 37–43. [DOI] [PubMed] [Google Scholar]
- Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW, Muller J. 2010. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 465: 243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EC, Devine SE. 2017. The role of somatic L1 retrotransposition in human cancers. Viruses 9: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott AF, Schmeckpeper BJ, Abdelrazik M, Comey CT, O'Hara B, Rossiter JP, Cooley T, Heath P, Smith KD, Margolet L. 1987. Origin of the human L1 elements: proposed progenitor genes deduced from a consensus DNA sequence. Genomics 1: 113–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EC, Gardner EJ, Masood A, Chuang NT, Vertino PM, Devine SE. 2016. A hot L1 retrotransposon evades somatic repression and initiates human colorectal cancer. Genome Res 26: 745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini FM, Radvinsky D. 2016. The pathways of genetic transformation in cholangiocarcinogenesis. Cancer Genet 209: 554–558. [DOI] [PubMed] [Google Scholar]
- Shukla R, Upton KR, Munoz-Lopez M, Gerhardt DJ, Fisher ME, Nguyen T, Brennan PM, Baillie JK, Collino A, Ghisletti S, et al. 2013. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell 153: 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer MF, Thayer RE, Grimaldi G, Lerman MI, Fanning TG. 1983. Homology between the KpnI primate and BamH1 (M1F-1) rodent families of long interspersed repeated sequences. Nucleic Acids Res 11: 5739–5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit JJ, Schinkel AH, Oude Elferink RP, Groen AK, Wagenaar E, van Deemter L, Mol CA, Ottenhoff R, van der Lugt NM, van Roon MA, et al. 1993. Homozygous disruption of the murine mdr2 P-glycoprotein gene leads to a complete absence of phospholipid from bile and to liver disease. Cell 75: 451–462. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. 2013. RepeatMasker Open-4.0. http://www.repeatmasker.org.
- Solyom S, Ewing AD, Rahrmann EP, Doucet T, Nelson HH, Burns MB, Harris RS, Sigmon DF, Casella A, Erlanger B, et al. 2012. Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res 22: 2328–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sookdeo A, Hepp CM, McClure MA, Boissinot S. 2013. Revisiting the evolution of mouse LINE-1 in the genomic era. Mob DNA 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara T, Ohsumi T, Kuromitsu J, Shibata K, Sasaki N, Okazaki Y, Shibata H, Sato S, Yoshiki A, Kusakabe M, et al. 1996. Dysfunction of the Orleans reeler gene arising from exon skipping due to transposition of a full-length copy of an active L1 sequence into the skipped exon. Hum Mol Genet 5: 989–993. [DOI] [PubMed] [Google Scholar]
- Tan J, Yu CY, Wang ZH, Chen HY, Guan J, Chen YX, Fang JY. 2015. Genetic variants in the inositol phosphate metabolism pathway and risk of different types of cancer. Sci Rep 5: 8473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Steranka JP, Ma S, Grivainis M, Rodic N, Huang CR, Shih IM, Wang TL, Boeke JD, Fenyo D, et al. 2017. Human transposon insertion profiling: analysis, visualization and identification of somatic LINE-1 insertions in ovarian cancer. Proc Natl Acad Sci 114: E733–E740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M. 2009. Polybromo-1: the chromatin targeting subunit of the PBAF complex. Biochimie 91: 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoolen B, Maronpot RR, Harada T, Nyska A, Rousseaux C, Nolte T, Malarkey DE, Kaufmann W, Kuttler K, Deschl U, et al. 2010. Proliferative and nonproliferative lesions of the rat and mouse hepatobiliary system. Toxicol Pathol 38: 5S–81S. [DOI] [PubMed] [Google Scholar]
- Tubio JMC, Li Y, Ju YS, Martincorena I, Cooke SL, Tojo M, Gundem G, Pipinikas CP, Zamora J, Raine K, et al. 2014. Mobile DNA in cancer. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science 345: 1251343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton KR, Gerhardt DJ, Jesuadian JS, Richardson SR, Sanchez-Luque FJ, Bodea GO, Ewing AD, Salvador-Palomeque C, van der Knaap MS, Brennan PM, et al. 2015. Ubiquitous L1 mosaicism in hippocampal neurons. Cell 161: 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. [DOI] [PubMed] [Google Scholar]
- Wei W, Morrish TA, Alisch RS, Moran JV. 2000. A transient assay reveals that cultured human cells can accommodate multiple LINE-1 retrotransposition events. Anal Biochem 284: 435–438. [DOI] [PubMed] [Google Scholar]
- Wilson BG, Roberts CW. 2011. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 11: 481–492. [DOI] [PubMed] [Google Scholar]
- Wu JN, Roberts CW. 2013. ARID1A mutations in cancer: another epigenetic tumor suppressor? Cancer Discov 3: 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie A, Jones AE, D'Brot A, Lu WJ, Kurtz P, Moran JV, Rakheja D, Chen KS, Hammer RE, Comerford SA, et al. 2016. p53 genes function to restrain mobile elements. Genes Dev 30: 64–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima I, Sato S, Kimura T, Yasumoto K, Shibahara S, Goding CR, Yamamoto H. 1999. An L1 element intronic insertion in the black-eyed white (Mitf[mi-bw]) gene: the loss of a single Mitf isoform responsible for the pigmentary defect and inner ear deafness. Hum Mol Genet 8: 1431–1441. [DOI] [PubMed] [Google Scholar]
- Yalcin B, Wong K, Bhomra A, Goodson M, Keane TM, Adams DJ, Flint J. 2012. The fine-scale architecture of structural variants in 17 mouse genomes. Genome Biol 13: R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. 2006. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J Biol Chem 281: 15694–15700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S, Li J, Zhou H, Frech C, Jiang X, Chu JS, Zhao X, Li Y, Li Q, Wang H, et al. 2014. Mutational landscape of intrahepatic cholangiocarcinoma. Nat Commun 5: 5696. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.