Abstract
Alternative pre-mRNA splicing plays a major role in expanding the transcript output of human genes. This process is regulated, in part, by the interplay of trans-acting RNA binding proteins (RBPs) with myriad cis-regulatory elements scattered throughout pre-mRNAs. These molecular recognition events are critical for defining the protein-coding sequences (exons) within pre-mRNAs and directing spliceosome assembly on noncoding regions (introns). One of the earliest events in this process is recognition of the 3′ splice site (3′ss) by U2 small nuclear RNA auxiliary factor 2 (U2AF2). Splicing regulators, such as the heterogeneous nuclear ribonucleoprotein A1 (HNRNPA1), influence spliceosome assembly both in vitro and in vivo, but their mechanisms of action remain poorly described on a global scale. HNRNPA1 also promotes proofreading of 3′ss sequences though a direct interaction with the U2AF heterodimer. To determine how HNRNPA1 regulates U2AF–RNA interactions in vivo, we analyzed U2AF2 RNA binding specificity using individual-nucleotide resolution crosslinking immunoprecipitation (iCLIP) in control and HNRNPA1 overexpression cells. We observed changes in the distribution of U2AF2 crosslinking sites relative to the 3′ss of alternative cassette exons but not constitutive exons upon HNRNPA1 overexpression. A subset of these events shows a concomitant increase of U2AF2 crosslinking at distal intronic regions, suggesting a shift of U2AF2 to “decoy” binding sites. Of the many noncanonical U2AF2 binding sites, Alu-derived RNA sequences represented one of the most abundant classes of HNRNPA1-dependent decoys. We propose that one way HNRNPA1 regulates exon definition is to modulate the interaction of U2AF2 with decoy or bona fide 3′ss.
Precursor messenger RNA (pre-mRNA) splicing is catalyzed by a large macromolecular complex composed of five uridine-rich small nuclear ribonucleoprotein particles (U snRNPs) and myriad protein factors (Wahl et al. 2009). This process is required to excise intervening sequences (introns) from the pre-mRNA and ligate protein-coding sequences (exons). The 5′ and 3′ ends of introns are defined by nearly invariant GU and AG dinucleotides as well as the branch point sequence, respectively. Combinatorial protein–RNA interactions play important roles in defining the splice sites and branch point during spliceosome assembly. The 5′ splice site (GU) is recognized by serine and arginine-rich splicing factors (SR Proteins) and through base-pairing interactions with the U1 snRNP (Eperon et al. 1993; Jamison et al. 1995). Similarly, the 3′ splice site (3′ss) is decoded by a combination of SR proteins and the U2 snRNP auxiliary factor (U2AF). After early (E) complex assembly, the branch point sequencing is specified through the interaction of U2AF, splicing factor 1 (SF1), and via base-pairing with the U2 snRNP (Ruskin et al. 1988; Berglund et al. 1997). The responsibility for 3′ss recognition is shared by the two subunits of U2AF (Merendino et al. 1999; Wu et al. 1999; Zorio and Blumenthal 1999). The small and large subunits of U2AF, encoded by U2AF1 and U2AF2, recognize the AG dinucleotide and the upstream polypyrimidine tract, respectively (Merendino et al. 1999; Wu et al. 1999; Zorio and Blumenthal 1999). These early steps in spliceosome assembly play critical roles in defining exon–intron boundaries.
HNRNPA1 is a well-characterized regulator of alternative splicing. One of the primary functions of HNRNPA1 is to prevent inclusion of specific exons (Mayeda and Krainer 1992; Caceres et al. 1994; Yang et al. 1994). Several models for HNRNPA1-dependent exon skipping have been described in the literature. For example, HNRNPA1 can compete with RBPs for binding to juxtaposed regulatory elements that would be normally occupied by splicing enhancer proteins such as SRSF1 (Eperon et al. 2000; Zahler et al. 2004). Another potential mechanism involves oligomerization and spreading of HNRNPA1 from an exonic splicing silencer across a regulated exon, thus antagonizing binding of splicing enhancers and spliceosomal factors, such as U2AF2 (Zhu et al. 2001; Okunola and Krainer 2009). Finally, HNRNPA1 binding to intronic splicing silencers on either side of an alternative exon can dimerize, causing “looping out” of an exon from the pre-mRNA, and promote its exclusion from the final mature transcript (Blanchette and Chabot 1999). A related alternative splicing factor, PTBP1, also promotes exon skipping via a looping mechanism (Chou et al. 2000; Lamichhane et al. 2010). Perhaps most relevant to this work, HNRNPA1 promotes proofreading of the 3′ss by U2AF2. In this case HNRNPA1 enables the U2AF heterodimer to reject suboptimal splice sites (Tavanez et al. 2012). In light of these diverse molecular mechanisms of HNRNPA1-dependent splicing regulation, there is a critical need to test their generality on a global scale.
In this study, we investigate how HNRNPA1 influences the association of U2AF2 with 3′ss on a transcriptome-wide scale. We used individual nucleotide resolution crosslinking immunoprecipitation (iCLIP) and high-throughput sequencing to map U2AF2–RNA interactions in control or HNRNPA1 overexpression cell lines. As HNRNPA1 has been shown to be overexpressed in a variety of cancer types (Ushigome et al. 2005; Boukakis et al. 2010; Zhou et al. 2013) and directly involved in specific splicing event that regulate disease phenotype (David et al. 2010; Chen et al. 2012; Bonomi et al. 2013; Loh et al. 2015), this in-cell model will allow us a global look at the effects of HNRNPA1 modulation on splicing and RBP distribution.
Results
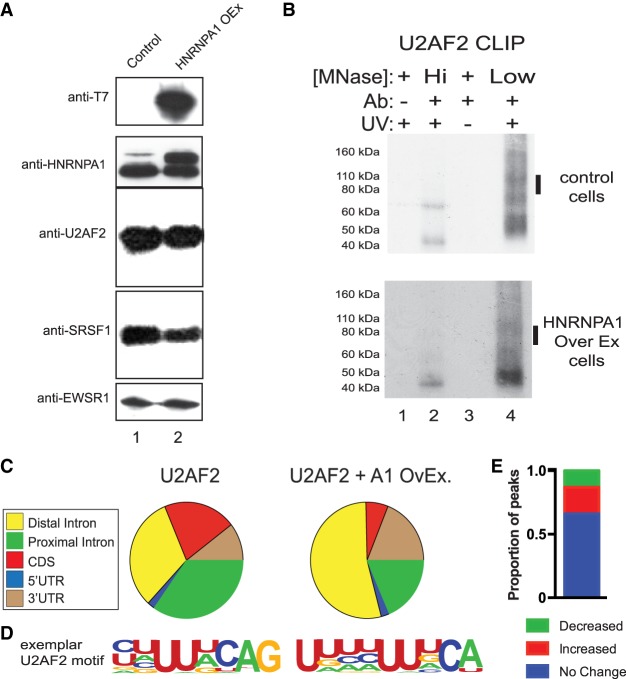
To determine how HNRNPA1 influences the association of U2AF2 with 3′ss on a global scale, we established an HNRNPA1-inducible expression system in HEK293 cells. We then assayed U2AF2 and HNRNPA1 protein–RNA interactions in control or HNRNPA1 overexpression cells using individual nucleotide resolution crosslinking immunoprecipitation and high throughput sequencing (iCLIP-seq) (Konig et al. 2010). We favored the overexpression approach because HNRNPA1 protein levels are elevated in many human cancers (Pino et al. 2003; Ushigome et al. 2005; Chen et al. 2010; Loh et al. 2015; Yu et al. 2015). Induction of HNRNPA1 results in an approximately twofold increase compared to the endogenous protein (and relative to EWSR1) and has no appreciable effect on SRSF1 or U2AF2 steady-state protein levels (Fig. 1A). We used iCLIP to purify HNRNPA1–, SRSF1–, and U2AF2–RNA complexes from control and HNRNPA1 overexpressing cells (Fig. 1B; Supplemental Fig. S2A,B). In all cases, the immunoprecipitated material was both UV- and antibody-dependent, nuclease-sensitive, and produced robust sequencing libraries (Supplemental Tables 1–3). After identification of binding site peaks using CLIPper (Supplemental Table 4; Lovci et al. 2013), we observed differences in the distribution of U2AF2 peaks across genomic locations between control and HNRNPA1 overexpression cell lines (Fig. 1C). Most notably, the proportion of peaks located in coding exons (CDS) or in exon-proximal intronic regions was reduced, whereas intronic peaks located >500 nucleotides (nt) from exons (distal intron) increased. A similar trend was observed for SRSF1 peaks (Supplemental Fig. 1). In both control cells and HNRNPA1 overexpression cells, intragenic, non-repeat-associated U2AF2 peaks are characterized by a pyrimidine-rich motif, closely resembling authentic 3′ss (Figs. 1D, 2E). Although the sequences are distinct from each other, SRSF1 and HNRNPA1 motifs also appear to be similar between control and HNRNPA1 overexpression cells (Supplemental Fig. 1A,B).
Figure 1.
Crosslinking immunoprecipitation of U2AF2 under HNRNPA1 modulation. (A) Western blot analysis of SRSF1, U2AF2, HNRNPA1, the T7 epitope, and EWSR1 in control (lane 1) and HNRNPA1 overexpression HEK293 cells. (B) Examples of iCLIP autoradiographs for U2AF2 either in control or following overexpression of HNRNPA1. Protein–RNA complex shifts are UV-, antibody-, and micrococcal-nuclease–sensitive. Bars denote the region of nitrocellulose blot excised for RNA isolation for iCLIP library preparation. Micrococcal nuclease treatment at 15 U (high) and 0.015 U (low). (C) CLIPper analysis of iCLIP RNA distribution for U2AF2 in control and HNRNPA1 overexpression conditions. (D) Top HOMER consensus binding motifs for U2AF2 in control and HNRNPA1 overexpression conditions in peaks that do not overlap repetitive elements. (E) Proportion of U2AF2 binding site peaks lost (green), gained (red), or unchanged (blue) in HNRNPA1 overexpression cells.
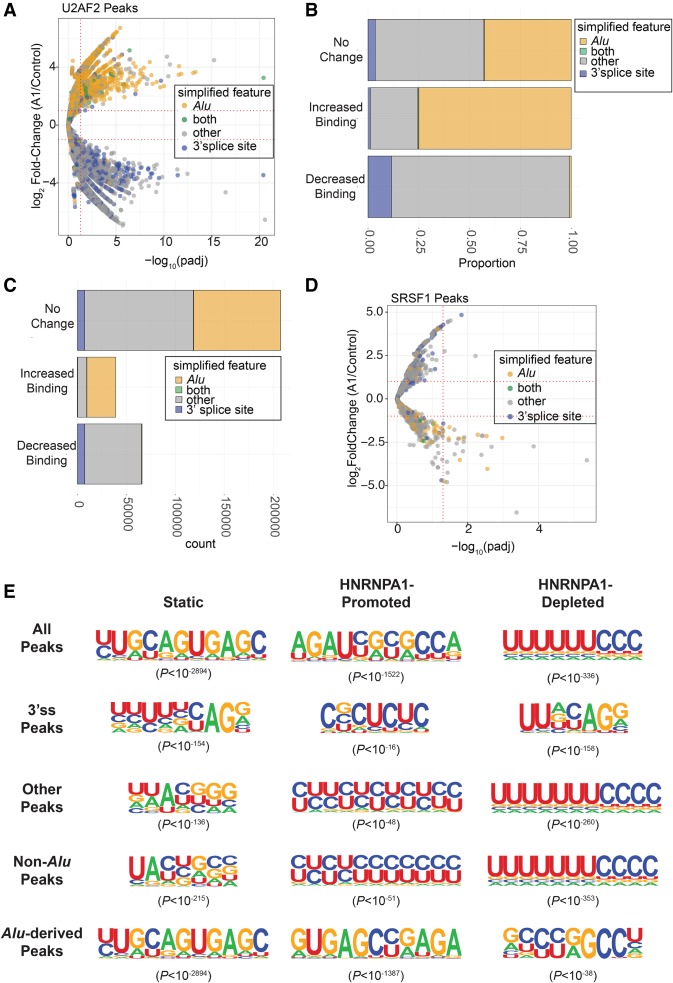
Figure 2.
Analysis of global changes in U2AF2 binding site and consensus motifs in control and HNRNPA1 overexpression cells. (A) Volcano plot representing the log2(fold-change) in U2AF2 crosslinking peaks (HNRNPA1 overexpression/control) found within Alu elements, 3′ splice site regions, both Alu and 3′ splice site regions (both), or other regions outside of these (other). These fold-changes are plotted against their corresponding –log10(adjusted P-value) associated with each fold-change. Horizontal dotted lines are associated with a log2(fold-change) of ±2 and between HNRNPA1 overexpression versus control samples. Vertical dotted lines are associated with −log10(adjusted P-value) corresponding to 0.05. (B) Stacked bar graph showing the proportion of U2AF2 crosslinking peaks showing no change (top), increased crosslinking peaks (Increased Binding), or decreased crosslinking peaks (Decreased Binding) found within Alu elements, 3′ splice site regions, both Alu and 3′ splice site regions (both), or other regions outside of these (other). (C) Stacked bar graph showing the total count of U2AF2 crosslinking peaks showing no change (top), increased crosslinking peaks (Increased Binding), or decreased crosslinking peaks (Decreased Binding) found within Alu elements, 3′ splice site regions, both Alu and 3′ splice site regions (both), or other regions outside of these (other). (D) Volcano plot representing the log2(fold-change) in SRSF1 crosslinking peaks found within Alu elements, 3′ splice site regions, both Alu and 3′ splice site regions (both), or other regions outside of these (other). (E) HOMER-generated exemplar U2AF2 motifs associated with static (left), HNRNPA1-disrupted (middle), and HNRNPA1-promoted (right) crosslinking peaks at 3′ splice sites with HNRNPA1 overexpression. Associated P-values for each motif are provided in parentheses.
To understand how HNRNPA1 overexpression alters U2AF2–RNA interactions, we identified differentially occupied binding sites in the two cell lines. The union of U2AF2 binding sites from control and HNRNPA1 overexpression cells consisted of nearly 307,000 peaks (Supplemental Tables 5, 6). The majority of these sites (66.3%) were unaffected by HNRNPA1 overexpression (Figs. 1E, 2D). In contrast, 21.5% and 12.2% of U2AF2 binding sites exhibited significant decreases or increases in crosslinking, respectively, upon HNRNPA1 overexpression (Supplemental Table 5). We used BEDTools along with GENCODE and RepeatMasker (Tarailo-Graovac and Chen 2009; Quinlan and Hall 2010; Wright et al. 2016) to annotate peaks with significantly different U2AF2 crosslinking in control and HNRNPA1 overexpression cells. U2AF2 peaks that were lost after HNRNPA1 overexpression were enriched for 3′ss annotations (P < 2.2 × 10−16, χ2 test of independence), whereas peaks gained showed a strong preference for Alu-derived RNA sequences (P < 2.2 × 10−16, χ2 test of independence) (Fig. 2A–C; Supplemental Fig. 2A). Like U2AF2 peaks, SRSF1 binding sites were also perturbed by HNRNPA1 overexpression, but they exhibit the opposite trend with respect to 3′ss (Supplemental Table 7) and Alu-derived RNA sequences (Fig. 2D; Supplemental Fig. 2B,C).
To determine if HNRNPA1 alters the sequence landscape of U2AF2–RNA interactions, we searched for overrepresented sequences in both stable and differential peaks in control and HNRNPA1 overexpression cells. We classified U2AF2 peaks into three groups: those overlapping Alu-derived sequences, those overlapping 3′ss and “other” sequences, for peaks not part of either class. For each group, we identified enriched sequences from lost, gained, or stable (static) peaks, resulting in nine different classes of U2AF2 binding sites. For most classes, we observe variations of pyrimidine-rich sequences that resemble 3′ss. For example, Figure 2E shows the top three exemplar motifs from stable, HNRNPA1-promoted, or HNRNPA1-disrupted U2AF2 peaks annotated as 3′ss. For the 13,986 stable peaks at 3′ss, ∼26% contained the most highly enriched motif shown in Figure 2E (P < 10−155). In contrast, at 3′ss peaks that were disrupted in HNRNPA1 overexpression cells, a distinct pyrimidine-rich motif was discovered in ∼31% of 13,318 peaks (P < 10−159). A much smaller subset of peaks exhibited increased U2AF2 crosslinking in the overexpression cells relative to control. Of these 867 binding sites, 9.6% contained the strong polypyrimidine motif (P < 10−16), which closely resembled the static sites. Although polypyrimidine-rich motifs are present in the majority of 3′ss and “other” binding sites, peaks annotated as overlapping Alu elements are less pyrimidine-rich (Fig. 2E). These data suggest that Alu-derived sequences are an abundant class of noncanonical binding motif that become accessible to U2AF2 in HNRNPA1 overexpression cells.
HNRNPA1 influences association of splicing factors such as U2AF2 with 3′ss on a case-by-case basis (Zhu et al. 2001; Tavanez et al. 2012). However, its effects on splicing-factor RNA targeting on a global scale are not well defined. In order to assess the effects of HNRNPA1 on transcriptome-wide splicing, we determined how titration of HNRNPA1 affected the distribution of SRSF1– and U2AF2–RNA crosslinks relative to 3′ss of constitutive or alternative cassette exons. As suggested by the peak analysis (Supplemental Fig. 3), there are no differences in HNRNPA1 crosslinking sites between control and overexpression cells (Supplemental Fig. 4A,B, left panels, blue and red lines, respectively). SRSF1 crosslinking to exonic sequences was modestly reduced in the HNRNPA1 overexpression cells compared to control, but the positional distribution of the SRSF1 sites relative to the 3′ss was largely unchanged for constitutive and skipped exons (Supplemental Fig. 4A,B, right panels, blue and red lines, respectively). U2AF2 crosslinking distribution relative to the 3′ss was substantially altered in HNRNPA1 overexpressing cells compared to the control, where a characteristic peak is observed over the 3′ss of both constitutive and skipped exons (Fig. 3A,B, blue line). In contrast, in cells overexpressing HNRNPA1, U2AF2 crosslinking density near alternative exons shifts downstream from the 3′ss and the peak is substantially reduced (Fig. 3B right panel, red line). To further determine if there is a direct relationship between HNRNPA1 binding and changes in U2AF2 or SRSF1 association with transcripts, we examined regions flanking the 3′ss of skipped exons with HNRNPA1 crosslinking in either condition (Fig. 3C; Supplemental Fig. 4C; Supplemental Tables 5–7). In regions with no detectable HNRNPA1 crosslinks, the change in U2AF2 crosslinking exhibits a bimodal distribution, which corresponds to regions flanking the 3′ss that show either increased or decreased U2AF2 crosslinking in HNRNPA1 overexpression cells relative to control cells (Fig. 3C, blue). In contrast, U2AF2 crosslinking to the vicinity of the 3′ss is significantly reduced when a direct association of HNRNPA1 is also evident (Fig. 3C, pink). For example, in both SRSF6 (serine/arginine-rich splicing factor 6 also known as SRP55) and PIEZO1 (piezo type mechanosensitive ion channel component 1), we found that U2AF2 crosslinking near the 3′ss is reduced in the cell lines overexpressing HNRNPA1 (Supplemental Fig. 5).
Figure 3.
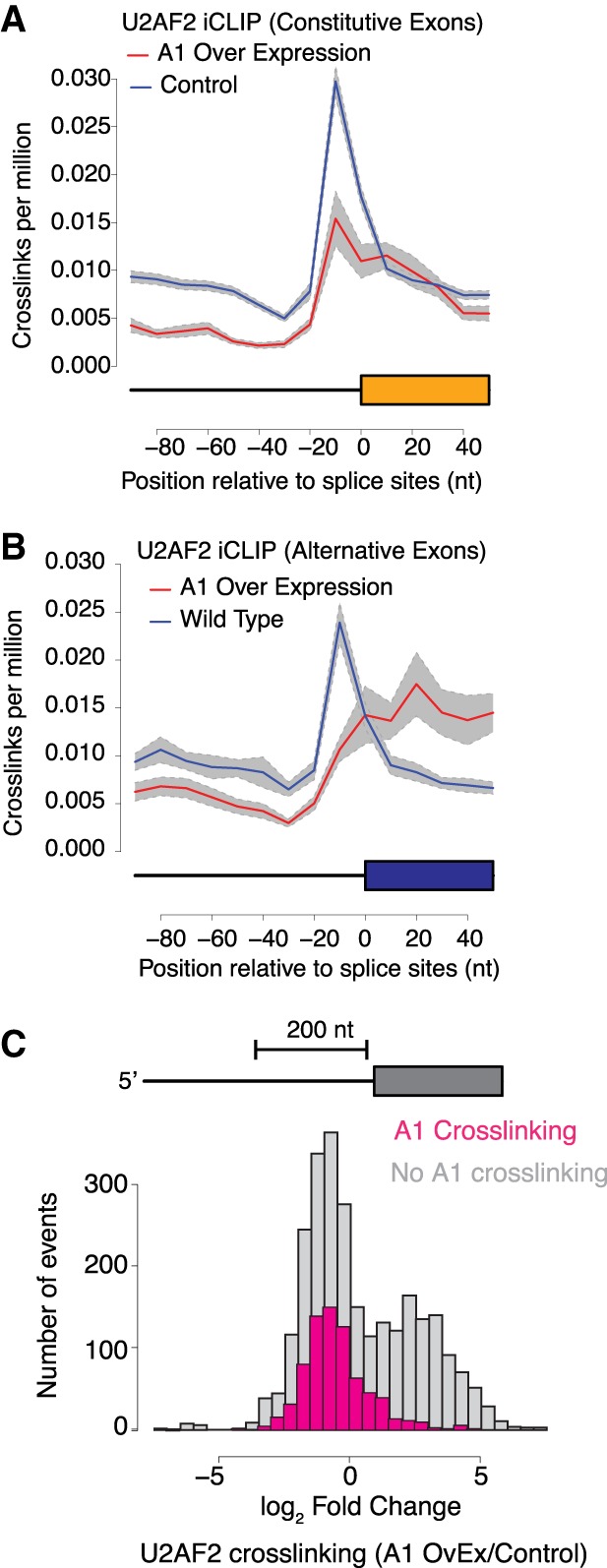
HNRNPA1 induced redistribution of U2AF2 crosslinking near 3′ss. (A,B) Normalized crosslinking distribution for U2AF2 in wild-type (blue line) and HNRNPA1 overexpression cell lines (red line) with 95% confidence interval (gray area). Data are divided between constitutive (A) and alternative cassette (B) exons. (C) Distribution of U2AF2 crosslinking changes within 200-bp intron regions near 3′ss of alternative cassette exons. Gray bars correspond to annotated alternative splicing events with no evidence of HNRNPA1 crosslinking in either condition, and pink represents annotated events with detectable HNRNPA1 crosslinking.
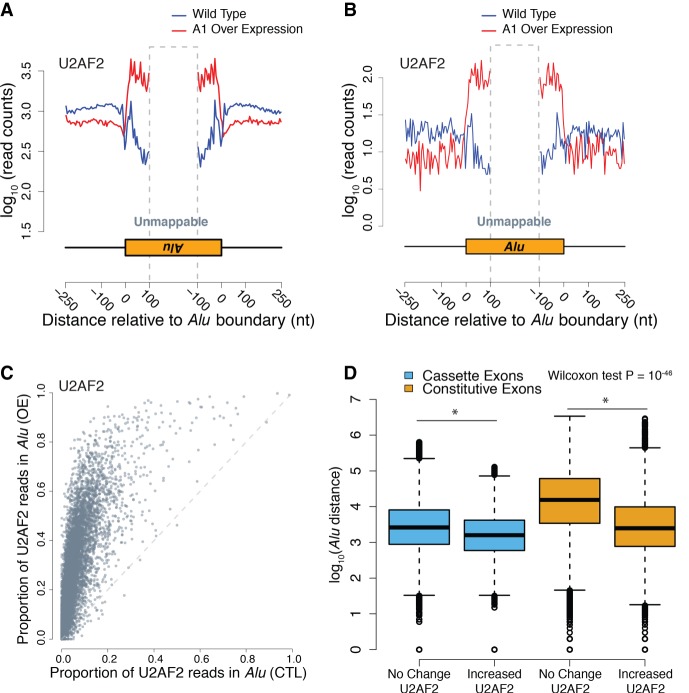
Previous work by Zarnack et al. (2013) demonstrated that HNRNP proteins, such as HNRNPC, compete with U2AF2 for binding to antisense Alu elements, in order to repress their exonization. We asked if HNRNPA1 similarly repressed U2AF2 crosslinking to Alu elements by measuring crosslink density across the boundaries of intronic Alu elements in control and HNRNPA1 overexpression cells. We observed a dramatic increase in U2AF2 crosslinking to Alu-containing RNA transcripts compared to control cells (Fig. 4A,B; Supplemental Tables 8–11). Like prior reports (Supplemental Table 8), U2AF2 crosslink density was nearly 10-fold greater in RNA derived from antisense Alu elements compared to sense (cf. Fig. 4A to B; Zarnack et al. 2013; Shao et al. 2014). To visualize HNRNPA1-dependent accumulation of U2AF2 in Alu elements, we calculated the coverage of all peaks across a consensus sequence assembled from all the Alu subtypes in the human genome (see Methods). For example, for subtype AluSc, HNRNPA1 overexpression induces U2AF2 crosslinking near the 3′ end of the element compared to control cells (Supplemental Fig. 6A). Conversely, HNRNPA1 crosslinking globally decreases over Alu elements with overexpression (Supplemental Fig. 6B,C; Supplemental Table 9). In contrast to U2AF2, crosslinking of SRSF1 to antisense Alu elements shows no appreciable changes (Supplemental Fig. 6D,E; Supplemental Table 9), suggesting that the effect of HNRNPA1 is specific to U2AF2. In order to asses any indirect effects of HNRNPA1 expression on known regulators of U2AF2-Alu binding, we also measured protein levels and cell localization of HNRNPC under control and overexpression of HNRNPA1, which showed no discernable changes (Supplemental Fig. 7).
Figure 4.
HNRNPA1 overexpression correlates with global redistribution of U2AF2 signal to Alu RNA elements. (A) Aggregated crosslinking sites on antisense Alu elements and nearby regions for U2AF2. Blue represents wild-type binding of the given RNA binding protein, and red represents HNRNPA1 overexpression of the log10 number of iCLIP read counts across all antisense-Alu elements. (B) Aggregated crosslinking sites on sense Alu elements and nearby regions for U2AF2. (C) Scatter plot of all human cassette exons measuring the proportion of U2AF2 iCLIP crosslinking sites found within Alu elements within the cassette exon event over the total number of crosslinks found within the event. Proportions from control and HNRNPA1 overexpression samples are compared for each individual cassette exon event. (D) Box plot representing the distance of Alu elements from cassette exons (blue) and constitutive exons (orange) that show no change in U2AF2 crosslinking versus those that show an increase in U2AF2 crosslinking.
To determine if Alu elements exhibiting HNRNPA1-dependent increases in U2AF2 crosslinking are located in cis relative to annotated skipped exons (Alu elements upstream of or downstream from a splicing event) or in trans (another locus) (Supplemental Fig. 8), we compared the proportion of U2AF2 crosslinks within Alu-elements relative to sequences flanking annotated as alternative cassette (skipped) exons in control or HNRNPA1 overexpression cells. The scatter plot shown in Figure 4C (data in Supplemental Tables 10 and 11) demonstrates that the proportion of U2AF2 crosslinks present in Alu elements increases significantly upon HNRNPA1 overexpression, whereas the proportion of HNRNPA1 crosslinks is decreased (Supplemental Fig. 6C; data in Supplemental Tables 12 and 13). Additionally, distribution of distal intronic U2AF2 peaks showing increases with HNRNPA1 overexpression show a significant enrichment for Alu elements compared to a null distribution (Supplemental Fig. 9). In contrast, the proportion of SRSF1 crosslinks to Alu elements is refractory to changes in HNRNPA1 expression levels (Supplemental Fig. 6E; data in Supplemental Tables 14 and 15). These data demonstrate a global change in U2AF2-Alu association and refute the hypothesis that a few spurious Alu elements are responsible for the signal observed in Figure 4A. To determine if cis-Alu elements are involved in HNRNPA1-regulated alternative splicing, we manually curated the list of 83 splicing events that also had detectable U2AF2 crosslinking. We found that 41% of HNRNPA1-dependent exon skipping events exhibited redistribution of U2AF2 to adjacent Alu elements (Table 1). Taken together, these data demonstrate that overexpression of HNRNPA1 influences the association of U2AF2 with both sense and antisense Alu elements.
Table 1.
Summary of HNRNPA1-dependent changes in exon skipping with detectable U2AF2 crosslinking
Alu elements influence alternative splicing, although the mechanisms are poorly understood (Sorek et al. 2002; Lev-Maor et al. 2008; Gal-Mark et al. 2009; Schwartz et al. 2009; Pastor and Pagani 2011). Splicing regulatory elements often exhibit position-dependent and context-dependent functions (Fu and Ares 2014). To determine if Alu-elements with HNRNPA1-dependent changes in U2AF2 crosslinking exhibit positional bias, we measured their distance relative to the 3′ss of constitutive or skipped exons. In general, we observed that Alu elements are closer to skipped exons than constitutive exons (P = 1.4 × 10−46, Wilcoxon rank-sum test) (Fig. 4D, cf. blue and orange boxes). But yet, those Alu elements with HNRNPA1-dependent increases in U2AF crosslinking are significantly closer to exons than those that are unchanged (P = 9.5 × 10−92) (Fig. 4D). Taken together, our data suggest that Alu-elements may function as cis-regulatory elements that compete with authentic exons for binding to splicing factors.
To determine if changes in U2AF2 crosslinking correlated with HNRNPA1-dependent splicing regulation, we sequenced poly(A)+ selected RNA libraries from control and HNRNPA1 overexpression cells (Supplemental Table 16). Of the 267 HNRNPA1-regulated cassette exons that exhibited at least a 33% change in splicing (Bayes factor >10), the majority exhibited increased levels of exon skipping upon HNRNPA1 overexpression (Fig. 5A; Supplemental Table 17). Eighty-three of the 267 differentially spliced exons also had detectable U2AF2 crosslinking proximal to 3′ss in either control or HNRNPA1-overexpressing cell lines. Of those 83 exons, ∼90% exhibited an HNRNPA1-dependent increase in exon skipping compared to ∼70% of exons with no detected U2AF2 crosslinking (P < 0.0025, Fisher's exact test) (Table 1). To investigate the relationship between exon skipping with loss or gain of U2AF2 crosslinking, we increased the sensitivity of our analysis by using less stringent thresholds for differential splicing (difference >1%, Bayes factor >5) (Supplemental Table 18). This resulted in 574 events, the majority of which favored exon skipping (424 events). Exon skipping events that also had evidence of HNRNPA1 crosslinking in either condition were enriched for loss of U2AF2 crosslinking as compared to events with no detectable HNRNPA1 crosslinking (P < 0.026 Fisher's exact test) (Table 2). In some cases, HNRNPA1-sensitive exons exhibited redistribution of U2AF2 signal from near 3′ss to distal, upstream crosslinking sites, suggesting their possible function in HNRNPA1-mediated splicing regulation (Fig. 5E; Supplemental Figs. 10–20). We use transient transfection of T7 epitope–tagged HNRNPA1 and RT-PCR to validate several endogenous splicing events exhibiting HNRNPA1-dependent reduction in U2AF2-3′ss crosslinking (Fig. 5B–D, upper panels). As expected, we observed a significant reduction in exon inclusion for ACIN1, SRSF6, and TRNAU1AP in HNRNPA1 overexpression cells relative to the control (Fig. 5B–D, lower panels).
Figure 5.
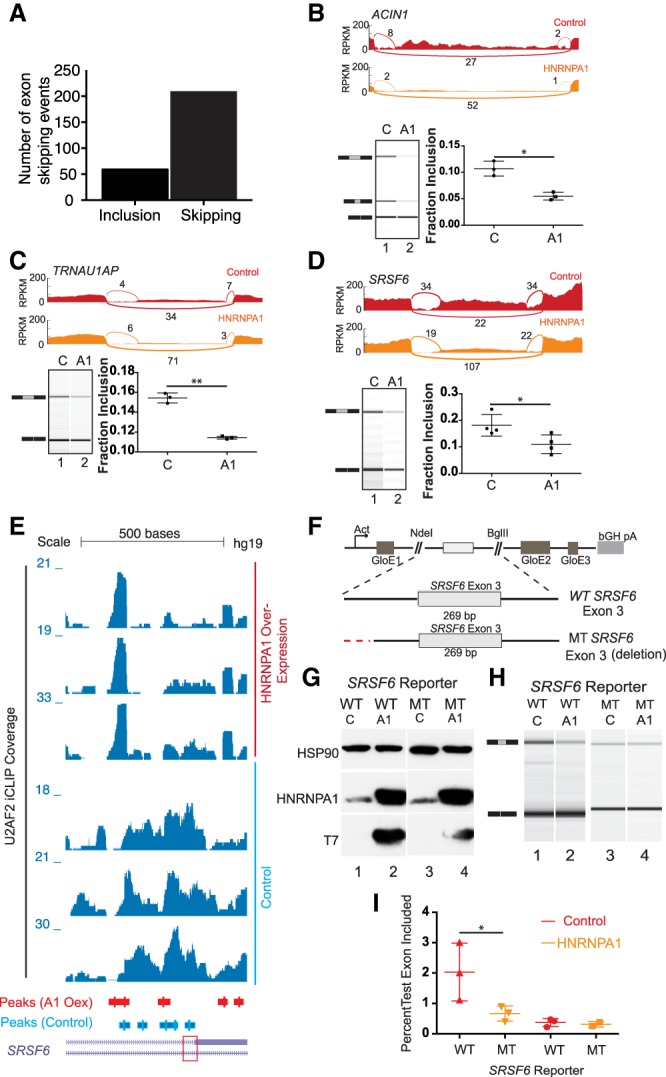
Noncanonical U2AF2 binding sites influence HNRNPA1-dependent exon skipping. (A) Bar graph of all HNRNPA1-dependent splicing changes as detected by MISO analysis. Events determined from the comparison of RNA-seq data derived from HNRNPA1 overexpressing HEK293T cell versus control HEK293T cells. (B–D) Sashimi plots showing read and junction coverage for three genes: ACIN1, SRSF6, and TRNAU1AP. Virtual gel representation of an Agilent 2100 Bioanalyzer DNA 1000 assay of RT-PCR products corresponding to endogenous ACIN1, SRSF6, and TRNAU1AP from control and HNRNPA1 overexpression cells (lower left panel). Bar graph depicting mean exon inclusion for TRNAU1AP and SRSF6 splicing reporters quantified using an Agilent 2100 Bioanalyzer with standard deviation bars. (*) P < 0.05; (**) P < 0.01 (lower right panel). (E) UCSC Genome Browser screen shot of U2AF2 iCLIP read coverage at the SRSF6 exon 3 locus in control or HNRNPA1 overexpression cells (bottom and top three tracks, respectively). Peaks called by CLIPper are depicted below the coverage tracks. Red box denotes the 3′ss of SRSF6 exon 3. (F) Splicing reporter constructs created representing wild-type (Wt) or mutant (Mt) versions of alternative exons in SRSF6. Noncanonical U2AF2 binding sites in SRSF6 were mutated (red lines) by deletion. GloE1, GloE2, and GloE3 designate exons 1–3 of beta-globin. The polyadenylation signal from the bovine growth hormone 1 gene is indicated by bGH pA. (G) Representative western blots probed with antibodies against HSP90, HNRNPA1, and the T7 epitope. Lysates were prepared from HEK293T cells transiently transfected with either wild-type (WT) or mutant (MT) SRSF6 reporter construct and control or T7 epitope–tagged HNRNPA1. (H) Virtual gel representation of an Agilent 2100 Bioanalyzer DNA 1000 assay of RT-PCR products from SRSF6 splicing construct transfections. (I) Bar graph depicting mean exon inclusion for SRSF6 splicing reporters quantified using an Agilent 2100 Bioanalyzer with standard deviation bars. (*) P < 0.05.
Table 2.
Summary of HNRNPA1-dependent changes in exon skipping and U2AF2 positioning
To determine if noncanonical U2AF2 binding sites are involved in HNRNPA1-dependent exon skipping, we created matched pairs of hemoglobin subunit beta (HBB) splicing reporter gene constructs (Rothrock et al. 2003) containing the wild-type or mutated intronic elements upstream of the HNRNPA1-sensitive exons found in the SRSF6 pre-mRNA (Fig. 5E). The SRSF6 splicing reporter constructs were cotransfected into HEK293T cells with epitope-tagged HNRNPA1 expression plasmid or a control plasmid (Fig. 5F). Because inclusion of the test exons may induce nonsense-mediated decay (NMD) by inducing an in-frame premature termination codon (PTC), as is the case with SRSF6, we assayed splicing in the presence of the translation inhibitor emetine dihydrochloride, a potent inhibitor of NMD in vivo (Noensie and Dietz 2001; Lareau et al. 2007; Ni et al. 2007). As shown in Figure 5H, RT-PCR of wild-type SRSF6 splicing reporters show expected decrease in exon inclusion in response to HNRNPA1 overexpression compared to endogenous levels (Fig. 5G). Deletion of the distal intronic U2AF2 binding site resulted in an attenuation of exon inclusion and a loss of HNRNPA1-dependent exon skipping. Quantification of amplicon ratios demonstrated that mutation of the putative HNRNPA1-induced, U2AF2-associated intronic elements disrupt splicing sensitivity to HNRNPA1 overexpression (P < 0.0029, respectively, ratio paired t-test) (Fig. 5I).
Discussion
In this study, we have used iCLIP to study the impact of perturbation of alternative splicing factor levels on core components of the spliceosome. These experiments allow for quantitative analysis of competitive RNA binding and splice site recognition on a global scale. In cells overexpressing HNRNPA1, we observed a pronounced shift in U2AF2 binding sites relative to control cells. Based on U2AF2 peak distributions, this HNRNPA1-dependent redistribution involves loss of proximal-intron and recognition of distal-intronic peaks (Figs. 1, 2A–C). A similar pattern was observed at the single nucleotide level, where U2AF2 crosslinking density near alternative 3′ss was reduced in HNRNPA1 overexpression cells and distributed across exons (Fig. 3). In addition to evidence for global changes in U2AF2 binding position, we also observed variation in sequence motifs enriched at U2AF2 peaks in both cell lines (Figs. 1, 2E). Splicing reporter assays revealed that upstream polypyrimidine tracks identified by iCLIP were involved in HNRNPA1-dependent splicing regulation (Fig. 5). These data also support the hypothesis that distant U2AF2 binding sites may function as splice site decoys that contribute to HNRNPA1-dependent splicing silencing.
HNRNPA1 represses splicing through diverse mechanisms. We think our data suggest that HNRNPA1 alters the competition between bona fide and decoy 3′ss. Because HNRNPA1 forms a ternary complex with the U2AF heterodimer and the 3′ss (Tavanez et al. 2012), HNRNPA1-dependent redistribution of U2AF2 from bona fide splice sites to “decoy” sites might arise from direct competition for these splicing factors at 3′ss. Tavanez et al. (2012) also demonstrated that HNRNPA1 is required for discrimination of AG and CG dinucleotides at 3′ss. HNRNPA1 displaces U2AF from noncanonical CG-containing splice sites but allows it to productively engage AG-containing splice sites. We observed a global decrease in U2AF2 crosslinking near alternative 3′ss, but little change near constitutive splice sites. Given that alternative exons are typically flanked by weak splice sites, it seems unlikely that this proofreading mechanism contributes to regulation of alternative splicing. Tavanez et al. (2012) also noted that depletion of HNRNPA1 from HeLa cells resulted in increased U2AF association at spurious sites, including within the 3′ UTRs of intronless messages when HNRNPA1 is knocked down. We also observed a global increase in U2AF2 crosslinking to 3′ UTRs but not CDS regions when HNRNPA1 is overexpressed (Supplemental Table 3). It will be important to understand if RNA determinants (sequence, structure, splice site strength) distinguish between a proofreading function for HNRNPA1 and a simple competition with U2AF (Jain et al. 2017).
Our findings are well aligned with a recent census of U2AF2 binding sites in HeLa cells, which documented a position-dependent code for U2AF2 in splicing regulation (Shao et al. 2014). Perhaps most relevant to the work presented here is their observation that noncanonical U2AF2 binding sites located upstream or within alternative exons correlate with exon skipping (Lim et al. 2011; Shao et al. 2014; Cho et al. 2015). We find strong evidence for an HNRNPA1-dependent shift in U2AF2 binding sites from exon-proximal to -distal intronic positions. We have demonstrated that a noncanonical U2AF2 binding site in SRSF6 intron 2 is required for HNRNPA1-dependent splicing regulation (Fig. 5E). Additionally, in the context of a splicing reporter, deletion of this element appears to dramatically reduce exon inclusion. It is possible that HNRNPA1-dependent eviction of U2AF2 at 3′ss could initiate remodeling at other functional RNA elements, such as nearby intronic splicing enhancers.
Our results suggest a role for HNRNPA1 in promoting recognition of noncanonical sites by U2AF2. One interesting observation is that although U2AF2 binding sites are predominantly pyrimidine-rich motifs (Singh et al. 1995), we observe variation in overrepresented sequence motifs at different classes of binding sites across the transcriptome, suggesting that U2AF2 occupies low-affinity binding sites when HNRNPA1 is overexpressed (Fig. 2E). This result is consistent with the hypothesis that U2AF2 and HNRNPA1 compete for closely related sequences (Zhu et al. 2001; Okunola and Krainer 2009; Jain et al. 2017). Alternatively, HNRNPA1 may modulate U2AF2 RNA binding specificity by altering the conformation of its RNA recognition motifs (Mackereth et al. 2011; Tavanez et al. 2012; Agrawal et al. 2016; Voith von Voithenberg et al. 2016). It is possible that the large-scale reorganization of U2AF2 binding to Alu-derived RNA sequences triggered by HNRNPA1 overexpression is an ultrasensitive response driven by molecular titration of U2AF–3′ss interactions by HNRNPA1 (Zhang et al. 2013).
Alu elements influence gene expression in diverse ways (Hasler and Strub 2006; Chen and Carmichael 2009; Gong and Maquat 2011; Pastor and Pagani 2011; Kelley et al. 2014; Tajnik et al. 2015). Recently, Zarnack et al. (2013) demonstrated that HNRNPC competes with U2AF2 to repress inclusion of antisense Alu-derived exons in mRNA. We find that HNRNPA1 overexpression correlates with increased U2AF2 association with both sense and antisense Alu-derived RNA sequences. We did not observe any change in HNRNPC protein expression or localization (Supplemental Fig. 7). Taken together, our data demonstrate that Alu-derived sequences may function as RNA regulatory elements that respond to changes to the intracellular concentration of splicing factors. It is possible that, as primate-specific elements, Alu elements may influence the evolution of splicing regulation by modulating recognition of bona fide exons. Our results suggest the intriguing hypothesis that Alu-derived RNA elements may contribute to species-specific differences in alternative splicing throughout the primate lineage.
Methods
iCLIP analysis of U2AF2, SRSF1, and HNRNPA1
iCLIP was performed as previously described (Konig et al. 2010; Huppertz et al. 2014). Briefly, Flp-In T-REx HEK293T cells (Invitrogen) lacking or containing a stable, inducible T7-tagged version of HNRNPA1 were treated with tetracycline for 24 h and then irradiated with UV-C light to form covalent crosslinks between proteins and nucleic acids in vivo. After cell lysis, RNA was partially fragmented using low concentrations of micrococcal nuclease, and U2AF65–, SRSF1–, or HNRNPA1–RNA complexes were immunopurified with U2AF65, (MC3; Santa Cruz Biotechnology), SRSF1 (96; Santa Cruz Biotechnology), and HNRNPA1 (4B10; Santa Cruz Biotechnology) antibodies immobilized on protein A–coated magnetic beads (Life Technologies), respectively. After stringent washing and dephosphorylation (Fast AP, Fermentas), RNAs were ligated at their 3′ ends with a preadenylated RNA adaptor (Bioo Scientific) and radioactively labeled to allow visualization. Samples were run using MOPS-based protein gel electrophoresis (in-house recipe) and transferred to a nitrocellulose membrane. Protein–RNA complexes migrating 15–80 kDa above free protein were cut from the membrane, and RNA was recovered from the membrane by proteinase K digestion under denaturing (3.5 M urea) conditions. The oligonucleotides for reverse transcription contained two inversely oriented adaptor regions adapted from the Bioo NEXTflex small RNA library preparation kit (Bioo Scientific), separated by a BamHI restriction site as well as a barcode region at their 5′ end containing a 4-nt experiment-specific barcode within a 5-nt random barcode to mark individual cDNA molecules. cDNA molecules were size-purified using denaturing PAGE gel electrophoresis, circularized by CircLigase II (Epicenter), annealed to an oligonucleotide complementary to the restriction site, and cut using BamHI (NEB). Linearized cDNAs were then PCR-amplified using ImmoMix PCR master mix (Bioline) with primers (Bioo) complementary to the adaptor regions and were subjected to high-throughput sequencing using Illumina HiSeq. A more detailed description of the iCLIP protocol has been published (Huppertz et al. 2014).
Mapping and analysis of iCLIP sequencing data
Single-end reads generated by Illumina HiSeq were inspected for the presence of adaptor sequences. Reads containing sequences corresponding to the 3′ RNA adaptor were retained if they were at least 30 bp long after the adaptor sequence was trimmed off. The first 9 bp in each read from the iCLIP library preparation, containing an internal barcode comprising 4 bp for replicate identification and 5 bp of random nucleotides for use in duplicate mapping removal, were also removed before mapping. Trimmed reads were checked for mapping to a repeat filter comprising RepeatMasker elements (Tarailo-Graovac and Chen 2009) in the human genome using Bowtie 2 (Langmead and Salzberg 2012). Reads that passed the repeat filter were mapped to the transcriptome and genome with TopHat2 (Kim et al. 2013). If reads mapped equally well to multiple loci, a single mapping was selected randomly by TopHat2. Duplicate mappings from each replicate were reduced to one per position if they had the same genomic endpoints and if they originated from reads with the same set of random 5-bp nucleotides. Following mapping and duplicate removal, individual reads were truncated to their 5′ ends to represent the site of crosslinking consistent with the iCLIP methodology. For all samples, only such crosslinking sites found to have nonzero mapping counts in two out of three replicates (or two out of two duplicates where applicable) were considered to be biologically reproducible candidates for further analysis. The counts at such reproducible crosslinking sites were summed over all replicates to create an aggregated data set for each cell condition and CLIP. To determine the background from the iCLIP data sets, the two cell conditions (control and HNRNPA1-overexpressing) were temporarily further aggregated for each CLIP (U2AF, SF2, A1), and those binding sites that had nonzero counts in all three temporary aggregate data sets were determined. A 41-nt mask was created by extending 20 nt upstream of and 20 nt downstream from each such three-way common binding site. The aggregated data set of binding sites for each cell condition and CLIP was then filtered using this mask, keeping only sites outside the mask that also had a mapping count of at least three in the aggregate data. These aggregated and filtered data were used for downstream analyses. This aggregation and filtering strategy was adapted from previously described iCLIP analysis pipelines (Friedersdorf and Keene 2014; Flynn et al. 2015). For use as input to CLIPper (Lovci et al. 2013), the filtered (single nucleotide) binding sites were expanded by 15 nt upstream and 15 nt downstream.
CLIPper (CLIP-seq peak enrichment; https://github.com/YeoLab/CLIPper) was used to determine genomic distribution of RNA crosslinking peaks as well as identify clusters representing binding sites for HNRNPA1, U2AF2, and SRSF1 for each condition as previously described (Lovci et al. 2013). For each condition, the resulting iCLIP peak data of the replicates were merged. The peaks were annotated to the human genome (hg19) and then divided into categories based on their genomic locations, including CDS, intron, and UTR. The peaks in each category were further subsetted based on whether they overlapped with Alu elements. For the motif analysis, 50-bp sequences were extracted from the peak regions (crosslinking site ± 25 bp). Strand-specific MEME-ChIP and HOMER analyses were performed on these sequences to find 6–10 bp long, or 5-, 7-, 9-, and 11-bp enriched motifs (Bailey et al. 2009; Heinz et al. 2010).
For differential crosslinking analysis, CLIPper peaks from both control and HNRNPA1-overexpressing cells were merged using BEDTools intersect, and peak coverage by crosslink-centered reads (i.e., reads constructed by expanding the 5′ end upstream and downstream by 15 nt) was assessed for all replicates using BEDTools multicov (Quinlan and Hall 2010). Differential status was subsequently determined using DESeq2 (Love et al. 2014). Peaks were determined to be increasing (FDR < 0.05, log2(fold-change) > 1) or decreasing (FDR < 0.05, log2(fold-change) < −1) based on FDR and log2(fold-change). These categories were further subdivided based on overlap of Alu elements, 3′ss (i.e., the 200-nt intronic region proximal to the 3′ss), and other. Enriched motifs were identified as described above using HOMER (Heinz et al. 2010).
RBP binding analysis
We extracted 40,769 cassette exons from MISO human genome (hg19) alternative events annotation version 2. We extracted 200,880 constitutive exons from RefSeq gene annotation by excluding the exons that overlap with cassette exons. Gene differential expression analysis was performed using edgeR (Robinson et al. 2010). We used 40,952 constitutive exons that were not significantly differentially expressed (FDR > 0.05) in further analysis.
For each RNA binding protein in each cell line, the iCLIP reads of all the replicates were merged together (Flynn et al. 2015). The start positions of the reads were considered as crosslinking sites. The number of reads near the 3′ss (100 bp into the intron, 50 bp into the exon) of each exon was calculated based on a 10-bp window. The raw read counts were normalized by the total library size.
The changes in binding of U2AF2 and SF2 near 3′ss were further analyzed with edgeR. Read counts were calculated for 200-bp intron regions near the 3′ss of the cassette exons. For each RBP, the regions with more than one count per million (CPM) in at least half of the replicates in either of the cell line were used for binding change analysis.
RBP binding near Alu elements
We extracted 315,974 antisense Alu elements from RepeatMasker. The merged iCLIP data for each condition were down-sampled to 1M reads. The total number of sense strand reads were calculated for Alu and nearby regions (250 bp from Alu boundary). For each cassette exon events (cassette exon + up-/downstream introns + up-/downstream exons), the number of reads in antisense Alu elements, and the total number of reads in the whole event were calculated separately. The proportion of reads that fall into antisense Alu elements for each event was used to represent the RBP binding change in Alu regions.
Randomization test for establishing significance of Alu-element overlap by distal intronic increasing peaks
U2AF2 iCLIP peaks exhibiting increasing binding following HNRNPA1 overexpression were extracted and subsetted for those found within distal intronic regions using a series of different distance constraints (minimum 200, 500, or 2000 bases from the nearest exon). Any intronic region found to overlap an alternative exon was removed from consideration (introns flanking the alternative exon were included). The peaks were then randomly shuffled within the same sets of distal intronic regions using BEDTools shuffle, and the number of shuffled peaks overlapping Alu elements was determined. The shuffling process was repeated 1000 times to construct a null distribution of expected Alu overlaps. Finally, the actual number of Alu-overlapping peaks was compared to the randomized null distribution. As the observed counts exceeded the entirety of the null distribution of counts for each definition of distal intron, the P-value has an upper bound of 0.001 for each comparison. The analysis was repeated for peaks found in corresponding proximal intronic regions and exons.
mRNA-seq of control or HNRNPA1-overexpressing HEK293 cells
RNA was isolated from whole-cell lysates of control and HNRNPA1-overexpressing Flp-In T-REx HEK 293T cells using TRI-Reagent LS (Sigma). Poly(A)+ sequencing libraries were prepared using the TrueSeq RNA library prep kit (Illumina). Each condition was analyzed in duplicate using the HiSeq 2000.
Quantification of alternative splicing by RNA-seq
poly(A)+ transcriptome sequencing reads were mapped to the human reference genome (hg19) with TopHat2. Mapped reads of duplicates were merged together for splicing analysis. Splicing change was analyzed with MISO (Katz et al. 2010). The MISO result was filtered with the following parameters: --num-inc 1 --num-exc 1 --num-sum-exc 10 --delta-psi 0.20 --bayes-factor 10. After filtering, 267 skipped exon events were left for further analysis. We chose to map our data to hg19 to facilitate analysis with the MISO alternative splicing event database, which is based on hg19 annotation. Because our study focuses on alternative splicing of protein-coding genes, remapping the data to GRCh38 will not have a significant impact on rigor or reproducibility.
Mapping to an Alu consensus sequence
Alu element annotations were obtained from the hg19 UCSC Genome Browser RepeatMasker track. A strategy similar to the one described in Jacobs et al. (2014) was used to show the CLIPper peak density over all Alu elements as follows: After removal of the longest 2%, the top 50 longest human Alu sequences were aligned with MUSCLE and used to construct a consensus sequence (Edgar 2004). CLIPper peaks were mapped from the genomic position to the consensus sequence position using a BLAT alignment of the repeat to the consensus, and the coverage of summits per bp of the consensus AluSc was plotted in Supplemental Figure 5c (Kent 2002). A Genome Browser session displaying the Repeat Masker Data can be found at the following URL: http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=Max&hgS_otherUserSessionName=pubRepeats2Sanford.
Data access
All iCLIP and RNA-seq data generated in this study have been submitted to the NCBI Gene Expression Omnibus GEO (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE83923. Access to other publicly available data sets from GEO used in this study is detailed in Supplemental Table S8.
Supplementary Material
Acknowledgments
We thank Professors Javier Caceres (MRC HGU, Edinburgh), Manny Ares (UCSC), and Al Zahler (UCSC) for discussion of the project. We thank Julia Philipp (UCSC) for thoughtful comments on the manuscript. This project was supported by National Institutes of Health grants GM1090146 (National Institute of General Medical Sciences; J.R.S.), HG007336 (National Human Genome Research Institute; M.T.), U41HG002371 (National Human Genome Research Institute; M.H.), GM008646 (National Institute of General Medical Sciences; NIH Training Grant Support for J.M.D.), and California Institute of Regenerative Medicine grant GC1R-06673-A (J.R.S.).
Author contributions: J.M.H., H.L., Y.L., and J.R.S. designed the experiments. J.M.H., H.L., A.J.W., G.K., J.M.D., M.H., and S.K. performed the experiments and data analysis. J.M.H., H.L., A.J.W., G.K., J.M.D., M.H., Y.L., and J.R.S. wrote and edited the manuscript. All authors read and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.229062.117.
References
- Agrawal AA, Salsi E, Chatrikhi R, Henderson S, Jenkins JL, Green MR, Ermolenko DN, Kielkopf CL. 2016. An extended U2AF65-RNA-binding domain recognizes the 3′ splice site signal. Nat Commun 7: 10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund JA, Chua K, Abovich N, Reed R, Rosbash M. 1997. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell 89: 781–787. [DOI] [PubMed] [Google Scholar]
- Blanchette M, Chabot B. 1999. Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J 18: 1939–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonomi S, di Matteo A, Buratti E, Cabianca DS, Baralle FE, Ghigna C, Biamonti G. 2013. HnRNP A1 controls a splicing regulatory circuit promoting mesenchymal-to-epithelial transition. Nucleic Acids Res 41: 8665–8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukakis G, Patrinou-Georgoula M, Lekarakou M, Valavanis C, Guialis A. 2010. Deregulated expression of hnRNP A/B proteins in human non-small cell lung cancer: parallel assessment of protein and mRNA levels in paired tumour/non-tumour tissues. BMC Cancer 10: 434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres JF, Stamm S, Helfman DM, Krainer AR. 1994. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science 265: 1706–1709. [DOI] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG. 2009. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell 35: 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Zhang J, Manley JL. 2010. Turning on a fuel switch of cancer: hnRNP proteins regulate alternative splicing of pyruvate kinase mRNA. Cancer Res 70: 8977–8980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, David CJ, Manley JL. 2012. Concentration-dependent control of pyruvate kinase M mutually exclusive splicing by hnRNP proteins. Nat Struct Mol Biol 19: 346–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Moon H, Loh TJ, Jang HN, Liu Y, Zhou J, Ohn T, Zheng X, Shen H. 2015. Splicing inhibition of U2AF65 leads to alternative exon skipping. Proc Natl Acad Sci 112: 9926–9931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou MY, Underwood JG, Nikolic J, Luu MH, Black DL. 2000. Multisite RNA binding and release of polypyrimidine tract binding protein during the regulation of c-src neural-specific splicing. Mol Cell 5: 949–957. [DOI] [PubMed] [Google Scholar]
- David CJ, Chen M, Assanah M, Canoll P, Manley JL. 2010. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 463: 364–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon IC, Ireland DC, Smith RA, Mayeda A, Krainer AR. 1993. Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO J 12: 3607–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon IC, Makarova OV, Mayeda A, Munroe SH, Caceres JF, Hayward DG, Krainer AR. 2000. Selection of alternative 5′ splice sites: role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol Cell Biol 20: 8303–8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RA, Martin L, Spitale RC, Do BT, Sagan SM, Zarnegar B, Qu K, Khavari PA, Quake SR, Sarnow P, et al. 2015. Dissecting noncoding and pathogen RNA–protein interactomes. RNA 21: 135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedersdorf MB, Keene JD. 2014. Advancing the functional utility of PAR-CLIP by quantifying background binding to mRNAs and lncRNAs. Genome Biol 15: R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD, Ares M Jr. 2014. Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet 15: 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Mark N, Schwartz S, Ram O, Eyras E, Ast G. 2009. The pivotal roles of TIA proteins in 5′ splice-site selection of Alu exons and across evolution. PLoS Genet 5: e1000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Maquat LE. 2011. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470: 284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasler J, Strub K. 2006. Alu elements as regulators of gene expression. Nucleic Acids Res 34: 5491–5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppertz I, Attig J, D'Ambrogio A, Easton LE, Sibley CR, Sugimoto Y, Tajnik M, Konig J, Ule J. 2014. iCLIP: protein–RNA interactions at nucleotide resolution. Methods 65: 274–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs FM, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR, Haussler D. 2014. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 516: 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N, Lin HC, Morgan CE, Harris ME, Tolbert BS. 2017. Rules of RNA specificity of hnRNP A1 revealed by global and quantitative analysis of its affinity distribution. Proc Natl Acad Sci 114: 2206–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamison SF, Pasman Z, Wang J, Will C, Luhrmann R, Manley JL, Garcia-Blanco MA. 1995. U1 snRNP-ASF/SF2 interaction and 5′ splice site recognition: characterization of required elements. Nucleic Acids Res 23: 3260–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB. 2010. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 7: 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DR, Hendrickson DG, Tenen D, Rinn JL. 2014. Transposable elements modulate human RNA abundance and splicing via specific RNA-protein interactions. Genome Biol 15: 537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT: the BLAST-like alignment tool. Genome Res 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. 2010. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 17: 909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane R, Daubner GM, Thomas-Crusells J, Auweter SD, Manatschal C, Austin KS, Valniuk O, Allain FH, Rueda D. 2010. RNA looping by PTB: evidence using FRET and NMR spectroscopy for a role in splicing repression. Proc Natl Acad Sci 107: 4105–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. 2007. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446: 926–929. [DOI] [PubMed] [Google Scholar]
- Lev-Maor G, Ram O, Kim E, Sela N, Goren A, Levanon EY, Ast G. 2008. Intronic Alus influence alternative splicing. PLoS Genet 4: e1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KH, Ferraris L, Filloux ME, Raphael BJ, Fairbrother WG. 2011. Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. Proc Natl Acad Sci 108: 11093–11098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh TJ, Moon H, Cho S, Jang H, Liu YC, Tai H, Jung DW, Williams DR, Kim HR, Shin MG, et al. 2015. CD44 alternative splicing and hnRNP A1 expression are associated with the metastasis of breast cancer. Oncol Rep 34: 1231–1238. [DOI] [PubMed] [Google Scholar]
- Lovci MT, Ghanem D, Marr H, Arnold J, Gee S, Parra M, Liang TY, Stark TJ, Gehman LT, Hoon S, et al. 2013. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat Struct Mol Biol 20: 1434–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackereth CD, Madl T, Bonnal S, Simon B, Zanier K, Gasch A, Rybin V, Valcarcel J, Sattler M. 2011. Multi-domain conformational selection underlies pre-mRNA splicing regulation by U2AF. Nature 475: 408–411. [DOI] [PubMed] [Google Scholar]
- Mayeda A, Krainer AR. 1992. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell 68: 365–375. [DOI] [PubMed] [Google Scholar]
- Merendino L, Guth S, Bilbao D, Martinez C, Valcarcel J. 1999. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402: 838–841. [DOI] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, Shiue L, Clark TA, Blume JE, Ares M Jr. 2007. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev 21: 708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noensie EN, Dietz HC. 2001. A strategy for disease gene identification through nonsense-mediated mRNA decay inhibition. Nat Biotechnol 19: 434–439. [DOI] [PubMed] [Google Scholar]
- Okunola HL, Krainer AR. 2009. Cooperative-binding and splicing-repressive properties of hnRNP A1. Mol Cell Biol 29: 5620–5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor T, Pagani F. 2011. Interaction of hnRNPA1/A2 and DAZAP1 with an Alu-derived intronic splicing enhancer regulates ATM aberrant splicing. PLoS One 6: e23349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino I, Pio R, Toledo G, Zabalegui N, Vicent S, Rey N, Lozano MD, Torre W, Garcia-Foncillas J, Montuenga LM. 2003. Altered patterns of expression of members of the heterogeneous nuclear ribonucleoprotein (hnRNP) family in lung cancer. Lung Cancer 41: 131–143. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothrock C, Cannon B, Hahm B, Lynch KW. 2003. A conserved signal-responsive sequence mediates activation-induced alternative splicing of CD45. Mol Cell 12: 1317–1324. [DOI] [PubMed] [Google Scholar]
- Ruskin B, Zamore PD, Green MR. 1988. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell 52: 207–219. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Gal-Mark N, Kfir N, Oren R, Kim E, Ast G. 2009. Alu exonization events reveal features required for precise recognition of exons by the splicing machinery. PLoS Comput Biol 5: e1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C, Yang B, Wu T, Huang J, Tang P, Zhou Y, Zhou J, Qiu J, Jiang L, Li H, et al. 2014. Mechanisms for U2AF to define 3′ splice sites and regulate alternative splicing in the human genome. Nat Struct Mol Biol 21: 997–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Valcarcel J, Green MR. 1995. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268: 1173–1176. [DOI] [PubMed] [Google Scholar]
- Sorek R, Ast G, Graur D. 2002. Alu-containing exons are alternatively spliced. Genome Res 12: 1060–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajnik M, Vigilante A, Braun S, Hanel H, Luscombe NM, Ule J, Zarnack K, Konig J. 2015. Intergenic Alu exonisation facilitates the evolution of tissue-specific transcript ends. Nucleic Acids Res 43: 10492–10505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo-Graovac M, Chen N. 2009. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics 25: 4.10.1–4.10.14. [DOI] [PubMed] [Google Scholar]
- Tavanez JP, Madl T, Kooshapur H, Sattler M, Valcarcel J. 2012. hnRNP A1 proofreads 3′ splice site recognition by U2AF. Mol Cell 45: 314–329. [DOI] [PubMed] [Google Scholar]
- Ushigome M, Ubagai T, Fukuda H, Tsuchiya N, Sugimura T, Takatsuka J, Nakagama H. 2005. Up-regulation of hnRNP A1 gene in sporadic human colorectal cancers. Int J Oncol 26: 635–640. [PubMed] [Google Scholar]
- Voith von Voithenberg L, Sanchez-Rico C, Kang HS, Madl T, Zanier K, Barth A, Warner LR, Sattler M, Lamb DC. 2016. Recognition of the 3′ splice site RNA by the U2AF heterodimer involves a dynamic population shift. Proc Natl Acad Sci 113: E7169–E7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718. [DOI] [PubMed] [Google Scholar]
- Wright JC, Mudge J, Weisser H, Barzine MP, Gonzalez JM, Brazma A, Choudhary JS, Harrow J. 2016. Improving GENCODE reference gene annotation using a high-stringency proteogenomics workflow. Nat Commun 7: 11778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Romfo CM, Nilsen TW, Green MR. 1999. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402: 832–835. [DOI] [PubMed] [Google Scholar]
- Yang X, Bani MR, Lu SJ, Rowan S, Ben-David Y, Chabot B. 1994. The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc Natl Acad Sci 91: 6924–6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Guo J, Liu Y, Jia J, Jia R, Fan M. 2015. Oral squamous cancer cell exploits hnRNP A1 to regulate cell cycle and proliferation. J Cell Physiol 230: 2252–2261. [DOI] [PubMed] [Google Scholar]
- Zahler AM, Damgaard CK, Kjems J, Caputi M. 2004. SC35 and heterogeneous nuclear ribonucleoprotein A/B proteins bind to a juxtaposed exonic splicing enhancer/exonic splicing silencer element to regulate HIV-1 tat exon 2 splicing. J Biol Chem 279: 10077–10084. [DOI] [PubMed] [Google Scholar]
- Zarnack K, Konig J, Tajnik M, Martincorena I, Eustermann S, Stevant I, Reyes A, Anders S, Luscombe NM, Ule J. 2013. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell 152: 453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Bhattacharya S, Andersen ME. 2013. Ultrasensitive response motifs: basic amplifiers in molecular signalling networks. Open Biol 3: 130031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZJ, Dai Z, Zhou SL, Fu XT, Zhao YM, Shi YH, Zhou J, Fan J. 2013. Overexpression of HnRNP A1 promotes tumor invasion through regulating CD44v6 and indicates poor prognosis for hepatocellular carcinoma. Int J Cancer 132: 1080–1089. [DOI] [PubMed] [Google Scholar]
- Zhu J, Mayeda A, Krainer AR. 2001. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol Cell 8: 1351–1361. [DOI] [PubMed] [Google Scholar]
- Zorio DA, Blumenthal T. 1999. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature 402: 835–838. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.