Abstract
High-quality meat is of great economic importance to the pig industry. The 1-acylglycerol-3-phosphate-O-acyltransferase 5 (AGPAT5) enzyme converts lysophosphatidic acid to phosphatidic acid in the mitochondrial membrane. In this study, we found that the porcine AGPAT5 gene was highly expressed in muscle tissue, influencing meat characteristics, and we also identified a non-synonymous single-nucleotide polymorphism (nsSNP) (rs196952262, c.673 A>G) in the gene, associated with a change of isoleucine 225 to valine. The presence of this nsSNP was significantly associated with meat color (lightness), lower cooking loss, and lower carcass temperatures 1, 4, and 12 h after slaughter (items T1, T4, and T12 on the recognized quality scale, respectively), and tended to increase backfat thickness and the water-holding capacity. These results suggest that nsSNP (c.673A>G) of the AGPAT5 gene is a potential genetic marker of high meat quality in pigs.
Keywords: AGPAT5, gene expression, non-synonymous SNP, meat quality, Berkshire pig
Introduction
For many years, production of high-quality meat has been the prime objective of the pork industry. Meat quality can be assessed from technological, nutritional, and sensory perspectives and may be influenced by multiple interacting factors before and after slaughter (Park et al., 2010). Many studies have focused on genetic factors affecting meat quality (Baby et al., 2014; Casiro et al., 2017; Gonzalez-Prendes et al., 2017; Hwang et al., 2017). These studies found that selective pig breeding and the use of DNA markers played important roles when seeking to enhance pork quality.
The 1-acylglycerol-3-phosphate O-acyltransferases (AGPATs), also known as lysophosphatidic acid acyltransferases, are key enzymes of phospholipid and triacylglyceride biosynthesis. To date, 11 AGPATs have been identified in both mouse and human; however, only the first five (AGPAT1-5) have been proven to catalyze phosphatidic acid synthesis from lysophosphatidic acid; phosphatidic acid is the precursor of all glycerolipids (including triacylglycerides) (Vance and Vance, 2008; Yamashita et al., 2014a). Therefore, AGPATs are important in terms of triacylglyceride biosynthesis because most fatty acids are incorporated into lipids by these enzymes (Coleman and Lee, 2004; Shindou and Shimizu, 2009; Yamashita et al., 2014b). Several studies have shown that fatty acid composition is associated with both meat quality and nutritional value (Choi et al., 2016; Kouba et al., 2003; Yu et al., 2013). However, no study has yet investigated how AGPAT5 affects pig meat quality.
In the present study, we identify a single-nucleotide polymorphism (SNP) in the AGPAT5 gene and explore the associations between this polymorphism and the meat quality traits of Berkshire pigs.
Materials and Methods
Animals
A total of 430 pigs of a pure Berkshire line (males, 210, females, 220), bred under similar conditions, were randomly selected and slaughtered at body weights of approximately 110 kg. The longissimus dorsi muscles were sampled immediately after slaughter and the samples were held at 4°C prior to the assessment of meat quality traits. Animal care and use, and all experimental protocols, conformed to the guidelines of the Animal Care and Use Committee of GNTECH, the Korean Animal Protection Act, and all related laws.
Analysis of AGPAT5 expression by RT-PCR
Total RNAs from various tissues (liver, stomach, lung, kidney, large and small intestines, spleen, and muscle) of three Berkshire pigs were isolated using the TRI-Reagent (Molecular Research Center, USA) and reverse-transcribed into cDNA with the aid of Superscript II Reverse Transcriptase (Invitrogen, USA), in accordance with the manufacturer’s protocol. The cDNAs were then subjected to RT-PCR for evaluation of the relative gene expression level of AGPAT5 and that of the gene encoding peptidylprolyl isomerase A (PPIA) (internal control), using appropriate primer pairs (Table 1). Amplifications proceeded on a Perkin Elmer 9700 system (Applied Biosystems, USA) under the following conditions: 95°C for 5 min; 30 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s; and final elongation for 7 min at 72°C. The amplification products were separated on 2% (w/v) TAE agarose gels and quantified using a Gel Logic model 200 imaging system (Kodak, USA).
Table 1. Oligonucleotides used for genotyping and RT-PCR.
| Application | Gene name | Sequence (5′ → 3′) | |
|---|---|---|---|
| Genotyping | AGPAT5 | Allele-specific Oligo1 | ACTTCGTCAGTAACGGACGTCGAAAGCCACTGTAACATCGTAAAT |
| Allele-specific Oligo2 | GAGTCGAGGTCATATCGTGTCGAAAGCCACTGTAACATCGTAAAC | ||
| Locus-specific Oligo | GCATCTAAATAACTCTTCATAGAATCCATGAGCGGGTTCGTACCAG | ||
| TCGTCTGCCTATAGTGAGTC | |||
| RT-PCR | AGPAT5 | Forward | TTTTCTCAGCATGGAGGGAT |
| Reverse | GGCCTTTTTGAGCAGCAAAT | ||
| PPIA | Forward | CACAAACGGTTCCCAGTTTT | |
| Reverse | TGTCCACAGTCAGCAATGGT | ||
AGPAT5 SNP detection and genotyping
An AGPAT5 nsSNP was detected in cDNAs synthesized from pooled liver RNAs of three Berkshire pigs using an Illumina GAII analyzer (Illumina, Inc., USA), as described previously (Jung et al., 2012). The nsSNP information was obtained with the aid of the NCBI dbSNP database. To explore AGPAT5 nsSNP genotypes, genomic DNAs were isolated from whole blood cells of 430 pigs and SNP genotypes were analyzed using an Illumina VeraCode GoldenGate Assay kit (Illumina, Inc.). The relevant oligonucleotide information is shown in Table 1.
Measurements of meat quality traits
The meat quality parameters examined included carcass weight (kg); backfat thickness (mm); meat colors (L* [lightness], a* [redness], and b* [yellowness]); cooking loss (%); water-holding capacity (%); carcass temperatures at 1, 4, and 12 h after slaughter (T1, T4, and T12, respectively); and the 24-h postmortem pH (pH24). Backfat thickness was measured at the 10th rib at a point 75% along the longissimus dorsi (toward the belly). Meat color was recorded by a Minolta Chromameter (CR-400; Minolta, Japan) after 30 min of blooming at 1°C. Cooking loss was the weight difference between before and after cooking. A slice 3 cm in thickness (weight 100±5 g) from the longissimus dorsi muscle was placed into a polypropylene bag (Dongbang Co., Korea), cooked for 40 min at 70°C in a water bath, and then cooled to room temperature. The pH24 was that at 24 h postmortem and was measured with the aid of a portable pH meter (Istek Inc., Korea) equipped with a glass electrode that could be inserted into muscle tissue. The water-holding capacity at 3 d postmortem was measured using a centrifugation method (Fan et al., 2010). Duplicate 10 g minced samples taken from one chop from each loin were placed into centrifuge tubes and spun for 10 min at 40,000 g. After centrifugation, the liquid was removed and the meat re-weighed. The percentage of water loss was measured and used to estimate the water-holding capacity.
Statistical analysis
The frequencies of the various AGPAT5 genotypes were calculated. To analyze associations between nsSNP genotypes and meat quality traits, we ran a general linear model using SAS software version 9.1.3 (SAS Institute Inc., USA). SNPs subjected to statistical analysis were characterized by a call rate < 0.90, a minor allele frequency > 0.01, and a Hardy-Weinberg equilibrium probability (the p value) > 0.05. The linear model employed was: yij =µ + Gi + Sj + eij, where yij is the phenotypic contribution of the target trait, μ the general mean, Gi the fixed effect of genotype i, Sj the fixed effect of sex j, and eij the random error. Significant differences (p<0.05) between the genotypic frequencies associated with various traits were sought with the aid of analysis of variance (featuring the Bonferroni correction) and the Kruskal-Wallis test.
Results and Discussion
AGPAT5 expression in various tissues of the Berkshire pig
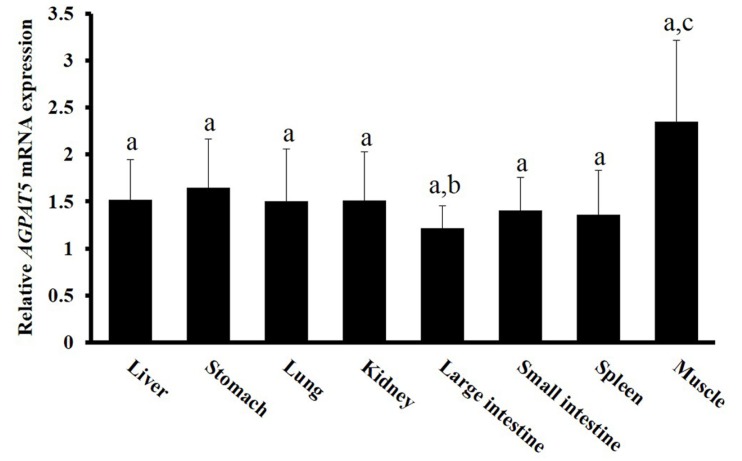
We used RT-PCR to evaluate AGPAT5 expression in various tissues of the Berkshire pig (Fig. 1). AGPAT5 was ubiquitously expressed in all tissues examined, but its expression was highest in muscle, as is the case for human AGPAT5 (Agarwal et al., 2006). Murine AGPAT5 is primarily expressed in skeletal muscle, brain, and heart; and is expressed at high levels in testis and prostate (Biao et al., 2005). Meat quality depends on physiological processes in muscle tissue, potentially involving many genes associated with muscle structure and metabolism. We assumed that AGPAT5 status would be a determinant of meat quality.
Fig. 1. The AGPAT5 mRNA expression was determined in various tissues by RT-PCR.
RNA was isolated from various tissues, including the liver, stomach, lung, kidney, large and small intestines, spleen and muscle. Letter a, b, and c above each bar indicate statistically significant differences among tissues (p<0.05). Values are mean±SD.
Association of the AGPAT5 nsSNP with meat quality traits
As variations in DNA sequence such as SNPs can enhance phenotypic diversity such as meat quality, we identified a new nsSNP (rs196952262, c.673A>G) in the AGPAT5 gene and investigated the contribution thereof to meat quality in Berkshire pigs. The nsSNP c.673A>G in AGPAT5 identified by RNA sequencing of liver tissue samples changes isoleucine 225 to valine in Berkshire pigs. To analyze the association between this nsSNP and meat quality, we genotyped 430 Berkshire pigs using the GoldenGate assay. The genotypic and allelic frequencies of the nsSNP are shown in Table 2. The GG genotype was much more common than the AG and AA genotypes. The frequencies of the G and A alleles were 0.792 and 0.206, respectively. The genotype frequencies were in Hardy-Weinberg equilibrium (p>0.05) (Falconer, 1996).
Table 2. Genotype and allele frequencies of non-synonymous SNP in AGPAT5 gene.
| SNP | Genotype | Genotype frequency | Allele | Allele frequency |
|---|---|---|---|---|
| AGPAT5 c.673A>G | GG (n=267) | 0.631 | G | 0.794 |
| AG (n=149) | 0.327 | A | 0.206 | |
| AA (n=14) | 0.042 |
χ2=1.55, 0.10 < p < 0.50
We investigated the association between the new nsSNP and meat quality traits (Table 3). All three genotypes (AA, AG, and GG) were detected in the pig population. The AGPAT5 nsSNP was significantly associated with lightness (the CIE L* value), less cooking loss, and lower carcass temperatures (T1, T4, and T12). The AG genotype was associated with higher meat quality than the AA and GG genotypes.
Table 3. Association between AGPAT5 nsSNP, c.677A>G, and meat quality traits.
| SNP | AGPAT5, c.673A>G | |||
|---|---|---|---|---|
| Genotype | GG (n=267) | AG (n=149) | AA (n=14) | |
| Carcass weight (kg) | 85.775±5.567 | 85.805±5.756 | 87.214±6.518 | |
| Backfat thickness (mm) | 24.738±5.337 | 25.624±5.220 | 23.429±3.857 | |
| Meat color | CIE L* | 48.510±2.894* | 48.758±2.816* | 50.371±0.611* |
| CIE a* | 6.149±1.058 | 6.131±0.977 | 6.228±1.332 | |
| CIE b* | 2.887±1.112 | 2.871±1.090 | 2.725±1.140 | |
| Cooking loss (%) | 27.574±3.545* | 26.615±4.241* | 28.121±3.159* | |
| Water holding capacity (%) | 58.213±2.774 | 58.413±2.704 | 56.830±1.692 | |
| T1 (°C) | 37.588±3.569* | 36.860±4.613* | 39.955±1.949* | |
| T4 (°C) | 26.588±4.132* | 26.151±5.174* | 30.955±3.567* | |
| T12 (°C) | 16.978±2.980* | 16.729±3.526* | 20.491±3.162* | |
| pH24 | 5.835±0.213 | 5.793±0.214 | 5.793±0.167 | |
Data is shown as Means±SD. Superscript indicates statistically significant differences among genotypes (p<0.05).
CIE L*, a* and b* respectively represent the meat color lightness, redness and yellowness.
T represents a postmortem temperature.
AGPAT-encoded enzymes convert lysophosphatidic acid to phosphatidic acid, a critical substrate for the synthesis of important lipid signaling molecules including phosphatidyl inositol (a second messenger of insulin signaling) and cardiolipin (a mitochondrial membrane phospholipid) (Yamashita et al., 2014a). Of the various AGPAT isoforms, several exhibit lysophospholipid acyltransferase activity, but only AGPAT4 and AGPAT5 are known to be located in mitochondria (Prasad et al., 2011). However, unlike AGPAT4, AGPAT5 is active on several lysophospholipid substrates, including lysophosphatidylinositol, lysophosphatidyl ethanolamine, lysophosphatidyl choline, and lysophosphatidyl serine (Prasad et al., 2011). Fats and fatty acids of adipose tissue and muscle are important contributors to various aspects of meat quality. Intramuscular fats are composed primarily of phospholipids located in the cell membranes and neutral lipids consisting of mainly triacylglycerols in the adipocytes (Smet et al., 2004). Fats vary greatly in melting point, and fat composition thus affects meat firmness/softness (Knothe and Dunn, 2009; Wood and Enser, 1997). Negative correlations were evident between various fatty acid profiles and meat quality traits (Razmaitė et al., 2009). Moreover, Kim et al. (2016) suggested that fat content affected meat quality by controlling the water-holding capacity and drip loss. Meat from heavy pigs (which were also fatter and faster growing) had lower Warner-Bratzler Shear Force and cooking loss than meat from light weight pigs (Magowan et al., 2011). The decrease in cooking loss with increased ultimate muscle pH is likely to be a reflection of improvements in water-holding capacity which are to be expected as the pH moves away from the average isoelectric point of muscle proteins (Monin et al., 1986). These suggest that AGPAT5 status may affect meat quality by regulating fatty acid synthesis.
In summary, we found that the porcine AGPAT5 gene was highly expressed in muscle and we explored the association between an AGPAT5 polymorphism and meat quality in the Berkshire pig. The AGPAT5 AG genotype reduced all of meat color, cooking loss, and carcass temperatures. Therefore, this nsSNP may help the breeding industry to select pigs of high meat quality.
Acknowledgments
This research was supported by Gyeongnam National University of Science and Technology Grant 2016.
References
- Agarwal A. K., Barnes R. I., Garg A. Functional characterization of human 1-acylglycerol-3-phosphate acyltransferase isoform 8: Cloning, tissue distribution, gene structure, and enzymatic activity. Archiv. Biochem. Biophys. 2006;449:64–76. doi: 10.1016/j.abb.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Baby S., Hyeong K. E., Lee Y. M., Jung J. H., Oh D. Y., Nam K. C., Kim T. H., Lee H. K., Kim J. J. Evaluation of genome based estimated breeding values for meat quality in a berkshire population using high density single nucleotide polymorphism chips. Asian-Australas. J. Anim. Sci. 2014;27:1540–1547. doi: 10.5713/ajas.2014.14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biao L., Jiang Y. J., Yaling Z., Hatch G. M. Cloning and characterization of murine 1-acyl-sn-glycerol 3-phosphate acyltransferases and their regulation by PPARα in murine heart. Biochem. J. 2005;385:469–477. doi: 10.1042/BJ20041348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casiro S., Velez-Irizarry D., Ernst C. W., Raney N. E., Bates R. O., Charles M. G., Steibel J. P. Genome-wide association study in an F2 Duroc × Pietrain resource population for economically important meat quality and carcass traits. J. Anim. Sci. 2017;95:545–558. doi: 10.2527/jas.2016.1003. [DOI] [PubMed] [Google Scholar]
- Coleman R. A., Lee D. P. Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 2004;43:134–176. doi: 10.1016/S0163-7827(03)00051-1. [DOI] [PubMed] [Google Scholar]
- Falconer D. S. Introduction to Quantitative Genetics. 4th Edition. Trudy F. C. Mackay; 1996. [Google Scholar]
- Fan B., Lkhagvadorj S., Cai W., Young J., Smith R. M., Dekkers J. C., Huff-Lonergan E., Lonergan S. M., Rothschild M. F. Identification of genetic markers associated with residual feed intake and meat quality traits in the pig. Meat Sci. 2010;84:645–650. doi: 10.1016/j.meatsci.2009.10.025. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Prendes R., Quintanilla R., Canovas A., Manunza A., Figueiredo Cardoso T., Jordana J., Noguera J. L., Pena R. N., Amills M. Joint QTL mapping and geneexpression analysis identify positional candidate genes influencing pork quality traits. Scientific Rep. 2017;7:39830. doi: 10.1038/srep39830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J. H., An S. M., Kwon S. G., Park D. H., Kim T. W., Kang D. G., Yu G. E., Kim I. S., Park H. C., Ha J., Kim C. W. Associations of the polymorphisms in DHRS4, SERPING1, and APOR genes with postmortem pH in berkshire pigs. Animal Biotechnol. 2017:1–6. doi: 10.1080/10495398.2017.1279171. [DOI] [PubMed] [Google Scholar]
- Jung W. Y., Kwon S. G., Son M., Cho E. S., Lee Y., Kim J. H., Kim B. W., Park D. H., Hwang J. H., Kim T. W., Park H. C., Park B. Y., Choi J. S., Cho K. K., Chung K. H., Song Y. M., Kim I. S., Jin S. K., Kim D. H., Lee S. W., Lee K. W., Bang W. Y., Kim C. W. RNA-Seq approach for genetic improvement of meat quality in pig and evolutionary insight into the substrate specificity of animal carbonyl reductases. PloS One. 2012;7:e42198. doi: 10.1371/journal.pone.0042198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knothe G., Dunn R. O. A comprehensive evaluation of the melting points of fatty acids and esters determined by differential scanning calorimetry. J. Am. Oil Chem. Soc. 2009;86:843–856. doi: 10.1007/s11746-009-1423-2. [DOI] [Google Scholar]
- Kouba M., Enser M., Whittington F. M., Nute G. R., Wood J. D. Effect of a high-linolenic acid diet on lipogenic enzyme activities, fatty acid composition, and meat quality in the growing pig. J. Anim. Sci. 2003;81:1967–1979. doi: 10.2527/2003.8181967x. [DOI] [PubMed] [Google Scholar]
- Magowan E., Moss B., Fearom A., Ball E. Effect of breed, finish weight and sex on pork meat and eating quality and fatty acid profile. Agri-Food Biosci. Inst. UK. 2011:28. [Google Scholar]
- Monin G., Talmant A., Laborde D., Zabari M., Sellier P. Compositional and enzymatic characteristics of the Longissimus Dorsi muscle from large white, halothane-positive and halothane-negative pietrain, and hampshire pigs. Meat Sci. 1986;16:307–316. doi: 10.1016/0309-1740(86)90041-0. [DOI] [PubMed] [Google Scholar]
- Park K. M., Pramod A. B., Kim J. H., Choe H. S., Hwang I. H. Molecular and biological factors affecting skeletal muscle cells after slaughtering and their impact on meat quality: A mini review. J. Muscle Foods. 2010;21:280–307. doi: 10.1111/j.1745-4573.2009.00182.x. [DOI] [Google Scholar]
- Prasad S. S., Garg A., Agarwal A. K. Enzymatic activities of the human AGPAT isoform 3 and isoform 5: localization of AGPAT5 to mitochondria. J. Lipid Res. 2011;52:451–462. doi: 10.1194/jlr.M007575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razmaitė V., Kerzienė S., Švirmickas G. Correlations between fatty acid composition in subcutaneous tissue and meat quality traits in hybrids from different genotype and gender. Veterinarija ir Zootechnika; 2009. pp. 67–72. [Google Scholar]
- Shindou H., Shimizu T. Acyl-CoA:lysophospholipid acyltransferases. J. Biol. Chem. 2009;284:1–5. doi: 10.1074/jbc.R800046200. [DOI] [PubMed] [Google Scholar]
- Smet S. D., Raes K., Demeyer D. Meat fatty acid composition as affected by fatness and genetic factors: A review. Anim. Res. 2004;53:81–98. doi: 10.1051/animres:2004003. [DOI] [Google Scholar]
- Vance D. E., Vance J. E. CHAPTER 8 - Phospholipid biosynthesis in eukaryotes, Biochemistry of Lipids, Lipoproteins and Membranes. Fifth Edition. Elsevier; San Diego: 2008. pp. 213–244. [Google Scholar]
- Wood J. D., Enser M. Factors influencing fatty acids in meat and the role of antioxidants in improving meat quality. Br. J. Nutr. 1997;78:S49–S60. doi: 10.1079/BJN19970134. [DOI] [PubMed] [Google Scholar]
- Yamashita A., Hayashi Y., Matsumoto N., Nemoto-Sasaki Y., Oka S., Tanikawa T., Sugiura T. Glycerophosphate/Acylglycerophosphate acyltransferases. Biology. 2014a;3:801–830. doi: 10.3390/biology3040801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A., Hayashi Y., Nemoto-Sasaki Y., Ito M., Oka S., Tanikawa T., Waku K., Sugiura T. Acyltransferases and transacylases that determine the fatty acid composition of glycerolipids and the metabolism of bioactive lipid mediators in mammalian cells and model organisms. Prog. Lipid Res. 2014b;53:18–81. doi: 10.1016/j.plipres.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Yu K., Shu G., Yuan F., Zhu X., Gao P., Wang S., Wang L., Xi Q., Zhang S., Zhang Y., Li Y., Wu T., Yuan L., Jiang Q. Fatty acid and transcriptome profiling of longissimus dorsi muscles between pig breeds differing in meat quality. Int. J. Biol. Sci. 2013;9:108–118. doi: 10.7150/ijbs.5306. [DOI] [PMC free article] [PubMed] [Google Scholar]