Abstract
Second-trimester amniotic fluid supernatant (AFS) contains cell-free fetal RNA (cffRNA) transcripts that can provide information about fetal gene expression. In a retrospective case–control study, we measured second-trimester fetal gene expression using cffRNA extracted from AFS in women who had spontaneous preterm birth (sPTB) <34 weeks and in women who delivered >37 weeks. We extracted cffRNA from AFS of women with singletons who had second-trimester genetic amniocenteses. Twenty-one gravidas who had sPTB and 21 term controls were matched 1:1 for maternal age, fetal sex, race, gestational age (GA) at the time of amniocentesis, and medication exposure. Cell-free fetal RNA was extracted and hybridized to a customized 65-gene NanoString panel containing genes related to oxidative stress, inflammation, and hypothalamic–pituitary–adrenal (HPA) axis and included 15 housekeeping genes. Two models were run, 1 examining sPTB in relation to case/control status and 1 examining sPTB in relation to GA as a continuous variable. Among cases, the gene expression of nitric oxide synthase 1 (NOS1), d-aspartate oxidase (DDO), and Beta-2-microglobulin (B2M) was higher than controls (P value < .05; false discovery rate–corrected Q value of ≤0.10). Nitric oxide synthase 1 and DDO are genes associated with oxidative stress; B2M is a marker of the fetal inflammatory response. Fetal HPA gene expression is not associated with GA at delivery or sPTB in second-trimester AFS. Alterations of fetal gene expression related to inflammation and oxidative stress antedate clinical symptoms and may be useful for early identification of patients at risk of having an sPTB.
Keywords: cell-free fetal RNA, preterm birth, inflammation, oxidative stress
Introduction
In the United States, preterm birth (PTB) is the most common cause of infant morbidity and mortality. Although prematurity rates have declined in the United States, 1 in 10 infants are born preterm, equivalent to 360 000 infants per year. In 2007, US medical care for preterm infants costs more than $51 billion.1 Primary prevention strategies such as antioxidant supplementation or screening and treatment of maternal infections have failed to reduce or eliminate PTB.2 Little is known about the fetal contribution to PTB, however, there are emerging data that variants in the DNA of the fetus, not the mother, may be the trigger for some PTBs.3
Amniotic fluid supernatant (AFS) offers the opportunity to study human fetal development and physiology via analysis of the cell-free fetal RNA (cffRNA). Although cell-free fetal DNA and RNA in maternal serum arise from the placenta,4 gene expression and epigenetic studies of cffRNA in amniotic fluid demonstrate that cell-free nucleic acids are mainly contributed by the fetus.5 Thus, analysis of cell-free nucleic acids in amniotic fluid provides critical information about fetal gene expression.
The purpose of the study was to measure second-trimester fetal gene expression related to common PTB pathways involving the fetal hypothalamic–pituitary–adrenal (HPA) axis, inflammation, and oxidative stress. These genes were chosen because they represent key genes in pathways related to spontaneous PTB (sPTB).6 We hypothesized that fetal gene expression from AFS at the time of a second-trimester genetic amniocentesis in women who eventually had sPTB would have differential gene expression related to common PTB pathways in fetuses of women who delivered preterm compared to those at term.
Materials and Methods
This was a retrospective case–control study of women who delivered via sPTB or term birth (TB) with singleton fetuses without structural anomalies and who underwent second-trimester (14-24 weeks) genetic amniocentesis at University of North Carolina (UNC) at Chapel Hill. Amniocenteses were performed for clinical indications (advanced maternal age, ultrasonographic soft markers of aneuploidy, abnormal serum screening results, maternal request). Since 2003, AFS has been banked at −80°C for research purposes. The study was approved by the institutional review board at the UNC at Chapel Hill (13-1515). Amniotic fluid supernatant from women who had sPTB (<37 weeks; n = 21) was matched to AFS from women who had a TB (>37 weeks; n = 21) by fetal sex, maternal age (within 2 years), gestational age (GA) at amniocentesis (within 1 week), race, year of amniocentesis (within 1 year), and medication exposure (Table 1). Patients with abnormal fetal karyotypes, structural abnormalities, or any symptom or sign of preterm delivery at the time of genetic amniocentesis (eg, cervical shortening) were excluded. Since there are many different phenotypes of sPTB (preterm premature rupture of membranes, placental abruption, chorioamnionitis), a retrospective chart review was conducted by the lead author and confirmed by another board-certified perinatologist to ensure that the cases were women who delivered via sPTB defined as labor without a precipitating factor (ie, women with preterm premature rupture of membranes or placental abruption were excluded) as our goal was to identify fetal gene expression only associated with sPTB.
Table 1.
Demographic Characteristics of Women With sPTB and TB.
| Characteristics | sPTB (n = 21) | TB (n = 21) | P Value |
|---|---|---|---|
| Mean maternal age in years (SD [range]) | 34.6 (5.04 [22-40]) | 35.8 (6.98 [21-44]) | .53 |
| Race | |||
| Caucasian | 12 (57.1%) | 12 (57.1%) | |
| African American | 5 (23.8%) | 5 (23.8%) | |
| Asian | 2 (9.5%) | 2 (9.5%) | |
| Unknown | 1 (4.8%) | 1 (4.8%) | |
| Hispanic | 1 (4.8%) | 1 (4.8%) | |
| Mean gestational age in days at amniocentesis (SD [range]) | 123.6 (15.2 [112-163]) | 122.9 (14.3 [106-141]) | .83 |
| Fetal sex (number of males/number of females) | 14/7 | 14/7 | |
| Psychiatry medications | 6 (28.6%) | 6 (28.6%) | |
Abbreviations: SD, standard deviation; sPTB, spontaneous preterm birth; TB, term birth.
Cell-free fetal RNA was extracted from 5 mL of AFS, according to previously described protocols.7 RNA quantity was determined by Nanodrop (Thermo Scientific, Wilmington, Delaware), and quality was determined using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, California). The RNA was hybridized to a custom NanoString panel comprising 65 genes of interest and 15 housekeeping genes (Supplemental Table 1). The NanoString was performed in the core preclinical genomic pathology laboratory at the UNC at Chapel Hill Lineberger Comprehensive Cancer Center. Genes related to PTB were selected based on previous publications and were chosen for their known biological roles in stress, inflammation, and oxidative stress.3,8,9 Briefly, the nCounter Analysis System probe library contains 2 sequence-specific probes, the capture probe and the reporter probe, for each gene of interest. Probe pairs are mixed with total RNA in 1 hybridization reaction and then imaged with the use of fluorescent microscopy.10,11 Expression is measured by counting the number of unique color tags within the gene probe tripartite structures and is reported as “counts.” This system is optimal for small quantities of RNA such as is found in AFS. This platform permits quantifying the expression of up to 800 targets, without the need to use enzymes (reverse transcriptase or Taq for polymerase chain reaction), which could introduce bias.
Raw expression data were extracted using the nSolver software (NanoString Technologies, Inc, Seattle, Washington) and normalized both to positive controls and housekeeping genes. A Grubbs test was used to test for outliers in the data set. Statistical analyses were performed using Partek Genomics Suite software (version 6.6; Partek Inc, St Louis, Missouri). Differences in cell-free fetal gene expression were evaluated using 2 methods. A t test was performed to examine differences in gene expression associated with case status, whereas linear regression was used to examine changes associated with GA as a continuous variable. Because 1:1 matching was performed for the following variables that can affect fetal gene expression (maternal medication use, race, fetal sex, and maternal age), these variables were not included in the analysis. Statistical significance was defined at level P < .05. The false discovery rate–corrected Q-value (FDR Q-val) was set at 10% (.10).
Results
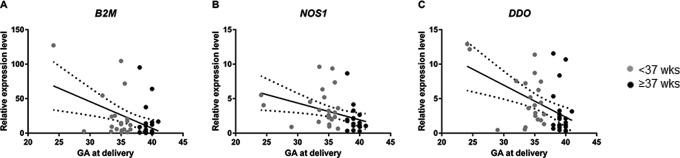
Table 1 summarizes selected demographic characteristics of the study participants. The sPTB and TB groups did not differ significantly with respect to maternal or GAs at the time of amniocentesis. Clinical indications for amniocentesis were the same in both groups (advanced maternal age, abnormal serum screen, soft marker on targeted anatomical survey). Although quality of RNA did vary among individual samples, those variations were not associated with year of collection (P = .45). Gene expression analysis was carried out on n = 65 genes selected based upon their relationship to PTB pathways with a focus on their biological roles in the HPA axis, inflammation, and oxidative stress; 15 housekeeping genes were also included (Supplemental Table 1). Although statistical analysis of dichotomous categories (sPTB/TB) showed no genes passed our FDR Q-val of ≤.10, there was a significant (P = .007) 2-fold increase in gene expression of nitric oxide synthase 1 (NOS1), a gene known to be associated with oxidative stress (Table 2). None of the other genes were differentially regulated other than NOS1 in the dichotomous (preterm/term) analysis (Table 2). Interestingly, there were differences in gene expression when GA was analyzed as a continuous variable. Effect size was also calculated between the 2 groups analyzing gene expression levels per week of gestation. The expression levels of beta-2-microglobulin (B2M) were significantly negatively associated with GA at delivery (FDR Q-val < 0.001), with a 60% decrease in gene expression with each increasing week of gestation. Additionally, the expression level of d-aspartate oxidase (DDO) was significant in relation to GA at delivery (FDR Q-val = 0.028) and showed 48% decrease in gene expression with each increasing week of gestation (Table 2; Figure 1). And although NOS1 (FDR Q val = 0.18) did not meet our FDR Q-val criteria, it does meet an accepted Q of 0.212,13 and showed a 38% decrease.
Table 2.
Cell-Free Fetal Gene Expression of Select Genes Related to Preterm Birth Pathways.a
| Pathway | Gene Name | Gestational Age | Case Status | ||||
|---|---|---|---|---|---|---|---|
| P Value | FDR Q Val | Effect Size per Week of Gestation | P Value | FDR Q Val | Fold Change (Up in Cases) | ||
| Oxidative stress | NOS1b,c | .014 | .183 | ↓ 38% | .007 | .59 | 2.11 |
| Oxidative stress | DDOb | .001 | .028 | ↓ 48% | .061 | .59 | 1.73 |
| Inflammation | B2Mb | <.001 | .001 | ↓ 60% | .12 | .59 | 2.82 |
| HPA axis | GRIA2b | .236 | .235 | ↓ 19% | .09 | .59 | 2.03 |
Abbreviations: B2M, beta-2 microglobulin; DDO, d-aspartate oxidase; FDR Q-val, false discovery rate–corrected Q value; GRIA2, glutamate ionotropic receptor AMPA type subunit 2; HPA, hypothalamic–pituitary–adrenal; NOS1, nitric oxide synthase 1; AMPA, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid.
aFour of 65 genes from various pathways were chosen to illustrate findings. Significance is defined as P < .05 with FDR Q-val of ≤0.10.
bGenes significantly negatively associated with gestational age at delivery.
cGenes significantly associated with case status.
Figure 1.
Association of beta-2-microglobulin (B2M; A), nitric oxide synthase 1 (NOS1; B), and d-aspartate oxidase (DDO; C) expression with gestational age at delivery (samples collected at 14-24 weeks’ gestation).
Comment
The goal of the study was to measure differences in gene expression in cffRNA extracted from AFS in women with sPTB compared to women delivered at term in second-trimester amniotic fluid. In contrast to our a priori hypothesis, the gene expression levels at the time of the second-trimester amniocentesis that are involved in the HPA axis were not associated with GA at delivery or sPTB. It is possible that changes in HPA axis fetal gene expression do occur, but do so in the third trimester or closer to the time of labor onset. We did find higher expression levels of B2M, a marker of the fetal inflammatory response,14 in fetuses that ultimately had sPTB compared to those at term. Fetal gene expression of NOS1 and DDO was also increased in fetuses that had sPTB. Nitric oxide synthase 1 and DDO are both associated with oxidative stress; NOS1 is also associated with smooth muscle contraction.15
Evidence for differential gene expression related to fetal inflammation and oxidative stress at the time of the second-trimester genetic amniocentesis in women who delivered at earlier GAs suggests that these are key pathways that initiate preterm labor. Beta-2-microglobulin is the noncovalently bound light chain of major histocompatibility class I, a protein found on the surface of all nucleated cells.16 Beta-2-microglobulin has been shown to have potent antibacterial properties in human amniotic fluid and exhibits antibacterial activity against pathogenic microbial strains. Beta-2-microglobulin has increased expression in amniotic cells during bacterial infection.14 Previous studies have shown subclinical infection at the time of genetic amniocentesis in women who deliver preterm17 or at the time of idiopathic preterm labor.18 It is possible that a subclinical infection stimulates a fetal inflammatory response that results in upregulation of B2M gene expression in the women who delivered via sPTB. However, given the small sample size, larger studies are needed to confirm results and further understand the mechanisms of the changes seen in fetal gene expression.
In this study, NOS1 and DDO gene expression was also higher in women who delivered at earlier GAs. Nitric oxide synthase 1, catalyzes the synthesis of nitric oxide from l-arginine, has been shown to have a relaxant effect on uterine smooth muscle cells in animal models.15,19 There is also other evidence that NOS1 has a role in sPTB as NOS1 isoforms are found in higher concentration in women in labor (term and preterm).20 We can speculate that the fetus is expressing increased amounts of NOS1 gene expression to relax the uterus because it is trying to counteract the inflammatory milieu caused by increased B2M gene expression. However, because of the exploratory nature of this work, further studies are needed to validate and understand the mechanism behind these findings.
This study is not without limitations. Although the sample size is small, the cases and controls were carefully matched. Since all of the amniocentesis samples were obtained because of routine indications, there is a possibility of introducing bias because of comorbid pregnancy conditions. To address this, we excluded any fetuses with chromosome abnormalities and structural abnormalities. There are numerous strengths of the study. First, the use of human amniotic fluid matched on factors known to alter fetal gene expression, including GA, is a strength as was limiting the phenotype of sPTB to women who had no known risk factor for sPTB (cervical shortening, infection, bleeding) in order to identify fetal gene expression changes prior to the onset of clinical symptoms. Additionally, the use of the highly sensitive NanoString technology for cffRNA assessment is a strength as it allows us the ability to target genes associated with PTB pathways. In addition, NanoString is a clinically validated tool, thus, there is high potential for it to be clinically useful in the prediction of PTB. This study sheds light on fetal transcripts and pathways that may be important in identifying women at risk of sPTB. These transcripts can potentially be identified noninvasively in maternal blood.21,22 Given the selected genes related to each PTB pathway, we believe this study provides novel data regarding molecular mechanisms related to a possible fetal contribution to sPTB. An alternative agnostic approach could also be performed to identify novel gene transcripts associated with PTB using a larger cohort of patients and a whole genome expression array.
In summary, targeted gene expression analysis was used to identify changes in cffRNA derived from AFS in women with sPTB and TB. Alterations of fetal gene expression related to inflammation and oxidative stress may antedate clinical symptoms and may be useful for early identification of patients at risk of having an sPTB. Future studies should confirm these findings in a larger cohort. Given the recent ability to monitor cell-free RNA noninvasively during human pregnancy to provide information about fetal tissue-specific transcription,21 biomarkers for sPTB prior to the onset of clinical symptoms using cffRNA in maternal serum may be identified in women at high risk for sPTB based on differentially expressed gene expression levels related to inflammatory and oxidative stress pathways.
Supplementary Material
Footnotes
Authors’ Note: Presented at 63rd annual meeting of the Society of Reproductive Investigation, Montreal, Québec, Canada.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: NICHD R03 HD080788; BIRCWH K12 HD001441; and Division of Maternal–Fetal Medicine: Cefalo Bowes Research Award.
Supplemental Material: The online Supplemental Table is available at http://journals.sagepub.com/doi/suppl/10.1177/1933719116670038.
References
- 1. The impact of premature birth on society. http://www.marchofdimes.org/mission/the-economic-and-societal-costs.aspx. 2015. Updated October 2015. Accessed March 2016.
- 2. Markham KB, Klebanoff M. Prevention of preterm birth in modern obstetrics. Clin Perinatol. 2014;41(4):773–785. [DOI] [PubMed] [Google Scholar]
- 3. Biggio JXF, Baldwin D, Bukowski R, et al. Neonatal, not maternal, copy number variants are associated with spontaneous preterm birth. American J Obstet Gynecol. 2015; Supplement to January 2015; 212(1):S8. [Google Scholar]
- 4. Flori E, Doray B, Gautier E, et al. Circulating cell-free fetal DNA in maternal serum appears to originate from cyto- and syncytio-trophoblastic cells. Case report. Hum Reprod. 2004;19(3):723–724. [DOI] [PubMed] [Google Scholar]
- 5. Lun FM, Chiu RW, Leung TY, Leung TN, Lau TK, Lo YM. Epigenetic analysis of RASSF1A gene in cell-free DNA in amniotic fluid. Clin Chem. 2007;53(4):796–798. [DOI] [PubMed] [Google Scholar]
- 6. Romero R, Dey SK, Fisher SJ. Preterm labor: one syndrome, many causes. Science. 2014;345(6198):760–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dietz JA, Johnson KL, Massingham LJ, et al. Comparison of extraction techniques for amniotic fluid supernatant demonstrates improved yield of cell-free fetal RNA. Prenat Diagn. 2011;31(6):598–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Behnia F, Parets SE, Kechichian T, et al. Fetal DNA methylation of autism spectrum disorders candidate genes: association with spontaneous preterm birth. Am J Obstet Gynecol. 2015;212(4):533.e531-e539. [DOI] [PubMed] [Google Scholar]
- 9. Uzun A, Dewan AT, Istrail S, Padbury JF. Pathway-based genetic analysis of preterm birth. Genomics. 2013;101(3):163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Geiss GK, Bumgarner RE, Birditt B, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26(3):317–325. [DOI] [PubMed] [Google Scholar]
- 11. Kulkarni MM. Digital multiplexed gene expression analysis using the NanoString nCounter system. Curr Protoc Mol Biol. 2011;Chapter 25:Unit25B.10. [DOI] [PubMed] [Google Scholar]
- 12. Sun Z, Asmann YW, Kalari KR, et al. Integrated analysis of gene expression, CpG island methylation, and gene copy number in breast cancer cells by deep sequencing. PloS One. 2011;6(2):e17490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yi Y, Li C, Miller C, George AL., Jr Strategy for encoding and comparison of gene expression signatures. Genome Biol. 2007;8(7):R133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim JY, Park SC, Lee JK, Choi SJ, Hahm KS, Park Y. Novel antibacterial activity of beta(2)-microglobulin in human amniotic fluid. PloS One. 2012;7(11):e47642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sladek SM, Regenstein AC, Lykins D, Roberts JM. Nitric oxide synthase activity in pregnant rabbit uterus decreases on the last day of pregnancy. Am J Obstet Gynecol. 1993;169(5):1285–1291. [DOI] [PubMed] [Google Scholar]
- 16. Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329(6139):506–512. [DOI] [PubMed] [Google Scholar]
- 17. Dortbudak O, Eberhardt R, Ulm M, Persson GR. Periodontitis, a marker of risk in pregnancy for preterm birth. J Clin Periodontol. 2005;32(1):45–52. [DOI] [PubMed] [Google Scholar]
- 18. Hitti J, Tarczy-Hornoch P, Murphy J, Hillier SL, Aura J, Eschenbach DA. Amniotic fluid infection, cytokines, and adverse outcome among infants at 34 weeks’ gestation or less. Obstet Gynecol. 2001;98(6):1080–1088. [DOI] [PubMed] [Google Scholar]
- 19. Riemer RK, Buscher C, Bansal RK, Black SM, He Y, Natuzzi ES. Increased expression of nitric oxide synthase in the myometrium of the pregnant rat uterus. Am J Physiol. 1997;272(6 pt 1):E1008–E1015. [DOI] [PubMed] [Google Scholar]
- 20. Natuzzi ES, Ursell PC, Harrison M, Buscher C, Riemer RK. Nitric oxide synthase activity in the pregnant uterus decreases at parturition. Biochem Biophys Res Commun. 1993;194(1):1–8. [DOI] [PubMed] [Google Scholar]
- 21. Koh W, Pan W, Gawad C, et al. Noninvasive in vivo monitoring of tissue-specific global gene expression in humans. Proc Natl Acad Sci U S A. 2014;111(20):7361–7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Curti A, Lapucci C, Berto S, et al. Maternal plasma mRNA species in fetal heart defects: a potential for molecular screening. Prenat Diagn. 2016;36(8):738–743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.