Abstract
The biosynthesis of phosphatidic acid, a key intermediate in the biosynthesis of lipids, is controlled by lysophosphatidic acid (LPA, or 1-acyl-glycerol-3-P) acyltransferase (LPAAT, EC 2.3.1.51). We have isolated a cDNA encoding a novel LPAAT by functional complementation of the Escherichia coli mutant plsC with an immature embryo cDNA library of oilseed rape (Brassica napus). Transformation of the acyltransferase-deficient E. coli strain JC201 with the cDNA sequence BAT2 alleviated the temperature-sensitive phenotype of the plsC mutant and conferred a palmitoyl-coenzyme A-preferring acyltransferase activity to membrane fractions. The BAT2 cDNA encoded a protein of 351 amino acids with a predicted molecular mass of 38 kD and an isoelectric point of 9.7. Chloroplast-import experiments showed processing of a BAT2 precursor protein to a mature protein of approximately 32 kD, which was localized in the membrane fraction. BAT2 is encoded by a minimum of two genes that may be expressed ubiquitously. These data are consistent with the identity of BAT2 as the plastidial enzyme of the prokaryotic glycerol-3-P pathway that uses a palmitoyl-ACP to produce phosphatidic acid with a prokaryotic-type acyl composition. The homologies between the deduced protein sequence of BAT2 with prokaryotic and eukaryotic microsomal LAP acytransferases suggest that seed microsomal forms may have evolved from the plastidial enzyme.
Phosphatidic acid is a key intermediate in the biosynthesis of phospho- and glycerolipids, essential components of all cellular membranes and of triacylglycerols. In most plant species, palmitoyl-ACP and oleoyl-ACP are the predominant products of de novo fatty acid biosynthesis in the chloroplast (Ohlrogge et al., 1993). These fatty acids may enter the prokaryotic pathway of lipid biosynthesis by transfer of the acyl group from ACP to glycerol-3-P, which is mediated by a stromal glycerol-3-P acyltransferase to form LPA or to position sn-2 of glycerol-3-P mediated by LPAAT (EC 2.3.1.51) to form phosphatidic acid. The phosphatidic acid produced by the prokaryotic pathway of the chloroplast is then used to produce phosphatidylglycerol (for review, see Ohlrogge and Browse, 1995) or is dephosphorylated to diacylglycerol, from which the glycodiacylglycerols characteristic of the thylakoid membrane are derived (for review, see Douce and Joyard, 1990).
Alternatively, the elongation of fatty acids may be terminated by the action of an acyl-ACP thioesterase, which hydrolyzes the acyl-ACP to release a free fatty acid, which then leaves the plastid. The fatty acid in the form of an acyl-CoA thioester may participate in the synthesis of glycerolipids via the eukaryotic pathway located at the ER. The phosphatidic acid of the cytoplasmic pathway is used to produce the phospholipids characteristic of extrachloroplastic lipids. The phosphatidylcholine produced by the eukaryotic pathway is a substrate for desaturation, after which the diacylglycerol moiety may be returned to the chloroplast. The contribution of each pathway to the synthesis of chloroplast lipids differs among species, and the relative amounts of hexadecatrienoic acid and linolenate present in the leaf galactolipids are an indication of the relative flux through each pathway (for review, see Browse and Somerville, 1991).
The important role that acyltransferases, and the LPAATs in particular, play in regulating lipid acyl composition is mediated through their substrate specificities (for review, see Frentzen, 1993). The plastidial LPAAT almost exclusively utilizes palmitoyl-ACP to produce a phosphatidic acid containing a saturated group at position sn-2, which is characteristic of lipids synthesized by the prokaryotic pathway. In contrast, the cytoplasmic LPAAT prefers linoleoyl to oleoyl groups; it discriminates strongly against saturated C16 and C18 acyl groups and further differs from the plastidial enzyme in that an unsaturated acceptor LPA is preferred. Thus, the phosphatidic acid produced by the eukaryotic pathway is highly enriched in C18:1 at position sn-2. In cytoplasmic LPAAT, additional seed-specific isozymes exist whose substrate specificity varies among species (Sun et al., 1988). These acyltransferases are of interest for engineering of seed oil composition.
Membrane-bound LPAATs have not been purified sufficiently to allow molecular cloning, except that of coconut (Davies et al., 1995). However, the complementation of the acyltransferase-deficient mutant of Escherichia coli (Coleman, 1990) using libraries derived from immature embryos of meadowfoam (Brown et al., 1995; Hanke et al., 1995) and from maize endosperm (Brown et al., 1994) has facilitated the isolation of cDNAs encoding LPAATs.
We are engaged in studies of structure and function among plant LPAATs and their role in determining lipid acyl composition. To this end we have isolated a novel LPAAT by functional complementation in an acyltransferase-deficient E. coli mutant. We describe the characterization of an oilseed rape (Brassica napus) cDNA encoding a prokaryotic-type LPAAT and provide evidence identifying this protein as the enzyme controlling phosphatidic acid biosynthesis in the plastid. We also compare the structural similarities among the plastidial, microsomal, and prokaryotic proteins and discuss evolutionary relationships among plant LPAATs.
MATERIALS AND METHODS
Plant Material
The progeny of a cross between the varieties B002 (<1% erucic acid) and Hokkaido (>50% erucic acid) of oilseed rape (Brassica napus) described in Barret et al. (1998) were used for the construction of cDNA libraries and the isolation of the BAT2 gene. For the mapping studies we used the double-haploid population, which was derived from the cross between the varieties Darmor-bzh (<1% erucic acid) and Yudal (>50% erucic acid) and was used for the construction of a genetic map of B. napus (Foisset et al., 1996).
Isolation of B. napus cDNAs by Heterologous Complementation
The protocol used for complementation was based on that of Delauney and Verma (1990). The construction of an immature embryo cDNA library of B. napus in the vector Lambda ZAP II (Stratagene) as described in Barret et al. (1998). An aliquot representative of the primary library was subjected to in vivo excision using the ExAssist system (Stratagene). Approximately 2 × 106 colonies were recovered after plasmid rescue and pooled for plasmid DNA preparation. The Escherichia coli mutant JC201 was used in the complementation experiments and has the following genotype: plsC thr-1 ara-14 d(gal-attl)-99 his G4 rpsL136 xyl-5 mtl-1 lacY1 tsx-78 eda-50 rfbD1 thi-1 (Coleman, 1990). E. coli JC201 was transformed via an electroporation protocol for bacteria based on that described by Ausubel et al. (1992).
Transformant colonies growing on Luria-Bertani agar containing 100 μg mL−1 ampicillin and 1 mm IPTG after incubation at 42°C were collected and plasmid DNA was isolated. An aliquot of this pooled DNA was used to retransform E. coli JC201 cells, and the procedure was repeated with pooled DNA from this second transformation. After three rounds of transformation, plasmid DNA was isolated from individual clones, which apparently corrected the temperature-sensitive phenotype. The ability of the individual clones to complement the temperature-sensitive phenotype of E. coli JC201 was tested by re-transformation with plasmid DNA isolated from individual cDNA clones, followed by incubation at 42°C. Plasmid DNA was isolated from individual transformant clones and sequenced to confirm identity with the original DNA.
DNA Sequencing and Analysis
Sequences were determined using cycle sequencing (DyeDeoxy Terminator, Applied Biosystems) on double-stranded DNA templates with a sequencer (model 373A, Applied Biosystems). Each strand was sequenced using oligonucleotide primers.
Southern and Northern Hybridization
DNA was extracted from young green leaf tissue according to the protocol of Doyle and Doyle (1990). A series of reactions containing 15 μg of DNA was digested with the appropriate restriction enzyme, and the resulting fragments were separated on a 0.75% agarose gel and transferred to nylon membrane (Hybond N+, Amersham). A 632-bp probe contained within the coding sequence was generated by PCR using the following primers: AT2.1, 5′-CAA GAA CTG TGA CTG TGA GAT CGG-3′; and AT2.2, 5′-CGT TAT TGG CAC CAC TGG AAC TCC-3′.
The resultant amplimer was radiolabeled by random priming (Rediprime, Amersham). Hybridization and washing at low and high stringency were according to the manufacturer's protocol.
Total RNA was extracted from various tissues by a protocol based on the method of Kay et al. (1987). Total RNA (20 μg) or 1 μg of poly(A+) RNA was fractionated on 1.2% agarose gels containing formaldehyde and transferred to nylon membrane (Hybond N, Amersham). The probe and the hybridization were as for the genomic Southern analysis. The filter was washed at 65°C in 0.125× SSC and 0.1% SDS and autoradiographed with intensifying screens at −80°C. The filters were subsequently stripped and reprobed with a 18S RNA probe to compare the relative amounts of RNA in each lane.
Chloroplast Import Studies
Plasmid DNA isolated from clone BAT2 was used to generate the precursor form of BAT2 by in vitro transcription using T3 polymerase, after which capped transcripts were translated in a wheat germ lysate in the presence of (35S)-Met. The translation product was incubated with intact, isolated pea chloroplasts, and the organelles were treated with thermolysin and then fractionated as described in Robinson (1993).
Assay of LPAAT Activity
E. coli JC201 cells were transformed in pBluescript SK vector (Stratagene) with the BAT2 sequence and in the vector only. Cells were sedimented at 3000g from a culture grown at 30°C to an optical density of 0.6, induced by the addition of IPTG to 1 mm final concentration, and grown for 3 h at 30°C. The cells were fractionated into periplasmic, cytoplasmic, and membrane fractions according to the protocol of Henderson and Macpherson (1986). The membrane pellet fraction was resuspended with 50 mm Tris-HCl, pH 8.0, 2 mm MgCl2, and 2 mm DTT, and stored at −80°C.
The assay was comprised of 50 mm Tris-HCl, pH 8, 1 mm MgCl2, 1 mm dithiotheitol, 55 μm 1-oleoyl-LPA, 10 μm [1-14C] acyl-CoA, and 15 μg of the membrane fraction and was incubated for 5 min at 30°C. This was essentially the protocol of Cao et al. (1990), as was the TLC analysis of products. The phosphatidic acid spot was visualized by autoradiography and collected for scintillation counting.
Mapping of the BAT2 Genes
The DNA insert contained within cDNA clone BAT2 was amplified by PCR and purified according to protocols described in Barret et al. (1998) using the following primers: BnAT2.4 = 5′-GTC ATC GGT TGG GCT ATG-3′ and BnAT2.5 = 5′-CCG ATC TCA CAG TCA CAG-3′.
RFLP analysis was performed as described in Sharpe et al. (1995). The BAT2/DraI markers were scored for each genotype, and linkage analysis was performed using Mapmaker (Lander et al., 1987) and Mapmaker QTL software, as described in Jourdren et al. (1996).
RESULTS
Heterologous Complementation of E. coli plsC
A population of plasmid clones excised from and representative of an immature embryo cDNA library of B. napus was transformed into the E. coli strain JC201 to identify clones that were capable of complementing the deficiency of LPAAT activity that is characteristic of this mutant (Coleman, 1990). This strain is unable to grow at 42°C but grows well at 30°C. Thus, the selection was based on the restoration of growth at the nonpermissive temperature in the presence of IPTG and ampicillin. We performed three successive rounds of transformation with pooled DNA obtained from colonies growing at the previous transformation step, with the expectation that each round of transformation would enrich the population with authentic complementing clones and eliminate revertants that apparently complement after acquisition of antibiotic resistance.
At the third round of transformation, approximately 1.5 × 106 colonies per microgram of DNA were obtained. An aliquot of this pooled DNA was propagated in E. coli XL-1-Blue (Stratagene) cells, and plasmid DNA was isolated from individual transformant colonies. When 100 clones were screened for insert size by PCR, approximately 85% contained an insert of 1.2 kb that shared homology with known LPAAT sequences. Four of these clones were sequenced at each extremity, and were found to be identical; these clones were designated BAT2 (Brassica acyltransferase 2).
A final transformation experiment was performed with plasmid DNAs of individual BAT2 clones. Plasmid DNA (50 ng) of one of the 1.2-kb clones was used to transform E. coli JC201 via heat shock, which resulted in the growth at 42°C of a minimum of 10,000 colonies in the presence of 1 mm IPTG and 100 μg mL−1 ampicillin. In contrast, 125 transformant colonies were present after transformation of E. coli JC201 with the vector pBluescript. We conclude that the expression of the 1.2-kb insert present in BAT2 resulted in the abolition of the temperature sensitivity of E. coli JC201 and restored growth at 42°C.
Sequence Analysis of a BAT2 cDNA Clone
The BAT2 cDNA (accession no. A111161) was composed of a 1155-bp sequence preceeding a poly(A+) tail of 18 bp. Translation of the cDNA sequence revealed the presence of a single ORF in the 5′ to 3′ direction of the cDNA, 351 amino acids (Mr 38,748), and a pI of 9.73. There were no in-frame stop codons present before the ORF, so it is not certain that the cDNA sequence isolated is complete. The first in-frame Met codon is positioned at nucleotides 24 to 26. The sequence surrounding this initiation codon does not correspond to the consensus for plants (Lutcke et al., 1987), so translation in the plant may not initiate from this codon. However, the BAT2 cDNA sequence, together with the EcoRI adaptor sequence, is in frame with and was probably expressed as a fusion protein with the β-galactosidase of the pBluescript vector. We used the technique of PCR walking (Devic et al., 1997) to determine the genomic sequence corresponding to the region 5′ to the cDNA coding sequence. This technique revealed the continuation of the ORF in the same reading frame as the BAT2 sequence and suggested that the BAT2 coding sequence may contain an additional eight amino acid residues to the nearest upstream AUG codon. Therefore, the BAT2 sequence may code for a protein of 359 amino acids with a Mr of 39,606 D. We conclude that the 1155-nt sequence of the pBAT2 clone contains sufficient information for the translation of a fusion protein in E. coli JC201 that eliminates the temperature-sensitive phenotype.
The deduced amino acid sequence of the ORF present in BAT2 was used to identify homologous sequences via an advanced BLASTX2 search (Gish and Slates, 1993). The BAT2 protein was found to share significant homologies with LPAAT sequences of eukaryotic and prokaryotic species (Fig. 1). The highest scores were obtained with a LPAAT of yeast, the SLC1 gene product (Nagiec et al., 1993), which revealed 32% identity and 51% similarity over a 204-amino acid overlap. A similar degree of homology was evident with the three sequences identified as microsomal LPAATs expressed in seeds of meadowfoam (Brown et al., 1995; Hanke et al., 1995; Lassner et al., 1995), which revealed 30% identity and 54% similarity within a 182-residue overlap, and with the coconut endosperm LPAAT (Knutzon et al., 1995), which revealed 31% identity and 47% similarity over 229 residues. Over a shorter sequence of 143 amino acids the BAT2 sequence shared a similar degree of homology (30% identity and 55% similarity with a putative ORF identified within the genome of Synechocystis sp. (accession no. P74498). Significant scores were obtained with bacterial LPAATs: The homology between the BAT2 protein and that of E. coli plsC was 31% identity and 50% similarity but within a shorter overlap of 115 residues. Absent from the BLAST alignments were the putative LPAAT sequences pMAT1 (Brown et al., 1994) and its B. napus homolog pBAT1 (accession no. Z49860).
Figure 1.
Comparison of the partial deduced amino acid sequences of B. napus BAT2 protein with LPAATs from meadowfoam (Limnanthes; accession no. Q42870), coconut (Cocos; accession no. Q42670), E. coli (accession no. P26647), yeast (Saccharomyces cerevisiae; accession no. P33333), and Synecocystis (Synechocys; accession no. P74498). Outlined boxes indicate identity; shaded boxes indicate conservative differences and unshaded boxes nonconservative differences.
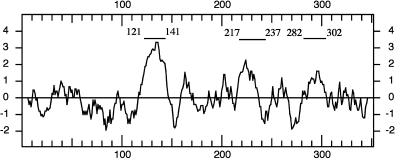
A comparison of the deduced amino acid sequence of BAT2 with each of the previously identified LPAAT sequences via the BLAST alignments suggested that the BAT2 protein was longer at the amino terminus by approximately 90 residues. This sequence is characterized by a paucity of acidic residues but is rich in Ser and enriched Ala and Val, and contains basic residues, all of which are characteristics of a chloroplast-targeting sequence (Keegstra et al., 1989). Sequence analysis by the PSORT program (Nakai and Horton, 1999) predicted that the BAT2 protein possessed an N-terminal chloroplast-targeting sequence together with an intrachloroplast sorting signal, so the BAT2 was predicted to be a thylakoid protein. The hydropathy profile of the deduced BAT2 protein sequence (Fig. 2) revealed the presence of several hydrophobic regions, three of which were predicted by the TOPRED2 program to be transmembrane domains. These putative membrane-spanning domains are located between residues 121 and 141, 217 and 237, and 282 and 302 with respect to the most N-terminal residue deduced from the cDNA sequence of BAT2.
Figure 2.
Hydropathy analysis of the BAT2 protein. Hydropathy profiles were determined using the deduced amino acid sequence of B. napus BAT2 using the Kyte-Doolittle algorithm. Horizontal numbers indicate amino acid positions in the protein sequence, and positive numbers on the vertical scale indicate a hydrophobic region. Three potential transmembrane domains are indicated by horizontal bars.
Subcellular Localization of the pBAT2 Protein
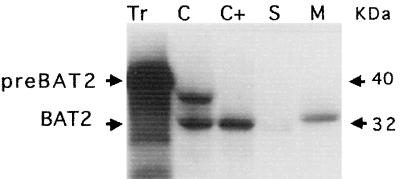
To establish whether the protein encoded by the cDNA clone pBAT2 may be located in the plastid, we tested the ability of isolated chloroplasts to import and process the putative precursor protein. The primary translation product was synthesized by in vitro transcription of the cDNA, followed by translation in a wheat germ cell-free system in the presence of radiolabeled Met. Figure 3 shows that this procedure generated a precursor protein of approximately 40 kD. Incubation of this polypeptide with intact pea chloroplasts resulted in the import and processing of the precursor protein to a mature protein of 32 kD, which was resistant to proteinase, indicating localization in the chloroplast. The preprotein present during the incubation with chloroplasts and evident in Figure 3, lane C, was sensitive to proteolysis, indicating that these molecules are associated with the external surface of the chloroplast, and are perhaps bound to the import receptors. Fractionation of the chloroplasts after the uptake incubation revealed the mature BAT2 protein (migrating with an apparent molecular mass greater than 32 kD in the presence of unlabeled thylakoid proteins) to be localized almost exclusively in the membrane fraction. We conclude from these data that the BAT2 protein is synthesized with a cleavable signal that is capable of targeting the preprotein to plastids and allowing compartmentalization of the mature protein into a membrane fraction.
Figure 3.
Import of BAT2 protein into isolated chloroplasts. 35S-Labeled proteins derived from an in vitro transcription-translation programmed by the BAT2 cDNA (lane Tr) were incubated with intact chlorplasts (lane C), treated with thermolysin (lane C+), and fractionated into stromal (lane S) and membrane (lane M) fractions. preBAT2, Preprotein; BAT2, mature protein. On the right are the apparent molecular masses in kD.
Mapping of the B. napus BAT2 Genes
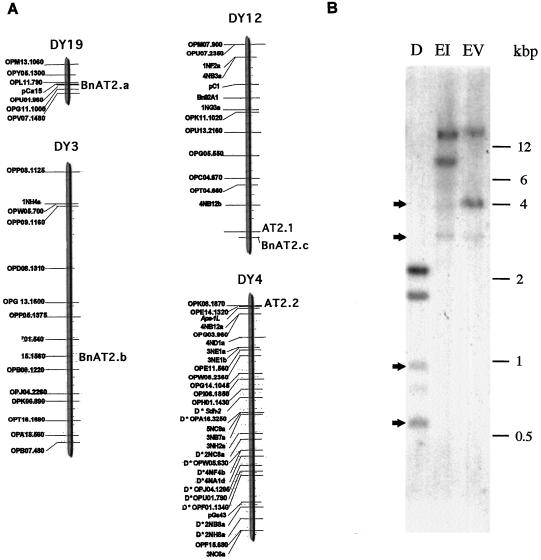
The BAT2 gene was positioned as a molecular marker on a B. napus genetic map (Foisset et al., 1996) using the BAT2 cDNA as a RFLP probe. Hybridization of the probe to Southern blots of the parental varieties Darmor-bzh and Yudal digested with DraI revealed at least two loci: AT2.1 (dominant) and AT2.2 (co-dominant). Examination of 90 progeny plants indicated that marker AT2.1 was positioned at the extremity of linkage group DY12, and AT2.2 was located at the extremity of group DY4 (Fig. 4A). A RFLP marker, 4NB12a, was located 14.3 cM from AT2.2, indicating either that DY4 and DY12 are homeologous chromosomes or that a fragment of DNA is duplicated between the two linkage groups. Using a cleaved amplified polymorphic sequences approach (Konieczny and Ausubel, 1993), a major band of 2.1 kb and several smaller bands were obtained after PCR, which gave rise to two markers. One 1.7-kb fragment, BnAT2a, was dominant and positioned on the DY19 linkage group. The second fragment was 1.2 kb and codominant. Digestion of the 2.1-kb fragment by HaeIII revealed a third marker, BnAT2b, which was positioned in the middle of the DY3 linkage group (Fig. 4A) 53 cM from locus L2, one of two loci controlling seed linolenic acid content. The marker BnAT2c was positioned on the DY12 group within 3.3 cM of the RFLP marker AT2.1 using 40 plants. The distance of 3.3 cM was obtained with a single recombinant plant between the two markers.
Figure 4.
A, Localization of BAT2 markers on a genetic map of B. napus: AT2.1 and AT2.2 were mapped by RFLP; BnAT2 (a, b, and c) were mapped by CAPS. B, Southern hybridization of B. napus BAT2 genes. Pooled, genomic DNA isolated from 20 HEAR segregant plants was digested with the enzymes DraI, EcoRI, and EcoRV, resolved on an agarose gel, and transferred to a nylon membrane. A truncated BAT2 coding sequence probe of 632 bp was hybridized, and the filter was washed to a stringency of 0.125× SSC and 0.1% SDS at 60°C and autoradiographed. Arrows indicate position of weakly hybridizing fragment.
Southern hybridization performed to confirm the number of genes encoding BAT2 revealed the presence of two strongly hybridizing fragments from each restriction digest of B. napus DNA. In addition, weakly hybridizing fragments were detected with the DNA digested with the enzymes DraI and EcoRI, as indicated by arrows in Figure 4B. Using the BAT2 sequence as a probe, we have subsequently isolated a highly homologous (97.2% amino acid identity) yet distinct sequence, BAT2.3. The heterogeneity between BAT2 and BAT2.3 cDNAs may be a consequence of the allotetraploid nature of B. napus and may reflect divergence between the parental BAT2 gene copy present in each donor A and C genome. This observation is consistent with the putative identification of pairs of homoeologous loci located in DY4 and DY12 linkage groups, a frequent feature of RFLP mapping in this amphidiploid species. The two strong hybridization signals detected on the Southern blot may correspond to the two cDNAs. The detection of additional, weakly hybridizing fragments may indicate the existence of structurally related genes. Thus, B. napus BAT2 protein is encoded by a minimum of two homologous genes and may be present in a small multigenic family of four members.
BAT2 Is Ubiquitously Expressed
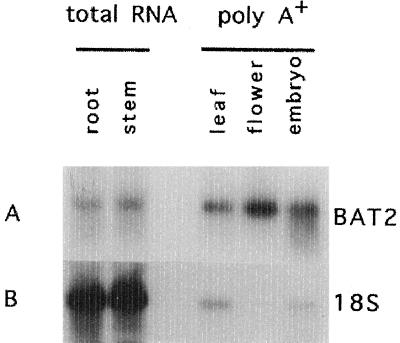
The pattern of B. napus BAT2 gene expression was determined to gain insight into its function. Northern hybridization was performed with total RNA isolated from root and stem tissues and with poly(A+) RNA isolated from leaf, flower, and embryos at 28 d after pollination (Fig. 5). A probe corresponding to 632 bp of the coding sequence was found to hybridize to at least one class of transcript of approximately 1.3 kb in each tissue examined. Apparent differences in the BAT2 transcript level among leaf, flower, and embryo were associated with a degree of contamination of the leaf and embryo poly(A+) RNA preparations by total RNA, as revealed by the use of the 18S rRNA probe as a quantitative control. We concluded that the BAT2 gene is expressed in a ubiquitous manner in all tissues, including nonphotosynthetic tissues where plastids are present.
Figure 5.
Northern hybridization of B. napus BAT2 expression. Total RNA was extracted from root and stem and poly(A+) RNA from leaf, flower, and immature embryos isolated at 28 d after pollination. The RNAs were resolved on formamide agarose gels, blotted to a nylon membrane, and hybridized with a 632-bp probe corresponding to the truncated BAT2 coding sequence cDNA insert and subsequently with an rRNA probe. The filters were washed to a stringency of 0.125× SSC and 0.1% SDS at 60°C and autoradiographed.
The BAT2 Protein Possesses LPAAT Activity
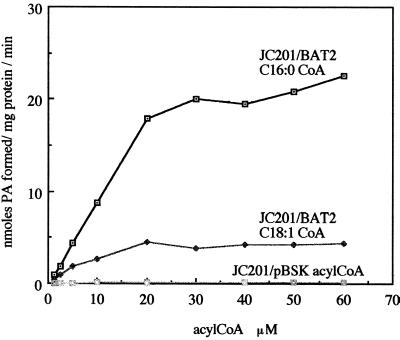
To confirm that the presence of the BAT2 cDNA sequence confers LPAAT activity, membranes were isolated from the acyltransferase-deficient E. coli mutant JC201 by expressing sequences present on the plasmid vector and from the pBAT2 cDNA, which were then assayed for the incorporation of acyl groups into phosphatidic acid using [1-14C]oleoyl-CoA or [1-14C]palmitoyl-CoA. Despite a preference for acyl-ACP thioesters, their physiological substrates, plastidial acyltransferases are capable of utilizing acyl-CoA thioesters for the synthesis of phosphatidic acid (Frentzen et al., 1983). LPAAT activity was determined in the presence of 55 μm 1-oleoyl-LPA, and varying concentrations of palmitoyl-CoA or oleoyl-CoA. The LPAAT activities of membrane preparations from E. coli JC201 transformants as a function of acyl-CoA concentration are shown in Figure 6.
Figure 6.
LPAAT activities in E. coli membrane preparations. E. coli JC201 cells were transformed in pBluescript SK vector (Stratagene) with the BAT2 sequence and in vector only. Membranes were obtained from cells induced by IPTG addition to cultures grown at 30°C. The membranes were assayed in the presence of 50 mm Tris-HCl, pH 8, 1 mm MgCl2, 55 μm LPA, 1 mm DTT, and varying concentrations of [1-14C]palmitoyl-CoA or [1-14C]oleoyl-oA, and the products of the reaction were resolved on TLC. The phosphatidic acid spot was visualized by autoradiography and collected for scintillation counting. Values are the means obtained with two individual membrane preparations. The values obtained with E. coli JC201/pBSK for palmitoyl-CoA and oleoyl-CoA were similar and are labeled as acyl-CoA.
The rate of formation of phosphatidic acid in the presence of membranes isolated from E. coli JC201 cells transformed with a vector containing the B. napus cDNA BAT2 was approximately 120-fold greater with [1-14C]palmitoyl-CoA and approximately 42-fold greater with [1-14C]oleoyl-CoA as substrates, compared with membranes isolated from E. coli JC201 cells transformed with the vector at acyl-CoA concentrations greater than 30 μm. Furthermore, the rate of formation of phosphatidic acid associated with the presence of the BAT2 cDNA sequence was significantly greater with [1-14C]palmitoyl-CoA (approximately 4.7-fold greater at saturating substrate concentrations) than with [1-14C]oleoyl-CoA. At 10 μm acyl-CoA the rate of formation of phosphatidic acid was approximately 2.2-fold greater with [1-14C]palmitoyl-CoA than with [1-14C]oleoyl-CoA. These data are consistent with a preference of the enzyme for the palmitoyl-CoA substrate when 1-oleoyl-LPA was the acyl acceptor.
DISCUSSION
Heterologous complementation in auxotrophic mutants of E. coli by eukaryotic cDNA expression libraries is a powerful approach for the isolation of genes not suitable for the conventional techniques of protein purification, and requires functional conservation of the mutated E. coli gene and its eukaryotic counterpart (Delauney and Verma, 1990). Although not all genes are suitable for this approach, LPAATs involved in the biosynthesis of triacylglycerols have been successfully isolated by complementation (Brown et al., 1995; Hanke et al., 1995). By adopting a similar approach, we have isolated a B. napus cDNA encoding a plastidial acyltransferase homologous to prokaryotic LPAATs. It is intriguing that our complementation screen resulted in the isolation of a plastidial LPAAT isoform only. This indicates, at least under our conditions of selection, that the plastidial enzyme was the most efficient in providing a functional substitution for the LPAAT deficiency, and is consistent with the sequence similarity between the plastidial and bacterial proteins. The abundance of cDNA does not seem to have been an important factor, since the BAT2 mRNA was not highly expressed (as evidenced by northern analysis and the absence of BAT2 homologs in expressed sequence tag databases).
The BAT2 cDNA Encodes a Plastidial LPAAT
Several lines of evidence confirm that the protein encoded by BAT2 is a LPAAT. A selection based on restoration of growth at 42°C of the temperature-sensitive, acyltransferase-deficient mutant of E. coli, JC201 (Coleman, 1990), allowed us to isolate a cDNA with a deduced protein sequence that revealed homologies with sequences known to possess LPAAT activity (Fig. 1). The strongest similarities were with the yeast SLC1 gene product (Nagiec et al., 1993) and plant microsomal LPAATs (Hanke et al., 1995; Knutzon et al., 1995; Lassner et al., 1995). We therefore conclude that the BAT2 cDNA encodes an activity that functionally compensates for the LPAAT deficiency, and thus complements the lesion associated with the temperature-sensitive phenotype of E. coli JC201. Because of the presence of a functional chloroplast-targeting sequence, the recovery of a corresponding genomic sequence from plant DNA, the isolation of pBAT2 from two independently constructed cDNA libraries of different populations of plants, and the strength of hybridizing signals on high-stringency genomic Southern blots, we believe that we have cloned an authentic plant cDNA and not an LPAAT from a contaminating endophyte, despite the sequence similarities among the BAT2, prokayotic, and yeast LPAATs.
Like the E. coli, yeast, and plant LPAATs (Lassner et al., 1995), the hydropathy profile of the deduced BAT2 protein indicates the presence of a minimum of three transmembrane-spanning domains. Such structural features of the BAT2 protein are consistent with results of protein purification confirming the LPAATs to be integral membrane proteins (Knutson et al., 1995). Protein-import assays (Fig. 3) suggest that the BAT2 protein may be imported into the chloroplast and localized in the total chloroplastic membrane fraction. Chloroplast glycerolipids are synthesized at the membranes of the plastid envelope, and chloroplastic LPAAT has been purified from inner and outer envelope membranes of spinach and from the inner envelope of pea chloroplasts (Douce and Joyard, 1990).
Taken together, these observations are consistent with the BAT2 protein being the LPAAT that incorporates an acyl group derived from an acyl-ACP thioester at the sn-2 position of a LPA-acceptor molecule during glycerolipid biosynthesis in the plastid. This enzyme, isolated from both C16:3 and C18:3 plants, has been shown to exhibit a nearly exclusive preference for palmitoyl groups (Frentzen et al., 1983). Consistent with this assignment, membrane fractions isolated from the acyltransferase-deficient E. coli mutant JC201 transformed with a plasmid containing the BAT2 sequence were demonstrated to possess LPAAT activity. Furthermore, the LPAAT activity was at least 4.7-fold greater with a palmitoyl-CoA substrate than with a oleoyl-CoA substrate (Fig. 6). The apparently ubiquitous expression of BAT2 and the presence of BAT2 transcripts in root tissues is consistent with the autonomy of nonphotosynthetically active tissues for glycerolipid biosynthesis (Browse and Slack, 1985; Stahl and Sparace, 1990).
BAT2 encodes the acyltransferase ultimately responsible for the production of hexadecatrienoic acid, which is present in substantial amounts at position sn-2 of galactolipids produced by the prokaryotic pathway and is characteristic of C16:3 plants. The availability of a plant deficient in chloroplastic LPAAT activity would confirm the assignment of the identity of BAT2. Such a mutant may be expected to have a phenotype similar to that of the Arabidopsis act1 mutant (Kunst et al., 1988), which is characterized by a deficiency in chloroplastic glycerol-3-P acyltransferase activity and results in a loss of the prokayotic pathway that is compensated for by the increased synthesis of lipids in the cytoplasmic pathway. The lesion effectively converts the C16:3 plant into a C18:3 plant. To our knowledge, no mutant of the chloroplastic LPAAT has been identified to date, and other Arabidopsis mutants with a similar fatty acid composition appear to be allelic to act1. It is possible that a lesion in chloroplastic LPAAT may be lethal because of accumulation of phosphatidic acid or depletion of available plastidial glycerol-3-P.
Two Classes of Higher Plant LPAAT
To date, three LPAAT activities have been characterized, appearing in the plastidial, mitochondrial, and cytoplasmic compartments of plant cells (Frentzen, 1993). In addition, cytoplasmic isozymes have been cloned from seed tissues (Brown et al., 1994; Hanke et al., 1995; Knutzon et al., 1995; Lassner et al., 1995). The isolation of the plastidial form confirms that at least two distinct classes of LPAAT sequences exist within higher plants. Despite the plastidial location and the difference in substrate preference, the protein encoded by BAT2 shares a strong identity to the microsomal LPAATs of seeds (Brown et al., 1995; Hanke et al., 1995). The similarity of the plastidial BAT2 sequence to that of a homologous sequence present in the genome of the blue-green algae Synechocystis sp. suggests a common progenitor gene and a direct line of descent to the ancestral chloroplastic gene (and its subsequent transfer to the nucleus).
The structural and functional similarity (evidenced by the complementation of the plsC lesion) of the plastidial sequence with that of microsomal LPAATs indicates that a duplication may have occurred, as well as the recruitment of the plastidial housekeeping gene for specialization of function in oilseeds. In this manner, enzymes with a substrate preference for the particular acyl group present at sn-2 of phosphatidic acid destined for triacylglycerol synthesis may have evolved. Molecular phylogenetic analysis using the DARWIN program (Gonnet et al., 1992) revealed the evolutionary proximity of the BAT2 protein to the Synechocystis sp. protein and that of the seed microsomal LPAATs to the prokaryotic LPAATs (data not shown); thus, the chloroplastic and ER forms involved in glycerolipid biosynthesis may constitute paralogous sequences. The precedent of the evolutionary relationships between seed-specific and ubiquitous acyl-ACP-thioesterases provides support for this hypothesis (Jones et al., 1995).
The second sequence type, represented by a LPAAT encoded by the BAT1 cDNA of B. napus (accession no. Z49860), a homolog of the maize MAT1 sequence (Brown et al., 1994), and the meadowfoam LAT1 protein (Brown et al., 1995), shares little identity with the protein encoded by BAT2. What, then, is the evolutionary relationship and role of the BAT1-type enzyme? The ubiquitously expressed transcript encoding the BAT1 protein is not imported into chloroplasts, but is probably located at the ER (data not shown), and thus may be involved in the production of phosphatidic acid by the eukaryotic pathway. The low sequence identity between BAT1 and BAT2 LPAATs of the same species suggest that these may be analogous sequences. The ubiquitous microsomal LPAAT of the eukaryotic pathway may have a different evolutionary origin, since no homologs of BAT1 are known to exist in the cyanobacterial genome, and molecular phylogenetic analysis reveals an evolutionary substantial distance from other LPAATs.
Biotechnological Applications for the Plastidial LPAAT
The acyl composition of seed oils is determined by the substrate specificities of the three microsomal acyltransferases of the Kennedy pathway, together with the composition of the donor acyl-CoA pool. The microsomal LPAATs that control the acylation at position sn-2 show species variation in substrate specificity. In B. napus, oleoyl-CoA is the preferred substrate, whereas in contrast, the LPAAT of meadowfoam incorporates C22:1 at position sn-2 of LPA. Similarly, the coconut enzyme displays a different specificity, accepting medium-chain acyl groups. It is suggested that expression of LPAAT with the appropriate substrate specificities in transgenic B. napus will alter the stereochemical composition of its seed oil. Transgenic plants expressing the plastidial enzyme redirected to the ER, together with a seed acyl-CoA pool enriched with C16:0 groups, may result in high-palmitoyl oils. The availability of the plastidial sequence is expected to provide insight into the structure and function relationships of LPAATs to aid protein engineering of enzymes possessing desired substrate and kinetic parameters.
ACKNOWLEDGMENTS
We are grateful to Dr. Jack Coleman of the Louisiana State University School of Medicine (New Orleans) for his generous gift of E. coli JC201. The authors also thank Dr. Martine Devic for critical appraisal of this manuscript and the Centre National de la Recherche Scientifique for financial support.
Abbreviations:
- ACP
acyl-carrier protein
- cM
centimorgan
- LPA
lysophosphatidic acid (1-acyl-glycerol-3-P)
- LPAAT
LPA acyltransferase
- IPTG
isopropyl-1-thio-β-d-galactopyranoside
- ORF
open reading frame
- RFLP
restriction fragment length polymorphism
The nucleotide sequence corresponding to the BAT2 cDNA reported in this article has been entered in the database under accession no. A111161.
Footnotes
This work was supported by a grant from the French Ministère de la Recherche et de la Technologie (no. 1994G0090) and the Organisation Nationale Interprofessionelle des Oléagineux, Paris.
LITERATURE CITED
- Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K, (1992) Short Protocols in Molecular Biology, Ed 2. Greene Publishing Associates and John Wiley, New York, Unit 1.8, pp 26–27
- Barret P, Delourme R, Renard M, Domergue F, Lessire R, Delseny M, Roscoe TJ. A B. napus FAE1 gene is linked to the E1 locus associated with variation in the content of erucic acid. Theor Appl Genet. 1998;96:177–186. [Google Scholar]
- Brown AP, Brough CL, Kroon JTM, Slabas AR. Identification of a cDNA that encodes a 1-acyl-sn-gylcerol-3-P acytransferase from Limnanthes douglasii. Plant Mol Biol. 1995;29:267–278. doi: 10.1007/BF00043651. [DOI] [PubMed] [Google Scholar]
- Brown AP, Coleman J, Tommy AM, Watson MD, Slabas AR. Isolation and characterisation of a maize cDNA that complements a 1-acyl-sn-glycerol-3-P acyltransferase mutant of Escherichia coli and encodes a protein which has similarities to other acyltransferases. Plant Mol Biol. 1994;26:211–223. doi: 10.1007/BF00039533. [DOI] [PubMed] [Google Scholar]
- Browse J, Slack CR. Fatty acid synthesis in plastids from maturing safflower and linseed cotyledons. Planta. 1985;166:74–80. doi: 10.1007/BF00397388. [DOI] [PubMed] [Google Scholar]
- Browse J, Somerville C. Glycerolipid synthesis: biochemistry and regulation. Annu Rev Plant Physiol Mol Biol. 1991;42:467–506. [Google Scholar]
- Cao Y, Oo KC, Huang AHC. Lysophosphatidate acyltransferase in the microsomal fraction from maturing seeds of meadowfoam (Limnanthes alba) Plant Physiol. 1990;9:1199–1206. doi: 10.1104/pp.94.3.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman J. Characterisation of Escherichia coli cells deficient in 1-acyl-sn-glycerol-3-P acyltransferase activity. J Biol Chem. 1990;265:17215–17221. [PubMed] [Google Scholar]
- Davies HM, Hawkins DJ, Nelson JS. Lysophosphatidic acid acyltransferase from coconut endosperm having medium chain length substrate specificity. Phytochemistry. 1995;39:989–996. [Google Scholar]
- Delauney AJ, Verma DPS (1990) Isolation of plant genes by heterologous complementation in Escherichia coli. In SB Gelvin, RA Schilperoort, DPS Verma, eds, Plant Molecular Biology Manual. Kluwer Academic Publishers, Dordrect, The Netherlands, Section A14, pp 1–23
- Devic M, Albert S, Delseny M, Roscoe TJ. Efficient PCR walking on plant genomic DNA. Plant Physiol Biochem. 1997;35:331–339. [Google Scholar]
- Douce R, Joyard J. Biochemistry and function of the plastid envelope. Annu Rev Cell Biol. 1990;6:173–216. doi: 10.1146/annurev.cb.06.110190.001133. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of Plant DNA from fresh tissues. Focus. 1990;12:13–15. [Google Scholar]
- Foisset N, Delourme R, Barret P, Hubert N, Landry BS, Renard M. Molecular-mapping analysis in Brassica napus using isozyme, RAPD and RFLP markers on a double haploid progeny. Theor Appl Genet. 1996;93:1017–1025. doi: 10.1007/BF00230119. [DOI] [PubMed] [Google Scholar]
- Frentzen M. Acyltransferases and triacylglycerols. In: Moore TS, editor. Lipid Metabolism in Plants. Boca Raton, FL: CRC Press; 1993. pp. 195–231. [Google Scholar]
- Frentzen M, Heinze E, McKeon TA, Stumpf PK. Specificities and selectivities of glycerol-3-P and 1-acyl-glycerol-3-P acyltransferases in pea and spinach chloroplasts. Eur J Biochem. 1983;129:629–636. doi: 10.1111/j.1432-1033.1983.tb07096.x. [DOI] [PubMed] [Google Scholar]
- Gish W, Slates DJ. Identitification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- Gonnet GM, Cohen MA, Benner SA. Exhaustive matching of the entire protein database. Science. 1992;256:1443–1445. doi: 10.1126/science.1604319. [DOI] [PubMed] [Google Scholar]
- Hanke C, Peterek G, Wolter FP, Frentzen M. A plant acyltransferase involved in triacylglycerol biosynthesis complements an Escherichia coli sn-1-acyl-glycerol-3-P acyltransferase mutant. Eur J Biochem. 1995;232:806–810. [PubMed] [Google Scholar]
- Henderson PJ, Macpherson AJ. Assay, genetics, proteins and reconstitution of proton linked galactose, arabinose and xylose transport systems of Escherichia coli. Methods Enzymol. 1986;125:387–429. doi: 10.1016/s0076-6879(86)25033-8. [DOI] [PubMed] [Google Scholar]
- Jones A, Davies HM, Volker TA. Palmitoyl-ACP thioesterase and the evolutionary origin of plant acyl-ACP thioesterases. Plant Cell. 1995;7:359–371. doi: 10.1105/tpc.7.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdren C, Barret P, Horvais R, Foisset N, Delourme R, Renard M, (1996) Identification of RAPD markers linked to the loci controlling erucic acid level in B. napus. Mol Breed 2: 61–71
- Kay R, Chan A, Daly M, McPherson J. Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science. 1987;236:1299–1302. doi: 10.1126/science.236.4806.1299. [DOI] [PubMed] [Google Scholar]
- Keegstra K, Olsen LJ, Theg M. Chloroplast precursors and their transport across the envelope. Annu Rev Plant Physiol Mol Biol. 1989;40:471–501. [Google Scholar]
- Knutzon DS, Lardizabal KD, Nelsen JS, Bleibaum JL, Davis HM, Metz JG. Cloning of a coconut endosperm cDNA encoding a 1-acyl-sn-1-acyl-glycerol-3-P acyltransferase that accepts medium chain length substrates. Plant Physiol. 1995;109:999–1006. doi: 10.1104/pp.109.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Kunst L, Browse J, Somerville C. Altered regulation of lipid biosynthesis in a mutant of Arabidopsis deficient in chloroplast glycerol-3-P acyltransferase activity. Proc Nat Acad Sci. 1988;85:4143–4147. doi: 10.1073/pnas.85.12.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Lassner MW, Levering CK, Davies HM, Knutzon DS. Lysophosphatidic acid acyltransferase from meadowfoam mediates insertion of erucic acid at the sn-2 position of triacylglycerol in transgenic B. napus oil. Plant Physiol. 1995;109:1389–1394. doi: 10.1104/pp.109.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutcke HA, Chow KC, Mickel FS, Moss KA, Kern HF, Scheele GA. Selection of AUG codons differs in plants and animals. EMBO J. 1987;6:43–48. doi: 10.1002/j.1460-2075.1987.tb04716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagiec MM, Wells GB, Lester RL, Dickson RC. A suppressor gene that enables Saccharomyces cerevisiae to grow without making phospholipids encodes a protein that resembles an Escherichia coli fatty acyltransferase. J Biol Chem. 1993;268:22156–22163. [PubMed] [Google Scholar]
- Nakai K, Horton P. PSORT: a programme for detecting the sorting signals of proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–35. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Ohlrogge J, Browse J. Lipid biosynthesis. Plant Cell. 1995;7:957–970. doi: 10.1105/tpc.7.7.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge JB, Jaworski JG, Post-Beittenmiller D (1993) De novo fatty acid biosynthesis. In TS Moore Jr, ed, Lipid Metabolism in Plants. CRC Press, Boca Raton, FL, pp 3–32
- Robinson C. Import of in vitro synthesised proteins into intact chloroplasts and isolated thylakoids. In: Bryant JA, editor. Methods in Plant Biochemistry. London: Academic Press; 1993. pp. 207–219. [Google Scholar]
- Sharpe AG, Parkin AP, Keith DJ, Lydiate DJ. Frequent non reciprocal translocations in the amphidiploid genome of B. napus (Brassica napus) Genome. 1995;38:1112–1121. doi: 10.1139/g95-148. [DOI] [PubMed] [Google Scholar]
- Stahl RJ, Sparace SA (1990) Fatty acid and glycerolipid biosynthesis in isolated pea root plastids. In PJ Quinn, JL Harwood, eds, Plant Lipid Biochemistry, Structure and Utilisation. Portland Press, London, pp 154–156
- Sun C, Cao YZ, Huang AHC. Acyl coenzymeA preference of the glycerol phosphate pathway in the microsomes from the maturing seeds of palm, maize and B. napus. Plant Physiol. 1988;88:56–60. doi: 10.1104/pp.88.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]