Abstract
Although endometrial adenocarcinoma is usually treated with surgery, patients with metastatic disease have a poor prognosis. To address the need for better treatment options, molecularly targeted drug therapies are being developed. These targeted therapies rely on accurate mutational profiling of the tumor, which is most often performed on DNA from the primary tumor. Our objective was to compare mutational concordance in primary tumors with their metastases. We genotyped 11 pairs of primary and metastatic endometrial adenocarcinomas using DNA from formalin-fixed paraffin-embedded tissue blocks and semiconductor-based next-generation sequencing. Five of these cases had multiple metastases for comparison. We sequenced 37 known cancer genes that are targets for new drug therapies. A total of 62 mutations were identified in 16 of these 37 genes. The most common mutations were in PIK3CA and PTEN. Overall, there was a 53% discordance in mutations between primary tumors and their paired (33 of 62). The absence of mutations in metastases (25 of 33, 76%) compared with the primary neoplasm was more common than gain of mutations (8 of 33, 24%). There was a 15% discordance rate between paired metastases within individuals (6 of 40), which was significantly less frequent than the rate between primary tumors and their metastases (Fisher exact P value <.0001). Although the sample size is relatively small, our data suggest it may be prudent to test metastases, rather than the primary neoplasm, when using molecularly targeted drug therapies, because isolated metastases may lack mutations detected in the heterogeneous mixture of the tumor’s origin.
Keywords: endometrial adenocarcinoma, Ion Torrent, mutational profiling
Introduction
Both the incidence and mortality of endometrial cancer are on the rise with an estimated 52 630 women diagnosed with endometrial cancer in 2014 and 8590 died of their disease.1 Women with early-stage disease are usually cured with surgery, but women with metastatic or recurrent disease have few therapeutic options.2 There is a clear need for new therapies for high-stage and recurrent disease, and molecularly targeted treatments are being explored in this setting.3–10 However, most molecular genetic studies of endometrial carcinoma have concentrated on readily available primary tumors and not on metastatic disease.
This is potentially a problem since tumor metastases are driven by mutations with genetic heterogeneity,11 which may lead to disparate genetic mutational profiles in primary and metastatic tumors.12–15 Our objective was to compare primary endometrial tumors with their paired metastases in a retrospective series of 11 cases and test for mutational discordance using a rapid cancer genotyping sequencing technology well suited for clinical testing.
Materials and Methods
Study Patients
In accordance with an institutional review board-approved protocol (IRB#8767) of Oregon Health & Science University (OHSU), we performed a retrospective review of all endometrioid endometrial adenocarcinomas with biopsy-proven metastatic disease diagnosed at our institution from 2005 to 2013. Cases of carcinosarcoma, clear cell carcinoma, uterine papillary serous carcinoma, and dedifferentiated carcinoma were excluded. Tumor grade was confirmed by surgical pathologist (T. K. M.), and clinical outcomes through November 2015 were documented from patient charts (Table 1). Archival formalin-fixed paraffin-embedded (FFPE) tissue blocks of the primary tumors and their metastasis(es) were used to core out tumor-rich areas (>90% of cells in coring field) for DNA extraction using the QIAamp DNA FFPE Tissue Kit in accordance with the manufacturer’s instructions (QIAGEN Inc, Valencia, California). For each sample, 2 to 3 cores were taken per tissue block for DNA extraction and the DNA was pooled to limit potential heterogeneity within the coring field. We did not test for genetic differences between independent cores within samples.
Table 1.
Characteristics of Patients With Primary Endometrial Adenocarcinoma.a
| Case | Age, years | Stage at Dx | Metastases/Recurrence | Outcome 2015 |
|---|---|---|---|---|
| 1 | 67 | II | Pelvic recurrence at 30 months | Persistent disease |
| 2 | 60 | IB | Pelvic lymph nodes at 4 months | Dead of disease |
| 3 | 72 | IIIA | Pelvic and periaortic lymph nodes | Persistent disease |
| 4 | 62 | IB | Vaginal cuff at 6 months | Persistent disease |
| 5 | 77 | IIIC | Right and left pelvic and periaortic | Persistent disease |
| 6 | 60 | IIIC | Common iliac lymph node | Persistent disease |
| 7 | 65 | IIIC | Pelvic and periaortic lymph nodes | Persistent disease |
| 8 | 63 | IIIC | Pelvic lymph node | Persistent disease |
| 9 | 50 | IVA | Omentum, pelvic, and periaortic nodes | Persistent disease |
| 10 | 59 | IIIC | Pelvic and periaortic lymph nodes | Persistent disease |
| 11 | 53 | IIIA | Ovary | Persistent disease |
Abbreviation: Dx, diagnosis.
aPrimary tumors were well to moderately differentiated at the time of diagnosis. There were no cases of uterine papillary serous carcinoma, clear cell carcinoma, or carcinosarcoma. There was no intervening chemotherapy in the 3 cases of low-stage disease with recurrence.
DNA Sequencing
We used the custom GeneTrails Solid Tumor Panel, which was sequenced on an Ion Torrent PGM (Thermo Fisher).16 The panel covers 37 genes (AKT1, AKT2, AKT3, ALK, BRAF, CDK4, CDKN2A, DDR2, EGFR, ERBB2 [HER2], FGFR1, FGFR3, GNA11, GNAQ, GNAS, HRAS, KDR, KIT, KRAS, MAP2K1 [MEK1], MET, NF1, NOTCH1, NRAS, NTRK2, NTRK3, PIK3CA, PIK3R1, PTEN, RAC1, RB1, RET, STK11, TP53, TSC1, TSC2, and VHL). For 20 of these genes, the presence of a mutation would justify the use of a Food and Drug Administration-approved therapeutic, either on-label or off-label. Alterations in 14 of the remaining 17 genes would support enrollment in a clinical trial using compounds currently in development. The other 3 genes are under investigation. The average read depth was 1000 reads, and the lower limit of detection for mutant alleles was 5%.16 The lower limit of detection is important to exclude potential polymerase chain reaction (PCR) errors registering as false positives, which may occur with Ion Torrent technology below a limit of 3%.
Statistical Analysis
Fisher exact test was performed using StatView software version 5.0 (SAS Institute, Cary, North Carolina) to compare the discordance frequency in paired primary and metastases in the cohort to paired metastases within individuals. P < .05 is significant.
Results
We identified 11 cases with paired metastases for genetic analysis. Three represented recurrent disease (within 4-30 months of primary tumor resection) and 8 were metastatic tumor collected at the same time as the primary tumor. All primary tumors were grade 1 to 2 endometrioid adenocarcinomas (Table 1). The metastases were similarly well to moderately differentiated without squamous differentiation (there were no histopathologic differences between the primary tumors and their corresponding metastases). The median age for the cohort was 62 years with a range of 50 to 77 years. For the patients with recurrent disease, no adjuvant therapy was given between the primary surgery and the recurrence. One patient died of disease and 10 patients are alive with disease with a median follow-up of 5 years. Six of the 11 had a single paired metastasis, 3 had 2 metastatic foci, and 2 patients had 3 metastatic foci for analysis.
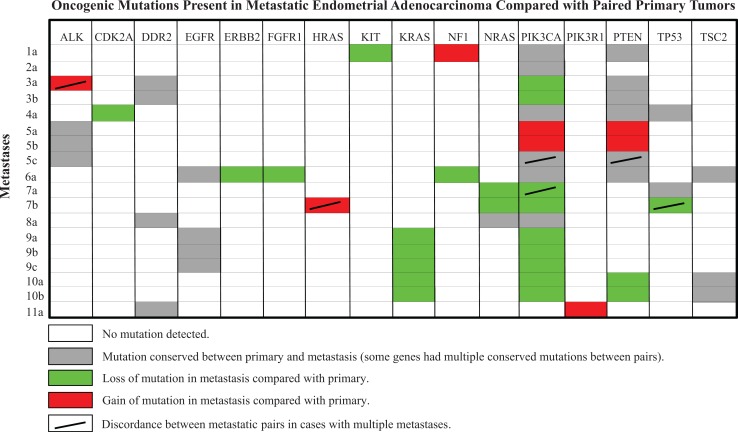
The most common mutations were in PIK3CA (10 of the 11 primary tumors) and PTEN (6 of the 11). Twenty-one of the 37 genes sequenced lacked mutations. Sixteen of the 37 had at least 1 mutation in the primary tumor, their metastases, or both, for a total of 62 mutations identified. Mutations in PIK3CA were the most discordant between primaries and metastases (5 of the 11 cases), being absent in metastatic lesions in 4 cases. Discrepancies in RAS gene mutations (KRAS, HRAS, and NRAS) were observed in 3 cases. The overall discordance rate in mutational profiles when comparing primary tumors to their metastases was 53% (33 of the 62). Compared with the primary neoplasm, a relative loss of mutations (25 of 33, 76%) was more common than a gain (8 of 33, 24%) in their metastases (Figure 1). Specific mutation sites within these genes are listed in Table 2. The discordance rate in mutations between metastases within individual patients was 15% (6 of 40), which was significantly less frequent than the discordance rate between primary tumors and their metastases (Fisher exact P value <.0001).
Figure 1.
Oncogenic mutations present in metastatic endometrial adenocarcinoma compared with paired primary tumors.
Table 2.
Specific Mutations in Paired Primary and Metastatic Foci.a
| Site | Gene | Exon | Mutation |
|---|---|---|---|
| Primary 1 | PIK3CA | 1, 4 | R88Q, V344M |
| PTEN | 4, 5 | Splice variant, R130G | |
| KIT | 17 | C788Y | |
| Metastasis 1a | PIK3CA | 1, 4 | R88Q, V344M |
| PTEN | 4, 5 | Splice variant, R130G | |
| NF1 | 37 | Y1639N | |
| Primary 2 | PIK3CA | 9 | E542K |
| Metastasis 2a | PIK3CA | 9 | E542K |
| Primary 3 | PIK3CA | 9 | R1212C |
| DDR2 | 14 | R752H | |
| PTEN | 5, 5 | R130G, frameshift | |
| Metastasis 3a | DDR2 | 14 | R752H |
| PTEN | 5, 5 | R130G, frameshift | |
| ALK | 23 | E545G | |
| Metastasis 3b | DDR2 | 14 | R752H |
| PTEN | 5, 5 | R130G, frameshift | |
| Primary 4 | PIK3CA | 18 | G914R |
| PTEN | 8 | 5′ Splice mutation | |
| TP53 | 8 | R273H | |
| CKDN2A | 2 | R138K | |
| Metastasis 4a | PIK3CA | 18 | G914R |
| PTEN | 8 | 5′ Splice mutation | |
| TP53 | 8 | R273H | |
| Primary 5 | ALK | 6 | Q459R |
| Metastasis 5a-b | ALK | 6 | Q459R |
| PIK3CA | 9 | E545K | |
| PTEN | 5 | C136R | |
| Metastasis 5c | ALK | 6 | Q459R |
| Primary 6 | PIK3CA | 20 | H1047Y |
| PTEN | 9 | L345P | |
| EGFR | 15 | T605A | |
| ERBB | 6 | S250Y | |
| FGFR | 6 | K256R | |
| NF1 | 21, 50 | Frameshift, T2475A | |
| TSC2 | 33 | S1431L | |
| Metastasis 6a | PIK3CA | 20 | H1047Y |
| PTEN | 9 | L345P | |
| EGFR | 15 | T605A | |
| NF1 | 50 | T2475A | |
| TSC2 | 33 | S1431L | |
| Primary 7 | PIK3CA | 20, 1, 1 | M1043I, R93W, R88Q |
| NRAS | 2 | Q61R | |
| TP53 | 5 | A159V | |
| Metastasis 7a | PIK3CA | 20 | M1043I |
| TP53 | 5 | A159V | |
| Metastasis 7b | PIK3CA | 20, 1 | M1043I, R93W |
| HRAS | 2 | E63 deletion | |
| Primary 8 | PIK3CA | 20 | H1047R |
| NRAS | 2 | Q61L | |
| DDR2 | 12 | E583D | |
| Metastasis 8a | PIK3CA | 20 | H1047R |
| NRAS | 2 | Q61L | |
| DDR2 | 12 | E583D | |
| Primary 9 | PIK3CA | 9 | Q546R |
| KRAS | 1 | G12A | |
| EGFR | 12 | I462M | |
| Metastasis 9a-c | EGFR | 12 | I462M |
| Primary 10 | PIK3CA | 20 | H1047R |
| PTEN | 6 | G165 | |
| KRAS | 1 | G12V | |
| TSC2 | 39 | R1706H | |
| Metastasis 10a-b | TSC2 | 39 | R1706H |
| Primary 11 | DDR2 | 15 | T782A |
| PIK3R1 | 12 | Q579Y | |
| Metastasis 11a | DDR2 | 15 | T782A |
| PIK3R1 | 12, 1 | Q579Y, frameshift |
aThe authors observed an overall 53% discordance rate (33 of 62) in mutational profiles when comparing primary tumors to their metastases with a relative loss (25 of 33, 76%) rather than a gain (8 of 33, 24%) of mutations in metastases. There was a 15% discordance rate in paired metastases (6 of 40), which was significantly less frequent than discordance between primaries and metastases (Fisher exact P value <.0001). Italics represent different mutation in same gene.
Discussion
In this study, we report a discordance rate of 53% in the mutational profile of 37 potentially actionable target genes between primary grade 1 to 2 endometrioid adenocarcinomas of the endometrium and their metastases. The predominant pattern was loss of 1 mutation in the metastatic site compared to the primary tumor. The discordance rate was significantly less between paired metastatic sites. Although the sample size is relatively small, our data suggest it may be best to use the genetic profile of metastases, rather than the primary neoplasm when designing mutation-based targeted drug therapies.
Genetic discordance between matched primary and metastatic tumors is a common phenomenon and is observed in a variety of cancers,12,13 but there are little data for endometrial adenocarcinoma.17 Krakstad et al used mass spectrometric-based mutation screening and found that PIK3CA and K-RAS were the most frequently mutated genes in primary endometrial adenocarcinomas.17 Of their 67 primary tumors, 7 had paired metastases. Three (43%) of the pairs had no change in mutation status, 3 (43%) showed a loss of a mutation, and 1 (14%) showed a gain. These percentages are similar to our observations in 11 paired cases, with predominantly a loss of mutations in the metastases. This somewhat unexpected loss of mutation has also been described for metastatic breast cancer.18
Nonetheless, it is interesting that our data suggest a relative absence of driver mutations in metastases compared with the primary neoplasm in some cases. The most likely explanation is the subset of tumor cells comprising the metastasis arose from a heterogeneous mixture of mutations identified in the primary, but by chance tumor cells comprising the metastasis did not have the mutations screened for in our panel. That is to say, the clonal expansion in the metastasis came from a cell within the primary that did not have the mutation in question in the first place. Certainly, metastatic tumor cells may have acquired other oncogenic driver mutations contributing to their metastatic potential that was not specifically tested for in our experiment.11 It is of course unlikely that the metastatic tumor cells reverted from mutation back to normal, but we did consider the possibility that tumor cell sampling for genetic analyses may have missed the tumor and solely sampled nonneoplastic elements of lymph nodes. Our experience and careful tissue sampling for DNA extraction,16 and the fact that all of the metastases tested had at least 1 oncogenic mutation, are reassuring.
Among 5 cases with multiple metastases within individuals, comparison between the metastases showed an overall concordance rate of 85% (34 of 40 mutations). Discordance in PIK3CA mutations occurred in 2 of these cases. Krakstad et al had 2 cases with multiple metastases and observed 80% concordance (4 of 5 mutations) with 1 loss of PIK3CA mutation.17 Thus, loss of PIK3CA mutations in metastases may be relatively common in endometrial cancer. This is important because PI3K inhibitors have generated great interest for the treatment of endometrial cancer with numerous registered trials. Notably, PTEN mutations were more likely to be conserved in metastases, with 1 additional case showing a gain of PTEN mutation. This is interesting because PTEN is accepted as the most commonly mutated gene in endometrial cancer and has been previously associated with metastatic endometrial cancer.19
The observation that PIK3CA mutations may be lost in metastatic or recurrent tumor sites may offer an explanation as to why PIK3CA mutations observed in the primary tumor do not reliably predict PI3K inhibitor response and are associated with a less favorable prognosis.8,9 In turn, our data support the assertion that mutational analysis of the recurrent or metastatic tumor—if at all possible—may be more informative to guide molecularly targeted therapies. Since the objective of targeted therapy is to treat the metastases and not the previously resected primary neoplasm, it is clear that the mutational profiles of metastatic tumors are required. A significant problem may be the oncogenic heterogeneity of independent metastases within an individual. Our data support the assertion that different metastases within a woman may show differences in loss and gain of oncogenic mutations compared with each other. In turn, what may be needed is more general assessment of the entire individual, perhaps free tumor DNA within plasma may provide better coverage of clinically relevant mutational profiles than sampling single metastatic foci.
Although the study sample size is relatively small, our results compare favorably with those reported by Krakstad et al17 and contribute to this growing body of literature. Moreover, the genes sequenced in our study are all potentially actionable in current regimens or clinical trials. In addition, we report the use of semiconductor-based sequencing of highly multiplexed PCR amplicons, which requires only 10 ng of DNA and it works well with DNA prepared from archived paraffin tissue blocks. Target preparation, sequencing, and data analysis may all be performed within 48 hours, which is well suited for rapid turnaround times and clinical testing.
Acknowledgments
The authors would like to thank the members of the Knight Diagnostic Laboratory for their expertise and support.
Authors’ Note: The article was presented at the Society of Reproductive Investigation (SRI) annual meeting in San Francisco, California, 2015.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to this research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was funded in part by the Oregon Clinical & Translational Research Institute (OCTRI), grant number TL1 RR024159, Robert L. Bacon Medical Education Enrichment Award, the N. L. Tartar Trust Fellowship 2014, and an American Medical Association Seed Grant 2015.
References
- 1. American Cancer Society. Cancer Facts & Figures 2014. Atlanta, GA: American Cancer Society; 2014. [Google Scholar]
- 2. Howlader N, Noone AM, Krapcho M, et al. eds. SEER Cancer Statistics Review, 1975-2013. Bethesda, MD: National Cancer Institute, http://seer.cancer.gov/csr/1975_2013/. [Google Scholar]
- 3. Levine RL, Cargile CB, Blazes MS, van Rees B, Kurman RJ, Ellenson LH. PTEN mutations and microsatellite instability in complex atypical hyperplasia, a precursor lesion to uterine endometrioid carcinoma. Cancer Res. 1998;58(15):3254–3258. [PubMed] [Google Scholar]
- 4. Oda K, Stokoe D, Taketani Y, McCormick F. High frequency of coexistant mutations of PIK3CA and PTEN genes in endometrial carcinoma. Cancer Res. 2005;65(23):10669–10673. [DOI] [PubMed] [Google Scholar]
- 5. Slomovitz BM, Broaddus RR, Burke TW, et al. Her-2/neu overexpression and amplification in uterine papillary serous carcinoma. J Clin Oncol. 2004;22(15):3126–3132. [DOI] [PubMed] [Google Scholar]
- 6. Konecny GE, Santos L, Winterhoff B, et al. HER2 gene amplification and EGFR expression in a large cohort of surgically staged patients with nonendometrioid (type II) endometrial cancer. Br J Cancer. 2009;100(1):89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lax SF, Kendall B, Tashiro H, Slebos RJ, Hedrick L. The frequency of p53, K-ras mutations, and microsatellite instability differs in uterine endometrioid and serous carcinoma: evidence of distinct molecular genetic pathways. Cancer. 2000;88(4):814–824. [PubMed] [Google Scholar]
- 8. Garcia-Dios DA, Lambrechts D, Coenegrachts L, et al. High-throughput interrogation of PIK3CA, PTEN, KRAS, FBXW7 and TP53 mutations in primary endometrial carcinoma. Gynecol Oncol. 2013;128(2):327–334. [DOI] [PubMed] [Google Scholar]
- 9. Dedes KJ, Wetterskog D, Ashworth A, Kaye SB, Reis-Filho JS. Emerging therapeutic targets in endometrial cancer. Nat Rev Clin Oncol. 2011;8(5):261–271. [DOI] [PubMed] [Google Scholar]
- 10. Mirkovic J, Sholl LM, Garcia E, Lindeman N, et al. Targeted genomic profiling reveals recurrent KRAS mutations and gain of chromosome 1q in mesonephric carcinomas of the female genital tract. Mod Pathol. 2015;28(11):1504–1514. [DOI] [PubMed] [Google Scholar]
- 11. Wan L, Pantel K, Kang Y. Tumor metastasis: moving new biological insights into the clinic. Nat Med. 2013;19(11):1450–1464. [DOI] [PubMed] [Google Scholar]
- 12. Stoecklein NH, Klein CA. Genetic disparity between primary tumours, disseminated tumour cells, and manifest metastasis. Int J Cancer. 2010;126(3):589–598. [DOI] [PubMed] [Google Scholar]
- 13. Aparicio S, Caldas C, The implications of clonal genome evolution for cancer medicine. N Engl J Med. 2013;368(9):842–851. [DOI] [PubMed] [Google Scholar]
- 14. Warth A, Macher-Gooppinger S, Mulley T, et al. Clonality of multifocal nonsmall cell lung cancer: implications for staging and therapy. Eur Respir J. 2012;39(6):1437–1442. [DOI] [PubMed] [Google Scholar]
- 15. Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor Heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Beadling C, Neff TL, Heinrich MC, et al. Combining highly multiplexed PCR with semiconductor-based sequencing for rapid cancer genotyping. J Mol Diagn. 2013;15(2):171–176. [DOI] [PubMed] [Google Scholar]
- 17. Krakstad C, Birkeland E, Seidel D, et al. High-throughput mutation profiling of primary and metastatic endometrial cancers identifies KRAS, FGFR2 and PIK3CA to be frequently mutated. PLoS One. 2012;7(12):e52795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Poplawski A, Jankowski M, Erickson S, et al. Frequent genetic differences between matched primary and metastatic breast cancer provide an approach to identification of biomarkers for disease progression. Eur J Hum Genet. 2010;18(5):560–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Salvesen HB, Stefansson I, Kalvenes MB, Das S, Akslen LA. Loss of PTEN expression is associated with metastatic disease in patients with endometrial carcinoma. Cancer. 2002;94(8):2185–2191. [DOI] [PubMed] [Google Scholar]