A robust working model delineates the main axes of the divergent metabolic strategies used by Arabidopsis thaliana in response to partial or total darkening treatments.
Abstract
In plants, an individually darkened leaf initiates senescence much more rapidly than a leaf from a whole darkened plant. Combining transcriptomic and metabolomic approaches in Arabidopsis (Arabidopsis thaliana), we present an overview of the metabolic strategies that are employed in response to different darkening treatments. Under darkened plant conditions, the perception of carbon starvation drove a profound metabolic readjustment in which branched-chain amino acids and potentially monosaccharides released from cell wall loosening became important substrates for maintaining minimal ATP production. Concomitantly, the increased accumulation of amino acids with a high nitrogen-carbon ratio may provide a safety mechanism for the storage of metabolically derived cytotoxic ammonium and a pool of nitrogen for use upon returning to typical growth conditions. Conversely, in individually darkened leaf, the metabolic profiling that followed our 13C-enrichment assays revealed a temporal and differential exchange of metabolites, including sugars and amino acids, between the darkened leaf and the rest of the plant. This active transport could be the basis for a progressive metabolic shift in the substrates fueling mitochondrial activities, which are central to the catabolic reactions facilitating the retrieval of nutrients from the senescing leaf. We propose a model illustrating the specific metabolic strategies employed by leaves in response to these two darkening treatments, which support either rapid senescence or a strong capacity for survival.
In the earliest stages of their development, emerging leaves are metabolic sinks that require nutrients from the rest of the plant to adequately sustain their growth. Once leaves are established, they progressively transition from a sink state to that of a metabolic source, in turn supporting the needs of the rest of the plant. As photoautotrophs, it is advantageous for plants to maintain leaves only for as long as they contribute to their survival, notably through the delivery of reduced carbon compounds. Thus, if the functional capacity of a leaf is deleteriously affected by developmental or external factors (abiotic or biotic), targeted leaf senescence can be initiated (Smart, 1994; Gan and Amasino, 1997; Thompson et al., 2004; Brouwer et al., 2012).
One of the most prevalent methods used to initiate a synchronized senescence under laboratory conditions is to grow plants in prolonged dark conditions. Research on this topic spans many decades and encompasses the myriad ways that darkness can influence plant phenotype, physiology, metabolism, and gene expression (Quinlan and Weaver, 1969; Stitt et al., 1985; Irving and Silsbury, 1988; Lee and Titus, 1992; Weaver and Amasino, 2001; van der Graaff et al., 2006; Keech et al., 2007; Sakuraba et al., 2012). Nonetheless, it appears that the term dark-induced senescence has long been used as a generic name in scientific studies that utilize darkness as a stress, leading to dramatically different results. In short, three experimental approaches are commonly employed: (1) the whole plant is placed in darkness (i.e. darkened plant [DP]); (2) one or a few leaves are darkened while the rest of the plant remains exposed to a normal photoperiodic cycle (i.e. individually darkened leaf [IDL]); and (3) a leaf or leaf disc is placed on flotation medium such as water and left in darkness. While the first two approaches maintain plant-to-organ communication, the third option clearly abolishes this connection, which may be problematic when studying traffic-dependent processes. In a set of studies comparing leaves subjected to DP or IDL conditions, Weaver and Amasino (2001) delineated these two treatments by establishing that IDL conditions triggered rapid leaf senescence while DP conditions actually inhibited the darkness-dependent induction of leaf senescence. Later, two additional studies demonstrated that, following 6 d of darkening treatment, plants from both experimental frameworks displayed major physiological and structural differences. Specifically, there were differential inductions of senescence, divergent photosynthetic and respiratory capacities (Keech et al., 2007), and pronounced differences in cytoskeletal dynamics (Keech et al., 2010), with the whole process being orchestrated by a complex, light-dependent signaling interplay (Liebsch and Keech, 2016). We proposed that, in response to the lack of photosynthetic activity, the leaves from DP entered a metabolically suppressed state, referred to as a standby mode, in which photosynthetic capacities were maintained while a state of reduced respiration rates was adopted. In contrast, IDL underwent accelerated senescence, maintained high mitochondrial respiration, while photosynthetic capacity decreased rapidly. While this comparison clearly indicated that DP and IDL leaves may have different metabolic strategies in their response to darkness, what remained unclear was the nature and basis of these strategies as well as their functional significance for the induction/suppression of senescence.
In order to tackle this question and improve our understanding of the metabolic adjustments occurring in leaves during either IDL or DP treatment, we conducted a comprehensive transcriptomic and metabolomic analysis of these two experimental frameworks in Arabidopsis (Arabidopsis thaliana). Additionally, to better understand the potential communication between the senescing leaf and the rest of the plant, we carried out a 13CO2-labeling time course and monitored the resulting 13C-labeled metabolites that were imported into the senescing leaf after 3 and 6 d of darkening. Ultimately, we focused on the differences observed between DP and IDL and proposed a working model that provides an overview of the different metabolic strategies employed by plants in response to these darkening treatments.
RESULTS
The Conserved Transcriptomic Responses to Darkness Accounted for More Than Half of All Significant Changes When Compared with Light
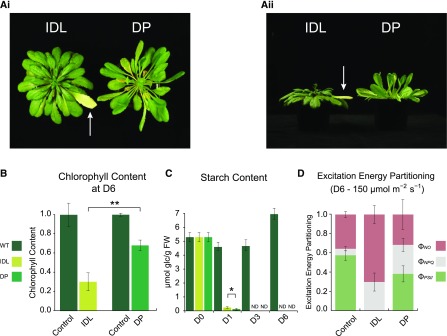
IDL and the leaves of DP showed a divergent phenotypical response to darkness. After 6 d of darkening, an individual leaf (leaves 14–20; see “Materials and Methods”) showed pronounced yellowing, while leaves of a similar age from a plant entirely darkened remained green, although paler than control leaves in the light (Fig. 1Ai). Interestingly, leaves from the DP also showed strong hyponasty with elongated petioles (Fig. 1Aii), features often characteristic of a shade-avoidance syndrome (Franklin and Whitelam, 2005). We observed that, in both darkening treatments, the relative water content (RWC) of the darkened leaves increased significantly throughout the time course as compared with light controls (Supplemental Fig. S1A). In addition, although the RWC was higher in DP samples than in IDL samples at each of the measured time points, it was only significantly higher at day 3 (P < 0.05; Student’s t test). After 6 d of darkening treatment, the chlorophyll content dropped by approximately 75% in IDL, whereas it only declined by approximately 30% in DP (Fig. 1B), when compared with control leaves in the light. These data corroborated our visual observations (Fig. 1A) as well as the results from previous studies (Keech et al., 2007; Brouwer et al., 2014). In response to darkness, starch content (from leaves sampled in the middle of their photoperiod) decreased substantially compared with light controls. Notably, at day 1 (D1), leaves exposed to IDL conditions contained significantly more starch than leaves from DP (P < 0.05; Student’s t test; Fig. 1C). In the subsequent time points (D3 and D6), starch levels in both samples from the two darkening treatments were no longer detectable. Finally, we estimated the photosynthetic efficiency of these leaves by measuring the partitioning of the excitation energy at two different light intensities. At 25 μmol m−2 s−1, a shade level slightly above the light compensation point, nearly 70% of the energy was photochemically utilized at the PSII centers, while the biggest fraction of energy dissipation occurred via nonphotochemical losses (ΦNO) rather than dissipation by down-regulation (ΦNPQ; Supplemental Fig. S1B). Following 6 d of IDL treatment, the energy quenched by the PSII centers (ΦPSII) dropped to approximately 20%, whereas it remained around 60% in DP leaves. When the actinic light was increased to 150 μmol m−2 s−1, standard light conditions for growth, an even greater loss of photosynthetic efficiency was measured. In control leaves, the ΦPSII declined slightly and the ΦNPQ increased correspondingly as compared with 25 μmol m−2 s−1. However, in IDL, no ΦPSII was recorded, whereas ΦPSII remained at nearly 40% in DP (Fig. 1D). Furthermore, as the photosynthetic photon flux density increased, a very high ΦNPQ was recorded in the two darkening treatments. Altogether, these results clearly demonstrated that, after 6 d of darkening, IDL lose their photosynthetic efficiency, whereas it is maintained at a functioning level in leaves from DP.
Figure 1.
Phenotypic and physiological analyses of Arabidopsis leaves following the darkening for 6 d of an individual leaf (IDL) and an entire plant (DP). A, Phenotypes of the leaves after 6 d of darkening treatment. The white arrows indicate the IDL. i, Top view; ii, side view. B, Chlorophyll content of leaves sampled at D6, grown under light, IDL, and DP conditions, measured in mg g−1 fresh weight and normalized to their respective wild-type (WT) control samples. Double asterisks indicate a significant (P < 0.01; Student’s t test) difference in chlorophyll abundance between the IDL and DP samples. Error bars indicate se. C, Starch content of sampled leaves grown under light, IDL, and DP conditions, measured in μmol Glc g−1 fresh weight (FW). The single asterisk indicates a significant (P < 0.05; Student’s t test) difference in starch abundance between the IDL and DP samples; ND indicates samples with nondetectable data. Error bars indicate se. D, Excitation energy partitioning at 150 μmol m−2 s−1 (growth conditions) of sampled leaves from control and after 6 d of IDL and DP conditions. Error bars indicate se.
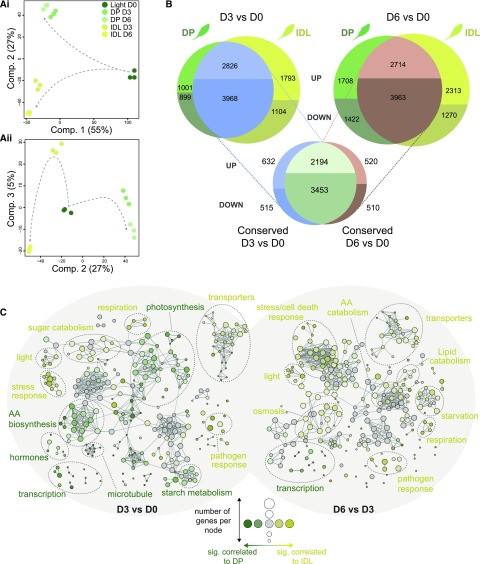
To gain a comprehensive insight into the differential regulation of metabolism between DP and IDL, we performed a microarray-based analysis using the Arabidopsis Gene 1.1 ST Array Strip technology from Affymetrix. For both experimental setups, we sampled at noon (i.e. the middle of the photoperiod) after D0, D3, and D6 of the respective darkening treatment (see “Materials and Methods”; Supplemental Table S1). This transcriptomic analysis revealed two distinct aspects in the response to darkness: (1) a conserved response arising from the two darkening treatments, and (2) a divergent, treatment-specific response. Plotting the first two components of a principal component analysis (PCA) from the normalized arrays, we observed that the first component, which accounted for 55% of the variance in the experiment, was explained by a conserved response to darkness (Fig. 2Ai). For an in-depth transcriptomic examination of this conserved response, see Supplemental Text S1. The second component, accounting for 27% of the variance, separated the contrasting responses of the two darkening treatments. In a second PCA, analyzing components 2 and 3 (Fig. 2Aii), component 3 accounted for 5% of the variance and reflected predominantly the temporal divergence in transcriptomes between D3 and D6. This time course-dependent difference was more pronounced in IDL than in DP, which indicates a more active transcriptomic reorganization in IDL, as suggested previously (Keech et al., 2007).
DP and IDL Also Showed Key Transcriptional Differences That Underpin the Differing Metabolic Strategies
As demonstrated above, DP and IDL share a common transcriptomic response to darkness. Nonetheless, nearly 20% of the genes down-regulated at D3 and 25% at D6 were differentially regulated in a treatment-specific manner (Fig. 2B). These figures rose dramatically for the transcripts increasing in abundance, with 26% for DP and 39% for IDL at D3, followed by 39% for DP and 46% in IDL at D6, that were significantly up-regulated in a treatment-specific manner (Fig. 2B). The identification of these treatment-specific transcriptomic responses formed the basis for further analysis. To visualize which biological functions contributed significantly to the overall response to darkness in either DP or IDL, we selected all the genes with an adjusted P value of less than 0.001 issued from the limma time-series analysis (Supplemental Methods S1) and established a biological network based on enriched Gene Ontology (GO) terms (P < 0.001) using the ClueGO plug-in (Bindea et al., 2009) for Cytoscape (Shannon et al., 2003). Two networks were created based on the differential expression between DP and IDL within the two time periods D3/D0 (Supplemental Fig. S2A) and D6/D3 (Supplemental Fig. S2B). Enriched GO terms associated with either IDL (palette of yellows) or DP (palette of greens) were clustered according to their functional categories. The size of a node is proportional to the number of genes contributing to this node, while the intensity of the color is proportional to the significance (the minimum significance being P < 0.05) of it belonging to a treatment (e.g. dark yellow = strongly associated with IDL, pale green = moderately associated with DP, and gray nodes = overrepresented in the gene subset but not significantly enriched in DP or IDL; the threshold of significance was set at 60% [see “Materials and Methods”]). Looking at the difference between D0 and D3, a rather balanced number of overrepresented GO terms were observed in DP and IDL (Fig. 2C; Supplemental Fig. S2A). In these first 3 d of darkening, responses that were enriched in IDL included respiration, transporter activities, sugar catabolism, pathogen, and stress. By contrast, the most significantly enriched GO terms in DP were amino acid biosynthesis, hormone metabolism, photosynthesis, starch metabolism, and regulation of transcription. Interestingly in the second part of the time course (from D3 to D6), the majority of the enriched GO terms were associated with IDL, including lipid and amino acid catabolic processes, transporters, respiration, responses to light, pathogen, starvation, and cell death mechanisms. Conversely, with few exceptions, only functional groups associated with the regulation of transcription were enriched in DP (Fig. 2C; Supplemental Fig. S2B). To summarize, this network analysis of GO terms indicated that, while IDL was supported by dynamic/differential transcriptomic changes throughout the process of senescence, DP underwent fewer additional transcriptomic changes leading to new metabolic functions.
Figure 2.
Transcriptomic analysis of the conserved and diverging responses in Arabidopsis leaves during two darkening treatments. A, PCA was used to reduce the dimensionality of the data and, thus, emphasize similarities and differences within the data set. i, Component 1 (55%) and component 2 (27%); ii, component 2 (27%) and component 3 (5%). B, Differential expression analysis was carried out comparing D3 versus D0 and D6 versus D0 for each darkening treatment. The resulting lists of significantly differentially expressed genes from each treatment were then matched to reveal the level of conservation in transcriptomic response. C, Biological network of enriched GO terms from significantly (P < 0.001) differentially expressed genes in DP versus IDL conditions at D3 and D6. Enriched GO terms associated with either DP (palette of greens) or IDL (palette of yellows) were clustered according to their functional categories. The size of a node is proportional to the number of genes contributing to this node, while the intensity of the color is proportional to the significance (the minimum significance being P < 0.05) of it belonging to a treatment (e.g. dark yellow = strongly associated with IDL, pale green = moderately associated with DP, and gray nodes = overrepresented in the gene subset but not enriched significantly in DP or IDL; the threshold of significance was set at 60%). AA, Amino acid.
As mentioned earlier, only the DP leaves displayed a strong hyponasty and elongation of the petiole (Fig. 1), traits generally observed during shade-avoidance responses. This analysis did not include the extent to which the signaling mechanisms controlling shade avoidance could overlap with the responses observed in DP or IDL. Thus, we conducted a complementary analysis by compiling and analyzing several publicly available microarrays performed with wild-type Arabidopsis leaves under control or enriched far-red light conditions. We identified a core subset of approximately 40 genes that displayed a conserved expression profile in response to shade-avoidance conditions (i.e. low red:far-red light ratio [R:FR]). Comparison of the expression of these genes under shade-avoidance response and DP conditions revealed that 76% of similarities were present at D3 but only 46% were observed at D6 (Supplemental Table S2). Interestingly, a corresponding comparison with IDL showed 65% similarities at D3 and only 46% at D6. This indicated that, in response to the onset of darkness and regardless of the treatment, a leaf will employ similar signaling mechanisms and metabolic strategies to those used in response to shade. When darkness is prolonged, the subsequent drastic shortage of available carbon prompts the strategy to shift into a more specialized, nuanced transcriptional response.
A Focus on the Transcriptomic Differences Associated with Metabolism Revealed Differing Strategies Used to Counter Extended Darkness
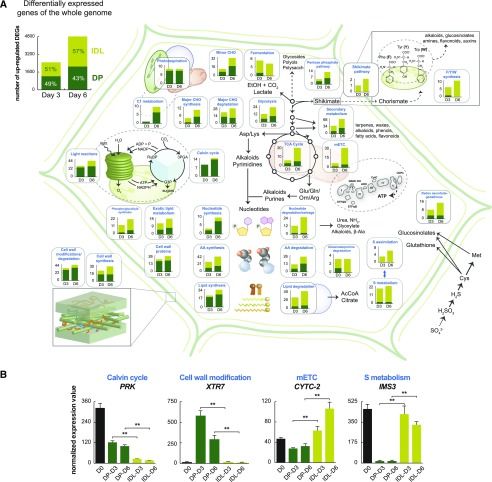
Our transcriptomic analyses revealed the increasing prevalence of distinct metabolic functions specific to each darkening treatment, which prompted a targeted examination of the major metabolic processes. To decipher the diverging metabolic strategies employed in DP and IDL, MapMan annotations (Usadel et al., 2005) were utilized to identify and isolate diverse metabolic functions present in the transcriptomic data. At both D3 and D6, the transcript abundance of genes belonging to specific metabolic functional groups was interrogated to determine which genes were significantly correlated with either DP or IDL (Fig. 3A). From this analysis, it was observed that functions linked to light-dependent and independent reactions of photosynthesis (light reactions, Calvin cycle, photorespiration, and C1 metabolism) were strongly correlated with DP. For instance, we have shown that genes encoding for proteins associated with photosynthesis were clearly down-regulated in both darkening treatments (Supplemental Fig. S3, A and C). However, the data presented here indicated that the amplitude of this down-regulation was significantly weaker in DP than IDL and could contribute to the final phenotype difference observed between the two darkening treatments. We also selected a few key genes from different metabolic or regulatory pathways that illustrated the divergence of the amplitude of the response between the two darkening treatments when compared with the illuminated controls (Fig. 3B; Supplemental Fig. S4). Furthermore, functions associated with lipid synthesis, cell wall modification/degradation, and cell wall proteins were all strongly correlated with DP (Supplemental Table S3). Nucleotides are nitrogen-rich compounds that have a diversity of roles in the cell, including as metabolites in bioenergetic processes and as precursors to nucleic acids and an array of macromolecules such as polysaccharides, phospholipids, and glycolipids (Stasolla et al., 2003). In this comparison, genes encoding proteins with nucleotide synthesis functions were strongly correlated with DP, while nucleotide degradation/salvage was more strongly associated with IDL.
Figure 3.
Transcriptomic analysis of leaf metabolism in DP and IDL. A, Differential expression analysis was carried out between DP and IDL samples at D3 and D6. The resulting genes that were defined as significant (adjusted P < 0.001; differential expression was carried out using limma) can be observed at top left. These genes were then cross referenced with MapMan annotations and sorted for metabolic functions. Differentially expressed genes (DEGs) that correlated to either DP or IDL conditions were recorded and graphed, with light green corresponding to correlation with IDL conditions and dark green corresponding to correlation with DP conditions. B, Histograms of the transcript abundance of representative genes to illustrate the different patterns of abundance present and differences of magnitude. Double asterisks indicate significant (P < 0.01; Student’s t test; three replicates) differences in transcript abundance between the IDL and DP samples. Error bars indicate se.
While many of the metabolic functions correlated with DP were associated with photosynthetic processes and cell wall/membrane metabolism, with IDL there was a pronounced correlation of transcripts encoding proteins that function in primary energy production, such as fermentation, β-oxidation, the tricarboxylic acid (TCA) cycle, and the mitochondrial electron transport chain (mETC). This fundamental divergence in metabolic programming indicated that, while in DP conditions, plants maintain basal photosynthetic processes with a reduced capacity for mitochondrial ATP production to outlast the deleterious darkened conditions. Furthermore, in IDL conditions, mitochondrial primary metabolism was maintained, likely to fuel nitrogen-yielding processes and to support the subsequent reallocation of nitrogen- and sulfur-rich nutrients (Fig. 3A; Chrobok et al., 2016). This implied the presence of both reducing power and carbon chains, either imported from other parts of the plant or derived from the degradation of low-Mr building blocks such as nucleotides, amino acids, and lipids, the catabolic processes of which were all strongly associated with IDL. Finally, functions associated with responding to and mitigating the effects of stress were highly correlated to IDL, such as redox regulation processes (e.g. the glutathione-ascorbate cycle) and sulfur metabolism and assimilation (Fig. 3A). While these metabolic pathways certainly contribute to the remobilization of sulfur from the senescing leaf to the rest of the plant (Abdallah et al., 2010), they also suggest that the stress status of these cells has become more severe and that steps are being taken to alleviate these effects (Supplemental Fig. S5). Additionally, of the differentially expressed components associated with the shikimate/chorismate pathway and the subsequent synthesis of the amino acids Phe, Tyr, and Trp, all but one gene was observed to correlate to IDL (Fig. 3A). These functions, which are linked to the biosynthesis of precursors to alkaloids, glucosinolates, amines, flavonoids, and auxins, could become essential for the protection of leaf cells in response to both biotic and abiotic stresses.
Metabolic Differences between Dark Treatments Increased over Time and Reflect the Diverging Carbon Status of the Leaves
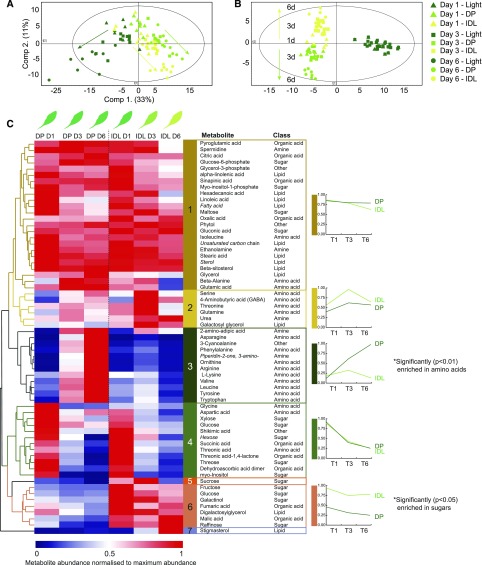
Having thoroughly examined the transcriptomic reprogramming of the metabolic machinery, a corresponding metabolomic analysis was undertaken. To achieve this, leaves were harvested after 1, 3, and 6 d of the respective darkening treatments, and metabolic profiling was conducted using gas chromatography-time of flight-mass spectrometry (GC-ToF-MS; Supplemental Table S4). PCA of the resulting data (Fig. 4A) greatly echoed the PCA of the transcriptomic data, as both darkening treatments diverged from the illuminated control samples in a similar manner at days 1, 3, and 6. This was better illustrated when a supervised method of dimensionality reduction was employed, such as orthogonal partial least squares-discriminant analysis (OPLS-DA; Fig. 4B). By categorizing the three treatments (light, DP, and IDL), within-group variance is revealed in starker contrast, and in this way, a clearer separation of the darkening treatments from the light samples was observed. Over the 6-d time course, the metabolomes of the two darkening treatments became progressively divergent (Fig. 4B). Hierarchical clustering of metabolites from the two darkening treatments and corresponding control samples (illuminated leaves from IDL plants) was utilized to analyze the abundance profiles of metabolites under these different conditions (Supplemental Fig. S6). Unsurprisingly, of the resulting six clusters, all but two were composed of metabolites that were reduced in abundance upon commencement of DP or IDL conditions (clusters 1, 2, 5, and 6). In contrast, cluster 3, which was significantly enriched in amino acids (P < 0.01; N-1 χ2 test), was characterized by metabolites that increased in abundance in DP conditions but not in IDL or control conditions (Supplemental Fig. S6). Since the focus of this study was on the diverging responses between leaves under DP and IDL conditions, an additional hierarchical cluster was prepared using only samples from the two darkening treatments in isolation (Fig. 4). This analysis resulted in seven clusters, of which two contained only single metabolites (Suc in cluster 5 and stigmasterol in cluster 7; Fig. 4C). Plotting the average abundance of the metabolites in each cluster quickly revealed conserved and divergent metabolite profiles in response to DP and IDL treatments. For example, cluster 1 displayed highly similar abundance patterns between DP and IDL at D1 and D3, but at D6, a reduction in abundance was observed for IDL while levels were maintained in DP. Notably, the polyamine spermidine and Glu were found in this cluster. Similar to the observations made when clustering the darkened samples with the illuminated control samples (Supplemental Fig. S6), cluster 3 was significantly (P < 0.01; N-1 χ2 test) enriched in amino acids. This cluster showed a marked divergence in metabolite abundance levels and notably contained the branched-chain amino acids (BCAA) Val and Leu; the aromatic amino acids Phe, Tyr, and Trp; as well as two amino acids with high N:C ratios, Asn and Arg. The metabolite abundance in IDL remained quite low, while in DP, the abundance increased substantially throughout the time course, reaching maximum levels at D6. In contrast, cluster 6 (enriched in sugars including Fru, Glc, galactinol, and raffinose) was characterized by low abundance levels in DP but much higher levels in IDL. Cluster 6 also contained two organic acids, malate and fumarate, which are central metabolites for primary metabolism. Digalactosylglycerol also accumulated in IDL, similar to galactosylglycerol in cluster 2, most likely as catabolic products of thylakoid degradation. Finally, cluster 4 was composed of several different metabolites (such as the amino acids Gly and Asp, the sugars Glc and an undefined hexose, and many organic acids) and was characterized by abundances that decreased at D3 and D6 in both darkening treatments.
Figure 4.
Metabolic profiling by GC-ToF-MS of leaves during a time course of 1, 3, and 6 d of DP or IDL conditions. A and B, PCA (A; component 1, 33%; component 2, 11%) and supervised OPLS-DA method (B; model with two predictive components [30.2%: 23.4% + 6.8%], three orthogonal components [32.1%], and Q2 [73.8%]), showing the progressive divergence of the respective metabolomes over time. C, Hierarchical clustering (Euclidean distance) of identified metabolites based on their abundance relative to internal standards. Seven clusters were determined. For each cluster (with the exception of clusters 5 and 7), the average metabolite abundance has been plotted to better visualize the changes in abundance that characterize these clusters. Italicized metabolites denote nonconfirmed metabolite identities. Clusters enriched significantly (using an N-1 x2 test) in amino acids (cluster 3) or sugars (cluster 6) are indicated with single asterisks.
From this metabolite analysis, clusters 3 and 6 could contribute to the understanding of the different metabolic strategies between DP and IDL. Therefore, we performed further experiments to analyze these amino acids and carbohydrates in more detail. Using leaves sampled after 6 d of darkening treatment, we analyzed by HPLC the abundance of 20 amino acids, γ-aminobutyric acid (a nonprotein amino acid associated with the modulation of a number of metabolic and stress processes), and free ammonium (a cytotoxic molecule that also is an important metabolic source of nitrogen; Table I). Corresponding leaves that remained illuminated also were sampled and analyzed. In agreement with the observations denoted in Figure 4C, this analysis revealed that the total amount of amino acids present in DP leaves was 3 and 5 times greater than in illuminated control leaves and IDL, respectively. Almost all amino acids, apart from Asp, Gly, and Pro, were accumulated in DP leaves. Asn alone accounted for nearly one-third of the total pool of amino acids with 14 µmol g−1 fresh weight, which represented a 40-fold accumulation compared with illuminated leaves. However, many more amino acids also accumulated from 20- to 100-fold in DP when compared with light samples. In IDL, fewer amino acids increased in abundance but not to the same extent as in DP, which fully supported our metabolomic data (Fig. 4C). The amount of Ala, Gln, and Glu decreased significantly in IDL when compared with illuminated and DP samples (Table I). This is in line with earlier data pertaining to the regulation of metabolism during developmental leaf senescence, where it was demonstrated that the abundance of Gln and Glu decreased significantly as a result of nitrogen export (Masclaux-Daubresse et al., 2010; Chrobok et al., 2016). Taken together, HPLC analysis confirmed a considerable accumulation of amino acids in DP, particularly those with a high N:C ratio (e.g. Asp and Arg) and, to a lesser extent, Gln, which could serve as nontoxic end products that scavenge free ammonium released from cellular metabolic activity. The 75% decrease of ammonium in DP leaves, albeit nonstatistically significant, also would support this explanation.
Table I. Amino acid content in leaves following darkening for 6 d of an individual leaf (IDL) and an entire plant (DP).
Results are means of three replicates ± se and are expressed in μmol g−1 fresh weight. Dark treatments were compared with light samples by Student’s t test (*, significant at P < 0.05; ns, not significant). n/a, Not applicable.
| Amino Acid | Light | DP | IDL |
|---|---|---|---|
| Ammonium | 0.80 ± 0.42 | 0.18 ± 0.05 ns | 0.54 ± 0.60 ns |
| Ala | 0.64 ± 0.11 | 1.03 ± 0.17* | 0.26 ± 0.04* |
| Arg | 0.03 ± 0.06 | 2.44 ± 0.06* | 0.33 ± 0.10* |
| Asn | 0.34 ± 0.04 | 14.44 ± 1.13* | 0.99 ± 0.62 ns |
| Asp | 1.85 ± 0.20 | 0.85 ± 0.25* | 0.22 ± 0.04* |
| Cys | n/a | n/a | n/a |
| γ-Aminobutyric acid | 0.00 ± 0.00 | 0.36 ± 0.03* | 0.17 ± 0.05* |
| Gln | 2.11 ± 0.19 | 2.93 ± 0.45 ns | 0.67 ± 0.27* |
| Glu | 2.15 ± 0.07 | 4.40 ± 0.34* | 1.86 ± 0.05* |
| Gly | 0.27 ± 0.07 | 0.00 ± 0.00* | 0.03 ± 0.03* |
| His | 0.00 ± 0.00 | 0.85 ± 0.03* | 0.05 ± 0.08 ns |
| Ile | 0.00 ± 0.00 | 0.99 ± 0.17* | 0.05 ± 0.05 ns |
| Leu | 0.11 ± 0.00 | 0.54 ± 0.07* | 0.22 ± 0.02* |
| Lys | 0.05 ± 0.09 | 0.47 ± 0.05* | 0.26 ± 0.03 ns |
| Met | n/a | n/a | n/a |
| Phe | 0.00 ± 0.00 | 2.06 ± 0.08* | 0.12 ± 0.02* |
| Pro | 0.79 ± 0.38 | 0.06 ± 0.02 ns | 0.07 ± 0.01 ns |
| Ser | 1.30 ± 0.22 | 2.04 ± 0.40 ns | 0.88 ± 0.15 ns |
| Thr | 0.60 ± 0.14 | 1.16 ± 0.38 ns | 0.37 ± 0.08 ns |
| Trp | 0.00 ± 0.00 | 0.88 ± 0.02* | 0.15 ± 0.04* |
| Tyr | 0.01 ± 0.02 | 0.40 ± 0.04* | 0.08 ± 0.01* |
| Val | 0.10 ± 0.00 | 2.05 ± 0.28* | 0.19 ± 0.02* |
| Total | 11.17 | 38.14 | 7.51 |
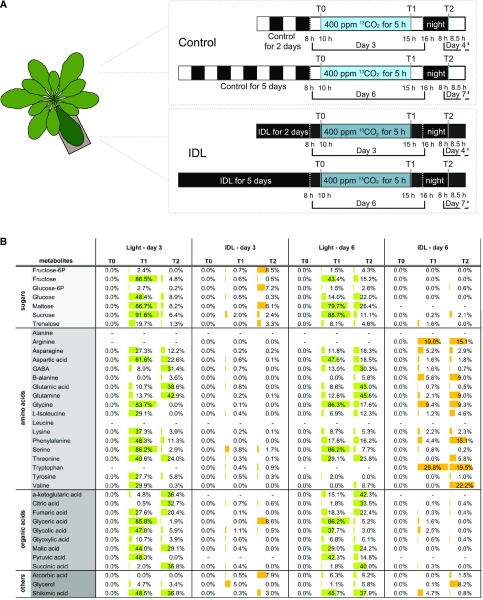
Interestingly, results from the leaf metabolite profiling suggest that the abundance of several sugars is initially reduced but then increases by day 6 in IDL (Fig. 4C). Since IDL leaves are not under photosynthesizing conditions, these sugars may have been imported from other parts of the plant. To test this, we conducted a 13CO2-labeling experiment with plants subjected to IDL treatment using a custom-made chamber that we developed recently (Lindén et al., 2016). Briefly, 6-week-old plants underwent 2 and 5 d of darkening treatment. In the morning of days 3 and 6, respectively, five plants with two individually covered leaves were exposed to 400 µL L−1 90% 13CO2-enriched CO2 for 5 h. Both IDL and illuminated control leaves were sampled at T0 (i.e. beginning of the experiment and a reference for natural 13C abundance), T1 (after 5 h of labeling), and T2 (the morning after the labeling experiment; Fig. 5A). We then analyzed the 13C enrichment in a set of metabolites that included mainly sugars, amino acids, and organic acids using GC-ToF-MS as described by Lindén et al. (2016) (total relative pools [12C + 13C] are found in Supplemental Table S5). At the end of the fumigation period, 80% to 90% of the pools of Suc, maltose, Gly, Ser, and glycerate were labeled in the light samples at either day 3 or day 6 (Fig. 5B). These values were as expected, since these metabolites are produced from and closely associated with the Calvin-Benson cycle. Importantly, Suc is a direct sink, maltose is a transition sugar from starch breakdown (note that similar labeling patterns have already been reported for maltose; Szecowka et al., 2013; Lindén et al., 2016), while Gly, Ser, and glycerate are direct photorespiratory products. Many other metabolites also were differentially labeled at T1 in the light samples. At T2, in both light samples, most of the labeling was found in amino and organic acids, while the percentage of labeling in sugar pools declined drastically. For IDL samples, the situation was considerably different compared with the light samples. For instance, the labeling pattern diverged markedly between leaves darkened for 3 and 6 d. At the end of the photoperiod (i.e. T1) for 3-d IDL samples, almost no metabolites were labeled. However, at T2, several sugar pools (Fru-6-P, Glc-6-P, trehalose, maltose, and Suc) showed significant 13C enrichment, suggesting the import of molecules containing 13C during the night. Intriguingly, the 13C enrichment pattern had diverged greatly for 6-d IDL samples. Labeling of the sugar pools was absent at T1 and minimal at T2; however, many amino acids were already significantly labeled by T1. Interestingly, Arg and Trp showed 20% enrichment, while the pool of these metabolites was not detectable in any of the light samples (Fig. 5B). At T2, a rather similar pattern was observed, with mostly pools of amino acids showing 13C enrichment. Also, a few amino acids, such as Gln, Ile, Phe, Thr, and Val, were labeled overnight. Together, these results indicated that, in IDL conditions, senescing leaves underwent an exchange of metabolites with other parts of the plant, and as this exchange occurs, the nature of the transported metabolites changes.
Figure 5.
13CO2 labeling of IDL after 2 and 5 d of darkening treatment. A, Experimental design of the labeling. Five plants with two individually covered leaves (2 or 5 d of treatment) were placed in a custom-made chamber and subjected to 5 h of 13CO2 at 400 µL L−1 and under similar controlled conditions to those in the growth chamber. Both IDL and leaves that remained in the light (Light) were sampled at T0, T1, and T2. B, Summary table of 13CO2 enrichment, expressed as a percentage of the total pool of a given metabolite. Only statistically relevant enrichments are shown (Lindén et al., 2016), and results are means of at least four independent biological replicates. To simplify the illustration of the results, the enrichment in 13CO2 is visualized as data bars, green for light and orange for IDL, and this enrichment was only compared between treatments (i.e. Light-D3 versus Light-D6 and IDL-D3 versus IDL-D6), as the labeling in light samples was so prominent compared with the labeling detected in IDL samples.
DISCUSSION
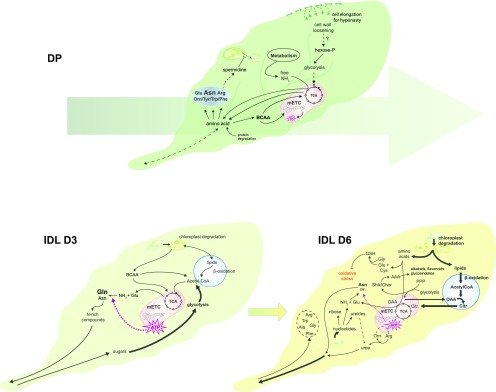
A plant’s ability to survive is based largely on its dependency to light. Hence, plants have evolved a variety of strategies to rapidly adapt to negative changes in light intensity and quality, arising from a decline in the irradiance of blue (400–500 nm) and red (600–700 nm) light. Additionally, low R:FR ratios occur at dusk as well as in environments of high plant density, which signal to the plant that it is being outcompeted for light by surrounding vegetation. If these conditions persist, a shade-avoidance response is triggered, often resulting in morphological changes such as the elongation of stems and petioles, leaf hyponasty, and increased apical dominance (Das et al., 2016; Kohnen et al., 2016). However, when an individual leaf or a whole plant is subjected to prolonged darkness, two unique divergent responses are observed: the IDL undergoes accelerated senescence, while the leaves from the whole DP adopt a metabolic strategy aimed at survival (Supplemental Fig. S4, senescence-associated genes [SAGs]). Understanding these different responses is essential to deepen our knowledge of the key mechanisms controlling the survival strategies used by plants as well as to identify novel targets for biotechnological approaches aimed at improving postharvest shelf life to reduce food waste. By complementing a thorough transcriptomic analysis with a set of metabolomic approaches, we propose a working model based on a cellular overview of the differing metabolic strategies employed by leaves under these two darkening conditions (Fig. 6).
Figure 6.
Model summarizing the different metabolic strategies employed by plants in response to partial or total darkening of the plant. The size and line weigh of the fonts and arrows are proportional to their implication in these metabolic processes. The large arrow behind the leaf in DP conditions depicts the conserved metabolic strategy maintained between 3 and 6 d of darkening. AAA, Aromatic amino acids; Citr, citrate; OAA, oxaloacetate; PPP, pentose phosphate pathway; Shik/Chor, shikimate/chorismate; TCA, tricarboxylic acid cycle.
Whole Darkened Plants Suppress Metabolism While Accumulating Nitrogen-Rich Amino Acids and Polyamines
When an entire plant is placed in darkness, all of its leaves are simultaneously compromised in their ability to produce the sugars required to support growth. In Arabidopsis, it has been shown that these debilitating conditions do not elicit leaf senescence but rather suppress it, most likely as a survival strategy resulting from a general developmental arrest (Weaver and Amasino, 2001; Keech et al., 2007, 2010; Fan et al., 2017). On a morphological level, the leaves undergo several changes, including strong hyponasty of the leaf blade and elongation of the petiole (Fig. 1), traits that are typical of shade-avoidance responses. The identification of a subset of genes differentially regulated specifically in response to shade avoidance and its comparison with DP-D3 conditions revealed a 76% similarity between the transcriptional responses of these two biological conditions (Supplemental Table S2). Unsurprisingly, when darkness was maintained for 6 d, this similarity dropped to 46%, suggesting a modified transcriptional response that likely resulted from the altered metabolic state. Interestingly, our transcriptomic data further revealed a mechanistic explanation for the phenotypic changes associated with the leaf hyponasty. At both D3 and D6 in DP, the genes associated with cell wall modification/degradation functions were significantly up-regulated compared with IDL (Figs. 3A and 6; Supplemental Table S3, cell wall metabolism). This group of genes contained a variety of pectin lyases, pectin esterases, β-galactosidases, and xyloglucan endotransglucosylases. Here, we propose a possible 2-fold outcome of this concerted cell wall modification/degradation. First, loosening of the cell wall while increasing cell turgency (as supported by the higher RWC under darkness; Supplemental Fig. S1A) would allow the plant to alter the aspect and posture of its leaves to better intercept light at a minor energy and carbon cost, since it avoids energy-consumptive processes such as cell division. Such rearrangement of the cell wall is strongly supported by earlier measurements from Keech et al. (2007), who reported that an increase in mesophyll cell size occurred in the leaves of plants that have been darkened for 6 d. This demonstrated that cells undergo a mechanical expansion in response to DP treatment. Second, we conjectured that the loosening of the cell wall could simultaneously participate in the release of valuable monosaccharides, which in turn would support primary metabolism. Indeed, several descriptive and experimental studies have suggested at least partial mobility of cell wall monosaccharides and polysaccharides in mature, nonreproductive plant tissues (for review, see Hoch, 2007). For example, experiments with barley (Hordeum vulgare) leaves suggested that cell wall (1→3,1→4)-β-d-glucans can be remobilized in nonelongating, dark-incubated leaves and the generated Glc could serve as an energy source under conditions of sugar depletion (Roulin et al., 2002).
Keech et al. (2007) also showed that, while leaves from DP preserved their photosynthetic capacity, mitochondrial respiration decreased. This corroborates the observations made in our work, as the transcript abundance of genes encoding proteins with functions associated with light-dependent and independent chloroplast functions (Fig. 3A, light reactions, Calvin cycle, photorespiration) was significantly less reduced in DP relative to IDL (Supplemental Fig. S4, RbcS2B), and the photosynthetic efficiency under growth light conditions was maintained at a higher level in DP compared with IDL (Fig. 1D). Similar patterns of expression were not observed for genes encoding proteins associated with mitochondrial primary metabolism (Fig. 3A, TCA cycle and mETC). Due to the maintenance of chloroplast integrity and photosynthetic efficiency observed in DP-treated leaves (Fig. 1; Supplemental Fig. S1B; Keech et al., 2007), it is notable that spermidine abundance was maintained at moderately high levels at all three time points in the leaves of DP, while it decreased in abundance in IDL (Fig. 4, cluster 1). The presence of spermidine under DP conditions likely contributes to the protection of chloroplast structures by preventing protein and chlorophyll degradation as well as by stabilizing numerous proteins such as D1 and D2 (photosystem b components), LHCII (light-harvesting complex), cytochrome f, and Rubisco (Sobieszczuk-Nowicka and Legocka, 2014), thus offering one explanation for the better survival strategy observed in DP leaves (Fig. 6).
In this metabolically suppressed state, plant cells must be resourceful in their utilization of various energy sources to yield ATP. In addition to the reduced carbon possibly liberated from cell wall modification/degradation, the enriched reserves of BCAA observed (Fig. 4, cluster 3; Table I) are slowly catabolized to yield Glu, which, in turn, can be converted to α-ketoglutarate via oxidative deamination and acetyl-CoA, both of which can feed directly into the TCA cycle (Fig. 6; Supplemental Table S3, BCAA). Concurrently, the electron transfer flavoprotein (ETF) and ETF ubiquinone oxide reductase (Supplemental Fig. S4, ETF-QO) can chaperone electrons released from BCAA catabolic pathways to the mETC, providing an additional source of ATP production (Ishizaki et al., 2006; Araújo et al., 2010).
In addition to the BCAA, we showed through HPLC (Table I) and GC-ToF-MS (Fig. 4) measurements that a large number of key amino acids accumulated in leaves during DP conditions, with the total amino acid pool increasing more than 3-fold compared with that of the light samples. The origins of these amino acids are likely varied, as has been highlighted previously (Soudry et al., 2005). During developmental leaf senescence, protein degradation is the primary source of free amino acids, particularly resulting from the degradation of Rubisco (Smart, 1994), and accounts for up to 35% of the nitrogen in a mature leaf (Makino et al., 1984; Soudry et al., 2005). However, in DP conditions, transcriptomic evidence suggests that this is not the case, as genes encoding functions associated with protein degradation were significantly underrepresented throughout the DP time course (Supplemental Fig. S7). During periods of prolonged stress, growth rates are reduced drastically, a feature that is often accompanied by increases in steady-state levels of ammonium, likely as a by-product of cellular metabolic activity (Rabe and Lovatt, 1984). Thus, the incorporation of accumulating ammonium into key amino acids would act to sequester valuable nitrogen while simultaneously reducing the levels of this cytotoxic molecule (Rabe and Lovatt, 1984; Gilbert et al., 1998). Many of these key amino acids with high N:C ratios accumulated under DP conditions, notably Asn (38% of the total amino acid pool; 40-fold increase compared with light samples) and Arg (6% of the total amino acid pool; 80-fold increase compared with light samples; Fig. 4, cluster 3; Table I), while concomitantly, the level of free ammonium decreased by 75%. Corresponding transcriptomic data supported these measurements. For example, Asn synthase (ASN1; AT3G47340) increased nearly 150-fold at D3 and D6 in DP compared with illuminated control samples (Supplemental Fig. S4). Asn, Gln, and Arg are ideally suited as transporter compounds of nitrogen, as each contains at least two amino groups tethered to a carbon backbone (Fig. 6; Supplemental Table S3, nitrogen metabolism). Preferential biosynthesis of amino acids with high N:C ratios would prove vital, as nitrogen sinks are limited during conditions where carbohydrate production and growth are impaired by different stresses (Urquhart and Joy, 1981; Lam et al., 1994; Dubay and Pessarakli, 1995; Gilbert et al., 1998). Ordinarily, these nitrogen-rich compounds (especially Asn) would be transported rapidly to sink tissues via phloem translocation, where they could be used to support growth and development (Masclaux-Daubresse et al., 2010). Conversely, a whole DP represents a unique situation whereby all organs and tissues have been compromised simultaneously and traditional sink/source relationships are altered. This would naturally lead to the accumulation of specific amino acid groups, likely compartmentalized in vacuoles, that would prove valuable upon reexposure to light conditions.
IDL Undergo Senescence, Using an Altered Source/Sink Relationship to Fuel the Export Process
It is clear that the leaves of DP and those that are individually darkened share many similarities in their response to darkness (Fig. 2, A and B), and the examination of the metabolic strategy employed by IDL does not deviate from this observation. Interestingly, we often noticed that, for a given gene or metabolic pathway, leaves from both treatments would respond in a similar way, but the amplitude of the response would be markedly different. For example, while BCAA catabolism is up-regulated in both darkening treatments, we clearly saw that the magnitude of the response was more pronounced in DP. A mirrored scenario was seen with β-oxidation, which appeared more pronounced in IDL (Figs. 2C and 3; Supplemental Figs. S3A and S4, KAT2). Nonetheless, when further scrutinizing the responses from DP and IDL to darkness, we identified numerous clear metabolic differences: fermentation, sulfur metabolism and assimilation, catabolic processes, and the shikimate pathway were among the biological functions strongly up-regulated in IDL. In addition, one of the most fundamental differences regarding primary metabolism we observed repeatedly throughout our data sets concerned mitochondrial functions, which became undoubtedly more central for the metabolism of IDL (Figs. 2C and 3A, TCA cycle and mETC; Figs. 4C and 5B; Supplemental Table S2). Regarding this aspect, IDL appears similar to developmental leaf senescence, as physiological studies of these two processes have demonstrated that the rapid decline in photosynthetic capacity is concomitant with a maintained mitochondrial respiration throughout the process of leaf senescence (Buchanan-Wollaston et al., 2005; Keech et al., 2007; Chrobok et al., 2016).
Besides providing the energy necessary for the degradation of metabolites such as nucleotides, amino acids, and lipids (Figs. 3A and 4C), mitochondria also orchestrate this process by hosting many catabolic pathways and supplying the requisite carbon backbones in the form of α-ketoglutarate or Glu. Chrobok et al. (2016) proposed that Glu originating from mitochondrial metabolism is the preferential carbon skeleton and that, upon export to the cytosol, can undergo amination via Gln synthetase1. The Gln that is produced can, in turn, provide an additional amine group to Asp, which, via the activity of ASN1, leads to the production of Asn. Here, the transcript abundance of these two enzymes was strongly up-regulated under IDL conditions (Supplemental Table S1). Finally, the resulting Gln and, to a lesser extent, Asn can be exported as stable stores of nitrogen (Masclaux-Daubresse et al., 2010). An additional source of mobile nitrogen arises from the urea derived from Arg and Orn metabolism (Fig. 4C) and from the degradation of nucleotides (Figs. 3 and 6; Supplemental Table S3, nucleotide degradation; Zrenner et al., 2006; Bohner et al., 2015). The catabolism of purines, specifically, leads to the production of xanthine, which can be degraded further to other ureides such as uric acid and allantoin, ultimately leading to the release of urea (Zrenner et al., 2006). In the context of senescence, ureides also have been demonstrated to have roles in reactive oxygen species scavenging (Brychkova et al., 2008). The transcript abundance of two key enzymes of this catabolic pathway, the xanthine dehydrogenases (AtXDH1 [AT4G34890] and AtXDH2 [AT4G34900]), which catalyze the conversion of xanthine to uric acid, is significantly higher in IDL samples than in DP at D6 (Supplemental Table S3, nucleotide degradation). In addition to supporting the production of nitrogen-rich molecules, nucleotide degradation also leads to the formation of ribose, a monosaccharide that is a critical component of many metabolites, such as NAD, S-Ado-Met, ATP, and cytokinins, as well as a participant in the nonoxidative phase of the pentose phosphate pathway (Riggs et al., 2016).
Interestingly, we also observed a switch in the substrates fueling mitochondria during the progression of senescence in IDL. Our experiments using 13CO2 labeling and subsequent GC-ToF-MS to monitor the 13C enrichment in metabolites demonstrated that, after 3 d, the IDL still received some sugars, notably Suc, from other parts of the plant (Figs. 4C and 5B). However, this import mostly dissipated after 6 d of darkening, where nearly no 13C labeling was found in sugars but many amino acids were labeled instead. Furthermore, both our transcriptomic and metabolomic analyses provided evidence for active lipid degradation, likely derived from chloroplast/thylakoid breakdown, as shown by the accumulation of galactosylglycerol, phytol, and unsaturated carbon chains (Fig. 3A, lipid degradation; Fig. 4C). Therefore, we propose a model in which sugars are imported at low levels into the darkened leaf until a threshold, likely predicated upon metabolic fluxes, is crossed. Following this junction, catabolic pathways come to dominate the metabolic landscape and provide enough alternative substrates to support cellular metabolism and the subsequent export of nutrients such as sulfur and nitrogen (Fig. 6). This metabolic shift occurs progressively, and by day 6, almost no sugars are imported, while the main bulk of Gln has already been exported (Fig. 4). Instead, the acetyl-CoA produced through β-oxidation is incorporated via the strongly up-regulated peroxisomal citrate synthases (CSY2 [AT3G58750] and CSY3 [AT2G42790]) to produce citrate (Supplemental Table S3, gluconeogenesis and the glyoxylate cycle). Consequently, organic acids produced in the peroxisome, mainly citrate and, to a lesser extent, malate and succinate produced via the glyoxylate cycle, can enter the Krebs cycle and support mitochondrial functions. Here, we favor citrate over malate/succinate as the main organic acid exported and fueling the TCA cycle for two reasons: (1) the export of citrate derived from β-oxidation to mitochondria is a metabolic mechanism known to occur during seed germination (Pracharoenwattana et al., 2005); and (2) the two key enzymes from the glyoxylate cycle (isocitrate lyase and malate synthase) are stably expressed in control and both darkened conditions (Fig. 6; Supplemental Table S3, gluconeogenesis and the glyoxylate cycle). That said, the partitioning between these two metabolic routes during leaf senescence remains to be challenged. We also believe that oxaloacetate would be preferred to malate as the organic acid imported into the peroxisomes (Fig. 6). Indeed, β-oxidation already produces copious amounts of reducing equivalents (e.g. NADH), and the conversion of malate to oxaloacetate by the peroxisomal malate dehydrogenase (pMDH) would only add even more reducing power within the peroxisome, which does not seem an efficient strategy. In line with this, both pMDH1 and pMDH2 are down-regulated in IDL (Supplemental Table S3, β-oxidation).
Such a metabolic switch between the import of carbohydrates and lipid degradation under darkening conditions is supported by Kunz et al. (2009), who utilized a loss-of-function mutation in the peroxisomal ABC-transporter1 (PXA1 [AT4G39850]; Supplemental Fig. S4, PXA1). The authors showed that, when pxa1 plants were subjected to DP conditions, severe leaf necrosis and death were observed; however, no corresponding phenotype was observed in the leaves of pxa1 subjected to IDL treatment. The authors hypothesized that this was due to the delivery of Suc from surrounding source leaves that alleviated the requirement for β-oxidation (Kunz et al., 2009). Furthermore, it has been shown that increased quantities of fatty acids can ultimately be deleterious for chloroplasts because of the detergent-like properties that derive from their amphipathic structure (Vernotte et al., 1983; Kunz et al., 2009). Thus, although the increased β-oxidation activity evidenced in IDL is coupled to mitochondrial activity, one can speculate that the increased β-oxidation also observed in DP aims primarily at minimizing the content of harmful fatty acids to preserve the chloroplasts while simultaneously providing small amounts of acetyl-CoA directly to the TCA cycle (Mackender and Leech, 1974; Dieuaide et al., 1992; Kunz et al., 2009; Fan et al., 2017).
Intriguingly, our transcriptomic analysis identified AtSIP2 (AT3G57520) as the 12th highest expressed gene in the entire transcriptome of IDL at D6, compared with being the 1,465th highest at T0 (Supplemental Fig. S4, AtSIP2). Originally thought to be a raffinose synthase, Peters et al. (2010) demonstrated that SIP2 was, in fact, an alkaline α-galactosidase, with a distinct substrate specificity for raffinose. GUS reporter assays showed that AtSIP2 was strongly expressed in sink tissues, and the authors suggested that it has a crucial physiological function in raffinose phloem unloading (Peters et al., 2010). Furthermore, it has been shown that galactinol and raffinose, which accumulated to significantly higher abundances in IDL than in DP (Fig. 4, cluster 6), can have important roles both as osmoprotectants under a range of stress conditions (Taji et al., 2002) and as scavengers of hydroxyl radicals to protect cells from oxidative damage and preserve redox homeostasis (Nishizawa et al., 2008). An earlier study, conducting spatiotemporal analysis of metabolomic shifts during developmental leaf senescence, revealed a localized accumulation of galactinol and raffinose to the basal region of leaves (Watanabe et al., 2013). Although we were unable to detect raffinose using a 13C-labeling approach, the combination of GC-ToF-MS data and pronounced up-regulation of AtSIP2 led us to hypothesize that, in IDL, imported raffinose could play a dual role of delivering reduced carbon to the darkened leaf and acting as a stress tolerance mechanism. Similarly, other genes with functions associated with hydroxyl scavenging, such as the glutathione-ascorbate cycle and sulfur metabolism and assimilation, were all strongly correlated with IDL (Figs. 3A and 6; Supplemental Table S3, secondary metabolites). Further evidence of the protective measures utilized by leaves under IDL conditions was the observed up-regulation of genes associated with the shikimate pathway and subsequent aromatic amino acid biosynthesis (Fig. 3A). The shikimate pathway is highly conserved across bacterial, fungal, and plant lineages, where it is one of the primary steps in the biosynthesis of aromatic amino acids and a multitude of aromatic secondary metabolites such as alkaloids, glucosinolates, amines, flavonoids, and auxins (Tohge et al., 2013). Under normal growth conditions, as much as 20% of the total fixed carbon in leaves is directed through the shikimate pathway (chiefly to Phe and Phe-derived products), while under stressful conditions, this proportion can increase markedly (Maeda and Dudareva, 2012; Tohge et al., 2013). This stress-induced increase in secondary metabolites is indicative of the diverse roles these molecules play in protecting the cell, primarily from biotic threats such as herbivory and bacterial and fungal pathogens. As Arabidopsis plants have lost the ability to abscise rosette leaves, this preferencing of secondary metabolites from the limited carbon pool available during IDL conditions provides an insight into how plants prophylactically protect their senescing tissues from becoming an entry point for pathogens, since a transcriptionally directed response would not be possible during the later stages of leaf senescence.
To conclude, our study establishes a robust working model that delineates the main axes of the divergent metabolic strategies employed by plants in response to two different darkening treatments. Furthermore, our work raises novel questions about the signaling pathways that coordinate these metabolic alterations. Two obvious families of molecules are considered key players in this regulation, sugars and phytohormones; although we do not rule out that additional molecules also could participate, such as mobile RNAs or even small peptides. The cross talk between these classes of signaling molecules is enormously complex. This will require innovative approaches from the scientific community to reveal the communication mechanisms and identify the key players orchestrating the metabolic response to prolonged darkness, which can lead leaf cells to an accelerated death or a surprising capacity for survival. Since our current food supply and distribution practices rely on the ability of harvested plants to repress senescence under storage conditions, which often require darkness, leaf metabolism under such conditions strongly merits further investigation.
MATERIALS AND METHODS
Plant Materials and Darkening Treatments
Arabidopsis thaliana (Arabidopsis) ecotype Columbia-0 was grown at 22°C under short-day conditions (8 h light and 16 h dark) with 180 µmol m−2 s−1 white light. Seven-week-old plants were used for darkening treatments. The individual covering of leaves (selected from among leaves 14–20), at most three per plant, was performed as described by Keech et al. (2007) and Brouwer et al. (2012). For the whole DP, plants were isolated from light by dark boxes and were returned to the same chamber to maintain the same temperature shift. For the IDL, one fully expanded leaf was covered on each side of the plant (a total of two covered leaves per plant) with plastic mittens coated with aluminum foil, while the rest of the rosette remained under the normal light/dark cycle (Weaver and Amasino, 2001; Keech et al., 2007). The progression of senescence was monitored at time points from 1 to 6 d of treatment. Illuminated leaves from plants with covered leaves were used as light control samples. Leaves of similar age were collected from dark-grown plants. At least eight to 10 individual plants were used for pool material.
RWC
The RWC was calculated using 100 × (FW − DW)/(TW − DW), where FW is the fresh matter, TW is the turgid matter after rehydrating the leaves for 24 h at 4°C in darkness, and DW is the dry matter after oven drying the leaves for 72 h at 65°C.
Photosynthetic Measurements
For chlorophyll measurements, samples from illuminated (control), IDL, and whole DP leaves were collected after 6 d of treatment, around midday (i.e. in the middle of the photoperiod), and immediately frozen in liquid nitrogen. Chlorophyll levels were estimated spectrophotometrically following an 80% (v/v) acetone extraction as described by Porra et al. (1989).
Steady-state fluorescence quenching was measured in air at 25 and 150 µmol m−2 s−1 at 22°C. The ΦPSII, ΦNPQ, and ΦNO were determined as described by Kramer et al. (2004). Measurements were performed as described by Keech et al. (2007) with detached leaves that were 45-min dark adapted prior to measurements. Each measurement was done in triplicate from three independent plants.
Microarrays
Samples were harvested at noon (i.e. in the middle of the photoperiod), at 0 d (Light D0), and after 3 and 6 d of the respective darkening treatments. For all conditions (Light D0, DP-D3, DP-D6, IDL-D3, and IDL-D6), three biological replicates were used, each of them being a pool of two leaves from independent plants. About 40 mg of frozen material was used for RNA isolation using the RNAeasy kit from Qiagen and followed by a DNase treatment (DNA-free; Ambion). RNA concentrations were determined using an ND-2000 spectrophotometer (NanoDrop Technologies), and RNA quality was assessed using the Agilent 2100 BioAnalyser 2100 and RNA 6000 Nano kits (Agilent). Microarrays were performed by NASCArrays (Affymetrix) using the Arabidopsis Gene 1.1 ST Array Strip technology from Affymetrix (Affymetrix).
Bioinformatics
Microarray Analysis
After reception of the CEL files, data were quality checked, preprocessed, and analyzed using R (R Development Core Team, 2013) and the Bioconductor (Huber et al., 2015), oligo (Carvalho and Irizarry, 2010), limma (Ritchie et al., 2015), and vsn (Huber et al., 2002) R packages (for the complete analysis transcript, see Supplemental Methods S1). Briefly, the array data were read into R using the oligo package that retrieved the pd.aragene.1.1.st array information, the features of which (i.e. probe set) were annotated using Affymetrix online resources through the getNetAffx function. The technical quality of the arrays was assessed using the arrayQualityMetrics R package (Kauffmann et al., 2009). Next, the resulting microarray data were subjected to a variance stabilizing transformation (within-array normalization) and to cyclic-loess between-array normalization. Following that step, the biological quality of the experiment was assessed by multidimensional scaling and PCA. Out of all 27,371 genes, 644 were targeted by several probe sets, in which case the probe set having recorded the highest fluorescence was kept. As seen from the PCA in Figure 2A and in Supplemental Methods S1, the biological replicates were highly similar, while the first and second dimensions (55% and 27% of the overall variance) were separating samples as expected, based on the experimental design (Light D0 versus DP versus IDL), both indicative of an adequate experimental control of the technical (microarray analysis) and biological variability. Finally, pairwise and time-series differential expression analyses were performed using limma. The results of the time-series differential expression analysis were converted into contingency tables and visualized as association plots.
The CEL files have been deposited to the Gene Expression Omnibus at the National Center for Biotechnology Information under accession number GSE98156.
Subset of Genes Associated with Shade Avoidance
To define a subset of genes typically responsive to the shade-avoidance syndrome, we compiled and analyzed several publicly available microarray analyses (GSE16333, the wild type in darkness, 1 h under far-red light, 24 h under far-red light; GSE17845, the wild type in white light, in dark, in far-red light; GSE35057, the wild type under high R/FR, under low R/FR; GSE35700, the wild type under high R/FR, under low R/FR). All raw intensity CEL files were imported into the Affymetrix expression console, and standard MAS5.0 normalization was initially conducted to determine present/absent/marginal calls for each probe set. Probe sets that were called present in two or more replicates were considered as expressed. Using R/Bioconductor, a GC-robust multiarray average normalization was carried out, and the resulting normalized intensities were used as the input for the differential expression analysis, which was carried out using the Cyber-T method, which implements a Bayesian method for the determination of probe sets showing significant changes in transcript abundance (Baldi and Long, 2001). We further manually curated a list of transcripts that have the same expression profile in response to low R/FR. The subset of approximately 40 genes is presented in Supplemental Table S2 and was used for comparison with DP and IDL treatments.
MapMan
Differential expression analysis was carried out between DP and IDL samples at D3 and D6, respectively and the resulting genes that were defined as significant (adjusted P < 0.001; differential expression carried out using limma) were cross referenced with Mapman annotations and sorted for metabolic functions. DEGs that correlated to either DP or IDL conditions were recorded and graphed, with light green corresponding to correlation with IDL conditions and dark green corresponding to correlation with DP conditions.
GO
Genes involved in the most significant expression profiles for DP and IDL were submitted to GO. This was performed using gProfiler (https://biit.cs.ut.ee/gprofiler/index.cgi; Reimand et al., 2007) with a moderate hierarchical filtering based on the best per parent, a manually uploaded input background, and a significance threshold determined by Benjamini-Hochberg false discovery rate (P < 0.05).
Building a Biological Network Based on GO Terms
A biological network was built using the Cytoscape (version 3.1.1; Shannon et al., 2003)-compatible plug-in ClueGO (version 2.1.6; Bindea et al., 2009). Genes issued from the comparison DP versus IDL using the limma package and being significantly differentially expressed at an adjusted P < 0.001 were used to create a biological network based on enriched GO terms and using the following parameters: min GO level = 1, max GO level = 19, number of genes per term = 4, and 20% genes per term; the minimum percentage for a cluster to be significant was 60%, both GO fusion and GO group were applied, Kappa score was fixed at 0.4, with an initial group size of 2 and a sharing group percentage set at 50%. The statistical test used for enrichment/depletion was a two-sided hypergeometric test with a Benjamini-Hochberg correction method applied.
Metabolomics
Starch Content
Starch was extracted from 20 mg fresh weight of material after ethanol extraction based on Smith and Zeeman (2006). After a digestion performed in 50 mm sodium acetate buffer, 0.45 units of α-amyloglucosidase, and 0.5 units of α-amylase overnight at 37°C, starch content was determined by quantifying Glc with the hexokinase/Glc-6-P dehydrogenase-based spectrophotometric assay.
Metabolite Extraction and Derivatization
Frozen and ground leaf material (15 ± 1 mg) was, with minor changes, extracted as described by Gullberg et al. (2004). Briefly, the samples were extracted in 1 mL of extraction mix (chloroform:methanol:water, 2:6:2 [v/v/v], including 7 ng μL−1 [13C3]myristic acid, [13C4]hexadecanoic acid, [2H7]cholesterol, [13C5]Pro, [2H4]putrescine, and [2H6]salicylic acid). A 3-mm tungsten bead was added to each sample before they were shaken at 30 Hz for 3 min in an MM 301 Vibration Mill (Retsch). The beads were removed prior to centrifugation at 15,000g for 10 min using a Mikro 220R instrument (Hettich, Zentrifugen). A 200μL supernatant was transferred to a GC-micro vial (Chromatol) and evaporated to dryness using a miVac quattro concentrator (Barnstead Genevac). Samples were derivatized by shaking with 30 μL of methoxyamine hydrochloride (15 mg mL−1) in pyridine for 10 min at 5°C before incubation for 16 h at room temperature. Samples were then trimethylsilylated with 30 μL of MSTFA with 1% (v/v) chlorotrimethylsilane and incubated 1 h at room temperature. Heptane (30 μL, including 15 ng μL−1 methylstearate) was added, and samples were vortexed before GC-MS analysis.
GC-MS Analysis
Samples were analyzed as described by Gullberg et al. (2004) using GC-ToF-MS together with blank control samples and a series of n-alkanes (C12–C40). Samples (1 μL) were injected into an Agilent Technologies 7890A GC system equipped with a 30 × 0.25-mm-diameter fused silica capillary column with a bonded 0.25-μm Durabond DB-5MSUI stationary phase (part no. 122-5222UI; Agilent J&W GC columns) by a CTC PAL Systems auto sampler. GC settings were as follows: injector temperature, 260°C; front inlet septum purge flow rate, 3 mL min−1; gas flow rate, 1 mL min−1; column temperature, 70°C for 2 min, then increased by 20°C min−1 to 320°C (held for 8 min). The effluent was introduced into the ion source of a Pegasus HT GC-high-throughput TOF-MS system (LECO). Electron impact was used for ionization. Transfer line temperature was 270°C, and ion source temperature was 200°C. Twenty spectra per second were recorded with a 50 to 800 mass-to-charge ratio range and 290-s solvent delay. All raw files were converted to NetCDF format by ChromaTOF software and exported to MATLAB version 8.1 (Mathworks). Peak detection and peak area calculations of both labeled and unlabeled features were done as described by Lindén et al. (2016). Data were normalized with respect to internal standards according to Redestig et al. (2009). Unlabeled metabolites were identified by comparing their retention indices and mass spectra with entries in commercial and in-house mass spectra libraries using NIST MS Search 2.0 (National Institute of Standards and Technology). For labeled metabolites, see “13C Labeling” below. Eight to 10 independent biological replicates were used for the metabolite analysis.
HPLC
HPLC was performed using an Elite La Chrome instrument (Hitachi) with Purosphere STAR RP-18 end-capped column (Merck). Samples were derivatized using the AccQTag Ultra Derivatization Kit (Waters). Separation was performed using a mixture of 0.1% HCOOH in ultrapure water (A) and 90% acetonitrile in 10% ultrapure water (B) as the mobile phase, at a flow rate of 2 mL min−1, and at the following gradients: 0 to 4.7 min, 99.9% A and 0.1% B; 4.7 to 8 min, 90.9% A and 9.1% B; 8 to 10 min, 80% A and 20% B; 10 to 12 min, 99.9% A and 0.1% B.
13C Labeling
The carbon dioxide used was 13CO2 (isotopic purity, 90 atom % 13C, 18O < 1.5 atom %) in a 25-L gas bottle purchased from Spectra Products and 12CO2 (AGA) in a 5-L gas bottle. The air-gas mix was tapped from the internal house gas supply. Labeling chamber construction was as described by Lindén et al. (2016). The temperature in the labeling cabinet was kept at 22°C, and the relative humidity was 80% to 85%. The 13CO2 gas pressure was 1 bar, and the air flow to the CO2 scrubber was 2 L min−1. During sampling, leaves were cut at the petiole, transferred to a 20-mL scintillator tube, and dropped into liquid nitrogen (whole procedure, ∼10 s).
In-house software, 13C_est, was used to correct for the natural abundance of 13C and isotope contributions from tetramethylsilane groups and to calculate the percentage of 13C incorporation for each identified metabolite pool. (Notably, the number of atoms labeled for a single molecule was not investigated.) Three to four independent biological replicates were used.
Statistical Analysis
Multivariate statistical investigations were performed using SIMCA 13.0 software (Umetrics). All variables were log10 transformed, centered, and scaled to unit variance before further analysis. PCA was used to obtain an overview, find clusters, and identify outliers in the data. OPLS-DA is a multivariate classification technique that is used to predict groupings for observations and to characterize the groups (Trygg and Wold, 2002; Bylesjö et al., 2006). This technique was used to find the combination of metabolites that provided the optimal discrimination between dark treatment (IDL and DL) and light. Normalized peak areas (metabolites) were used as descriptors (X), and treatment (light, IDL, DL) was used as the response (Y). R2X is the cumulative modeled variation in X, R2Y is the cumulative modeled variation in Y, and Q2Y is the cumulative predicted variation in Y, using cross-validation.
Hierarchical Clustering for Metabolomics
The relative abundance of metabolites was obtained by normalizing the abundance value to their maximum, in this case set to 1. Hierarchical clustering was performed using the MultiExperiment Viewer (version 4.8.1; Saeed et al., 2003) with the following parameters: average linkage, leaf order optimization for metabolites, and Euclidean distance.
Accession Numbers
The Arabidopsis Genome Initiative numbers of featured genes are as follows: ADC1 (AT2G16500), ADC2 (AT4G34710), ASN1 (AT3G47340), AtSIP2 (AT3G57520), CYTC-2 (AT4G10040), ETF-QO (AT2G43400), HFR1 (AT1G02340), IMS3 (AT5G23010), KAT2 (AT2G33150), PAO (AT3G44880), PHYA (AT1G09570), PHYB (AT2G18790), PIF3 (AT1G09530), PRK (AT1G32060), PXA1 (AT4G39850), RbcS2B (AT5G38420), SAG2 (AT5G60360), SAG12 (AT5G45890), SAG21 (AT5G45890), SAMDC (AT3G02470), SGR1 (AT4G22920), SPDS2 (AT1G70310), and XTR7 (AT1G32060).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Physiological analysis of Arabidopsis leaves following 6 d of IDL and DP conditions.
Supplemental Figure S2. Full biological network of enriched GO terms from significantly differentially expressed genes in DP versus IDL conditions at D3 and D6.
Supplemental Figure S3. Transcriptomic analysis of the conserved and diverging responses to darkness in DP and IDL conditions.
Supplemental Figure S4. Transcript abundance of selected genes at D0, D3, and D6 under DP or IDL conditions.
Supplemental Figure S5. Transcriptomic analysis of markers of stress in leaves under DP and IDL conditions.
Supplemental Figure S6. Hierarchical clustering of metabolite abundances extracted from illuminated control, DP, and IDL leaves.
Supplemental Figure S7. Transcriptomic analysis of protein metabolism in leaves under DP and IDL conditions.
Supplemental Table S1. Total present set of transcriptomic data of leaves sampled after 0, 3, and 6 d of DP or IDL conditions.
Supplemental Table S2. Subset of genes specifically regulated in response to shade avoidance.
Supplemental Table S3. List of genes associated with cell wall metabolism and gluconeogenesis functions, as defined by MapMan annotations.
Supplemental Table S4. Processed data from GC-ToF-MS for relative abundance of metabolites.
Supplemental Table S5. Relative abundance (12C + 13C) of identified metabolites under IDL and light conditions at D3 and D6.
Acknowledgments
We thank Dr. Daniela Liebsch for critical comments on the article and Hans Stenlund for help with the 13C-labeling determination.
Footnotes
This work was supported by the Swedish Research Council “VetenskapsRådet” (grant: 621-2014-4688) as well as by the Kempe Foundations (Gunnar Öquist Fellowship) and the Carl Tryggers Stiftelse (CTS14-247 & CTS15-262).
Articles can be viewed without a subscription.
References
- Abdallah M, Dubousset L, Meuriot F, Etienne P, Avice JC, Ourry A (2010) Effect of mineral sulphur availability on nitrogen and sulphur uptake and remobilization during the vegetative growth of Brassica napus L. J Exp Bot 61: 2635–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo WL, Ishizaki K, Nunes-Nesi A, Larson TR, Tohge T, Krahnert I, Witt S, Obata T, Schauer N, Graham IA, et al. (2010) Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22: 1549–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi P, Long AD (2001) A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17: 509–519 [DOI] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J (2009) ClueGO: a Cytoscape plug-in to decipher functionally grouped Gene Ontology and pathway annotation networks. Bioinformatics 25: 1091–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohner A, Kojima S, Hajirezaei M, Melzer M, von Wirén N (2015) Urea retranslocation from senescing Arabidopsis leaves is promoted by DUR3-mediated urea retrieval from leaf apoplast. Plant J 81: 377–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer B, Gardeström P, Keech O (2014) In response to partial plant shading, the lack of phytochrome A does not directly induce leaf senescence but alters the fine-tuning of chlorophyll biosynthesis. J Exp Bot 65: 4037–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer B, Ziolkowska A, Bagard M, Keech O, Gardeström P (2012) The impact of light intensity on shade-induced leaf senescence. Plant Cell Environ 35: 1084–1098 [DOI] [PubMed] [Google Scholar]
- Brychkova G, Alikulov Z, Fluhr R, Sagi M (2008) A critical role for ureides in dark and senescence-induced purine remobilization is unmasked in the Atxdh1 Arabidopsis mutant. Plant J 54: 496–509 [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, et al. (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42: 567–585 [DOI] [PubMed] [Google Scholar]
- Bylesjö M, Rantalainen M, Cloarec O, Nicholson JK, Holmes E, Trygg J (2006) OPLS discriminant analysis: combining the strengths of PLS-DA and SIMCA classification. J Chemometr 20: 341–351 [Google Scholar]
- Carvalho BS, Irizarry RA (2010) A framework for oligonucleotide microarray preprocessing. Bioinformatics 26: 2363–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrobok D, Law SR, Brouwer B, Lindén P, Ziolkowska A, Liebsch D, Narsai R, Szal B, Moritz T, Rouhier N, et al. 2016) Dissecting the metabolic role of mitochondria during developmental leaf senescence. Plant Physiol 172: 2132–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D, St Onge KR, Voesenek LA, Pierik R, Sasidharan R (2016) Ethylene- and shade-induced hypocotyl elongation share transcriptome patterns and functional regulators. Plant Physiol 172: 718–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieuaide M, Brouquisse R, Pradet A, Raymond P (1992) Increased fatty acid beta-oxidation after glucose starvation in maize root tips. Plant Physiol 99: 595–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubay R, Pessarakli M (1995) Physiological mechanisms of nitrogen absorption and assimilation in plants under stressful conditions. In Pessarakli M, ed, Handbook of Plant and Crop Physiology. Marcel Dekker, New York, pp 605–626 [Google Scholar]
- Fan J, Yu L, Xu C (2017) A central role for triacylglycerol in membrane lipid breakdown, fatty acid β-oxidation, and plant survival under extended darkness. Plant Physiol 174: 1517–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Whitelam GC (2005) Phytochromes and shade-avoidance responses in plants. Ann Bot 96: 169–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan S, Amasino RM (1997) Making sense of senescence (molecular genetic regulation and manipulation of leaf senescence). Plant Physiol 113: 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert GA, Gadush MV, Wilson C, Madore MA (1998) Amino acid accumulation in sink and source tissues of Coleus blumei Benth. during salinity stress. J Exp Bot 49: 107–114 [Google Scholar]
- Gullberg J, Jonsson P, Nordström A, Sjöström M, Moritz T (2004) Design of experiments: an efficient strategy to identify factors influencing extraction and derivatization of Arabidopsis thaliana samples in metabolomic studies with gas chromatography/mass spectrometry. Anal Biochem 331: 283–295 [DOI] [PubMed] [Google Scholar]
- Hoch G. (2007) Cell wall hemicelluloses as mobile carbon stores in non-reproductive plant tissues. Funct Ecol 21: 823–834 [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, et al. (2015) Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 12: 115–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, von Heydebreck A, Sültmann H, Poustka A, Vingron M (2002) Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics (Suppl 1) 18: S96–S104 [DOI] [PubMed] [Google Scholar]
- Irving DE, Silsbury JH (1988) The respiration of mature field bean (Vicia faba L.) leaves during prolonged darkness. Ann Bot (Lond) 62: 473–479 [Google Scholar]
- Ishizaki K, Schauer N, Larson TR, Graham IA, Fernie AR, Leaver CJ (2006) The mitochondrial electron transfer flavoprotein complex is essential for survival of Arabidopsis in extended darkness. Plant J 47: 751–760 [DOI] [PubMed] [Google Scholar]
- Kauffmann A, Gentleman R, Huber W (2009) arrayQualityMetrics: a Bioconductor package for quality assessment of microarray data. Bioinformatics 25: 415–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech O, Pesquet E, Ahad A, Askne A, Nordvall D, Vodnala SM, Tuominen H, Hurry V, Dizengremel P, Gardeström P (2007) The different fates of mitochondria and chloroplasts during dark-induced senescence in Arabidopsis leaves. Plant Cell Environ 30: 1523–1534 [DOI] [PubMed] [Google Scholar]
- Keech O, Pesquet E, Gutierrez L, Ahad A, Bellini C, Smith SM, Gardeström P (2010) Leaf senescence is accompanied by an early disruption of the microtubule network in Arabidopsis. Plant Physiol 154: 1710–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohnen MV, Schmid-Siegert E, Trevisan M, Petrolati LA, Sénéchal F, Müller-Moulé P, Maloof J, Xenarios I, Fankhauser C (2016) Neighbor detection induces organ-specific transcriptomes, revealing patterns underlying hypocotyl-specific growth. Plant Cell 28: 2889–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer DM, Johnson G, Kiirats O, Edwards GE (2004) New fluorescence parameters for the determination of QA redox state and excitation energy fluxes. Photosynth Res 79: 209–218 [DOI] [PubMed] [Google Scholar]
- Kunz HH, Scharnewski M, Feussner K, Feussner I, Flügge UI, Fulda M, Gierth M (2009) The ABC transporter PXA1 and peroxisomal beta-oxidation are vital for metabolism in mature leaves of Arabidopsis during extended darkness. Plant Cell 21: 2733–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HM, Peng SSY, Coruzzi GM (1994) Metabolic regulation of the gene encoding glutamine-dependent asparagine synthetase in Arabidopsis thaliana. Plant Physiol 106: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Titus JS (1992) Nitrogen accumulation and nitrate reductase activity in MM. 106 apple trees as affected by nitrate supply. J Hortic Sci Biotechnol 67: 273–281 [Google Scholar]
- Liebsch D, Keech O (2016) Dark-induced leaf senescence: new insights into a complex light-dependent regulatory pathway. New Phytol 212: 563–570 [DOI] [PubMed] [Google Scholar]
- Lindén P, Keech O, Stenlund H, Gardeström P, Moritz T (2016) Reduced mitochondrial malate dehydrogenase activity has a strong effect on photorespiratory metabolism as revealed by 13C labelling. J Exp Bot 67: 3123–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackender RO, Leech RM (1974) The galactolipid, phospholipid, and fatty acid composition of the chloroplast envelope membranes of Vicia faba L. Plant Physiol 53: 496–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Dudareva N (2012) The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu Rev Plant Biol 63: 73–105 [DOI] [PubMed] [Google Scholar]
- Makino A, Mae T, Ohira K (1984) Relation between nitrogen and ribulose-1,5-bisphosphate carboxylase in rice leaves from emergence through senescence. Plant Cell Physiol 25: 429–437 [Google Scholar]
- Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A (2010) Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann Bot 105: 1141–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Shigeoka S (2008) Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol 147: 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters S, Egert A, Stieger B, Keller F (2010) Functional identification of Arabidopsis ATSIP2 (At3g57520) as an alkaline α-galactosidase with a substrate specificity for raffinose and an apparent sink-specific expression pattern. Plant Cell Physiol 51: 1815–1819 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Pracharoenwattana I, Cornah JE, Smith SM (2005) Arabidopsis peroxisomal citrate synthase is required for fatty acid respiration and seed germination. Plant Cell 17: 2037–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan JD, Weaver RJ (1969) Influence of benzyladenine, leaf darkening, and ringing on movement of C-labeled assimilates into expanded leaves of Vitis vinifera L. Plant Physiol 44: 1247–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2008) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Rabe E, Lovatt CJ (1984) De novo arginine biosynthesis in leaves of phosphorus-deficient citrus and poncirus species. Plant Physiol 76: 747–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redestig H, Fukushima A, Stenlund H, Moritz T, Arita M, Saito K, Kusano M (2009) Compensation for systematic cross-contribution improves normalization of mass spectrometry based metabolomics data. Anal Chem 81: 7974–7980 [DOI] [PubMed] [Google Scholar]
- Reimand J, Kull M, Peterson H, Hansen J, Vilo J (2007) g:Profiler: a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res 35: W193–W200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs JW, Rockwell NC, Cavales PC, Callis J (2016) Identification of the plant ribokinase and discovery of a role for Arabidopsis ribokinase in nucleoside metabolism. J Biol Chem 291: 22572–22582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulin S, Buchala AJ, Fincher GB (2002) Induction of (1→3,1→4)-β-D-glucan hydrolases in leaves of dark-incubated barley seedlings. Planta 215: 51–59 [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Sakuraba Y, Balazadeh S, Tanaka R, Mueller-Roeber B, Tanaka A (2012) Overproduction of chl B retards senescence through transcriptional reprogramming in Arabidopsis. Plant Cell Physiol 53: 505–517 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart C. (1994) Gene expression during leaf senescence. New Phytol 126: 419–448 [DOI] [PubMed] [Google Scholar]
- Smith AM, Zeeman SC (2006) Quantification of starch in plant tissues. Nat Protoc 1: 1342–1345 [DOI] [PubMed] [Google Scholar]
- Sobieszczuk-Nowicka E, Legocka J (2014) Plastid-associated polyamines: their role in differentiation, structure, functioning, stress response and senescence. Plant Biol (Stuttg) 16: 297–305 [DOI] [PubMed] [Google Scholar]
- Soudry E, Ulitzur S, Gepstein S (2005) Accumulation and remobilization of amino acids during senescence of detached and attached leaves: in planta analysis of tryptophan levels by recombinant luminescent bacteria. J Exp Bot 56: 695–702 [DOI] [PubMed] [Google Scholar]
- Stasolla C, Katahira R, Thorpe TA, Ashihara H (2003) Purine and pyrimidine nucleotide metabolism in higher plants. J Plant Physiol 160: 1271–1295 [DOI] [PubMed] [Google Scholar]
- Stitt M, Wirtz W, Gerhardt R, Heldt HW, Spencer C, Walker D, Foyer C (1985) A comparative study of metabolite levels in plant leaf material in the dark. Planta 166: 354–364 [DOI] [PubMed] [Google Scholar]
- Szecowka M, Heise R, Tohge T, Nunes-Nesi A, Vosloh D, Huege J, Feil R, Lunn J, Nikoloski Z, Stitt M, et al. (2013) Metabolic fluxes in an illuminated Arabidopsis rosette. Plant Cell 25: 694–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K (2002) Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J 29: 417–426 [DOI] [PubMed] [Google Scholar]
- Thompson JE, Hopkins MT, Taylor C, Wang TW (2004) Regulation of senescence by eukaryotic translation initiation factor 5A: implications for plant growth and development. Trends Plant Sci 9: 174–179 [DOI] [PubMed] [Google Scholar]
- Tohge T, Watanabe M, Hoefgen R, Fernie AR (2013) Shikimate and phenylalanine biosynthesis in the green lineage. Front Plant Sci 4: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trygg J, Wold S (2002) Orthogonal projections to latent structures (O-PLS). J Chemometr 16: 119–128 [Google Scholar]
- Urquhart AA, Joy KW (1981) Use of phloem exudate technique in the study of amino acid transport in pea plants. Plant Physiol 68: 750–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, et al. (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol 138: 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff E, Schwacke R, Schneider A, Desimone M, Flügge UI, Kunze R (2006) Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol 141: 776–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernotte C, Solis C, Moya I, Maison B, Briantais JM, Arrio B, Johannin G (1983) Multiple effects of linolenic acid addition to pea thylakoids. Biochim Biophys Acta 725: 376–383 [Google Scholar]
- Watanabe M, Balazadeh S, Tohge T, Erban A, Giavalisco P, Kopka J, Mueller-Roeber B, Fernie AR, Hoefgen R (2013) Comprehensive dissection of spatiotemporal metabolic shifts in primary, secondary, and lipid metabolism during developmental senescence in Arabidopsis. Plant Physiol 162: 1290–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LM, Amasino RM (2001) Senescence is induced in individually darkened Arabidopsis leaves, but inhibited in whole darkened plants. Plant Physiol 127: 876–886 [PMC free article] [PubMed] [Google Scholar]
- Zrenner R, Stitt M, Sonnewald U, Boldt R (2006) Pyrimidine and purine biosynthesis and degradation in plants. Annu Rev Plant Biol 57: 805–836 [DOI] [PubMed] [Google Scholar]