Biotin attachment domain-containing proteins contribute to irreversible inhibition of ACCase under normal growth conditions and under conditions of fatty acid oversupply in Arabidopsis thaliana.
Abstract
The first committed step in fatty acid synthesis is mediated by acetyl-CoA carboxylase (ACCase), a biotin-dependent enzyme that carboxylates acetyl-CoA to produce malonyl-CoA. ACCase can be feedback regulated by short-term or long-term exposure to fatty acids in the form of Tween 80 (predominantly containing oleic acid), which results in reversible or irreversible ACCase inhibition, respectively. Biotin attachment domain-containing (BADC) proteins are inactive analogs of biotin carboxyl transfer proteins that lack biotin, and their incorporation into ACCase down-regulates its activity by displacing active (biotin-containing) biotin carboxyltransferase protein subunits. Arabidopsis (Arabidopsis thaliana) lines containing T-DNA insertions in BADC1, BADC2, and BADC3 were used to generate badc1 badc2 and badc1 badc3 double mutants. The badc1 badc3 mutant exhibited normal growth and development; however, ACCase activity was 26% higher in badc1 badc3 and its seeds contained 30.1% more fatty acids and 32.6% more triacylgycerol relative to wild-type plants. To assess whether BADC contributes to the irreversible phase of ACCase inhibition, cell suspension cultures were generated from the leaves of badc1 badc3 and wild-type plants and treated with 10 mm Tween 80. Reversible ACCase inhibition was similar in badc1 badc3 and wild-type cultures after 2 d of Tween 80 treatment, but irreversible inhibition was reduced by 50% in badc1 badc3 relative to wild-type plants following 4 d of Tween 80 treatment. In this study, we present evidence for two important homeostatic roles for BADC proteins in down-regulating ACCase activity: by acting during normal growth and development and by contributing to its long-term irreversible feedback inhibition resulting from the oversupply of fatty acids.
Acetyl-CoA carboxylase (ACCase; EC 6.4.1.2) is the enzyme responsible for the first committed step in fatty acid (FA) synthesis, the carboxylation of acetyl-CoA to produce malonyl-CoA (Ohlrogge and Jaworski, 1997; Cronan and Waldrop, 2002). The reaction comprises two sequential partial reactions (Guchhait et al., 1974a, 1974b; Polakis et al., 1974; Blanchard and Waldrop, 1998; Cronan and Waldrop, 2002): the first involves the ATP-dependent carboxylation of a biotin moiety, and the second involves transfer of the activated carboxyl group to acetyl-CoA to produce malonyl-CoA.
There are two distinct classes of ACCase. Most plant chloroplasts and bacteria contain a multisubunit, or heteromeric, ACCase that is readily dissociated into its component proteins (Kondo et al., 1991; Li and Cronan, 1992a, 1992b; Choi et al., 1995). By contrast, the plant cytosol, mammals, and fungi contain a single large multifunctional, homomeric, polypeptide (Wei and Tong, 2015; Hunkeler et al., 2016). In this study, we use ACCase to refer to the dissociable heteromeric ACCase. In plants, the plastid-localized heteromeric ACCase primarily contributes malonyl-CoA to de novo FA synthesis, whereas the cytosolic homomeric form contributes malonyl-CoA to a number of other processes, including FA elongation and polyketide biosynthesis.
The heteromeric ACCase is composed of four distinct subunits: biotin carboxyl transfer protein (BCCP), biotin carboxylase (BC), and α- and β-carboxyltransferases (CT). These subunits are organized in two separable subcomplexes, one comprising BCCP and BC, which mediates the carboxylation of biotin, and a second α-CT/β-CT complex that catalyzes the carboxyltransferase reaction.
Since it catalyzes the rate-limiting step in FA synthesis, ACCase is tightly regulated by a variety of mechanisms (Salie and Thelen, 2016). For example, the BCCP2 subunit is transcriptionally regulated by the WRINKLED1 transcription factor (Maeo et al., 2009). ACCase also has been shown to be biochemically regulated by multiple independent mechanisms, including pH change and thioredoxin activity, which results in the reduction of a disulfide bond between the α- and β-CT subunits (Kozaki and Sasaki, 1999; Kozaki et al., 2001). A small protein, PII, which exists as a homotrimer, has been shown to reversibly bind to the BCCP biotin cofactor in an ATP-dependent manner and to down-regulate ACCase activity. The PII-BCCP association can be destabilized by 2-oxoglutarate (Feria Bourrellier et al., 2010; Rodrigues et al., 2014; Gerhardt et al., 2015; Hauf et al., 2016).
Another class of proteins, annotated as biotin attachment domain-containing (BADC) proteins, also associate with ACCase (Olinares et al., 2010). All three Arabidopsis (Arabidopsis thaliana) BADC isoforms interact with BCCP isoforms 1 and 2 (Salie et al., 2016). While BADC proteins share 25% to 30% sequence similarity to BCCP subunits, their sequences show significant divergence with respect to the biotinylation motif such that they are unable to bind biotin, rendering them inactive (Salie et al., 2016). The displacement of competent BCCP subunits in ACCase with inactive BADC subunits thereby reduces the enzyme’s capacity to perform the carboxylation half reaction (Shintani and Ohlrogge, 1995).
Feedback inhibition of ACCase was initially demonstrated in tobacco (Nicotiana tabacum) suspension cell cultures fed with Tween-oleic acid (18:1; Shintani and Ohlrogge, 1995). We subsequently used a Brassica napus embryo-derived cell culture to characterize the feedback inhibition of ACCase and demonstrated that it occurs in two distinct phases (Andre et al., 2012). The first phase is the reversible inhibition of ACCase, which occurs in response to short-term exposure of cells to Tween-18:1 and results from the allosteric inhibition of ACCase by 18:1-acyl carrier protein. The second phase of inhibition occurs upon exposure to Tween-18:1 for several days and is not reversible upon transfer to Tween-free medium.
In this work, we characterize the physiological consequences of null mutations in various combinations of the three individual BADC isoforms using available Arabidopsis T-DNA insertion lines. We crossed these lines to create badc1 badc2 and badc1 badc3 double mutants and used the single and double mutants to quantify the effect of BADC on seed oil accumulation. These studies revealed that badc1 badc3, which grows similarly to the wild type, produces seeds containing 15% to 20% higher total fatty acids (TFAs) and triacylglycerol (TAG) than the wild type. To test the hypothesis that BADC isoforms contribute to the feedback inhibition of ACCase, cell suspension cultures were prepared from wild-type and badc1 badc3 leaves. While the reversible phase of inhibition did not differ between wild-type and badc1 badc3 lines, irreversible inhibition was mitigated significantly in the double mutant, demonstrating a role for BADC1 and BADC3 in the irreversible feedback inhibition of ACCase.
RESULTS
[14C]Acetate Incorporation into Total Lipids of Arabidopsis Cell Suspension Culture Demonstrated Reversible and Irreversible Inhibition of ACCase by Tween 80
Tween 80, which contains primarily 18:1, was utilized as a delivery system for FAs into T87 Arabidopsis cell suspension culture. The rate of [14C]acetate incorporation into FAs was determined by in vivo labeling using [14C]acetate, followed by total lipid extraction and scintillation counting. Supplementation of the NT-1 medium to 10 mm with Tween 80 resulted in a reduction in the rate of [14C]acetate incorporation within 3 h.
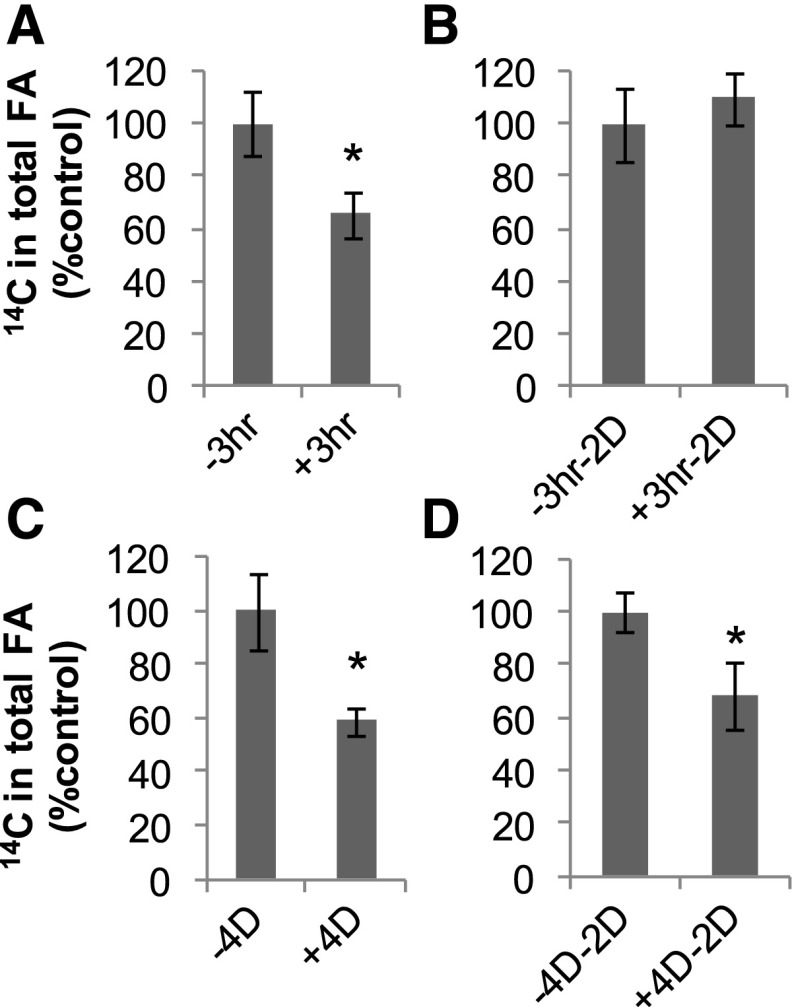
Cell cultures were maintained in NT-1 medium supplemented to 10 mm Tween 80 for either 3 h to induce short-term reversible ACCase inhibition or 4 d to induce long-term irreversible ACCase inhibition (Fig. 1). The rates of [14C]acetate incorporation into FAs were reduced by approximately 25% to 40% after both short-term (Fig. 1A) and long-term (Fig. 1C) exposure to Tween 80. Two days after the transfer of cells to Tween 80-free medium following a short-term exposure to Tween 80, rates of ACCase returned to those of the untreated controls (Fig. 1B). In contrast, cells transferred to Tween 80-free medium for 2 d after a 4-d exposure to Tween 80 displayed irreversible inhibition (Fig. 1D).
Figure 1.
[14C]Acetate incorporation into total lipids demonstrated reversible and irreversible inhibitions of ACCase by Tween 80 in Arabidopsis T87 cell suspension culture. Following short-term treatment (A), the inhibition of [14C]acetate incorporation was completely reversible in the absence of Tween 80 (B), but following long-term treatment (C), it persisted in the absence of Tween 80 (D). + or − indicates with or without Tween 80 treatment for the number of hours (hr)/days (D) specified. Asterisks indicate significant differences compared with the Tween 80-free controls, which were determined by Student’s t test (P < 0.05). Values are presented as means ± sd of three biological replicates.
Irreversible Inhibition of ACCase by Tween 80 Is Not the Result of Transcriptional Control
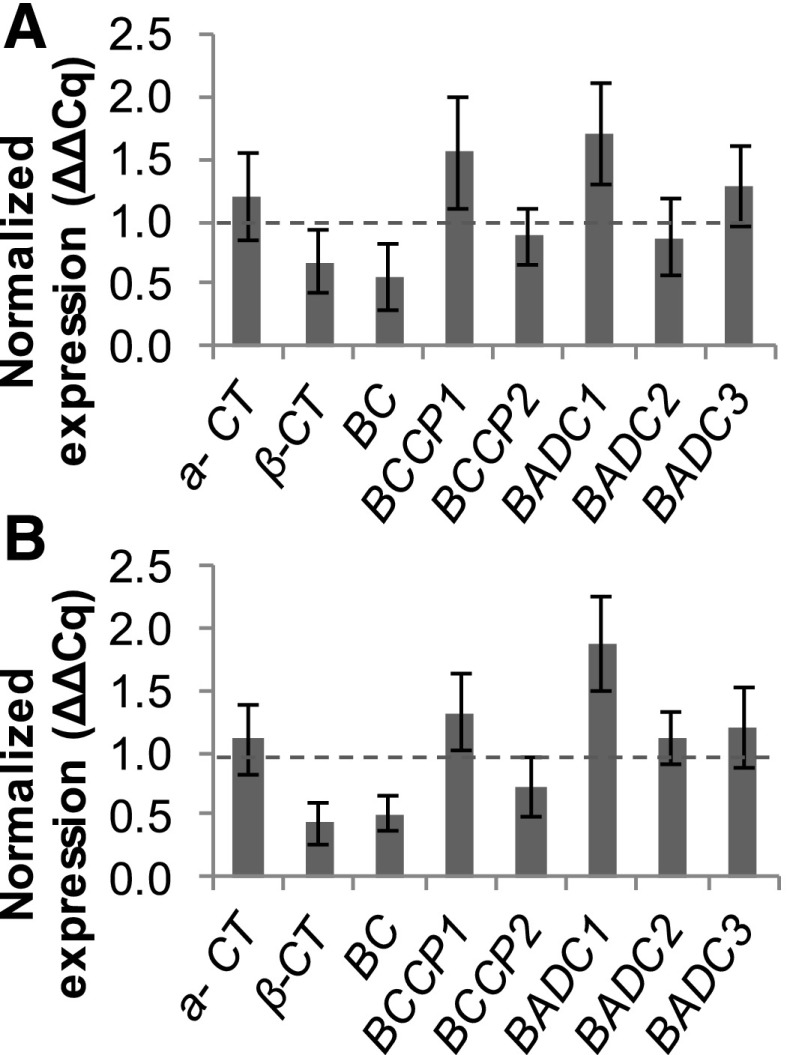
To determine if irreversible inhibition resulting from long-term treatment of Tween 80 is regulated through transcriptional control, RNAs were extracted from T87 cells exposed to long-term (4 d) Tween 80 treatment (Fig. 2A) and from those exposed to Tween 80 for 4 d followed by a 2-d incubation in Tween 80-free medium (Fig. 2B). This RNA was subjected to quantitative reverse transcription PCR (RT-qPCR) to estimate the transcript abundance of ACCase subunit-encoding genes, along with those of BADC1, BADC2, and BADC3. Despite the significant reduction in ACCase activity observed after 4 d of Tween 80 exposure, and after 4 d of Tween 80 exposure followed by 2 d of incubation in Tween 80-free medium, transcript levels of individual ACCase subunit and individual BADC subunits did not differ significantly (P < 0.05) from those of non-Tween 80-exposed controls using relative expression (REST)-specific analysis (Pfaffl et al., 2002). The levels of transcripts corresponding to these genes were not significantly affected, suggesting that irreversible inhibition is not the result of transcriptional control.
Figure 2.
Expression levels of ACCase-encoding genes in Arabidopsis T87 cells were not significantly affected 4 d after exposure to 10 mm Tween 80 (A) or after 4 d of exposure to Tween 80 followed by 2 d of incubation in Tween 80-free medium (B). RT-qPCR values are presented as percentages of untreated controls normalized as described in “Materials and Methods.” All data are means ± sd (n = 3), and no values were found to differ significantly (P < 0.05) from controls using mean crossing point deviation analysis computed by the relative expression (REST) software algorithm (Pfaffl et al., 2002).
Molecular Characterization of badc1, badc2, and badc3 T-DNA Insertion Mutants and Combinations Thereof
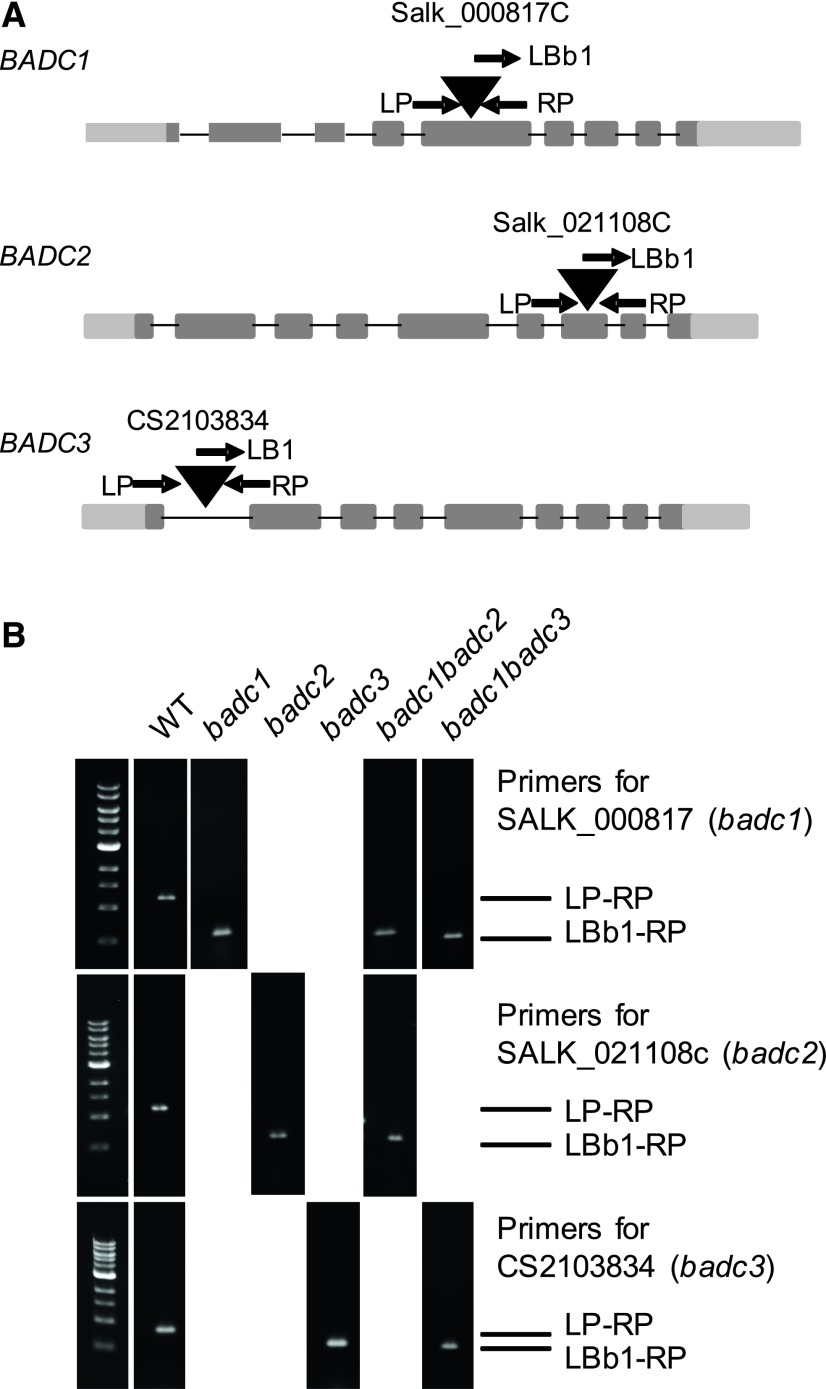
T-DNA insertion mutants of BADC1, BADC2, and BADC3 were genetically characterized. We confirmed that badc1 (Salk 000817C), badc2 (Salk 021108C), and badc3 (CS2103834) contain T-DNAs inserted in the fifth exon, seventh exon, and first intron, respectively (Fig. 3A). RT-qPCR of total RNA extracted from 14-d-old leaves confirmed that these T-DNA insertion lines were null mutants for the corresponding genes (Supplemental Fig. S1). In an attempt to create double mutants, homozygous badc1, badc2, and badc3 lines were crossed using pollen donors in both directions. The genotypes of the F3 lines were determined using gene-specific primer pairs in combination with T-DNA-specific primers. Two of the three possible homozygous double mutant lines (i.e. badc1 badc2 and badc1 badc3) were identified (Fig. 3B), while a homozygous badc2 badc3 line was not identified despite extensive attempts. However, the genotyping did identify multiple lines that were homozygous for one mutation and heterozygous for the other mutation. For example, badc2 badc3 line 21 was homozygous for badc2 and heterozygous for badc3, and badc2 badc3 line 79 was homozygous for badc3 and heterozygous for badc2 (Fig. 4A).
Figure 3.
Genotyping of BADC1, BADC2, and BADC3. A, Genomic organization of the T-DNA insertion knockout line in the Columbia-0 (Col-0) background. The positions of T-DNA insertions are indicated by triangles. The positions of primers used for genotyping are indicated by arrows. B, Individual plants were determined to be either heterozygous or homozygous using PCR with the indicated gene-specific primer pairs and combinations with T-DNA-specific primers. Wild-type (WT) Col-0 was used as a positive control for the gene-specific primer pairs and as a negative control for T-DNA-specific primer pairs.
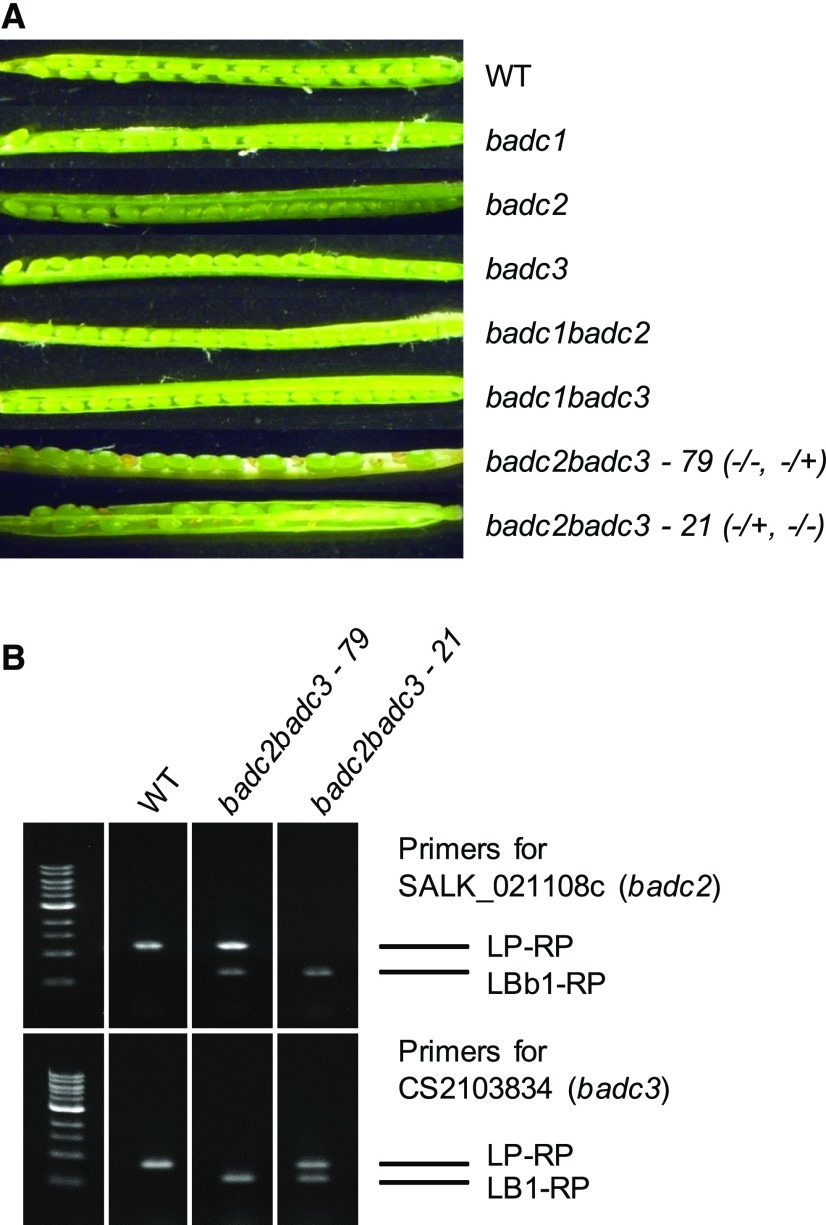
Figure 4.
Phenotypes of developing siliques of badc mutants. A, No aborted seeds were observed in siliques of the wild type (WT) or single badc mutants, while the double mutant badc2 badc3 resulted in embryo abortion. B, Individual plants were determined to be either heterozygous or homozygous using PCR with the indicated gene-specific primer pairs and combinations with T-DNA-specific primers. Wild-type Col-0 was used as a positive control for the gene-specific primer pairs and as a negative control for T-DNA-specific primer pairs. No badc2 badc3 homozygous line was obtained.
A possible explanation for the failure to identify homozygous badc2 badc3 plants (Fig. 4B) is that they are embryo lethal. To test this hypothesis, we visually inspected siliques from the homozygous wild type, badc single and double mutants, along with badc2 badc3 line 21 and badc2 badc3 line 79. Analysis of the seeds in developing siliques of badc2 badc3 line 79 and badc2 badc3 line 21 (for representative siliques, see Fig. 4A) revealed viable:aborted seed ratios of 699:213 and 489:157, respectively, under conditions where no aborted seeds were observed in siliques of single badc mutants or the wild type (Fig. 4A). The observed values for viable:aborted seed ratios are consistent with the expected 3:1 ratio predicted for the embryo lethality of homozygous badc2 badc3 (using a χ2 test and 0.05 level of significance; Supplemental Table S1).
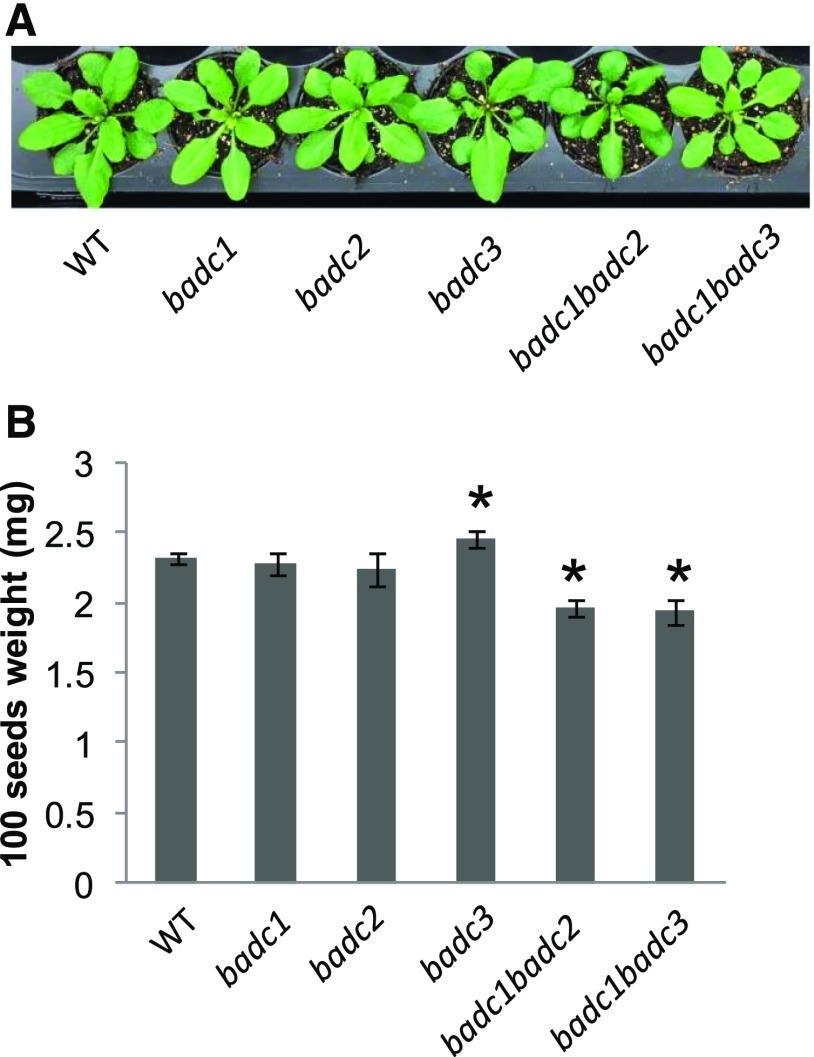
Phenotypes of the Homozygous badc Single and Available Double Mutant Lines
Homozygous badc1, badc2, badc3, badc1 badc2, and badc1 badc3 plants exhibited growth and development similar to those of wild-type plants grown under equivalent conditions (Fig. 5A). We next investigated the impact of these mutations on seed fill and observed that desiccated seeds harvested from the badc1 and badc2 mutants showed no significant differences in weight relative to the wild type. However, badc3 showed a small significant increase in desiccated seed weight (2.46 mg per 100 seeds) compared with the wild type (2.32 ± 0.04 mg per 100 seeds), whereas the two homozygous double mutants (badc1 badc2 and badc1 badc3) showed small significant (Student’s t test, P < 0.05) decreases in weight, 1.96 ± 0.06 mg per 100 seeds for badc1 badc2 and 1.94 ± 0.09 mg per 100 seeds for badc1 badc3, relative to the wild type (2.32 ± 0.04 mg per 100 seeds; Fig. 5B).
Figure 5.
The badc mutants grew similarly to wild-type plants. A, Phenotypes of 3-week old badc mutants and wild-type (WT) plants. B, Seeds of double mutants (badc1 badc2 and badc1 badc3) were significantly smaller than those of the wild type. Asterisks indicate significant differences compared with wild-type plants, which were determined by Student’s t test (P < 0.05). Values are presented as means ± sd of five biological replicates.
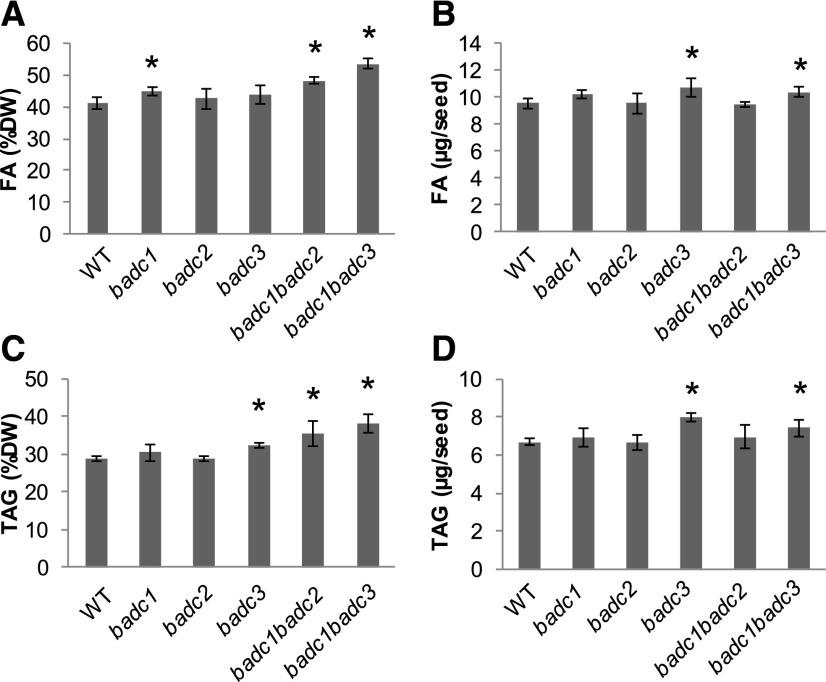
TFA and TAG Levels Increased in badc1 badc2 and badc1 badc3
Previous work provided biochemical evidence for the negative regulation of ACCase by BADC and demonstrated a significant elevation in oil content in BADC1 RNA interference transgenic Arabidopsis (Salie et al., 2016). Here, we quantify the effect of homozygous null mutations in each individual BADC gene, and in homozygous badc1 badc2 and badc1 badc3 lines, on TFA and TAG. Our data showed that the badc1 mutation, either alone or in combination with the badc2 or badc3 mutation, resulted in significant increases of 15% to 30% dry weight for FA (Fig. 6, A and B) and 18% to 30% dry weight for TAG (Fig. 6, C and D). Wild-type TFA, wild-type TAG, badc1 badc3 TFA, badc1 badc3 TAG, badc1 badc2 TFA, and badc1 badc2 TAG are 41.2% dry weight, 28.9% dry weight, 53.6% dry weight, 38.3% dry weight, 48.3% dry weight, and 35.5% dry weight, respectively. TFA in badc1 badc3 also was significantly higher than in wild-type plants (Student’s t test, P < 0.05). We also observed that, despite the smaller size in seeds of badc1 badc3 compared with the wild type, TFA and TAG per seed were significantly higher than in wild-type plants (Student’s t test, P < 0.05; Fig. 6, B and D).
Figure 6.
TFA and TAG increases in badc1 badc2 and badc1 badc3 double mutant seeds. A, TFA contents per dry weight. B, TFA contents per seed. C, Total TAG contents per dry weight. D, Total TAG contents per seed. Asterisks indicate significant differences from the wild type at the level of P < 0.05 as determined by Student’s t test. Values are presented as means ± sd of five biological replicates.
Arabidopsis Cell Suspension Cultures of Wild-Type Col-0 and badc1 badc3 as Models for Irreversible Inhibition by Tween 80
Ten-day-old wild-type (Col-0) and badc1 badc3 leaves were used to create callus cultures. These were transferred to liquid NT-1 medium to initiate cell suspension cultures. Both cell cultures were subcultured simultaneously and maintained in NT-1 medium in the absence or presence of 10 mm Tween 80 for 8 d to evaluate their relative growth rates. Cells were collected and weighed at the beginning of the experiment (day 0) and on days 2, 4, 6, and 8. Both wild-type and badc1 badc3 cell cultures grew at similar rates in the absence and presence of Tween 80 (Supplemental Fig. S2). In addition, wild-type and badc1 badc3 cell suspension cultures grown in NT-1 medium in the absence of Tween 80 for 6 d showed no significant difference in their FA profiles (Supplemental Fig. S3), validating both wild-type and badc1 badc3 genotypes of cell suspension culture as suitable models for a long-term Tween 80 treatment under these conditions.
BADC Contributes to the Irreversible Phase of Inhibition of ACCase
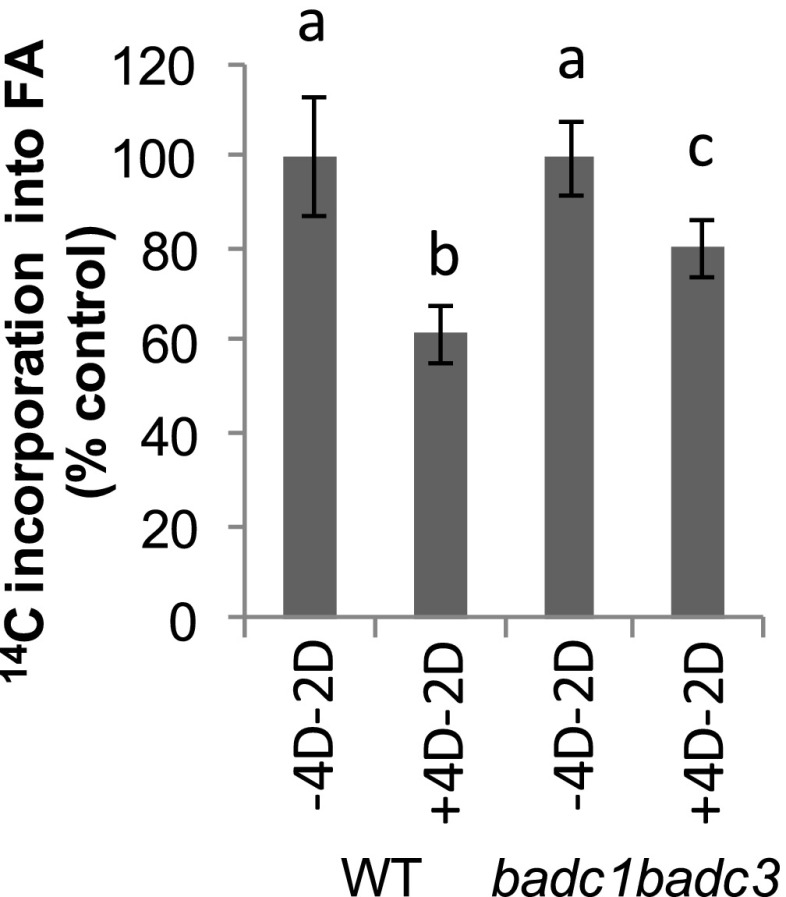
Wild-type and badc1 badc3 suspension culture cells were maintained in NT-1 medium containing 10 mm Tween 80 for 4 d and subsequently transferred to fresh medium lacking Tween 80 for an additional 2 d. During the experiments, medium with and without Tween 80 was refreshed every 48 h. The rate of [14C]acetate incorporation into FAs was determined by in vivo labeling using [14C]acetate followed by scintillation counting of lipid extracts. Wild-type culture cells showed approximately 39% irreversible inhibition compared with control cells in Tween 80-free medium (Fig. 7). In contrast, badc1 badc3 showed only 19% inhibition compared with the respective control cells in Tween 80-free medium (Fig. 7). These results suggest that BADC1 and BADC3 contributed approximately 50% of the observed Tween 80-induced irreversible inhibition observed in wild-type cultures. The overall irreversible inhibition of ACCase observed in the badc1 badc3 double mutant resulted in changes in the FA profile, with larger decreases in wild-type cultures (Supplemental Fig. S4A) relative to badc1 badc3 cultures as determined by Student’s t test (P < 0.05; Supplemental Fig. S4B).
Figure 7.
[14C]Acetate incorporation into total lipids showed that the persistence of inhibitions of ACCase by Tween 80 is stronger in the wild type (WT) than in badc1 badc3 in cell suspension culture. + or − indicate with or without Tween 80 treatment for the number of days (D) specified. Different letters indicate significant differences at the level of P < 0.05 as determined by Student’s t test. Values are presented as means ± sd of three biological replicates. A level of 100% corresponds to the wild type, 11.3k cpm; badc1 badc3, 14.3k cpm.
DISCUSSION
In this study, we quantified the effects of T-DNA knockouts of BADC1, BADC2, and BADC3, as well as the badc1 badc2 and badc1 badc3 double mutants, on the seed FA and TAG contents in Arabidopsis. The BADC1 knockout resulted in a significant increase in seed FA as a percentage dry weight, which has been observed previously in BADC1 RNA interference lines (Salie et al., 2016), whereas the badc2 and badc3 mutants showed smaller increases in FA relative to the wild type. However, the seed weights of badc1 and badc2 were similar to that of wild-type plants, whereas badc3 showed a significant increase in seed weight. In contrast, badc1 badc2 and badc1 badc3 showed significant decreases in weight per seed. The increase in seed weight for badc3 seeds was reflected in its significant increase in FA per seed relative to those of badc1, badc2, and the wild type. The large increase in percentage FA dry weight in badc1 badc3 is sufficient to overcome the reduction in seed weight, and the FA per seed is significantly higher than that of wild-type plants. TAG, on the other hand, is significantly higher as a percentage dry weight in badc3 than in badc1 and badc2, as it is in both badc1 badc2 and badc1 badc3. However, the dry weight of TAG per seed was significantly higher than in the wild type for badc3 compared with badc1 and badc2, again due to the increase in seed weight for badc3. Only badc3 and badc1 badc3 contained more TAG per seed than wild-type seeds.
At present, we do not have an explanation for the increased seed weight of badc3; however, the decreased weight of badc1 badc2 and badc1 badc3 along with our inability to obtain badc2 badc3 seeds suggest that BADC proteins play a currently undefined role in normal seed development. The consistently higher TAG and TFA in badc1 badc2 and badc1 badc3 seeds compared with wild-type seeds indicate the presence of constitutive BADC-dependent inhibition of ACCase under normal physiological conditions. Indeed, this is consistent with the identification of BADC subunits from ACCase pull-down experiments (Olinares et al., 2010). Constitutive down-regulation of ACCase by BADC is further supported by the observation that rates of FA synthesis in the double mutant were 26% higher in badc1 badc3 than in wild-type plants. Alleviation of ACCase suppression by BADC could allow free FAs in excess of cellular needs to accumulate to toxic levels, which have well-documented negative metabolic consequences, including alterations in longitudinal cell growth (Li et al., 2011) and cell death (Fan et al., 2013; Yang et al., 2015). It is also possible that unchecked FA synthesis could critically reduce the levels of ATP and reductant necessary for other critical cellular processes.
Previously, tobacco and B. napus somatic embryo cell suspension lines were used to characterize the inhibition of the heteromeric plastidial ACCase when cells were cultured in Tween-containing medium (Shintani and Ohlrogge, 1995; Andre et al., 2012). Here, we extend this approach to include Arabidopsis leaf suspension cell lines to exploit the available Arabidopsis T-DNA gene disruption lines. Similar to badc1 badc3 mutant plants, badc1 badc3 cell suspension cultures exhibited normal growth compared with wild-type cultures. Furthermore, the results presented here show that, when grown on Tween 80-containing medium, Arabidopsis leaf-derived cell lines mirror the responses of B. napus somatic embryo cell lines in displaying two phases of inhibition of ACCase: a short-term reversible phase and a longer-term irreversible phase. This allowed us to test the hypothesis that BADC is involved in the long-term irreversible phase of ACCase regulation. The observation that long-term inhibition of ACCase was reduced by approximately 50% in the badc1 badc3 cell line establishes a role for BADC in the long-term irreversible ACCase feedback inhibition. The absence of significant changes in the transcription of the BADC genes suggests the existence of other mechanisms for the inclusion of BADC into ACCase, possibly involving posttranslational modification.
ACCase activity did not fully recover to non-Tween 80-fed control levels in the badc1 badc3 mutant, suggesting that a factor other than BADC1 and BADC3 is responsible for the remainder of the irreversible inhibition. A likely candidate is BADC2, because the individual badc2 mutant showed elevation in the levels of FA dry weight in seeds and badc1 badc2 showed significantly elevated FA dry weight relative to both wild-type and badc1 plants. However, our inability to obtain a homozygous badc2 badc3 and, therefore, badc1 badc2 badc3 triple mutants precludes our ability to test this hypothesis directly. Another formal candidate is the PII protein, which inhibits ACCase under low-2-oxoglutarate conditions; however, this is unlikely because this interaction is reversible, whereas the inhibition under study is irreversible (Gerhardt et al., 2015; Hauf et al., 2016). However, PII could potentially play a role in the displacement of BCCP subunits by BADC, because it has been shown to directly bind BCCP, possibly favoring subunit exchange. Alternatively, another proposed, but yet to be identified, factor could contribute to a portion of the irreversible inhibition by altering ACCase partitioning between the stroma and envelope fractions of the plastid (Thelen and Ohlrogge, 2002).
CONCLUSION
In this work, we show that T-DNA insertions into each of the three BADC genes resulted in increased FA accumulation in seeds and that badc1 badc2 and badc1 badc3 seeds show FA increases of 15% to 30% and 18% to 30% for TAG, whereas the double mutant badc2 badc3 results in embryo abortion. Under normal physiological conditions, ACCase activity is partially repressed by BADC. BADC1 and BADC3 account for approximately 50% of the irreversible inhibition of ACCase resulting from long-term exposure to Tween 80. These data provide direct evidence for the involvement of BADC in the homeostatic feedback regulation of ACCase.
MATERIALS AND METHODS
Plant Growth Conditions
Wild-type Arabidopsis (Arabidopsis thaliana; ecotype Col-0), badc1 (Salk 000817C), badc2 (Salk 021108C), and badc3 (CS2103834) were used in this study. For growth on solid medium, seeds were surface sterilized with 70% (v/v) ethanol, followed by 30% (v/v) bleach containing 0.01% (v/v) Tween 20, and rinsed three times with sterile water. Seeds were stratified for 3 d at 4°C in the dark and germinated on one-half-strength Murashige and Skoog (MS) medium supplemented with 1% (w/v) Suc in an incubator with a light/dark cycle of 18 h/6 h at 23°C, photosynthetic photon flux density of 250 μmol m−2 s−1, and 75% relative humidity.
Generation of Double Mutants and Genotyping Analysis
Homozygous single mutants were confirmed by PCR using gene-specific primer pairs and a combination of T-DNA-specific primers. The genotyping primers (5′ to 3′) were as follows: BADC1 LP, TTCATTTTGCTGGTTAATGCC; BADC1 RP, TTCTGCTCCTCTGAAACTTGC; BADC2 LP, GTGGGAGGAGCTCCATTAATC; BADC2 RP, CAAAGGTAATGGTACGGACAAAG; BADC3 LP, GTTGTCCAGCCTTTTCACC; BADC3 RP, CTCATCAATTAGATTAGCTTGGC; LBb1, GCGTGGACCGCTTGCTGCAACT; and LB1, GTAAGGTAATGGGCTACACTG. Double mutants were generated by crossing the corresponding mutant lines. Homozygous double mutant lines were identified by genotyping using gene-specific primers.
RNA Isolation and RT-qPCR
Total RNA was isolated from 14-d-old leaves using the RNeasy Plant Mini Kit (Qiagen), and cDNA was prepared using SuperScript IV VILO Master Mix (with ezDNase exzyme; Invitrogen). SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) was used in the reaction mix. RT-qPCR was carried out on the CFX96 Real-time PCR Detection System (Bio-Rad). Gene-specific primers (5′ to 3′) used in the analysis were as follows: α-CT forward, GGTTGCACGTGAAGTAGCTG, and α-CT reverse, TTTTGTTGTTTCAAGCGAAGTGG; β-CT forward, ATCTCCTACTACCGGTGGGG, and β-CT reverse, CTTGTGAACCTTCAGGCACG; BC forward, TATGGGATCCGTAGTAGGCG, and BC reverse, ACCAAAACTTGCTGTCACCC; BCCP1 forward, AGAAACTTCCTAGGATTGAAAGAGG, and BCCP1 reverse, CAGAGGCTGTAGTCACACCA; BCCP2 forward, TCTATCTCTATCCTCGCCGGT, and BCCP2 reverse, TGACGCTTCAAGGCACGAT; BADC1 forward, CAGTTGGGAACAGAACTTCCA, and BADC1 reverse, GCAACCAGAGGATCTCCATAAC; BADC2 forward, CGACTCTGCGATCTGTGAAA, and BADC2 reverse, TGCTACATACAGGCGGAAAC; and BADC3 forward, CGAATGGATCAGCTTCCTCTAC, and BADC3 reverse, GGATCTCCTGAAGAACCCTACT. The primers used for reference genes were as follows: F-box forward, GGCTGAGAGGTTCGAGTGTT, and F-box reverse, GGCTGTTGCATGACTGAAGA; and UBQ10 forward, ACCATCACTTTGGAGGTGGA, and UBQ10 reverse, GTCAATGGTGTCGGAGCTTT. Statistical analysis of RT-qPCR data was carried out with REST2009 (Pfaffl et al., 2002).
Arabidopsis T87 Cell Culture
Arabidopsis ecotype Col-0 cell line T87 was obtained from the Arabidopsis Biological Resource Center at Ohio State University. Cells were maintained as a suspension culture in NT-1 medium (4.3 g L−1 MS salt, 30 g L−1 Suc, 0.18 g L−1 KH2PO4, 1 mg L−1 thiamine, 0.44 mg L−1 2,4-dichlorophenoxyacetic acid [2,4-D], and 100 mg L−1 myoinositol, pH adjusted to 5.8 with NaOH). The cell suspension culture was then maintained at 24°C on a rotary shaker (120 rpm) under constant light. Cell suspensions were subcultured into 100-mL Erlenmeyer flasks containing fresh NT-1 medium (1:20, v/v) every 7 d.
Callus Production from Leaves
Leaves harvested from wild-type and badc1 badc3 double mutant seedlings 10 d after germination were lightly abraded with sterile surgical forceps and placed onto solid callus growth medium. Plates were incubated at 22°C in the dark until calluses formed. Callus growth medium consisted of MS medium supplemented with 30 g L−1 Suc, 0.2 g L−1 myoinositol, 0.5 mg mL−1 nebzylaminopurine, 1 mg mL−1 naphthaleneacetic acid, 1 mg mL−1 indole acetic acid, and 1 mg mL−1 2,4-D (Encina et al., 2001).
Cell Culture Growth Conditions
Calluses obtained from solid callus growth medium plates were transferred to and maintained as suspension cultures in NT-1 medium. The cell suspension culture was then maintained at 24°C on a rotary shaker (120 rpm) under constant light. Approximately 1 mL of suspension culture cells were subcultured into 20 mL of medium in 100-mL Erlenmeyer flasks containing fresh NT-1 medium every 7 d. For Tween 80 treatment experiments, a stock solution of 150 nm Tween 80 (Sigma-Aldrich) was filter sterilized (Andre et al., 2012) and added to NT-1 medium at room temperature. The medium was refreshed every 48 h by replacing half of the volume with fresh medium. For lipid extraction and RNA isolation, cells were rinsed with water, harvested by filtration, flash frozen, and stored at −80°C.
[14C]Acetate Incorporation Assay
[1-14C]Acetic acid, sodium salt, was purchased from PerkinElmer. Suspension culture cells were labeled 1 week after subculturing with a cell weight of approximately 40 mg. Cells were labeled by incubating in 0.2 μCi of [14C]acetate for 20 min at room temperature with constant shaking. Cells were subsequently rinsed three times with water. Total lipids were extracted with 500 μL of methanol:chloroform:formic acid (20:10:1, v/v). The organic phase was then extracted with 370 μL of 1 m KCl and 0.2 m H3PO4 and suspended in 2 mL of Ultima Gold liquid scintillation cocktail (PerkinElmer). The incorporated radioactivity was measured in cpm with a scintillation counter (Packard BioScience).
Lipid Extraction and Quantification
Total lipids were extracted from approximately 100 mg fresh weight of frozen suspension culture cells or 100 desiccated seeds. Materials were homogenized in 350 μL of methanol:chloroform:formic acid (20:10:1, v/v) and mixed repeatedly by shaking at room temperature for 1 h. The organic phase was added to 370 μL of 1 m KCl and 0.2 m H3PO4 and centrifuged at 12,000g for 1 min for phase separation. The organic phase was dried under an N2 stream and resuspended in chloroform. For TFA analysis, total lipid extracts were transmethylated into fatty acid methyl esters (FAMEs) by incubation in 1 mL of 1 n methanolic HCl at 80°C for 60 min (for suspension culture cells) or incubated in 1 mL of boron trichloride-methanol at 80°C to 85°C for 2.5 h (for desiccated seeds). For TAG quantification, total lipid extracts were separated by thin-layer chromatography using a hexane:diethyl ether:acetic acid (70:30:1, v/v/v) solvent system on Silica Gel 60 plates (EMD Millipore). Lipids were visualized by spraying 0.05% (w/v) primuline (in 80% [v/v] acetone). TAG fractions were identified under UV light, excised from the plate, and transmethylated into FAMEs by incubation in 1 mL of boron trichloride-methanol. FAMEs were extracted into hexane and dried under an N2 stream prior to resuspending in 100 μL of hexane. The quantification of FAMES was carried out on Agilent GC 7890A/MSD 5975C system using a capillary DB23 column (60 m × 0.250 mm; Agilent Technologies) in full mass scan mode (Nguyen et al., 2010). Heptadecanoic acid (5 μg) was used as an internal standard.
Statistics
Pairwise comparisons were performed using Student’s t test at P < 0.05 significance level. Phenotypic proportions of mutant seeds of F3 plants resulting from crossing between badc2 and badc3 were analyzed using χ2 goodness of fit at P < 0.05 significance level. Gene expression calculated from RT-qPCR data was analyzed using REST2009 software at P < 0.05 significance level.
Accession Numbers
Accession numbers for the genes employed in this study are as follows: BADC1, AT3G56130; BADC2, AT1G52670; BADC3, AT3G15690; α-CT, AT2G38040; β-CT, ATCG00500; BC, AT5G35360; BCCP1, AT5G16390; and BCCP2, AT5G15530.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. RT-qPCR analysis of 10-d-old leaves confirming that the T-DNA insertion lines were null mutants for the corresponding genes.
Supplemental Figure S2. Addition of Tween 80 did not affect the growth rates of both wild-type and badc1 badc3 cell suspension cultures.
Supplemental Figure S3. FA profile of badc1 badc3 cell cultures did not differ significantly from the wild type, which was determined by Student’s t test (P < 0.05).
Supplemental Figure S4. FA synthesis inhibition in badc1 badc3 was less compared with the wild type.
Supplemental Table S1. Mutant seeds of F3 plants resulting from crossing between badc2 and badc3.
Acknowledgments
We thank Carl Andre and Jay Thelen for critical reading of the article.
Footnotes
Initial aspects of the work were conceived by J.S. and Jay Thelen under the National Science Foundation (grant DBI 1117680). The work was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under contract number DE-SC0012704, specifically through the Physical Biosciences program of the Chemical Sciences, Geosciences, and Biosciences Division.
Articles can be viewed without a subscription.
References
- Andre C, Haslam RP, Shanklin J (2012) Feedback regulation of plastidic acetyl-CoA carboxylase by 18:1-acyl carrier protein in Brassica napus. Proc Natl Acad Sci USA 109: 10107–10112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard CZ, Waldrop GL (1998) Overexpression and kinetic characterization of the carboxyltransferase component of acetyl-CoA carboxylase. J Biol Chem 273: 19140–19145 [DOI] [PubMed] [Google Scholar]
- Choi JK, Yu F, Wurtele ES, Nikolau BJ (1995) Molecular cloning and characterization of the cDNA coding for the biotin-containing subunit of the chloroplastic acetyl-coenzyme A carboxylase. Plant Physiol 109: 619–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan JE Jr, Waldrop GL (2002) Multi-subunit acetyl-CoA carboxylases. Prog Lipid Res 41: 407–435 [DOI] [PubMed] [Google Scholar]
- Encina C, Constantin M, Botella J (2001) An easy and reliable method for establishment and maintenance of leaf and root cell cultures of Arabidopsis thaliana. Plant Mol Biol Rep 19: 245–248 [Google Scholar]
- Fan J, Yan C, Xu C (2013) Phospholipid:diacylglycerol acyltransferase-mediated triacylglycerol biosynthesis is crucial for protection against fatty acid-induced cell death in growing tissues of Arabidopsis. Plant J 76: 930–942 [DOI] [PubMed] [Google Scholar]
- Feria Bourrellier AB, Valot B, Guillot A, Ambard-Bretteville F, Vidal J, Hodges M (2010) Chloroplast acetyl-CoA carboxylase activity is 2-oxoglutarate-regulated by interaction of PII with the biotin carboxyl carrier subunit. Proc Natl Acad Sci USA 107: 502–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt EC, Rodrigues TE, Müller-Santos M, Pedrosa FO, Souza EM, Forchhammer K, Huergo LF (2015) The bacterial signal transduction protein GlnB regulates the committed step in fatty acid biosynthesis by acting as a dissociable regulatory subunit of acetyl-CoA carboxylase. Mol Microbiol 95: 1025–1035 [DOI] [PubMed] [Google Scholar]
- Guchhait RB, Polakis SE, Dimroth P, Stoll E, Moss J, Lane MD (1974a) Acetyl coenzyme A carboxylase system of Escherichia coli: purification and properties of the biotin carboxylase, carboxyltransferase, and carboxyl carrier protein components. J Biol Chem 249: 6633–6645 [PubMed] [Google Scholar]
- Guchhait RB, Polakis SE, Hollis D, Fenselau C, Lane MD (1974b) Acetyl coenzyme A carboxylase system of Escherichia coli: site of carboxylation of biotin and enzymatic reactivity of 1′-N-(ureido)-carboxybiotin derivatives. J Biol Chem 249: 6646–6656 [PubMed] [Google Scholar]
- Hauf W, Schmid K, Gerhardt EC, Huergo LF, Forchhammer K (2016) Interaction of the nitrogen regulatory protein GlnB (PII) with Biotin Carboxyl Carrier Protein (BCCP) controls acetyl-CoA levels in the cyanobacterium Synechocystis sp. PCC 6803. Front Microbiol 7: 1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunkeler M, Stuttfeld E, Hagmann A, Imseng S, Maier T (2016) The dynamic organization of fungal acetyl-CoA carboxylase. Nat Commun 7: 11196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo H, Shiratsuchi K, Yoshimoto T, Masuda T, Kitazono A, Tsuru D, Anai M, Sekiguchi M, Tanabe T (1991) Acetyl-CoA carboxylase from Escherichia coli: gene organization and nucleotide sequence of the biotin carboxylase subunit. Proc Natl Acad Sci USA 88: 9730–9733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozaki A, Mayumi K, Sasaki Y (2001) Thiol-disulfide exchange between nuclear-encoded and chloroplast-encoded subunits of pea acetyl-CoA carboxylase. J Biol Chem 276: 39919–39925 [DOI] [PubMed] [Google Scholar]
- Kozaki A, Sasaki Y (1999) Light-dependent changes in redox status of the plastidic acetyl-CoA carboxylase and its regulatory component. Biochem J 339: 541–546 [PMC free article] [PubMed] [Google Scholar]
- Li M, Bahn SC, Guo L, Musgrave W, Berg H, Welti R, Wang X (2011) Patatin-related phospholipase pPLAIIIβ-induced changes in lipid metabolism alter cellulose content and cell elongation in Arabidopsis. Plant Cell 23: 1107–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SJ, Cronan JE Jr (1992a) The gene encoding the biotin carboxylase subunit of Escherichia coli acetyl-CoA carboxylase. J Biol Chem 267: 855–863 [PubMed] [Google Scholar]
- Li SJ, Cronan JE Jr (1992b) The genes encoding the two carboxyltransferase subunits of Escherichia coli acetyl-CoA carboxylase. J Biol Chem 267: 16841–16847 [PubMed] [Google Scholar]
- Maeo K, Tokuda T, Ayame A, Mitsui N, Kawai T, Tsukagoshi H, Ishiguro S, Nakamura K (2009) An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J 60: 476–487 [DOI] [PubMed] [Google Scholar]
- Nguyen HT, Mishra G, Whittle E, Pidkowich MS, Bevan SA, Merlo AO, Walsh TA, Shanklin J (2010) Metabolic engineering of seeds can achieve levels of omega-7 fatty acids comparable with the highest levels found in natural plant sources. Plant Physiol 154: 1897–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge JB, Jaworski JG (1997) Regulation of fatty acid synthesis. Annu Rev Plant Physiol Plant Mol Biol 48: 109–136 [DOI] [PubMed] [Google Scholar]
- Olinares PD, Ponnala L, van Wijk KJ (2010) Megadalton complexes in the chloroplast stroma of Arabidopsis thaliana characterized by size exclusion chromatography, mass spectrometry, and hierarchical clustering. Mol Cell Proteomics 9: 1594–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakis SE, Guchhait RB, Zwergel EE, Lane MD, Cooper TG (1974) Acetyl coenzyme A carboxylase system of Escherichia coli: studies on the mechanisms of the biotin carboxylase- and carboxyltransferase-catalyzed reactions. J Biol Chem 249: 6657–6667 [PubMed] [Google Scholar]
- Rodrigues TE, Gerhardt EC, Oliveira MA, Chubatsu LS, Pedrosa FO, Souza EM, Souza GA, Müller-Santos M, Huergo LF (2014) Search for novel targets of the PII signal transduction protein in bacteria identifies the BCCP component of acetyl-CoA carboxylase as a PII binding partner. Mol Microbiol 91: 751–761 [DOI] [PubMed] [Google Scholar]
- Salie MJ, Thelen JJ (2016) Regulation and structure of the heteromeric acetyl-CoA carboxylase. Biochim Biophys Acta 1861: 1207–1213 [DOI] [PubMed] [Google Scholar]
- Salie MJ, Zhang N, Lancikova V, Xu D, Thelen JJ (2016) A family of negative regulators targets the committed step of de novo fatty acid biosynthesis. Plant Cell 28: 2312–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani D, Ohlrogge J (1995) Feedback inhibition of fatty acid synthesis in tobacco suspension cells. Plant J 7: 577–587 [Google Scholar]
- Thelen JJ, Ohlrogge JB (2002) Metabolic engineering of fatty acid biosynthesis in plants. Metab Eng 4: 12–21 [DOI] [PubMed] [Google Scholar]
- Wei J, Tong L (2015) Crystal structure of the 500-kDa yeast acetyl-CoA carboxylase holoenzyme dimer. Nature 526: 723–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Munz J, Cass C, Zienkiewicz A, Kong Q, Ma W, Sanjaya, Sedbrook J, Benning C (2015) Ectopic expression of WRINKLED1 affects fatty acid homeostasis in Brachypodium distachyon vegetative tissues. Plant Physiol 169: 1836–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]