Evolutionary analyses suggest that plant R-genes originated in charophytes and plant R-proteins evolved in a modular fashion through frequent gain or loss of protein domains.
Abstract
During plant-pathogen interactions, plants use intracellular proteins with nucleotide-binding site and Leu-rich repeat (NBS-LRR) domains to detect pathogens. NBS-LRR proteins represent a major class of plant disease resistance genes (R-genes). Whereas R-genes have been well characterized in angiosperms, little is known about their origin and early diversification. Here, we perform comprehensive evolutionary analyses of R-genes in plants and report the identification of R-genes in basal-branching streptophytes, including charophytes, liverworts, and mosses. Phylogenetic analyses suggest that plant R-genes originated in charophytes and R-proteins diversified into TIR-NBS-LRR proteins and non-TIR-NBS-LRR proteins in charophytes. Moreover, we show that plant R-proteins evolved in a modular fashion through frequent gain or loss of protein domains. Most of the R-genes in basal-branching streptophytes underwent adaptive evolution, indicating an ancient involvement of R-genes in plant-pathogen interactions. Our findings provide novel insights into the origin and evolution of R-genes and the mechanisms underlying colonization of terrestrial environments by plants.
Plants rely on two branches of innate immunity system to prevent or eliminate microbial infections: one involves cell surface receptors to respond to pathogen- or microbe-associated molecular patterns, and the other acts inside plant cells by using proteins with nucleotide-binding site (NBS) and Leu-rich repeat (LRR) domains (Dangl and Jones, 2001; Jones and Dangl, 2006; Jones et al., 2016). NBS-LRR proteins confer recognition of pathogen effectors either directly or indirectly and trigger disease resistance (Jones and Dangl, 2006; Jones et al., 2016; van der Hoorn and Kamoun, 2008). Most of the plant disease-resistance genes (R-genes) cloned so far encode NBS-LRR proteins (we use the term R-genes to refer to NBS-LRR genes hereafter; Jones and Dangl, 2006; Jones et al., 2016).
The architectures of plant R-proteins are characterized by an N-terminal signaling domain, a nucleotide-binding adaptor shared by APAF-1, certain R gene products and CED-4 (NB-ARC) domain, and a series of LRRs (McHale et al., 2006; Urbach and Ausubel, 2017). The NB-ARC domain belongs to the STAND (signal transduction ATPases with numerous domain) superfamily and might function as a molecular switch in disease-resistance signaling (McHale et al., 2006). The LRR domain plays an important role in the negative regulation of NBS-mediated oligomerization (Jones et al., 2016). NBS-LRR proteins are typically classified into two major subfamilies based on the presence or absence of an N-terminal signaling domain, namely the Toll/IL receptor (TIR) domain, and are referred to as TIR-NBS-LRR proteins (TNLs) and non-TIR-NBS-LRR proteins (nTNLs; McHale et al., 2006; Shao et al., 2016). Many nTNLs also possess a coiled-coil motif at the N terminus and are thus known as CC-NBS-LRR proteins (Meyers et al., 2003). Moreover, some nTNLs encode a resistance to the powdery mildew8 (RPW8) domain and are thus designated RPW8-NBS-LRR proteins (RNLs; Xiao et al., 2001; Shao et al., 2014).
Metazoan innate immunity also involves intracellular receptors known as Nod-like receptors (NLRs; Ausubel, 2005). Similar to plant NBS-LRR proteins, NLR proteins are characterized by a domain architecture with a STAND domain (NACHT [NAIP, CIIA, HET-E, and TEP1]) and a series of LRRs. The NB-ARC domain shares certain similarity with the NACHT domain (Urbach and Ausubel, 2017). Phylogenetic analyses suggest plant NBS-LRRs and metazoan NLRs originated independently by convergent evolution (Yue et al., 2012; Urbach and Ausubel, 2017).
Comparative genomic analyses show that R-genes are widely distributed in land plants (Meyers et al., 2002, 2003; McHale et al., 2006; Yue et al., 2012; Shao et al., 2014, 2016; Jones et al., 2016; Urbach and Ausubel, 2017; Xue et al., 2012). However, no R-genes have been reported in algae to date (Yue et al., 2012; Urbach and Ausubel, 2017). Thus, plant R-genes were proposed to originate in land plants (Yue et al., 2012; Urbach and Ausubel, 2017). Based on the observations that TNLs were first observed in Physcomitrella patens and nTNLs were first found in Selaginella moellendorffii, TNLs were thought to have an earlier origin than nTNLs (Yue et al., 2012). It currently remains uncertain about whether R-genes are present in basal-branching streptophytes, such as charophytes (a group of firewater algae from which land plants originated; Qiu et al., 1998, 2007; McCourt et al., 2004; Wickett et al., 2014; Delwiche and Cooper, 2015; Lang et al., 2018; Puttick et al., 2018).
In this study, we performed comparative genomic and phylogenetic analyses of R-genes in a wide variety of plants, with an emphasis on basal-branching plants. We identified the presence of R-genes in the genomes of basal-branching streptophytes, including charophytes, liverworts, and mosses. Moreover, we found R-proteins diversified into TNLs and nTNLs in charophytes. Our evolutionary analyses provide many novel insights into the early evolution and diversification of R-genes in plants.
RESULTS
Identification of R-Genes in Plants
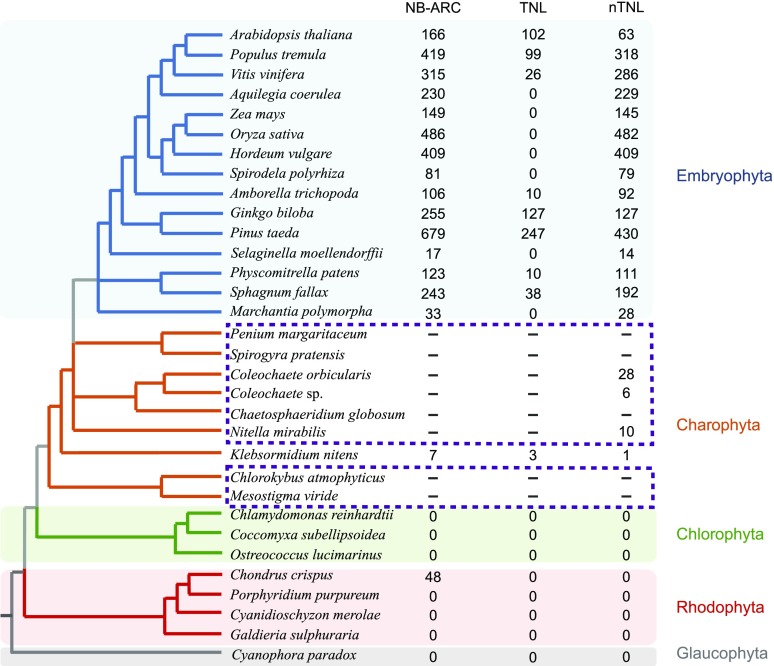
We performed comparative genomic and phylogenetic analyses of R-genes in the genomes of 24 representative plant species that cover a broad diversity of plants (Fig. 1; Supplemental Table S1). Given plant NBS-LRR proteins have been extensively reported in angiosperms, we focused our study on basal-branching plants. Using a Markov model of the NB-ARC domain generated from representative NB-ARC sequences as seeds to mine those plant proteomes, we identified a total of 3,766 proteins with the NB-ARC domain in the genomes of 17 plants (Fig. 1; Supplemental Table S2; Supplemental Data Set S1). No protein with the NB-ARC domain was identified in chlorophytes, rhodophytes (except Chondrus crispus), or glaucophytes.
Figure 1.
The identification and distribution of R-proteins in plants. The distribution and number of NB-ARC-domain proteins and R-proteins, including those with TIR-NBS-LRR domains (TNLs) and non-TIR-NBS-LRR proteins (nTNLs), in various plants is shown in the right columns. For species in the dashed-line boxes, a similarity search was performed against transcriptome data. Because the absence of one gene in the transcriptome does not guarantee its absence in the genome, only the number of gene sequences we identified was labeled. The phylogenetic relationships among plants are based on Qiu et al. (1998), Qiu et al. (2007), Wickett et al. (2014), Delwiche and Cooper (2015), and Puttick et al. (2018).
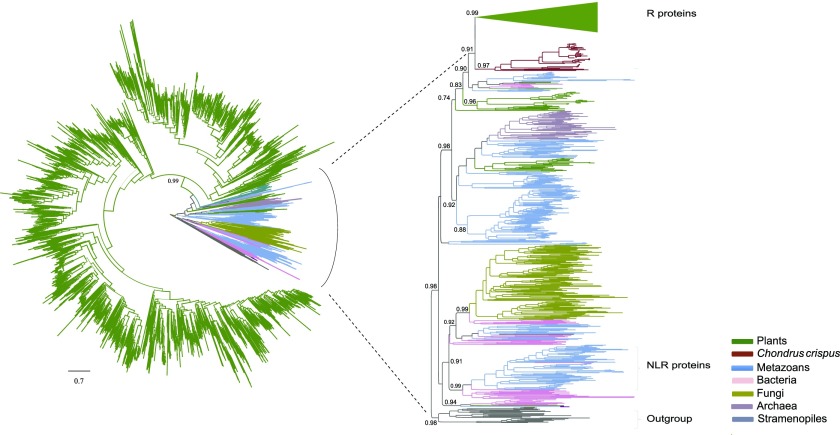
We performed a large-scale phylogenetic analysis of these proteins with the NB-ARC domain and other representative proteins with the NB-ARC or NACHT domains (Urbach and Ausubel, 2017). We found plant R-proteins form a monophyletic group with high support value and identified a total of 3,712 R-proteins within the genomes of 16 plant species (Fig. 2; Supplemental Table S2; Supplemental Data Set S1). Although some plant R-proteins lack a LRR domain (Fig. 3; Supplemental Table S3), these proteins cluster with NBS-LRR proteins, indicating that they might arise from NBS-LRR proteins by deleting LRRs (for convenience, we still called them NBS-LRR proteins or R-proteins). The number of R-genes identified in this study is roughly the same as that reported previously (Shao et al., 2016); for example, for Arabidopsis (Arabidopsis thaliana), 165 R-genes were identified in both this study and Shao et al. (2016). The majority of the Arabidopsis R-genes (157/165) were annotated as disease-resistance genes in the Arabidopsis genome release 10. All the other eight genes were confirmed to contain the NB-ARC domain, and some of them contain the TIR domain and/or LRRs, which might arise from R-proteins as aforementioned and represent annotation problems (Supplemental Table S3). Interestingly, R-proteins were identified in charophytes and all the land plants. The copy number of R-genes was found to vary greatly among species, from 4 in the charophyte Klebsormidium nitens to 677 in the gymnosperm Pinus taeda. No NBS-LRR proteins were found in chlorophytes, rhodophytes, or glaucophytes (Fig. 1; Supplemental Table S2). Although proteins with the NB-ARC domain were identified in the rhodophyte C. crispus, these proteins exhibit TIR-NB-ARC-WD40 or NB-ARC-WD40 domain organization and are thus not authentic NBS-LRR proteins.
Figure 2.
Phylogenetic analysis of NB-ARC-domain proteins. Phylogenetic analysis of the NB-ARC-domain proteins identified in this study and other representative proteins with NB-ARC or NAT domains was performed using an approximate maximum likelihood method. The SH-like values are depicted near the selected nodes. The plant R-proteins and animal NLR proteins are labeled. Proteins with MalT and SWACOS domains were used as outgroups. The expanded section represents a part of the whole tree.
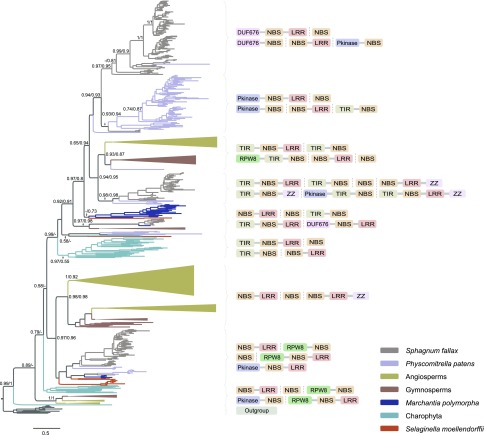
Figure 3.
Phylogenetic analysis of plant R-proteins. The angiosperm and gymnosperm clades are collapsed. The full tree is available in Supplemental Figure S1. The aLRT/SH-like support values are depicted near the selected nodes. The domain organizations are shown near the corresponding clades.
To further study R-genes in charophytes, we performed similarity searches and phylogenetic analyses to identify R-genes within eight charophyte transcriptomes (Timme et al., 2012; Ju et al., 2015). We found that R-genes are present in two other charophyte genera (Coleochaete and Nitella; Fig. 1; Supplemental Data Set S2). It should be noted that the absence of one gene in the transcriptome of one species does not guarantee its absence in that species (Wang et al., 2015). Nevertheless, our results further demonstrate that R-genes are widely present in charophytes. Taken together, our pan-Plantae comparative genomic analyses provide evidence that R-genes are ubiquitous in streptophytes.
Diversification of R-Genes in Streptophytes
To further explore the evolutionary relationship among R-proteins in streptophytes, we performed phylogenetic and domain architecture analyses of R-proteins of basal-branching streptophytes (charophytes, liverworts, and mosses) and representative R-proteins of seed plants (Fig. 3; Supplemental Fig. S1). Phylogenetic analyses were performed using two different methods (see “Materials and Methods” for details). We found there is no significant difference between two trees (P = 1.00 for the Kishino-Hasegawa test and P = 0.82 for the Shimodaira-Hasegawa test). Phylogenetic analyses show that charophyte R-proteins cluster into multiple independent groups (Supplemental Fig. S1). Both TNLs and nTNLs are present in the charophyte genomes. We found that R-proteins of land plants nest within the diversity of R-proteins of charophytes. Therefore, both the distribution and phylogenetic analyses suggest that plant R-genes originated and diversified in charophytes.
We found the proteins with a single NBS domain frequently cluster with the proteins with the NBS-LRR domain organization or proteins with the TIR-NBS-LRR domain organization, and proteins with the NBS-LRR domain organization frequently cluster with the proteins with the TIR-NBS-LRR domain organization (Fig. 3; Supplemental Table S4). This absence of the N- and C-terminal domains indicates that domain loss at the N or C terminus occurred frequently in the evolution of plant NBS-LRR proteins, which could also explain why neither TNLs nor nTNLs form a monophyletic group (Yue et al., 2012). Phylogenetic analysis showed that TNLs fall into the diversity of nTNLs and thus TNLs might have originated via insertion of the TIR domain into NBS-LRR proteins. RNLs are widespread in land plants and appear to originate from NBS-LRR via gain of the additional RPW8 domain (Supplemental Fig. S1).
Interestingly, we also found several additional domain architectures for R-proteins, such as Pkinase-NBS-LRR (PNL; Xue et al., 2012) and DUF676-NBS-LRR (DNL) proteins. The PNL proteins were identified in mosses (P. patens and Sphagnum fallax), angiosperms (Oryza sativa, Zea mays, Hordeum vulgare, and Aquilegia coerulea), and gymnosperms (P. taeda). However, DNL proteins were only identified in S. fallax (Supplemental Table S3).
Adaptive Evolution of R-Genes in Plants
To test whether R-genes underwent adaptive evolution in basal-branching streptophytes, we performed positive selection analysis in 34 groups of R-genes of K. nitens, Marchantia polymorpha, P. patens, and S. fallax (Supplemental Fig. S1). We detected amino acid residues under positive selection for the majority of these R-gene groups (26/34; Supplemental Table S5). The number of positively selected sites varies among the 26 R-gene groups in which positive selection was detected. The positively selected sites are dispersed along these proteins but appear to be not enriched in particular domains (Fig. 4). Our results show most of the R-gene clusters in basal-branching streptophytes underwent adaptive divergence.
Figure 4.
Positive selection in R-proteins of basal-branching streptophytes. The domain organization for each gene group is shown. A vertical line indicates a positively selected site. The detail for each gene group is available in Supplemental Figure S1. Cor, C. orbicularis; Mpo, M. polymorpha; Ppa, P. patens; Sfa, S. fallax.
DISCUSSION
In this study, we performed comparative genomic and phylogenetic analyses of R-genes in 24 plant species. We identified the presence of R-genes in basal-branching streptophytes (charophytes, liverworts, and mosses) and all the land plants. Phylogenetic analysis shows that TNLs fall into the diversity of nTNLs, indicating that nTNLs may be ancestral. TNLs in charophytes, mosses, and seed plants did not cluster together, suggesting the possibility that TNLs originated multiple times. On the other hand, the pattern that TNLs of seed plants are closely related that but no TNLs were identified in A. coerulea and the monocots (as reported in Tarr and Alexander, 2009) potentially indicates multiple losses of TNLs in plant lineages. Nevertheless, our evolutionary analyses reveal the complex evolutionary history of R-proteins in streptophytes.
Some proteins with the NB-ARC domain were identified in the rhodophyte C. crispus. These C. crispus proteins have WD40 repeats but not LRRs at the N terminus and thus are not R-proteins. It is possible that R-proteins originated in the last common ancestor of rhodophytes and charophytes. However, this scenario requires the loss of R-proteins in Chlorophyta and the replacement of LRRs by WD repeats, which seems less parsimonious than the hypothesis that R-proteins originated in charophytes.
Unusual domains were previously found to integrate into R-proteins, such as the heavy metal-associated domain in the rice (Oryza sativa) R-proteins RGA5 and Pik-1 (Césari et al., 2014; Maqbool et al., 2015). Some integrated domains play an essential role in pathogen effector recognition, which lays the foundation of the so-called integrated decoy model (Cesari, 2017). We found R-proteins are also associated with several novel domains, such as Pkinase and DUF676. The Pkinase domain might function in recognition of pathogen ligands (Song et al., 1995; Shiu and Bleecker, 2001; Afzal et al., 2008), raising the possibility that the integrated Pkinase domain acts as baits for the pathogen effectors. Potential decoy domain integration into R-proteins occurred frequently in flowering plants (Kroj et al., 2016; Sarris et al., 2016). Our study shows that domain integration also took place in mosses, indicating that domain integration is commonplace for the evolution of plant R-genes and might play an important role in the diversification of plant R-proteins. Our results suggest that recruiting new protein domains occurred multiple times during the evolution of plant R-proteins. Moreover, we found R-proteins frequently lost TIR and/or LRR domains at the N and/or C terminus, which might be due to either deletion or partial gene duplication. Taken together, our analyses reveal a highly dynamic nature of the domain architectures of R-proteins and suggest that plant R-proteins evolved in a modular fashion by frequent gains and losses of protein domains.
Comparative studies of genes belonging to evolutionary clusters suggest that adaptive evolution was commonplace for R-genes of dicots and monocots, which is thought to be associated with host-pathogen interaction (Bergelson et al., 2001; Mondragón-Palomino et al., 2002). We found most of the R-gene groups of basal-branching streptophytes, namely K. nitens, M. polymorpha, P. patens, and S. fallax, underwent adaptive evolution, implying that R-genes might have been involved in plant-pathogen interaction in basal-branching streptophytes.
The copy number of R-genes varies greatly in basal-branching streptophytes. Whereas charophytes, liverworts, and lycopods possess relatively low numbers of R-genes, the number of R-genes in the two moss species (P. patens and S. fallax) is comparable to or even greater than that of seed plants. This finding indicates that the high number of R-genes in mosses might be due to lineage-specific expansion of certain R-genes, such as PNLs and DNLs. The expansion of R-gene repertories might be caused by both adaption and drift. For the R-gene clusters that mainly underwent adaptive divergence, the increase in R-gene number might also be caused by the process of adaptation.
The colonization of the terrestrial environment by plants is one of the most important events in the history of life (Kenrick and Crane, 1997; Delwiche and Cooper, 2015). Land plants originated from charophytes, some of which were already living on land (Stebbins and Hill, 1980; McCourt et al., 2004; Delwiche and Cooper, 2015; Harholt et al., 2016). During terrestrial environment colonization, plants had to overcome novel pathogen species and levels of infection (Rensing et al., 2008; Hori et al., 2014). The emergence of a novel R-protein-associated strategy to eliminate pathogen infections might have helped charophytes and the earliest land plants to adapt to their new terrestrial environments. Indeed, the R-genes of charophytes mainly underwent adaptive evolution, which is likely the result of plant-pathogen interactions. Coincidently, the phenylpropanoid biosynthesis pathway, which plays an important role in response to phytopathogens, was also found to originate in streptophyte algae (de Vries et al., 2017). The establishment of symbiotic interactions with fungi is thought to have facilitated terrestrial environment colonization by early plants (Delaux et al., 2015; Field et al., 2015; de Vies and Archibald, 2018). R-proteins have been suggested to control the specificity of legume-rhizobia symbiosis (Yang et al., 2010). It is likely that R-proteins might contribute toward the symbiosis of fungi and early land plants. Taken together, we propose that the emergence of R-genes provided early land plants with the genetic toolkit needed for survival in a terrestrial environment and might play an important role in plant land colonization.
MATERIALS AND METHODS
Plant Species
To explore the origin and early diversification of R-genes in plants, we used the genome sequences of 24 representative plants (one gluacophyte, four rhodophytes, three chlorophytes, one charophyte, one liverwort, two mosses, one lycopodiophyte, two gymnosperms, and nine angiosperms; Supplemental Table S1). Moreover, we also used eight charophyte transcriptomes (Timme et al., 2012; Ju et al., 2015; Supplemental Table S1). These species cover a broad diversity of plants (Supplemental Table S1).
Identification of NB-ARC Domains
A hidden Markov model (HMM) of NB-ARC was generated using nine seed sequences of the NB-ARC domain (Pfam: PF00931) and hmmbuild from the HMMER package 3.1b2 (Eddy, 2011). The seed sequences of the NB-ARC domain (Pfam: PF00931) were retrieved from Pfam 31.0 (http://pfam.xfam.org). The HMM profile was used to search proteins with the NB-ARC domain against the protein sequences of 24 genomes with a cutoff E-value of 10−3 (Fig. 1; Supplemental Table S1). The homologs of R-genes in the transcriptomes of charophytes were identified using the tblastn algorithm with K. nitens proteins as queries and a cutoff value of 10−5. The NB-ARC domain sequences of plant R-proteins identified here and 964 representative proteins with the NB-ARC domain from both eukaryotes and prokaryotes (Urbach and Ausubel, 2017) were aligned using hmmalign from the HMMER package and improved by manual editing. Large-scale phylogenetic analysis was performed using FastTree 2.1 with the WAG+CAT model (Price et al., 2009). Support for each node in the phylogenetic tree was assessed using the SH-like value (Price et al., 2009). Proteins with MalT and SWACOS domains were used as outgroups (Urbach and Ausubel, 2017). The protein domain architecture was annotated using PHMMER from the EMBL-EBI database (Finn et al., 2011) and CD-Search (Marchler-Bauer et al., 2017).
Phylogenetic Analysis of Plant R-Genes
The NB-ARC domain sequences of the R-proteins of charophytes, Marchantia polymorpha, Physcomitrella patens, Sphagnum fallax, and Selaginella moellendorffii as well as representative R-proteins of seed plants were aligned using hmmalign from the HMMER package and improved by manual editing. Phylogenetic analysis was performed using a maximum likelihood method implemented in PhyML 3.0 (Guindon et al., 2010) with the VT+G+I+F model, the best fitting model selected by smart model selection implemented in PhyML 3.0 (Lefort et al., 2017), and an approximately maximum likelihood method implemented in FastTree 2.1 (Price et al., 2009) with the WAG+CAT model. Branch support was assessed using the approximate likelihood ratio test (aLRT) in PhyML, because conventional bootstrap analysis is extremely time-consuming for large trees and aLRT is a fast, accurate, and powerful alternative (Anisimova and Gascuel, 2006). Branch support was also assessed using the Shimodaira-Hasegawa-like value implemented in FastTree 2.1. To examine whether there is difference between trees inferred by two different methods, the Kishino-Hasegawa and Shimodaira-Hasegawa tests was performed using PAUP* 4.0a (Swofford 2003).
Positive Selection Analysis
Because R-genes are too divergent for alignment and positive selection analysis, we partitioned them and chose 34 groups based on the phylogenetic analysis (Supplemental Fig. S1). Some sequences are too divergent or fragmented (for transcriptome sequences) and thus were excluded for analysis. The nucleotide sequences were aligned using MAFFT (Katoh and Toh, 2008) and manually edited. Likelihood ratio test of positive selection was performed by comparing M7 (beta) and M8 (beta&ω) models using PAML4.9 software package (Yang et al., 2000; Yang, 2007). For gene groups with evidence of positive selection based on the likelihood ratio test, sites under positive selection were identified using the Bayes empirical Bayes (BEB) procedure.
Accession Numbers
Sequence data from this article can be found in data libraries under accession numbers detailed in Supplemental Data Sets S1 and S2 and Supplemental Tables S1 and S4.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogenetic analysis of plant R-proteins.
Supplemental Table S1. Reference information for the genomes and transcriptomes used in this study.
Supplemental Table S2. The distribution of NB-ARC-domain proteins and R-proteins in green plants.
Supplemental Table S3. The distribution of different nTNLs in green plants.
Supplemental Table S4. The annotation of Arabidopsis R-proteins identified in this study.
Supplemental Table S5. Positive selection analysis in earlier branching streptophytes.
Supplemental Data Set S1. Sequence information of proteins with an NB-ARC domain identified in plant genomes.
Supplemental Data Set S2. Sequence information of R-genes identified in charophyte transcriptomes.
Footnotes
This work was supported by the National Natural Science Foundation of China (31701091), the Natural Science Foundation of Jiangsu Province (BK20161016), the Program for Jiangsu Excellent Scientific and Technological Innovation Team (17CXTD00014), and the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions.
References
- Afzal AJ, Wood AJ, Lightfoot DA (2008) Plant receptor-like serine threonine kinases: Roles in signaling and plant defense. Mol Plant Microbe Interact 21: 507–517 [DOI] [PubMed] [Google Scholar]
- Anisimova M, Gascuel O (2006) Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol 55: 539–552 [DOI] [PubMed] [Google Scholar]
- Ausubel FM. (2005) Are innate immune signaling pathways in plants and animals conserved? Nat Immunol 6: 973–979 [DOI] [PubMed] [Google Scholar]
- Bergelson J, Kreitman M, Stahl EA, Tian D (2001) Evolutionary dynamics of plant R-genes. Science 292: 2281–2285 [DOI] [PubMed] [Google Scholar]
- Cesari S. (2017) Multiple strategies for pathogen perception by plant immune receptors. New Phytol [DOI] [PubMed] [Google Scholar]
- Césari S, Kanzaki H, Fujiwara T, Bernoux M, Chalvon V, Kawano Y, Shimamoto K, Dodds P, Terauchi R, Kroj T (2014) The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J 33: 1941–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Jones JD (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Delwiche CF, Cooper ED (2015) The evolutionary origin of a terrestrial flora. Curr Biol 25: R899–R910 [DOI] [PubMed] [Google Scholar]
- Delaux PM, Radhakrishnan GV, Jayaraman D, Cheema J, Malbreil M, Volkening JD, Sekimoto H, Nishiyama T, Melkonian M, Pokorny L, et al. (2015) Algal ancestor of land plants was preadapted for symbiosis. Proc Natl Acad Sci USA 112: 13390–13395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J, Archibald JM (2018) Plant evolution: Landmarks on the path to terrestrial life. New Phytol 217: 1428–1434 [DOI] [PubMed] [Google Scholar]
- de Vries J, de Vries S, Slamovits CH, Rose LE, Archibald JM (2017) How embryophytic is the biosynthesis of phenylpropanoids and their derivatives in streptophyte algae? Plant Cell Physiol 58: 934–945 [DOI] [PubMed] [Google Scholar]
- Eddy SR. (2011) Accelerated profile HMM searches. PLOS Comput Biol 7: e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field KJ, Pressel S, Duckett JG, Rimington WR, Bidartondo MI (2015) Symbiotic options for the conquest of land. Trends Ecol Evol 30: 477–486 [DOI] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR (2011) HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res 39: W29–W37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Harholt J, Moestrup Ø, Ulvskov P (2016) Why plants were terrestrial from the beginning. Trends Plant Sci 21: 96–101 [DOI] [PubMed] [Google Scholar]
- Hori K, Maruyama F, Fujisawa T, Togashi T, Yamamoto N, Seo M, Sato S, Yamada T, Mori H, Tajima N, et al. (2014) Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat Commun 5: 3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Jones JD, Vance RE, Dangl JL (2016) Intracellular innate immune surveillance devices in plants and animals. Science 354: aaf6395. [DOI] [PubMed] [Google Scholar]
- Ju C, Van de Poel B, Cooper ED, Thierer JH, Gibbons TR, Delwiche CF, Chang C (2015) Conservation of ethylene as a plant hormone over 450 million years of evolution. Nat Plants 1: 14004. [DOI] [PubMed] [Google Scholar]
- Katoh K, Toh H (2008) Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 9: 286–298 [DOI] [PubMed] [Google Scholar]
- Kenrick P, Crane PR (1997) The origin and early evolution of plants on land. Nature 389: 33–39 [Google Scholar]
- Kroj T, Chanclud E, Michel-Romiti C, Grand X, Morel JB (2016) Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. New Phytol 210: 618–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D, Ullrich KK, Murat F, Fuchs J, Jenkins J, Haas FB, Piednoel M, Gundlach H, Van Bel M, Meyberg R, et al. (2018) The Physcomitrella patens chromosome-scale assembly reveals moss genome structure and evolution. Plant J 93: 515–533 [DOI] [PubMed] [Google Scholar]
- Lefort V, Longueville JE, Gascuel O (2017) SMS: Smart model selection in PhyML. Mol Biol Evol 34: 2422–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, et al. (2017) CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45: D200–D203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maqbool A, Saitoh H, Franceschetti M, Stevenson CEM, Uemura A, Kanzaki H, Kamoun S, Terauchi R, Banfield MJ (2015) Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. eLife 4: e08709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCourt RM, Delwiche CF, Karol KG (2004) Charophyte algae and land plant origins. Trends Ecol Evol 19: 661–666 [DOI] [PubMed] [Google Scholar]
- McHale L, Tan X, Koehl P, Michelmore RW (2006) Plant NBS-LRR proteins: Adaptable guards. Genome Biol 7: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Morgante M, Michelmore RW (2002) TIR-X and TIR-NBS proteins: Two new families related to disease resistance TIR-NBS-LRR proteins encoded in Arabidopsis and other plant genomes. Plant J 32: 77–92 [DOI] [PubMed] [Google Scholar]
- Mondragón-Palomino M, Meyers BC, Michelmore RW, Gaut BS (2002) Patterns of positive selection in the complete NBS-LRR gene family of Arabidopsis thaliana. Genome Res 12: 1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP (2009) FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26: 1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttick MN, Morris JL, Williams TA, Cox CJ, Edwards D, Kenrick P, Pressel S, Wellman CH, Schneider H, Pisani D, et al. (2018) The interrelationships of land plants and the nature of the ancestral embryophyte. Curr Biol 28: 733–745 [DOI] [PubMed] [Google Scholar]
- Qiu YL, Cho Y, Cox JC, Palmer JD (1998) The gain of three mitochondrial introns identifies liverworts as the earliest land plants. Nature 394: 671–674 [DOI] [PubMed] [Google Scholar]
- Qiu YL, Li LB, Wang B, Chen ZD, Dombrovska O, Lee J, Kent L, Li RQ, Jobson RW, Hendry TA (2007) A non-flowering land plant phylogeny inferred from nucleotide sequences of seven chloroplast, mitochondrial and nuclear genes. Int J Plant Sci 168: 691–708 [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Sarris PF, Cevik V, Dagdas G, Jones JD, Krasileva KV (2016) Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biol 14: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao ZQ, Xue JY, Wu P, Zhang YM, Wu Y, Hang YY, Wang B, Chen JQ (2016) Large-scale analyses of angiosperm nucleotide-binding site-leucine-rich repeat genes reveal three anciently diverged classes with distinct evolutionary patterns. Plant Physiol 170: 2095–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao ZQ, Zhang YM, Hang YY, Xue JY, Zhou GC, Wu P, Wu XY, Wu XZ, Wang Q, Wang B, et al. (2014) Long-term evolution of nucleotide-binding site-leucine-rich repeat genes: Understanding gained from and beyond the legume family. Plant Physiol 166: 217–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB (2001) Plant receptor-like kinase gene family: Diversity, function, and signaling. Sci STKE 2001: re22. [DOI] [PubMed] [Google Scholar]
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, et al. (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270: 1804–1806 [DOI] [PubMed] [Google Scholar]
- Stebbins GL, Hill GJC (1980) Did multicellular plants invade the land? Am Nat 115: 342–353 [Google Scholar]
- Swofford DL. (2003). PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4 Sinauer Associates, Sunderland, MA [Google Scholar]
- Tarr DE, Alexander HM (2009) TIR-NBS-LRR genes are rare in monocots: Evidence from diverse monocot orders. BMC Res Notes 2: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme RE, Bachvaroff TR, Delwiche CF (2012) Broad phylogenomic sampling and the sister lineage of land plants. PLoS One 7: e29696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach JM, Ausubel FM (2017) The NBS-LRR architectures of plant R-proteins and metazoan NLRs evolved in independent events. Proc Natl Acad Sci USA 114: 1063–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn RA, Kamoun S (2008) From guard to decoy: A new model for perception of plant pathogen effectors. Plant Cell 20: 2009–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Liu Y, Li SS, Han GZ (2015) Insights into the origin and evolution of the plant hormone signaling machinery. Plant Physiol 167: 872–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett NJ, Mirarab S, Nguyen N, Warnow T, Carpenter E, Matasci N, Ayyampalayam S, Barker MS, Burleigh JG, Gitzendanner MA, et al. (2014) Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci USA 111: E4859–E4868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Ellwood S, Calis O, Patrick E, Li T, Coleman M, Turner JG (2001) Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291: 118–120 [DOI] [PubMed] [Google Scholar]
- Xue J-Y, Wang Y, Wu P, Wang Q, Yang L-T, Pan X-H, Wang B, Chen J-Q (2012) A primary survey on bryophyte species reveals two novel classes of nucleotide-binding site (NBS) Genes. PLoS ONE 7: e36700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2007) PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Pedersen AMK (2000) Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155: 431–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Tang F, Gao M, Krishnan HB, Zhu H (2010) R gene-controlled host specificity in the legume-rhizobia symbiosis. Proc Natl Acad Sci USA 107: 18735–18740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue JX, Meyers BC, Chen JQ, Tian D, Yang S (2012) Tracing the origin and evolutionary history of plant nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes. New Phytol 193: 1049–1063 [DOI] [PubMed] [Google Scholar]