A chromatin regulatory complex displays two modes of action in regulating the gene expression of a subclade of a transcription factor family during early endosperm development in Arabidopsis.
Abstract
Early endosperm development presents a unique system in which to uncover epigenetic regulatory mechanisms because the contributing maternal and paternal genomes possess differential epigenetic modifications. In Arabidopsis (Arabidopsis thaliana), the initiation of endosperm coenocytic growth upon fertilization and the transition to endosperm cellularization are regulated by the FERTILIZATION-INDEPENDENT SEED (FIS)-Polycomb Repressive Complex 2 (PRC2), a putative H3K27 methyltransferase. Here, we address the possible role of the FIS-PRC2 complex in regulating the type I MADS-box gene family, which has been shown previously to regulate early endosperm development. We show that a subclass of type I MADS-box genes (C2 genes) was expressed in distinct domains of the coenocytic endosperm in wild-type seeds. Furthermore, the C2 genes were mostly up-regulated biallelically during the extended coenocytic phase of endosperm development in the FIS-PRC2 mutant background. Using allele-specific expression analysis, we also identified a small subset of C2 genes subjected to FIS-PRC2-dependent maternal or FIS-PRC2-independent paternal imprinting. Our data support a dual role for the FIS-PRC2 complex in the regulation of C2 type I MADS-box genes, as evidenced by a generalized role in the repression of gene expression at both alleles associated with endosperm cellularization and a specialized role in silencing the maternal allele of imprinted genes.
The endosperm is a nutritive tissue within the seeds of angiosperms that supports the growth of the embryo and germinating seedlings (Lopes and Larkins, 1993; Olsen, 2004; Li and Berger, 2012; Lafon-Placette and Köhler, 2014). During double fertilization, two haploid sperm cells fuse with a haploid egg cell and a homodiploid central cell to give rise to a diploid embryo and a triploid endosperm, respectively (Yadegari and Drews, 2004; Berger et al., 2008). In most angiosperms, including Arabidopsis (Arabidopsis thaliana), endosperm development follows a “nuclear”-type program composed of a coenocytic phase and a subsequent cellularized phase (Olsen, 2004). The coenocytic phase is characterized by a series of mitoses without cytokinesis. In Arabidopsis, the coenocytic endosperm differentiates into micropylar, peripheral, and chalazal domains (Boisnard-Lorig et al., 2001; Brown et al., 2003). Cellularization begins after the eighth mitosis in the micropylar domain that surrounds the embryo, then extends across the peripheral domain toward the chalazal domain (Boisnard-Lorig et al., 2001; Sørensen et al., 2002; Olsen, 2004). After cellularization, the Arabidopsis endosperm gradually degenerates except for an endosperm epidermal layer (Brown et al., 1999; Olsen, 2004; Li and Berger, 2012).
Epigenetic processes have been implicated in controlling the duration of the coenocytic phase. Global DNA hypomethylation in methyltransferase1 (met1) antisense mutant causes either a prolonged or an abbreviated coenocytic phase depending on hypomethylation of the maternal or paternal genome, respectively (Adams et al., 2000). A similar phenomenon has been observed in interploidy and interspecific crosses, where the parental genome dosages are imbalanced and seed size is positively associated with the extent of coenocytic endosperm development (Scott et al., 1998; Adams et al., 2000; Lafon-Placette and Köhler, 2016; Gehring and Satyaki, 2017). The sensitivity of the endosperm to parental genome dosage suggests that some regulators of endosperm development are subjected to imprinting. This notion is further supported by the fact that the majority of the imprinted genes (MEGs, for maternally expressed imprinted genes, and PEGs, for paternally expressed imprinted genes) reported to date are expressed in the endosperm (Gehring et al., 2011; Hsieh et al., 2011; Luo et al., 2011; Waters et al., 2011; Wolff et al., 2011; Zhang et al., 2011, 2016; Xin et al., 2013; Xu et al., 2014; Hatorangan et al., 2016; Klosinska et al., 2016; Gehring and Satyaki, 2017). In addition to DNA methylation, histone modification likely contributes to the epigenetic mechanisms regulating endosperm development. The coenocytic program is repressed prior to fertilization and at the onset of cellularization through the activity of the FERTILIZATION-INDEPENDENT SEED (FIS)-Polycomb Repressive Complex2 (PRC2), which is a putative H3K27 methyltransferase composed primarily of FIS2, MEDEA (MEA), FERTILIZATION-INDEPENDENT ENDOSPERM (FIE), and the Arabidopsis homolog of MULTICOPY SUPPRESSOR OF IRA1 (MSI1; Ohad et al., 1996, 1999; Chaudhury et al., 1997; Grossniklaus et al., 1998; Luo et al., 1999; Cao et al., 2002; Czermin et al., 2002; Köhler et al., 2003a; Mozgova and Hennig, 2015; Pu and Sung, 2015). The prominent role played by the epigenetic machineries during endosperm development suggests that the key regulators of the coenocytic phase, such as transcription factors, likely interact with epigenetic control mechanisms. Although some transcription factors, such as AGAMOUS-LIKE62 (AGL62) and components of the MINISEED3-HAIKU pathway, have been shown to promote coenocytic development (Garcia et al., 2003; Luo et al., 2005; Kang et al., 2008; Zhou et al., 2009; Wang et al., 2010a), their interaction with the epigenetic machinery has not yet been investigated fully.
Type I MADS-box transcription factor genes may constitute a regulatory circuitry that links the above-mentioned epigenetic mechanisms with the regulation of genes expressed during early endosperm development. Type I MADS-box genes are preferentially expressed in the female gametophyte and seed (Day et al., 2008; Bemer et al., 2010). A member of this gene family, AGL62, has been shown to regulate coenocytic endosperm development and seed size (Kang et al., 2008; Hehenberger et al., 2012; Kradolfer et al., 2013a). In addition, pheres1 (phe1; also known as agl37), agl62, and agl90 mutants alleviate endosperm-mediated postzygotic barriers between Arabidopsis and Arabidopsis arenosa (Josefsson et al., 2006; Walia et al., 2009). Several type I MADS-box genes are expressed in coenocytic endosperm, up-regulated in FIS-PRC2 mutants, and up-regulated in crosses with excess paternal genome (Day et al., 2008; Walia et al., 2009; Bemer et al., 2010; Tiwari et al., 2010; Gehring and Satyaki, 2017). These observations suggest that this gene family is partly required for regulating coenocytic development. However, the function of type I MADS-box genes remains elusive, since nearly all single-gene mutants have no discernible phenotype in the endosperm, the only exception being agl62, which exhibits precocious cellularization (Kang et al., 2008). Several type I MADS-box genes are regulated by FIS-PRC2-mediated histone modification and MET1- or polymerase IV-dependent small interfering RNAs (p4-siRNAs; Walia et al., 2009; Tiwari et al., 2010; Shirzadi et al., 2011; Lu et al., 2012). Some of these epigenetic machineries maintain imprinting at AGL36 and PHE1 loci, which are paternally and maternally imprinted type I MADS-box genes, respectively (Köhler et al., 2005; Shirzadi et al., 2011).
To further understand the mechanism by which FIS-PRC2 regulates early endosperm development, we systematically identified a subset of the type I MADS-box genes that are regulated by FIS-PRC2 and analyzed the mode of this regulation during early endosperm development in Arabidopsis. We conducted a comprehensive expression analysis of type I MADS-box genes in the wild type and a FIS-PRC2 double mutant (mea;swn) and identified a subclade of the type I MADS-box gene family (C2) to be repressed by the FIS-PRC2 complex, which is necessary to restrict C2 gene expression to the coenocytic phase of endosperm development. We further showed that a few of these genes were subjected to genomic imprinting. Our observations support a dual role for FIS-PRC2 regulation of early endosperm development through a generalized repression of the C2 subclade of type I MADS-box genes associated with endosperm cellularization and a more specialized role in imprinting of the maternal allele.
RESULTS
FIS-PRC2 Is Required for the Down-Regulation of a Subset of Type I MADS-Box Genes during Endosperm Cellularization
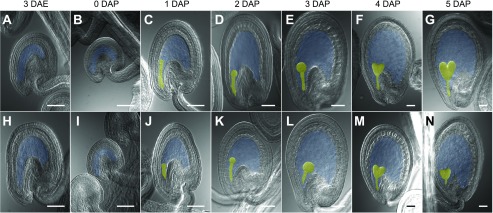
To identify type I MADS-box genes regulated by the FIS-PRC2 complex during endosperm development, we analyzed the mRNA levels using reverse transcription-quantitative PCR (RT-qPCR) for 69 MADS-box genes that included all type I MADS-box genes and all other MADS-box genes not belonging to the MIKCC subfamily (Supplemental Table S1; Parenicová et al., 2003; de Folter et al., 2005; Gramzow and Theissen, 2010; Dreni and Kater, 2014). We analyzed the mRNA levels in carpels or siliques of the wild-type Columbia-0 (Col-0) accession and swn-3/−;mea-3/+ double-mutant plants (Col-0 background) at seven time points corresponding to three developmental phases of endosperm development in wild-type seeds, including the unfertilized central cell 3 d after emasculation (DAE) and 0 d after pollination (DAP), the coenocytic endosperm (1, 2, and 3 DAP), and the cellularized endosperm (4 and 5 DAP; Fig. 1). swn-3/−;mea-3/+ plants produce an enhanced autonomous endosperm phenotype in comparison with mea single-mutant plants, indicating that SWN and MEA FIS-PRC2 complexes perform partially redundant roles in regulating the initiation of endosperm development upon fertilization (Wang et al., 2006). Unlike in the wild type (Col-0), mutant plants produced an autonomous endosperm at 3 DAE and a persistent coenocytic, noncellularized-type endosperm in seeds 4 and 5 DAP (Fig. 1; Supplemental Fig. S1). Of the 69 genes analyzed, 16 did not show expression at any stage analyzed and were not considered further (Supplemental Fig. S2).
Figure 1.
Stages of unfertilized seeds and early fertilized seeds encompassing the three phases of endosperm development used in the RT-qPCR assays. Cleared ovules or seeds at 3 DAE (A and H), 0 DAP (B and I), 1 DAP (C and J), 2 DAP (D and K), 3 DAP (E and L), 4 DAP (F and M), and 5 DAP (G and N) from Col-0 (A–G) and mea-3;swn-3 mutant (H–N) plants are shown. The embryo is false colored in yellow. The embryo sac or the part of the embryo sac excluding the embryo is false colored in blue. Bars = 50 µm.
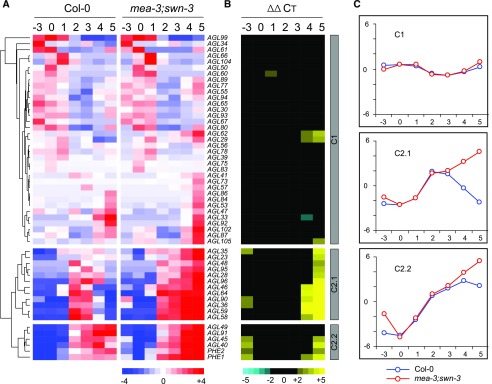
Using a hierarchical clustering approach, we identified two major expression clusters, termed C1 (35 genes) and C2 (18 genes), that differed in both the temporal expression profile in Col-0 and the degree of dysregulation in the mutant (Fig. 2). In Col-0, C1 genes exhibited diverse patterns of expression, with most showing lower mRNA levels during coenocytic endosperm development (1–3 DAP) compared with other stages (Fig. 2A). Many C1 genes, including central cell-expressed AGL61 and AGL80 (Bemer et al., 2008; Steffen et al., 2008), showed higher relative expression levels either before fertilization (0 DAP) or upon endosperm cellularization (4 and 5 DAP; Fig. 2A). In contrast, all C2 genes were distinguished by their up-regulation after fertilization within 1 to 2 DAP (Fig. 2A). They were further divided into two subclusters based on the onset of their down-regulation in Col-0: C2.1 genes showed a reduction in relative levels of mRNA at 4 to 5 DAP, whereas C2.2 genes maintained high relative levels of mRNA at 4 to 5 DAP (Fig. 2, A and C).
Figure 2.
mRNA profiles of 53 MADS-box genes during early seed development in Col-0 and swn-3/−;mea-3/+ double-mutant plants. A, Centered average CT values obtained from RT-qPCR analysis of mRNA levels in 3-DAE carpels (−3), 0-DAP carpels (0), and 1- to 5-DAP siliques (1–5). Values were subjected to hierarchical cluster analysis and are presented as a heat map to identify the three expression clusters C1, C2.1, and C2.2 based on the relative mRNA levels in both genetic backgrounds. Red denotes relatively high and blue denotes relatively low mRNA levels. B, Heat map depicting the differences in mRNA levels (ΔΔCT) between Col-0 and swn-3/−;mea-3/+ mutant backgrounds. Genes are arranged in the same order as in A. Yellow denotes up-regulation in the mutant, and blue denotes down-regulation in the mutant. ΔΔCT values below the statistical threshold are in black (|ΔΔCT| > 2; P < 0.05, Student’s t test). C, Graphic representation of average mRNA profiles of C1, C2.1, and C2.2 genes.
In addition to their distinct temporal mRNA profiles in wild-type Col-0, C1 and C2 genes were distinguished based on the extent to which they showed altered mRNA levels in the mutant. Overall, C1 genes did not show any significant change in mRNA levels (|ΔΔCT| > 2; P < 0.05, Student’s t test) in the mutant as compared with Col-0 (Fig. 2B; Supplemental Table S1), except for five genes, including the previously described AGL62 that is expressed during early endosperm development (Kang et al., 2008; Bemer et al., 2010). In contrast, 16 of 18 C2 genes (all except AGL49 and AGL91) showed a dramatically up-regulated expression pattern in the mutant, as both C2.1 and C2.2 genes showed a similar, continuous increase in mRNA levels (|ΔΔCT| > 2; P < 0.05, Student’s t test) at 3 DAE, 4 DAP, and 5 DAP (Fig. 2B; Supplemental Table S1). As noted above, both C2 subclusters displayed distinct expression kinetics in Col-0 (Fig. 2). This up-regulated pattern corresponded to the presence of the autonomous (3 DAE) and noncellularized endosperm (4–5 DAP) phenotypes observed in the FIS-PRC2 mutant combination (Figs. 1 and 2). Therefore, our data indicate that, although the loss of FIS-PRC2 activity does not affect the fertilization-induced activation of the C2 subset of type I MADS-box genes, FIS-PRC2 is likely required for the proper down-regulation of these genes at the onset of endosperm cellularization.
C2.1 and C2.2 Type I MADS-Box Genes Are Expressed in Distinct Domains of Coenocytic Endosperm
To confirm the association of C2 gene expression with coenocytic endosperm development and to further characterize the expression patterns of the designated expression clusters, we carried out a series of gene fusion analyses in transgenic plants for 38 selected genes representing the main expression clusters (20 C1 and 16 C2 genes) and control genes (two denoted as nonexpressors; Table I; Supplemental Tables S2–S4). Using promoter and full-length gene fusions, all C2 genes tested were shown to be expressed primarily in the endosperm following fertilization, during the coenocytic phase, and were mostly down-regulated (see below) as the endosperm became cellularized (Table I; Fig. 3; Supplemental Figs. S4 and S5). Of these, only five genes were shown to be expressed earlier in the female gametophyte, with three displaying weak expression in the central cell (Table I; Supplemental Figs. S6 and S7). In contrast, most C1 genes tested were either not expressed (nine genes) or expressed in both the female gametophyte and the endosperm (four genes: AGL29, AGL84, AGL87, and AGL99; Supplemental Table S3; Supplemental Figs. S6 and S8); endosperm-specific expression was observed in only three C1 genes (AGL33, AGL102, and AGL104) tested (Supplemental Table S3). No GFP activity in ovules or seeds was detected for the two control genes (AGL26 and AGL74; Supplemental Table S3).
Table I. Expression analysis of C2 type I MADS-box genes in ovules and seeds.
A, Antipodal cells; C, central cell; ENC, signal is stronger in the chalazal region than in the peripheral and micropylar regions; ENM, signal is stronger in the micropylar region and not detected or weak in the chalazal region; ENM&C, signal is stronger in the micropylar and chalazal regions than in the peripheral region; ENM&P, signal is present only in the micropylar and peripheral regions; FG, female gametophyte; ND, not determined; ne, no detectable expression; s, synergid cells; SBE, single-base extension with fluorescently labeled dideoxyribonucleotide triphosphates (ddNTPs).
| Expression Cluster |
Reporter Analysisa |
Imprinting Analysisb |
||||
|---|---|---|---|---|---|---|
| Gene | Arabidopsis Genome Initiative No. | qPCRc | FG | Endosperm | Reporterc | SBE |
| AGL23 | AT1G65360 | C2.1 | C, s | ENM&C | Biallelic | Paternal |
| AGL28 | AT1G01530 | C2.1 | c | ENM&C | Biallelic | Biallelic |
| AGL35 | AT5G26630 | C2.1 | ne | ENM&C | Biallelic (biallelic) | Biallelic |
| AGL36 | AT5G26650 | C2.1 | ne | ENM | Maternal (maternal) | Maternal |
| AGL46 | AT2G28700 | C2.1 | c | ENM&P | Biallelic | Biallelic |
| AGL48 | AT2G40210 | C2.1 | ne | ENM | Biallelic | Biallelic |
| AGL58 | AT1G28450 | C2.1 | ne | ENM | Biallelic | ND |
| AGL59 | AT1G28460 | C2.1 | ne | ENM | Biallelic | ND |
| AGL64 | AT1G29962 | C2.1 | ne | ENM | ND | Biallelic |
| AGL96 | AT5G06500 | C2.1 | ne | ENM | Maternal | Maternal |
| AGL95 | AT2G15660 | C2.1 | ND | ND | ND | Biallelic |
| AGL90 | AT5G27960 | C2.1 | ne | ENM | Maternal | Maternal |
| PHE1 | AT1G65330 | C2.2 | ne | ENM&C | Biallelic (paternal) | Paternal |
| PHE2 | AT1G65300 | C2.2 | c | ENM&C | Biallelic | Biallelic |
| AGL40 | AT4G36590 | C2.2 | ne | ENM&C | Biallelic | Biallelic |
| AGL45 | AT3G05860 | C2.2 | A | ENC | Biallelic | Biallelic |
| AGL49 | AT1G60040 | C2.2 | ND | ND | ND | ND |
| AGL91 | AT3G66656 | C2.2 | ne | ENC | Biallelic | ND |
GFP/YFP reporter signals were analyzed 1 DAE (corresponding to the mature female gametophyte) and at flower stage 16 (corresponding to coenocytic endosperm stages V and VI). Consistent (uppercase) or sporadic (lowercase) GFP expression was observed in the female gametophyte, while expression in the endosperm was consistent.
Imprinting of paternal or maternal allele-specific expression was determined using reporter lines and/or SBE analysis.
The genes significantly up-regulated at 5 DAP in the swn;mea mutant are in boldface.
All genes were analyzed in promoter fusion reporter lines except for three genes also analyzed in full-length gene fusion (indicated by parentheses).
Figure 3.
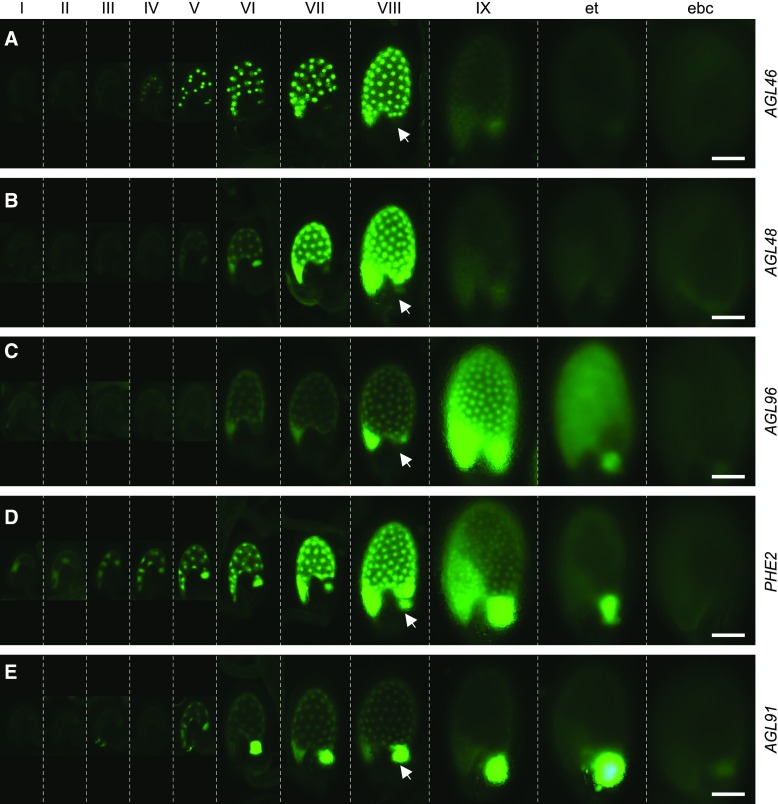
Expression of C2 type I MADS-box gene reporter lines during endosperm development. Promoter-GFP activities were analyzed at 11 stages (separated by dashed lines) during endosperm development, including the coenocytic phase (endosperm stages I–VIII) and cellularized phases (endosperm stage IX, early torpedo stage [et], and early bent-cotyledon stage [ebc]). Five distinct expression patterns are represented by AGL46 (A), AGL48 (B), AGL96 (C), PHE2 (D), and AGL91 (E). All seeds are oriented with the micropylar end toward the bottom left. Arrows indicate the position of the chalazal domain at endosperm stage VIII. Bars = 100 µm.
Further examination of the reporter-gene expression patterns (Table I; Fig. 3; Supplemental Figs. S4 and S5) indicated differences in both temporal and spatial patterns of C2.1 and C2.2 transcriptional activities during endosperm development. Prior to endosperm cellularization (stages VII and VIII), C2.1 genes (eight of 11; i.e. all except AGL23, AGL28, and AGL35) generally displayed higher activity in the micropylar and peripheral regions of the coenocytic endosperm as compared with the chalazal region (Table I; Fig. 3, A–C; Supplemental Fig. S4), whereas all five C2.2 genes typically displayed higher GFP activity in both the micropylar and chalazal regions or were expressed predominantly in the chalazal region of the coenocytic endosperm (Table I; Fig. 3, D and E; Supplemental Fig. S5). Shortly after the initiation of endosperm cellularization (stage IX), C2.1 genes (seven of 11) were rapidly down-regulated in all three endosperm compartments, except for four genes (AGL23, AGL28, AGL35, and AGL96) that showed a more delayed down-regulation pattern (Fig. 3, A–C; Supplemental Fig. S4). In contrast, all five C2.2 genes displayed reduced activity in the micropylar and peripheral endosperm by stage IX but maintained high activity in the chalazal endosperm (Fig. 3, D and E; Supplemental Fig. S5). Therefore, the micropylar-peripheral-preferred expression of C2.1 genes distinguishes this subcluster from the C2.2 genes, which show a chalazal-preferred expression pattern. This observation is in agreement with our RT-qPCR data showing a delayed down-regulation of C2.2 genes upon cellularization of the micropylar-peripheral endosperm (Fig. 2A). Notably, the chalazal compartment cellularizes late as compared with other endosperm compartments (Belmonte et al., 2013). Furthermore, an analysis of the publicly available microarray (Hruz et al., 2008) and RNA sequencing (Liu et al., 2012) data indicated that C2 gene expression was highly specific to the endosperm as compared with C1 gene expression (Supplemental Fig. S9). The latter showed a broad pattern of expression during the plant life cycle, whereas the C2.1 and C2.2 genes exhibited differential mRNA accumulation in the endosperm compartments consistent with our gene fusion patterns. Taken together, our data indicate that the C2 genes represent an endosperm-specific subclade of type I MADS-box genes and that the C2.1 and C2.2 genes are programmed for differential expression in distinct domains of coenocytic endosperm prior to cellularization.
C2 Type I MADS-Box Genes Exhibit Both Nonimprinted and Imprinted Expression Patterns during Early Seed Development
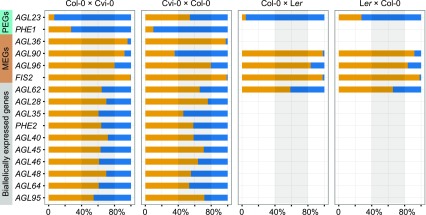
Components of the FIS-PRC2 complex have been shown to maintain the locus-specific, paternal or maternal imprinting of some endosperm-expressed genes, including MEA (Gehring et al., 2006) and PHE1 (Köhler et al., 2005; Hsieh et al., 2011). To determine whether C2 genes are regulated through FIS-PRC2-mediated imprinting, we first investigated the imprinting status of C2 genes in the wild-type background. Single-base extension with fluorescently labeled ddNTPs (SBE) was employed to measure the ratio of maternal to paternal transcripts in 3-DAP seeds from reciprocal crosses between Col-0 and Cvi-0 (Col-0 × Cvi-0 and Cvi-0 × Col-0) based on the presence of single-nucleotide polymorphisms (SNPs) between the pair of accessions (Fig. 4). These analyses indicated that the majority of the C2 genes (nine of 14) were biallelically expressed in both reciprocal crosses (Fig. 4; Supplemental Table S5). Five genes, AGL23, AGL36, PHE1, AGL90, and AGL96, showed preferential accumulation of either maternal or paternal transcripts in at least one of the reciprocal crosses and, therefore, were identified as a MEG or a PEG (Fig. 4; Supplemental Table S5). These data are in agreement with previous reports that identified AGL36 as a MEG and PHE1 as a PEG (Köhler et al., 2005; Shirzadi et al., 2011). Interestingly, AGL96 transcripts exhibited high maternal frequency but not at levels that would suggest a nearly complete silencing of the paternal allele, as detected for FIS2 (Fig. 4). Of all the genes assayed, only AGL23 and AGL90 showed preferential accumulation of paternal and maternal transcripts, respectively, in only one of the crosses (Col-0 × Cvi-0; Fig. 4). Therefore, we tested allelic transcript levels for AGL23 and AGL90, along with three control genes, AGL62, AGL96, and FIS2, in reciprocal crosses between Col-0 and Landsberg erecta (Ler; Col-0 × Ler and Ler × Col-0) and showed that AGL90 mRNA was derived primarily from the maternal allele in both crosses while AGL23 transcripts were primarily paternal (Fig. 4). The results of SBE analysis were further supported by allele-specific RT-PCR experiments using cleaved-amplified polymorphic sequence (CAPS) markers (Supplemental Fig. S10). These results suggested that an accession-specific regulatory program (Wolff et al., 2011; Waters et al., 2013; Pignatta et al., 2014; Gehring and Satyaki, 2017) drives the allelic expression of AGL23 and AGL90. Therefore, with the exception of the MEGs AGL36, AGL90, and AGL96 and the PEGs AGL23 and PHE1, C2 genes generally exhibited a biallelic expression pattern during coenocytic endosperm development.
Figure 4.
Differential imprinted expression patterns of C2 type I MADS-box genes. The allele-specific expression of 14 C2 genes and control genes was analyzed in reciprocal crosses between Col-0 and Cvi-0 or Ler by single-base extension with fluorescently labeled ddNTPs (SBE) based on SNPs. Orange denotes maternal allele expression frequency, and blue denotes paternal allele expression frequency. In cross descriptions, the maternal parent is denoted first. FIS2 represents a MEG, and AGL62 represents a biallelically expressed gene. Three independent biological replicates were used. The threshold for determining MEGs and PEGs was set at 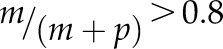 and
and 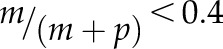 (where m denotes maternal transcripts and p denotes paternal transcripts measured by SBE) in both reciprocal crosses, as described previously (Hsieh et al., 2011). P values calculated using Student’s t test are provided in Supplemental Table S5.
(where m denotes maternal transcripts and p denotes paternal transcripts measured by SBE) in both reciprocal crosses, as described previously (Hsieh et al., 2011). P values calculated using Student’s t test are provided in Supplemental Table S5.
We tested for the activity of the upstream regulatory sequences and further confirmed both spatiotemporal and parental expression of the C2 genes (15 plus two C1 genes) at 2 and 5 DAP using promoter fusion reporter lines in reciprocal crosses (Table I; Supplemental Table S3). These stages corresponded to a coenocytic and an early-cellularized phase of endosperm development, respectively (Fig. 1). We also tested a set of full-length gene fusions for three representative genes, AGL35 (biallelic), AGL36 (MEG), and PHE1 (PEG), which included the 3′-flanking sequences in addition to the entire transcribed region and the 5′-flanking sequences for each gene (Table I; Supplemental Fig. S11). In agreement with our SBE analysis, only AGL36, AGL90, and AGL96 promoter reporter genes showed maternally restricted activity at 2 DAP (Table I; Fig. 5A; Supplemental Fig. S12), whereas AGL23 and PHE1 promoter reporter genes showed a biallelic pattern of expression at the same stage of development (Table I; Supplemental Fig. S12). Full-length gene fusions for AGL36 and PHE1 displayed the expected patterns of expression (Table I; Fig. 5B; Supplemental Fig. S12) as described previously (Makarevich et al., 2008; Shirzadi et al., 2011), indicating a functional role for 5′- and 3′-flanking sequences, respectively, in locus-specific regulation of the two genes. However, the majority of the C2 genes tested, including AGL48 and AGL91, were biallelically expressed as promoter reporters (Table I; Fig. 5, C and D; Supplemental Fig. S12). Taken together, for the small subset of the imprinted C2 genes, our data indicate that 5′- or 3′-flanking sequences are required to drive the allele-specific expression of MEGs or PEGs, respectively.
Figure 5.
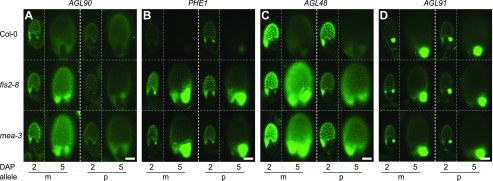
Allele-specific expression of C2 type I MADS-box genes in the endosperm. GFP or YFP signals were analyzed at 2 and 5 DAP. The AGL reporter transgenes, including the pAGL90:GFP promoter fusion (A), the PHE1:YFP full-length gene fusion (B), the pAGL48:GFP fusion (C), and the pAGL91:GFP fusion (D), were inherited either from the maternal (m) or paternal (p) parent, while the mutant allele (mea-3 and fis2-8) was always inherited from the maternal parent. Bars = 100 µm.
The Active Alleles of C2 Genes Are Repressed by the FIS-PRC2 Complex upon Cellularization through Their 5′-Flanking Regions
The results of our RT-qPCR analysis indicated that the C2 genes were up-regulated in the mea-3;swn-3 mutant corresponding to the period of endosperm cellularization in wild-type seeds (Figs. 1 and 2). In order to address whether either or both alleles are controlled by FIS-PRC2 during early endosperm development, we assayed the parent-specific activity of promoter and/or full-length gene fusions for 15 C2 genes (all C2 genes except AGL49, AGL64, and AGL95) as well as two C1 genes (AGL33 and AGL87) in the fis2-8 and mea-3 mutant backgrounds at 2 and 5 DAP (Fig. 5; Supplemental Fig. S12). Our analysis indicated that the promoter reporters of the MEGs AGL36, AGL90, and AGL96 (as well as the AGL36 full-length gene fusion) displayed an exclusively maternal up-regulation in fis2 and mea mutants at 5 DAP, whereas their paternal alleles remained silenced at 2 and 5 DAP (Fig. 5A; Supplemental Fig. S12). This suggests that the paternal silencing of C2 MEGs can be maintained exclusively through the 5′-flanking region of the gene by a FIS-PRC2-independent mechanism. In contrast, both the maternal and the paternal alleles of the PEG PHE1 full-length gene fusion were up-regulated at 5 DAP in fis2 and mea mutant seeds (Fig. 5B). A similar up-regulation pattern was detected with the PHE1 promoter reporter lines (although the absolute levels of expression were higher than for the full-length gene fusion; Supplemental Fig. S12), suggesting that the 5′-flanking sequences are sufficient to mediate FIS-PRC2-dependent repression of either allele during the transition to cellularization. Interestingly, an up-regulation of both the paternal and maternal alleles was observed at 5 DAP in the mutants for most of the biallelically expressed C2 genes when tested as promoter reporters (11 of 12 [i.e. all except AGL91]; Fig. 5C; Supplemental Fig. S12). As FIS-PRC2 mutants lack proper endosperm cellularization (Chaudhury et al., 1997; Grossniklaus et al., 1998; Köhler et al., 2003a), the observed up-regulation of the C2 genes at 5 DAP may result from an extended coenocytic phase. However, this is unlikely, as AGL91 and AGL33, two coenocytic-endosperm-expressed genes, were not observed to be up-regulated in the mutants at 5 DAP (Fig. 5D; Supplemental Fig. S12). Therefore, our data support a dual role for the FIS-PRC2 complex in the regulation of AGL genes: it maintains the maternal but not the paternal imprinting of the imprinted AGL genes, and it likely functions through their 5′-flanking sequences to down-regulate the actively expressed AGL genes prior to endosperm cellularization irrespective of their imprinting status.
DISCUSSION
To understand how FIS-PRC2 regulates early endosperm development, we analyzed the expression of the type I MADS-box genes in both the wild type and a mea;swn double mutant that produces persistent coenocytic endosperm. A subset of type I MADS-box genes (C2 genes) was found to be up-regulated in the mutant (Fig. 2). Further analysis revealed that the C2 genes were expressed primarily in distinct domains of the coenocytic endosperm and that this expression pattern was regulated by FIS-PRC2 acting through the C2 5′-flanking sequences of both alleles (Figs. 3 and 5; Supplemental Figs. S4 and S5). A subset of C2 genes was subjected to FIS-PRC2-dependent maternal imprinting or FIS-PRC2-independent paternal imprinting (Table I; Figs. 4 and 5; Supplemental Fig. S12). These data indicate a dual role for FIS-PRC2 in the regulation of its target genes during early endosperm development.
FIS-PRC2 Regulates a Subset of Endosperm-Expressed Type I MADS-Box Genes That Are Expressed in Distinct Domains of the Coenocytic Endosperm
Many type I MADS-box genes were expressed preferentially in distinct domains of the coenocytic endosperm: C2.1 genes were expressed primarily in the micropylar domain; C2.2 genes were expressed primarily in both the micropylar and chalazal domains; and two C2.2 genes (AGL45 and AGL91) and several C1 genes were expressed primarily in the chalazal domain (Table I; Fig. 3; Supplemental Table S3; Supplemental Figs. S4, S5, and S8). Our data differ from a study in which several C2.1 genes were reported as being expressed in the embryo rather than the micropylar endosperm that surrounds the embryo (Bemer et al., 2010). In agreement with our observation, an analysis of the available expression data (Hruz et al., 2008; Liu et al., 2012) indicated that C2.1 genes are expressed primarily in the endosperm (Supplemental Fig. S9). We observed several reporter lines in which the GFP signal intensity differed dramatically between neighboring endosperm domains (Fig. 3; Supplemental Figs. S4, S5, and S8), which suggests that the cytoplasmic contents of the coenocytic endosperm are not freely diffusible. Our data support the notion that the coenocytic endosperm domains are metabolically and functionally distinct (Boisnard-Lorig et al., 2001; Ingram, 2010).
In addition to generating domain-specific markers that can be used to monitor endosperm differentiation, our study also provides an entry point to unravel the genetic network controlling coenocytic endosperm differentiation. Because C2.1 genes encode transcription factors that are enriched in micropylar endosperm (Table I; Fig. 3; Supplemental Fig. S4), they may belong to gene regulatory networks for micropylar endosperm differentiation. It remains to be tested whether C2.1 genes act upstream of other micropylar-specific genes, such as SUCROSE-PROTON SYMPORTER5 (SUC5), ABNORMAL LEAF-SHAPE1 (ALE1), ZHOUPI (ZOU; also known as RETARDED GROWTH OF EMBRYO1, RGE1), and EMBRYO SURROUNDING FACTOR1 (ESF1), which were reported to regulate nutrient transfer into the embryo, the degeneration of micropylar endosperm, or early embryo patterning (Tanaka et al., 2001; Baud et al., 2005; Kondou et al., 2008; Yang et al., 2008; Xing et al., 2013; Costa et al., 2014). In light of the recent genome-wide discovery of genes expressed in specific endosperm domains (Le et al., 2010; Belmonte et al., 2013), it is possible to address if they are, in fact, regulated by type I MADS-box genes that are expressed in the same endosperm domains.
FIS-PRC2 Regulation of C2 Genes Reflects the Complexity of Epigenetic Regulatory Mechanisms That Control Early Seed Development
The expression of C2 genes is subjected to at least two levels of epigenetic control. First, FIS-PRC2-mediated histone modification through the 5′-flanking sequences of the gene is likely involved in repressing C2 genes upon endosperm cellularization (Supplemental Fig. S13). Second, a number of C2 genes are subjected to either maternal imprinting (the PEGs AGL23 and PHE1) or paternal imprinting (the MEGs AGL36, AGL90, and AGL96; Supplemental Fig. S13).
Our data revealed that FIS-PRC2 activity is required to repress the expression of the majority of C2 genes (16 of 18 genes, i.e. all except AGL49 and AGL91) before endosperm initiation and during endosperm cellularization (Figs. 2 and 5; Supplemental Figs. S7 and S12). The repression of C2 genes is likely achieved through direct binding of the FIS-PRC2 complex to C2 loci, as reported previously for PHE1, PHE2 (also known as AGL38), and AGL62 (Köhler et al., 2003b; Villar et al., 2009; Hehenberger et al., 2012; Moreno-Romero et al., 2016). Since both FIS-PRC2 and C2 genes are expressed in coenocytic endosperm (Luo et al., 2000; Yadegari et al., 2000; Wang et al., 2006; Table I; Fig. 3; Supplemental Figs. S4 and S5), it is not clear how FIS-PRC2-mediated repression is circumvented during the coenocytic endosperm phase to allow C2 gene expression. Using the available whole-genome profiling of H3K27me3-modified chromatin of 4-DAP endosperm nuclei (Moreno-Romero et al., 2016), we found that the promoters of 10 of the 16 FIS-PRC2-regulated C2 genes lack any detectable H3K27me3 modification in the regions used in the promoter fusions (Supplemental Fig. S14). These include eight of 11 biallelically expressed C2 genes (AGL28, AGL35, PHE2, AGL40, AGL45, AGL46, AGL64, and AGL95) and two of three MEGs (AGL36 and AGL90). Among these, PHE2 has been reported as a direct target of FIS-PRC2, with H3K27me3 modifications detected at its promoter sequence from closed flowers (Villar et al., 2009). These observations suggest a dynamic change in the pattern of H3K27me3 modifications at FIS-PRC2-regulated C2 genes during early endosperm development (Supplemental Fig. S13). These may occur via dissociation of the FIS-PRC2 complex from C2 loci during the coenocytic endosperm phase and a subsequent reassociation during endosperm cellularization. Alternatively, the FIS-PRC2 complex may remain associated with C2 loci in the form of bivalent chromatin domains, which are able to adopt either active or silenced states due to the presence of antagonistic histone modifications (Ku et al., 2008; Chakravarthy et al., 2011; Finnegan et al., 2011; Mozzetta et al., 2011; Mozgova and Hennig, 2015).
Several endosperm-expressed type I MADS-box genes, including C1 genes, are unlikely to be regulated by the FIS-PRC2 complex in the same manner. Genes expressed in coenocytic endosperm were identified from both the C1 and C2 clusters (Table I; Supplemental Table S3), which is in agreement with previous reports showing that type I MADS-box genes are expressed primarily in the endosperm and the female gametophyte (Köhler et al., 2003b; Portereiko et al., 2006; Bemer et al., 2008, 2010; Colombo et al., 2008; Day et al., 2008; Kang et al., 2008; Steffen et al., 2008). However, unlike the C2 genes, most endosperm-expressed C1 genes were not up-regulated in FIS-PRC2 mutants (Table I; Fig. 2; Supplemental Table S3), suggesting that persistent coenocytic endosperm produced in the FIS-PRC2 mutants does not cause indiscriminate up-regulation of all the genes expressed in coenocytic endosperm.
The FIS-PRC2 complex is required to maintain maternal imprinting in all reported cases, including PHE1 (Fig. 5; Köhler et al., 2005; Hsieh et al., 2011; Huang et al., 2017). Whole-genome H3K27me3 profiling has shown that PEGs including PHE1 are marked by maternal-specific H3K27me3 in 4-DAP endosperm (Moreno-Romero et al., 2016), consistent with the results of reciprocal crosses and gene fusion studies indicating a requirement of 3′-flanking sequences in the maintenance of PHE1 imprinting (Makarevich et al., 2008; Villar et al., 2009). Using the same data set, we found the SBE-typed PEG AGL23 to display a similar parentally asymmetric H3K27me3 modification both at its 5′- and 3′-flanking sequences (Supplemental Fig. S15). Because our AGL23 promoter or protein reporter lines did not display a PEG pattern consistent with our SBE results (Table I; Fig. 4; Supplemental Figs. S12 and S16), the available H3K27me3 profile suggests that the maintenance of the maternal imprinting pattern of AGL23 would require either distal 5′-flanking sequences or 3′-flanking sequences, as is the case for PHE1 (Makarevich et al., 2008; Villar et al., 2009). However, unlike PHE1, which is not expressed detectably in the female gametophyte before fertilization (Table I; Supplemental Fig. S7), AGL23 functions both in the female gametophyte and during seed development (Colombo et al., 2008). Accordingly, it is expressed within the central cell (weak) and the synergid cells (very weak) before fertilization, and it is expressed predominantly paternally within the endosperm after fertilization (Table I; Fig. 4; Supplemental Figs. S7 and S10). Therefore, as compared with PHE1, the regulation of AGL23 involves a more complex process of switching from a prefertilization maternal to a postfertilization paternal pattern. Nevertheless, the regulation of PEGs during early endosperm development, including maintenance of the silenced state of the maternal allele during coenocytic growth and initiation of repression of the paternal allele in transition to endosperm cellularization, would require the activity of the FIS-PRC2 complex.
On the other hand, multiple mechanisms have been proposed for paternal imprinting, including MET1-dependent DNA methylation, FIS-PRC2-dependent histone modification, and an unknown mechanism independent of MET1 or FIS-PRC2 that likely involves RNA-directed DNA methylation (Kinoshita et al., 2004; Gehring et al., 2006; Jullien et al., 2006, 2012; Tiwari et al., 2008; Fitz Gerald et al., 2009; Hsieh et al., 2011; Luo et al., 2011; Raissig et al., 2011; Vu et al., 2013; Du et al., 2014; Gehring and Satyaki, 2017). Our data indicate that paternally imprinted C2 MEGs are regulated by a FIS-PRC2-independent imprinting mechanism with the requirement of the 5′-flanking region of the gene but not the 3′-flanking region (Table I; Fig. 5; Supplemental Fig. S12), which agrees with a report showing that the paternal imprinting of AGL36 relies on MET1 instead of FIS-PRC2 (Shirzadi et al., 2011).
Potential Role of FIS-PRC2-Regulated Type I MADS-Box C2 Genes in Regulating Endosperm Development and Hybridization
There is an apparent link between imprinting and C2 genes: C2 genes are either imprinted themselves, or regulated by paternally imprinted FIS-PRC2 genes, or both (Table I; Figs. 4 and 5; Supplemental Figs. S12 and S13). Imprinting has been suggested as the underlying cause of abnormal endosperm development in crosses with imbalanced parental genome dosage (Lin, 1984; Feil and Berger, 2007; Berger and Chaudhury, 2009; Lafon-Placette and Köhler, 2016; Gehring and Satyaki, 2017). It has been proposed that imprinted genes act in macromolecular complexes that are sensitive to changes in stoichiometric ratio (Dilkes and Comai, 2004). Such complexes may form between FIS-PRC2 proteins and the C2 loci as part of the chromatin structure (Köhler et al., 2003b; Villar et al., 2009). Alternatively, they may form between C2 proteins as α-γ heterodimers (de Folter et al., 2005), since most C2 genes belong to either the α or γ subfamily (Parenicová et al., 2003). The existence of imprinted components in the complex, such as FIS-PRC2 or C2 proteins, renders the complex sensitive to imbalanced parental genome dosage. Therefore, imbalanced crosses may affect the performance of the complex, leading to abnormal expression or function of C2 genes. In line with these observations, C2 genes constitute the majority of the type I MADS-box genes that are either up-regulated or down-regulated in paternal-excess or maternal-excess interploidy and interspecific crosses (Walia et al., 2009; Tiwari et al., 2010; Shirzadi et al., 2011; Kradolfer et al., 2013a, 2013b; Burkart-Waco et al., 2015; Wolff et al., 2015). This suggests that altered C2 gene expression contributes to the abnormal endosperm phenotype observed in crosses with imbalanced parental genome dosage.
Although direct genetic evidence is limited (likely due to extensive genetic redundancy), the potential role of FIS-PRC2-regulated type I MADS-box C2 genes in regulating endosperm development can be gleaned from the functions of key interacting protein(s). MADS-box proteins are known to act as dimers or in higher order complexes (Davies et al., 1996; Egea-Cortines et al., 1999; Honma and Goto, 2001; Smaczniak et al., 2012). An analysis of the Plant Interactome Database for potential binding partners of C2 proteins suggests that the protein interaction network is based mainly on Mα-Mβ and Mα-Mγ interactions (Supplemental Fig. S17). Most of the C2 proteins are participants of the Mα-Mγ subnetwork. Interestingly, the Mα-Mγ subnetwork also includes AGL62, a gene known to control endosperm cellularization (Kang et al., 2008). Since AGL62 directly binds four C2 proteins (AGL36, PHE1, PHE2, and AGL90; Supplemental Fig. S17), the observation further supports the notion that at least some of the C2 genes regulate endosperm development.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) accession Cvi-0 (CS22682) was obtained from the Arabidopsis Biological Resource Center (Berardini et al., 2015). The mea-3/+, fis2-8/+, and mea-3/+;swn-3/− mutants in the Col-0 background were described previously (Wang et al., 2006). Plants were grown under a 16-h-light/8-h-dark photoperiod at 22°C as described previously (Wang et al., 2006).
Tissue Collection and RNA Isolation
Tissues used in RT-qPCR analysis were collected from Col-0 and the mea-3/+;swn-3/− double-mutant plants. Unfertilized carpels at 0 DAP and 3 DAE were harvested from stage 13 flowers containing differentiated female gametophytes at stages FG6 to FG7 (Smyth et al., 1990; Christensen et al., 1997) and at 3 DAE of stage 12 flowers (Smyth et al., 1990), respectively. Flower stage 14 (Smyth et al., 1990) is considered to be the time of pollination. Self-fertilized siliques were harvested every day from 1 to 5 DAP.
Tissues used in imprinting analysis were collected from reciprocal crosses between Col-0 and Cvi-0 or Col-0 and Ler. Flowers from one accession were emasculated at flower stage 12 (Smyth et al., 1990) and pollinated with pollen from a different accession at 1 DAE. Seeds were then harvested at 3 DAP.
RNA was isolated from carpels, siliques, or seeds in three biological replicates, each collected from multiple plants, using the hot-borate method (Wilkins and Smart, 1996), treated with TURBO DNase (Ambion), and purified with the RNeasy MinElute Cleanup Kit (Qiagen). RT was carried out with oligo(dT) primers using the RETROscript Kit (Ambion) following the manufacturer’s instructions. First-strand cDNA was treated with RNase H (New England Biolabs) and purified with the MinElute PCR Purification Kit (Qiagen).
RT-qPCR
RT-qPCR was performed using a LightCycler 1.5 instrument and the LightCycler FastStart DNA MasterPLUS SYBR Green I Kit (Roche) as described previously (Wang et al., 2010b). In brief, the PCR program consisted of a denaturing step at 95°C for 5 min followed by 45 cycles at 95°C for 10 s, 60°C for 5 s, and 72°C for 10 s. Each RT-qPCR run consisted of one pair (Col-0 and mea-3/+;swn-3/−) of reactions for ACTIN2 (ACT2) and 15 pairs of reactions for MADS-box genes. Primers used in RT-qPCR analysis are described in Supplemental Table S1. The CT values were calculated using the standard approach provided in the LightCycler software 4.0 package (Roche) and normalized as described previously (normalized CT, target = CT, target − CT, ACT2 +19; Wang et al., 2010b). The normalized CT value was cut off at greater than 35, which was set as an arbitrary threshold for negligible levels of mRNA. Differences in mRNA levels between Col-0 and the mea-3/+;swn-3/− double mutant were calculated using the −ΔΔCT method (Livak and Schmittgen, 2001), where ΔΔCT = normalized CT, wt − normalized CT, mutant. Calculation of the Pearson correlation coefficients and Student’s t test were performed using the corresponding functions in Excel (Microsoft).
Sixty-nine MADS-box genes were analyzed initially at 0, 3, and 5 DAP in Col-0 and the swn-3;mea-3 mutant. Sixteen AGLs with negligible mRNA level were not included in further analyses. The remaining 53 genes were analyzed with RT-qPCR using three biological replicates at 3 DAE and 0 to 5 DAP in Col-0 and the swn-3;mea-3 mutant (Supplemental Fig. S2). The RT-qPCR data were highly reproducible between biological replicates (r ∼ 0.875–0.99).
Reporter Constructs and Plant Transformation
Promoter regions containing 1- to 3-kb 5′ flanking sequences and the first one to seven codons were amplified with Phusion (Finnzymes) or ExTaq (Takara) DNA polymerases from Col-0 genomic DNA using the primers described in Supplemental Table S4, restriction digested, and cloned into pBI-GFP[S65T] (Yadegari et al., 2000), pBN-YFP/GFP (Wang et al., 2006), or pBI-n2GFP (nucleus-localized GFP; Wang et al., 2010b). Shorter 5′-flanking sequences were employed in cases where the upstream intergenic region was less than 1 kb. All promoter-vector junctions were verified by sequencing.
Full-length gene fusions were amplified using triple-template PCR with Phusion DNA polymerase (Finnzymes), restriction digested, and cloned into pBI101 (Jefferson et al., 1987). The procedure for triple-template PCR (Shevchuk et al., 2004) was as follows for each gene. Fragment 1 containing ∼3 kb of 5′-flanking sequences and the coding region without a translation stop codon was amplified from Col-0 genomic DNA using primers P1F and P1R, chimeric-YFP fragment 2 was amplified from the pBN-YFP vector using the chimeric primers P2F and P2R, and fragment 3 containing ∼3 kb of 3′-flanking sequences after the translation stop codon was amplified from Col-0 genomic DNA using primers P3F and P3R. The primary product was amplified from equal moles of the above three fragments using 11 cycles of PCR under low annealing temperature without primers. The final triple-template PCR product was amplified from the purified primary product using 30 cycles of PCR under high annealing temperature with nested primers P4F and P4R. Primer sequences are provided in Supplemental Table S6. All full-length gene-vector insertions were verified by sequencing.
Plant transformations were performed with the Arabidopsis accession Col-0 following the standard floral dip method using Agrobacterium tumefaciens strain GV3101 pMP90 or LBA4404 containing the binary vector as described previously (Wang et al., 2010b). The presence of the transgene in T1 plants was confirmed using PCR.
Promoter Reporter Gene and Full-Length Gene Fusion Expression Analysis
Ovules and seeds from transgenic plants carrying promoter or protein fusions were excised and imaged as described previously (Wang et al., 2006). GFP/YFP activity within the mature female gametophyte was analyzed at 1 DAE, while activity within the endosperm was initially analyzed in self-fertilized seeds. The expression of the transgene was determined in the T1 or T2 generation. The number of T1 lines analyzed per construct is provided in Supplemental Table S2.
Allele-specific expression within the endosperm was determined at 2 and 5 DAP in the wild-type, mea-3, and fis2-8 backgrounds. Allele-specific expression in the wild-type background was analyzed in reciprocal crosses between the reporter line and Col-0. Paternal allele expression in the mutant background was determined by crossing the mutant to the reporter line as the pollen donor, while maternal allele expression in the mutant background was determined by crossing the mutant carrying the transgene to Col-0 as the pollen donor.
Image acquisition and processing were performed as described previously (Wang et al., 2006). In brief, bright-field and epifluorescence images were captured with a MicroFire CCD camera (Optronics) attached to an Axiophot compound epifluorescence microscope (Carl Zeiss) equipped with an enhanced GFP bandpass filter (filter set 38 HE EGFP, exciter 450–490 nm, dichroic 495 nm, emitter 500–550 nm; Carl Zeiss) and a YFP bandpass filter (exciter 490–510, dichroic 515, emitter 520–550; Chroma Technology). Image processing was carried out using Photoshop and Illustrator CS (Adobe Systems).
Confocal Laser Scanning Microscopy
The swn-3/−;mea-3/+ double mutant plants were allowed to self-pollinate. The resulting siliques were fixed according to a published protocol (Christensen et al., 1997). Briefly, pistils were cut open along the replum and fixed for 2 h at room temperature in a fixative containing 4% (v/v) glutaraldehyde and 12.5 mm cacodylate (pH 6.9). Fixed siliques were dehydrated in an ethanol series (20% (v/v) steps for 10 min each). After dehydration, the siliques were cleared in a 2:1 mixture of benzyl benzoate and benzyl alcohol and mounted in immersion oil.
An MRC 1024 laser scanning confocal system (Bio-Rad) equipped with a 568-nm laser line and YHS filter block was used to illuminate glutaraldehyde-fixed seeds. Images were collected using LaserSharp2000 software (Bio-Rad).
Allele-Specific Expression Analysis Based on SNPs
SNPs between Col-0 and Cvi-0 or Col-0 and Ler were identified using the TAIR GBrowser (Berardini et al., 2015). Primers were designed to amplify each gene fragment from cDNA that contained at least one SNP between Col-0 and Cvi-0 or Col-0 and Ler (Supplemental Table S7). The amplicons from Cvi-0 and Ler genomic DNA were confirmed by sequencing the associated SNPs. When an insertion or deletion between accessions was found, a new SNP would be selected to exclude the insertion or deletion. Allele-specific expression was analyzed using single-base extension with fluorescently labeled ddNTPs (SBE) and further confirmed by CAPS.
SBE was generated using the ABI PRISM SNaPshot Multiplex Kit (Applied Biosystems) and analyzed with the ABI PRISM 3730 DNA analyzer at the University of Arizona Genetics Core facility. Primers used for SBE are described in Supplemental Table S7. Relative transcript levels for the two alleles were assessed by peak height, which was analyzed with STRand (Veterinary Genetics Laboratory). Since different ddNTPs with specific fluorescent labels give unequal fluorescence intensity, the estimation of allelic transcript frequency based directly on the peak height (H) of fluorescence intensity has a systematic bias and requires a calibration process (Shifman et al., 2002). Genomic DNA from Cvi-0 × Col-0 F1 hybrid seedlings is a natural pool with equal copies of DNA for the two alleles Col-0 and Cvi-0. Here, we used the observed fluorescence intensity ratio of two alleles from genomic DNA as an adjustment parameter to remove the systematic bias from different fluorescent labels for each gene with each SNP, 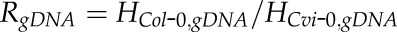 . The adjustment parameter was obtained from an average of two replicates (Supplemental Table S8). In the calculation of the allelic transcript frequency in reciprocal crosses between Col-0 and Cvi-0, the adjusted intensity for Cvi-0 was
. The adjustment parameter was obtained from an average of two replicates (Supplemental Table S8). In the calculation of the allelic transcript frequency in reciprocal crosses between Col-0 and Cvi-0, the adjusted intensity for Cvi-0 was 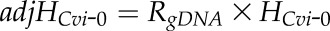 , and the estimated frequency of Col-0 intensity to the total intensity was
, and the estimated frequency of Col-0 intensity to the total intensity was 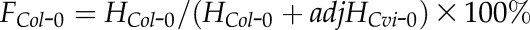 . A similar procedure was applied to calculate the allelic transcript frequency in reciprocal crosses between Col-0 and Ler (Supplemental Table S8). To assess the accuracy of allele frequency estimation, we constructed artificial DNA pools of PHE1 and AGL96 with different ratios of amplicons from Col-0 and Cvi-0 cDNA (in pools, the allele frequency of Col-0 was 10%, 25%, 50%, 75%, and 90%, while the corresponding allele frequency of Cvi-0 was 90%, 75%, 50%, 25%, and 10%). The measured allele frequency was not significantly different from the expected allele frequency (the paired Student’s t test P ∼ 0.721–0.806 > 0.05). We set the threshold for determining MEGs and PEGs at
. A similar procedure was applied to calculate the allelic transcript frequency in reciprocal crosses between Col-0 and Ler (Supplemental Table S8). To assess the accuracy of allele frequency estimation, we constructed artificial DNA pools of PHE1 and AGL96 with different ratios of amplicons from Col-0 and Cvi-0 cDNA (in pools, the allele frequency of Col-0 was 10%, 25%, 50%, 75%, and 90%, while the corresponding allele frequency of Cvi-0 was 90%, 75%, 50%, 25%, and 10%). The measured allele frequency was not significantly different from the expected allele frequency (the paired Student’s t test P ∼ 0.721–0.806 > 0.05). We set the threshold for determining MEGs and PEGs at 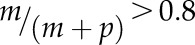 and
and 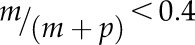 (where m denotes maternal transcripts and p denotes paternal transcripts measured by SBE; P < 0.05, Student’s t test) in both reciprocal crosses, as described previously (Hsieh et al., 2011). Analysis and graphing of the SBE data were carried out in R (Wickham, 2009; R Development Core Team, 2016).
(where m denotes maternal transcripts and p denotes paternal transcripts measured by SBE; P < 0.05, Student’s t test) in both reciprocal crosses, as described previously (Hsieh et al., 2011). Analysis and graphing of the SBE data were carried out in R (Wickham, 2009; R Development Core Team, 2016).
For CAPS, the products were first amplified from RT-PCR fragments using a two-step PCR that included regular PCR for 16 cycles followed by reconditional PCR (Thompson et al., 2002) with a 10-fold diluted regular PCR product as a template for three additional cycles to decrease heteroduplex formation. The products were digested subsequently with a restriction endonuclease that differentially cleaved Col-0 and Cvi-0 or Ler alleles (Supplemental Table S9), and the digested products were run through a 2% to 3% (w/v) MetaPhor agarose (Cambrex) gel. Primers used for CAPS are shown in Supplemental Table S9.
Literature-Curated Protein-Protein Interactions
A total of 111 pairs of literature-curated protein-protein interactions that exist among the 69 AGLs were extracted from the Plant Interactome Database (http://interactome.dfci.harvard.edu/A_thaliana/index.php; Arabidopsis Interactome Mapping Consortium, 2011). The majority of the interactions were based on a published yeast two-hybrid study for Arabidopsis MADS-box genes (de Folter et al., 2005). The interaction map was visualized using the organic layout option from Cytoscape version 3.6.0 (Shannon et al., 2003).
Accession Numbers
The accession numbers for all genes integral to this study are indicated in Supplemental Table S1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Seed phenotype in swn-3;mea-3 double mutants.
Supplemental Figure S2. Flow chart of the analyses carried out in this study.
Supplemental Figure S3. Individual mRNA profiles of MADS-box genes during early seed development from Col-0 and swn-3/−;mea-3/+ double-mutant plants.
Supplemental Figure S4. Expression of an additional set of C2.1 type I MADS-box gene reporter lines during endosperm development.
Supplemental Figure S5. Expression of an additional set of C2.2 type I MADS-box gene reporter lines during endosperm development.
Supplemental Figure S6. Expression of C1 and C2 gene reporter lines in the mature female gametophyte.
Supplemental Figure S7. Expression of C2 type I MADS-box gene reporter lines in unfertilized seeds from Col-0 and the mea-3 and fis2-8 mutants.
Supplemental Figure S8. Expression of C1 gene reporter lines during endosperm development.
Supplemental Figure S9. Global analyses of mRNA levels of MADS-box genes using the available expression data.
Supplemental Figure S10. Identification of allele-specific expression using.
Supplemental Figure S11. Expression analysis of the full-length gene and promoter fusions for AGL35, AGL36, and PHE1.
Supplemental Figure S12. Allele-specific expression of an additional set of C1 and C2 MADS-box genes in the endosperm.
Supplemental Figure S13. Models for regulation of C2 type I MADS-box genes by FIS-PRC2 during endosperm development.
Supplemental Figure S14. Total H3K27me3 profiles of C2 type I MADS-box genes in Col-0 × Ler and Ler × Col-0.
Supplemental Figure S15. Parental-specific H3K27me3 profiles of AGL23 and PHE1 in the Col-0 and Ler accessions.
Supplemental Figure S16. Allele-specific expression of the AGL23 protein reporter line in the endosperm.
Supplemental Figure S17. Protein-protein interaction map of the 69 AGLs.
Supplemental Table S1. RT-qPCR analysis of MADS-box genes.
Supplemental Table S2. Expression analysis of MADS-box genes.
Supplemental Table S3. Expression analysis of non-C2 MADS-box genes in ovules and seeds.
Supplemental Table S4. Construction of promoter and gene fusions for MADS-box genes.
Supplemental Table S5. Single-base extension analysis of C2 type I MADS-box genes.
Supplemental Table S6. Primers used in triple-template PCR for constructing the full-length gene fusions.
Supplemental Table S7. Single-base extension analysis-related information of C2 type I MADS-box genes.
Supplemental Table S8. Single-base extension data postcalibration.
Supplemental Table S9. Information on C2 type I MADS-box genes for cleaved amplified polymorphic sequence analysis.
Acknowledgments
We thank Adele Zhou for help with CAPS experiments.
Footnotes
This work was supported by National Science Foundation Grants IOS-0520008 (to K.S.S., G.N.D. and R.Y.) and IOS-0923880 (to G.N.D. and R.Y.).
Articles can be viewed without a subscription.
References
- Adams S, Vinkenoog R, Spielman M, Dickinson HG, Scott RJ (2000) Parent-of-origin effects on seed development in Arabidopsis thaliana require DNA methylation. Development 127: 2493–2502 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Wuillème S, Lemoine R, Kronenberger J, Caboche M, Lepiniec L, Rochat C (2005) The AtSUC5 sucrose transporter specifically expressed in the endosperm is involved in early seed development in Arabidopsis. Plant J 43: 824–836 [DOI] [PubMed] [Google Scholar]
- Belmonte MF, Kirkbride RC, Stone SL, Pelletier JM, Bui AQ, Yeung EC, Hashimoto M, Fei J, Harada CM, Munoz MD, et al. (2013) Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc Natl Acad Sci USA 110: E435–E444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemer M, Heijmans K, Airoldi C, Davies B, Angenent GC (2010) An atlas of type I MADS box gene expression during female gametophyte and seed development in Arabidopsis. Plant Physiol 154: 287–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemer M, Wolters-Arts M, Grossniklaus U, Angenent GC (2008) The MADS domain protein DIANA acts together with AGAMOUS-LIKE80 to specify the central cell in Arabidopsis ovules. Plant Cell 20: 2088–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini TZ, Reiser L, Li D, Mezheritsky Y, Muller R, Strait E, Huala E (2015) The Arabidopsis Information Resource: making and mining the “gold standard” annotated reference plant genome. Genesis 53: 474–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F, Chaudhury A (2009) Parental memories shape seeds. Trends Plant Sci 14: 550–556 [DOI] [PubMed] [Google Scholar]
- Berger F, Hamamura Y, Ingouff M, Higashiyama T (2008) Double fertilization: caught in the act. Trends Plant Sci 13: 437–443 [DOI] [PubMed] [Google Scholar]
- Boisnard-Lorig C, Colon-Carmona A, Bauch M, Hodge S, Doerner P, Bancharel E, Dumas C, Haseloff J, Berger F (2001) Dynamic analyses of the expression of the HISTONE:YFP fusion protein in Arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 13: 495–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE, Nguyen H (2003) Events during the first four rounds of mitosis establish three developmental domains in the syncytial endosperm of Arabidopsis thaliana. Protoplasma 222: 167–174 [DOI] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE, Nguyen H, Olsen OA (1999) Development of endosperm in Arabidopsis thaliana. Sex Plant Reprod 12: 32–42 [Google Scholar]
- Burkart-Waco D, Ngo K, Lieberman M, Comai L (2015) Perturbation of parentally biased gene expression during interspecific hybridization. PLoS ONE 10: e0117293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Chakravarthy H, Ormsbee BD, Mallanna SK, Rizzino A (2011) Rapid activation of the bivalent gene Sox21 requires displacement of multiple layers of gene-silencing machinery. FASEB J 25: 206–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury AM, Ming L, Miller C, Craig S, Dennis ES, Peacock WJ (1997) Fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA 94: 4223–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen CA, King EJ, Jordan JR, Drews GN (1997) Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex Plant Reprod 10: 49–64 [Google Scholar]
- Colombo M, Masiero S, Vanzulli S, Lardelli P, Kater MM, Colombo L (2008) AGL23, a type I MADS-box gene that controls female gametophyte and embryo development in Arabidopsis. Plant J 54: 1037–1048 [DOI] [PubMed] [Google Scholar]
- Costa LM, Marshall E, Tesfaye M, Silverstein KA, Mori M, Umetsu Y, Otterbach SL, Papareddy R, Dickinson HG, Boutiller K, et al. (2014) Central cell-derived peptides regulate early embryo patterning in flowering plants. Science 344: 168–172 [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185–196 [DOI] [PubMed] [Google Scholar]
- Davies B, Egea-Cortines M, de Andrade Silva E, Saedler H, Sommer H (1996) Multiple interactions amongst floral homeotic MADS box proteins. EMBO J 15: 4330–4343 [PMC free article] [PubMed] [Google Scholar]
- Day RC, Herridge RP, Ambrose BA, Macknight RC (2008) Transcriptome analysis of proliferating Arabidopsis endosperm reveals biological implications for the control of syncytial division, cytokinin signaling, and gene expression regulation. Plant Physiol 148: 1964–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Folter S, Immink RG, Kieffer M, Parenicová L, Henz SR, Weigel D, Busscher M, Kooiker M, Colombo L, Kater MM, et al. (2005) Comprehensive interaction map of the Arabidopsis MADS box transcription factors. Plant Cell 17: 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilkes BP, Comai L (2004) A differential dosage hypothesis for parental effects in seed development. Plant Cell 16: 3174–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreni L, Kater MM (2014) MADS reloaded: evolution of the AGAMOUS subfamily genes. New Phytol 201: 717–732 [DOI] [PubMed] [Google Scholar]
- Du M, Luo M, Zhang R, Finnegan EJ, Koltunow AM (2014) Imprinting in rice: the role of DNA and histone methylation in modulating parent-of-origin specific expression and determining transcript start sites. Plant J 79: 232–242 [DOI] [PubMed] [Google Scholar]
- Egea-Cortines M, Saedler H, Sommer H (1999) Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J 18: 5370–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Berger F (2007) Convergent evolution of genomic imprinting in plants and mammals. Trends Genet 23: 192–199 [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Bond DM, Buzas DM, Goodrich J, Helliwell CA, Tamada Y, Yun JY, Amasino RM, Dennis ES (2011) Polycomb proteins regulate the quantitative induction of VERNALIZATION INSENSITIVE 3 in response to low temperatures. Plant J 65: 382–391 [DOI] [PubMed] [Google Scholar]
- Fitz Gerald JN, Hui PS, Berger F (2009) Polycomb group-dependent imprinting of the actin regulator AtFH5 regulates morphogenesis in Arabidopsis thaliana. Development 136: 3399–3404 [DOI] [PubMed] [Google Scholar]
- Garcia D, Saingery V, Chambrier P, Mayer U, Jürgens G, Berger F (2003) Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol 131: 1661–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL (2006) DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124: 495–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Missirian V, Henikoff S (2011) Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds. PLoS ONE 6: e23687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Satyaki PR (2017) Endosperm and imprinting, inextricably linked. Plant Physiol 173: 143–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramzow L, Theissen G (2010) A hitchhiker’s guide to the MADS world of plants. Genome Biol 11: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB (1998) Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280: 446–450 [DOI] [PubMed] [Google Scholar]
- Hatorangan MR, Laenen B, Steige KA, Slotte T, Köhler C (2016) Rapid evolution of genomic imprinting in two species of the Brassicaceae. Plant Cell 28: 1815–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehenberger E, Kradolfer D, Köhler C (2012) Endosperm cellularization defines an important developmental transition for embryo development. Development 139: 2031–2039 [DOI] [PubMed] [Google Scholar]
- Honma T, Goto K (2001) Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 525–529 [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TF, Shin J, Uzawa R, Silva P, Cohen S, Bauer MJ, Hashimoto M, Kirkbride RC, Harada JJ, Zilberman D, et al. (2011) Regulation of imprinted gene expression in Arabidopsis endosperm. Proc Natl Acad Sci USA 108: 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Zhu QH, Zhu A, Wu X, Xie L, Wu X, Helliwell C, Chaudhury A, Finnegan EJ, Luo M (2017) Mutants in the imprinted PICKLE RELATED 2 gene suppress seed abortion of fertilization independent seed class mutants and paternal excess interploidy crosses in Arabidopsis. Plant J 90: 383–395 [DOI] [PubMed] [Google Scholar]
- Ingram GC. (2010) Family life at close quarters: communication and constraint in angiosperm seed development. Protoplasma 247: 195–214 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsson C, Dilkes B, Comai L (2006) Parent-dependent loss of gene silencing during interspecies hybridization. Curr Biol 16: 1322–1328 [DOI] [PubMed] [Google Scholar]
- Jullien PE, Kinoshita T, Ohad N, Berger F (2006) Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting. Plant Cell 18: 1360–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien PE, Susaki D, Yelagandula R, Higashiyama T, Berger F (2012) DNA methylation dynamics during sexual reproduction in Arabidopsis thaliana. Curr Biol 22: 1825–1830 [DOI] [PubMed] [Google Scholar]
- Kang IH, Steffen JG, Portereiko MF, Lloyd A, Drews GN (2008) The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20: 635–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, Jacobsen SE, Fischer RL, Kakutani T (2004) One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303: 521–523 [DOI] [PubMed] [Google Scholar]
- Klosinska M, Picard CL, Gehring M (2016) Conserved imprinting associated with unique epigenetic signatures in the Arabidopsis genus. Nat Plants 2: 16145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C, Hennig L, Bouveret R, Gheyselinck J, Grossniklaus U, Gruissem W (2003a) Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J 22: 4804–4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C, Hennig L, Spillane C, Pien S, Gruissem W, Grossniklaus U (2003b) The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev 17: 1540–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C, Page DR, Gagliardini V, Grossniklaus U (2005) The Arabidopsis thaliana MEDEA Polycomb group protein controls expression of PHERES1 by parental imprinting. Nat Genet 37: 28–30 [DOI] [PubMed] [Google Scholar]
- Kondou Y, Nakazawa M, Kawashima M, Ichikawa T, Yoshizumi T, Suzuki K, Ishikawa A, Koshi T, Matsui R, Muto S, et al. (2008) RETARDED GROWTH OF EMBRYO1, a new basic helix-loop-helix protein, expresses in endosperm to control embryo growth. Plant Physiol 147: 1924–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kradolfer D, Hennig L, Köhler C (2013a) Increased maternal genome dosage bypasses the requirement of the FIS polycomb repressive complex 2 in Arabidopsis seed development. PLoS Genet 9: e1003163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kradolfer D, Wolff P, Jiang H, Siretskiy A, Köhler C (2013b) An imprinted gene underlies postzygotic reproductive isolation in Arabidopsis thaliana. Dev Cell 26: 525–535 [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. (2008) Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon-Placette C, Köhler C (2014) Embryo and endosperm, partners in seed development. Curr Opin Plant Biol 17: 64–69 [DOI] [PubMed] [Google Scholar]
- Lafon-Placette C, Köhler C (2016) Endosperm-based postzygotic hybridization barriers: developmental mechanisms and evolutionary drivers. Mol Ecol 25: 2620–2629 [DOI] [PubMed] [Google Scholar]
- Le BH, Cheng C, Bui AQ, Wagmaister JA, Henry KF, Pelletier J, Kwong L, Belmonte M, Kirkbride R, Horvath S, et al. (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA 107: 8063–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Berger F (2012) Endosperm: food for humankind and fodder for scientific discoveries. New Phytol 195: 290–305 [DOI] [PubMed] [Google Scholar]
- Lin BY. (1984) Ploidy barrier to endosperm development in maize. Genetics 107: 103–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Jung C, Xu J, Wang H, Deng S, Bernad L, Arenas-Huertero C, Chua NH (2012) Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24: 4333–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lopes MA, Larkins BA (1993) Endosperm origin, development, and function. Plant Cell 5: 1383–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Zhang C, Baulcombe DC, Chen ZJ (2012) Maternal siRNAs as regulators of parental genome imbalance and gene expression in endosperm of Arabidopsis seeds. Proc Natl Acad Sci USA 109: 5529–5534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Dennis ES, Peacock WJ, Chaudhury A (2000) Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc Natl Acad Sci USA 97: 10637–10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Koltunow A, Dennis ES, Peacock WJ, Chaudhury AM (1999) Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA 96: 296–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Dennis ES, Berger F, Peacock WJ, Chaudhury A (2005) MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc Natl Acad Sci USA 102: 17531–17536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Taylor JM, Spriggs A, Zhang H, Wu X, Russell S, Singh M, Koltunow A (2011) A genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm. PLoS Genet 7: e1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevich G, Villar CB, Erilova A, Köhler C (2008) Mechanism of PHERES1 imprinting in Arabidopsis. J Cell Sci 121: 906–912 [DOI] [PubMed] [Google Scholar]
- Moreno-Romero J, Jiang H, Santos-González J, Köhler C (2016) Parental epigenetic asymmetry of PRC2-mediated histone modifications in the Arabidopsis endosperm. EMBO J 35: 1298–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozgova I, Hennig L (2015) The polycomb group protein regulatory network. Annu Rev Plant Biol 66: 269–296 [DOI] [PubMed] [Google Scholar]
- Mozzetta C, Consalvi S, Saccone V, Forcales SV, Puri PL, Palacios D (2011) Selective control of Pax7 expression by TNF-activated p38α/polycomb repressive complex 2 (PRC2) signaling during muscle satellite cell differentiation. Cell Cycle 10: 191–198 [DOI] [PubMed] [Google Scholar]
- Ohad N, Margossian L, Hsu YC, Williams C, Repetti P, Fischer RL (1996) A mutation that allows endosperm development without fertilization. Proc Natl Acad Sci USA 93: 5319–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad N, Yadegari R, Margossian L, Hannon M, Michaeli D, Harada JJ, Goldberg RB, Fischer RL (1999) Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11: 407–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen OA. (2004) Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell (Suppl) 16: S214–S227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenicová L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, et al. (2003) Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15: 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatta D, Erdmann RM, Scheer E, Picard CL, Bell GW, Gehring M (2014) Natural epigenetic polymorphisms lead to intraspecific variation in Arabidopsis gene imprinting. eLife 3: e03198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portereiko MF, Lloyd A, Steffen JG, Punwani JA, Otsuga D, Drews GN (2006) AGL80 is required for central cell and endosperm development in Arabidopsis. Plant Cell 18: 1862–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu L, Sung ZR (2015) PcG and trxG in plants: friends or foes. Trends Genet 31: 252–262 [DOI] [PubMed] [Google Scholar]
- Raissig MT, Baroux C, Grossniklaus U (2011) Regulation and flexibility of genomic imprinting during seed development. Plant Cell 23: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RJ, Spielman M, Bailey J, Dickinson HG (1998) Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125: 3329–3341 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchuk NA, Bryksin AV, Nusinovich YA, Cabello FC, Sutherland M, Ladisch S (2004) Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res 32: e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifman S, Pisanté-Shalom A, Yakir B, Darvasi A (2002) Quantitative technologies for allele frequency estimation of SNPs in DNA pools. Mol Cell Probes 16: 429–434 [DOI] [PubMed] [Google Scholar]
- Shirzadi R, Andersen ED, Bjerkan KN, Gloeckle BM, Heese M, Ungru A, Winge P, Koncz C, Aalen RB, Schnittger A, et al. (2011) Genome-wide transcript profiling of endosperm without paternal contribution identifies parent-of-origin-dependent regulation of AGAMOUS-LIKE36. PLoS Genet 7: e1001303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczniak C, Immink RG, Angenent GC, Kaufmann K (2012) Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 139: 3081–3098 [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen MB, Mayer U, Lukowitz W, Robert H, Chambrier P, Jürgens G, Somerville C, Lepiniec L, Berger F (2002) Cellularisation in the endosperm of Arabidopsis thaliana is coupled to mitosis and shares multiple components with cytokinesis. Development 129: 5567–5576 [DOI] [PubMed] [Google Scholar]
- Steffen JG, Kang IH, Portereiko MF, Lloyd A, Drews GN (2008) AGL61 interacts with AGL80 and is required for central cell development in Arabidopsis. Plant Physiol 148: 259–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Onouchi H, Kondo M, Hara-Nishimura I, Nishimura M, Machida C, Machida Y (2001) A subtilisin-like serine protease is required for epidermal surface formation in Arabidopsis embryos and juvenile plants. Development 128: 4681–4689 [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2016) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Thompson JR, Marcelino LA, Polz MF (2002) Heteroduplexes in mixed-template amplifications: formation, consequence and elimination by ‘reconditioning PCR’. Nucleic Acids Res 30: 2083–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S, Schulz R, Ikeda Y, Dytham L, Bravo J, Mathers L, Spielman M, Guzmán P, Oakey RJ, Kinoshita T, et al. (2008) MATERNALLY EXPRESSED PAB C-TERMINAL, a novel imprinted gene in Arabidopsis, encodes the conserved C-terminal domain of polyadenylate binding proteins. Plant Cell 20: 2387–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S, Spielman M, Schulz R, Oakey RJ, Kelsey G, Salazar A, Zhang K, Pennell R, Scott RJ (2010) Transcriptional profiles underlying parent-of-origin effects in seeds of Arabidopsis thaliana. BMC Plant Biol 10: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar CB, Erilova A, Makarevich G, Trösch R, Köhler C (2009) Control of PHERES1 imprinting in Arabidopsis by direct tandem repeats. Mol Plant 2: 654–660 [DOI] [PubMed] [Google Scholar]
- Vu TM, Nakamura M, Calarco JP, Susaki D, Lim PQ, Kinoshita T, Higashiyama T, Martienssen RA, Berger F (2013) RNA-directed DNA methylation regulates parental genomic imprinting at several loci in Arabidopsis. Development 140: 2953–2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia H, Josefsson C, Dilkes B, Kirkbride R, Harada J, Comai L (2009) Dosage-dependent deregulation of an AGAMOUS-LIKE gene cluster contributes to interspecific incompatibility. Curr Biol 19: 1128–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Garcia D, Zhang H, Feng K, Chaudhury A, Berger F, Peacock WJ, Dennis ES, Luo M (2010a) The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J 63: 670–679 [DOI] [PubMed] [Google Scholar]
- Wang D, Tyson MD, Jackson SS, Yadegari R (2006) Partially redundant functions of two SET-domain polycomb-group proteins in controlling initiation of seed development in Arabidopsis. Proc Natl Acad Sci USA 103: 13244–13249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zhang C, Hearn DJ, Kang IH, Punwani JA, Skaggs MI, Drews GN, Schumaker KS, Yadegari R (2010b) Identification of transcription-factor genes expressed in the Arabidopsis female gametophyte. BMC Plant Biol 10: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters AJ, Bilinski P, Eichten SR, Vaughn MW, Ross-Ibarra J, Gehring M, Springer NM (2013) Comprehensive analysis of imprinted genes in maize reveals allelic variation for imprinting and limited conservation with other species. Proc Natl Acad Sci USA 110: 19639–19644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters AJ, Makarevitch I, Eichten SR, Swanson-Wagner RA, Yeh CT, Xu W, Schnable PS, Vaughn MW, Gehring M, Springer NM (2011) Parent-of-origin effects on gene expression and DNA methylation in the maize endosperm. Plant Cell 23: 4221–4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2009) ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag, New York [Google Scholar]
- Wilkins TA, Smart LB (1996) Isolation of RNA from plant tissue. In Krieg PA, ed, A Laboratory Guide to RNA: Isolation, Analysis, and Synthesis. Wiley-Liss, New York, pp 21–41 [Google Scholar]
- Wolff P, Jiang H, Wang G, Santos-González J, Köhler C (2015) Paternally expressed imprinted genes establish postzygotic hybridization barriers in Arabidopsis thaliana. eLife 4: e10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff P, Weinhofer I, Seguin J, Roszak P, Beisel C, Donoghue MT, Spillane C, Nordborg M, Rehmsmeier M, Köhler C (2011) High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm. PLoS Genet 7: e1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Yang R, Li G, Chen H, Laurie J, Ma C, Wang D, Yao Y, Larkins BA, Sun Q, et al. (2013) Dynamic expression of imprinted genes associates with maternally controlled nutrient allocation during maize endosperm development. Plant Cell 25: 3212–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Q, Creff A, Waters A, Tanaka H, Goodrich J, Ingram GC (2013) ZHOUPI controls embryonic cuticle formation via a signalling pathway involving the subtilisin protease ABNORMAL LEAF-SHAPE1 and the receptor kinases GASSHO1 and GASSHO2. Development 140: 770–779 [DOI] [PubMed] [Google Scholar]
- Xu W, Dai M, Li F, Liu A (2014) Genomic imprinting, methylation and parent-of-origin effects in reciprocal hybrid endosperm of castor bean. Nucleic Acids Res 42: 6987–6998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadegari R, Drews GN (2004) Female gametophyte development. Plant Cell (Suppl) 16: S133–S141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadegari R, Kinoshita T, Lotan O, Cohen G, Katz A, Choi Y, Katz A, Nakashima K, Harada JJ, Goldberg RB, et al. (2000) Mutations in the FIE and MEA genes that encode interacting polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell 12: 2367–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Johnston N, Talideh E, Mitchell S, Jeffree C, Goodrich J, Ingram G (2008) The endosperm-specific ZHOUPI gene of Arabidopsis thaliana regulates endosperm breakdown and embryonic epidermal development. Development 135: 3501–3509 [DOI] [PubMed] [Google Scholar]
- Zhang M, Li N, He W, Zhang H, Yang W, Liu B (2016) Genome-wide screen of genes imprinted in sorghum endosperm, and the roles of allelic differential cytosine methylation. Plant J 85: 424–436 [DOI] [PubMed] [Google Scholar]
- Zhang M, Zhao H, Xie S, Chen J, Xu Y, Wang K, Zhao H, Guan H, Hu X, Jiao Y, et al. (2011) Extensive, clustered parental imprinting of protein-coding and noncoding RNAs in developing maize endosperm. Proc Natl Acad Sci USA 108: 20042–20047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhang X, Kang X, Zhao X, Zhang X, Ni M (2009) SHORT HYPOCOTYL UNDER BLUE1 associates with MINISEED3 and HAIKU2 promoters in vivo to regulate Arabidopsis seed development. Plant Cell 21: 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]