Mutagenesis reveals which CESA protein classes have the greatest influence on the regulation of cellulose microfibril biosynthesis in the Arabidopsis secondary cell wall.
Abstract
Cellulose microfibrils are the basic units of cellulose in plants. The structure of these microfibrils is at least partly determined by the structure of the cellulose synthase complex. In higher plants, this complex is composed of 18 to 24 catalytic subunits known as CELLULOSE SYNTHASE A (CESA) proteins. Three different classes of CESA proteins are required for cellulose synthesis and for secondary cell wall cellulose biosynthesis these classes are represented by CESA4, CESA7, and CESA8. To probe the relationship between CESA proteins and microfibril structure, we created mutant cesa proteins that lack catalytic activity but retain sufficient structural integrity to allow assembly of the cellulose synthase complex. Using a series of Arabidopsis (Arabidopsis thaliana) mutants and genetic backgrounds, we found consistent differences in the ability of these mutant cesa proteins to complement the cellulose-deficient phenotype of the cesa null mutants. The best complementation was observed with catalytically inactive cesa4, while the equivalent mutation in cesa8 exhibited significantly lower levels of complementation. Using a variety of biophysical techniques, including solid-state nuclear magnetic resonance and Fourier transform infrared microscopy, to study these mutant plants, we found evidence for changes in cellulose microfibril structure, but these changes largely correlated with cellulose content and reflected differences in the relative proportions of primary and secondary cell walls. Our results suggest that individual CESA classes have similar roles in determining cellulose microfibril structure, and it is likely that the different effects of mutating members of different CESA classes are the consequence of their different catalytic activity and their influence on the overall rate of cellulose synthesis.
Plant cells are surrounded by a multilayered cell wall. This composite structure is composed of a primary cell wall that undergoes significant remodeling during cell expansion. After reaching their final size and shape, many cell types deposit the secondary cell wall, which is considerably thicker and frequently more rigid. Consequently, the secondary cell wall provides structural support for the plant and also constitutes the majority of plant biomass (Burton et al., 2010). Cellulose makes up the largest component of the secondary cell wall that also frequently consists of lignin and hemicellulose, making cellulose, xylan, and lignin the three most abundant natural biopolymers on Earth. Increasing interest in the use of plant biomass as renewable feedstocks for the production of biofuels and other specialty chemicals and biomaterials has provided the impetus for understanding the synthesis and breakdown of these polymers (Pauly and Keegstra, 2008; Carroll and Somerville, 2009; Kumar et al., 2016).
Cellulose is composed of linear chains of β-1,4-linked Glc units. These chains align to form cellulose microfibrils, the basic structure of cellulose found in the cell wall. In vascular plants, current models of cellulose microfibrils estimate that there are 18 to 24 cellulose chains in each microfibril (Fernandes et al., 2011; Newman et al., 2013; Thomas et al., 2013; Wang and Hong, 2016). Cellulose is synthesized at the plasma membrane by the cellulose synthase complex (CSC). The CSC can be visualized by electron microscopy and forms a rosette structure with 6-fold symmetry (Mueller and Brown, 1980; Kimura et al., 1999). Although the rosette structure appears to be conserved among the green plants, there is variation in the size of the cellulose synthesized, and the microfibrils deposited during primary wall synthesis generally are smaller than those in the secondary cell wall (Cosgrove and Jarvis, 2012). These differences in size have been proposed to result from the products of more than one CSC aggregating and being incorporated into a single microfibril (Salmén, 2004; Li et al., 2016).
The catalytic components of the CSC are the CESA proteins, although a number of other proteins have been shown to be associated with the complex (Gu et al., 2010; Vain et al., 2014; Endler et al., 2015; Liu et al., 2016). An analysis of higher plant CESA protein sequences suggests that they can be categorized into six main classes (Carroll and Specht, 2011; Kumar and Turner, 2015b). Three of these classes, CESA1, CESA3, and CESA6, are responsible for producing cellulose in the primary cell walls (Desprez et al., 2007; Persson et al., 2007), while the other three classes, CESA4, CESA7, and CESA8, make cellulose in secondary cell walls (Taylor et al., 2003). However, a recent analysis of CESA proteins from the moss Physcomitrella patens suggests that separate CESAs are required for both primary and secondary cell walls, but these CESA proteins do not form separate classes and it seems likely that a single CESA class is sufficient to make cellulose (Norris et al., 2017). This implies that the requirement for multiple CESA proteins in higher plants is not essential for cellulose synthesis per se but may be a means of fine-tuning the regulation of cellulose synthesis in response to various external cues. Several studies have shown that phosphorylation is important for regulating CESA catalytic activity, movement, and response to external signals and that the modification of a single class of CESAs can affect the rates of cellulose synthesis (Chen et al., 2010; Sánchez-Rodríguez et al., 2017). The influence of a single CESA class on the activity of the entire CSC has been illustrated by a recent study that overexpressed different members of the CESA6 class (Hu et al., 2017), which resulted in significant changes in cell shape, cellulose content, and cell wall thickness.
Higher plants use three classes of CESAs, and each CESA appears to occupy a unique position within the rosette (Kumar et al., 2017); consequently, there is an opportunity to probe the relationship between CESA subunit position within the CSC and cellulose microfibril ultrastructure. Mutants defective in cellulose deposition in the secondary cell wall exhibit an irregular xylem (irx) phenotype (Turner and Somerville, 1997). When first described, it was noted that CESA8irx1-1 contains significantly less cellulose than the wild type but significantly more than knockout mutants in other CESA proteins, such as cesa7irx3-1. Subsequent analysis has shown that the mutation in CESA8irx1-1 is a point mutation that alters Asp-683 of CESA8 to Asn (Taylor et al., 2000). Comparison of the sequence of CESA8 with other characterized glycosyltransferase family 2 members, and particularly the crystal structure of the cellulose synthase gene from Rhodobacter sphaeroides bacterial cellulose synthase A (BCSA; Morgan et al., 2013; Kumar and Turner, 2015b), indicates that Asp-683 forms part of the invariant TED motif found in the catalytic site, with the Asp residue likely to be the catalytic base, making it essential for catalysis (Morgan et al., 2013). Consequently, mutating this Asp may largely retain the structure of the CESA protein, but it is unlikely to be able to catalyze the formation of cellulose. The assumption is that, since an individual complex involved in secondary cell wall synthesis is made of three different CESA proteins (CESA8, CESA7, and CESA4), the residual cellulose in the walls of the cesa8irx1-1 mutant plants results from the catalytic activity of CESA4 and CESA7 (Turner and Somerville, 1997; Taylor et al., 2003). This result provides an opportunity to examine whether all CESA proteins within the CSC are equivalent or whether the cesa8irx1-1 mutant revealed something particular to only CESA8. Available data suggest that the stoichiometry of CESA4, CESA7, and CESA8 is 1:1:1 (Hill et al., 2014); however, if they occupy different positions within the rosette, then mutating individual CESAs may have a different effect on cellulose synthesis.
In this study, equivalent Asp-to-Asn mutations were introduced into putative catalytic motifs of Arabidopsis (Arabidopsis thaliana) CESA4, CESA7, and CESA8 and then transformed back into the corresponding cesa null mutant. These mutations had different effects on both cellulose content and plant growth. Mutations in CESA4 had the smallest effect on cellulose content, while mutations in CESA8 had the largest effect. Furthermore, we found that, although equivalent mutations in CESA7 and CESA4 exhibited only small differences in cellulose content, the CESA4 mutants are able to grow healthier and more closely resemble wild-type plants. Solid-state NMR (ssNMR) and Fourier transform infrared (FTIR) microspectroscopy were used to examine the structure of the cellulose in these mutants in order to improve our understanding of the relationship between the CSC and cellulose microfibril production in the secondary cell wall.
RESULTS
Mutations in the TED, DXD, and DDX Motifs of Secondary Wall CESAs
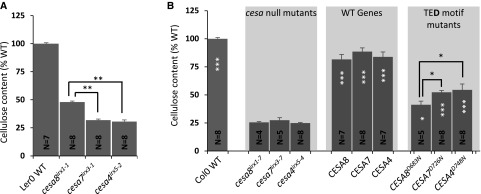
We analyzed the cellulose content of plants containing single point mutations in the three CESA genes essential for secondary cell wall-specific cellulose synthesis: cesa8irx1-1 (D683N), cesa7irx3-1 (W859STOP), and cesa4irx5-2 (W995STOP). Consistent with previous observations (Turner and Somerville, 1997), the cellulose content of cesa8irx1-1 plants was found to be significantly higher than that of cesa7irx3-1 and cesa4irx5-2 plants (Fig. 1A). We then made the cesa8irx1-1 equivalent TED motif mutants in CESA8, CESA7, and CESA4 (Supplemental Table S1), transformed them into the corresponding null mutant, and measured the stem cellulose content. Columbia-0 (Col-0) wild-type plants had a cellulose content of about 33% of alcohol-insoluble residue (AIR), while the cesa T-DNA mutants contained about 9% cellulose in their cell walls. To allow for easy comparisons between different experiments, all cellulose content data throughout this article are expressed as percentages of wild-type data, which results in the cesa T-DNA mutants exhibiting about 25% of the wild-type cellulose content. The TED motif mutants of CESA8 (CESA8D683N), CESA7 (CESA7D726N), and CESA4 (CESA4D748N) all had significantly higher cellulose contents than their background cesa T-DNA mutants, exhibiting cellulose contents of 41%, 52%, and 54% of wild-type values, respectively (Fig. 1B). The CESA8D683N mutant that would be equivalent to cesa8irx1-1 actually showed the poorest complementation, while those transformed with CESA4D748N exhibited the highest complementation (Fig. 1B).
Figure 1.
Cellulose content of the plants with point mutations in the secondary wall CESA genes. A, Data for three cesa mutants, cesa8irx1-1 (D683N), cesa7irx3-1 (W859STOP), and cesa4irx5-2 (W995STOP). B, Secondary wall CESA T-DNA null mutants (cesa8irx1-7, cesa7irx3-7, and cesa4irx5-4) were transformed with wild-type (WT) genes (CESA8, CESA7, and CESA4, respectively) or TED motif Asp-to-Asn mutants (CESA8D683N, CESA7D726N, and CESA4D748N, respectively). Measurements were taken from single plants at the T1 stage. N refers to the number of individual transformants analyzed. Cellulose content is expressed as a percentage of the wild-type value. Error bars indicate se. Significance levels from univariate ANOVA are indicated for comparison between the genotype and the background mutant (white asterisks inside each bar) or between particular genotypes (black asterisks and lines above the bars): ***, significant at 0.001; **, significant at 0.01; and *, significant at 0.05.
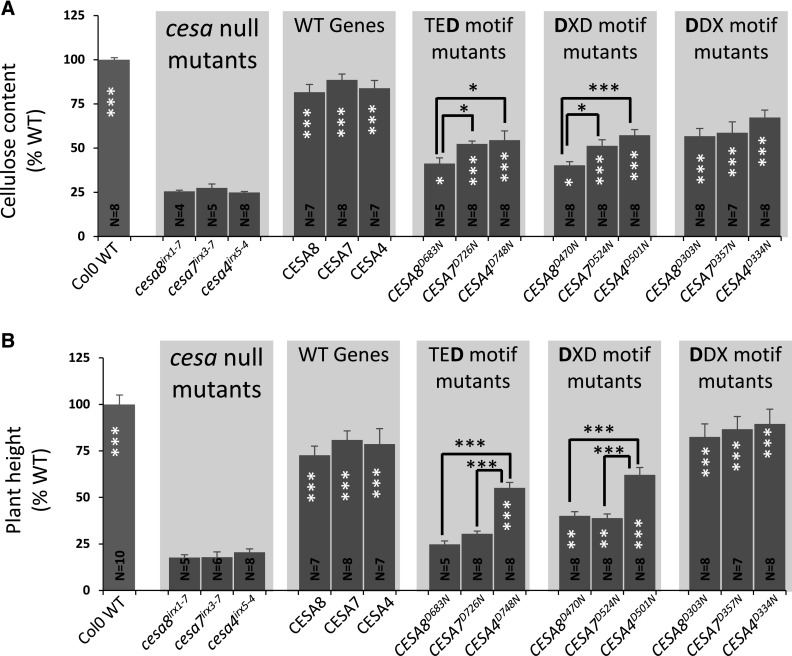
It is possible that differences in the level of complementation for the CESA8/7/4 TED mutants result from variation in the effects of these mutations on the catalytic activity of the CSC. Consequently, we made two further series of mutations in two other highly conserved motifs that also are believed to be essential for CESA activity. We mutated the first Asp of the DXD motif that is required for binding of the UDP-Glc substrate and the highly conserve DDX motif that also is found in the catalytic site (Morgan et al., 2013; Supplemental Table S1). Throughout this article, these mutations are referred to as TED, DXD, and DDX, with the mutated Asp highlighted in boldface. These Asp residues were mutated to Asn, and the mutated genes were placed under the control of their native promoters and transformed into the corresponding null cesa T-DNA mutants. All these mutants had significant increases in cellulose content compared with their background T-DNA mutants (Fig. 2A). The cellulose content of the DXD motif mutants was very similar to that of the TED motif mutants and constituted 40%, 51%, and 57% of wild-type values for CESA8, CESA7, and CESA4, respectively (Fig. 2A). Again, we found that there was significantly more cellulose in the CESA4 mutant than in the CESA8 mutant, as we had observed previously for the TED mutants. By contrast, we found that the DDX mutants had a much smaller effect on cellulose content, with cellulose contents of 57%, 59%, and 67% of wild-type values for CESA8, CESA7, and CESA4, respectively (Fig. 2A).
Figure 2.
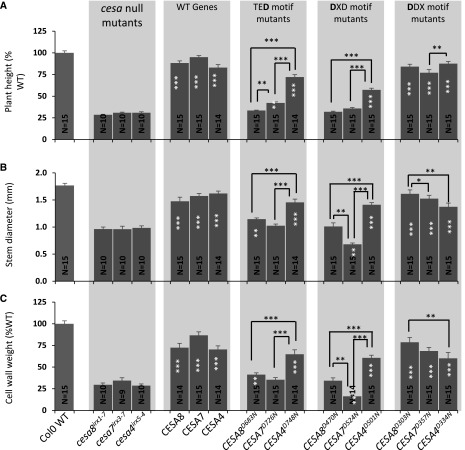
Cellulose content of the plants containing Asp-to-Asn mutation in the TED, DXD, and DDX motifs of secondary wall CESAs. A, Cellulose content. B, Plant height. CESA T-DNA null mutants (cesa8irx1-7, cesa7irx3-7, and cesa4irx5-4) were transformed with wild-type (WT) genes (CESA8, CESA7, and CESA4, respectively), TED motif Asp-to-Asn mutants (CESA8D683N, CESA7D726N, and CESA4D748N, respectively), DXD motif Asp-to-Asn mutants (CESA8D470N, CESA7D524N, and CESA4D501N, respectively), or DDX motif Asp-to-Asn mutants (CESA8D303N, CESA7D357N, and CESA4D334N, respectively). N refers to the number of individual T1 transformants analyzed. Cellulose content and plant height are expressed as percentages of the wild-type values. The cellulose data for the TED motif are the same as in Figure 1B. Error bars indicate se. Significance levels from univariate ANOVA are indicated for comparison between the genotype and the background mutant (white asterisks inside each bar) or between particular genotypes (black asterisks and lines above the bars): ***, significant at 0.001; **, significant at 0.01; and *, significant at 0.05.
We analyzed up to six independent lines for the cesa T-DNA mutants transformed with wild-type CESAs and TED/DXD/DDX motif mutants for protein expression using qualitative western blotting. The analysis showed that the CESA proteins could be detected in all lines (Supplemental Fig. S1). Although the level of expression was variable, the variation in expression levels exhibited no obvious correlation with the level of complementation for plant height or cellulose content.
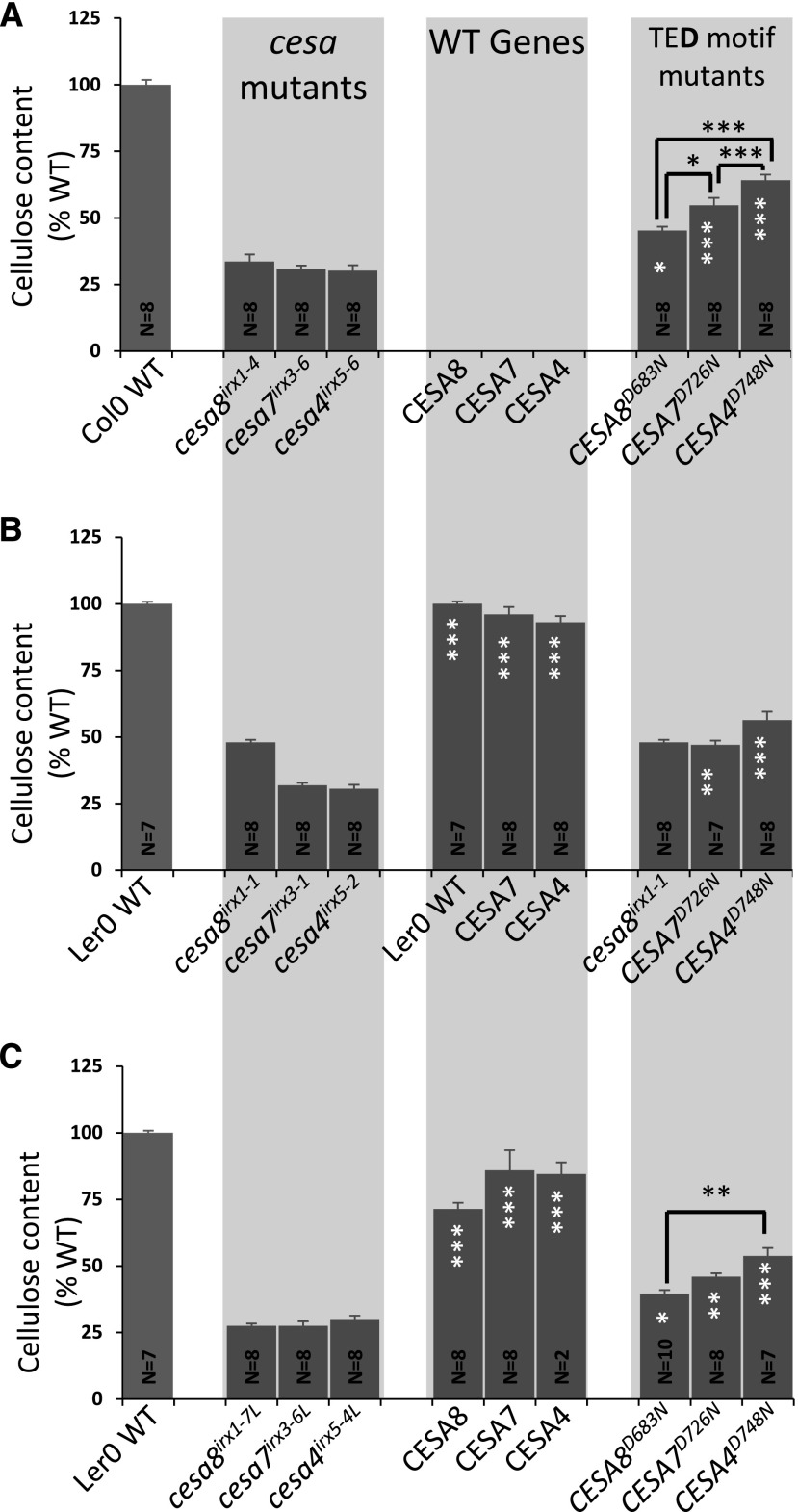
Variation in the Effects of Mutating Individual CESA Proteins Is Independent of the Genetic Background
The original cesa8irx1-1 allele was isolated in the Landsberg erecta (Ler) ecotype, and we consistently observed that mutants in the Ler background grew better than equivalent mutants in the Col-0 background. Furthermore, cesa8irx1-1 appeared to have slightly more cellulose (48%) than the CESA8D683N mutant transformed into cesa8irx1-7, a T-DNA allele in the Col-0 ecotype (41%; Fig. 1). To test whether some of the differences observed may vary between backgrounds or alleles, we transformed the TED motif Asp mutants into three different sets of alternative mutant backgrounds (Fig. 3). We first used an alternative set of T-DNA alleles in the Col-0 background. These three mutants, cesa8irx1-4, cesa7irx3-6, and cesa4irx5-6, all contain T-DNA insertions in exons of the genes and are likely to be complete knockouts. The second alternative set of background mutants consisted of cesa7irx3-1 and cesa4irx5-2, which are both nonsense mutations in the Ler background. To complete this set, we used cesa8irx1-1 and compared it with the wild type. Finally, as there were no T-DNA insertions in CESA genes in the Landsberg ecotype available, cesa8irx1-7, cesa7irx3-6, and cesa4irx5-4 mutants were backcrossed with the Ler wild type for five generations, selecting for the characteristic Ler morphology while retaining the T-DNA insertions. For all three sets of alternative mutant alleles, we obtained a pattern of complementation similar to that observed for the original T-DNA mutant alleles (Fig. 3). Thus, all three sets of the TED Asp mutants had significantly more cellulose than the background mutants (cesa8irx1-4, cesa7irx3-6, and cesa4irx5-6; cesa7irx3-1 and cesa4irx5- 2; and cesa8irx1-7L, cesa7irx3-6L, and cesa4irx5-4L), and the order of complementation was CESA8 < CESA7 < CESA4, with the plants transformed with catalytically inactive CESA8 mutants having significantly less cellulose than those transformed with corresponding CESA4 mutants. Apart from the Ler cesa8irx1-1 allele, which had similar levels of cellulose to CESA7D726N (transformed into cesa7irx3-1; Fig. 3B), the results are consistent between different alleles and genetic backgrounds.
Figure 3.
Cellulose content of the plants containing an Asp-to-Asn mutation in the TED motif of secondary wall CESAs in alternative cesa backgrounds. A, Secondary wall CESA T-DNA null mutants (cesa8irx1-4, cesa7irx3-6, and cesa4irx5-6) were transformed with TED Asp-to-Asn mutants (CESA8D683N, CESA7D726N, and CESA4D748N, respectively). Transformation of wild-type (WT) genes was not performed for this set of mutants. B, cesa7irx3-1 and cesa4irx5-2 were transformed with either the wild-type genes (CESA7 and CESA4, respectively) or TED Asp-to-Asn mutants (CESA7D726N and CESA4D748N, respectively) and compared with the cesa8irx1-1 mutant. For CESA8, Ler wild-type measurements have been duplicated in the middle graph in order to make easy comparisons. C, cesa8irx1-7, cesa7irx3-6, and cesa4irx5-4 were backcrossed into Ler five times to create cesa8irx1-7L, cesa7irx3-6L, and cesa4irx5-4L, respectively. These were then transformed with either the wild-type genes (CESA8, CESA7, and CESA4, respectively) or TED Asp-to-Asn mutants (CESA8D683N, CESA7D726N, and CESA4D748N, respectively). N refers to the number of individual T1 transformants analyzed. Cellulose content is expressed as a percentage of the wild-type value. Error bars indicate se. Significance levels from univariate ANOVA are indicated for comparison between the genotype and the background mutant (white asterisks inside each bar) or between particular genotypes (black asterisks and lines above the bars): ***, significant at 0.001; **, significant at 0.01; and *, significant at 0.05.
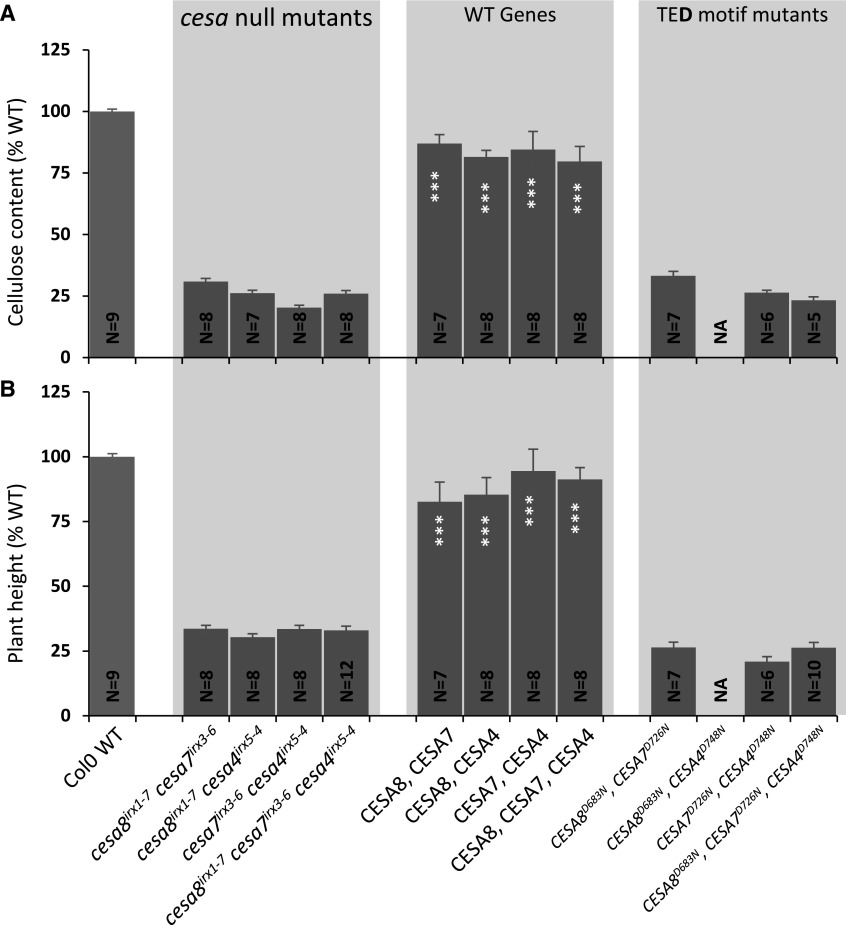
Mutating the TED Asp of Multiple Secondary Cell Wall CESAs
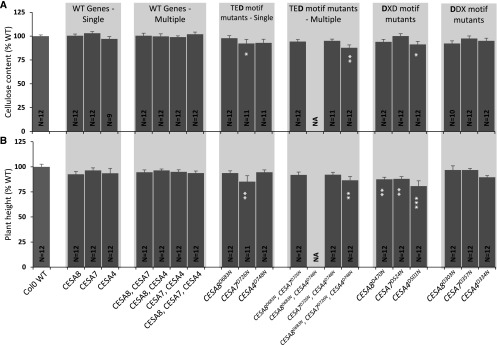
There are two possibilities that explain how plants in which only a single class of CESA is mutated are still able to make some cellulose. Either the cellulose is synthesized by the two remaining CESA classes or mutating the TED and DXD motifs is not sufficient to completely abolish their activity and they are able to synthesize some cellulose, albeit at a reduced rate. In the latter case, the rate of cellulose synthesis by the CSC is likely determined by the rate of synthesis of the defective CESA. Consequently, introducing a TED mutation into more than one CESA class should result in some cellulose, with the rate determined by the CESA class displaying the lowest activity. To investigate this idea, we made two double combinations (CESA8/CESA7 and CESA7/CESA4) and a triple combination (CESA8/CESA7/CESA4) of the TED Asp mutants. The multi-CESA mutants were cloned within the same plasmid and transformed into their respective double and triple cesa T-DNA mutant combinations. None of the lines containing more than one defective CESA showed evidence of any complementation and had a cellulose content that was not significantly different from the T-DNA mutant controls (Fig. 4). This supports the idea that residual cellulose synthesis in TED single mutants is supported by cellulose synthesis by the two remaining active classes of CESA proteins.
Figure 4.
Analysis of the plants containing TED motif Asp-to-Asn mutations in multiple secondary wall CESAs. A, Cellulose content. B, Plant height. Data are shown for all possible double (cesa8irx1-7 cesa7irx3-6, cesa8irx1-7 cesa4irx5-4, and cesa7irx3-6 cesa4irx5-4) and triple (cesa8irx1-7 cesa7irx3-6 cesa4irx5-4) mutant combinations of secondary wall CESAs transformed with plasmids containing the corresponding wild-type (WT) CESA genes or the TED motif Asp-to-Asn mutants. N refers to the number of individual T1 transformants analyzed. Cellulose content and plant height are expressed as percentages of the wild-type values. The cellulose data for the TED motif are the same as in Figure 1B. Error bars indicate se. Significance levels from univariate ANOVA are indicated for comparison between the genotype and the background mutant (white asterisks inside each bar): ***, significant at 0.001. NA indicates no data available.
Cellulose Content in CESA Mutants Does Not Always Correlate with Plant Morphology
Plant height is often a useful proxy for the degree of complementation of the cellulose content (Kumar et al., 2017), and this relationship largely holds true for the plants analyzed in this study (Fig. 2B). We found that cesa mutants complemented with the DDX motif mutants had sufficient cellulose to completely restore plant height, while the mutants complemented with the DXD and TED mutants that had less cellulose exhibited only partial restoration of plant height (Fig. 2B). This relationship between cellulose content and plant height does not always hold, and both the TED and DXD mutants of CESA7 (CESA7D726N and CESA7D524N) exhibited only marginally less cellulose than the corresponding mutants in CESA4 (CESA4D748N and CESA4D334N; Fig. 1A), but the complementation of plant height was always much greater in the CESA4 mutant plants (Fig. 2B).
In order to investigate the apparent discrepancy of the CESA7/CESA4 DXD and TED mutants in terms of cellulose content and growth phenotypes, the T2 generation of all mutants was grown as part of a single experiment, where 15 replicates of each genotype, consisting of five plants from three independent lines, were grown side by side. The growth differences between the genotypes were apparent from an early stage. While there were no differences between CESA8, CESA7, and CES4 for the DDX mutants, rosettes of 4-week-old plants of the CESA7 DXD mutant (CESA7D524N) appeared considerably smaller than those of CESA8 (CESA8D470N) or CESA4 (CESA4D501N; Supplemental Fig. S2). Similarly, the rosettes of the CESA7 TED Asp mutant were smaller than those of CESA4. These differences in growth between CESA7 and CESA4 continued throughout development (Fig. 5). When the plant height, stem diameter, and weight of cell wall material were compared, we observed the same trend as we observed with our initial plant height observations (Figs. 2B and 5A). The level of complementation was higher for cesa4irx5-4 than for cesa7irx3-7 when using either corresponding DXD or TED mutants. In addition, complementation of the CESA7 DXD mutant (CESA7D524N) was particularly poor, suggestive of some kind of dominant negative effect (Fig. 5B).
Figure 5.
Growth characteristics of the plants containing an Asp-to-Asn mutation in the TED, DXD, and DDX motifs. A, Plant height. B, Stem diameter. C, Weight of the cell wall preparation. Error bars indicate se. N refers to the number of T2 plants analyzed. For each genotype, up to 15 plants (five plants each for three selected lines) were analyzed. Significance levels from univariate ANOVA are indicated for comparison between the genotype and the background mutant (white asterisks inside each bar) or between particular genotypes (black asterisks and lines above the bars): ***, significant at 0.001; **, significant at 0.01; and *, significant at 0.05. WT, Wild type.
Defective CESA Proteins Exhibit Little or No Detectable Effects on Cellulose Synthesis in the Wild Type
To test more directly whether the CESA Asp mutants might have a dominant effect on plant growth, and in particular whether the mutation in CESA7 might cause a particularly severe dominant negative phenotype, we transformed the three sets of CESA Asp mutants as well as the double and triple mutant combinations of the TED Asp mutants into wild-type Col-0. Even with the most severe phenotypes, we observed only relatively modest reductions in cellulose content and plant height. As might be expected, the TED CESA triple combination caused the largest decrease in cellulose content and a significant decrease in plant height. However, there was no consistent trend. Although the CESA7 TED mutant showed a significant decrease in cellulose content and plant height, all the DXD mutant plants grew slightly worse than the wild-type plants, but there was little effect on cellulose (Fig. 6).
Figure 6.
Analysis of all constructs used in this study in the Col-0 wild-type (WT) background. A, Cellulose content. B, Plant height. Data are shown for Col-0 transformed with wild-type CESA genes, single, double, and triple mutant combinations of CESA genes carrying the TED Asp mutants, and single CESA genes containing the DXD or DDX Asp mutants. N refers to the number of individual T1 transformants analyzed. Cellulose content and plant height are expressed as percentages of the wild-type values. The cellulose data for the TED motif are the same as in Figure 1B. Error bars indicate se. Significance levels from univariate ANOVA are indicated for comparison between the genotype and the background mutant (white asterisks inside each bar): ***, significant at 0.001; **, significant at 0.01; and *, significant at 0.05. NA indicates no data available.
Chemical Analysis of Cell Walls in the DDX, DXD, and TED Motifs of Secondary Cell Wall CESAs
Since the Updegraff method only measures the crystalline cellulose and our Asp mutants appeared to show varying levels of crystalline cellulose, it is possible that differences in the cellulose content of the Asp mutants are offset by an increase in noncrystalline cellulose or other forms of β-glucan. Consequently, the noncellulosic sugars were quantified. Consistent with previous studies (Brown et al., 2005), Xyl was the most abundant sugar and constituted more than 50% of the total cell wall sugars, but it was found at similar levels in all genotypes (Table I). In comparison with the Col-0 wild type, all three cesa T-DNA mutants had higher levels of the minor sugars Rha, Ara, and Gal. The relative proportion of Glc was reduced from 6% in Col-0 to only ∼3% in the cesa T-DNA mutants. Man content also was reduced in the cesa T-DNA mutants (Table I). For the various Asp mutants, the sugar composition followed the trends shown by the complementation for the cellulose content. Therefore, where cellulose complementation was good, such as in the DDX mutants, the noncellulosic sugar composition more closely matched that of the wild type, while in the poorly complemented lines, the profile looked similar to the composition of the cesa T-DNA mutants (Table I). Overall, there was no evidence to suggest that any of the CESA Asp mutants exhibit any increase in the amount of noncrystalline glucan that may compensate for or form in response to the reduction in cellulose found in these lines.
Table I. Noncellulosic sugar analysis of the plants containing an Asp-to-Asn mutation in the DDX, DXD, and TED motifs of secondary wall CESAs.
Relative amounts of six sugars expressed as percentages of total noncellulosic sugars are shown. Negligible amounts of Fuc also were detected but are not shown here. For each genotype, stem powder from three independent lines was analyzed. For each line, up to 36 T2 plants were pooled. Gas chromatography (GC) analysis was performed in duplicate for each sample. Mean values of six replicates (three biological × two technical) are shown ± se.
| Type | Genotype | Sugar |
|||||
|---|---|---|---|---|---|---|---|
| Rha | Ara | Gal | Glu | Man | Xyl | ||
| Controls | Col-0 wild type | 6.54 ± 0.16 | 4.48 ± 0.15 | 6.74 ± 0.19 | 21.74 ± 0.9 | 7.67 ± 0.15 | 52.43 ± 0.65 |
| cesa8irx1-7 | 11.61 ± 1.1 | 11.56 ± 0.24 | 11.36 ± 0.35 | 9.97 ± 0.76 | 5.48 ± 0.05 | 49.61 ± 2.18 | |
| cesa7irx3-7 | 10.42 ± 0.45 | 11.29 ± 0.43 | 11.47 ± 0.57 | 9.95 ± 0.77 | 5.85 ± 0.09 | 50.58 ± 1.71 | |
| cesa4irx5-4 | 11.36 ± 0.43 | 11.56 ± 0.38 | 11.51 ± 0.16 | 10.1 ± 0.2 | 5.5 ± 0.25 | 49.51 ± 0.5 | |
| Wild-type genes | CESA8_wild type | 5.85 ± 1.2 | 4.17 ± 0.06 | 7.5 ± 0.34 | 22.07 ± 0.27 | 8.2 ± 0.16 | 52.06 ± 0.84 |
| CESA7_wild type | 6.97 ± 0.38 | 3.95 ± 0.05 | 6.63 ± 0.23 | 24.81 ± 0.42 | 8.45 ± 0.08 | 49.05 ± 0.81 | |
| CESA4_wild type | 7.34 ± 0.34 | 5.16 ± 0.21 | 7.19 ± 0.12 | 14.67 ± 0.96 | 8.05 ± 0.1 | 57.35 ± 0.72 | |
| TED motif | CESA8D683N | 11.19 ± 0.28 | 8.57 ± 0.2 | 9.21 ± 1.65 | 13.82 ± 0.85 | 6.02 ± 1.07 | 51.19 ± 1.16 |
| CESA7D726N | 9.5 ± 0.79 | 7.04 ± 0.23 | 8.88 ± 0.13 | 18.04 ± 0.16 | 7.64 ± 0.23 | 48.41 ± 0.46 | |
| CESA4D748N | 8.09 ± 0.44 | 6.59 ± 0.57 | 9.4 ± 1.42 | 16.49 ± 0.97 | 8.24 ± 0.16 | 51.19 ± 0.86 | |
| DXD motif | CESA8D470N | 9.12 ± 0.14 | 10.47 ± 0.39 | 12.33 ± 0.33 | 11.83 ± 0.62 | 6.67 ± 0.08 | 48.62 ± 1.41 |
| CESA7D524N | 10.86 ± 0.13 | 10.15 ± 0.18 | 12.96 ± 0.34 | 13.53 ± 0.2 | 6.72 ± 0.13 | 44.6 ± 0.83 | |
| CESA4D501N | 8.99 ± 0.52 | 6.69 ± 0.16 | 9.95 ± 0.37 | 15.5 ± 1.59 | 7.29 ± 0.12 | 50.6 ± 2.11 | |
| DDX motif | CESA8D303N | 7.18 ± 0.06 | 4.4 ± 0.11 | 6.72 ± 0.27 | 20.53 ± 1.11 | 8.21 ± 0.05 | 52.95 ± 0.81 |
| CESA7D357N | 6.65 ± 0.11 | 4.93 ± 0.14 | 8.2 ± 0.1 | 15.97 ± 0.5 | 8.07 ± 0.05 | 55.45 ± 0.44 | |
| CESA4D334N | 5.96 ± 0.18 | 4.55 ± 0.06 | 7.18 ± 0.12 | 19.51 ± 0.34 | 7.96 ± 0.02 | 54.19 ± 0.36 | |
ssNMR or FTIR Showed No Alteration in Cellulose Structure in the CESA Asp Mutants
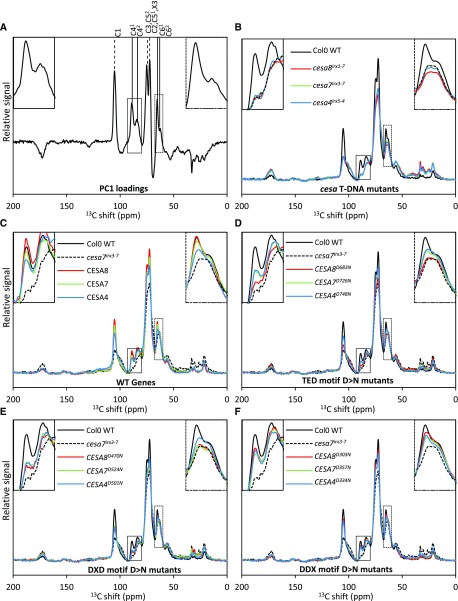
By incorporating a defective CESA protein into the CSC, it is possible that the product of each CSC will be an abnormal microfibril with fewer than normal cellulose chains. Since we know that CESA proteins exhibit high class specificity and likely occupy unique sites within the complex (Carroll and Specht, 2011; Kumar et al., 2017), the position within the CSC occupied by a defective class of CESA proteins may influence the ability of the remaining chains to form microfibrils and, consequently, may explain why mutations in particular classes of CESA proteins have differing effects on cellulose synthesis. To address this question, both ssNMR and FTIR were used to examine cellulose structure in plants from the wild type, cesa T-DNA null mutants, and the three CESA T-DNA mutants with wild-type genes or with TED, DXD, and DDX Asp mutants. A principal component analysis (PCA) plot of the C13 CP-MAS ssNMR data reveals that the majority of variability (89%) is explained by PC1 (Supplemental Fig. S3). The samples separate along PC1 more or less in order of their cellulose content (i.e. Col-0 wild-type genes, DDX mutants, DXD/TED mutants, and cesa T-DNA mutants). Furthermore, the loading plot of PC1 is dominated by peaks attributed to cellulose (Fig. 7A). Two regions of the spectra were of particular interest: the 80- to 93-ppm area that contains the C41 (89 ppm) and C42 (84 ppm) peaks and the 60- to 67-ppm area containing the C61 (65 ppm) and C62 (63 ppm) peaks. The C41 and C61 signals are attributed to the interior region of the cellulose domain or crystalline cellulose, while C42 and C62 have been attributed to surface or amorphous cellulose (Atalla and Vanderhart, 1984; Ha et al., 1998). Differences in microfibril size between primary and secondary cell walls are likely to manifest themselves in differences in the ratio of these two pairs of peaks (Ha et al., 1998). The full spectra (0–200 ppm) of all 16 genotypes are shown in Figure 7, B to F. Consistent with an absence of cellulose in the secondary cell wall mutants, there were large differences between the Col-0 wild type and cesa T-DNA mutants in both the 80- to 93-ppm and 60- to 67-ppm regions (Fig. 7B). The wild-type genes and DDX motif Asp mutants showed a high degree of complementation in both regions (Fig. 7, C and F), consistent with their relatively normal cellulose content. For the DXD mutants, CESA8, CESA7, and CESA4 all showed very similar profiles in the C4 region, but in the C6 region, the CESA4 DXD mutant showed far greater complementation (Fig. 7E). For the TED Asp, CESA7 and CESA4 were very similar to each other, while CESA8 showed a much lower level of complementation in both the C4 and C6 regions (Fig. 7D). Together, these data suggest that the spectra are dominated by the amount of secondary cell wall cellulose and reflect the degree of complementation. The fact that the spectral differences observed between the different DXD Asp mutants is not reflected in the TED Asp mutants makes further interpretation of these subtle differences difficult.
Figure 7.
NMR analysis of the plants containing an Asp-to-Asn mutation in the DDX, DXD, and TED motifs of secondary wall CESAs. A, Loadings for PC1 of the PCA that accounts for 89% data variability. B to F, ssNMR data for 16 genotypes. For each genotype, stem powder from up to 36 T2 plants (one selected line) was analyzed. Each graph shows the total signal-normalized individual spectra for the five groups of genotypes. Col-0 wild-type (WT) and cesa7irx3-7 data are repeated in C to F for comparison. The positions of cellulose peaks are indicated by vertical lines above the peaks in A. In each graph, the inset at top left shows the C4 peak region (80–93 ppm), while the inset at top right shows the C6 peak region (60–67 ppm).
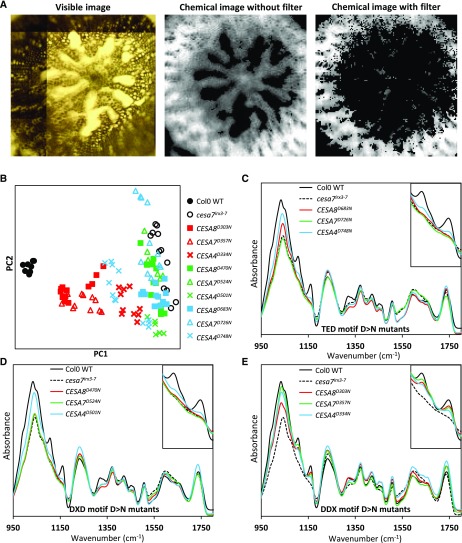
The ssNMR spectra were obtained from whole stem powder, which constitutes a mixture of primary and secondary cell walls. In our FTIR microspectroscopy analysis, we attempted to enrich for signals derived from the secondary cell walls. First, FTIR spectra were recorded from thin hypocotyl sections using a focal plane array detector that collects 128 × 128 = 16,384 pixels (spectra) from a 700- × 700-µm window that was sufficiently large to cover all or most of the section. We then filtered the pixels based on signal intensity at wave number 1,508 cm−1, which has been assigned to an aromatic c=c stretch indicative of lignin (Gorzsás et al., 2011). Using this approach, the remaining spectra were largely from the region of the hypocotyl that contains cells with a secondary cell wall (Fig. 8A).
Figure 8.
FTIR microspectroscopy analysis of the plants containing an Asp-to-Asn mutation in the DDX, DXD, and TED motifs of secondary wall CESAs. A, Visible image as seen with the FTIR microscope and chemical images based on signal intensity at 1,035 cm−1. The filtered chemical image was obtained after removing pixels that had 1,109 cm−1 signal intensity less than the mean 1,109 cm−1 intensity of all pixels in the image. B, PCA based on the filtered data set. Groups of genotypes are color coded: black, control genotypes; red, DDX motif mutants; green, DXD motif mutants; and blue, TED motif mutants. Each data point refers to an FTIR tile. For each genotype, up to 12 tiles were generated from T2 generation plants (three independent lines, one hypocotyl per line, four sections per hypocotyl, and one tile per section). C to E, Normalized mean (of all tiles for a genotype) spectra for the three groups of genotypes. Col-0 wild-type (WT) and cesa7irx3-7 data are included in all graphs for comparison. In each graph, the inset at top right shows the 1,085 to 1,185 cm−1 peaks associated with cellulose.
PCA of enriched data showed that, similar to the ssNMR, PC1 separated the samples essentially in order of their cellulose content complementation levels (Fig. 8B). The FTIR spectra showed considerable variation among genotypes in the 1,085- to 1,185-cm−1 region (Fig. 8, C–E) that contains peaks attributed to the signal from cellulose. Similar to the ssNMR data, the FTIR spectra in these regions correlated with the crystalline cellulose content, and the TED and DXD mutants had the lowest levels of complementation (Fig. 8, C–E).
Microfibril Angle and Cell Wall Crystallinity Correlate with the Amount of Cellulose in the Secondary Cell Wall
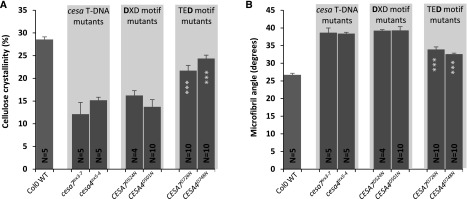
Since ssNMR and FTIR did not reveal any obvious differences in cellulose structure, we used wide-angle x-ray diffraction to examine the cell wall crystallinity and microfibril angle. The analysis was focused on the TED and DXD mutants of CESA4 and CESA7, since these plants exhibit only small differences in cellulose content but large differences in plant height (Fig. 2). We observed significant differences between the wild type and the cesa T-DNA mutants, with the wild type exhibiting a higher cell wall crystallinity but lower microfibril angle compared with the mutants. While the wall crystallinity and microfibril angle of the DXD mutants were similar to those of the cesa mutants, the TED motif mutants exhibited a level of cell wall crystallinity and a microfibril angle that were intermediate between the cesa mutants and the wild type (Fig. 9). There were no significant differences between the equivalent CESA4 and CESA7 TED or DXD Asp mutants for either microfibril angle or cell wall crystallinity (Fig. 9).
Figure 9.
Physical properties of the plants containing an Asp-to-Asn mutation in the DXD and TED motifs of secondary wall CESAs. A, Cellulose crystallinity. B, Microfibril angle. Error bars indicate se. N refers to the number of T2 plants analyzed. For each genotype, up to 10 plants (five plants each for two selected lines) were analyzed. Significance levels from univariate ANOVA are indicated for comparison between the genotype and the background mutant (white asterisks inside each bar): ***, significant at 0.001. WT, Wild type.
DISCUSSION
The fact that the CSC of the plant secondary cell wall is composed of three different CESAs and that these CESAs exhibit a high degree of class specificity (Kumar et al., 2017) suggests that each CESA occupies a particular position within the complex. Consequently, this provides an opportunity to investigate how altering the synthesis of individual cellulose chains at any given position affects cellulose microfibril structure. As demonstrated in this study (Fig. 4) and has been documented previously (Turner and Somerville, 1997; Taylor et al., 2000; Brown et al., 2005), both T-DNA knockout mutants and mutants that cause premature termination of the CESA protein result in plants with reduced secondary cell wall cellulose. This cellulose-deficient phenotype is assumed to result from the remaining two CESA isoforms being unable to form a functional complex (Ha et al., 2002; Gardiner et al., 2003; Taylor et al., 2003; Kumar and Turner, 2015a). By introducing CESA genes with specific Asp residues mutated to Asn, it should be possible to block cellulose synthesis specifically within a single CESA isoform, resulting in a catalytically inactive CESA isoform within a CSC that is still able to assemble correctly. Three different catalytic Asp mutants were tested (TED, DXD, and DDX). CESA proteins with the first DDX motif Asp mutation all still exhibited reasonable levels of complementation (Fig. 2) and are assumed to still display some catalytic activity. Even though this motif is highly conserved in plant CESA proteins (Pear et al., 1996) and in the bacterial BCSA structure, it maps close to the catalytic site (Morgan et al., 2013) and it may not participate directly in catalysis. In contrast, the Asp in the TED motif is believed to be the catalytic base (Morgan et al., 2013), and mutating this residue is likely to abolish activity. This assertion is supported by the fact that, when we mutate the highly conserved DXD motif essential for substrate binding (Morgan et al., 2013), we obtain essentially the same results as with mutating the TED motif. This result also holds when we examine a range of different alleles and different ecotypes (Figs. 2 and 3). A consistent pattern is observed across all these complementation experiments with the mutated DXD and TED motifs: CESA4 always gives the best complementation, while CESA8 gives the worst complementation and CESA7 is intermediate. If these different levels of complementation were the result of the residual catalytic activity, when we introduce mutants in two or more CESA isoforms, we would anticipate that the level of cellulose complementation would be comparable to that of the most severe mutant. However, we find that the lines in which two or more CESAs are mutated exhibit no significant complementation (Fig. 4). This result supports the idea that the residual catalytic activity of the mutated CESAs is not the cause of the partial complementation seen with the CESA DXD and TED mutants.
A more plausible explanation of the partial complementation exhibited by the DXD and TED CESA mutants is that the complex can assemble and be transported to the plasma membrane, where the two remaining CESA isoforms are still able to synthesize cellulose chains. This poses the questions of why mutations in different CESA isoforms give different levels of complementation. The available data indicate that CESA proteins in the secondary cell wall CSC are in a 1:1:1 stoichiometry (Hill et al., 2014) and, therefore, cannot explain the differences between how the DXD and TED mutants of CESA4 are better than CESA8 in their ability to complement the cesa T-DNA mutants. The data presented here support the idea that, if one CESA is inactive, the ability of the remaining two CESAs to continue to synthesize cellulose microfibrils depends upon which sites are active or inactive and the positions of the active CESAs.
Previous work on CESA class specificity suggests that each CESA protein may occupy a particular position within the CSC (Kumar et al., 2017). Consequently, we hypothesized that the differential effect of the inactivation of CESA8, CESA7, and CESA4 was a consequence of the differential effects these positions may have on cellulose microfibril assembly, which would be reflected in alteration in the structure of cellulose produced. Our analysis using ssNMR, FTIR, and x-ray diffraction was not able to identify changes in cellulose microfibril structure to support this hypothesis. An alternative explanation is based upon the differential catalytic activity of individual CESA proteins that may be a consequence of their inherent structure or a result of regulation by posttranslational modification or interaction with other accessory proteins. It was shown that phosphorylating CESA1 can regulate the bidirectional movement of primary wall CSCs and, hence, cellulose deposition (Chen et al., 2010). Furthermore, altering the levels of a particular member of the CESA6 class has a big effect on cellulose synthesis (Hu et al., 2017). Together, these two studies provide an example for how one particular CESA isoform can exert an effect on cellulose synthesis by an entire complex and how one isoform can have a greater effect on cellulose synthesis than others in certain circumstances. Furthermore, it has been proposed that the polymerization and crystallization of the cellulose chains provide the energy for the movement of the CSC through the plasma membrane (Diotallevi and Mulder, 2007). A CSC generating only two-thirds of the normal cellulose chains is likely to synthesize cellulose more slowly, since less energy will be generated to force the CSC through the plasma membrane. If some classes of CESA proteins were less active than others, either inherently from their structure or as a result of posttranslational modification, mutating the least active CESA would have the smallest effect on cellulose synthesis. The data from this study suggest that the least active CESA in secondary cell wall synthesis is CESA4, while the most active is CESA8.
In order to examine how different active CESA combinations might affect microfibril structure, we initially used ssNMR to examine the structure of cellulose. As expected, we found large differences in the spectra when we compared the wild type with the cesa T-DNA mutants. PCA of the data shows that PC1 separates on the basis of cellulose content and accounts for 89% of the variation (Supplemental Fig. S3). The loadings plot shows much more variation in the C41 and C61 peaks, which correspond to the interior domains, than it does for the C42 and C62 peaks, which are attributed to the outer domains. While frequently overlooked, the loadings plot reflects not only the differences in cellulose content between the wild type and the cesa mutants but also changes in cellulose structure that are inevitably the consequence of changes in the relative proportion of the signal derived from cellulose in either the primary or secondary cell walls. It is generally accepted that cellulose microfibrils in secondary cell walls are frequently composed of aggregates and contain more crystalline cellulose compared with those in the primary cell walls (Salmén, 2004; Terashima et al., 2009; Cosgrove and Jarvis, 2012). Comparing the wild type with the cesa T-DNA mutants also reveals big differences between the microfibril angle and crystallinity that also could reflect differences between the primary and secondary cell walls (Fig. 9). Consequently, it is likely that the crystallinity of many mutants with reduced secondary cell wall cellulose will be somewhere between that of the wild type and the cesa T-DNA mutants, independent of whether they show additional changes in microfibril structure. Small differences were observed in the ssNMR spectra of lines with similar cellulose content, but these differences do not follow a consistent trend that would easily allow us to explain the data, and the data do not reveal large changes in microfibril structure that might explain the different levels of complementation found in the mutants.
While studying cellulose biosynthesis in secondary cell walls offers many experimental advantages and practical applications, the large secondary wall microfibrils synthesized by CSC clusters (Li et al., 2016) may hinder the detection of subtle changes in microfibril structure. For example, if a primary cell wall microfibril is a product of one CSC, it would contain ∼18 chains. If one-third of the CESA proteins were defective in the CSC, then it would be reduced to only 12 chains, resulting in a very large change in the ratio of interior to exterior chains. In contrast, if a secondary wall microfibril is a product of four CSCs and contains 72 chains, reducing this to a microfibril of 48 chains is likely to have a much smaller effect on the ratio of interior to exterior cellulose chains.
Plants in which the DXD and TED Asp residues of CESA7 are mutated only have marginally less cellulose than the equivalent mutations in CESA4. However, the CESA4 DXD and TED mutant plants grow substantially larger and can reach almost twice the height of the corresponding CESA7 mutants (Fig. 2). The reasons for this difference are not clear, but we frequently observe better complementation of the plant height defect compared with the cellulose defect. For example, the DDX mutants show partial complementation of the cellulose defect but grow normally (Fig. 2). The CESA4 DXD and TED mutants consistently have slightly more cellulose that the corresponding CESA7 mutants. It is possible that the CESA4 DXD and TED mutants make just enough cellulose to get them over a certain threshold that allows the plants to make at least a partially functioning secondary wall that is able to support better plant growth, while the corresponding CESA7 mutants do not.
In summary, we have shown that the effects of mutating individual isoforms of the CESA proteins that make up the CSC of the secondary cell walls are not equivalent. Most of the techniques we have used to look at microfibril structure are affected by the relative proportions of primary and secondary cell wall cellulose in the samples. This is likely to mask more subtle changes in microfibril structure that may exist within the various classes of mutants we have identified. In the future, it may be necessary to use more sensitive techniques, such as two-dimensional ssNMR, to closely examine both microfibril structure and also the interaction between the microfibrils and the xylan that is bound to them (Dupree et al., 2015; Grantham et al., 2017).
MATERIALS AND METHODS
Plant Material
The Arabidopsis (Arabidopsis thaliana) secondary cell wall cesa mutants used in this study and their main properties are listed in Supplemental Table S2. Some have been described previously: cesa8irx1-1 (Turner and Somerville, 1997; Taylor et al., 2000), cesa7irx3-1 (Turner and Somerville, 1997; Taylor et al., 1999), cesa4irx5-2 (Taylor et al., 2003), cesa8irx1-7 and cesa7irx3-7 (Kumar and Turner, 2015a), cesa4irx5-4 and cesa8irx1-5 (Brown et al., 2005), and cesa7irx3-6 (Kumar and Turner, 2015a). Double and triple mutant combinations of secondary cell wall cesa mutants are described by Kumar and Turner (2015a). cesa4irx5-6, cesa8irx1-7L, cesa7irx3-6L, and cesa4irx5-4L are new alleles used in this study. For all T-DNA mutants, seeds were obtained from the Nottingham Arabidopsis Stock Centre, and homozygous plants were identified by genotyping using a PCR-based method. To create cesa8irx1-7L, cesa7irx3-6L, and cesa4irx5-4L T-DNA mutants in the Col-0 background (cesa8irx1-7, cesa7irx3-6, and cesa4irx5-4, respectively), they were crossed with the Ler wild type. The F1 plants were then crossed with the Ler wild type again, and the process was repeated another three times, after which the plants were selfed and homozygous plants were isolated from the segregating population and genotyped.
Plasmid Constructs
The full schematics for constructing the plasmids used in this study are shown in Supplemental Figure S4. Three Gateway destination vectors were constructed on pCambia1300 (hygromycin resistance). These vectors were VX21 (proCESA8::Gateway_casette:term35S), VX22 (proCESA7::RGS-6xHIS-STREP-Gateway_casette:term35S), and VX23 (proCESA4::Gateway_casette:term35S; Supplemental Fig. S4, A–C). The component fragments of these vectors (promoters, Gateway cassette, and terminators) were PCR amplified and cloned into a pJET vector using the CloneJET PCR Cloning Kit (Thermo Fisher Scientific). All primers included appropriate restriction sites to allow concatenation of the fragments. The inserts in the pJET vector were fully sequenced before assembling to create the final destination vectors. The three promoter::Gateway_casette:terminator cassettes also were designed to have unique restriction sites at the ends to allow two or three cassettes to be put together in a single plasmid.
The cloning of the coding sequences of wild-type CESA8, CESA7, and CESA4 in a Gateway entry vector, pDONOR/pZEO, has been described previously (Kumar et al., 2017). These entry clones were used as templates in PCR-based site-directed mutagenesis (Atanassov et al., 2009). For each mutant, two PCR fragments (A and B) were amplified. For fragment A, primers MF (5′-GTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAG-3′) and GSR (gene-specific reverse) were used, while for fragment B, GSF (gene-specific forward) and MR (5′-CAGGAAACAGCTATGACCATGTAATACGACTCACTA-3′) were used. The sequences of the different GSF primers used are listed in Supplemental Table S3. The GSR primers had a complementary sequence to the GSF primers. PCR fragments A and B were gel extracted with a gel extraction kit (Qiagen) and then combined in an overlap extension reaction (Atanassov et al., 2009), which was then purified. The overlap extension product was then used in an LR reaction (Thermo Fisher Scientific) with Gateway destination vectors described to drive the expression of CESA8, CESA7, and CESA4 with native promoters. The positive selection of Gateway technology meant that only the clones containing both fragments in the correct order would survive. Plasmids were extracted with a plasmid miniprep kit (Qiagen) and sequenced fully to verify that no mutations had occurred. Double and triple CESA constructs were made using the unique restriction sites at the ends of the promoter::Gateway:terminator cassettes (Supplemental Fig. S4, D–F).
Plant Growth and Analysis
Sequenced plasmids were transformed into the Agrobacterium tumefaciens strain pGV3101, which was then transformed into Arabidopsis plants using the floral dip method (Clough and Bent, 1998). Plants were analyzed at T1 and T2 stages. Seed was surface sterilized with 10 fold dilution of sodium hypochlorite solution (∼30% free chlorine), followed by three washes with water and stratification for 48 h. The seed was then selected on one-half-strength Murashige and Skoog plates containing 35 µg mL−1 hygromycin. After growing for 7 d on plates in an incubator, eight to 10 independent lines for each construct were transplanted into a 6:1:1 mixture of compost, perlite, and vermiculite. Plants were grown for another 6 weeks on soil under long-day conditions (16-h/8-h day/night, 22°C/18°C temperature, and 80% humidity). Col-0 wild-type and cesa mutant plants were grown on plates without any selection before being transplanted. A vector-only control for the Col-0 wild type was included in one of the experiments, and no differences were found in the growth patterns or cellulose content as compared with the Col-0 wild type grown on nonselection plates. Plant height measurements were taken when plants were 7 weeks old, after which 50-mm pieces from the primary inflorescence stem, starting at 5 mm above the base, were harvested and stored in 70% ethanol for the analysis of cellulose content.
Cellulose Assays
Stem material was harvested from 7-week-old plants. A 5-cm piece from the primary shoot was collected 5 mm above the base. Cellulose content was determined using a medium-throughput adaptation of Updegraff’s method (Kumar and Turner, 2015a).
ssNMR Analysis
For each genotype, 27 T2 plants were grown on plates and compost as described above. Stem material was harvested when plants were 9 weeks old and stripped of their leaves and siliques. Stems were freeze dried for 72 h, after which they were ground into a coarse powder using a pestle and mortar. The coarse powder was further ground into a fine powder using the Retch MM301 mixer mill. For each grinding run, about 500 mg of coarse powder was put into a 20-mL-capacity steel tube along with four 10-mm-diameter steel balls and ground for 3 min at a vibrational frequency of 24 Hz. About 100 mg of fine powder was used for the preparation of AIR followed by destarching in 2-mL screw-cap tubes using a modification of the method of Foster et al. (2010). Briefly, the fine powder was extracted twice with 1 mL of 70% ethanol (at 70°C) and once with 1 mL of a 1:1 mixture of chloroform:methanol. After the addition of each solvent, the samples were centrifuged at 14,000 relative centrifugal force for 10 min and the supernatant was discarded. Finally, 1 mL of acetone was added, and the samples were left to dry in a fume hood overnight. For destarching, the AIR was rehydrated in 1.5 mL of a 0.1 m sodium acetate buffer, pH 5, and heated for 20 min at 80°C in a heating block. After cooling the suspension on ice, 35 μL of amylase (50 μg mL−1 solution in water; from Bacillus spp.; Sigma) and 17 μL of pullulanase (17.8 units; from Bacillus acidopullulyticus; Sigma) were added and the tubes were vortexed thoroughly. The tubes were incubated in a 37°C shaker overnight. The enzyme mix was inactivated by incubating the samples at 100°C for 10 min, and the supernatant was discarded following centrifugation. The remaining pellet was washed three times by adding 1.5 mL of water, vortexing, centrifuging, and decanting. Finally, the pellet was resuspended in 1 mL of acetone and air dried in a fume hood overnight. Dried samples were stored at room temperature until needed for further processing.
Up to 50 mg of powder was analyzed by ssNMR in C13 CP-MAS experiments. Solid-state 13C spectra were recorded at 100.56 MHz using a Varian VNMRS spectrometer and a 4-mm (rotor o.d.) magic-angle spinning probe. Spectra were obtained using cross-polarization with a 2-s recycle delay, a 3-ms contact time, an ambient probe temperature of ∼25°C, and a sample spin rate of 10 kHz. Between 1,000 and 1,600 repetitions were accumulated. Spectral referencing was performed with respect to an external sample of neat tetramethylsilane (carried out by setting the high-frequency signal from adamantane to 38.5 ppm). Recorded spectra were baseline corrected using MNOVA software (Mestrelab Research) and exported to Microsoft Excel, where they were normalized to make the total signal for each spectrum 100%. Further plotting of the spectra was performed in Excel. PCA was performed using Matlab R2016a (MathWorks).
Analysis of Noncellulosic Sugars
AIR was prepared as described above for ssNMR analysis. Approximately 10 mg of AIR powder was transferred to prenumbered 2-mL screw-cap vials. Weak acid hydrolysis was performed by adding 500 µL of 1 m H2SO4 to each vial and heating at 121°C for 60 min. Before heating, 20 µL of a 20 mg mL−1 myoinositol solution was added to each vial as an internal control. After letting the tubes cool to room temperature, they were centrifuged at 3,000 rpm for 5 min, and a 250-µL aliquot of the supernatant was transferred to a 15-mL screw-cap glass tube. The released monosaccharides were converted into their respective alditol acetate derivatives as described previously (Blakeney et al., 1983; Reiter et al., 1993).
Derivatized sugars were transferred to GC vials for analysis on a Supelco SP-2330 (Sigma; product code 24073) fused silica capillary column (dimensions: 30-m length, 0.32-mm i.d., and 0.2-µm film thickness) mounted on an Agilent 6850 GC instrument. Analysis was performed in splitless mode using helium as the carrier gas with a flow rate of 25 mL min−1. The temperature program was as follows: 2 min at 160°C, 40°C min−1 gradient to 200°C, held at 200°C for 4 min, 20°C min−1 gradient to 220°C, and held at 220°C for 17 min. Peak assignment was performed by analyzing a run of seven individual sugars: Rha, Fuc, Ara, Xyl, Man, Gal, and Glc, as well as myoinositol. The myoinositol signal was used to normalize the signal for every peak detected. The detector response to each sugar was established by analyzing a sugar mix containing equal amounts of all seven sugars. Sugar peaks in the sample runs were normalized with myoinositol and corrected for the detector response. Relative sugar content was then calculated as a percentage of total sugars. All calculations were performed in Microsoft Excel.
Analysis of Protein Expression
Up to six independent lines for each genotype were tested for protein expression analysis. A total of 16 plants for each line were grown until 5 weeks old, when stems were harvested and stripped of their leaves and flowers. Stems were ground in liquid nitrogen into a fine powder using a pestle and mortar. Protein extracts were analyzed by quantitative western blotting as described previously (Kumar et al., 2017) using anti-CESA8, anti-CESA7, and anti-CESA4 antibodies. An anti-HSP73 antibody was used as a control for sample loading.
X-Ray Diffraction
Microfibril angle and cell wall crystallinity were determined by x-ray diffraction using the Bruker D8 Discover x-ray diffraction unit. Both the x-ray source and the detector were set to θ = 0° for microfibril angle determination. The average T-value of the two 002 diffraction arc peaks was used for microfibril angle calculations, according to the method of Ukrainetz et al. (2008), with the exception of a calibration curve developed specifically for Arabidopsis. In contrast, the 2θ (source) was set to 17° for wood crystallinity determination. Crystallinity was determined by mathematically fitting the data using the method of Vonk (1973). Crystallinity measures were precalibrated by capturing diffractograms of pure Acetobacter xylinum bacterial cellulose known to be 87% crystalline.
FTIR
Hypocotyls were harvested from 7-week-old plants, frozen in liquid nitrogen, and stored at −80°C until further use. For sectioning, the hypocotyls were mounted in OCT embedding medium (Thermo Fisher Scientific) and 10-µm thin cryosections were obtained using a cryomicrotome (Leica CM3050). Sections were air dried between two glass slides to keep them flat and stored at room temperature until use. The FTIR protocol as described previously (Pilling et al., 2015) was used. Briefly, dried sections were scraped off and placed on CaF2 discs (20 × 1 mm; Crystran) and analyzed using a Varian 670 infrared spectrometer coupled to a Varian 620-IR imaging microscope equipped with a 128- × 128-pixel liquid nitrogen-cooled mercury cadmium telluride focal plane array detector. The infrared microscope consisted of a 15× magnification objective with a resultant field of view of 704 μm. The process of recording FTIR spectra on the Varian 620-IR imaging microscope was controlled by Resolutions Pro software (Agilent). Within the program, a mosaic of multiple visible images, each covering a 500- × 500-µm area, was created to cover the whole hypocotyl section. An area was then selected to record FTIR spectra so that a single infrared window of 704 × 704 µm was able to cover a majority of the hypocotyl. This constituted an FTIR tile. Part of the mosaic corresponding to the infrared tile was produced by the program as a single .jpg image. No further processing of the visible image was performed. An example of such a mosaic is shown in Figure 8A. For each section, a single tile of 16,384 pixels (128 × 128) was recorded. For each genotype, three plants (one each for three independent transgenic lines) were analyzed, and for each plant, four sections were analyzed, making a total of 12 tiles per genotype.
The interferograms were processed using triangular apodization with one level of zero filling giving a data point spacing of approximately 4 cm−1, with the spectral range 900 to 4,000 cm−1 retained. All preprocessing and data analysis were performed using Matlab 2012a (MathWorks) and the ProSpect Toolbox (London Spectroscopy). Matlab scripts were used for background correction and quality control to filter out bad-quality pixels. In order to enrich for pixels coming from secondary cell walls, we then filtered the pixels based on signal intensity at wave number 1,508 cm−1, which has been assigned to an aromatic c=c stretch indicative of lignin (Gorzsás et al., 2011). Mean spectra for each tile were calculated using these pixels, and the data were exported to Microsoft Excel, where all tiles for each genotype were averaged to produce the spectra shown in Figure 8.
Accession Numbers
Accession numbers are as follows: CESA4, AT5G44030; CESA7, AT5G17420; and CESA8, AT4G18780.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Protein expression analysis of the plants containing Asp-to-Asn mutation in the DDX, DXD, and TED motifs.
Supplemental Figure S2. Growth characteristics of the plants containing Asp-to-Asn mutation in the DDX, DXD, and TED motifs.
Supplemental Figure S3. PCA plot of ssNMR data.
Supplemental Figure S4. Schematics for constructing the plasmids
Supplemental Table S1. Amino acid residue numbers of the mutated Asp residues in DDX, DXD, and TED motifs of Arabidopsis secondary cell wall CESA proteins
Supplemental Table S2. List of cesa mutant alleles used in this study.
Supplemental Table S3. List of gene-specific forward primers used for constructing the Asp mutants.
Acknowledgments
We thank Dr. Liam Campbell, Holly Allen, and Sophie Mogg for critical reading of the article. We are grateful to Mike Jarvis and Paul Dupree for helpful discussion about FTIR and NMR, respectively. Solid-state NMR spectra were obtained at the EPSRC UK National Solid-State NMR Service at Durham.
Footnotes
This work was funded by Biotechnology and Biological Sciences Research Council Grants BB/H012923/1 and BB/P01013X/1.
Articles can be viewed without a subscription.
References
- Atalla RH, Vanderhart DL (1984) Native cellulose: a composite of two distinct crystalline forms. Science 223: 283–285 [DOI] [PubMed] [Google Scholar]
- Atanassov II, Atanassov II, Etchells JP, Turner SR (2009) A simple, flexible and efficient PCR-fusion/Gateway cloning procedure for gene fusion, site-directed mutagenesis, short sequence insertion and domain deletions and swaps. Plant Methods 5: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeney AB, Harris PJ, Henry RJ, Stone BA (1983) A simple and rapid preparation of alditol acetates for monosaccharide analysis. Carbohydr Res 113: 291–299 [Google Scholar]
- Brown DM, Zeef LAH, Ellis J, Goodacre R, Turner SR (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Gidley MJ, Fincher GB (2010) Heterogeneity in the chemistry, structure and function of plant cell walls. Nat Chem Biol 6: 724–732 [DOI] [PubMed] [Google Scholar]
- Carroll A, Somerville C (2009) Cellulosic biofuels. Annu Rev Plant Biol 60: 165–182 [DOI] [PubMed] [Google Scholar]
- Carroll A, Specht CD (2011) Understanding plant cellulose synthases through a comprehensive investigation of the cellulose synthase family sequences. Front Plant Sci 2: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Ehrhardt DW, Somerville CR (2010) Mutations of cellulose synthase (CESA1) phosphorylation sites modulate anisotropic cell expansion and bidirectional mobility of cellulose synthase. Proc Natl Acad Sci USA 107: 17188–17193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ, Jarvis MC (2012) Comparative structure and biomechanics of plant primary and secondary cell walls. Front Plant Sci 3: 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diotallevi F, Mulder B (2007) The cellulose synthase complex: a polymerization driven supramolecular motor. Biophys J 92: 2666–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupree R, Simmons TJ, Mortimer JC, Patel D, Iuga D, Brown SP, Dupree P (2015) Probing the molecular architecture of Arabidopsis thaliana secondary cell walls using two- and three-dimensional 13C solid state nuclear magnetic resonance spectroscopy. Biochemistry 54: 2335–2345 [DOI] [PubMed] [Google Scholar]
- Endler A, Kesten C, Schneider R, Zhang Y, Ivakov A, Froehlich A, Funke N, Persson S (2015) A mechanism for sustained cellulose synthesis during salt stress. Cell 162: 1353–1364 [DOI] [PubMed] [Google Scholar]
- Fernandes AN, Thomas LH, Altaner CM, Callow P, Forsyth VT, Apperley DC, Kennedy CJ, Jarvis MC (2011) Nanostructure of cellulose microfibrils in spruce wood. Proc Natl Acad Sci USA 108: E1195–E1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster CE, Martin TM, Pauly M (2010) Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass). Part II. Carbohydrates. J Vis Exp 1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner JC, Taylor NG, Turner SR (2003) Control of cellulose synthase complex localization in developing xylem. Plant Cell 15: 1740–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorzsás A, Stenlund H, Persson P, Trygg J, Sundberg B (2011) Cell-specific chemotyping and multivariate imaging by combined FT-IR microspectroscopy and orthogonal projections to latent structures (OPLS) analysis reveals the chemical landscape of secondary xylem. Plant J 66: 903–914 [DOI] [PubMed] [Google Scholar]
- Grantham NJ, Wurman-Rodrich J, Terrett OM, Lyczakowski JJ, Stott K, Iuga D, Simmons TJ, Durand-Tardif M, Brown SP, Dupree R, et al. (2017) An even pattern of xylan substitution is critical for interaction with cellulose in plant cell walls. Nat Plants 3: 859–865 [DOI] [PubMed] [Google Scholar]
- Gu Y, Kaplinsky N, Bringmann M, Cobb A, Carroll A, Sampathkumar A, Baskin TI, Persson S, Somerville CR (2010) Identification of a cellulose synthase-associated protein required for cellulose biosynthesis. Proc Natl Acad Sci USA 107: 12866–12871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha MA, Apperley DC, Evans BW, Huxham IM, Jardine WG, Viëtor RJ, Reis D, Vian B, Jarvis MC (1998) Fine structure in cellulose microfibrils: NMR evidence from onion and quince. Plant J 16: 183–190 [DOI] [PubMed] [Google Scholar]
- Ha MA, MacKinnon IM, Sturcová A, Apperley DC, McCann MC, Turner SR, Jarvis MC (2002) Structure of cellulose-deficient secondary cell walls from the irx3 mutant of Arabidopsis thaliana. Phytochemistry 61: 7–14 [DOI] [PubMed] [Google Scholar]
- Hill JL Jr, Hammudi MB, Tien M (2014) The Arabidopsis cellulose synthase complex: a proposed hexamer of CESA trimers in an equimolar stoichiometry. Plant Cell 26: 4834–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Zhang R, Feng S, Wang Y, Wang Y, Fan C, Li Y, Liu Z, Schneider R, Xia T, et al. (2017) Three AtCesa6-like members enhance biomass production by distinctively promoting cell growth in Arabidopsis. Plant Biotechnol J (in press) [DOI] [PMC free article] [PubMed]
- Kimura S, Laosinchai W, Itoh T, Cui X, Linder CR, Brown RM Jr (1999) Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis. Plant Cell 11: 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Atanassov I, Turner S (2017) Functional analysis of cellulose synthase (CESA) protein class specificity. Plant Physiol 173: 970–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Campbell L, Turner S (2016) Secondary cell walls: biosynthesis and manipulation. J Exp Bot 67: 515–531 [DOI] [PubMed] [Google Scholar]
- Kumar M, Turner S (2015a) Protocol: a medium-throughput method for determination of cellulose content from single stem pieces of Arabidopsis thaliana. Plant Methods 11: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Turner S (2015b) Plant cellulose synthesis: CESA proteins crossing kingdoms. Phytochemistry 112: 91–99 [DOI] [PubMed] [Google Scholar]
- Li S, Bashline L, Zheng Y, Xin X, Huang S, Kong Z, Kim SH, Cosgrove DJ, Gu Y (2016) Cellulose synthase complexes act in a concerted fashion to synthesize highly aggregated cellulose in secondary cell walls of plants. Proc Natl Acad Sci USA 113: 11348–11353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Schneider R, Kesten C, Zhang Y, Somssich M, Zhang Y, Fernie AR, Persson S (2016) Cellulose-microtubule uncoupling proteins prevent lateral displacement of microtubules during cellulose synthesis in Arabidopsis. Dev Cell 38: 305–315 [DOI] [PubMed] [Google Scholar]
- Morgan JLW, Strumillo J, Zimmer J (2013) Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SC, Brown RM Jr (1980) Evidence for an intramembrane component associated with a cellulose microfibril-synthesizing complex in higher plants. J Cell Biol 84: 315–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman RH, Hill SJ, Harris PJ (2013) Wide-angle x-ray scattering and solid-state nuclear magnetic resonance data combined to test models for cellulose microfibrils in mung bean cell walls. Plant Physiol 163: 1558–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris JH, Li X, Huang S, Van de Meene AML, Tran ML, Killeavy E, Chaves AM, Mallon B, Mercure D, Tan HT, et al. (2017) Functional specialization of cellulose synthase isoforms in a moss shows parallels with seed plants. Plant Physiol 175: 210–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly M, Keegstra K (2008) Cell-wall carbohydrates and their modification as a resource for biofuels. Plant J 54: 559–568 [DOI] [PubMed] [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM (1996) Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA 93: 12637–12642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, Khitrov N, Auer M, Somerville CR (2007) Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci USA 104: 15566–15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilling MJ, Bassan P, Gardner P (2015) Comparison of transmission and transflectance mode FTIR imaging of biological tissue. Analyst (Lond) 140: 2383–2392 [DOI] [PubMed] [Google Scholar]
- Reiter WD, Chapple CCS, Somerville CR (1993) Altered growth and cell walls in a fucose-deficient mutant of Arabidopsis. Science 261: 1032–1035 [DOI] [PubMed] [Google Scholar]
- Salmén L. (2004) Micromechanical understanding of the cell-wall structure. C R Biol 327: 873–880 [DOI] [PubMed] [Google Scholar]
- Sánchez-Rodríguez C, Ketelaar K, Schneider R, Villalobos JA, Somerville CR, Persson S, Wallace IS (2017) BRASSINOSTEROID INSENSITIVE2 negatively regulates cellulose synthesis in Arabidopsis by phosphorylating cellulose synthase 1. Proc Natl Acad Sci USA 114: 3533–3538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100: 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Laurie S, Turner SR (2000) Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12: 2529–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Scheible WR, Cutler S, Somerville CR, Turner SR (1999) The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11: 769–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima N, Kitano K, Kojima M, Yoshida M, Yamamoto H, Westermark U (2009) Nanostructural assembly of cellulose, hemicellulose, and lignin in the middle layer of secondary wall of Ginkgo tracheid. J Wood Sci 55: 409–416 [Google Scholar]
- Thomas LH, Forsyth VT, Sturcová A, Kennedy CJ, May RP, Altaner CM, Apperley DC, Wess TJ, Jarvis MC (2013) Structure of cellulose microfibrils in primary cell walls from collenchyma. Plant Physiol 161: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SR, Somerville CR (1997) Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9: 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukrainetz NK, Ritland K, Mansfield SD (2008) Identification of quantitative trait loci for wood quality and growth across eight full-sib coastal Douglas-fir families. Tree Genet Genomes 4: 159–170 [Google Scholar]
- Vain T, Crowell EF, Timpano H, Biot E, Desprez T, Mansoori N, Trindade LM, Pagant S, Robert S, Höfte H, et al. (2014) The cellulase KORRIGAN is part of the cellulose synthase complex. Plant Physiol 165: 1521–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonk CG. (1973) Computerization of Ruland’s x-ray method for determination of crystallinity in polymers. J Appl Cryst 6: 148–152 [Google Scholar]
- Wang T, Hong M (2016) Solid-state NMR investigations of cellulose structure and interactions with matrix polysaccharides in plant primary cell walls. J Exp Bot 67: 503–514 [DOI] [PMC free article] [PubMed] [Google Scholar]