Arabidopsis expresses four ALY family nuclear RNA-binding proteins that interact with the RNA helicase UAP56, and their depletion causes nuclear mRNA accumulation and developmental defects.
Abstract
The regulated transport of mRNAs from the cell nucleus to the cytosol is a critical step linking transcript synthesis and processing with translation. However, in plants, only a few of the factors that act in the mRNA export pathway have been functionally characterized. Flowering plant genomes encode several members of the ALY protein family, which function as mRNA export factors in other organisms. Arabidopsis (Arabidopsis thaliana) ALY1 to ALY4 are commonly detected in root and leaf cells, but they are differentially expressed in reproductive tissue. Moreover, the subnuclear distribution of ALY1/2 differs from that of ALY3/4. ALY1 binds with higher affinity to single-stranded RNA than double-stranded RNA and single-stranded DNA and interacts preferentially with 5-methylcytosine-modified single-stranded RNA. Compared with the full-length protein, the individual RNA recognition motif of ALY1 binds RNA only weakly. ALY proteins interact with the RNA helicase UAP56, indicating a link to the mRNA export machinery. Consistently, ALY1 complements the lethal phenotype of yeast cells lacking the ALY1 ortholog Yra1. Whereas individual aly mutants have a wild-type appearance, disruption of ALY1 to ALY4 in 4xaly plants causes vegetative and reproductive defects, including strongly reduced growth, altered flower morphology, as well as abnormal ovules and female gametophytes, causing reduced seed production. Moreover, polyadenylated mRNAs accumulate in the nuclei of 4xaly cells. Our results highlight the requirement of efficient mRNA nucleocytosolic transport for proper plant growth and development and indicate that ALY1 to ALY4 act partly redundantly in this process; however, differences in expression and subnuclear localization suggest distinct functions.
In eukaryotic cells, pre-mRNAs are synthesized in the nucleus by RNA polymerase II and cotranscriptionally processed by diverse events, including 5′ capping, splicing, and 3′ polyadenylation. Only when these processes are completed are the mature mRNAs exported from the nucleus to the cytosol for translation. In yeast and metazoa, a plethora of proteins have been identified that mediate the transport of export-competent mRNAs across the nuclear envelope through nuclear pore complexes (Köhler and Hurt, 2007; Wickramasinghe and Laskey, 2015). The export of mRNAs is an integrated step in mRNA biogenesis, and the TREX (for transcription-export) multiprotein complex plays a central role in the coupling of transcription, processing, and export (Katahira, 2012; Heath et al., 2016). TREX consists of the THO core complex that associates with various other components, including the RNA helicase UAP56 (also known as DDX39B; Sub2 in yeast) and mRNA export factors such as CIP29/SARNP (Tho1 in yeast) and ALY (Yra1 in yeast; Strässer et al., 2002; Masuda et al., 2005; Dufu et al., 2010). In metazoa, TREX is recruited to mRNAs in a capping- and splicing-dependent manner (Katahira, 2012; Heath et al., 2016). In addition to its function in splicing, UAP56 serves an important role in initiating the export of mRNAs by loading mRNA export adaptors such as ALY (Ally of AML-1 and LEF-1 [Bruhn et al., 1997], also termed REF) onto both spliced and intronless mRNAs (Luo et al., 2001; Taniguchi and Ohno, 2008). mRNA adaptors are recruited to the RNA molecule cotranscriptionally through various processing events to license mRNA for export (Walsh et al., 2010). ALY (and other TREX components) recruit the mRNA export receptor NXF1/NXT1 (Mex67/Mtr2 in yeast) and hand the mRNA over to NXF1/NXT1 (Viphakone et al., 2012). Finally, NXF1/NTF1 interacts with TREX-2 at the nuclear pore and directs the mRNA through the nuclear pore by binding to FG repeats of nucleoporins (Katahira, 2012; Wickramasinghe and Laskey, 2015; Heath et al., 2016).
In plants, only a few factors that are involved in the above-mentioned mRNA export pathway have been functionally characterized to date (Xu and Meier, 2008; Merkle, 2011; Gaouar and Germain, 2013). The composition of the Arabidopsis (Arabidopsis thaliana) THO core complex of TREX (consisting of HPR1, THO2, THO5A/B, THO6, THO7A/B, and TEX1) resembles metazoan THO rather than that of yeast (Yelina et al., 2010). Likewise, THO associates with UAP56, ALYs, and MOS11 (the ortholog of CIP29) in Arabidopsis cells (Sørensen et al., 2017). MOS11 and ALY2 interact directly with the ATP-dependent RNA helicase UAP56 (Kammel et al., 2013). Four putative mRNA export adaptors of the ALY family (ALY1–ALY4) are encoded by the Arabidopsis genome, and in transient assays, it was observed that they display a distinct subnuclear distribution (Uhrig et al., 2004; Pendle et al., 2005). They were identified as interactors of the tomato bushy stunt virus P19 protein, and the association with P19 resulted in a relocation of ALY2 and ALY4 from the nucleus to the cytosol (Uhrig et al., 2004; Canto et al., 2006). Surprisingly, some ALY proteins were identified along with components of the exon junction complex in Arabidopsis nucleoli (Uhrig et al., 2004; Pendle et al., 2005). ALY4 (and a closely related ortholog from Nicotiana benthamiana) was reported to be involved in a variety of plant responses upon pathogen infection, including stomatal closure (Teng et al., 2014).
THO and TREX-2 subunits as well as MOS11 were demonstrated experimentally to be involved in plant mRNA export, as mos11, hpr1, tex1 mos11, and thp1 mutants exhibit nuclear mRNA accumulation (Germain et al., 2010; Lu et al., 2010; Pan et al., 2012; Xu et al., 2015; Sørensen et al., 2017), but so far, none of the ALY mRNA export adaptor candidates have been shown to function in the nucleocytosolic transport of mRNAs in plants.
In this study, we systematically studied the Arabidopsis ALY proteins, including their expression and subcellular localization. Moreover, we examined their interactions with RNA and the RNA helicase UAP56. Mutant plants deficient in the expression of all four ALY genes exhibit nuclear mRNA accumulation and show various developmental defects in both the sporophyte and the female gametophyte. Our results indicate that Arabidopsis ALY1 to ALY4 play a role in mRNA export and that the efficient nucleocytosolic transport of mRNAs is a requirement for proper plant growth and development.
RESULTS
ALY Proteins in Arabidopsis and Other Plants
First, we compared the amino acid sequences of the four Arabidopsis ALY proteins (ALY1–ALY4) as well as of putative orthologs from several other plants. The Arabidopsis ALY proteins (245–295 amino acid residues; 25.8–31.3 kD) are generally rich in the amino acid residues Arg and Lys; accordingly, they are basic proteins with theoretical pI values of 9.8 to 11.6. An alignment of the amino acid sequences revealed that the four Arabidopsis ALY proteins (like mammalian ALY proteins; Walsh et al., 2010) have in common a centrally positioned conserved RNA recognition motif (RRM; Burd and Dreyfuss, 1994) including characteristic RNP sequences (Supplemental Fig. S1A). The RRM is flanked by more variable Arg/Gly-rich N- and C-terminal domains. ALY1 and ALY2 (54% amino acid sequence identity) as well as ALY3 and ALY4 (70% amino acid sequence identity) share a high degree of sequence similarity, whereas the similarity of ALY1/2 versus ALY3/4 is clearly lower (less than 42% amino acid sequence identity; Supplemental Fig. S1B). Arabidopsis ALY1/2 and ALY3/4 show ∼44% and ∼38% amino acid sequence identity with human ALY, respectively (Supplemental Fig. S1B). Searching protein sequences of several other plant model species using Arabidopsis ALY1 as a query demonstrated that ALY-like proteins are present in flowering plants (monocots, dicots, and Amborella spp.) as well as in Selaginella and Physcomitrella spp. Monocot and dicot plants generally encode several ALY proteins (four or five different genes), whereas there seem to be only two ALY-like proteins in Amborella spp. and one in Selaginella spp. A multiple sequence alignment of these sequences revealed that the ALY amino acid sequences of other flowering plants are diversified similarly to those of Arabidopsis (Supplemental Fig. S2).
RNA Binding of ALY1
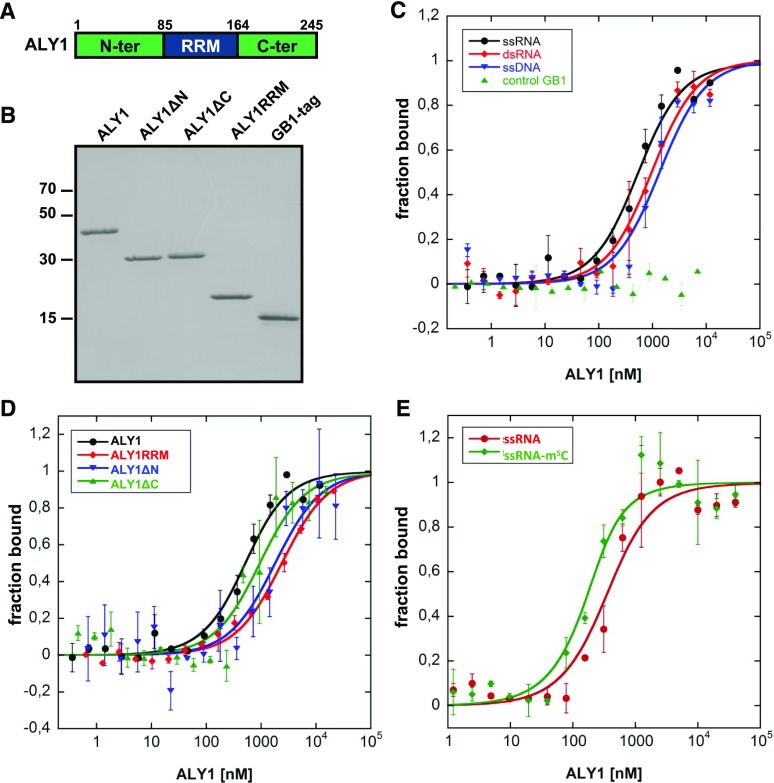
A multitude of proteins interacting with RNA have been identified in Arabidopsis (Köster et al., 2017), and a critical feature of mRNA export adaptors is their ability to bind RNA (Walsh et al., 2010). However, RNA interactions of plant ALY proteins have not been characterized thus far, and we selected the ALY1 protein for RNA-binding analyses. In addition to full-length ALY1 (amino acids 1–245), we examined the predicted RRM (ALY1RRM; amino acids 85–164) and ALY1 lacking either the N-terminal domain (ALY1ΔN; amino acids 85–245) or the C-terminal domain (ALY1ΔC; amino acids 1–164; Fig. 1A). ALY1 and truncated versions of the protein were expressed in Escherichia coli as 6xHis-GB1 fusion proteins, purified by two-step chromatography, and examined by SDS-PAGE (Fig. 1B). For comparison, we also used the unfused 6xHis-GB1 tag. The purified recombinant proteins were analyzed for RNA binding in solution using microscale thermophoresis (MST). First, binding affinities were measured comparatively for fluorescently labeled 25-nucleotide single-stranded RNA (ssRNA), double-stranded RNA (dsRNA), and single-stranded DNA (ssDNA) probes by incubation with increasing concentrations of ALY1. Compared with dsRNA and ssDNA (Kd = 918 ± 201 and 1,300 ± 479 nm, respectively), ALY1 exhibited a preference for interaction with ssRNA (Kd = 481 ± 109 nm; Fig. 1C; Student’s t test, P < 0.05 and P < 0.001, respectively). The unfused 6xHis-GB1 tag did not exhibit affinity for ssRNA. To examine which domains of ALY1 contribute to the RNA interactions, the binding to ssRNA of the different recombinant ALY1 versions was measured (Fig. 1D; Student’s t test, P < 0.005). Full-length ALY1 bound the RNA probe with more than 5-fold higher affinity compared with that of the individual RRM (Kd = 414 ± 99 versus 2,285 ± 315 nm). For the ALY1ΔN and ALY1ΔC proteins, intermediate affinities (Kd = 1,700 ± 741 and 830 ± 388 nm, respectively) were determined. Therefore, for efficient RNA binding of ALY1, the RRM and the terminal domains are required, and the contribution of the N-terminal domain seems to be greater than that of the C-terminal domain. Recently, it was reported that 5-methylcytosine (m5C) is a common and important posttranscriptional modification of mRNAs and other RNAs in Arabidopsis (David et al., 2017) and that mammalian ALY preferentially binds to m5C-modified RNA (Yang et al., 2017). Therefore, we comparatively analyzed the interaction of Arabidopsis ALY1 with an m5C-modified 19-nucleotide ssRNA oligonucleotide relative to the unmodified control RNA. The MST measurements demonstrated that ALY1 bound the m5C-modified RNA (Kd = 77 ± 32 nm) with about 3-fold higher affinity than that for the unmodified RNA (Kd = 249 ± 88 nm; Fig. 1E; Student’s t test, P < 0.01).
Figure 1.
Production of recombinant ALY1 proteins and MST analysis of their interactions with nucleic acids. A, Schematic representation of ALY1 depicting the different domains of the protein. The RRM and the relatively variable N- and C-terminal domains (compare with Supplemental Fig. S1) are indicated. Numbers indicate amino acid sequence positions that delimit the domains. B, SDS-PAGE analysis of purified full-length and truncated ALY1 expressed in E. coli, alongside the unfused 6xHis-GB1 tag. C, MST analysis of the interaction of full-length ALY1 with different nucleic acids. Incubation of the unfused 6xHis-GB1 tag with ssRNA was included as a control. D, MST analysis of the interaction of full-length and truncated versions of ALY1 with ssRNA. E, MST analysis of the interaction of full-length ALY1 with m5C-modified 19-nucleotide ssRNA and with the unmodified control RNA. Each data point represents two biological and two technical replicates, and error bars indicate sd of the biological replicates.
The ALY Proteins Interact Directly with the RNA Helicase UAP56
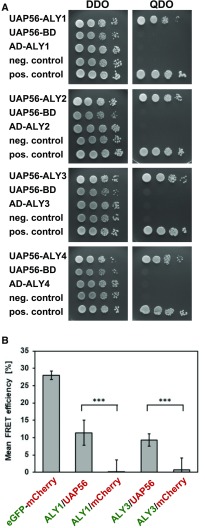
The ALY proteins copurify with the THO/TREX components TEX1 and UAP56 from Arabidopsis cells (Sørensen et al., 2017), and ALY2 interacts with the RNA helicase UAP56 in yeast cells and in vitro (Kammel et al., 2013). We examined whether all four ALY proteins can interact directly with UAP56 using the yeast two-hybrid assay. UAP56 fused to the DNA-binding domain (BD) of GAL4 was expressed in yeast cells, whereas the ALY sequences were fused to the activation domain (AD) of GAL4. Yeast cells coexpressing both types of fusion proteins were scored for their growth. Whereas all strains grew on the double dropout (DDO) medium, only the strains expressing both the UAP56-BD and the AD-ALY fusion proteins (along with a positive control) grew on the high-stringency, selective quadruple dropout (QDO) medium (Fig. 2A), demonstrating that ALY1 to ALY4 interact with UAP56 in yeast cells. Strains expressing only one of the two fusion proteins and the negative control showed no growth on the QDO medium. The interaction between ALY1 and ALY3 with UAP56 was analyzed further by Förster resonance energy transfer (FRET) in Agrobacterium tumefaciens-infiltrated N. benthamiana leaves, introducing plasmids that drive the expression of eGFP and mCherry fusion proteins as donor/acceptor pairs. The recovery of donor fluorescence after acceptor photobleaching (APB) was quantified (Fig. 2B). An eGFP-mCherry fusion protein (27.9% ± 1.3% FRET-APB efficiency) served as a positive control for the experiment, while cells expressing ALY1/3-eGFP along with unfused mCherry provided background levels (0.2% ± 3.7%/0.7% ± 3.5% FRET-APB efficiency). FRET-APB efficiencies of 11.4% ± 3.6% and 9.3% ± 1.8% were recorded for the combinations ALY1-eGFP/UAP56-mCherry and ALY3-eGFP/UAP56-mCherry, respectively. Therefore, according to yeast two-hybrid assays and FRET-APB analyses, ALY proteins interact directly with UAP56.
Figure 2.
ALY proteins interact with UAP56 in yeast two-hybrid assays and FRET analyses. A, Yeast two-hybrid assays with cells harboring the indicated constructs grown on DDO (left) or QDO (right) medium. B, Protein interactions analyzed by FRET-APB. N. benthamiana leaves were coinfiltrated with vectors expressing donor (eGFP, ALY1, and ALY3; green)/acceptor (mCherry and UAP56; red) combinations as indicated, alongside positive and negative controls. Transiently transformed cells were analyzed using two biological replicates by FRET-APB. Mean FRET-APB efficiencies (±sd, eight analyzed nuclei each) are shown. ***, Statistically significant difference by Student’s t test (P < 0.001).
Expression and Subcellular Localization of ALY Proteins
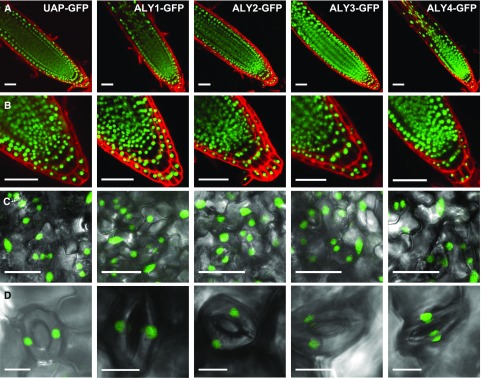
The subnuclear distribution of ALY1- to ALY4-GFP fusion proteins was analyzed previously in infiltrated N. benthamiana leaves (Uhrig et al., 2004) and transiently transformed suspension cultured Arabidopsis cells (Pendle et al., 2005). Since we intended to analyze the expression and subcellular localization of ALY-GFP fusion proteins in stably transformed plants, ideally in the absence of the respective endogenous protein, we started out characterizing T-DNA insertion lines of the four ALY genes. According to PCR-based genotyping and sequencing of the corresponding genomic regions, plants homozygous for T-DNA insertions within exon sequences were obtained for each gene (Supplemental Fig. S3, A and B). Reverse transcription (RT)-PCR analyses revealed that the respective ALY transcript was not detectable in the mutant plants, while the levels of the other ALY transcripts appeared unaltered (Supplemental Fig. S3C). The four aly mutant lines had essentially a wild-type appearance (see below for details) and were transformed with constructs driving the expression of the respective ALY-GFP fusion protein under the control of its promoter. Based on RT-PCR analysis of the different ALY-GFP-expressing plants, the ALY transcript levels in the transformed plant lines were comparable to those in Columbia-0 (Col-0; Supplemental Fig. S4). Root tips of the plants expressing ALY-GFP fusion proteins (and, for comparison, UAP56-GFP) were stained with propidium iodide and analyzed by confocal laser scanning microscopy (CLSM). In agreement with immunofluorescence analyses (Kammel et al., 2013), UAP56-GFP was detected exclusively in the cell nuclei (Fig. 3, A and B). The GFP signal of the ALY proteins also was restricted to the cell nuclei, which is in line with previous studies (Uhrig et al., 2004; Pendle et al., 2005). The expression of the four ALY proteins and of UAP56 was detected in all root cells. Examination of the leaf surfaces of these plants revealed that the ALY-GFP and UAP56-GFP fluorescence was observed in the nuclei of epidermal cells and guard cells (Fig. 3, C and D). Therefore, the four ALY proteins and UAP56 are widely expressed in sporophytic cells of Arabidopsis plants.
Figure 3.
Localization of ALY1- to ALY4-GFP and UAP56-GFP fusion proteins in nuclei of root and leaf cells. A, Root tips of plants at 8 d after stratification (DAS) expressing the indicated GFP fusion proteins under the control of the respective native promotors. GFP fluorescence is visible in green, while propidium iodide staining is in red. B, Root tips as in A shown at higher magnification. C, Abaxial surfaces of the second leaf pair from 14-DAS plants depicting GFP fluorescence in nuclei of epidermal cells and of stomatal guard cells. D, Magnified images of guard cells with nuclear GFP fluorescence. Bars = 50 µm (A and B), 25 µm (C), and 10 µm (D).
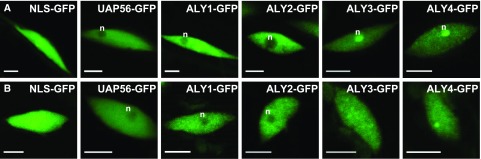
The subnuclear localization of the ALY-GFP fusions was inspected in more detail by CLSM in comparison with the UAP56-GFP and GFP-NLS controls. GFP-NLS (Antosch et al., 2015) was distributed evenly throughout root nuclei, and UAP56 occurs in the nucleoplasm with no clear signal in nucleoli (Fig. 4A), in agreement with the findings of an immunofluorescence microscopy analysis (Kammel et al., 2013). In comparison, the subnuclear localization of the ALY proteins is more variable, generally showing a less uniform distribution in the nucleoplasm. Strikingly, in many root cells, ALY3 and ALY4 were strongly enriched in nucleoli, whereas ALY1 and ALY2 were clearly less prominent in nucleoli relative to the nucleoplasm (Supplemental Table S1). In leaf cells, the nucleoplasmic distribution of the ALY proteins appeared more heterogenous than in root cells, particularly for ALY4, which partially localized to nucleoplasmic foci (Fig. 4B). Notably, the nucleolar enrichment of ALY3 and ALY4 seen in root cells was not observed in leaf cells, and the proteins were distributed similarly in nucleoplasm and nucleoli. As in root cells, ALY1 and ALY2 were detected primarily in the nucleoplasm of leaf cells and less in nucleoli. Thus, in accordance with their amino acid sequence similarities (Supplemental Fig. S1), the subnuclear distribution of ALY1/2 differs to some extent from that of ALY3/4. Recently, the motif WxHD was identified to play a role in the subnuclear localization of mammalian ALY (Gromadzka et al., 2016). Since this motif also occurs in ALY1 and a similar WxxD motif is found in ALY2, but not in ALY3 and ALY4 (Supplemental Fig. S1), we considered whether this motif might cause the distinct subnuclear localization of Arabidopsis ALY proteins. However, the analysis of plants expressing ALY1 and ALY3 versions with the WxxD motif swapped between the two proteins (Supplemental Fig. S5A) did not reveal an altered subnuclear distribution of these proteins when compared with control proteins (Supplemental Fig. S5B).
Figure 4.
Subnuclear localization of ALY1- to ALY4-GFP and UAP56-GFP fusion proteins in nuclei of root and leaf cells. A, Root cell nuclei from 8-DAS plants expressing the indicated GFP fusion proteins under the control of the respective ALY native promotors. The images depicted represent the most commonly observed subnuclear distributions of fusion protein. B, Cell nuclei from the second leaf pair of 14-DAS plants of the same GFP fusion protein lines depicted in A. n, Nucleoli. Bars = 5 μm.
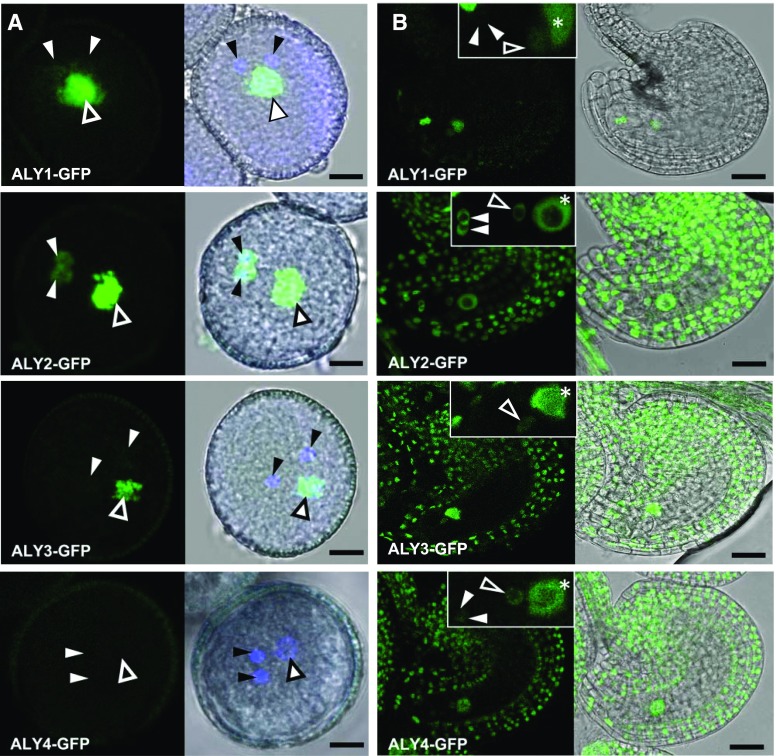
In view of the apparently ubiquitous expression of the four ALY-GFP proteins in sporophytic cells, their occurrence was analyzed in male and female gametophytes by CLSM. In mature pollen grains (Fig. 5A), the fluorescent signal of ALY1-GFP and ALY2-GFP was clearly visible in the vegetative nucleus. ALY2-GFP fluorescence was readily detectable in sperm cells, whereas the sperm cell-derived signal of ALY1-GFP was much weaker. ALY3-GFP fluorescence was present in the vegetative nucleus but not detectable in sperm cells. Nevertheless, the lack of ALY3-GFP detection in sperm cells by CLSM may be due to the overall weaker fluorescence of ALY3-GFP. By contrast, no fluorescence was detectable in transgenic ALY4-GFP mature pollen grains, even when the pollen grains were not counterstained with 4′,6-diamidin-2-phenylindol (DAPI) to exclude the possibility of DAPI-induced attenuated GFP fluorescence. Comparable results were obtained after in vitro pollen germination, when the fluorescence of the ALY-GFP fusion proteins was monitored in growing pollen tubes (Supplemental Fig. S6). Detailed analysis of mature unfertilized ovules (Fig. 5B) demonstrated that ALY1-GFP was detected specifically within the female gametophyte. The nuclei of the synergid cells and the homodiploid central cell nucleus showed the strongest ALY1-GFP signals, whereas the fluorescence was much weaker in the egg cell nucleus. No fluorescence was detected in the sporophytic tissues of ALY1-GFP-expressing ovules. Compared with ALY1-GFP, ALY2-GFP and ALY4-GFP displayed similar expression patterns in the nuclei of female gametophytic cells, with overall weaker fluorescence for ALY4-GFP. By contrast, ALY3-GFP was expressed in the central cell nucleus and the egg cell nucleus but was not detectable in the synergid cells. Furthermore, ALY2-GFP, ALY3-GFP, and ALY4-GFP also were expressed in the sporophytic cells of the ovule, such as the inner and outer integuments, the nucellus, and the funiculus. Therefore, in contrast to root and leaf cells, the analysis of pollen grains and ovules established clear differences in the expression pattern and the expression strength of the four ALY proteins. Moreover, unlike in root cells, we did not observe any nucleolar enrichment of ALY3-GFP and ALY4-GFP in the cells of the female gametophyte.
Figure 5.
Localization of ALY-GFP fusion proteins in pollen grains and ovules. A, Mature pollen grains. Closed and open arrowheads indicate sperm cell nuclei and vegetative nuclei, respectively. DNA is counterstained with DAPI (blue). GFP fluorescence (left images) and the corresponding bright-field images, merged with fluorescent signals (right images), are shown. B, Mature unfertilized ovules. GFP fluorescence (left images) and the corresponding bright-field images, merged with GFP signals (right images), are shown. Insets depict ALY-GFP signals in the nuclei of female gametophytic cells. Closed and open arrowheads and asterisks indicate synergid nuclei, egg cell nuclei, and central cell nuclei, respectively. Bars = 5 µm (A) and 25 µm (B).
Plants Lacking ALY Expression Exhibit Various Developmental Defects
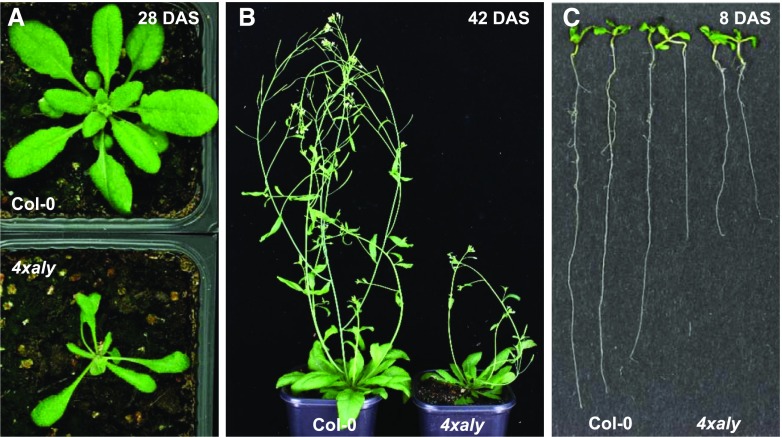
As mentioned above, plants of the four individual aly mutant lines are phenotypically essentially unaffected. Therefore, we generated double mutants combining the single mutations of the more closely related aly1 and aly2 as well as aly3 and aly4. Like single aly mutants, the aly1 aly2 and aly3 aly4 double mutant plants displayed basically a wild-type appearance (Supplemental Fig. S7A), including normal flower architecture (Supplemental Fig. S7B) and unaffected fertility (Supplemental Fig. S7C). The only noteworthy variance was that the aly3 mutant (and consistently the aly3 aly4 double mutant) bolted slightly earlier than Col-0 (Supplemental Fig. S7A). In view of the very mild impairment of these plants under standard growth conditions, we subsequently crossed the aly1 aly2 and aly3 aly4 double mutants. Plants homozygous for all four T-DNA insertions were obtained, and these aly1 aly2 aly3 aly4 quadruple mutants are referred to herein as 4xaly. The quadruple mutant plants were phenotypically severely affected, as evident from the reduced size of leaves and rosette diameter as well as from the decreased number of primary inflorescences (Fig. 6, A and B; Supplemental Fig. S8, A and B). At the seedling stage, the hypocotyls of 4xaly plants were remarkably elongated relative to those of Col-0 (Supplemental Fig. S8C). The mutant plants had fewer leaves than Col-0 when they started bolting (Supplemental Fig. S8D), and the bolting of 4xaly plants was delayed when compared with that of Col-0 (Supplemental Fig. S8E). In addition to the defects in the aerial parts, the growth of the primary root was clearly decreased (Fig. 6C; Supplemental Fig. S8F).
Figure 6.
Phenotype of the quadruple aly mutant plants (4xaly) relative to Col-0. A and B, Representative images of plants at 28 DAS (A) and 42 DAS (B). Plants were grown under long-day conditions in soil. C, Roots of 8-DAS plants grown on solid Murashige and Skoog (MS) medium.
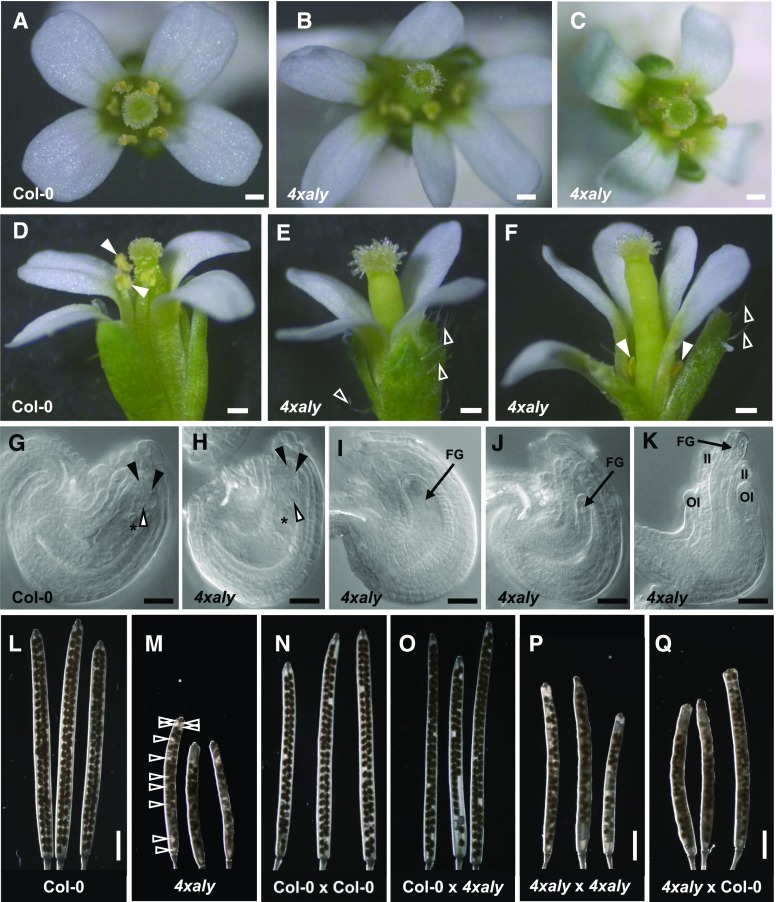
When we analyzed the inflorescences of 4xaly plants we observed that approximately 25% of the flowers showed an abnormal morphology, including altered number of petals (ranging from three to seven; Fig. 7, A–C), markedly increased number of trichomes on the sepals, and severely reduced length of the stamen filaments (Fig. 7, D–F). Detailed microscopy examination of cleared 4xaly ovules demonstrated that a fraction of the ovules (28%; n = 317 ovules) showed diverse developmental defects, whereas others were indistinguishable from Col-0 ovules (Fig. 7, G–K). The 4xaly ovule phenotype was composed of arrested female gametophyte development or collapsed female gametophytes (16% ± 1.12% [mean ± se]) and abnormal integument development (12% ± 0.37% [mean ± se]). Notably, the observed ovule phenotypes did not correlate with the abnormal flower morphology. In contrast to the severe developmental defects observed for a portion of the ovules, no altered phenotypes were detected for pollen grains and germinated pollen tubes. Based on light microscopy analyses and DAPI staining, the pollen grains released from 4xaly anthers had a wild-type appearance (Supplemental Fig. S9). The aly1 T-DNA insertion line is in the quartet (qrt1-2) background; therefore, 4xaly anthers produce microspores that fail to separate during pollen development, due to the lack of a pectin methylesterase in qrt1-2 (Francis et al., 2006). In vitro germination of 4xaly pollen tetrads revealed that their germination is as efficient as that of Col-0 pollen, indicating that they are fully viable (Supplemental Fig. S9). However, while performing the pollen germination assays, we observed that the pollen is not as easily shed from dehiscent 4xaly anthers as the pollen of dehiscent Col-0 anthers. Furthermore, the phenotype of compromised pollen dispersal is independent from the severely reduced preanthesis filament elongation observed in a portion of 4xaly flowers (Fig. 7F).
Figure 7.
4xaly plants produce a number of flowers with altered morphology and abnormal ovules. A to F, Flower morphology. A, Col-0 flower depicting normal petal number. B, 4xaly flower depicting altered petal number. C, Normal 4xaly flower as observed in the majority of cases. D, Col-0 flower depicting normal trichome number on sepals and stamen filament length (closed arrowheads). E and F, Abnormal 4xaly flower depicting increased trichome number on sepals (open arrowheads) and reduced stamen filament length (closed arrowheads). G to K, Differential contrast microscopy of cleared ovules. G, Col-0 ovule depicting normal morphology. H, 4xaly ovule depicting normal morphology. I, 4xaly ovule depicting arrested female gametophyte development. J, 4xaly ovule depicting collapsed female gametophyte. K, 4xaly ovule depicting abnormal integument development. Closed and open arrowheads and asterisks indicate synergid cell nuclei, egg cell nuclei, and central cell nuclei, respectively. FG, Female gametophyte; II, inner integuments; OI, outer integuments. L and M, Morphology of self-pollinated siliques of the indicated genotypes (pistil × pollen). N to Q, Morphology of cross-pollinated siliques of the indicated genotypes (pistil × pollen). Open arrowheads in M indicate gaps with undeveloped seeds in a 4xaly silique. Bars = 0.2 mm (A–F), 20 µm (G–K), and 2 mm (L–Q).
Compared with siliques of self-pollinated Col-0 flowers (Fig. 7L), 4xaly siliques were shorter and the seed set was reduced, which was readily identified by gaps between developed seeds (Fig. 7M). Counting the gaps in almost mature siliques derived from self-pollinated flowers revealed 22.85% ± 3.63% (mean ± se; n = 11) nondeveloped seeds in 4xaly siliques compared with 2.62% ± 0.97% (mean ± se; n = 10) nondeveloped seeds in Col-0 siliques. Subsequently, we performed hand-pollination experiments to determine whether the male or the female or both contribute to the observed reproductive defects in 4xaly siliques. When we hand pollinated Col-0 pistils with 4xaly pollen, the seed set was comparable to that of Col-0 pistils that were hand pollinated with Col-0 pollen (Fig. 7, N and O). This suggests that the reduced stamen length of 4xaly anthers and/or the compromised pollen dispersal but not a reduced fertility of 4xaly pollen contribute to the reproductive defects in 4xaly plants. Furthermore, the hand pollination of 4xaly pistils either with Col-0 or 4xaly pollen showed that the reduced seed set in 4xaly siliques mainly originated from ovules with abortive female gametophytes. Independent of the pollen donor plant, both the length of 4xaly siliques and the seed set were comparable (Fig. 7, P and Q). As expected, the reciprocal crossings of Col-0 pistils with 4xaly pollen and 4xaly pistils with Col-0 pollen produced uniformly heterozygous progeny (n = 15 each) for all four ALY genes.
The Absence of Conserved ALY Factors in 4xaly Plants Affects Nucleocytosolic mRNA Transport
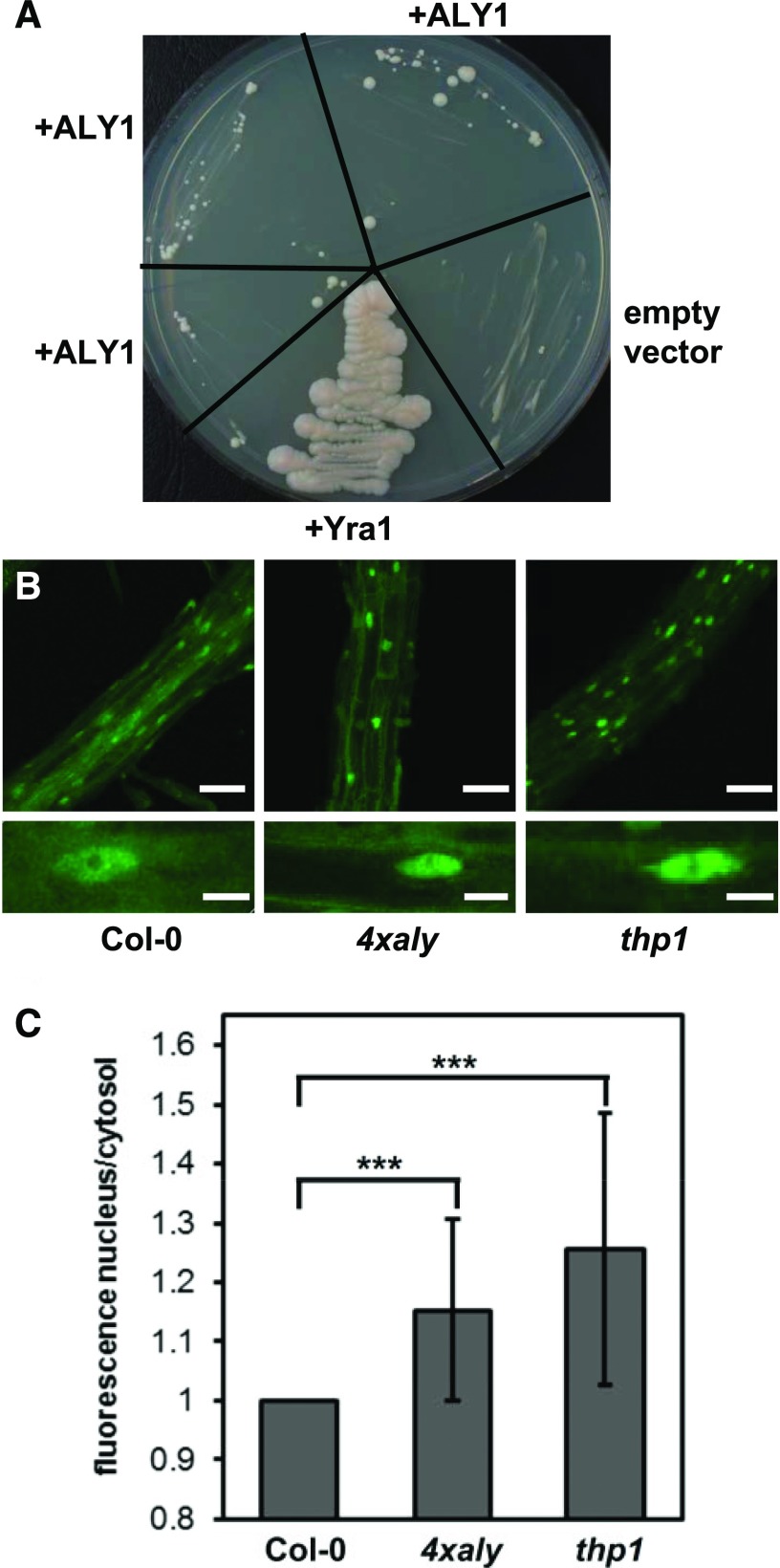
To examine whether an Arabidopsis ALY protein can functionally replace the yeast ortholog Yra1, a vector driving the expression of ALY1 under the control of the GPD promoter was introduced into a Yra1 shuffle strain (yra1::HIS3, pURA3-Yra1; Strässer and Hurt, 2000). Colonies formed after several days when transformants were grown on 5′-fluoroorotic acid (5-FOA)-containing plates (to select for colonies that had lost the pURA3-Yra1 plasmid), whereas the shuffle strain transformed with the empty plasmid did not grow (Fig. 8A). Therefore, the expression of ALY1 can restore the growth of the otherwise nonviable yra1 strain. However, as seen with the similar complementation experiment performed using mouse ALY (Strässer and Hurt, 2000), the complementation appears relatively inefficient (formation of small, slow-growing colonies) compared with the strain expressing Yra1 under the control of the GPD promoter (Fig. 8A).
Figure 8.
Complementation of the yeast yra1 mutant and analysis of the nucleocytosolic distribution of polyadenylated mRNA in 4xaly plants. A, Growth of the YRA1 shuffle strain (yra1::HIS3, pURA3-Yra1) following complementation with Arabidopsis ALY1 or yeast YRA1 alongside an empty-vector negative control. For ALY1, three individual transformants were analyzed. Cells were grown on 5-FOA plates, thus only allowing growth of yra1::HIS3 cells that have lost the pURA3-Yra1 plasmid. The plate was photographed after 18 d at 24°C to better visualize small, slow-growing colonies. B, Root cells of 6-DAS Col-0, 4xaly, and thp1 seedlings examined using whole-mount in situ hybridization with a fluorescently labeled 48-nucleotide oligo(dT) probe. CLSM images (taken using identical microscope settings) of representative regions of the analyzed roots (top row) and individual cells (bottom row) are shown. Bars = 60 μm (top row) and 10 μm (bottom row). C, The fluorescent hybridization signal of nuclei relative to cytosol as quantified for 58 or more nuclei from three roots of each genotype. The ratios are depicted relative to Col-0 (ratio of 1), with error bars indicating sd. ***, Statistically significant difference by two-tailed Student’s t test (P < 0.001).
Finally, to analyze whether mRNA export is affected in 4xaly plants, in situ hybridization with a fluorescently labeled oligo(dT) probe was used to detect bulk poly(A) mRNA. Using this method (Gong et al., 2005), the nucleocytosolic distribution of polyadenylated mRNA was examined in seedling root cells. The fluorescent signals were comparatively analyzed by CLSM for Col-0, 4xaly, and thp1 plants, since plants deficient in the THP1 subunit of the TREX-2 complex show severe defects in mRNA export (Lu et al., 2010); thus, thp1 served as a reference for the experiment. When compared with Col-0, stronger fluorescent signals were observed in the nuclei of thp1 cells (Fig. 8A). With 4xaly cells, the nuclear fluorescence relative to the cytosolic signal also was clearly enhanced, albeit not quite to the same level as for thp1 cells (Fig. 8B). These in situ hybridization experiments revealed an increased nuclear retention of polyadenylated mRNAs in 4xaly (and thp1) relative to that in Col-0. Therefore, the ALY proteins (like THP1; Lu et al., 2010) play a role in the export of mRNAs from plant nuclei.
DISCUSSION
The export of mRNAs from the nucleus to the cytosol for translation is a critical step in the expression of protein-coding genes, and this process is poorly characterized in plants. In yeast and metazoa, mRNA export adaptors such as ALY proteins play a central role in nucleocytosolic mRNA transport. A characteristic feature of mRNA export adaptors is their RNA-binding activity (Walsh et al., 2010). In MST assays, Arabidopsis ALY1 bound in solution with higher affinity to ssRNA when compared with its binding to dsRNA and ssDNA. The interaction of mammalian ALY and yeast Yra1 with ssRNA was demonstrated using UV cross-linking, electrophoretic mobility shift assays (EMSAs) and NMR spectroscopy (Strässer and Hurt, 2000; Stutz et al., 2000; Zenklusen et al., 2001; Golovanov et al., 2006; Katahira et al., 2009). Consistent with our MST measurements, preferential binding to ssRNA also was seen with murine ALY, as its binding was competed more efficiently by the addition of tRNAs than by ssDNA (Stutz et al., 2000). In EMSAs, the variable N- and C-terminal domains of mammalian ALY and yeast Yra1 were found to bind ssRNA, whereas no protein-RNA interactions were detected for the individual RRM (Rodrigues et al., 2001; Zenklusen et al., 2001). Using NMR chemical shift mapping and UV cross-linking, it was observed that the isolated RRM of mammalian ALY weakly bound to ssRNA, but high-affinity RNA interaction required the variable N- and C-terminal domains (Golovanov et al., 2006). In another EMSA study, the minimal RNA-binding site was mapped to a stretch of 21 amino acid residues within the variable N-terminal domain of murine ALY (Hautbergue et al., 2008), but this region is not well conserved in the Arabidopsis ALY sequences. Our MST analyses of ALY1 RNA binding revealed that the individual RRM interacts with ssRNA weakly and that both the N- and C-terminal domains increase the affinity of the protein for RNA, whereby the N-terminal domain has a greater effect. However, apparently, RNA-binding sites scattered over the length of ALY1 are required for its full capacity to interact with RNA. The presence of separate RNA-binding sites would provide ALY proteins with the potential to interact with different parts of an RNA molecule, which may play an important role in the packaging of mRNPs (Heath et al., 2016). Like mammalian ALY (Yang et al., 2017), Arabidopsis ALY1 exhibits a significantly higher affinity for m5C-modified RNA. The m5C modification is suggested to play an essential role in mRNA export and other posttranscriptional processes in mammals (Yang et al., 2017), a mechanism that may be conserved in plants (David et al., 2017).
Metazoan ALY and yeast Yra1 can be recruited to mRNAs cotranscriptionally by the THO/TREX complex, and the interaction is mediated by the DEAD-box RNA helicase UAP56/Sub2. Additional ways of ALY loading onto mRNAs are through the 5′ cap-binding complex and the polyadenylation machinery (Wickramasinghe and Laskey, 2015; Heath et al., 2016). ALY proteins were affinity purified from Arabidopsis suspension culture cells together with the THO core component TEX1 and with UAP56 (Sørensen et al., 2017), and ALY2 bound directly to UAP56 in vitro (Kammel et al., 2013). Here, ALY proteins interacted with UAP56 in the yeast two-hybrid assay and FRET-APB experiments. Taken together, these findings indicate that UAP56-mediated recruitment through the THO/TREX complex is conserved for the four Arabidopsis ALY proteins. The Arabidopsis genome contains two genes encoding identical UAP56 proteins but likely no additional paralog (Kammel et al., 2013). Hence, the THO/TREX interaction with ALY proteins in Arabidopsis might be mediated exclusively by the RNA helicase UAP56.
The fluorescence microscopy analysis of plants expressing ALY-GFP fusion proteins under the control of their respective native promoters in an aly mutant background revealed that the four ALY proteins are widely expressed throughout the plant. ALY1 to ALY4 were detected in the nuclei of all root tip cells and leaf epidermis cells as well as stomatal guard cells. Metazoan ALY and yeast Yra1 also are exclusively nuclear proteins, as they are displaced from the mRNA before nucleocytosolic translocation (Walsh et al., 2010; Wickramasinghe and Laskey, 2015). Closer inspection of the subnuclear localization of ALY proteins in leaf cells, root cells, and haploid cells of the female gametophyte revealed some differences. In many root cells, ALY3/4 were detected weakly in the nucleoplasm with a strong enrichment in nucleoli, whereas ALY1/2 were prominently present in the nucleoplasm and appear to be excluded from nucleoli. Whereas ALY1/2 displayed a comparable subnuclear distribution in root and leaf cells, the nucleolar enrichment of ALY3/4 was observed neither in leaf cells nor in cells of the female gametophyte. Nucleolar localization of the ALY mRNA export factors may seem surprising, but, in addition to rRNA synthesis/processing, nucleoli play important roles in the processing of other types of RNA, including mRNA splicing and export (Shaw and Brown, 2012; Meier et al., 2017). It was demonstrated that, in metazoa, and particularly in the best-studied model yeast, RNAs other than mRNAs are exported by different mechanisms involving exportins (Crm1 in yeast; Köhler and Hurt, 2007). rRNAs are exported from the nucleus in the form of large preribosomes (pre-60S and pre-40S), which, in yeast, is essentially mediated by Crm1 and the adaptor protein Nmd3 (Peña et al., 2017). Both exportins and Nmd3-like proteins are conserved in plants, and Arabidopsis NMD3 is required for the export of ribosomal subunits (Merkle, 2011; Chen et al., 2012). Therefore, it is unlikely that plant ALY proteins act in the export of preribosomes. Previous analyses addressing the subcellular localization of Arabidopsis ALY-GFP fusion proteins utilized transient fusion protein expression driven by viral promoters either in N. benthamiana leaves (Uhrig et al., 2004) or in Arabidopsis suspension culture cells (Pendle et al., 2005), yielding somewhat different results especially for ALY1, ALY3, and ALY4. The most striking differences include a remarkable enrichment of ALY1 in nucleoli, a very prominent ring-shaped staining of the nucleolar periphery particularly with ALY3/4, and generally that the ALY proteins often are localized to speckles and foci within the nucleoplasm. The comparable subnuclear distribution of ALY1 and ALY2 versus ALY3 and ALY4 observed in root and leaf cell nuclei of stably transformed Arabidopsis plants is in line with the degree of amino acid sequence similarity of ALY1 and ALY2 versus ALY3 and ALY4 (Supplemental Figs. S1 and S2), which also is reflected by the conservation of their respective gene structures (Supplemental Fig. S3).
Whereas the ALY-GFP fusion proteins appeared to be expressed ubiquitously in roots and leaves, the analysis of reproductive tissues revealed interesting differences regarding their expression pattern and expression level. In pollen grains, the expression of ALY3 is weaker and differs from that of ALY1/2, since it was detectable only in the nucleus of the pollen vegetative cell, whereas ALY4 was not expressed at all. However, despite the expression of ALY1 to ALY3 in the pollen vegetative cell, neither pollen development nor pollen germination, nor pollen tube growth, was noticeably affected when 4xaly pollen were grown in vitro or when they were used for hand-pollination experiments. These observations suggest either that ALY-mediated mRNA export does not play a critical role in pollen or that mRNA export from the nucleus of the pollen vegetative cell is achieved primarily by additional, yet uncharacterized, mRNA export factors (see below). Furthermore, our hand-pollination experiments using Col-0 pistils also demonstrate that 4xaly sperm cells are fully functional. Although ALY1 and ALY2 are expressed in sperm cells, the seed development of ovules fertilized with 4xaly sperm cells was not reduced. Thus, male reproductive 4xaly phenotypes are caused only by a significantly reduced length of stamen filaments and the compromised release of pollen from dehiscent anthers. In the ovule, ALY1 was restricted to the female gametophyte, whereas the other ALY proteins also were expressed in the surrounding sporophytic tissues. Around 16% of 4xaly ovules showed developmental defects in the female gametophyte, whereas the development of the integuments was affected in 12% of the mutant ovules. Considering these phenotypes, the expression pattern of ALY proteins suggests that all four ALY proteins act redundantly during female gametogenesis. On the other hand, the joint expression of ALY2, ALY3, and ALY4 in the sporophytic tissues of the ovule implies their contribution to the anatomically correct development of the integuments. Nevertheless, the phenotypic changes in 4xaly ovules occurred with low penetrance, suggesting that ALY proteins act redundantly with additional mRNA export factors during female gametophyte development. Since we also found filament elongation and pollen dispersal to be compromised in 4xaly stamens, cross-pollination experiments were necessary to clarify the reason(s) for the reduced seed set of self-pollinated 4xaly pistils. These experiments clearly showed that the ovules with abortive female gametophytes provoke the reduced seed set in 4xaly siliques, whereas 4xaly pollen is fully fertile when all physical restraints are overcome.
In metazoa, the number of genes encoding different ALY proteins ranges from one in humans to two in Drosophila melanogaster to three in Caenorhabditis elegans (Gatfield and Izaurralde, 2002; Longman et al., 2003; Katahira et al., 2009). The ALY genes of flowering plants seem to be more diversified, as the analyzed monocot and dicot plant genomes encode four or five different ALY proteins. Inactivation of the human ALY in HeLa cells, and similarly the depletion of ALY in D. melanogaster SL2 cells, resulted in a mild defect in the export of bulk mRNAs (Gatfield and Izaurralde, 2002; Katahira et al., 2009), whereas the simultaneous down-regulation of the three ALY proteins in C. elegans did not cause a detectable nuclear accumulation of polyadenylated mRNAs (Longman et al., 2003). In agreement with these findings, in Arabidopsis 4xaly plants, the nuclear accumulation of polyadenylated mRNAs was observed, but not a complete block of mRNA export. Therefore, as in metazoa, there is some redundancy among mRNA export adaptors (Wickramasinghe and Laskey, 2015; Heath et al., 2016), and, in addition to ALYs, other proteins may serve this function in Arabidopsis. Possible candidates for additional mRNA export adaptors include certain SR proteins and/or UIF-like proteins (Heath et al., 2016). D. melanogaster cells lacking ALY proteins show a mild proliferation defect, and simultaneous depletion of the three ALYs in C. elegans resulted only in a slight decrease of larval mobility (Gatfield and Izaurralde, 2002; Longman et al., 2003). Compared with these findings, 4xaly plants exhibited various defects in vegetative and reproductive development, whereas aly single mutants as well as the aly1 aly2 and aly3 aly4 double mutants displayed essentially a wild-type appearance. These results indicate that there is functional redundancy between the different ALY proteins and that the phenotypic defects of 4xaly plants may be caused by cumulative effects on mRNA export that are not detectable in the single and double mutants. However, the fact that flowering plants generally express several ALY proteins argues against simple redundancy and suggests that there is (partial) functional specialization. This is supported by the observations that Arabidopsis ALY proteins are differently expressed, at least in reproductive tissue, and that ALY1/2 display subnuclear localization that is distinct from that of ALY3/4 in root and leaf cells. Moreover, there is the possibility that various ALY proteins are involved in the nucleocytosolic transport of different subsets of mRNAs. Since this could be combined with tissue- and/or developmental stage-specific effects, analyzing these options will be a challenge for future investigations.
CONCLUSION
In plants, only a few components of the mRNA export machinery have been functionally characterized to date. These include some nucleoporins of the nuclear pore complex (Parry, 2015; Meier et al., 2017) and the TREX-2 complex that localizes to the nucleoplasmic side of nuclear pores (Lu et al., 2010). Moreover, subunits of the THO/TREX core complex as well as the associated MOS11 protein are involved in nucleocytosolic mRNA transport in Arabidopsis (Germain et al., 2010; Pan et al., 2012; Xu et al., 2015; Sørensen et al., 2017). The data presented here demonstrate that the ALY proteins of flowering plants are more diversified than those in other organisms and that they are part of the plant mRNA export pathway, likely acting as mRNA export adaptors. This notion is supported by their nuclear localization, their RNA-binding activity, their UAP56-mediated interaction with the THO/TREX complex, that the expression of ALY1 can complement the growth of the nonviable yeast yra1 mutant, and, most importantly, the nuclear mRNA accumulation in the absence of the four ALY proteins. However, it remains to be seen whether ALY proteins interact functionally with the thus far unidentified plant mRNA export receptor. Finally, the phenotype of plants lacking ALY proteins illustrates that impaired mRNA export can cause severe defects in vegetative and reproductive development.
MATERIALS AND METHODS
Plant Material and Documentation
According to standard protocol, Arabidopsis (Arabidopsis thaliana) Col-0 was grown at 21°C in a growth chamber under long-day conditions (16-h photoperiod per day) on soil (Antosz et al., 2017), while for some analyses, plants were grown on MS medium (Murashige and Skoog, 1962). After sowing, seeds were stratified in darkness for 48 h at 4°C prior to incubation in the plant growth chamber. The age of plants is stated as days after stratification (DAS). For root analyses, plants were grown on vertically oriented MS plates in a plant incubator (Percival Scientific) under long-day conditions as described before (Dürr et al., 2014). Seeds of the aly T-DNA insertion lines were obtained from the European Arabidopsis Stock Centre (http://www.arabidopsis.info/), and those of thp1-3 (Lu et al., 2010) were a gift from Yuhai Cui. Mutant plants were characterized by PCR-based genotyping in combination with sequencing of the insertion region and by RT-PCR (Antosz et al., 2017). By crossing the parental lines as described previously (Lolas et al., 2010), higher order mutant lines were generated. Using plasmids harboring different ALY-GFP (or UAP56-GFP) fusion constructs under the control of the respective native promoters (Supplemental Table S2) and Agrobacterium tumefaciens-mediated floral dip transformation as described previously (Pedersen et al., 2010; Antosch et al., 2015), plants were generated that express ALY-GFP (or UAP56-GFP). Plant phenotypes were documented and quantified as described previously (Dürr et al., 2014; Antosz et al., 2017). All phenotypic analyses were performed independently at least twice, and representative examples of the reproduced experiments are shown.
Preparation of Pollen Grains and Pollen Germination
Mature pollen grains were mounted in DAPI solution (6 µg mL−1 DAPI in 0.1 m PBS, pH 7.4) and imaged by fluorescence microscopy. Pollen grains were germinated as described previously (Vogler et al., 2014).
Preparation of Ovules and Siliques
Ovules were dissected and mounted as described previously (Sprunck et al., 2012) using unpollinated pistils at floral stage 12 (Smyth et al., 1990). Ovules were dissected on glass slides, either in chloral hydrate:glycerol:water (8:1:2, w/v/v) for differential interference contrast microscopy or in 50 mm sodium phosphate buffer (pH 7.5) for CLSM. Emasculated pistils at floral stage 12 were used for cross-pollination experiments. The seed set of self-pollinated and 11-d-old cross-pollinated siliques was investigated as described (Sprunck et al., 2012).
Plasmid Construction
The required gene or cDNA sequences were amplified by PCR with KAPA DNA polymerase (PeqLab) using Arabidopsis genomic DNA or cDNA as template and the primers (providing also the required restriction enzyme cleavage sites) listed in Supplemental Table S2. The PCR fragments were inserted into suitable plasmids using standard methods. All plasmid constructs were checked by DNA sequencing, and details of the plasmids generated in this work are summarized in Supplemental Table S2.
Production of Recombinant Proteins
For MST experiments, full-length ALY1 and truncated versions of the protein (Supplemental Table S2) were expressed in Escherichia coli using the plasmid pET24b-GB1 (Hautbergue et al., 2008) for the expression of the ALY1 proteins fused to a 6xHis tag for metal-chelate purification and a GB1 tag to increase protein stability/solubility (Gronenborn et al., 1991). The proteins were isolated from E. coli lysates by metal-chelate affinity chromatography using Ni-NTA beads (Qiagen) as described previously (Kammel et al., 2013). Eluted ALY1 proteins were further purified by FPLC ion-exchange chromatography using a Resource Q column (GE Healthcare) equilibrated in buffer D (10 mm sodium phosphate, pH 9, 1 mm EDTA, 1 mm DTT, and 0.5 mm PMSF; Grasser et al., 1996). Proteins were eluted with a linear gradient of 0 to 1 m NaCl in buffer D, and ALY1-containing fractions were pooled and passed through PD-10 columns (GE Healthcare) changing to protein buffer (50 mm Tris-HCl, pH 8, 100 mm NaCl, 10 mm 2-mercaptoethanol, 10% [v/v] glycerol, 0.5% [v/v] Triton X-100, 0.5 mm phenylmethylsulfonyl fluoride, 50 mm l-Arg, and 50 mm l-Glu; Golovanov et al., 2004). Finally, proteins were characterized by SDS-PAGE in combination with Coomassie Blue staining and by mass spectrometry.
Yeast Two-Hybrid and Complementation Assays
Yeast two-hybrid assays were performed essentially according to the manufacturer’s instructions (Clontech). In brief, yeast cells of the strain AH109 were cotransformed with pGBKT7 and pGADT7 plasmids (Supplemental Table S2) and grown at 30°C on SD/-Leu/-Trp (DDO) medium. For interaction assays, cells were grown on SD/-Ade/-Leu/-Trp/-His (QDO) medium for 2 d (Kammel et al., 2013). The interactions between p53 and the SV40 large T-antigen, and between lamin and the SV40 large T-antigen, served as positive and negative controls, respectively, as provided by the manufacturer. Using the YRA1 shuffle strain (yra1::HIS3, pURA3-Yra1; Strässer and Hurt 2000), we tested whether ALY1 expressed under the control of the GPD promoter using the 2µ p425-GPD vector (Mumberg et al., 1995) can replace Yra1, which was performed as described previously (Strässer and Hurt, 2000) by ultimately growing the complemented cells on 5-FOA plates.
PCR-Based Genotyping and RT-PCR
To distinguish between plants being wild type, heterozygous, or homozygous for the T-DNA insertions, genomic DNA was isolated from leaves. The genomic DNA was used for PCR analysis with Taq DNA polymerase (PeqLab) and primers specific for T-DNA insertions and the target genes (Supplemental Table S2). For RT-PCR, total RNA was extracted from ∼100 mg of frozen plant tissue using the TRIzol method (Invitrogen) before the RNA samples were treated with DNase. RT was performed using 2 μg of RNA and Revert Aid H minus M-MuLV reverse transcriptase (Thermo Scientific). The obtained cDNA was amplified by PCR using Taq DNA polymerase (PeqLab) as described previously (Dürr et al., 2014).
Fluorescent MST Binding Assay
MST binding experiments were carried out essentially as described previously (Kammel et al., 2013) with 200 nm 25-nucleotide Cy3-labeled ssRNA, dsRNA, or ssDNA oligonucleotides or 19-nucleotide Cy3-labeled ssRNA with or without m5C modification (Eurofins Genomics; Yang et al., 2017; Supplemental Table S2). MST measurements were performed in protein buffer with a range of concentrations of ALY1 proteins at 40% MST power and 50% LED power in standard capillaries at 25°C on a Monolith NT.115 device (NanoTemper Technologies). The fluorescence was monitored and used for data analysis in Thermophoresis and TJump. To calculate the fraction bound, the ΔFnorm value of each point is divided by the amplitude of the fitted curve, resulting in values from 0 to 1 (0 = unbound and 1 = bound), and processed using the KaleidaGraph 4.5 software and fitted using the Kd fit formula derived from the law of mass action. Differences between data sets were statistically evaluated using an unpaired Student’s t test.
Light Microscopy
For the analysis of GFP fusion protein expression, plants of at least three independent transgenic lines each harboring the different ALY-GFP fusion constructs in the respective mutant background were selected for further analysis based on RT-PCR analysis indicating uniform expression levels of ALY similar to Col-0 (Antosch et al., 2015). The imaging of GFP fluorescence was performed on an inverted Leica SP8 CLSM device with a 40×/1.3 numerical aperture and a 60×/1.3 numerical aperture oil-immersion objective. GFP was excited using the 488-nm argon laser line, and emission was detected from 495 to 545 nm (roots and leaves) or 500 to 535 nm (ovules and pollen) by a hybrid detector. Propidium iodide staining was performed as described previously (Dürr et al., 2014). Flowers and cleared siliques were imaged using a SteREO Discovery.V8 stereomicroscope (Zeiss). Cleared ovules and DAPI-stained pollen grains were investigated at the APOTOME FL (Zeiss) with a differential interference contrast 40×/1.4 oil objective and a 49DAPI reflector for DAPI fluorescence. Germinated pollen tubes were analyzed using an inverse Nikon Eclipse 2000-S microscope equipped with a Zeiss Axio-Cam MRm CCD camera using a 209/0.4 numerical aperture air objective.
FRET
FRET-APB was performed essentially as described previously (Weidtkamp-Peters and Stahl, 2017) using an SP8 microscope (Leica). Briefly, a square (0.5 × 0.5 cm) of an infiltrated Nicotiana benthamiana leaf was mounted in water on an objective slide with the abaxial side facing up. A circular area of 12 μm was bleached at 100% laser power for 80 iterations. Fifteen prebleach and 15 postbleach images were analyzed by ImageJ. The mean FRET efficiency was calculated as follows: ((IPOST - IPRE))/IPRE) × 100, where IPOST - IPRE = mean fluorescence intensity of 15 postbleached - mean fluorescence intensity of 15 prebleached frames.
Whole-Mount in Situ Hybridization
To determine the relative distribution of bulk mRNA in nuclei and cytosol, a previously described protocol was adopted (Gong et al., 2005) using 6-d-old seedlings grown on solid MS medium. Hybridization was performed in PerfectHyb plus solution (Sigma) with an Alexa Fluor 488-labeled 48-nucleotide oligo(dT) probe. Fluorescent signals of seedling roots were analyzed using CLSM with a Leica SP8 microscope (Sørensen et al., 2017).
Accession Numbers
Sequence data for the genes described in this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ALY1 (At5g59950), ALY2 (At5g02530), ALY3 (At1g66260), ALY4 (At5g37720), UAP56A (At5g11170), and UAP56B (At5g11200).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Amino acid sequence identity and alignment of Arabidopsis ALY proteins.
Supplemental Figure S2. Sequence similarity of ALY sequences.
Supplemental Figure S3. Molecular characterization of aly T-DNA insertion mutants.
Supplemental Figure S4. ALY transcript levels in aly mutant plants expressing ALY-GFP fusion proteins.
Supplemental Figure S5. Subnuclear distribution of mutant ALY1 and ALY3.
Supplemental Figure S6. ALY-GFP signals in growing pollen tubes, analyzed by CLSM.
Supplemental Figure S7. Phenotypes of aly single and double mutant plants.
Supplemental Figure S8. Phenotypic characterization of 4xaly plants relative to Col-0.
Supplemental Figure S9. The pollen grain phenotype and pollen germination efficiency of the 4xaly mutant are similar to those of the wild type.
Supplemental Table S1. Relative subnuclear localization of the different ALY-GFP proteins in root cells.
Supplemental Table S2. Oligonucleotide primers used in this study and construction of plasmids.
Acknowledgments
We thank Anna Geiger, Valentin Bergér, and Dominik Fiegle for contributions during early stages of the project, Benedikt Müller for help with CLSM imaging, Astrid Bruckmann and Eduard Hochmuth for mass spectrometry analyses, Antje Böttinger for technical assistance, Stuart Wilson for providing plasmid pET24b-GB1, Ed Hurt for the yeast YRA1 shuffle strain (yra1::HIS3, pURA3-YRA1), Yuhai Cui for the thp1-3 mutant line, and the Nottingham Arabidopsis Stock Centre for providing Arabidopsis T-DNA insertion lines.
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft through grant SFB960 to K.D.G.
Articles can be viewed without a subscription.
References
- Antosch M, Schubert V, Holzinger P, Houben A, Grasser KD (2015) Mitotic lifecycle of chromosomal 3xHMG-box proteins and the role of their N-terminal domain in the association with rDNA loci and proteolysis. New Phytol 208: 1067–1077 [DOI] [PubMed] [Google Scholar]
- Antosz W, Pfab A, Ehrnsberger HF, Holzinger P, Köllen K, Mortensen SA, Bruckmann A, Schubert T, Längst G, Griesenbeck J, et al. (2017) The composition of the Arabidopsis RNA polymerase II transcript elongation complex reveals the interplay between elongation and mRNA processing factors. Plant Cell 29: 854–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhn L, Munnerlyn A, Grosschedl R (1997) ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRalpha enhancer function. Genes Dev 11: 640–653 [DOI] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science 265: 615–621 [DOI] [PubMed] [Google Scholar]
- Canto T, Uhrig JF, Swanson M, Wright KM, MacFarlane SA (2006) Translocation of Tomato bushy stunt virus P19 protein into the nucleus by ALY proteins compromises its silencing suppressor activity. J Virol 80: 9064–9072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MQ, Zhang AH, Zhang Q, Zhang BC, Nan J, Li X, Liu N, Qu H, Lu CM, Sudmorgen, et al. (2012) Arabidopsis NMD3 is required for nuclear export of 60S ribosomal subunits and affects secondary cell wall thickening. PLoS ONE 7: e35904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David R, Burgess A, Parker B, Li J, Pulsford K, Sibbritt T, Preiss T, Searle IR (2017) Transcriptome-wide mapping of RNA 5-methylcytosine in Arabidopsis mRNAs and noncoding RNAs. Plant Cell 29: 445–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufu K, Livingstone MJ, Seebacher J, Gygi SP, Wilson SA, Reed R (2010) ATP is required for interactions between UAP56 and two conserved mRNA export proteins, Aly and CIP29, to assemble the TREX complex. Genes Dev 24: 2043–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürr J, Lolas IB, Sørensen BB, Schubert V, Houben A, Melzer M, Deutzmann R, Grasser M, Grasser KD (2014) The transcript elongation factor SPT4/SPT5 is involved in auxin-related gene expression in Arabidopsis. Nucleic Acids Res 42: 4332–4347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis KE, Lam SY, Copenhaver GP (2006) Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methylesterase gene. Plant Physiol 142: 1004–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaouar O, Germain H (2013) mRNA export: threading the needle. Front Plant Sci 4: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield D, Izaurralde E (2002) REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J Cell Biol 159: 579–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain H, Qu N, Cheng YT, Lee E, Huang Y, Dong OX, Gannon P, Huang S, Ding P, Li Y, et al. (2010) MOS11: a new component in the mRNA export pathway. PLoS Genet 6: e1001250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovanov AP, Hautbergue GM, Tintaru AM, Lian LY, Wilson SA (2006) The solution structure of REF2-I reveals interdomain interactions and regions involved in binding mRNA export factors and RNA. RNA 12: 1933–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovanov AP, Hautbergue GM, Wilson SA, Lian LY (2004) A simple method for improving protein solubility and long-term stability. J Am Chem Soc 126: 8933–8939 [DOI] [PubMed] [Google Scholar]
- Gong Z, Dong CH, Lee H, Zhu J, Xiong L, Gong D, Stevenson B, Zhu JK (2005) A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 17: 256–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasser KD, Grimm R, Ritt C (1996) Maize chromosomal HMGc: two closely related structure-specific DNA-binding proteins specify a second type of plant high mobility group box protein. J Biol Chem 271: 32900–32906 [DOI] [PubMed] [Google Scholar]
- Gromadzka AM, Steckelberg AL, Singh KK, Hofmann K, Gehring NH (2016) A short conserved motif in ALYREF directs cap- and EJC-dependent assembly of export complexes on spliced mRNAs. Nucleic Acids Res 44: 2348–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronenborn AM, Filpula DR, Essig NZ, Achari A, Whitlow M, Wingfield PT, Clore GM (1991) A novel, highly stable fold of the immunoglobulin binding domain of streptococcal protein G. Science 253: 657–661 [DOI] [PubMed] [Google Scholar]
- Hautbergue GM, Hung ML, Golovanov AP, Lian LY, Wilson SA (2008) Mutually exclusive interactions drive handover of mRNA from export adaptors to TAP. Proc Natl Acad Sci USA 105: 5154–5159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath CG, Viphakone N, Wilson SA (2016) The role of TREX in gene expression and disease. Biochem J 473: 2911–2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammel C, Thomaier M, Sørensen BB, Schubert T, Längst G, Grasser M, Grasser KD (2013) Arabidopsis DEAD-box RNA helicase UAP56 interacts with both RNA and DNA as well as with mRNA export factors. PLoS ONE 8: e60644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J. (2012) mRNA export and the TREX complex. Biochim Biophys Acta 1819: 507–513 [DOI] [PubMed] [Google Scholar]
- Katahira J, Inoue H, Hurt E, Yoneda Y (2009) Adaptor Aly and co-adaptor Thoc5 function in the Tap-p15-mediated nuclear export of HSP70 mRNA. EMBO J 28: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler A, Hurt E (2007) Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 8: 761–773 [DOI] [PubMed] [Google Scholar]
- Köster T, Marondedze C, Meyer K, Staiger D (2017) RNA-binding proteins revisited: the emerging Arabidopsis mRNA interactome. Trends Plant Sci 22: 512–526 [DOI] [PubMed] [Google Scholar]
- Lolas IB, Himanen K, Grønlund JT, Lynggaard C, Houben A, Melzer M, Van Lijsebettens M, Grasser KD (2010) The transcript elongation factor FACT affects Arabidopsis vegetative and reproductive development and genetically interacts with HUB1/2. Plant J 61: 686–697 [DOI] [PubMed] [Google Scholar]
- Longman D, Johnstone IL, Cáceres JF (2003) The Ref/Aly proteins are dispensable for mRNA export and development in Caenorhabditis elegans. RNA 9: 881–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Tang X, Tian G, Wang F, Liu K, Nguyen V, Kohalmi SE, Keller WA, Tsang EW, Harada JJ, et al. (2010) Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J 61: 259–270 [DOI] [PubMed] [Google Scholar]
- Luo ML, Zhou Z, Magni K, Christoforides C, Rappsilber J, Mann M, Reed R (2001) Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413: 644–647 [DOI] [PubMed] [Google Scholar]
- Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R (2005) Recruitment of the human TREX complex to mRNA during splicing. Genes Dev 19: 1512–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier I, Richards EJ, Evans DE (2017) Cell biology of the plant nucleus. Annu Rev Plant Biol 68: 139–172 [DOI] [PubMed] [Google Scholar]
- Merkle T. (2011) Nucleo-cytoplasmic transport of proteins and RNA in plants. Plant Cell Rep 30: 153–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Pan H, Liu S, Tang D (2012) HPR1, a component of the THO/TREX complex, plays an important role in disease resistance and senescence in Arabidopsis. Plant J 69: 831–843 [DOI] [PubMed] [Google Scholar]
- Parry G. (2015) The plant nuclear envelope and regulation of gene expression. J Exp Bot 66: 1673–1685 [DOI] [PubMed] [Google Scholar]
- Pedersen DS, Merkle T, Marktl B, Lildballe DL, Antosch M, Bergmann T, Tönsing K, Anselmetti D, Grasser KD (2010) Nucleocytoplasmic distribution of the Arabidopsis chromatin-associated HMGB2/3 and HMGB4 proteins. Plant Physiol 154: 1831–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña C, Hurt E, Panse VG (2017) Eukaryotic ribosome assembly, transport and quality control. Nat Struct Mol Biol 24: 689–699 [DOI] [PubMed] [Google Scholar]
- Pendle AF, Clark GP, Boon R, Lewandowska D, Lam YW, Andersen J, Mann M, Lamond AI, Brown JW, Shaw PJ (2005) Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol Biol Cell 16: 260–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues JP, Rode M, Gatfield D, Blencowe BJ, Carmo-Fonseca M, Izaurralde E (2001) REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc Natl Acad Sci USA 98: 1030–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P, Brown J (2012) Nucleoli: composition, function, and dynamics. Plant Physiol 158: 44–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen BB, Ehrnsberger HF, Esposito S, Pfab A, Bruckmann A, Hauptmann J, Meister G, Merkl R, Schubert T, Längst G, et al. (2017) The Arabidopsis THO/TREX component TEX1 functionally interacts with MOS11 and modulates mRNA export and alternative splicing events. Plant Mol Biol 93: 283–298 [DOI] [PubMed] [Google Scholar]
- Sprunck S, Rademacher S, Vogler F, Gheyselinck J, Grossniklaus U, Dresselhaus T (2012) Egg cell-secreted EC1 triggers sperm cell activation during double fertilization. Science 338: 1093–1097 [DOI] [PubMed] [Google Scholar]
- Strässer K, Hurt E (2000) Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J 19: 410–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strässer K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondón AG, Aguilera A, Struhl K, Reed R, et al. (2002) TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304–308 [DOI] [PubMed] [Google Scholar]
- Stutz F, Bachi A, Doerks T, Braun IC, Séraphin B, Wilm M, Bork P, Izaurralde E (2000) REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA 6: 638–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi I, Ohno M (2008) ATP-dependent recruitment of export factor Aly/REF onto intronless mRNAs by RNA helicase UAP56. Mol Cell Biol 28: 601–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng W, Zhang H, Wang W, Li D, Wang M, Liu J, Zhang H, Zheng X, Zhang Z (2014) ALY proteins participate in multifaceted Nep1Mo-triggered responses in Nicotiana benthamiana and Arabidopsis thaliana. J Exp Bot 65: 2483–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrig JF, Canto T, Marshall D, MacFarlane SA (2004) Relocalization of nuclear ALY proteins to the cytoplasm by the tomato bushy stunt virus P19 pathogenicity protein. Plant Physiol 135: 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viphakone N, Hautbergue GM, Walsh M, Chang CT, Holland A, Folco EG, Reed R, Wilson SA (2012) TREX exposes the RNA-binding domain of Nxf1 to enable mRNA export. Nat Commun 3: 1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler F, Schmalzl C, Englhart M, Bircheneder M, Sprunck S (2014) Brassinosteroids promote Arabidopsis pollen germination and growth. Plant Reprod 27: 153–167 [DOI] [PubMed] [Google Scholar]
- Walsh MJ, Hautbergue GM, Wilson SA (2010) Structure and function of mRNA export adaptors. Biochem Soc Trans 38: 232–236 [DOI] [PubMed] [Google Scholar]
- Weidtkamp-Peters S, Stahl Y (2017) The use of FRET/FLIM to study proteins interacting with plant receptor kinases. Methods Mol Biol 1621: 163–175 [DOI] [PubMed] [Google Scholar]
- Wickramasinghe VO, Laskey RA (2015) Control of mammalian gene expression by selective mRNA export. Nat Rev Mol Cell Biol 16: 431–442 [DOI] [PubMed] [Google Scholar]
- Xu C, Zhou X, Wen CK (2015) HYPER RECOMBINATION1 of the THO/TREX complex plays a role in controlling transcription of the REVERSION-TO-ETHYLENE SENSITIVITY1 gene in Arabidopsis. PLoS Genet 11: e1004956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XM, Meier I (2008) The nuclear pore comes to the fore. Trends Plant Sci 13: 20–27 [DOI] [PubMed] [Google Scholar]
- Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY, Li A, Wang X, Bhattarai DP, Xiao W, et al. (2017) 5-Methylcytosine promotes mRNA export: NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res 27: 606–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelina NE, Smith LM, Jones AME, Patel K, Kelly KA, Baulcombe DC (2010) Putative Arabidopsis THO/TREX mRNA export complex is involved in transgene and endogenous siRNA biosynthesis. Proc Natl Acad Sci USA 107: 13948–13953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Vinciguerra P, Strahm Y, Stutz F (2001) The yeast hnRNP-Like proteins Yra1p and Yra2p participate in mRNA export through interaction with Mex67p. Mol Cell Biol 21: 4219–4232 [DOI] [PMC free article] [PubMed] [Google Scholar]