Physcomitrella patens RECX maintains mitochondrial genome stability by interacting with the mitochondrial recombinase RECA1.
Abstract
The chloroplast and mitochondrial genomes are essential for photosynthesis and respiration, respectively. RECA and RECG, which are plant-specific homologs of the bacterial homologous recombination repair proteins RecA and RecG, maintain organelle genome stability by suppressing aberrant recombination between short dispersed repeats (SDRs) in the moss Physcomitrella patens. In this study, we analyzed the plant-specific factor RECX, a homolog of bacterial RecX that regulates RecA. RECX fused to GFP colocalized with mitochondrial RECA1 and chloroplast RECA2 on mitochondrial and chloroplast nucleoids, respectively. Knockout (KO) and overexpression (OEX) of RECX did not alter the P. patens morphological phenotype. Analysis of mitochondrial DNA, however, showed that products from recombination between SDRs increased significantly in RECX OEX mitochondria and modestly in RECX KO mitochondria. By contrast, analysis of chloroplast DNA revealed no substantial alteration in the number of products from recombination between SDRs in RECX KO and OEX chloroplasts. Yeast two-hybrid analysis revealed interactions between RECX and RECA1 and between RECX and RECA2. Expression profiles showed a positive correlation between RECX and factors maintaining the stability of both organelle genomes and RECA1. Collectively, these results suggest that RECX maintains mitochondrial genome stability, likely by modulating RECA1 activity, and that the compromised function of RECX induces mitochondrial genome instability.
Plant chloroplasts and mitochondria are energy-producing organelles that possess their own genomic DNA, ∼100 to 200 kb and 100 to 500 kb in size, respectively, although mitochondrial DNA (mtDNA) is more variable, ranging from ∼13 kb in some algae to ∼11 Mb in some expanded angiosperm mtDNA genomes (Smith and Keeling, 2015). As a result of gene transfer from organelles to the nucleus during evolution, most genes encoding proteins involved in organelle functions are encoded by nuclear DNA. However, organellar genomes contain genes essential for photosynthesis and respiration as well as for gene expression in the organelle. In particular, organelle genomes encode genes for redox regulation according to the colocation of redox regulation as proposed by Allen (2015). Although organelle DNA generally forms single or multiple circular structures, it can exist as a mixture of circular and linear molecules that sometimes exceeds the concatemeric genome size (Bendich, 2004; Morley and Nielsen, 2017). Like bacterial chromosomal DNA, organelle DNA is packed into DNA-protein complexes called nucleoids, in which proteins involved in transcription, translation, and DNA metabolism are associated in addition to core nucleoid proteins (Powikrowska et al., 2014).
Homologous recombination repair (HRR), a ubiquitous mechanism conserved from bacteria to eukaryotes, maintains genome integrity by repairing DNA double strand breaks and stalled or collapsed replication forks. In bacterial HRR, RecA recombinase binds to a processed single-stranded DNA and then recombines with a homologous sequence to perform strand exchange (Lusetti and Cox, 2002). The resulting branched structures are the target of RecG and other branch-specific DNA helicases (Whitby et al., 1993). Plant nuclear DNA encodes orthologs of bacterial RecA and RecG proteins (RECA and RECG, respectively) that are not found in animal or fungal nuclear DNA (Cerutti et al., 1992; Khazi et al., 2003; Lin et al., 2006; Odahara et al., 2015b). Land plants have a few copies of RECA and a single copy of RECG. Along with most of the proteins functioning in chloroplasts and mitochondria, plant RECA and RECG are targeted to the chloroplast and/or mitochondria.
In the moss Physcomitrella patens, a model organism for studying plant functions with efficient nuclear gene targeting (Schaefer, 2002), two RECA genes, namely RECA1 and RECA2, are found in the nuclear genome. RECA1 is targeted to mitochondria, and RECA1 knockout (KO) mutants display a severe delay in growth and development as well as defective mitochondria. RECA1 KO mutants also exhibit gross mtDNA rearrangements due to induced recombination between short dispersed repeats (SDRs; less than 100 bp) ranging from 13 to 90 bp (Odahara et al., 2009, 2015b), and they show remarkable variation in gene dosage in mtDNA (Odahara et al., 2015b). RECA2 is a chloroplast-targeted protein (Inouye et al., 2008), and RECA2 KO mutants display a modest delay in growth (Odahara et al., 2015a). Although the P. patens chloroplast DNA (cpDNA) contains only a few pairs of relatively long repeats (more than 40 bp), in contrast to P. patens mtDNA, which contains 20 pairs of relatively long repeats (Odahara et al., 2009), RECA2 KO leads to chloroplast genome instability. This is due to the induction of recombination between these relatively long cpDNA repeats as well as between repeats shorter than 30 bp (Odahara et al., 2015a).
RECG is a dual-targeted protein, and a similar growth and developmental delay is observed in P. patens RECG KO strains compared with RECA1 KO strains, albeit to a lesser extent (Odahara et al., 2015b). Similar to KO of RECA1 or RECA2, KO of RECG leads to instability of both organelle genomes due to the induction of recombination between SDRs, and the repeats involved in organelle genome instability are highly similar to those in RECA1 or RECA2 KO mutants (Odahara et al., 2015a, 2015b). The fact that RECA and RECG KO similarly lead to organelle genome instability through aberrant recombination suggests that the HRR pathway might maintain organelle genome stability by suppressing recombination between SDRs.
MutS homolog1 (MSH1) is another factor that helps to maintain organelle genome stability in Arabidopsis (Arabidopsis thaliana; Abdelnoor et al., 2003). Arabidopsis has a single MSH1 gene, and its mutation results in variegated phenotypes and the rearrangement of organelle DNA attributed to aberrant recombination between SDRs (Abdelnoor et al., 2003; Xu et al., 2011). P. patens is unique in that it possesses two MSH1 genes, MSH1A and MSH1B, and MSH1A lacks the C-terminal GIY-YIG endonuclease domain. Interestingly, the P. patens MSH1A and MSH1B double KO mutant and both single mutants show no obvious differences in morphological phenotype. However, MSH1 double and MSH1B single mutants exhibit organelle genome instability due to recombination between SDRs. Double mutation analysis showed that MSH1B interacts genetically with the HRR factors RECA2 and RECG in chloroplasts, and with RECG in mitochondria, to suppress recombination between SDRs (Odahara et al., 2017).
In bacteria, recX often is located downstream of recA (Sano, 1993; De Mot et al., 1994; Papavinasasundaram et al., 1997) and is cotranscribed with recA (Pagès et al., 2003). Moreover, recX is largely conserved among bacteria but is absent in some bacterial organisms, such as α-proteobacteria, ε-proteobacteria, cyanobacteria, Mollicutes, and Chlamydiae species (Rocha et al., 2005). Loss of recX modestly increases sensitivity to UV light and other agents that damage DNA and decreases RecA-dependent genetic recombination (Stohl et al., 2003; Cárdenas et al., 2012). Overproduction of RecX protein affects RecA-dependent phenomena such as SOS induction and P1 transduction (Stohl et al., 2003). RecX binds to RecA and RecA nucleofilaments in vitro, inhibits filament extension, and promotes filament disassembly to regulate RecA activity, including strand exchange (Venkatesh et al., 2002; Drees et al., 2004; Ragone et al., 2008). This bacterial RecX acts in the HRR pathway by modulating the activity of RecA.
In this work, we identified homologs of bacterial RecX in plant nuclear genomes. Using P. patens, we characterized RECX by generating RECX KO and overexpression (OEX) strains. Our results demonstrate the role of RECX in maintaining mitochondrial genome stability, probably by modulating the activity of a plant mitochondrial RecA homolog.
RESULTS
RECX Is a Plant-Specific Homolog of Bacterial RecX
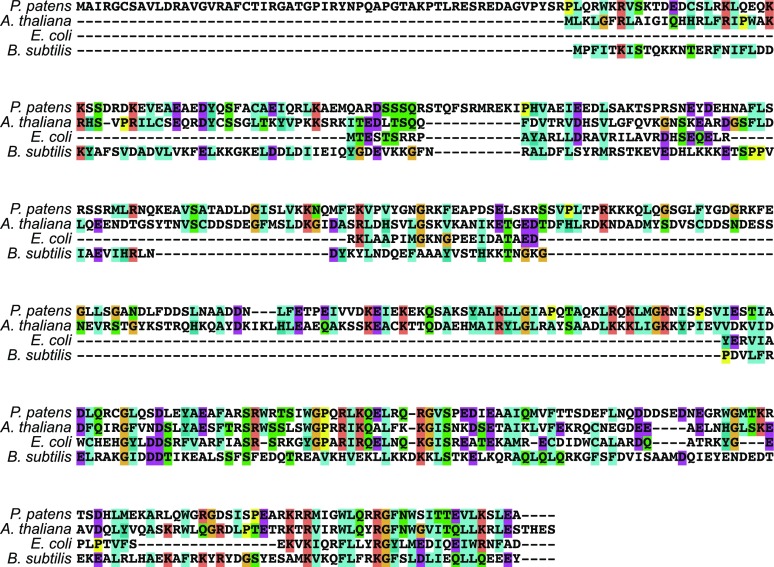
Using the Escherichia coli RecX protein sequence, a BLAST search identified genes encoding similar proteins in plant genomes but not in animal or fungal genomes. These RECX sequences were present as single-copy genes in plants from green algae to angiosperms but not in the red alga Cyanidioschyzon meloraeas (Supplemental Table S1). Multiple alignment showed that the C-terminal region of plant RECX and bacterial RecX proteins is highly conserved, but the N-terminal region varies in terms of the amino acid sequence and length (Fig. 1). No DNA-binding motifs or domains were found in plant RECX proteins. Plant RECA genes are derived from their cyanobacteria and α-proteobacteria ancestors (Lin et al., 2006), and none of the bacteria investigated possess recX genes except for Agrobacterium tumefaciens (Rocha et al., 2005). It was difficult to generate a reliable phylogenetic tree of plant RECX and bacterial RecX proteins due to variations in amino acid sequences and structures (Fig. 1), but the analysis indicates a single clade of plant RECX proteins (Supplemental Fig. S1). Unlike bacterial RecX proteins, the extended N-terminal region of plant RECX proteins indicates the presence of a transit peptide and/or a presequence for chloroplast localization and/or mitochondrial localization. In silico prediction of the subcellular localization of plant RECX proteins showed that most appear to be found in mitochondria (Supplemental Table S1).
Figure 1.
Multiple alignment of plant RECX and bacterial RecX protein sequences. Protein sequences of RECX from P. patens and Arabidopsis, alongside RecX from E. coli and Bacillus subtilis, were aligned using ClustalX2. Identical or similar amino acids are colored according to their biochemical properties.
RECX Associates with Organelle Nucleoids and with RECA
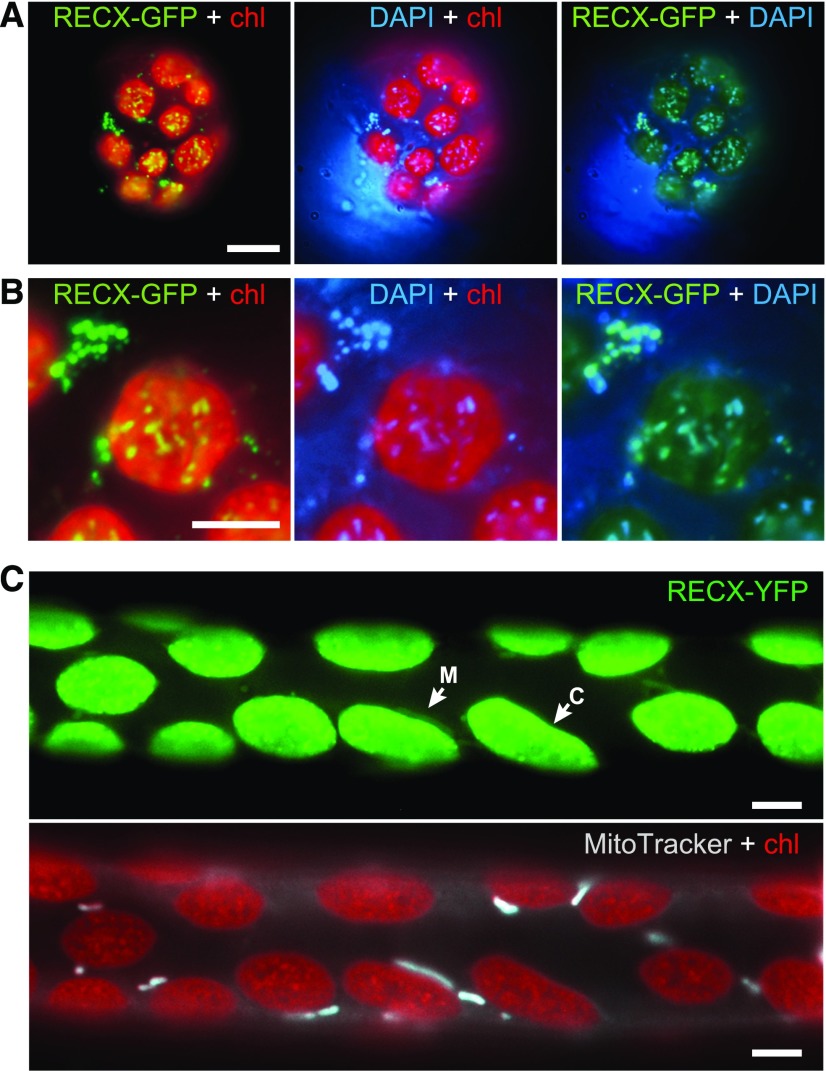
To investigate the intracellular localization of RECX, RECX tagged with GFP was transiently expressed under the control of a modified 35S promoter in P. patens protoplast cells. Fluorescence microscopy showed that RECX-GFP formed multiple punctate structures both inside and outside chloroplasts (Fig. 2, A and B). RECX-GFP in chloroplasts corresponded to chloroplast nucleoids, which were dispersed throughout the chloroplast as small granular structures following staining with DAPI, a DNA-specific dye (Fig. 2B). Similarly, the GFP signal outside chloroplasts corresponded to mitochondrial nucleoids, which appeared as extrachloroplastic cytosolic small granules following DAPI staining (Fig. 2B). These results suggest that RECX associates with organelle nucleoids in P. patens. Next, we analyzed the intracellular localization of RECX in RECX-YFP knock-in plants generated by gene targeting (Supplemental Fig. S2), which is achieved via efficient homologous recombination in nuclei of P. patens cells (Schaefer, 2002). Analysis of RECX-YFP knock-in protonemal cells confirmed that RECX was a dual-targeted protein and formed punctate structures in chloroplasts (Fig. 2C).
Figure 2.
Intracellular localization of RECX. A and B, Fluorescence microscopy of RECX-GFP in protoplast cells. RECX fused in frame with GFP was expressed under the control of the P7113 promoter in P. patens protoplast cells. Cells were stained with the 4,6-diamidino-2-phenylindole (DAPI) DNA-specific dye and observed using fluorescence microscopy. Chlorophyll autofluorescence (chl) was observed to discriminate chloroplasts. Overview images (A) and localization of RECX-GFP in chloroplast and mitochondrial nucleoids (B) are shown. C, Fluorescence microscopy of RECX-YFP knock-in cells. Protonemal cells stained with MitoTracker Orange were observed using fluorescence microscopy. Arrows labeled with C and M indicate the localization of YFP signals in chloroplasts and mitochondria, respectively. Bars = 10 μm in A and 5 μm in B and C.
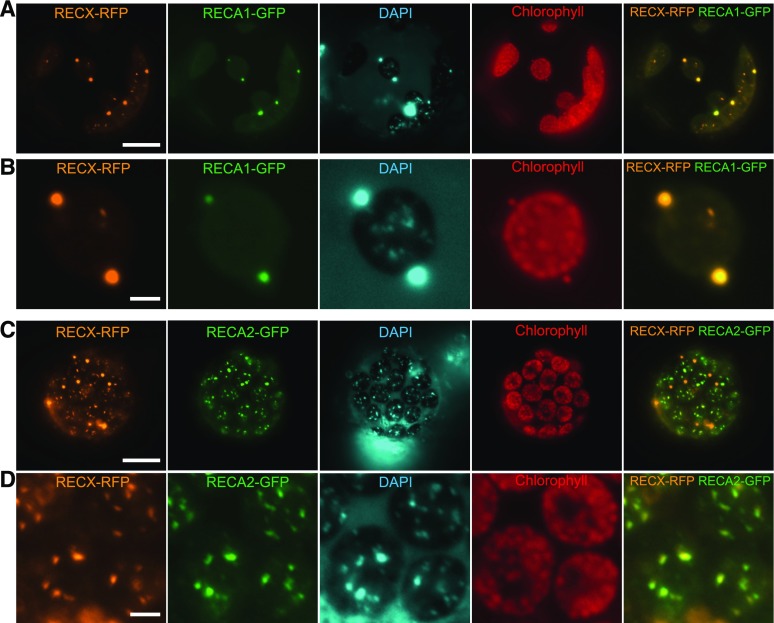
RECA2 was localized to chloroplast nucleoids, as demonstrated by the localization of its full-length product tagged to GFP in protoplast cells (Fig. 3, C and D; Inouye et al., 2008). A similar analysis of full-length RECA1 tagged with GFP showed that RECA1-GFP signals corresponded with DAPI-stained mitochondrial nucleoids (Fig. 3A), suggesting that RECA1 associates with mitochondrial nucleoids. Next, we investigated the localization of RECX in protoplast cells expressing GFP-tagged RECA1 or RECA2. Fluorescence microscopy showed that RECX tagged with RFP was colocalized exclusively with both RECA1 and RECA2 foci formed on mitochondrial and chloroplast nucleoids, respectively (Fig. 3). These results suggest that RECX is an organelle nucleoid-associated protein that potentially functions with RECA1 and RECA2 on organelle nucleoids.
Figure 3.
Colocalization of RECX with RECA1 and RECA2 on organelle nucleoids. RECX tagged with RFP was coexpressed with RECA1-GFP (A and B) or RECA2-GFP (C and D) in P. patens protoplast cells. Individual RFP and GFP fluorescence signals (two left columns) are depicted alongside the DAPI DNA-specific dye, chlorophyll autofluorescence, and merged RFP/GFP signals (three right columns). A and C, Whole cell images. B and D, Closeup images of cells. Fluorescence leakage from the strong RFP signal can be seen in the chlorophyll fluorescence. Bars = 10 μm in A and C and 5 μm in B and D.
KO and OEX of RECX
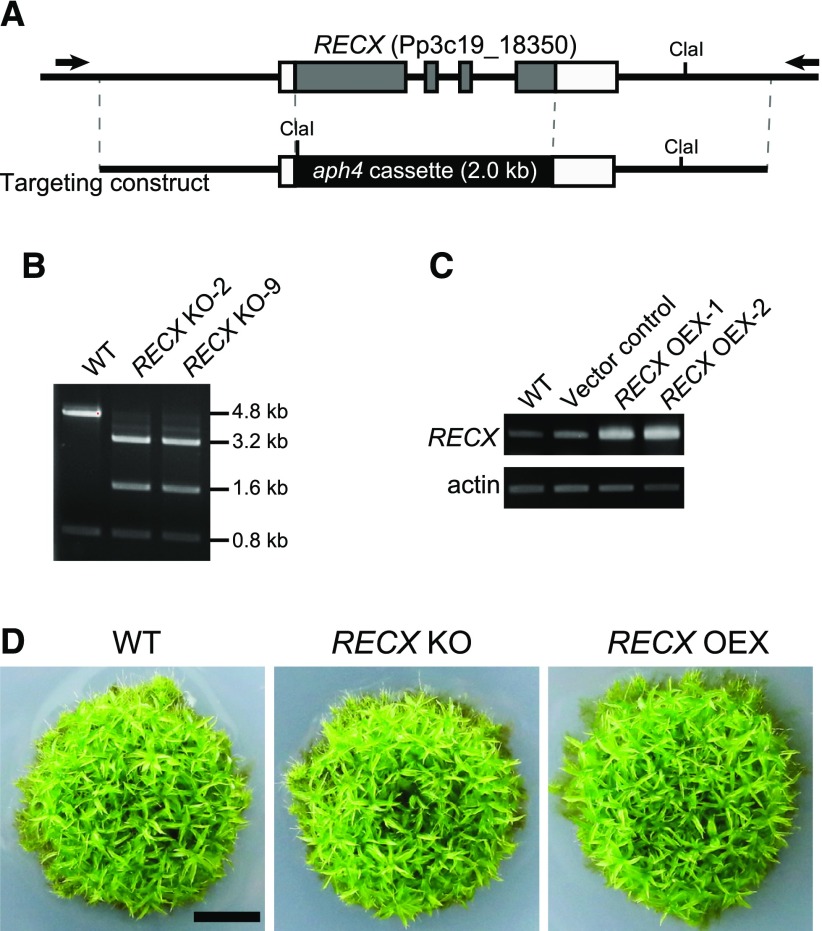
We knocked out and overexpressed RECX in P. patens to investigate the role of RECX in plants. RECX KO was carried out by replacing the RECX coding region with an aph4 gene cassette, and RECX OEX was achieved by expressing RECX cDNA under the control of a modified 35S promoter from the ectopic neutral site (Fig. 4). P. patens initially forms a colony composed of protonemal cells on an agar plate, then develops gametophores. We obtained RECX KO and OEX plants and found that both grew normally and developed gametophores on agar plates, comparable with the wild type (Fig. 4). Thus, RECX KO and OEX plants displayed no morphological phenotypic differences.
Figure 4.
KO and OEX of RECX. A, Scheme showing KO of the RECX gene. The P. patens genomic RECX locus (top) and the targeting construct (bottom) are shown. Gray boxes indicate coding regions, white boxes indicate untranslated regions, and lines indicate intergenic regions. Primers (black arrows) and ClaI restriction enzyme sites used in B are indicated. B, PCR genotyping of RECX KO mutants. PCR amplicons of the RECX locus from the wild type (WT) and RECX KO mutants were digested with ClaI. C, Reverse transcription-PCR analysis of RECX OEX strains. Actin was amplified as an internal control. D, Comparison of growth and morphology. Wild-type, RECX KO, and RECX OEX strains were grown on BCDATG agar plates for 4 weeks. Bar = 5 mm.
Mitochondrial Genome Instability in RECX KO and OEX Plants
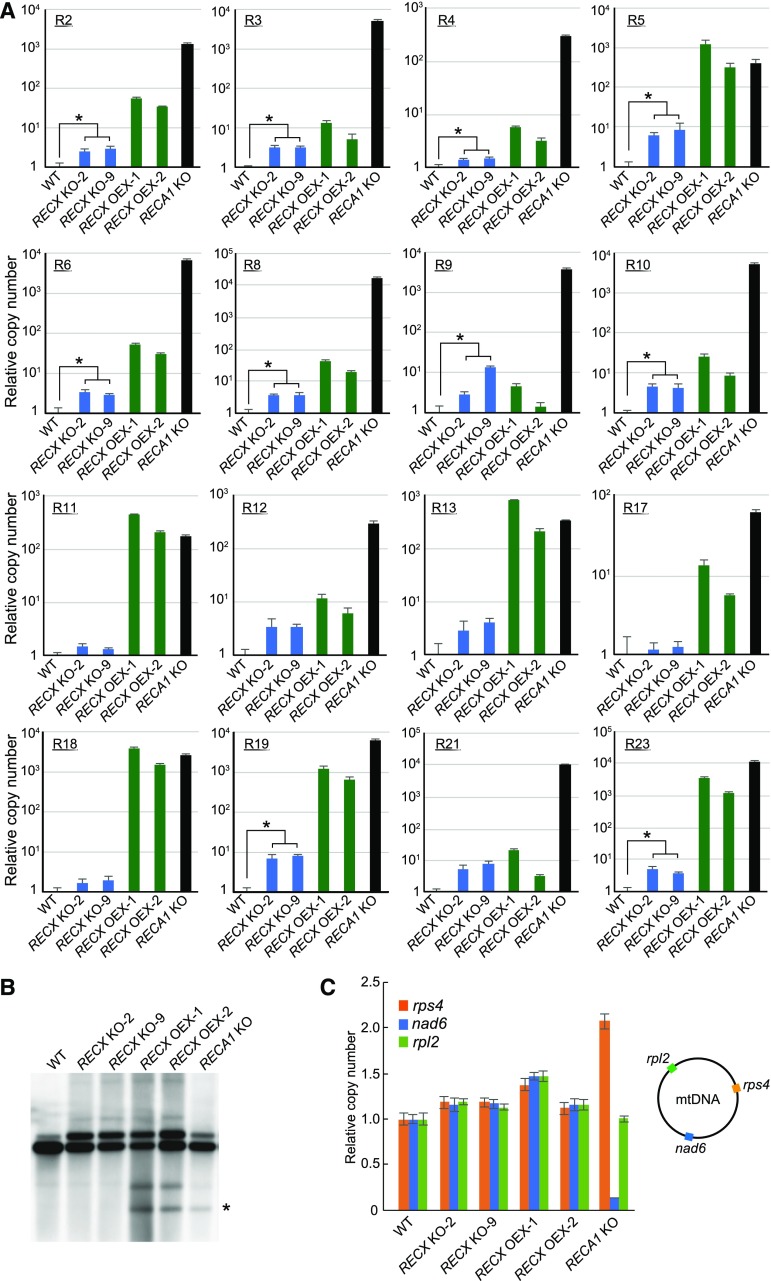
We analyzed mitochondrial genome stability in RECX KO and OEX plants, focusing on recombination between SDRs. In P. patens mtDNA, 20 pairs of SDRs of 46 to 90 bp in length were identified and named R1 to R20 (Odahara et al., 2009). KO of RECA1 induced the accumulation of products from recombination between most of these repeats (Odahara et al., 2009, 2015b). Repeats were redefined as those longer than 30 bp with a maximum 1-bp mismatch. We searched for such repeats in P. patens mtDNA (Table I; Supplemental Fig. S3) and evaluated the accumulation of recombination products between all these repeats, including R1 to R20, by performing qPCR and DNA gel blotting. qPCR analysis of products from recombination between repeats R2 to R23 revealed significantly increased accumulation of these recombination products in RECX OEX plants to amounts comparable to that in RECA1 KO in some cases (Fig. 5A). By contrast, the amounts of these recombination products were not changed significantly in RECX KO plants; there was no decrease in any of the products and only a modest increase in several repeats in RECX KO plants (Fig. 5A). We also observed an accumulation of products from recombination between nad9-nad7 repeats, a hallmark of mtDNA instability resulting from KO of RECA1, in RECX OEX and RECA1 KO plants, by DNA gel blotting (Fig. 5B). These results collectively show that recombination between mitochondrial SDRs was markedly induced in RECX OEX plants, and this recombination was similar to that induced in RECA1 KO plants in terms of both sites and efficiency; however, recombination was increased modestly rather than decreased in RECX KO plants.
Table I. Repeated sequences in P. patens mtDNA (at least 30 bp, 1-bp mismatch permitted).
Repeated sequences were identified by REPuter, and palindromic sequences and tandem repeats were omitted from the list. R1 to R20 were described by Odahara et al. (2009), and R21 to R24 are newly identified. Recombination products between the repeats were analyzed by quantitative real-time PCR (qPCR) or DNA gel blotting (Gel blot). DR, Direct repeats; IR, inverted repeats; N/A, not applicable for qPCR due to insufficient amplification of products.
| Repeats | Assay | Repeat 1 |
Repeat 2 |
Direction | Mismatch | E Value | ||
|---|---|---|---|---|---|---|---|---|
| Length | Position | Length | Position | |||||
| bp | bp | |||||||
| R1 | Gel blot | 79 | 79,450 | 79 | 90,132 | DR | 0 | 8.54E-39 |
| R2 | qPCR | 78 | 43,155 | 78 | 79,454 | DR | 1 | 3.17E-35 |
| R3 | qPCR | 75 | 43,155 | 75 | 90,136 | DR | 1 | 1.95E-33 |
| R5 | qPCR | 57 | 43,453 | 57 | 70,208 | DR | 1 | 1.02E-22 |
| R6 | qPCR | 56 | 50,034 | 56 | 73,796 | DR | 1 | 4.01E-22 |
| R8 | qPCR | 50 | 50,375 | 50 | 74,201 | DR | 0 | 2.46E-21 |
| R9 | qPCR | 47 | 20,077 | 47 | 102,360 | DR | 0 | 1.58E-19 |
| R11 | qPCR | 51 | 70,214 | 51 | 80,507 | DR | 1 | 3.74E-19 |
| R12 | qPCR | 48 | 944 | 48 | 9,303 | DR | 1 | 2.25E-17 |
| R4 | qPCR | 44 | 49,773 | 44 | 73,534 | DR | 1 | 5.28E-15 |
| R17 | qPCR | 41 | 58,284 | 41 | 104,768 | IR | 1 | 3.15E-13 |
| R4 | qPCR | 40 | 49,804 | 39 | 73,565 | DR | 1 | 1.23E-12 |
| R13 | qPCR | 38 | 43,472 | 38 | 80,520 | DR | 1 | 1.87E-11 |
| R21 | qPCR | 35 | 2,314 | 35 | 26,780 | DR | 1 | 1.10E-09 |
| R22 | N/A | 34 | 65,453 | 34 | 72,964 | DR | 1 | 4.28E-09 |
| R23 | qPCR | 34 | 29,149 | 34 | 58,287 | DR | 1 | 4.28E-09 |
| R20 | N/A | 33 | 12,923 | 33 | 101,036 | IR | 1 | 1.66E-08 |
| R10 | qPCR | 31 | 52,628 | 31 | 103,632 | DR | 1 | 2.50E-07 |
| R15 | N/A | 31 | 1,671 | 31 | 41,363 | IR | 1 | 2.50E-07 |
| R16 | N/A | 31 | 43,479 | 31 | 90,573 | DR | 1 | 2.50E-07 |
| R19 | qPCR | 30 | 27,052 | 30 | 29,571 | DR | 1 | 9.67E-07 |
| R24 | N/A | 30 | 37,364 | 30 | 61,690 | IR | 1 | 9.67E-07 |
| R14 | Gel blot | 58 | 80,499 | 58 | 90,545 | DR | 4 | 9.81E-17 |
| R18 | qPCR | 53 | 70,213 | 53 | 90,552 | DR | 5 | 4.59E-12 |
Figure 5.
Mitochondrial genome instability in RECX KO and OEX strains. A, qPCR analyses of products from recombination between mitochondrial SDRs in RECX KO and OEX strains. The relative copy numbers of products from recombination between repeats (R2–R23) are normalized against nuclear actin. Data are means of three technical replicates ± sd. Asterisks indicate significant differences (P < 0.05, Student’s t test). B, DNA gel-blot analysis of the configuration at the mitochondrial nad7 locus. SacII-digested total DNAs from wild-type (WT), RECX KO, and RECX OEX strains, and the RECA1 KO mutant, were probed with labeled nad7 DNA. The asterisk denotes products from recombination between nad9-nad7 repeats comprising R1 and R14. C, qPCR analysis of gene dosage at three mitochondrial loci. The relative copy numbers of mitochondrial rps4, nad6, and rpl2 are normalized against nuclear actin. Data are means of three technical replicates ± sd.
Besides the increased accumulation of products from recombination between SDRs in mitochondria, KO of RECA1 led to a dosage imbalance of mitochondrial genes that appeared as a decrease and an increase in nad6 and rps4 loci, respectively (Fig. 5C; Odahara et al., 2015b). qPCR evaluation of the copy number at three mitochondrial loci, namely rps4, nad6, and rpl2, revealed a modest increase at all loci in RECX KO and OEX plants but not in RECA1 KO plants (Fig. 5C), showing that RECX KO or OEX had a mild effect on the dosage imbalance of mitochondrial genes.
Stability of cpDNA in RECX KO and OEX Plants
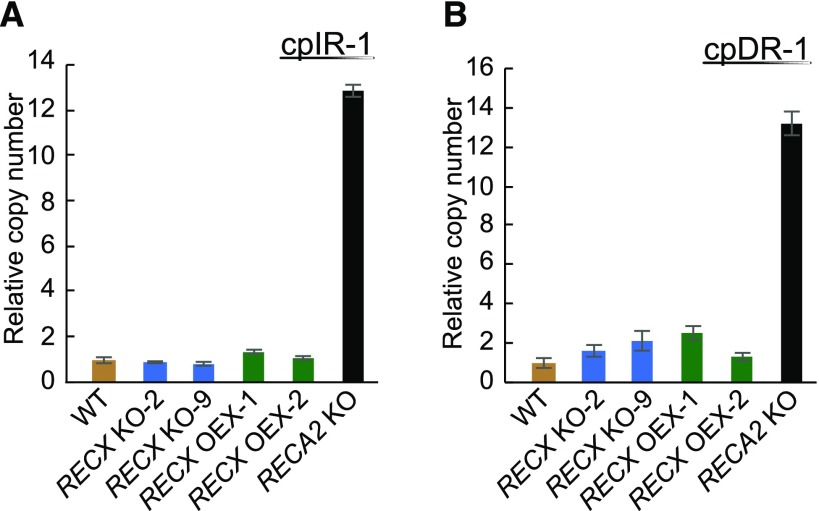
Compared with mtDNA, P. patens cpDNA possesses fewer relatively long (40–100 bp) repeats, except for palindromic sequences (Odahara et al., 2015a). Among the repeats, products from recombination between cpIR-1, located in rpl16 and trnG, and cpDR-1, located in psaA and psaB, are increased by KO of RECA2 (Odahara et al., 2015a) as well as by KO of RECG (Odahara et al., 2015b). Therefore, we measured the levels of these recombination products to test the involvement of RECX in the maintenance of chloroplast genome stability. qPCR analysis of cpDNA showed that neither KO nor OEX of RECX led to a significant alteration in the amount of cpIR-1 recombination products, compared with an ∼13-fold increase in products resulting from KO of RECA2 (Fig. 6). Similarly, qPCR analysis showed that KO or OEX of RECX had only a minor effect on the amount of cpDR-1 recombination products that were increased slightly in the RECA2 KO mutant (Fig. 6). These results suggest that RECX KO and OEX had only a minimal effect on the stability of cpDNA.
Figure 6.
Analysis of recombination between SDRs in chloroplasts. qPCR analyses of products from recombination between chloroplast SDRs is shown. The relative copy numbers of products from recombination between cpIR-1 (A) and cpDR-1 (B) are normalized against nuclear actin. Data are means of three technical replicates ± sd. WT, Wild type.
RECX Interacts with RECA1 and RECA2
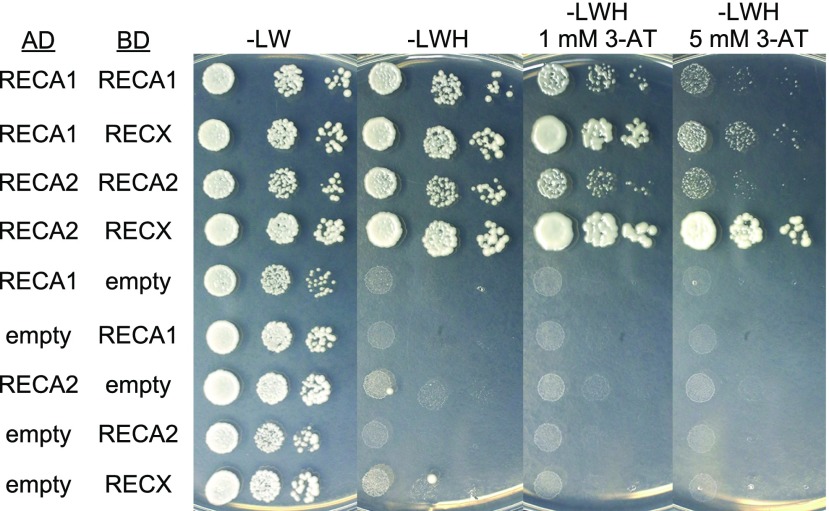
The colocalization of RECX with RECA proteins and the comparable mitochondrial genome instability in RECX OEX and RECA1 KO plants raise the possibility of protein-protein interaction between RECX and RECA. Thus, we used the yeast two-hybrid system to test protein-protein interactions between RECX and RECA1 or RECA2. Full-length proteins of RECX, RECA1, and RECA2 were produced in yeast cells. The results showed moderate interaction between RECA1 and RECA1 and between RECA2 and RECA2 (Fig. 7). The monomer-monomer interactions observed for RECA1 and RECA2 indicate polymerization, a characteristic feature of RecA family proteins (Lusetti and Cox, 2002). We also observed a strong protein-protein interaction between RECX and RECA1, which was retained in the presence of 1 mm 3-AT, a competitive inhibitor of the reporter HIS3 (Fig. 7). Moreover, we found an even stronger protein-protein interaction between RECX and RECA2 that was retained in the presence of 5 mm 3-AT (Fig. 7). These results indicate robust protein-protein interactions between RECX and RECA1 and between RECX and RECA2, in addition to the monomer-monomer interaction between RECA1 and RECA1 and between RECA2 and RECA2.
Figure 7.
RECX interacts with RECA1 and RECA2. Protein-protein interactions were investigated by yeast two-hybrid assay with HIS3 as a reporter gene. Yeast strains harboring genes fused to the activation domain (AD) and the DNA-binding domain (BD), or individual AD/BD gene fusions alongside empty vector controls, were spotted onto standard medium lacking Leu and Trp (-LW) or selective medium lacking Leu, Trp, and His (-LWH) following 10-fold serial dilution. 3-Amino-1,2,4-triazole (3-AT) was added to -LWH medium at two different concentrations (1 and 5 mm).
Expression Profiles of RECX and Other Genes
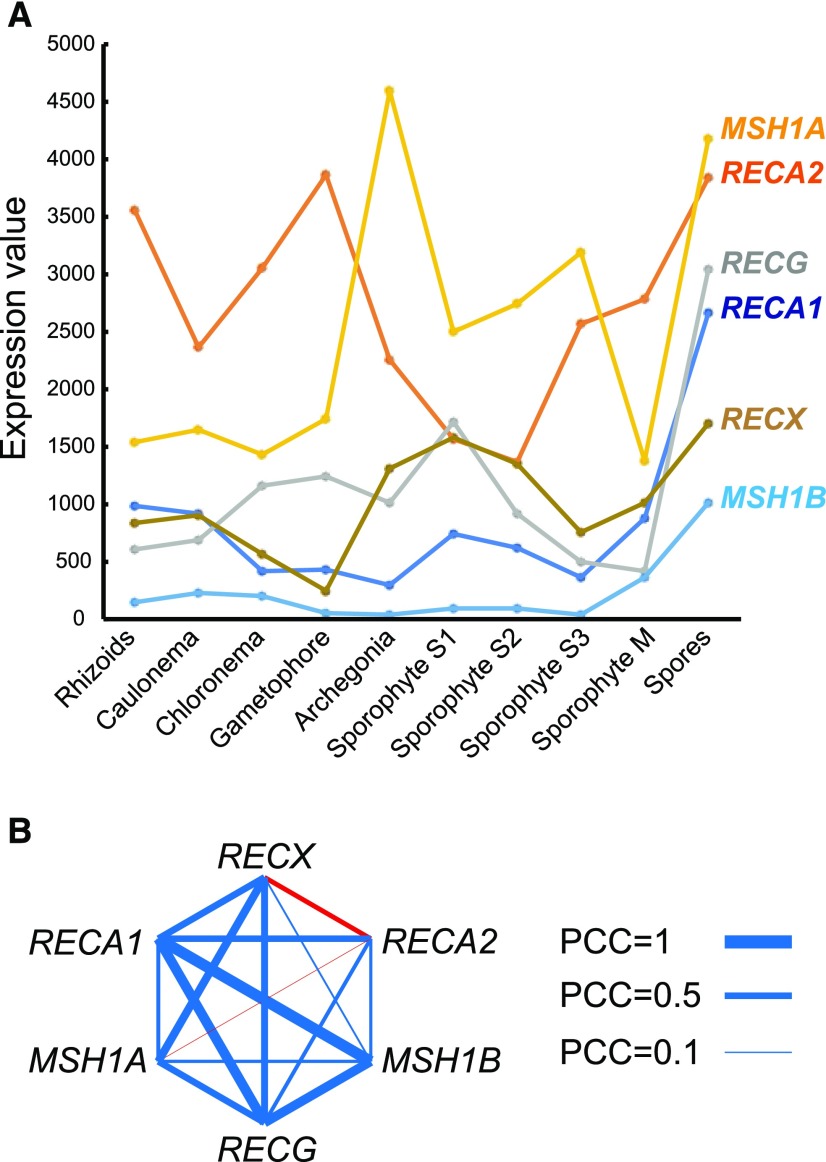
Expression profiles of RECX and other genes involved in the maintenance of organelle genome stability were retrieved from the Physcomitrella eFP Browser that consists of microarray data from different tissue types (Ortiz-Ramírez et al., 2016), and correlations in expression were analyzed. The expression of RECX was widespread from vegetative growth tissues to reproductive tissues but lower in gametophores (Fig. 8A). RECA1 exhibited a similar expression profile and a moderate positive correlation with RECX expression (Fig. 8). By contrast, the expression of RECA2 was higher in rhizoids, gametophores, and spores, which correlates negatively with RECX expression (Fig. 8). However, the expression of MSH1B and RECG is correlated highly with RECA1 and moderately with RECX, with a marked increase in expression in spores (Fig. 8). Collectively, the analysis of expression profiles revealed a positive correlation between the expression of RECX and other genes, except for RECA2, and a positive correlation between RECA1, RECG, and MSH1B expression.
Figure 8.
Expression profiles and correlation analysis of RECX and other factors. A, Expression profiles of genes based on microarray data from the Physcomitrella eFP Browser. B, Expression correlation between genes using the Pearson’s correlation coefficient (PCC), indicated by lines. Line width denotes the PCC value as exemplified at right. Blue and red lines denote positive and negative interactions, respectively.
DISCUSSION
In this article, we describe a RECX in plants that, upon OEX, induces mitochondrial genome instability due to recombination between SDRs. However, the results suggest that its actual role is to maintain mitochondrial genome stability, probably by interacting with mitochondrial RECA1 and modulating RECA1 protein activity. RECX also interacts with chloroplast RECA2; however, no significant morphological phenotypic differences were observed in cpDNA following RECX KO or OEX.
Among eukaryotes, homologs of bacterial RecX are present in plant genomes but not in animal or fungal genomes, and RECA also is only found in plants and the slime mold Dictyostelium discoideum (Lin et al., 2006). Although phylogenetic analysis suggests that plant RECA genes originated from the cyanobacteria and α-proteobacteria ancestors of chloroplasts and mitochondria, respectively (Lin et al., 2006), recX genes are absent in these bacteria except for A. tumefaciens (Rocha et al., 2005). This suggests that plant RECX genes were acquired via horizontal gene transfer from bacteria other than cyanobacteria or α-proteobacteria or were derived from ancestral bacteria, but related extant cyanobacteria and α-proteobacteria have now lost their recX genes. The high sequence and structural variations in plant RECX and bacterial RecX protein sequences make it difficult to predict the origin of the plant RECX gene.
RNA and proteins involved in DNA and RNA metabolism are associated with organelle nucleoids (Powikrowska et al., 2014). Along with other proteins involved in organelle genome stability, RECX is associated with chloroplast and mitochondrial nucleoids even though it has no typical domains or motifs related to DNA binding. Our results showed that RECA1 and RECA2 were associated with mitochondrial and chloroplast nucleoids, respectively, and that RECX colocalized and interacted with RECA1 and RECA2 proteins. Thus, RECX appears to associate with organelle nucleoids by interacting with RECA1 and RECA2.
RECA1 and RECG KO mutants exhibit growth and developmental defects as well as mitochondrial defects (Odahara et al., 2009, 2015b). These phenotypes most likely are caused by gross mtDNA rearrangements attributed to recombination between SDRs, because the amount of most recombination products is increased in cells displaying severe phenotypes (Odahara et al., 2015b). Although RECX KO and OEX did not alter the morphological phenotype, mutants displayed increased mitochondrial genome instability, as evidenced by the accumulation of DNA products from recombination between SDRs. In particular, RECX OEX plants exhibited an ∼1,000-fold increase in recombination products from several mitochondrial repeats (R5, R11, R13, R18, R19, and R23) comparable with increases in RECA1 KO plants (Fig. 5A). However, the accumulation of products from recombination between other repeats in RECX OEX plants was much lower than that in the RECA1 KO mutant (Fig. 5A). Moreover, the severe dosage imbalance seen in the three mtDNA loci of RECA1 KO plants was not observed in RECX OEX mtDNA (Fig. 5C). Collectively, the level of mtDNA defects due to increased recombination between SDRs and the accompanying mtDNA dosage imbalance in RECX OEX plants likely failed to exceed the threshold for the appearance of visible phenotypes.
The increased accumulation of products from recombination between mitochondrial repeats in RECX OEX plants raises the possibility that RECX induces aberrant recombination in the wild type. Importantly, however, our results showed no significant decrease in mitochondrial recombination products in RECX KO plants (Fig. 5A). In the case of mitochondrial repeats R2, R3, R4, R6, R8, R9, R10, R19, and R23, products from these repeats were increased significantly in the RECX KO mutant, suggesting that RECX does not induce, but rather suppresses, such aberrant recombination in mitochondria in the wild type. In some bacteria, the RecX protein binds to the RecA filament formed on DNA, inhibiting filament extension and promoting filament disassembly, thereby modulating recombination activity (Drees et al., 2004; Ragone et al., 2008; Cárdenas et al., 2012). Our results showed strong protein-protein interaction between the RecA homolog RECA1 and RECX. Moreover, comprehensive analysis of the mtDNA products from recombination between repeats suggested that RECX OEX plants shared common recombination sites with RECA1 KO plants (Fig. 5A). Given the evidence from bacterial RecX and the model for the suppression of aberrant recombination by RECA1 (Odahara et al., 2009), our results suggest that RECX maintains mitochondrial genome stability, probably by inhibiting RECA1 activity to facilitate the optimal homologous recombination repair of stalled or collapsed replication forks.
The tagged RECX protein was localized not only to mitochondrial nucleoids but also to chloroplast nucleoids, where it was colocalized with RECA2 foci. Our yeast two-hybrid assay revealed a protein-protein interaction between RECX and RECA2 and indicates that the interaction between RECX and RECA2 is stronger than that between RECX and RECA1. Thus, based on the effects of RECX OEX on mitochondria, OEX of RECX would be expected to lead to a phenotype that is similar to that caused by KO of RECA2 in chloroplasts. Our analysis showed that RECX KO and OEX did not lead to significant increases in products from recombination between chloroplast SDRs, a characteristic and important phenotype of RECA2 KO (Odahara et al., 2015a). However, because RECA2 KO caused a milder phenotype consistent with a minor induction of aberrant recombination between chloroplastic SDRs than occurred following RECA1 KO (Odahara et al., 2009, 2015a), the phenotype caused by RECX OEX would be expected to be modest in chloroplasts if RECX inhibits RECA2 activity.
The eFP expression profiles of RECX and other genes involved in the maintenance of organelle genome stability revealed a positive relationship between their expression, except for RECA2. RECG, MSH1A, MSH1B, and RECX function in both chloroplasts and mitochondria (Odahara et al., 2015b, 2017), but their gene expression increased with the increasing expression of mitochondrial RECA1 rather than chloroplast RECA2. Given that the effects of RECG, MSH1B, or RECX KO were more prominent in mtDNA than in cpDNA (Odahara et al., 2015b, 2017), this stronger relationship with mitochondrial RECA1 suggests that the regulation of RECG, MSH1A, MSH1B, and RECX expression may be essential in mitochondria. The eFP data also revealed a broad expression of RECX and other genes in different kinds of tissues and increased expression of these genes in reproductive tissues, especially spores (Fig. 8A). Thus, the maintenance of organelle genome stability appears to be more active in these tissues and may be altered in different tissues and developmental stages.
Thus, in this study, we describe the RECX gene in plants and demonstrate that RECX OEX induces mitochondrial genome instability due to aberrant recombination between SDRs. In general, mutation of genes in mtDNA by recombination, when fixed, causes defective mitochondria, but rarely does it correlate with cytoplasmic male sterility (CMS) traits in angiosperms. CMS is phenotypically normal but is defective in pollen and, thus, important for plant breeding (Hanson and Bentolila, 2004). Sandhu et al. (2007) showed that modulation of MSH1 expression by RNA interference results in heritable CMS in tobacco (Nicotiana tabacum) and tomato (Solanum lycopersicum) plants. Our results here showed that RECX OEX led to higher induction of recombination in mtDNA than occurred upon MSH1 mutation in P. patens (Odahara et al., 2017). RECX OEX, which should be easier than RNA interference, may be another way to induce CMS in angiosperms.
In summary, our analysis identified the role of the plant RECX protein in the maintenance of mitochondrial genome stability: RECX suppresses aberrant recombination between SDRs, probably by interacting with and regulating RECA1 activity. Although RECX interacts with RECA2, our analyses could not identify any phenotypic differences in RECX KO plants or alterations in cpDNA. Future analysis of RECX KO and OEX mutants in conditions in which RECA2 KO results in severe phenotypic differences and/or in tissues in which RECA2 is highly expressed (as revealed by eFP) may determine the role of RECX in chloroplasts.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Physcomitrella patens ssp. patens was used in this study, and protonemata were cultivated on BCDAT agar medium (Nishiyama et al., 2000) supplemented with 0.5% (w/v) Glc (BCDATG) at 25°C under constant white light. For growth comparisons, small pieces of protonemal cells were inoculated onto BCDATG agar plates and cultivated for 4 weeks.
Extraction of DNA and RNA
Total genomic DNA and total RNA were extracted using the cetyl-trimethyl-ammonium bromide method (Murray and Thompson, 1980) and the RNeasy Plant Mini Kit (Qiagen), respectively. Protonemal cells cultivated on BCDATG agar medium for 4 d after homogenization and inoculation were used for the extraction of genomic DNA and total RNA. Reverse transcription was performed with total RNA treated with TURBO DNase (Thermo Fisher Scientific) and ReverTra Ace (Toyobo) to prepare first-strand cDNAs.
Sequence Analysis of Plant RECX and Bacterial RecX
Protein sequences of plant RECX and bacterial RecX were aligned with ClustalX2 (Larkin et al., 2007) and subjected to Molecular Evolutionary Genetic Analysis (Tamura et al., 2007) for construction of a phylogenetic tree.
Protoplast Transformation
Protonemal cells cultivated on BCDATG agar medium for 3 d were used for polyethylene glycol (PEG)-mediated protoplast transformation according to Nishiyama et al. (2000).
Generation of RECX-YFP Knock-In Strains
To construct the RECX-YFP knock-in plasmid, 5′ fragments amplified with primers P1 and P2 (primer sequences are listed in Supplemental Table S2) and 3′ fragments amplified with primers P3 and P4 were inserted on either side of the nptII cassette of pCTRN-NPTII 2 (accession no. AB697058). The plasmid was linearized and introduced into P. patens cells by PEG-mediated protoplast transformation. Transformants were selected on BCDAT agar medium containing G418. Primers P5 and P6 were used for PCR genotyping.
Fluorescence Microscopy
For tagging of proteins with sGFP or TagRFP, cDNA sequences amplified with primers P7 and P8 for RECX, P9 and P10 for RECA1, and P11 and P12 for RECA2 were fused in frame to sGFP or TagRFP and expressed under the control of the E7113 promoter (Mitsuhara et al., 1996). These plasmids were introduced into P. patens protoplast cells by PEG-mediated transformation. For analysis of the colocalization of tagged proteins, plasmids were mixed and introduced into protoplast cells, and cells were cultured and stained with 1 μg mL−1 DAPI. Mitochondria were analyzed by staining protonemal cells with 200 nm MitoTracker Orange (Thermo Fisher Scientific). Cells were observed with an epifluorescence microscope (Axio Imager 2; Zeiss).
KO and OEX of RECX
A construct for KO of RECX was prepared by inserting RECX 5′ fragments amplified with primers P13 and P14, and RECX 3′ fragments amplified with primers P15 and P16, into either side of the aph4 cassette of pTN186 (Addgene). A construct for OEX of RECX was prepared by inserting RECX cDNA fragments amplified with primers P17 and P18 into pPpMADS2-7113, which targets the nuclear MADS2 locus and expresses genes under the control of the E7113 promoter (Mitsuhara et al., 1996). These plasmids were linearized and introduced into P. patens cells by PEG-mediated protoplast transformation. Transformants were selected on BCDAT agar medium containing hygromycin B for RECX OEX or G418 for RECX KO. For PCR genotyping of RECX KO plants, PCR products amplified with primers P5 and P6 were digested with the restriction enzyme ClaI. Reverse transcription-PCR analysis of RECX transcripts was performed using first-strand cDNA with primers P19 and P20 for RECX and primers P21 and P22 for actin.
Analysis of mtDNA and cpDNA
qPCR analysis of organelle DNA was performed using genomic DNA extracted by the cetyl-trimethyl-ammonium bromide method, PowerUp SYBR Green Master Mix (Thermo Fisher Scientific), a 7500 Fast Real-Time PCR System (Thermo Fisher Scientific), and primers listed in Supplemental Table S3. For DNA gel-blot analysis, total genomic DNA digested with SacII was separated, transferred to a nylon membrane, and hybridized with digoxigenin-labeled nad7 probe prepared by PCR using primers P23 and P24. Hybridization of probes was performed at 37°C, and membranes were washed in 2× SSC containing 0.1% (w/v) SDS at 25°C and in 0.5× SSC with 0.1% (w/v) SDS at 65°C. The detection of digoxigenin-labeled probes was performed with Anti-DIG-Alkaline Phosphatase (Roche) and AttoPhos (Promega).
Yeast Two-Hybrid Analysis
cDNA was amplified with primers P25 and P26 for RECX, P27 and P28 for RECA1, and P29 and P30 for RECA2, and products were introduced into pGADT7 and pGBKT7 vectors (Clontech). The resulting plasmids were introduced into yeast strain PJ-69-4A (James et al., 1996). Yeast strains harboring plasmids were cultivated at 30°C in synthetic defined medium without Leu or Trp until the A600 (OD600) reached 0.15, at which point they were spotted onto synthetic defined medium without Leu, Trp, or His but containing 1 or 5 mm 3-AT.
eFP Analysis
Expression levels of genes were retrieved from the Physcomitrella eFP Browser (http://bar.utoronto.ca/efp_physcomitrella/cgi-bin/efpWeb.cgi) for the following gene identifiers: Pp1s172_79V6.1 for RECX, Pp1s98_157V6.1 for RECA1, Pp1s412_17V6.1 for RECA2, Pp1s290_9V6.1 for RECG, Pp1s222_41V6.1 for MSH1A, and Pp1s134_115V6.1 for MSH1B.
Accession Numbers
Sequence data from this article can be found in the GenBank data library under accession numbers LC333045 for P. patens RECX, BAA16560 for Escherichia coli RecX, and NP_388733 for Bacillus subtilis RecX as well as in the TAIR data library under accession number AT3G13226 for Arabidopsis RECX.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogenetic analysis of plant RECX and bacterial RecX proteins.
Supplemental Figure S2. Generation of RECX-YFP knock-in strains.
Supplemental Figure S3. Map of repeats in P. patens mtDNA.
Supplemental Table S1. In silico prediction of intracellular localization.
Supplemental Table S2. Oligomeric DNA primers used in this study.
Supplemental Table S3. Oligomeric DNA primers used for quantitative PCR.
Acknowledgments
We thank K. Tachikawa for technical assistance and T. Nishiyama, T. Fujita, and M. Hasebe for technical advice and providing plasmids.
Footnotes
This work was supported by the Japan Society for the Promotion of Science (16K18588 to M.O.) and the Strategic Research Foundation Grant-Aided Project for Private Universities from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (S1201003 to Y.S.).
References
- Abdelnoor RV, Yule R, Elo A, Christensen AC, Meyer-Gauen G, Mackenzie SA (2003) Substoichiometric shifting in the plant mitochondrial genome is influenced by a gene homologous to MutS. Proc Natl Acad Sci USA 100: 5968–5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JF. (2015) Why chloroplasts and mitochondria retain their own genomes and genetic systems: colocation for redox regulation of gene expression. Proc Natl Acad Sci USA 112: 10231–10238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendich AJ. (2004) Circular chloroplast chromosomes: the grand illusion. Plant Cell 16: 1661–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas PP, Carrasco B, Defeu Soufo C, César CE, Herr K, Kaufenstein M, Graumann PL, Alonso JC (2012) RecX facilitates homologous recombination by modulating RecA activities. PLoS Genet 8: e1003126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti H, Osman M, Grandoni P, Jagendorf AT (1992) A homolog of Escherichia coli RecA protein in plastids of higher plants. Proc Natl Acad Sci USA 89: 8068–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mot R, Schoofs G, Vanderleyden J (1994) A putative regulatory gene downstream of recA is conserved in gram-negative and gram-positive bacteria. Nucleic Acids Res 22: 1313–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees JC, Lusetti SL, Chitteni-Pattu S, Inman RB, Cox MM (2004) A RecA filament capping mechanism for RecX protein. Mol Cell 15: 789–798 [DOI] [PubMed] [Google Scholar]
- Hanson MR, Bentolila S (2004) Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell (Suppl) 16: S154–S169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye T, Odahara M, Fujita T, Hasebe M, Sekine Y (2008) Expression and complementation analyses of a chloroplast-localized homolog of bacterial RecA in the moss Physcomitrella patens. Biosci Biotechnol Biochem 72: 1340–1347 [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazi FR, Edmondson AC, Nielsen BL (2003) An Arabidopsis homologue of bacterial RecA that complements an E. coli recA deletion is targeted to plant mitochondria. Mol Genet Genomics 269: 454–463 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lin Z, Kong H, Nei M, Ma H (2006) Origins and evolution of the recA/RAD51 gene family: evidence for ancient gene duplication and endosymbiotic gene transfer. Proc Natl Acad Sci USA 103: 10328–10333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusetti SL, Cox MM (2002) The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu Rev Biochem 71: 71–100 [DOI] [PubMed] [Google Scholar]
- Mitsuhara I, Ugaki M, Hirochika H, Ohshima M, Murakami T, Gotoh Y, Katayose Y, Nakamura S, Honkura R, Nishimiya S, et al. (1996) Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol 37: 49–59 [DOI] [PubMed] [Google Scholar]
- Morley SA, Nielsen BL (2017) Plant mitochondrial DNA. Front Biosci 22: 1023–1032 [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Hiwatashi Y, Sakakibara I, Kato M, Hasebe M (2000) Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res 7: 9–17 [DOI] [PubMed] [Google Scholar]
- Odahara M, Inouye T, Nishimura Y, Sekine Y (2015a) RECA plays a dual role in the maintenance of chloroplast genome stability in Physcomitrella patens. Plant J 84: 516–526 [DOI] [PubMed] [Google Scholar]
- Odahara M, Kishita Y, Sekine Y (2017) MSH1 maintains organelle genome stability and genetically interacts with RECA and RECG in the moss Physcomitrella patens. Plant J 91: 455–465 [DOI] [PubMed] [Google Scholar]
- Odahara M, Kuroiwa H, Kuroiwa T, Sekine Y (2009) Suppression of repeat-mediated gross mitochondrial genome rearrangements by RecA in the moss Physcomitrella patens. Plant Cell 21: 1182–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odahara M, Masuda Y, Sato M, Wakazaki M, Harada C, Toyooka K, Sekine Y (2015b) RECG maintains plastid and mitochondrial genome stability by suppressing extensive recombination between short dispersed repeats. PLoS Genet 11: e1005080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Ramírez C, Hernandez-Coronado M, Thamm A, Catarino B, Wang M, Dolan L, Feijó JA, Becker JD (2016) A transcriptome atlas of Physcomitrella patens provides insights into the evolution and development of land plants. Mol Plant 9: 205–220 [DOI] [PubMed] [Google Scholar]
- Pagès V, Koffel-Schwartz N, Fuchs RP (2003) recX, a new SOS gene that is co-transcribed with the recA gene in Escherichia coli. DNA Repair (Amst) 2: 273–284 [DOI] [PubMed] [Google Scholar]
- Papavinasasundaram KG, Movahedzadeh F, Keer JT, Stoker NG, Colston MJ, Davis EO (1997) Mycobacterial recA is cotranscribed with a potential regulatory gene called recX. Mol Microbiol 24: 141–153 [DOI] [PubMed] [Google Scholar]
- Powikrowska M, Oetke S, Jensen PE, Krupinska K (2014) Dynamic composition, shaping and organization of plastid nucleoids. Front Plant Sci 5: 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragone S, Maman JD, Furnham N, Pellegrini L (2008) Structural basis for inhibition of homologous recombination by the RecX protein. EMBO J 27: 2259–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP, Cornet E, Michel B (2005) Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genet 1: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu AP, Abdelnoor RV, Mackenzie SA (2007) Transgenic induction of mitochondrial rearrangements for cytoplasmic male sterility in crop plants. Proc Natl Acad Sci USA 104: 1766–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano Y. (1993) Role of the recA-related gene adjacent to the recA gene in Pseudomonas aeruginosa. J Bacteriol 175: 2451–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer DG. (2002) A new moss genetics: targeted mutagenesis in Physcomitrella patens. Annu Rev Plant Biol 53: 477–501 [DOI] [PubMed] [Google Scholar]
- Smith DR, Keeling PJ (2015) Mitochondrial and plastid genome architecture: reoccurring themes, but significant differences at the extremes. Proc Natl Acad Sci USA 112: 10177–10184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohl EA, Brockman JP, Burkle KL, Morimatsu K, Kowalczykowski SC, Seifert HS (2003) Escherichia coli RecX inhibits RecA recombinase and coprotease activities in vitro and in vivo. J Biol Chem 278: 2278–2285 [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Venkatesh R, Ganesh N, Guhan N, Reddy MS, Chandrasekhar T, Muniyappa K (2002) RecX protein abrogates ATP hydrolysis and strand exchange promoted by RecA: insights into negative regulation of homologous recombination. Proc Natl Acad Sci USA 99: 12091–12096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby MC, Ryder L, Lloyd RG (1993) Reverse branch migration of Holliday junctions by RecG protein: a new mechanism for resolution of intermediates in recombination and DNA repair. Cell 75: 341–350 [DOI] [PubMed] [Google Scholar]
- Xu YZ, Arrieta-Montiel MP, Virdi KS, de Paula WB, Widhalm JR, Basset GJ, Davila JI, Elthon TE, Elowsky CG, Sato SJ, et al. (2011) MutS HOMOLOG1 is a nucleoid protein that alters mitochondrial and plastid properties and plant response to high light. Plant Cell 23: 3428–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]