In Arabidopsis thaliana, scopoletin 8-hydroxylase mediates the final step in the biosynthesis of fraxetin, a coumarin with a catechol moiety, which is secreted by roots of iron-deficient plants at elevated pH to mobilize iron.
Abstract
Iron (Fe) is an essential mineral nutrient and an important factor for the composition of natural plant communities. Low Fe availability in aerated soils with neutral or alkaline pH has led to the evolution of elaborate mechanisms that extract Fe from the soil solution. In Arabidopsis (Arabidopsis thaliana), Fe is acquired by an orchestrated strategy that comprises mobilization, chelation, and reduction of Fe3+ prior to its uptake. Here, we show that At3g12900, previously annotated as scopoletin 8-hydroxylase (S8H), participates in Fe acquisition by mediating the biosynthesis of fraxetin (7,8-dihydroxy-6-methoxycoumarin), a coumarin derived from the scopoletin pathway. S8H is highly induced in roots of Fe-deficient plants both at the transcript and protein levels. Mutants defective in the expression of S8H showed increased sensitivity to growth on pH 7.0 media supplemented with an immobile source of Fe and reduced secretion of fraxetin. Transgenic lines overexpressing S8H exhibited an opposite phenotype. Homozygous s8h mutants grown on media with immobilized Fe accumulated significantly more scopolin, the storage form of scopoletin, supporting the designated function of S8H in scopoletin hydroxylation. Fraxetin exhibited Fe-reducing properties in vitro with higher rates being observed at neutral relative to acidic pH. Supplementing the media containing immobile Fe with fraxetin partially rescued the s8h mutants. In natural Arabidopsis accessions differing in their performance on media containing immobilized Fe, the amount of secreted fraxetin was highly correlated with growth and Fe and chlorophyll content, indicating that fraxetin secretion is a decisive factor for calcicole-calcifuge behavior (i.e. the ability/inability to thrive on alkaline soils) of plants.
As a key edaphic factor, soil pH has a strong impact on the availability of mineral nutrients and the distribution of species in natural plant communities. Iron (Fe) solubility decreases dramatically with increasing pH, excluding so-called calcifuge (“chalk-fleeing”) species from carbonate-rich, alkaline soils due to their inability to acquire sufficient Fe under such conditions. Calcicole behavior, i.e. the ability to thrive on alkaline soils, has been attributed to the efficiency of Fe acquisition of a cultivar or species, a trait that strongly contributes to the ability to survive and reproduce on such soils (Zohlen and Tyler, 2004).
Plants have been categorized into two phylogenetically distinct groups that have evolved mutually exclusive mechanisms to increase the solubility of Fe (Römheld and Marschner, 1986). In nongraminaceous species, such as Arabidopsis (Arabidopsis thaliana), Fe is mobilized by net proton extrusion into the rhizosphere mediated by the P-type ATPase AHA2 (Santi and Schmidt, 2009) and reduced through the root surface Fe chelate reductase FERRIC REDUCTION OXIDASE2 (FRO2; Robinson et al., 1999) before Fe2+ is taken up via the high-affinity Fe2+ transporter IRON REGULATED TRANSPORTER1 (IRT1; Eide et al., 1996; Vert et al., 2002). This mechanism has been referred to as strategy I (Römheld and Marschner, 1986; Brumbarova et al., 2015). Grasses (Poales) have adopted a system in which secretion of high-affinity Fe chelators of the mugineic acid family, generically referred to as phytosiderophores, precedes uptake of the Fe3+-phytosiderophore complex without prior reduction (strategy II; Kobayashi and Nishizawa, 2012). However, recent studies suggest that secretion of Fe-chelating compounds is not unique to graminaceous species. In Arabidopsis, the scopoletin pathway is reprogrammed upon Fe deficiency to produce and secrete coumarins with Fe-mobilizing properties, which is of particular importance in alkaline soils in which the free ion activity of Fe is extremely low (Rodríguez-Celma et al., 2013; Schmid et al., 2014; Fourcroy et al., 2014; Schmidt et al., 2014). Expression profiling, genetic, and biochemical approaches implicated the 2-oxoglutarate and Fe(II)-dependent dioxygenase (2OGD) FERULOYL-COA 6'-HYDROXYLASE1 (F6′H1) and the ABC transporter PLEIOTROPIC DRUG RESISTANCE9 (PDR9) as key nodes in the biosynthesis and secretion of Fe-mobilizing coumarins, respectively (Kai et al., 2008; Yang et al., 2010; Lan et al., 2011; Rodríguez-Celma et al., 2013; Schmid et al., 2014; Fourcroy et al., 2014). F6'H1 mediates the ortho-hydroxylation of feruloyl CoA to generate scopoletin, which, together with its β-glucoside scopolin, constitutes the most abundant coumarin in Arabidopsis (Kai et al., 2008; Döll et al., 2018). Subsequent studies revealed that aglycones of coumarins containing a catechol moiety, such as esculetin and fraxetin, were the dominant active compounds in Arabidopsis (Schmidt et al., 2014; Strehmel et al., 2014; Sisó-Terraza et al., 2016a). However, the final step(s) of the biosynthetic pathway of these compounds has not yet been annotated.
The assimilation of Fe is under a surprisingly complex, multifaceted control, which is required to keep Fe within a tight cellular concentration. Similar to other key processes in Fe acquisition, the biosynthesis and secretion of coumarins is regulated by the basic helix-loop-helix (bHLH)-type transcription factor FIT (Ivanov et al., 2012; Colangelo and Guerinot, 2004; Schmidt et al., 2014). FIT dimerizes with the subgroup Ib bHLH proteins, bHLH38, bHLH39, bHLH100, and bHLH101, to regulate its target genes (Yuan et al., 2008; Sivitz et al., 2012; Wang et al., 2013). The transcription of FIT and its partners is responsive to Fe and directly activated by three other bHLH proteins, bHLH34, bHLH104, and bHLH105 (Li et al., 2016; Zhang et al., 2015). These three proteins also activate the expression of POPEYE, a negative regulator of the Fe deficiency response (Long et al., 2010), which regulates a subset of genes that do not overlap with the FIT regulatory network. The abundance of bHLH104 and bHLH105 is likely regulated by the Fe-binding hemerythrin domain-containing E3 ligase BRUTUS in a proteasomal-dependent manner (Long et al., 2010; Selote et al., 2015; Li et al., 2016; Zhang et al., 2015). Coregulation of all components of the reduction-based Fe uptake system suggests high cooperativity of the various processes. It is therefore reasonable to speculate that the final step(s) of the biosynthesis of Fe-mobilizing coumarins is part of the same regulon.
Iron deficiency-induced production of Fe-mobilizing compounds by nongraminaceous species and their subsequent release into the rhizosphere has been observed over several decades (Dakora and Phillips, 2002; Tsai and Schmidt, 2017). Since their discovery, the role of root exudates in Fe acquisition was subject to a long-standing debate. Both reductive release of Fe from Fe3+ chelates and chelation of Fe3+ to provide the substrate for an unidentified ferric chelate reductase were suggested as functions of Fe deficiency-induced phenolic secretion (Brown and Ambler, 1973; Hether et al., 1984; Römheld and Marschner, 1984). After the molecular identification of the Fe chelate reductase FRO2 in Arabidopsis (Robinson et al., 1999), the so-called “reductants” were considered as being of minor importance in Fe acquisition. Several aspects collectively led to a gross underestimation of the ecological significance of Fe-mobilizing compounds of strategy I species. First, in the laboratory, plants are generally grown at optimal, i.e. slightly acidic pH at which the Fe acquisition machinery works most efficiently. However, Fe chlorosis is most frequently observed at alkaline pH, conditions under which FRO2 activity is compromised (Susín et al., 1996). As a second factor, experiments are mostly conducted with highly soluble sources of Fe, such as Fe3+ complexed by aminopolycarboxylates, at concentrations that well exceed the Km of FRO2, a situation that is not likely to occur in situ. Under such conditions, the activity of FRO2 is presumably rate-limiting for Fe uptake, which makes the contribution of the secretion of Fe-scavenging compounds to Fe uptake insignificant.
The question as to the traits that allow calcicole plants to thrive on neutral or alkaline soil has puzzled plant biologists for more than two centuries (Link, 1789; Unger, 1836; Grime and Hodgson, 1968). Attempts to single out specific factors have been largely unsuccessful, an approach that is also hampered by the difficulties inherent to interspecies comparisons (De Silva, 1934; Lee, 1998; Schmidt and Fühner, 1998). The huge variety of the secreted substances and a lack of knowledge regarding the exact chemical nature and precise role of the secreted compounds have excluded targeted approaches and have rendered attempts to associate secreted secondary metabolites with the autecology of species difficult. In this study, we addressed the biosynthesis and ecological role of Fe-mobilizing coumarins secreted by Fe-deficient plants. We report that besides the ortho-hydroxylation of feruloyl CoA by F6’H1, the introduction of a second hydroxyl group in the ortho position, a reaction catalyzed by scopoletin 8-hydroxylase (S8H; At3g12900), is critical for producing the Fe-mobilizing coumarin fraxetin. Scopoletin hydroxylation at the C8 position by At3g12900 was shown previously (Siwinska et al., 2018). By comparing diverse Arabidopsis accessions differing in the ability to grow on alkaline media supplemented with an immobile Fe source, we further show that the abundance of fraxetin in the media determines the ability to acquire Fe at elevated pH.
RESULTS
At3g12900 Is a Putative S8H
Up-regulation of general phenylpropanoid and scopoletin pathway genes upon Fe deficiency has been implicated with the secretion of Fe-mobilizing phenolic compounds, aiding in the acquisition of sparingly soluble Fe pools (Rodríguez-Celma et al., 2013; Schmidt et al., 2014; Fourcroy et al., 2014). Scopoletin biosynthesis and secretion are increased upon Fe starvation (Lan et al., 2011; Schmid et al., 2014), but the structural features of scopoletin do not support a function for Fe chelation or mobilization and rather suggest a role as an antimicrobial repellent or as a precursor for such a compound. To bind Fe efficiently, two vicinal hydroxyl groups are required (Mladenka et al., 2010; Schmid et al., 2014; Schmidt et al., 2014), implying the generation of novel substances in response to Fe deficiency by extension of the scopoletin pathway. Introduction of a hydroxyl group can be catalyzed by 2OGDs. Referring to our previously published transcriptomic and proteomic data, the 2OGD At3g12900 is a promising candidate for mediating this reaction (Lan et al., 2011; Rodríguez-Celma et al., 2013). The gene is the closest paralog of F6’H1, which is highly Fe-responsive and was shown to be essential for the generation of Fe-mobilizing coumarins (Rodríguez-Celma et al., 2013; Schmid et al., 2014). F6’H1 and At3g12900 belong to the same clade of the 2OGD family predicted to be involved in coumarin biosynthesis (Kawai et al., 2014), indicative of a close evolutionary and, possibly, functional relationship of the two proteins. Two adjacent hydroxyl groups in the ortho-position are found in Fe deficiency-induced coumarins secreted by Arabidopsis, such as fraxetin (7,8-dihydroxy-6-methoxycoumarin), which has been consistently detected in Fe-deficient plants (Schmid et al., 2014; Schmidt et al., 2014; Fourcroy et al., 2014; Sisó-Terraza et al., 2016a). Based on its function, we designated At3g12900 as S8H. Hydroxylation of scopoletin by At3g12900 has recently been demonstrated by heterologous expression of the protein in Escherichia coli (Siwinska et al., 2018).
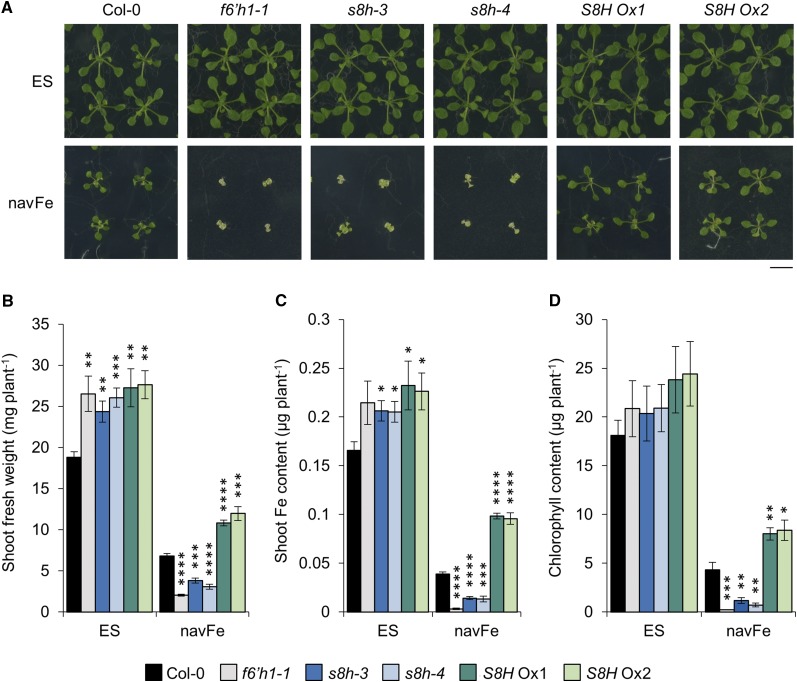
S8H Is Crucial for Mobilizing Fe
To elucidate the role of S8H in Fe acquisition, we generated S8H overexpression (S8H Ox) lines using the cauliflower mosaic virus (CaMV) 35S promoter and obtained two independent T-DNA insertion mutants designated s8h-3 (SALK_041283) and s8h-4 (SALK_073361), both of which harbor a tandem two-copy inverted T-DNA insertion in the promoter region (Supplemental Fig. S1). To investigate whether changes in the expression of S8H will affect the tolerance of plants to low Fe availability at circumneutral pH, the performance of Col-0 (wild type), s8h mutants, and S8H Ox lines was assessed after 14 d of growth on either Estelle and Somerville (ES) media supplemented with 40 µm readily available Fe in the form of Fe3+-EDTA adjusted to pH 5.5 or unavailable Fe (navFe) media supplemented with insoluble Fe in the form of FeCl3 adjusted to pH 7.0. The f6’h1-1 mutant, which is defective in the biosynthesis of scopoletin, was included as a negative control (Fig. 1A). When grown under control conditions, s8h mutants and S8H Ox lines had significantly more fresh weight than wild-type plants (Fig. 1B). Higher fresh weight relative to the wild type was also observed for the f6’h1-1 mutant, and increased growth in these lines was accompanied by a higher shoot Fe content (Fig. 1C), which might be due to a reduced demand for Fe required as a cofactor for F6’H1 and S8H in the mutants and improved Fe uptake due to increased production of Fe-mobilizing compounds in the S8H Ox lines. When cultivated on navFe media, growth of wild-type plants was severely restricted and accompanied by a dramatic reduction in shoot Fe and chlorophyll content (Fig. 1, A–D). This effect was even more pronounced in all three mutant lines under investigation (i.e. f6’h1-1 and the two s8h alleles). By contrast, S8H Ox plants performed considerably better on navFe media than wild-type plants. Both S8H Ox lines not only showed significantly higher shoot fresh weight, but also increased chlorophyll and Fe content when compared to Col-0, indicative of improved mobilization of Fe in the media (Fig. 1, B–D). These experiments show that S8H is of critical importance for the uptake of Fe from recalcitrant Fe pools.
Figure 1.
Effects of Fe availability on the growth of Col-0, f6’h1-1, s8h-3, s8h-4, S8H Ox1, and S8H Ox2 plants. A, Phenotypes of plants grown for 14 d on either ES or navFe media. Representative images of five independent experiments are shown. Bar = 1 cm. B, Shoot fresh weight. C, Shoot Fe content. D, Chlorophyll content. Each bar represents the mean ± se of five independent experiments. Statistical testing was carried out using Student’s t test. Asterisks indicate significant differences from the wild type in each treatment: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
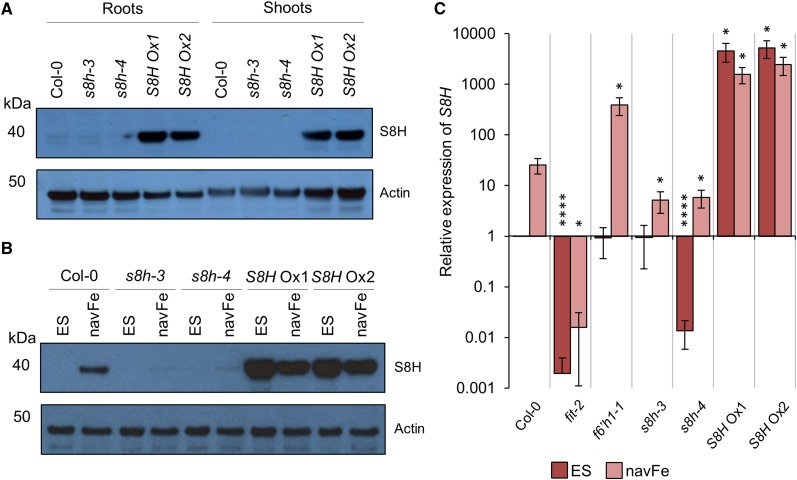
S8H Is Induced by Low Fe Availability
To investigate the pattern and regulation of S8H expression, we raised polyclonal antibodies against the synthetic peptide CGVTLEEEKMNGLMG and used the affinity-purified antibodies to detect S8H in protein extracts from 14-d-old Arabidopsis seedlings by western-blot analysis (Fig. 2, A and B; Supplemental Fig. S2). To validate the antibody and to investigate the expression pattern of S8H, protein extracts from roots of Col-0, s8h mutants, and S8H Ox lines grown under control conditions were compared to extracts from shoots. Bands of the expected mass of approximately 40 kD were only detected in protein extracts from roots of wild-type plants and s8h mutants, suggesting that S8H expression is root specific (Fig. 2A). In S8H Ox lines, S8H was detected in protein extracted from both roots and shoots. Next, we investigated the effect of growth conditions in which Fe was immobilized and not readily available for uptake on the expression of S8H. As show in Figure 2B, S8H expression in roots was strongly induced under navFe conditions in wild-type plants; a weak induction was observed in the s8h mutants. S8H protein abundance in S8H Ox lines was slightly less pronounced when plants were grown under navFe conditions compared to those grown on ES media (Fig. 2B).
Figure 2.
Expression of S8H protein and transcripts. A and B, Detection of S8H in protein extracted from 14-d-old Col-0, s8h-3, s8h-4, S8H Ox1, and S8H Ox2 plants by western-blot analysis. Actin abundance is shown in each panel as a control for equal protein loading. Representative images of three independent experiments are shown. A, Total protein extracted from roots and shoots of plants grown on ES media. Per lane, 10 μg of total protein was loaded. B, Total protein extracted from roots of plants grown on either ES or navFe media. Per lane, 2.5 μg of total protein was loaded. C, RT-qPCR analysis of S8H expression in root samples from Col-0, fit-2, f6’h1-1, s8h-3, s8h-4, S8H Ox1, and S8H Ox2 plants grown for 14 d on either ES or navFe media. The ΔΔCT method was used to determine relative gene expression, and expression of elongation factor 1 alpha was used as an internal control. Each bar represents the mean ± se of three independent experiments. Statistical testing was carried out using Student’s t test. Asterisks indicate significant differences from the wild type (Col-0) in each treatment: *, P ≤ 0.05; ****, P ≤ 0.0001.
Consistent with the western-blot results, analysis of S8H transcript abundance by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) confirmed strong induction of S8H expression in roots of the wild type upon growth on navFe; an ∼80% decrease in S8H transcript levels was observed in both s8h mutants. In s8h-4, the expression level of S8H was also significantly (P < 0.0001) reduced relative to Col-0 when grown on ES medium. Interestingly, in f6’h1-1 plants, induction of S8H was much more pronounced by growth under navFe conditions than in Col-0, possibly due to a lacking end-product inhibition on F6’H1 expression. The absence of detectable S8H expression in the fit mutant confirmed FIT-dependent regulation of S8H expression (Fig. 2C; Colangelo and Guerinot, 2004). S8H Ox lines showed massive (>4,000-fold) constitutive induction of S8H expression when grown on ES media. Although mRNA levels of S8H Ox lines decreased when plants were grown on navFe media, transcript levels remained >1,500-fold higher than in the wild type (Fig. 2C). These observations are consistent with the proposed role of S8H in the biosynthesis of root-borne compounds secreted into the rhizosphere. Furthermore, the Fe-dependent and root-specific expression of S8H is reminiscent of F6’H1 (Rodríguez-Celma et al., 2013; Schmid et al., 2014). Although both the f6’h1-1 and s8h mutants exhibited severely defective phenotypes when grown on navFe media, the expression of S8H was massively increased in f6’h1-1 plants grown on navFe conditions (Fig. 2C). Similar to the wild type, expression of F6’H1 was induced in s8h mutants by growth on navFe media (Supplemental Fig. S3). This shows that induction of S8H alone is insufficient to rescue the f6’h1-1 mutant phenotype, suggesting that S8H and F6’H1 are both essential to plants for the production and secretion of Fe-mobilizing compounds at elevated pH.
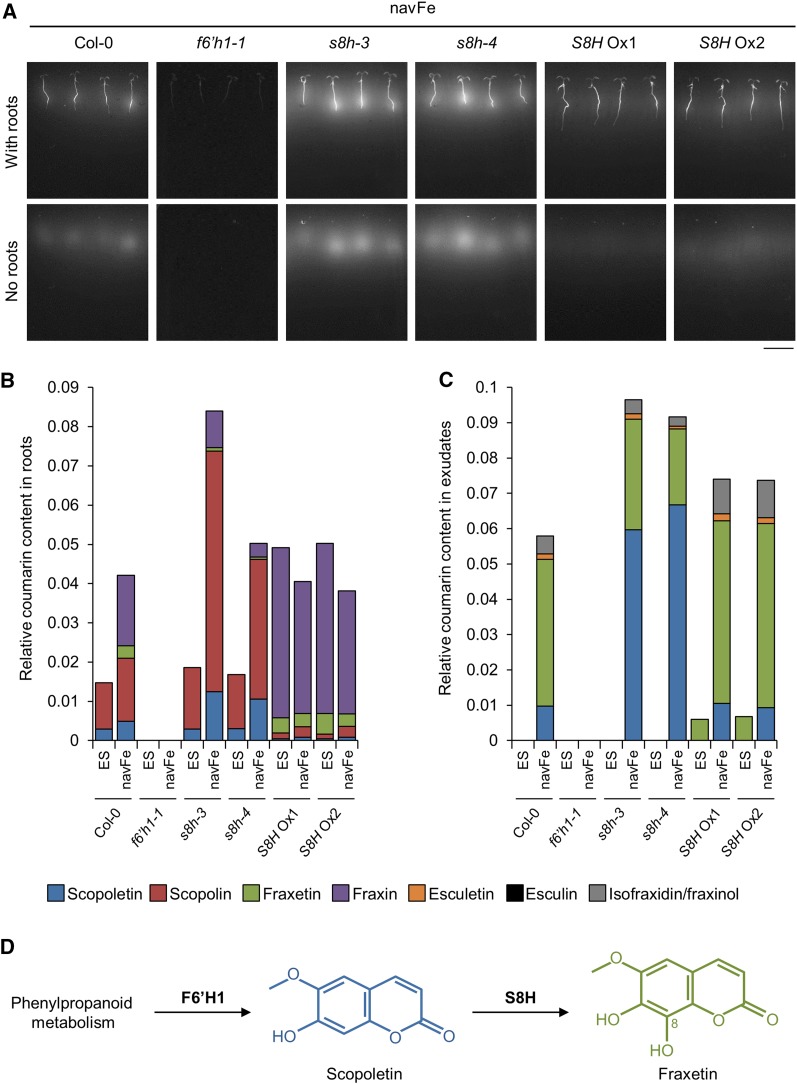
S8H Activity Alters the Production of Fluorescent Coumarins
Autofluorescence of coumarins has been used as a noninvasive method to detect and assess the amount of root-secreted Fe-mobilizing coumarins (Rodríguez-Celma et al., 2013; Schmid et al., 2014; Döll et al., 2018). To determine whether S8H is involved in the biosynthesis of coumarins, autofluorescence was observed in roots and root exudates of the genotypes under investigation (Fig. 3A). The fluorescence signal was monitored before and after plants were removed from the media. As expected, under control conditions where Fe is available and coumarins are not required, no fluorescence was observed in roots or in the growth media of the wild type and s8h and f6’h1-1 mutants. Roots of S8H Ox lines grown on ES media emitted a weak fluorescence signal in the roots (Supplemental Fig. S4). Except for the f6’h1-1 mutant, fluorescence was dramatically increased in the roots of all other lines investigated when plants were grown on navFe media, a response that was particularly pronounced in the s8h mutants. Fluorescence remained detectable after removing plants from the media, indicative of secretion of the compounds. Interestingly, a markedly higher fluorescence relative to wild-type plants was detected in the media of the s8h-3 and s8h-4 mutants, while the S8H Ox lines produced slightly weaker signals than the wild type (Fig. 3A). It has been previously shown that fluorescence of fraxetin is exceedingly low in comparison to scopoletin and its derivatives (Sisó-Terraza et al., 2016a). Therefore, the strong fluorescence signal in the media of the s8h mutants could be explained by the secretion of (fluorescent) scopoletin, the hydroxylation of which is compromised in the mutant. In the S8H Ox lines, high hydroxylation rates may have led to increased secretion of nonfluorescent fraxetin and concomitant lower levels of scopoletin.
Figure 3.
Accumulation of coumarins in roots and root exudates. A, Visualization of fluorescent compounds produced and secreted by 7-d-old plants grown vertically on navFe media. Photos were taken before (with roots) and after (no roots) removing plants from the media using 365 nm as the excitation wavelength. Representative images of three independent experiments are shown. Bar = 1 cm. B and C, Relative abundance of selected coumarin compounds in root extracts (B) and root exudates (C) of Col-0, f6’h1-1, s8h-3, s8h-4, S8H Ox1, and S8H Ox2 plants grown on ES and navFe media for 14 d. Bars represent the relative content of different coumarin compounds presented as peak area normalized to the number of roots and internal standard peak area. Values are mean relative coumarin content obtained from three independent experiments. D, Generation of fraxetin via S8H-mediated hydroxylation of scopoletin at the C8 position.
Coumarin Profiling Designates At3g12900 as Scopoletin 8-Hydroxylase
To validate the proposed function of S8H, we carried out a targeted-coumarin profile analysis of the methanolic extracts of the roots and media from the Col-0, f6’h1-1, s8h, and S8H Ox lines grown on either control ES or navFe media for 14 d (Fig. 3, B and C). Ultra-performance liquid chromatography (UPLC)/ESI-QTOF-MS was used to detect and measure the relative abundance of coumarins shown in Supplemental Figure S5. Peaks were compared to authentic standards. In order to obtain a signal that can be reliably quantified, peaks with a signal-to-noise ratio below 10 were excluded. Under control conditions, the major coumarins detected in roots of wild-type plants were scopolin and, to a lesser extent, its corresponding aglycone scopoletin (Fig. 3B). Growing the plants under navFe conditions caused a relatively small increase in scopolin and doubled the scopoletin level. Fraxin and its aglycone fraxetin were detected in amounts comparable to scopolin/scopoletin, together constituting the major Fe deficiency-inducible coumarins. Under control conditions, the composition and amounts of coumarins in the s8h mutants did not deviate greatly from the wild type. When grown on navFe media, the biosynthesis of both scopolin and scopoletin was massively induced in the mutants. Fraxin and fraxetin levels were lower in both mutants than in the wild type, but clearly detectable, probably due to residual gene activity in the knockdown mutants. In S8H Ox lines, the amount of scopolin/scopoletin was markedly reduced relative to Col-0, and the major coumarins were fraxin and its aglycone fraxetin. No major differences between the two Fe regimes were observed for the S8H Ox lines except for a slightly reduced fraxin and fraxetin production under navFe conditions. As anticipated, no coumarins were produced in f6’h1-1 plants. In summary, consistent with the proposed function of S8H as a scopoletin 8-hydroxylase, S8H activity appears to mediate the biosynthesis of the fraxetin/fraxin pair at the expense of scopoletin and scopolin.
Regarding the exudates, no coumarins were detected in the media of Col-0 under control conditions; fraxetin was the predominant secreted compound under navFe condition (Fig. 3C). Smaller amounts of scopoletin, esculetin, and isofraxidin/fraxinol were also detected in the wild type. Similar to Col-0, no coumarins were detected in the root exudate of the s8h mutants under control conditions. As anticipated from the decreased activity of S8H, Fe-deficient s8h mutants secreted significantly more scopoletin and less fraxetin than the wild type. The high amount of scopoletin corresponds to the bright fluorescence detected in the media of these mutants (Fig. 3A). Increased levels of scopoletin in the s8h mutant growth media are consistent with previous reports (Siwinska et al., 2018). Intriguingly, under control conditions, S8H Ox lines secreted exclusively fraxetin. When grown on navFe media, S8H Ox lines secreted more fraxetin than Col-0. Isofraxidin/fraxinol and esculetin were also secreted into the media. This observation can be explained by the up-regulation of the general phenylpropanoid pathway and strong induction of F6’H1 under Fe-deficient conditions. Together, these observations suggest that S8H hydroxylates scopoletin at the C8 position to generate fraxetin, the aglycone of the storage form fraxin, at the expense of scopoletin and scopolin (Fig. 3D). Although theoretically scopoletin can be hydroxylated at C5, data from Siwinska et al. (2018) strongly support that S8H catalyzes C8 hydroxylation of scopoletin. Only coumarin aglycones were secreted into the media, supporting a scenario in which the sugar moiety of glycosylated coumarins is removed prior to secretion (Le Roy et al., 2016).
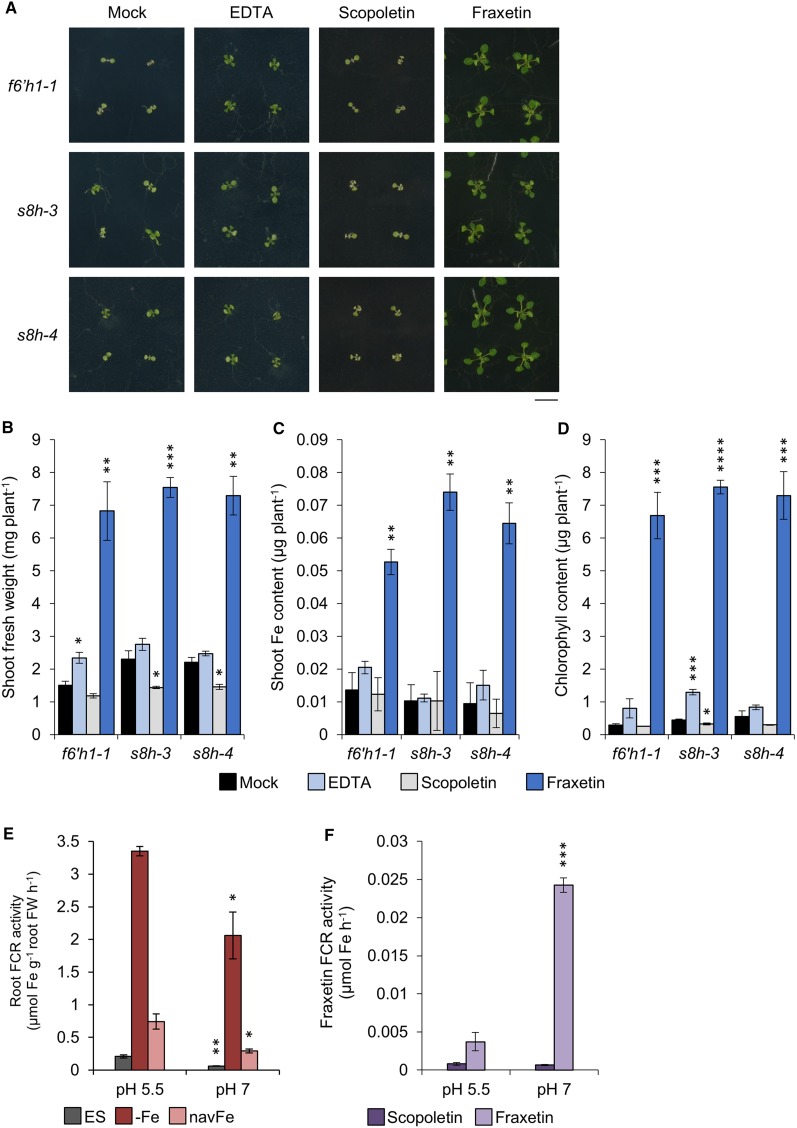
Fraxetin Supplementation Complements the Phenotypes of the f6’h1-1 and s8h Mutants
To demonstrate that fraxetin is the active compound participating in the acquisition of Fe at elevated pH, we investigated the growth of the s8h mutants in navFe media supplemented with fraxetin. The phenotypes of the f6’h1-1, s8h-3, and s8h-4 mutants were assessed after 14 d of growth. The synthetic Fe chelator EDTA and scopoletin were added to the media as positive and negative controls, respectively. Growth and shoot Fe and chlorophyll content were dramatically increased in all three mutants by adding fraxetin to the media (Fig. 4, A–D). For example, chlorophyll content was increased by ∼30-fold in the presence of fraxetin when compared to the mock treatment. Addition of EDTA had minor and inconsistent effects on growth and Fe content; scopoletin did not improve growth of any of the tested lines. These observations suggest that fraxetin is a bioactive compound in the acquisition of immobile Fe.
Figure 4.
Rescue of mutants by exogenous application of fraxetin in navFe media. A, Phenotypes of f6’h1-1, s8h-3, and s8h-4 mutants grown for 14 d on navFe media supplemented with 120 μm of either EDTA, scopoletin, or fraxetin. Representative images from three independent experiments are shown. Bar = 1 cm. B, Shoot fresh weight. C, Shoot Fe content. D, Chlorophyll content. Each bar represents the mean ± se of three independent experiments. Statistical testing was carried out using Student’s t test. Asterisks indicate significant differences from the mock treatment for each mutant: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. E and F, FCR activity of roots (E) and fraxetin (F) was assayed at pH 5.5 (MES) and 7.0 (MOPS). Col-0 plants were precultivated in ES media for 10 d and transferred to either ES, −Fe (ES with no Fe3+-EDTA, containing 100 μm ferrozine), or navFe media for 3 d before conducting the FCR assay. For measuring Fe3+ reduction by coumarin compounds, 100 μm of either fraxetin or scopoletin was added in the FCR assay instead of roots and normalized to blanks with DMSO. Each bar represents the mean ± se of three independent experiments. Statistical testing was carried out using Student’s t test. Asterisks indicate significant differences from the assay buffered at pH 5.5: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Fraxetin Extends the pH Range of Fe Chelate Reduction
Chelation of sparingly soluble Fe is an unequivocal function of catechol-type coumarins and was previously demonstrated (Mladenka et al., 2010; Schmid et al., 2014; Sisó-Terraza et al., 2016a). Reduction of Fe3+ by secreted coumarins remains more ambiguous. Fraxetin was shown to reductively mobilize Fe from Fe3+ oxide at neutral pH (Sisó-Terraza et al., 2016a). Under the present conditions, ferric chelate reduction by fraxetin was significantly more efficient at pH 7.0 when compared to pH 5.5 (Fig. 4F). This stands in contrast to the pH dependence of root-mediated ferric chelate reduction. Col-0 precultivated for 10 d and subsequently grown on ES, −Fe (ES with no Fe3+-EDTA, containing 100 µm ferrozine), or navFe conditions for 3 d was assayed at pH 5.5 and 7.0. At pH 5.5, root ferric-chelate reductase (FCR) activity of Fe-deficient and navFe-treated plants was strongly induced relative to Fe-sufficient plants. However, this induction was significantly decreased when the pH of the assay was buffered at pH 7.0. Furthermore, it was shown that the FCR of plants treated with navFe condition is much lower than that of plants treated with –Fe condition (Fig. 4F). This observation confirms the previously reported mildly acidic pH optimum of FRO2-mediated Fe chelate reduction (Römheld and Marschner, 1984; Susín et al., 1996). It thus appears that secretion of fraxetin extends the pH range of efficient Fe3+ reduction, thereby reducing the sensitivity of plants to neutral or alkaline conditions.
Fraxetin Determines the Efficiency of Fe Uptake from an Immobile Fe Source
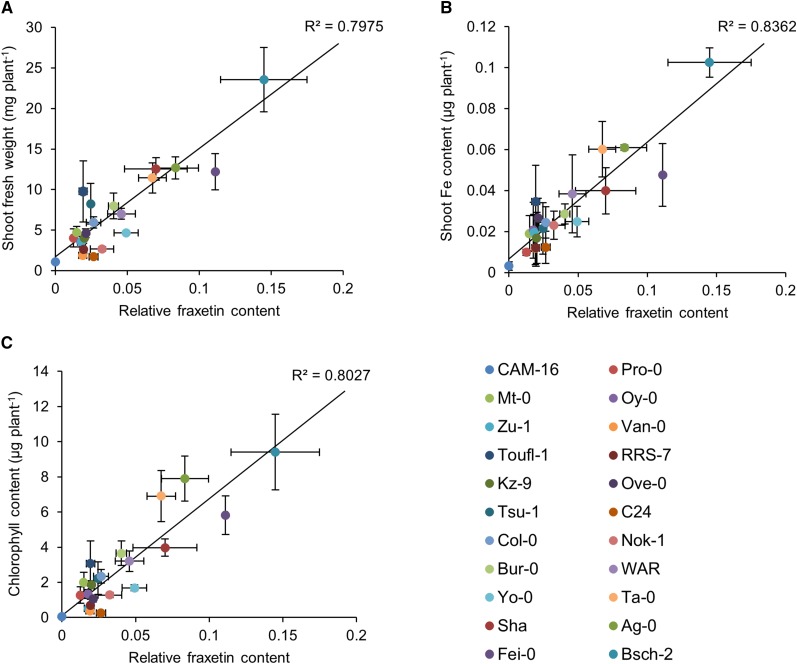
To investigate whether secretion of fraxetin determines Fe uptake from immobilized Fe pools and the performance of the plant at circumneutral pH, we investigated the growth of 22 Arabidopsis accessions, selected for their differing growth response to navFe media (Supplemental Fig. S6). Since the secretion of fraxetin can be affected by both production and secretion rates, determination of fraxetin was chosen over quantification of transcript or protein abundance to determine its contribution to the calcicole-calcifuge behavior of the various accessions. The selected accessions differed widely in their rosette growth and shoot Fe and chlorophyll content (Supplemental Fig. S6). The amount of fraxetin in the media was highly correlated with these three parameters, with the highest correlation being observed between fraxetin and shoot Fe content (Fig. 5). This shows that fraxetin secretion is a decisive and growth-restricting parameter for Fe uptake from immobile Fe pools, which putatively defines the amplitude of requirements for successful establishment of a genotype to a given set of edaphic parameters.
Figure 5.
Correlation analysis between the growth performance and the relative fraxetin content in 22 different Arabidopsis accessions. Plants were grown on navFe media for 14 d. Linear regression analysis was carried out by plotting shoot fresh weight (A), shoot Fe content (B), and chlorophyll content (C) of each accession against their relative fraxetin content.
DISCUSSION
While secretion of Fe-mobilizing compounds has been acknowledged as part of the Fe deficiency response of strategy I plants for several decades, their biosynthesis, ecological significance, and specific role in Fe acquisition have just begun to be elucidated. Congruous with a recent study (Siwinska et al., 2018), we show here that the 2OGD S8H is a critical component in the synthesis of fraxetin, a coumarin with a catechol moiety and a methoxy substituent derived from scopoletin. While mutants with partial loss of S8H activity were more severely impacted when grown on media that restrict the solubility of Fe than wild-type plants, transgenic plants overexpressing S8H showed better performance and had higher Fe and chlorophyll content than the wild type under these conditions. Targeted analysis of coumarins secreted by s8h mutants and S8H Ox lines demonstrated that At3g12900 is a scopoletin 8-hydroxylase, a function that has been validated recently for this protein based on biochemical evidences (Siwinska et al., 2018). Fraxetin, but not its precursor scopoletin, was able to ameliorate the phenotypes of s8h and f6’h1 mutant plants grown on navFe media, indicating that fraxetin is necessary and sufficient to mobilize Fe from immobile sources at circumneutral pH. We further show that fraxetin can reduce Fe3+, a property that was more pronounced at neutral pH conditions where the activity of the FRO2/IRT1-dependent Fe uptake system is compromised. Importantly, fraxetin is not only able to reduce Fe3+ in synthetic chelates, but also in Fe3+ oxides (Sisó-Terraza et al., 2016a), which is the form Fe is likely to be present in neutral or alkaline soils. However, the FRO2/IRT1 system still appears to be required, since fro2 and irt1 mutants are not rescued by root exudates from the wild type (Fourcroy et al., 2016). Most importantly, we show here that the amount of secreted fraxetin is tightly correlated with the growth and Fe and chlorophyll content of accessions differing in their ability to thrive on media with low Fe availability. Thus, the rate of fraxetin secretion appears to constitute a decisive, genetically determined factor for the calcicole/calcifuge behavior of a genotype.
While alkaline pH triggers a partial Fe-deficiency response also in the presence of Fe (Sisó-Terraza et al., 2016a), it appears that the balance between the two phenotypic readouts, i.e. root-mediated reduction and fraxetin secretion, is tipped by an additional, pH-dependent control mechanism. High media pH values shift the allocation of energetic and metabolic resources toward the biosynthesis and secretion of Fe-mobilizing compounds, a strategy that is more important at alkaline pH than enzymatic Fe reduction because of decreased substrate concentration for the reductase. Secretion of fraxetin remains low at acidic pH even under conditions of Fe deficiency (Sisó-Terraza et al., 2016a). Moreover, both reduction and mobilization of Fe by fraxetin is more efficient at neutral or alkaline pH (this study; Sisó-Terraza et al., 2016a); the pronounced (∼30-fold) stimulation of fraxetin secretion caused by a pH shift from 5.5 to 7.5 (Sisó-Terraza et al., 2016a) and a similarly pronounced decrease in enzymatic Fe3+ reduction supports that pH induces an “efficiency control” on the response to Fe deficiency. Such a regulatory node may be provided by processing of glycosylated coumarins prior to secretion. PDR9-mediated secretion of coumarins is dependent on the activity of the β-glucosidase BGLU42; Fe-deficient bglu42 mutant plants do not secrete coumarins (Zamioudis et al., 2014). This inhibition is thought to be due to compromised removal of the sugar moiety of glycosylated coumarins, a necessary step prior to secretion. Expression of BGLU42 is dependent on the MYB-type transcription factors MYB10 and MYB72, direct targets of FIT (Palmer et al., 2013). BGLU42 expression is misregulated in myb72 myb10 double mutants, which are sensitive to growth under alkaline conditions (Palmer et al., 2013). Expression of MYB72 is affected by beneficial soil bacteria, suggesting that MYB72 provides a gate for additional signals derived from the induced systemic resistance pathway (Zamioudis et al., 2014). However, how pH is sensed and alters the secretion rate of coumarins remains to be elucidated.
While habitat requirements and ecological fitness are complex traits that are defined by a multitude of factors, research into the factors that confer calcicole behavior has identified the ability to acquire Fe as a key determinant for the efficient establishment of a genotype on calcareous sites (Zohlen and Tyler, 2000, 2004; von Wandruszka, 2006). Up to 40% of photosynthetically fixed carbon is secreted into the rhizosphere (Whipps, 1990; Marschner, 1995), a costly strategy with many facets. Root exudates can act as chemical attractants or repellants, influence the growth of competing plant species, and change the chemical and physical properties of the soil (Nardi et al., 2000). The role of root exudates in the acquisition of mineral nutrients and successful establishment to a given set of soil conditions remains understudied. In Arabidopsis, fraxetin appears to be the most prominent and most robustly detected coumarin secreted by Fe-deficient plants (this study; Schmidt et al., 2014; Fourcroy et al., 2014; Sisó-Terraza et al., 2016a). Given the diversity of secondary metabolites, other species-dependent substances with similar features may extend the list of putative Fe-mobilizing compounds. An example for a genus- or genera-dependent strategy is the secretion of riboflavin sulfates by Fe-deficient sugar beet (Beta vulgaris) roots, which was suggested to provide a redox shuttle between intracellular reducing equivalents and soil Fe3+ compounds (Sisó-Terraza et al., 2016b). It should also be noted that S8H may be important for the biosynthesis of other putative Fe-mobilizing compounds downstream of fraxetin. An example is hydroxyfraxetin, which was shown to accumulate upon Fe deficiency (Ziegler et al., 2017). This investigation emphasizes the possibly underrated importance of root exudates for defining habitat requirements. Systematic approaches to catalog changes in rates and composition of secreted compounds in response to limitations of a particular mineral nutrient have been hampered by the complexity of such analyses and the general difficulties of comparing species that differ in a multitude of aspects such as root architecture, growth pattern, resource allocation, or reproductive strategies. The wide geographical distribution of Arabidopsis accessions and the concomitant adaptation of secondary metabolism to a given set of environmental conditions set the stage to link the composition of root exudates with habitat requirements.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) mutants and transgenic lines used in this study are in the Col-0 background. Seeds of the accession Col-0 and the T-DNA insertion mutants s8h-3 (SALK_041283), s8h-4 (SALK_073361), and f6’h1-1 (SALK_132418C) were obtained from the Arabidopsis Biological Resource Center (Ohio State University). Homozygous s8h-3 and s8h-4 T-DNA mutants were selected from PCR-based genotyping, and the positions of the T-DNA insertions were mapped by sequencing using primers shown in Supplemental Table S1. Seeds of the fit-2 mutant (SALK_126020) were provided by Dr. Catherine Curie (INRA Montpellier, France). The fit-2 and f6’h1-1 mutants were characterized previously (Sivitz et al., 2011; Kai et al., 2008).
For the generation of S8H Ox lines, the full-length At3g12900 cDNA was amplified from Arabidopsis root cDNA using the primers indicated in Supplemental Table S1, followed by gel purification and cloning into the BamHI-digested and dephosphorylated expression vector pROK2 containing the CaMV 35S promoter. The final construct was verified by sequencing. Agrobacterium tumefaciens-mediated transformation was used to transform wild-type plants (Col-0). Transformants were selected based on kanamycin resistance. Plants with a single insertion of the transgene were selected for self-pollination to generate T3 homozygous lines for further analysis. Overexpression of S8H in the transgenic plants was confirmed by RT-qPCR and western-blot analyses.
The Arabidopsis accessions used in this study were either purchased from the Arabidopsis Biological Resource Center or were provided by Dr. Paul E. Verslues (Institute of Plant and Microbial Biology, Academia Sinica, Taiwan).
Growth Conditions
Arabidopsis plants were grown under sterile conditions in a growth chamber on agar-based media as described by Estelle and Somerville (1987). Seeds were surface-sterilized by immersing in 30% (v/v) commercial bleach containing 6% NaClO (Clorox) and 70% (v/v) absolute ethanol containing 0.1% (v/v) Tween 20 for 6 min, followed by three rinses with absolute ethanol. Seeds were sown on petri dishes and kept for 2 d at 4°C in the dark before the plates were transferred to a growth chamber and grown at 21°C under continuous illumination (50 µmol m−2 s−2). The growth medium comprised 5 mm KNO3, 2 mm MgSO4, 2 mm Ca(NO3)2, 2.5 mm KH2PO4, 70 µm H3BO3, 14 µm MnCl2, 1 µm ZnSO4, 0.5 µm CuSO4, 0.01 µm CoCl2, and 0.2 µm Na2MoO4, supplemented with 1.5% (w/v) Suc, and solidified with 0.4% Gelrite pure (Kelco). For control ES media, 40 µm Fe3+-EDTA and 1 g/L MES were added and the pH was adjusted to 5.5 with KOH. For navFe media, 40 µm FeCl3 and 1 g/L MOPS were added and pH was adjusted to 7.0 with KOH. Unless stated otherwise, plants were grown directly on media for 14 d.
Protein Extraction and Western-Blot Analysis
Total protein was extracted from frozen root and shoot samples using glass beads in a TissueLyzer II (Qiagen) with 150 µL of extraction buffer (100 mm Tris-HCl, pH 7.4, 4% SDS, 200 mm DTT, 2× protease inhibitor cocktail, and 1 mm PMSF). The slurry was kept on ice for 10 min (vortexed every 2 min) before centrifugation at 13,200 rpm for 15 min at 4°C. The supernatant was collected and protein concentration was determined using the Pierce 660 nm Protein Assay Kit in the presence of 50 mm Ionic Detergent Compatible Reagent (Thermo Fisher Scientific). Equal amounts of total protein were heated at 100°C for 10 min in a solution containing 1× NuPAGE LDS sample buffer and 1× NuPAGE sample reducing agent prior to separation on NuPAGE 4-12% Bis-Tris Gels with NuPAGE MES SDS Running buffer (Invitrogen) at 200 V for 55 min using the XCell SureLock Mini Cell electrophoresis system (Life Technologies). Proteins were then transferred onto PVDF membranes (Bio-Rad) using the Trans-Blot Turbo Transfer System (Bio-Rad). Membranes were blocked in 1× NAP-Blocker in TBS solution (G-Biosciences) for 1 h before sequential probing with primary and secondary antibodies. Polyclonal anti-S8H antibody was raised in rabbits against the synthetic peptide CGVTLEEEKMNGLMG (generated by Genscript). Membranes were incubated with anti-S8H antibody diluted 1:2,000 in blocking buffer for 3 h and then the secondary antibody at a 1:10,000 dilution (GE Healthcare; NA934-100UL) for 45 min at room temperature. The membranes were washed three times using 1× PBS (Invitrogen; AM9625) with 0.5% Tween 20 and the signal detection was performed using Pierce ECL western Blotting Substrate (Thermo Fisher Scientific). Monoclonal antiactin (plant) antibody was used as a loading control (Sigma-Aldrich; A0480). The antiactin (plant) antibody diluted 1:4,000 was incubated for 45 min at room temperature followed by the secondary antibody (GE Healthcare; NA931-1ML) at a 1:10,000 dilution incubated for 45 min at room temperature.
RNA Extraction and Reverse Transcription Quantitative PCR
Total RNA was isolated from root samples using the RNeasy mini kit (Qiagen) and treated with DNase using the TURBO DNA-free kit (Ambion) according to the manufacturer’s instructions. Nucleic acid concentration was determined with a NanoDrop ND-1000 UV-Vis spectrophotometer (NanoDrop Technologies). First-strand cDNA was synthesized from equal amounts of DNA-free RNA using oligo(dT) primer and Superscript II reverse transcriptase (Invitrogen) as indicated by the manufacturer. The first-strand cDNA was used as a template for RT-qPCR in a 25-µL reaction mix using the SYBR Green PCR Master Mix (Applied Biosystems) with programs recommended by the manufacturer in the QuantStudio 12K Flex Real-Time PCR System (Applied Biosystems). Three independent replicates were performed for each sample. The ΔΔCT method was used to determine the relative gene expression (Livak and Schmittgen, 2001), with the expression of elongation factor 1 alpha (EF1α; At5g60390) used as an internal control. Primers used for RT-qPCR analyses are given in Supplemental Table S1.
Chlorophyll Content
Chlorophyll was extracted and measured following a protocol from Abadía and Abadía (1993), with some modifications. The equation used to calculate chlorophyll concentrations is described by Lichtenthaler (1987). Frozen shoot samples (five per sample) were disrupted using stainless steel beads in a TissueLyzer II (Qiagen). Chlorophyll was extracted with 500 μL 80% (v/v) acetone in a TissueLyzer II, centrifuged, and the supernatant was collected in Eppendorf tubes. Extraction was carried out two more times and extracts were pooled to give a total volume of 1.5 mL. Absorbance was measured at 663-, 647-, and 750-nm wavelengths in a PowerWave XS2 microplate spectrophotometer (BioTek). Absorbance at 663 and 647 nm was corrected for scattering with the 750-nm absorbance measurements.
Shoot Fe Content
Total Fe in shoots was determined by the BPDS method described by Pan et al. (2015), with slight modifications. Shoot samples (20 plants per sample) were dried in an oven at 50°C for 2 d and incubated in 225 μl of 65% (v/v) nitric acid (HNO3) at 96°C for 6 h, followed by 150 μL of 30% (v/v) H2O2 at 56°C for 2 h. Dilution of each sample was made by the addition of 225 μL of sterile water. Samples were mixed in assay solution that contained 1 mm BPDS, 0.6 m sodium acetate, and 0.48 m hydroxylamine hydrochloride. The concentration of Fe2+-BPDS3 complexes were measured by A535 in a PowerWave XS2 microplate spectrophotometer (BioTek). Fe concentrations were determined against a standard curve made with FeCl3 that was treated in the same way as the plant materials.
Detection of Fluorescent Compounds in Roots and Media
The accumulation of fluorescent compounds was visualized in the BioSpectrum 600 imaging system (UVP). Photographs were taken before and after removing plants from the growth media using 365 nm as the excitation wavelength, SYBR Gold 485 to 655 nm as the emission filter, and 9 s as the exposure time.
Collection of Methanolic Extracts from Roots and Growth Media
To extract coumarins from Arabidopsis roots, samples (20 roots per sample) were pulverized using stainless steel beads in a TissueLyzer II (Qiagen) and 500 μL of 80% (v/v) methanol containing 0.5 ppm 4-methylumbelliferone as an internal standard. The extracts were centrifuged and supernatant was collected in Eppendorf tubes. Extraction was carried out one more time, and extracts were pooled to give a total volume of 1 mL. To extract root exudates from growth media, media were dried in an oven at 50°C for 2 d and extraction was done by incubating the media in 10 mL of 80% (v/v) methanol containing 0.1 ppm 4-methylumbelliferone as an internal standard for 30 min. The extract was collected and extraction was repeated twice. Extracts were pooled to give a total volume of 30 mL. Samples were centrifuged at 13,200 rpm for 10 min before transferring into appropriate vials for further analysis.
Targeted Coumarin Profile Analysis
Coumarins were analyzed following a protocol adapted from Schmid et al. (2014). UPLC-QTOF-MS analysis was performed on an Acquity UPLC system (Waters) and a SYNAPT G2 high-definition mass spectrometry system (Waters) with an electrospray ionization interface, ion mobility, and time-of-flight system. Samples were separated on an Acquity BEH phenyl column (100 mm × 2.1 mm, 1.7 μm; Waters). The column temperature was set to 35°C. The mobile phase consisted of 0.1% (v/v) formic acid in 2% (v/v) acetonitrile (solution A) and 0.1% (v/v) formic acid in 100% acetonitrile (solution B). The gradient duration was 13 min at a flow rate of 0.4 mL/min. The elution gradient was as follows: starting with 1% B for 1 min, 1 to 99% B within 10 min, holding at 99% B for 0.5 min, 99 to 1% B in 0.01 min, and then holding at 1% B for another 1.49 min. The injected volume was 5 μL. Spectra were collected in the positive ionization (ES+) mode. The electrospray capillary voltage was set to 3 kV and the cone voltage was set to 30 V. The source and desolvation temperatures were 80°C and 350°C, respectively. The desolvation and cone gas flow rates were 700 and 20 L/h, respectively. A lock mass calibration of Leu encephalin at a concentration of 1 ppm in water:methanol (50:50, v/v) was infused at a flow rate of 10 μL/min via a lock spray. The MS range acquired was 50 to 1,200 m/z with 0.2 s/scan in the centroid mode. For data analysis, the acquired mass data were imported to Markerlynx (Waters) within the Masslynx software (version 4.1) for peak detection and alignment. The retention time and m/z data for each peak were determined by the software and compared to authentic standards. Coumarin compounds studied are indicated in Supplemental Figure S5. Data were processed using TargetLynx application package within the MassLynx software (Waters). Relative coumarin levels were quantified by normalizing the metabolite peak area to the internal standard peak area and the number of roots. Peaks with a signal-to-noise ratio below 10 were excluded.
Determination of Root and Fraxetin FCR Activity
For the root FCR experiment, plants were precultivated for 10 d in ES media and then transferred to either ES, navFe, or –Fe (ES with no Fe3+-EDTA, containing 100 μm ferrozine) media for 3 d. The protocol for measuring root FCR activity was adapted from Grillet et al. (2014). Whole-root systems from five Arabidopsis plants were excised, rinsed with either 10 mm MES (pH 5.5) or MOPS (pH 7.0) buffer solution, and incubated in 2 mL of assay solution with shaking in the dark for 1 h. The assay solution was buffered at pH 5.5 (with 10 mm MES) or pH 7.0 (with 10 mm MOPS), containing 300 µm BPDS and 100 µm Fe3+-EDTA. Blanks without roots were included to correct for any FCR activity not attributable to roots. The concentration of Fe2+-BPDS3 complexes was measured by A535 in a PowerWave XS2 microplate spectrophotometer (BioTek). A standard curve was prepared by dilution of a stock FeSO4 solution. The FCR activity of fraxetin and scopoletin dissolved in DMSO was measured in similar ways as described above. Instead of incubating root samples, 100 µm fraxetin and scopoletin was added. Blanks including DMSO were included to correct for any FCR activity not associated to the tested compounds.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: S8H, At3g12900; F6’H1, At3g13610; FIT, At2g28160; FRO2, At1g01580; and IRT1, At4g19690.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. T-DNA insertion alleles of s8h.
Supplemental Figure S2. Full scans of western blots.
Supplemental Figure S3. F6’H1 transcript levels.
Supplemental Figure S4. Visualization of fluorescent compounds.
Supplemental Figure S5. Structures of coumarin compounds.
Supplemental Figure S6. Effects of Fe availability on the growth of 22 Arabidopsis accessions.
Supplemental Table S1. Primers used in this study.
Acknowledgments
We thank Dr. Catherine Cure (INRA Montpellier, France) for kindly providing the fit-2 mutant seeds, Dr. Paul E. Verslues (Institute of Plant and Microbial Biology, Academia Sinica, Taiwan) for kindly providing seeds of different Arabidopsis accessions, and Dr. Thomas J. Buckhout for critical comments on the manuscript. We acknowledge the Small Molecule Metabolomics core facility, sponsored by IPMB and the Scientific Instrument Center, Academia Sinica, for providing technical assistance, useful discussion, and support with data acquisition and analysis of Synapt HDMS experiments. We also acknowledge the use of the QuantStudio 12K Flex Real-Time PCR System at the Genomic Technology Core Facility at the Institute of Plant and Microbial Biology, Academia Sinica. We also thank Ms. Ya-Tan Cheng (Schmidt laboratory) for technical assistance.
Footnotes
This work was supported by a grant from the Ministry of Science and Technology to W.S. (grant 104-2311-B-001-039-MY3).
Articles can be viewed without a subscription.
References
- Abadía J, Abadía A (1993) Iron and plant pigments. In Barton LL, Hemming BC, eds, Iron Chelation in Plants and Soil Microorganisms, Academic Press, San Diego, CA, pp 327–343 [Google Scholar]
- Brown J, Ambler J (1973) “Reductants” released by roots of Fe-deficient soybeans. Agron J 65: 311–314 [Google Scholar]
- Brumbarova T, Bauer P, Ivanov R (2015) Molecular mechanisms governing Arabidopsis iron uptake. Trends Plant Sci 20: 124–133 [DOI] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML (2004) The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakora FD, Phillips DA (2002) Root exudates as mediators of mineral acquisition in low-nutrient environments. Plant Soil 245: 35–47 [Google Scholar]
- Döll S, Kuhlmann M, Rutten T, Mette MF, Scharfenberg S, Petridis A, Berreth DC, Mock HP (2018) Accumulation of the coumarin scopolin under abiotic stress conditions is mediated by the Arabidopsis thaliana THO/TREX complex. Plant J 93: 431–444 [DOI] [PubMed] [Google Scholar]
- De Silva B. (1934) The distribution of “calcicole” and “calcifuge” species in relation to the content of the soil in calcium carbonate and exchangeable calcium, and to soil reaction. J Ecol 22: 532–553 [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93: 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estelle MS, Somerville C (1987) Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol Gen Genet 206: 200–206 [Google Scholar]
- Fourcroy P, Sisó-Terraza P, Sudre D, Savirón M, Reyt G, Gaymard F, Abadía A, Abadia J, Alvarez-Fernández A, Briat JF (2014) Involvement of the ABCG37 transporter in secretion of scopoletin and derivatives by Arabidopsis roots in response to iron deficiency. New Phytol 201: 155–167 [DOI] [PubMed] [Google Scholar]
- Fourcroy P, Tissot N, Gaymard F, Briat JF, Dubos C (2016) Facilitated Fe nutrition by phenolic compounds excreted by the Arabidopsis ABCG37/PDR9 transporter requires the IRT1/FRO2 high-affinity root Fe2+ transport system. Mol Plant 9: 485–488 [DOI] [PubMed] [Google Scholar]
- Grillet L, Ouerdane L, Flis P, Hoang MT, Isaure MP, Lobinski R, Curie C, Mari S (2014) Ascorbate efflux as a new strategy for iron reduction and transport in plants. J Biol Chem 289: 2515–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grime J, Hodgson J (1968) Investigation of the ecological significance of lime-chlorosis by means of largescale comparative experiments. In IH Rorison, ed, Ecological Aspects of the Mineral Nutrition of Plants. Blackwell, Oxford, UK, pp 67–99 [Google Scholar]
- Hether N, Olsen R, Jackson L (1984) Chemical identification of iron reductants exuded by plant roots. J Plant Nutr 7: 667–676 [Google Scholar]
- Ivanov R, Brumbarova T, Bauer P (2012) Fitting into the harsh reality: regulation of iron-deficiency responses in dicotyledonous plants. Mol Plant 5: 27–42 [DOI] [PubMed] [Google Scholar]
- Kai K, Mizutani M, Kawamura N, Yamamoto R, Tamai M, Yamaguchi H, Sakata K, Shimizu B (2008) Scopoletin is biosynthesized via ortho-hydroxylation of feruloyl CoA by a 2-oxoglutarate-dependent dioxygenase in Arabidopsis thaliana. Plant J 55: 989–999 [DOI] [PubMed] [Google Scholar]
- Kawai Y, Ono E, Mizutani M (2014) Evolution and diversity of the 2-oxoglutarate-dependent dioxygenase superfamily in plants. Plant J 78: 328–343 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Nishizawa NK (2012) Iron uptake, translocation, and regulation in higher plants. Annu Rev Plant Biol 63: 131–152 [DOI] [PubMed] [Google Scholar]
- Lan P, Li W, Wen TN, Shiau JY, Wu YC, Lin W, Schmidt W (2011) iTRAQ protein profile analysis of Arabidopsis roots reveals new aspects critical for iron homeostasis. Plant Physiol 155: 821–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA. (1998) The calcicole-calcifuge problem revisited. Adv in Bot Res 29: 1–30 [Google Scholar]
- Le Roy J, Huss B, Creach A, Hawkins S, Neutelings G (2016) Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants. Front Plant Sci 7: 735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang H, Ai Q, Liang G, Yu D (2016) Two bHLH transcription factors, bHLH34 and bHLH104, regulate iron homeostasis in Arabidopsis thaliana. Plant Physiol 170: 2478–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Functional organization of carotenoids and prenylquinones in the photosynthetic membrane. In PK Stumpf, JB Mudd, WD Nes, eds, The Metabolism, Structure, and Function of Plant Lipids. Springer, Boston, pp 63–74 [Google Scholar]
- Link HF. (1789) Florae Göttingensis specimen, sistens vegetabilia saxo calcareo propria: Diss inaug botan
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN (2010) The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22: 2219–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H. (1995) Mineral Nutrition of Higher Plants. Academic Press, London [Google Scholar]
- Mladenka P, Macáková K, Zatloukalová L, Reháková Z, Singh BK, Prasad AK, Parmar VS, Jahodár L, Hrdina R, Saso L (2010) In vitro interactions of coumarins with iron. Biochimie 92: 1108–1114 [DOI] [PubMed] [Google Scholar]
- Nardi S, Pizzeghello D, Reniero F, Rascio N (2000) Chemical and biochemical properties of humic substances isolated from forest soils and plant growth. Soil Sci Soc Am J 64: 639–645 [Google Scholar]
- Palmer CM, Hindt MN, Schmidt H, Clemens S, Guerinot ML (2013) MYB10 and MYB72 are required for growth under iron-limiting conditions. PLoS Genet 9: e1003953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan IC, Tsai HH, Cheng YT, Wen TN, Buckhout TJ, Schmidt W (2015) Post-transcriptional coordination of the Arabidopsis iron deficiency response is partially dependent on the E3 ligases RING DOMAIN LIGASE1 (RGLG1) and RING DOMAIN LIGASE2 (RGLG2). Mol Cell Proteomics 14: 2733–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römheld V, Marschner H (1984) Plant‐induced pH changes in the rhizosphere of “Fe‐efficient” and “Fe‐inefficient” soybean and corn cultivars. J Plant Nutr 7: 623–630 [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Celma J, Lin W-D, Fu G-M, Abadía J, López-Millán A-F, Schmidt W (2013) Mutually exclusive alterations in secondary metabolism are critical for the uptake of insoluble iron compounds by Arabidopsis and Medicago truncatula. Plant Physiol 162: 1473–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römheld V, Marschner H (1986) Evidence for a specific uptake system for iron phytosiderophores in roots of grasses. Plant Physiol 80: 175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi S, Schmidt W (2009) Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol 183: 1072–1084 [DOI] [PubMed] [Google Scholar]
- Schmid NB, Giehl RF, Döll S, Mock HP, Strehmel N, Scheel D, Kong X, Hider RC, von Wirén N (2014) Feruloyl-CoA 6′-Hydroxylase1-dependent coumarins mediate iron acquisition from alkaline substrates in Arabidopsis. Plant Physiol 164: 160–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Günther C, Weber M, Spörlein C, Loscher S, Böttcher C, Schobert R, Clemens S (2014) Metabolome analysis of Arabidopsis thaliana roots identifies a key metabolic pathway for iron acquisition. PLoS One 9: e102444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt W, Fühner C (1998) Sensitivity to and requirement for iron in Plantago species. New Phytol 138: 639–651 [Google Scholar]
- Selote D, Samira R, Matthiadis A, Gillikin JW, Long TA (2015) Iron-binding E3 ligase mediates iron response in plants by targeting basic helix-loop-helix transcription factors. Plant Physiol 167: 273–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisó-Terraza P, Luis-Villarroya A, Fourcroy P, Briat JF, Abadía A, Gaymard F, Abadía J, Álvarez-Fernández A (2016a) Accumulation and secretion of coumarinolignans and other coumarins in Arabidopsis thaliana roots in response to iron deficiency at high pH. Front Plant Sci 7: 1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisó-Terraza P, Rios JJ, Abadía J, Abadía A, Álvarez-Fernández A (2016b) Flavins secreted by roots of iron-deficient Beta vulgaris enable mining of ferric oxide via reductive mechanisms. New Phytol 209: 733–745 [DOI] [PubMed] [Google Scholar]
- Sivitz A, Grinvalds C, Barberon M, Curie C, Vert G (2011) Proteasome-mediated turnover of the transcriptional activator FIT is required for plant iron-deficiency responses. Plant J 66: 1044–1052 [DOI] [PubMed] [Google Scholar]
- Sivitz AB, Hermand V, Curie C, Vert G (2012) Arabidopsis bHLH100 and bHLH101 control iron homeostasis via a FIT-independent pathway. PLoS One 7: e44843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwinska J, Siatkowska K, Olry A, Grosjean J, Hehn A, Bourgaud F, Meharg AA, Carey M, Lojkowska E, Ihnatowicz A (2018) Scopoletin 8-hydroxylase: a novel enzyme involved in coumarin biosynthesis and iron-deficiency responses in Arabidopsis. J Exp Bot [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehmel N, Böttcher C, Schmidt S, Scheel D (2014) Profiling of secondary metabolites in root exudates of Arabidopsis thaliana. Phytochemistry 108: 35–46 [DOI] [PubMed] [Google Scholar]
- Susín S, Abadía A, Gonzalez-Reyes JA, Lucena JJ, Abadia J (1996) The pH requirement for in vivo activity of the iron deficiency-induced “Turbo” ferric chelate reductase - A comparison of the iron-deficiency-induced iron reductase activities of intact plants and isolated plasma membrane fractions in sugar beet. Plant Physiol 110: 111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HH, Schmidt W (2017) Mobilization of iron by plant-borne coumarins. Trends Plant Sci 22: 538–548 [DOI] [PubMed] [Google Scholar]
- Unger F. (1836) Über den Einfluss des Bodens auf die Vertheilung der Gewächse, nachgewiesen in der Vegetation des nordoestlichen Tirols. Rohrmann & Schweigert, Wien, Germany [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wandruszka R. (2006) Phosphorus retention in calcareous soils and the effect of organic matter on its mobility. Geochem Trans 7: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Cui Y, Liu Y, Fan H, Du J, Huang Z, Yuan Y, Wu H, Ling HQ (2013) Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol Plant 6: 503–513 [DOI] [PubMed] [Google Scholar]
- Whipps J. (1990) The Rhizosphere, Lynch JM, ed. Wiley, Chichester, UK [Google Scholar]
- Yang JW, Lin W, Schmidt W (2010) Transcriptional Profiling of the Arabidopsis Iron Deficiency Response Reveals Conserved Transition Metal Homeostasis Networks. Plant Physiol 152: 2130–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ (2008) FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res 18: 385–397 [DOI] [PubMed] [Google Scholar]
- Zamioudis C, Hanson J, Pieterse CMJ (2014) β-Glucosidase BGLU42 is a MYB72-dependent key regulator of rhizobacteria-induced systemic resistance and modulates iron deficiency responses in Arabidopsis roots. New Phytol 204: 368–379 [DOI] [PubMed] [Google Scholar]
- Zhang X, Gou M, Guo C, Yang H, Liu CJ (2015) Down-regulation of Kelch domain-containing F-box protein in Arabidopsis enhances the production of (poly)phenols and tolerance to ultraviolet radiation. Plant Physiol 167: 337–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler J, Schmidt S, Strehmel N, Scheel D, Abel S (2017) Arabidopsis Transporter ABCG37/PDR9 contributes primarily highly oxygenated Coumarins to Root Exudation. Sci Rep 7: 3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohlen A, Tyler G (2000) Immobilization of tissue iron on calcareous soil: differences between calcicole and calcifuge plants. Oikos 89: 95–106 [Google Scholar]
- Zohlen A, Tyler G (2004) Soluble inorganic tissue phosphorus and calcicole-calcifuge behaviour of plants. Ann Bot 94: 427–432 [DOI] [PMC free article] [PubMed] [Google Scholar]