In the wheat Rht18 semidwarf, increased expression of GA2oxA9 metabolizes GA12 precursor to inactive GA110, thereby reducing flux through the GA biosynthetic pathway, resulting in a lower content of bioactive GA and reduced growth.
Abstract
Semidwarfing genes have improved crop yield by reducing height, improving lodging resistance, and allowing plants to allocate more assimilates to grain growth. In wheat (Triticum aestivum), the Rht18 semidwarfing gene was identified and deployed in durum wheat before it was transferred into bread wheat, where it was shown to have agronomic potential. Rht18, a dominant and gibberellin (GA) responsive mutant, is genetically and functionally distinct from the widely used GA-insensitive semidwarfing genes Rht-B1b and Rht-D1b. In this study, the Rht18 gene was identified by mutagenizing the semidwarf durum cultivar Icaro (Rht18) and generating mutants with a range of tall phenotypes. Isolating and sequencing chromosome 6A of these “overgrowth” mutants showed that they contained independent mutations in the coding region of GA2oxA9. GA2oxA9 is predicted to encode a GA 2-oxidase that metabolizes GA biosynthetic intermediates into inactive products, effectively reducing the amount of bioactive GA (GA1). Functional analysis of the GA2oxA9 protein demonstrated that GA2oxA9 converts the intermediate GA12 to the inactive metabolite GA110. Furthermore, Rht18 showed higher expression of GA2oxA9 and lower GA content compared with its tall parent. These data indicate that the increased expression of GA2oxA9 in Rht18 results in a reduction of both bioactive GA content and plant height. This study describes a height-reducing mechanism that can generate new genetic diversity for semidwarfism in wheat by combining increased expression with mutations of specific amino acid residues in GA2oxA9.
Gibberellins (GAs) are a class of plant hormones involved in many aspects of growth and development, including seed germination, leaf growth, floral induction, and stem elongation (Yamaguchi, 2008). The regulation of plant height, either by lowering the bioactive GA content or by inhibiting GA signaling, formed the basis of the Green Revolution that was associated with major yield increases in wheat (Triticum aestivum) and rice (Oryza sativa; Hedden, 2003). The wheat semidwarfing genes Rht-B1b and Rht-D1b encode DELLA proteins that lack a functional DELLA domain, preventing normal GA signaling and resulting in reduced responsiveness to GA and reduced elongation (Peng et al., 1999). The major semidwarfing gene sd-1 in rice encodes a nonfunctional GA 20-oxidase, a key enzyme in the GA biosynthetic pathway, and causes a lower content of bioactive GA (Sasaki et al., 2002; Spielmeyer et al., 2002). Mutations in GA 20- and GA 3-oxidase biosynthetic genes that reduce plant height have also been identified in other crop species, including maize (Zea mays; dwarf1) and barley (Hordeum vulgare; sdw1/denso; Spray et al., 1996; Jia et al., 2015). The GA metabolic enzymes are encoded by multiple genes, often with distinct tissue and developmental specific expression patterns (Pearce et al., 2015). Mutations in stem-expressed GA 20-oxidase, such as in the rice sd-1 mutant, reduce plant height without affecting other important GA-dependent processes, such as reproductive development (Spielmeyer et al., 2002). However, unlike the DELLA GA signaling mutants, GA biosynthetic mutants are responsive to the application of GA.
GA biosynthesis in cereals proceeds through a common intermediate GA12 that is hydroxylated at C-13 to form GA53 before being converted to bioactive GA1 in a multistep process catalyzed by dioxygenase enzymes (Yamaguchi, 2008; Hedden and Thomas, 2012). The content of GA1 depends on its rate of synthesis and on the rate at which it is inactivated by 2-oxidation. Different GA 2-oxidase isoforms metabolize GA1 and its immediate precursor GA20 to the inactive GA8 and GA29, respectively (C19 isoforms), or convert the earlier GA biosynthetic intermediates GA12 and GA53 to the inactive GA110 and GA97, respectively (C20 isoforms; Sakamoto et al., 2003; Schomburg et al., 2003). Metabolism of biosynthetic intermediates to these inactive products reduces flux through the biosynthetic pathway, leading to a lower GA1 content and reduced growth (Schomburg et al., 2003; Lo et al., 2008).
A new strategy for reducing height and achieving semidwarf phenotypes has been to overexpress GA 2-oxidase transgenes in a range of species (Busov et al., 2003; Elias et al., 2012; Wuddineh et al., 2015). In wheat, ectopic expression of the bean (Phaseolus coccineus) PcGA2ox1 gene decreased GA content and strongly reduced plant height, but also had pleiotropic effects on growth habit, tillering, and ear size (Appleford et al., 2007). In rice, overexpression of the C19 OsGA2ox1 gene also resulted in severe height reduction, whereas expression of the same transgene using the promoter of a native GA biosynthesis gene yielded plants with semidwarf phenotypes and fewer undesirable characteristics (Sakamoto et al., 2001, 2003). Overexpressing the C20 OsGA2ox6 or OsGA2ox9 genes in rice resulted in moderate height reductions without affecting time to anthesis or grain production (Lo et al., 2008; Huang et al., 2010). Taken together, these studies demonstrate the agronomic potential of overexpressing GA 2-oxidase transgenes to achieve semidwarf lines with varying degrees of height reduction.
Rht18 is a dominant, semidwarfing gene that was identified following fast neutron mutagenesis of the tall durum wheat variety Anhinga and released as the commercial semidwarf durum cultivar Icaro (Konzak, 1988). Rht18 has also been introgressed into hexaploid bread wheat, and initial field studies have demonstrated its agronomic potential (Yang et al., 2015; Tang, 2015). In a recent mapping study of two durum RIL (recombinant inbred line) populations, Rht18 was mapped within an interval of 1.8 centimorgan (cM) on chromosome 6A that contained a GA 2-oxidase gene, GA2oxA9 (Vikhe et al., 2017). An association mapping study of hexaploid winter wheats identified a major quantitative trait locus for reduced height on chromosome 6A that overlapped with the Rht18 region and included GA2oxA9 (Würschum et al., 2017). Rht18 is GA responsive, as coleoptile length and seedling leaf elongation rates both increased in Rht18 lines following GA application (Ellis et al., 2004; Vikhe et al., 2017). Given its GA responsiveness and its distinct location from Rht-B1 and Rht-D1 (chromosome group 4), Rht18 represents a semidwarfing gene in wheat that is unrelated to mutant DELLA genes.
In this study, we identified the Rht18 gene by mutagenizing the semidwarf line Icaro with sodium azide and screening for overgrowth mutations that suppressed dwarfism and produced tall lines. Thirteen such mutants contained independent mutations in the coding region or splice sites of GA2oxA9. The transcript levels of GA2oxA9 were higher, and the GA1 content lower in semidwarf Icaro compared with its tall parent Anhinga. We also found that the GA2oxA9 gene encodes a functional enzyme that metabolizes a C20 GA precursor (GA12) and that enzyme activity was greatly reduced in one of the overgrowth mutants. The data indicate that the Rht18 phenotype is caused by increased expression of GA2oxA9, resulting in a lower content of bioactive GA. This study highlights how an allelic variant in a GA 2-oxidase gene can regulate growth in a manner that previously has only been observed in transgenic plants.
RESULTS
Rht18 Maps to the Centromeric Region of Chromosome 6A
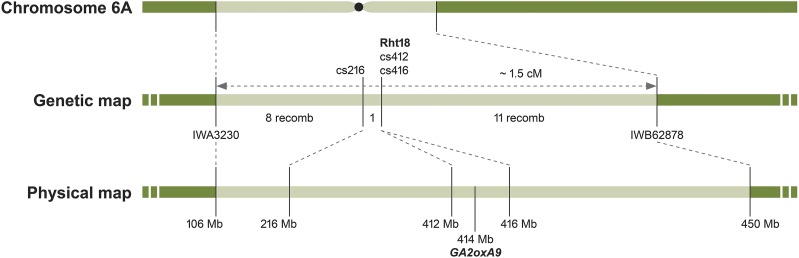
The Rht18 semidwarfing gene was previously mapped to chromosome 6A in a region flanked by the single-nucleotide polymorphism (SNP)-based markers IWA3230 and IWB62878 in durum wheat (Fig. 1; Tang, 2015). In this study, the flanking markers were used to screen an F2 population derived from a cross between Icaro (semidwarf) and Langdon (tall) durum wheat cultivars to define a genetic interval of approximately 1.5 cM, which contained Rht18. IWA3230 was located at 106 Mb on the short arm and IWB62878 at 450 Mb on the long arm of chromosome 6A, spanning a centromeric region of approximately 344 Mb on the Chinese Spring wheat genome sequence (Fig. 1). The unfavorable relationship of genetic to physical distance in this region (1 cM/230 Mb) precluded a map-based cloning approach to characterize the Rht18 locus.
Figure 1.
Genetic and physical mapping of Rht18 region on chromosome 6A. Genetic map developed from Icaro x Langdon F2:F3 mapping population. Markers were mapped to the IWGSC Ref Seq v1.0 wheat genome assembly to determine their physical locations.
Identification of Rht18 Overgrowth Mutants in Tetraploid and Hexaploid Wheat
Rht18 behaves as a dominant gene in durum wheat (Tang, 2015) and is a presumed gain-of-function mutant. We predicted that sodium azide induced mutations in the Rht18 gene would result in loss-of-function (tall) overgrowth derivatives, similar to the strategy recently used to identify second site mutations within the severe DELLA dwarfing allele Rht-B1c (Chandler and Harding, 2013). Chromosome 6A could then be sequenced from a set of multiple, independent, allelic mutants and analyzed using recently developed bioinformatics tools to define the gene responsible for the dwarfing phenotype (Sánchez-Martín et al., 2016).
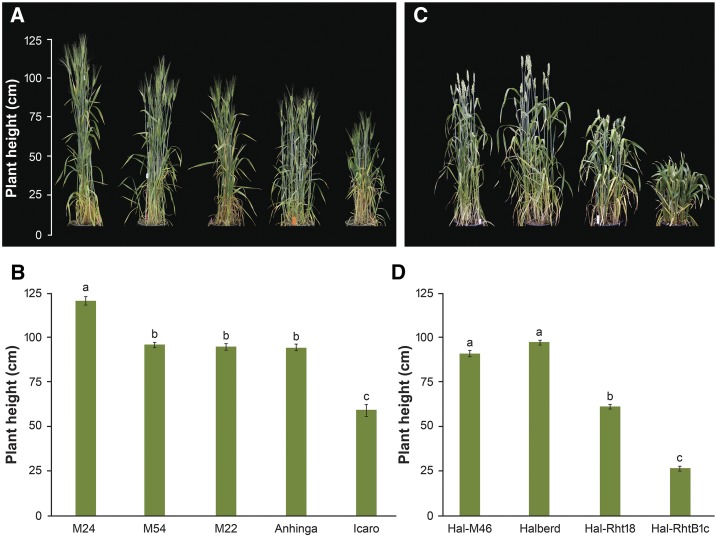
Icaro was mutagenized with sodium azide and a bulk M2 population was screened for height in the field. Putative overgrowth mutants were then selected and progeny tested in the glasshouse. Fourteen mutants (ranging in height from 73–121 cm) were confirmed to be taller than Icaro (59 cm) and closer in height to the tall parent Anhinga (94 cm; Fig. 2). Five overgrowth mutants, M12, M17, M22, M24, and M54, which were most likely to represent independent events based on the structure of the M2 population, were selected for further study. Overgrowth mutants were also identified in a hexaploid bread wheat background by screening a sodium azide M2 population of Halberd-Rht18 for height in the field. Eighty plants judged to be taller than their semidwarf parent were selected, and 30 were subsequently confirmed by progeny testing in the glasshouse. The mean height of the Halberd-Rht18 overgrowth mutants ranged between 86 and 116 cm, which was taller than the Halberd-Rht18 parent (61 cm) and closer to the tall Halberd isogenic line (97 cm; Fig. 2).
Figure 2.
Plant height is increased in Rht18 overgrowth mutants in tetraploid (A and B) and hexaploid (C and D) wheat. Plant height was measured at maturity and data presented as mean values ± se (n = 12 for each line). Different letters indicate significant differences in plant height of tetraploid (B) and hexaploid (D) wheats (ANOVA, P < 0.05).
Mutations in the GA2oxA9 Gene Are Responsible for the Overgrowth Phenotype
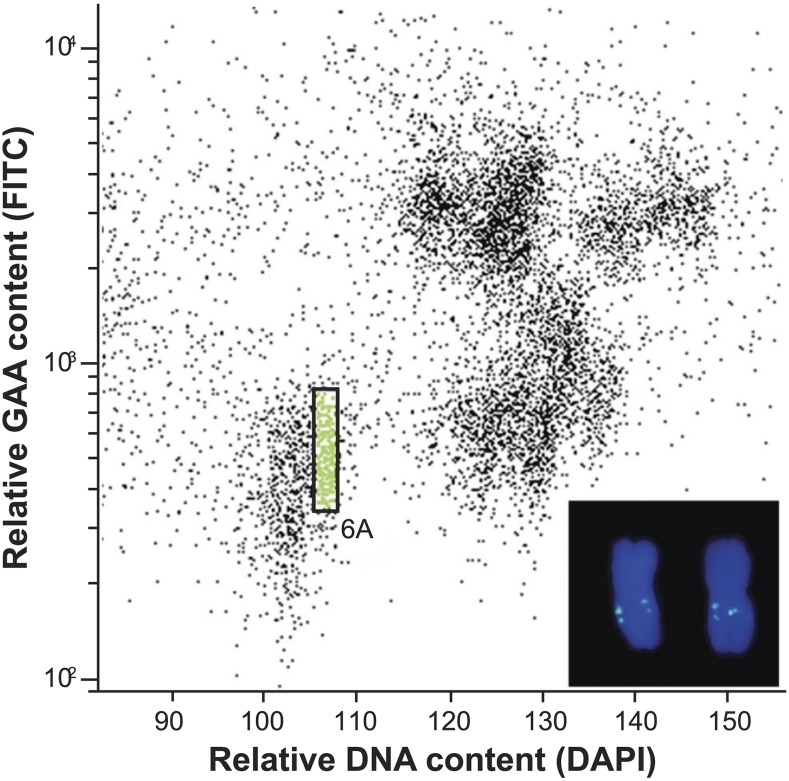
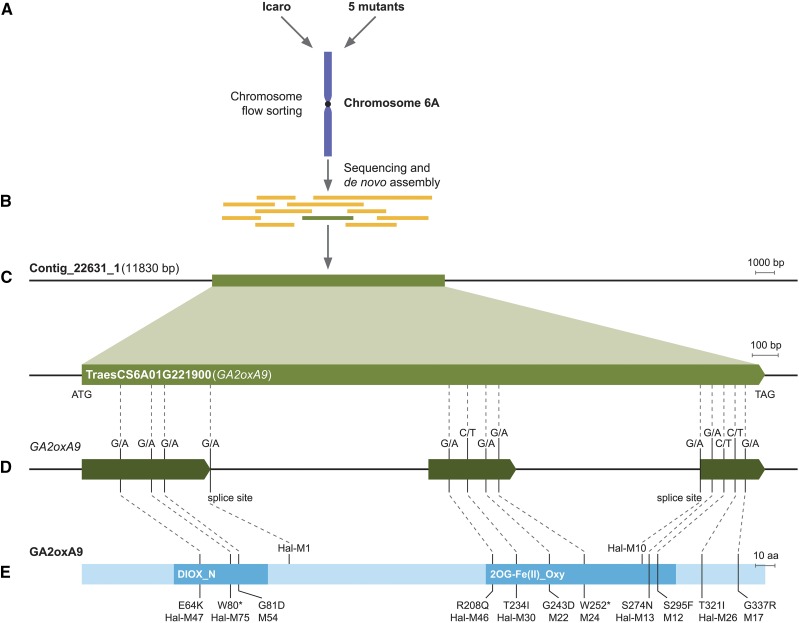
To test the hypothesis that overgrowth mutants carried loss-of-function mutations in Rht18, three durum overgrowth mutants (M12, M24, and M54) were intercrossed and outcrossed to Icaro and Anhinga, generating F2 populations for height measurement. Genetic analysis showed that the overgrowth mutations were most likely allelic and either tightly linked to, or within, the Rht18 gene (see “Materials and Methods”). We hypothesized that overgrowth lines would carry mutations in the Rht18 gene. We therefore isolated and sequenced chromosome 6A from mutants and Icaro to identify mutations that may be responsible for the overgrowth phenotype. Chromosome 6A was isolated by flow cytometry sorting from Icaro, Anhinga, and the five durum overgrowth mutants, M12, M17, M22, M24, and M54 (Fig. 3). DNA prepared from isolated chromosomes was sequenced at greater than 80× coverage (Fig. 4). Illumina sequence reads from Icaro were assembled de novo, producing an assembly with a total length of 444 Mb (∼70% of the total length of 6A) that was predicted to cover most of the genic regions. Raw reads from the overgrowth mutants and Icaro were then mapped to the Icaro assembly and single-nucleotide variants (SNVs) identified (Fig. 4). Contigs that contained at least one independent SNV in each of the five overgrowth mutants were identified; there was only a single contig, from a total of >120,000, which contained five independent SNVs, one in each mutant (Fig. 4; Supplemental Fig. S1). This contig (contig_22631_1) contained a single predicted open reading frame (ORF) that encoded a GA 2-oxidase gene (GA2oxA9; Fig. 4). The D subgenome homeolog GA2oxD9 has been demonstrated to encode an enzyme that metabolizes GA biosynthetic intermediates, potentially lowering the content of bioactive GAs. Thus, GA2oxA9 was presumed to have a similar function (Pearce et al., 2015).
Figure 3.
Biparametric flow karyotype of Triticum durum cv Icaro. The population of target chromosome 6A is highlighted in green. Inset: Images of two flow-sorted chromosomes 6A. The chromosome was identified using fluorescence in situ hybridization with a probe for GAA microsatellite (yellow-green). Chromosome were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; blue).
Figure 4.
Identification of GA2oxA9 as a candidate gene for Rht18. A, Chromosome 6A from Icaro and overgrowth mutants isolated and Illumina-sequenced at greater than 80× coverage. B, Icaro Illumina reads de novo assembled and sequence of the overgrowth mutants mapped back to the assembly. C, Contig (22631_1) was identified which contained gene TraesCS6A01G221900 (GA2oxA9). D, Gene model of GA2oxA9 showing three exons and five independent mutations (G/A or C/T transitions) in durum Icaro background. Additional mutations were found in eight Halberd overgrowth mutants (Hal-M47, Hal-M75, Hal-M13, Hal-M46, Hal-M30, Hal-M10, Hal-M1, and Hal-M26). E, Predicted amino acid sequence of GA2oxA9 protein showing single amino acid substitutions in durum and hexaploid wheat mutants except in M24 and Hal-M75 where mutations resulted in early termination codons and Hal-M1 and Hal-M10 where mutations resulted changing spliceosome recognition site. Predicted functional domains include nonhaem dioxygenase (DIOX_N) domain and 2-oxoglutarate and Fe(II)-dependent oxygenase [2OGFe(II)_Oxy] domain.
Nucleotide sequences of the predicted GA2oxA9 ORF from the five mutants were translated and compared with Icaro and other wheat and rice GA 2-oxidases (Supplemental Fig. S2). All mutations occurred within the predicted coding region of GA2oxA9; four mutations involved changes in conserved amino acids, M12 (S295F), M17 (G337R), M22 (G243D), and M54 (G81D), whereas in the fifth mutant, M24 (W252*) a premature termination codon was introduced that was predicted to result in a nonfunctional protein (Fig. 4; Supplemental Fig. S2). Based on these results in durum, we sequenced the GA2oxA9 gene from Halberd-Rht18 and eight of the Halberd-Rht18 overgrowth mutants. Five mutants contained SNVs in the predicted coding region of GA2oxA9, resulting in changes in a conserved amino acid, Hal-M13 (S274N), Hal-M26 (T321I), Hal-M30 (T234I), Hal-M46 (R208Q), and Hal-M47 (E64K). One introduced a premature termination codon, Hal-M75 (W80*), and the remaining two mutants Hal-M1 and Hal-M10 contained a SNV that was predicted to change a spliceosome recognition site (Fig. 4; Supplemental Figs. S3 and S4). These data indicate that 13 independent mutations in the GA2oxA9 gene, in both tetraploid and hexaploid overgrowth lines, are responsible for their enhanced growth through either a loss or reduction in GA2oxA9 protein amount or activity.
Rht18 Semidwarfism Is Associated with Increased Expression of GA2oxA9
In the Icaro x Langdon mapping population, SNP-based markers cs_412 and cs_416 that flank GA2oxA9 in the Chinese Spring physical map cosegregated with Rht18. This was consistent with the possibility that GA2oxA9 is also responsible for the semidwarf phenotype of Rht18 (Fig. 1). However, the nucleotide sequences of the predicted ORF of GA2oxA9 were identical in Icaro and Anhinga. We therefore hypothesized that increased gene expression of GA2oxA9 in Icaro relative to Anhinga could result in a lower GA1 content and reduced height. This idea was supported by previous studies that have shown that increased expression of closely related GA 2-oxidase genes in wheat, rice, poplar (Populus tremula × Populus alba) and Arabidopsis (Arabidopsis thaliana) result in dwarf phenotypes (Busov et al., 2003; Schomburg et al., 2003; Appleford et al., 2007; Lo et al., 2008).
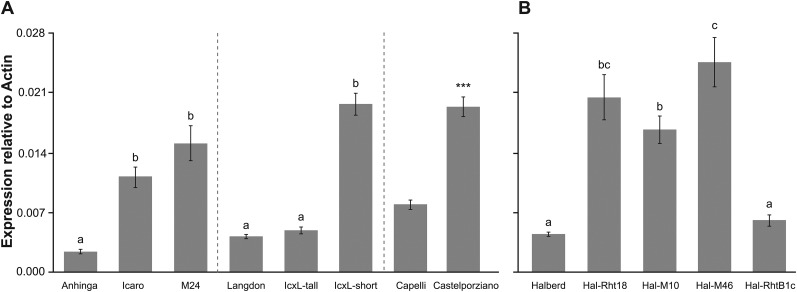
GA2oxA9 expression was assayed by quantitative, real-time reverse transcriptase PCR (qPCR) in elongating peduncles, as most of the height reduction in Icaro was due to reduced peduncle length. Expression of GA2oxA9 was ∼4-fold higher in the Icaro and the M24 overgrowth mutant compared to the tall parent Anhinga (Fig. 5A). GA2oxA9 expression was also determined in fixed short and tall segregants from the Icaro x Langdon mapping population. The expression level of GA2oxA9 was higher in Icaro and the fixed short progeny compared to the Langdon and fixed tall progeny (Fig. 5A). Previously, another semidwarf mutant Castelporziano (Rht14) was identified following the treatment of the tall durum wheat Capelli with fast neutron radiation (Gale et al., 1985). Genetic studies have shown that Rht14 is probably allelic to Rht18 (Haque et al., 2011; Tang, 2015). The nucleotide sequences of the predicted GA2oxA9 ORF from Castelporziano and Capelli were identical to Icaro and Anhinga. Like Icaro, GA2oxA9 expression was also increased in Castelporziano compared to its tall parent Capelli (Fig. 5A). As both Icaro and Castelporziano are tetraploid wheats, the expression of the homeologous gene on the B genome (GA2oxB9) was also assayed in peduncle tissue. In Icaro, M24, Anhinga, Castelporziano, and Capelli, GA2oxB9 expression was below the limit of detection by qPCR.
Figure 5.
GA2oxA9 expression is increased in Rht18 and Rht14 wheat isolines. GA2oxA9 expression relative to actin in elongating peduncle tissue from tetraploid (A) and hexaploid (B) wheat. Data are mean values of four biological replicates ± se. Different letters indicate significant differences between Anhinga, Icaro (Rht18), and M24, between Langdon, fixed tall, and fixed short segregants from Icaro x Langdon mapping population, and between Halberd-Rht18 isolines (ANOVA, P < 0.05). Significant differences between Castelporziano (Rht14) and Capelli are indicated by asterisks (Student’s t test,***P < 0.001).
Expression of GA2oxA9 was also assayed in the Halberd bread wheat (hexaploid) background. Similar to the results seen in the tetraploid lines, expression of GA2oxA9 was increased in Halberd-Rht18 and the Halberd-Rht18 overgrowth mutants Hal-M10 and Hal-M46 compared to Halberd (Fig. 5B). The fold change in expression was similar in magnitude across tetraploid and hexaploid isolines. GA2oxA9 expression was also determined in a severe Halberd dwarf carrying the mutant DELLA Rht-B1c allele (Fig. 2). The expression of GA2oxA9 was similar in Halberd-Rht-B1c compared to Halberd (Fig. 5B), indicating that the increased expression of GA2oxA9 in Halberd-Rht18 is not simply a consequence of short plant stature.
Expression of GA2oxA9 was also determined in elongating first leaves, where the rate of leaf elongation was lower in Icaro compared with Anhinga and M24 (Supplemental Fig. S5). Similar to the results observed in peduncle tissue, expression of GA2oxA9 was increased in Icaro, M24, and Castelporziano compared to the tall parents Anhinga and Capelli (Supplemental Fig. S5). Taken together, these data show a consistent increase in GA2oxA9 expression in elongating peduncle and leaf tissue of semidwarf lines relative to tall parents in both tetraploid and hexaploid wheat.
Rht18 Semidwarfism Is Associated with a Lower Content of Bioactive GA
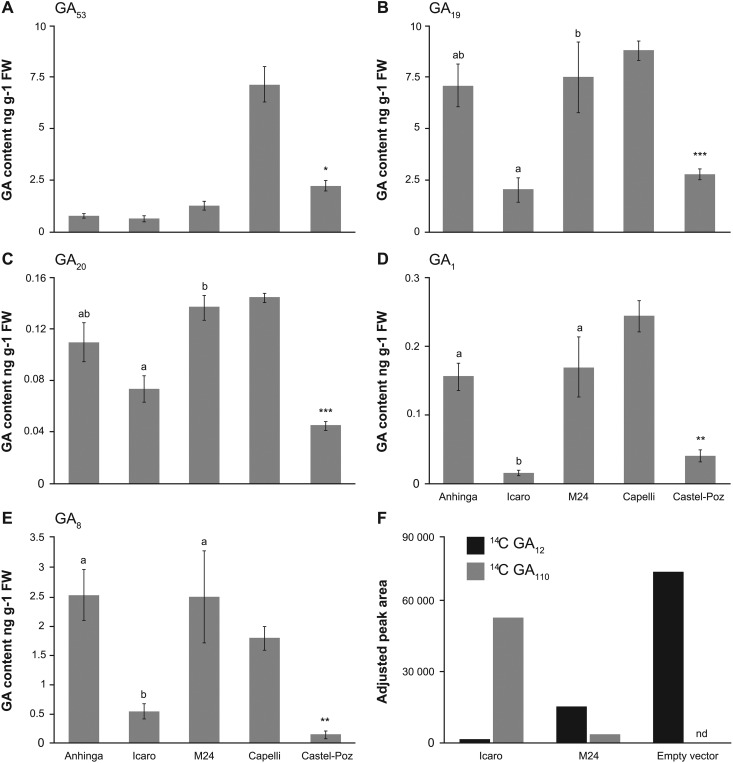
The increase in GA2oxA9 expression observed in Rht18 and Rht14 relative to tall lines is consistent with the hypothesis that increased expression of this gene leads to semidwarfism associated with a lower content of bioactive GA. GA contents were determined in elongating peduncle tissue of both sets of tall and semidwarf isogenic lines (Fig. 6). The content of bioactive GA1 was substantially reduced in the semidwarf lines Icaro and Castelporziano compared to tall lines Anhinga and Capelli (Fig. 6D). The levels of GA precursors were also lower in Castelporziano (GA53, GA19, and GA20), as was the GA1 catabolite GA8 in both Icaro and Castelporziano, suggesting that the lower content of GA1 was the result of reduced flux through the GA biosynthetic pathway (Fig. 6). The content of several endogenous GAs in the M24 overgrowth mutant was greater than in Icaro and similar to the tall isoline Anhinga, consistent with a nonfunctional GA2oxA9 protein resulting from a newly introduced termination codon (Figs. 4 and 6).
Figure 6.
GA content in Icaro (Rht18), Castelporziano (Rht14), and respective tall isolines and enzyme activity of GA2oxA9 from Icaro (Rht18) and tall isoline (M24). A to E, Content of GA precursors GA53, GA19, and GA20 (A–C), bioactive GA1 (D), and inactive catabolite GA8 (E; ng/g FW) in rapidly elongating peduncle tissue from tetraploid wheat lines. Data are mean values of three biological replicates ± se. Different letters indicate significant differences between Icaro (Rht18), M24, and Anhinga (ANOVA, P < 0.05). Significant differences between Castelporziano (Rht14) compared to Capelli are indicated by asterisks (Student’s t test,*P < 0.05, **P < 0.01, and ***P < 0.001). F, Adjusted peak areas of 14C-GA12 and product 14C-GA110 following 1 h incubation of 14C-GA12 with cell-free extracts from E. coli expressing GA2oxA9 from Rht18 (Icaro), tall mutant (M24), and empty vector control monitored by liquid chromatography-tandem mass spectrometry. Extracts from E. coli with empty vector control had no detectable (nd) 14C-GA110.
GA2oxA9 Encodes a Functional GA 2-Oxidase That Inactivates GA12
GA2oxA9 is a member of the C20 GA 2-oxidase family, which have been shown to catalyze the conversion of the C20 GA biosynthetic intermediates GA12 and GA53 to inactive GA110 and GA97, respectively (Schomburg et al., 2003; Lo et al., 2008). The biochemical function of GA2oxA9 was evaluated by expressing the GA2oxA9 gene from Icaro and the M24 overgrowth mutant in Escherichia coli (see “Materials and Methods”). The GA2oxA9 protein from Icaro converted most of the radiolabeled [14C1]GA12 substrate to [14C1]GA110 within 1 h of incubation (Fig. 6F). In comparison, in a parallel assay with the protein from M24, much less GA110 was formed and more of the substrate was retained, indicating that the mutant protein was able to catalyze [14C1]GA12 to [14C1]GA110 but with greatly reduced efficiency compared with Icaro (Fig. 6F). When similar assays were carried out with [14C1]GA1 and [14C1]GA20, no [14C1]GA8 or [14C1]GA29 products were detected, respectively (data not shown), indicating the enzyme may be specific for C20 2-oxidation steps. Although the mutation in M24 was predicted to result in a premature termination codon and a nonfunctional protein in planta, some functional protein must have been expressed in E. coli to enable the catalysis of substrate, since no activity was detected in the empty vector control. The result of the functional analysis was consistent with some predicted full-length protein being detected on the SDS-PAGE gel separating protein extracted from E. coli expressing the M24 construct (Supplemental Fig. S6).
GA2oxA9 Promoter and Upstream Sequences Are Highly Conserved in Icaro and Anhinga
To elucidate DNA polymorphisms that could be responsible for the differential expression of GA2oxA9, we compared 100 kb of Icaro sequence immediately upstream from the predicted transcription start site of GA2oxA9 with Anhinga using the Chinese Spring IWGSC RefSeq v1.0 reference genome assembly (https://wheat-urgi.versailles.inra.fr/). The GA2oxA9 promoter and upstream sequence were highly conserved (Supplemental Fig. S7) with only 13 SNPs detected between Icaro and Anhinga and the closest SNP being 7,314 bp upstream of the transcriptional start site (Supplemental Table S1). The 100-kb sequence of the IWGSC RefSeq 1.0 assembly was identical to the recently released sequence in the Triticum 3.1 assembly (Zimin et al., 2017). The sequences of the 3′ untranslated regions of Anhinga and Icaro were also identical, indicating that transcript stability was unlikely to play a role in the differential expression of the gene. As mutations induced by fast neutron radiation are characterized by the introduction of small deletions, we identified all deletions 5 Mb upstream of the predicted transcription start site of GA2oxA9 in Icaro compared to Anhinga (Supplemental Table S2). Eight deletions ranging in size from 1 to 32 bp were identified, the closest being a 1 bp deletion ∼0.8 Mb upstream of GA2oxA9 (Supplemental Table S2). It is possible that sequence divergence between Anhinga and Chinese Spring prevented the identification of causal deletions in Icaro. Increased expression of GA2oxA9 is likely to have resulted from the fast neutron mutagenesis, and the results from the fine mapping showed that the causal change must be tightly linked to the Rht18 locus. Future work will focus on identifying this causal mutation.
DISCUSSION
The GA2oxA9 gene was identified through a combination of genome complexity reduction, next-generation sequencing, and the application of bioinformatics tools to find causal SNVs in 13 independent mutants. Recent technological and scientific advances in all these areas allow researchers to isolate wheat genes while bypassing lengthy and laborious recombination-based mapping and cloning procedures. In particular, the ability to isolate individual wheat chromosomes in most genetic backgrounds constitutes a paradigm shift in complexity reduction of the wheat genome (Giorgi et al., 2013). A key advantage of whole-chromosome sequencing and analysis using the MutChromSeq protocol compared to other strategies such as RNA-seq or exome capture is that it provides a nonbiased approach to target causal SNVs in protein coding and regulatory sequences alike (Sánchez-Martín et al., 2016). The MutChromSeq approach is particularly useful for target regions close to centromeres, which are difficult to analyze using recombination-based approaches. The mutations in GA2oxA9 were identified without any prior knowledge of detailed map information of the gene on chromosome 6A.
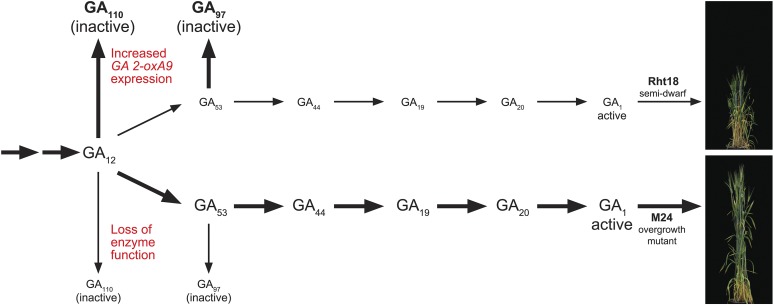
We propose the following model for GA2oxA9 regulation of plant height (Fig. 7): In the tall parent Anhinga, the production of GAs proceeds through the intermediates GA12, GA53, GA44, GA19, and GA20 to bioactive GA1 (Hedden and Thomas, 2012). In the semidwarf Icaro, there is increased expression of GA2oxA9 and enhanced conversion of GA12 to GA110, resulting in decreased flux through the biosynthetic pathway and ultimately lower bioactive GA1 (Fig. 7). This lower content of bioactive GA1 results in reduced plant height in Icaro (Fig. 7). In the overgrowth mutant M24, the function of the GA2oxA9 protein is impaired and the conversion of GA12 to GA110 is greatly reduced, allowing normal flux through the GA biosynthetic pathway and thereby restoring normal GA1 content and plant height (Fig. 7). Thus, a single gene, GA2oxA9, is most likely responsible for both the semidwarf Rht18 and the overgrowth phenotypes in durum and bread wheat.
Figure 7.
GA2oxA9 is responsible for semidwarf and overgrowth phenotypes. Increased expression of GA2oxA9 stimulates conversion of GA12 and GA53 to inactive GA110 and GA97, respectively. This reduces flux in the GA biosynthetic pathway, resulting in lower contents of bioactive GA1 and semidwarfism. Mutations in GA2oxA9 cause loss of protein function and impair the conversion of GA12 and GA53 to inactive GA110 and GA97. This restores flux through the GA biosynthetic pathway, increasing GA1 content and restoring plant height.
The mutation in the M24 overgrowth mutant introduced an early termination codon and the line was taller than wild-type Anhinga, indicating that GA2oxA9 may play a role in the regulation of plant height. However, in our study, we could only detect very low expression of GA2oxA9 in the expanding peduncle tissue of Anhinga (Fig. 5). GA2oxA9 was expressed in bread wheat during the early stages of stem elongation (Pearce et al., 2015), indicating that the GA2oxA9 gene may regulate plant height, but at an earlier time point than that assessed in our study.
An increase in expression of a GA metabolic gene resulted in a lower bioactive GA content and the Rht18 semidwarf phenotype. This mechanism is distinct from mutant DELLA genes Rht-B1b and Rht-D1b that disrupt GA signaling in wheat and from height-reducing mechanisms in other crop species (Peng et al., 1999; Sasaki et al., 2002; Spielmeyer et al., 2002). In barley and rice, semidwarfism is also caused by a reduced bioactive GA content, but this is achieved through loss of function mutations in key GA biosynthetic genes (Sasaki et al., 2002; Spielmeyer et al., 2002; Jia et al., 2015). Such loss-of-function mutants have not been isolated in wheat, most likely due to the presence of closely related, functional gene members in the two (durum) or three (bread wheat) subgenomes (Borrill et al., 2015). In contrast, mutations that increase expression of a GA 2-oxidase gene like Rht18 are dominant gain-of-function mutants that are largely unaffected by gene redundancy in polyploid wheat.
The regulation of plant height by increased expression of GA 2-oxidase has been reported in numerous studies that overexpressed members of this gene family ectopically or by activation-tagged mutants (Sakamoto et al., 2001; Sakai et al., 2003; Appleford et al., 2007; Lo et al., 2008). However, constitutive expression of GA 2-oxidases often results in severe height reduction and reduced fertility (Sakamoto et al., 2001; Sakai et al., 2003; Appleford et al., 2007; Lo et al., 2008). Here, we report nontransgenic wheat mutants with changes in both expression and coding sequence in a GA 2-oxidase gene that controls plant height. A combination of increased gene expression and mutations in conserved amino acid residues generates a range of semidwarf heights in durum and hexaploid wheat. This strategy was used in transgenic rice to generate height variants with improved agronomic performance (Lo et al., 2017). Constitutive expression of mutant C20 OsGA2ox6 isoforms carrying single amino acid changes generated transgenic rice lines with increased yield potential and stress tolerance compared to lines that express the wild-type C20 OsGA2ox6 gene under the control of the endogenous promoter (Lo et al., 2017). The increased expression of the C20 GA2oxA9 in Rht18 has also generated semidwarfs with agronomic potential and provides a nontransgenic approach to further increase genetic diversity by combining with mutations in conserved amino acid residues (Yang et al., 2015). In this study, we have generated more than 30 overgrowth mutants in hexaploid and tetraploid wheat that may prove to be a valuable source of new semidwarfing alleles.
Rht14 and Rht24 Are likely Alleles of Rht18
Rht14 and Rht18 were previously reported to be linked to the same simple sequence repeat marker on chromosome 6A and the analysis of progeny from intercrosses showed that these genes were likely to be alleles (Haque et al., 2011; Tang, 2015). In Castelporziano (Rht14), GA2oxA9 expression was increased and GA content decreased relative to a tall isogenic line mirroring the changes observed between Rht18 isogenic lines (Figs. 5 and 6). These data support the hypothesis that Rht14 and Rht18 are allelic and that the semidwarf phenotype of Rht14 is the result of increased expression of GA2oxA9. The Rht14 and Rht18 semidwarfs were identified by separate groups in fast neutron mutagenized populations of two different tall durum genetic backgrounds. The predicted ORFs of GA2oxA9 were identical between mutants and wild types, so changes in gene expression most probably were responsible for the mutant phenotype. What is the molecular basis that led to changes in gene expression in these mutants? These changes may have arisen because of either mutagen treatment or by spontaneous events. Comparison of sequences upstream of the predicted transcription start sites between mutant and the wild type did not reveal any characteristic small deletions that are typical following treatment with fast neutron radiation (Supplemental Table S1; Koornneef et al., 1982). Changes in chromatin structure were recently linked to the regulation of GA 2-oxidase gene in Arabidopsis root meristems (Li et al., 2017b). Future work will investigate the molecular basis of increased expression of GA2oxA9 in Icaro and Castelporziano. Another major quantitative trait locus for reduced height (Rht24) was recently reported in an association mapping study of European winter wheats (Würschum et al., 2017). The map location of Rht24 overlapped the Rht18 region on chromosome 6A that included GA2oxA9. Natural variation in gene expression and/or allelic diversity in the ORF of GA2oxA9 may be responsible for reducing height in winter wheat cultivars, suggesting that this locus is already playing a role in wheat improvement.
CONCLUSION
Characterizing Rht18 has provided insight into a new height reducing mechanism for wheat by linking an induced mutant to an increase in expression of a GA 2-oxidase gene. We anticipate that this discovery will generate additional variation for height by combining increased expression with specific amino acid changes in GA 2-oxidase to develop a range of semidwarfs independent of mutant DELLA genes in one of the most important food crops.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The tetraploid wheat (Triticum aestivum) Icaro (Rht18) and Castelporziano (Rht14) were derived from independent fast neutron treated populations (Gale et al., 1985; Konzak, 1988). Tetraploid wheats and the hexaploid wheat Halberd were obtained from the Australian Winter Cereals Collection. Halberd-Rht18 and Halberd-Rht-B1c are BC4F4 fixed near isogenic lines that were developed at CSIRO. Plants used for assessment of plant height, GA2oxA9 expression, and GA content were grown in glasshouse conditions at 21°C day/18°C night with daylength extended to 16 h by artificial lighting. Four plants were grown in 20-cm pots containing a soil/compost mix supplemented with slow release NPK fertilizer. Plant height was recorded at maturity by measuring the main stem from the surface of the soil to the tip of the spike. The method for determining leaf elongation rates has been previously described (Chandler and Robertson, 1999).
Genetic and Physical Mapping
A F2:F3 mapping population was generated by crossing Icaro with the tall cultivar Langdon. DNA was extracted from the endosperm half of 700 F2 seeds and screened with SNP markers IWA3230 and IWB62878 previously shown to flank the Rht18 locus (Tang, 2015). Twenty recombinants were identified and phenotyped by measuring heights on 4 to 16 F3 progeny from each recombinant in the glasshouse. A Langdon short read cDNA data set (Trick et al., 2012) was downloaded from the Sequence Read Archive, and reads were quality trimmed using Trimmomatic V0.32 (Bolger et al., 2014). Icaro and Langdon reads were mapped with Bowtie2 to the Chinese Spring IWGSC RefSeq v1.0 genome assembly (https://wheat-urgi.versailles.inra.fr/). The mapped reads within the Rht18 region of interest were visually inspected with the Integrative Genomics Viewer (Robinson et al., 2011) and three SNVs identified and converted to KASP markers (cs216, cs412, and cs416). The 20 recombinants were genotyped, and only one recombination event was identified between cs216 and cs416. The physical location of GA2oxA9, the IWA3230 and IWB62878 markers from the 90K SNP array, and the cs216, cs412, and cs416 markers generated in this study were determined by using their sequences as a query for a BLASTn search of the Chinese Spring IWGSC RefSeq v1.0 genome assembly (https://wheat-urgi.versailles.inra.fr/). Sequences of KASP markers used in this study are shown in Supplemental Table S3.
Mutagenesis and Identification of Overgrowth Mutants
Approximately 4,000 Icaro grains were treated with sodium azide (Chandler and Harding, 2013) and M1 plants grown in an outdoor birdcage area in Canberra in 2013. To evaluate independent mutant events, M2 subpopulations were created by harvesting seed in bulk rows. Approximately 135,000 M2 seeds were sown in plots at the Leeton NSW trial site in 2014. Fourteen overgrowth plants were identified that were 10 to 30 cm taller than Icaro (approximately one overgrowth mutant per 10,000 M2 seed). Five of these lines were selected from different M2 subpopulations and were therefore likely to represent independent events. M3 seed from the 14 overgrowth lines were progeny tested in the glasshouse in 2015 and five overgrowth mutants M12, M17, M22, M24, and M54, which were likely to represent independent events, were selected for further study. M12, M24, and M54 were intercrossed and F2 populations scored for height in the glasshouse. The lack of dwarf lines in the F2 generation indicated that the overgrowth mutations were probably allelic (data not shown). M12 and M24 were also crossed to Icaro and Anhinga. F1 plants from crosses with Icaro were short, indicating that the overgrowth mutation was recessive, and F2 populations segregated for a single dominant dwarfing gene (data not shown). F1 plants from crosses with Anhinga were tall, and the lack of dwarf F2 progeny indicated that the dominant Rht18 dwarfing effect was lost in these mutants (data not shown).
Overgrowth mutants were also generated in a hexaploid background. Approximately 6,000 grains of the BC4F4 Halberd-Rht18 seed was treated with sodium azide as above and sown in the field in 2014. M2 seed was harvested from three bulked plots and ∼300,000 grains were sown in 2015. Screening of the M2 population from one bulked plot identified 110 lines judged to be taller than the bulk population (approximately three overgrowth mutants per 10,000 M2 seed). Eighty of these lines were progeny tested in glasshouse conditions, and 30 have so far been confirmed as overgrowth mutants.
Chromosome Flow Sorting
Suspensions of intact mitotic metaphase chromosomes were prepared from synchronized root tip meristem of young seedlings (Vrána et al., 2000). Prior to flow cytometry, chromosome GAA microsatellite loci were fluorescently labeled using 5′-FITC-GAA7-FITC-3′ and chromosomal DNA was stained by 4′,6-diamidino-2-phenylindole at 2 µg/mL (Vrána et al., 2016). Chromosome samples were analyzed by FASCAria II SORP flow sorter (BD Biosciences). Approximately 35,000 copies of chromosome 6A (equivalent of ∼50 ng DNA) were flow sorted from each line into a 0.25-mL PCR tube containing 40 µL of sterile distilled water. Contamination of the sorted 6A fractions by other chromosomes ranged from 3% to 11% (6% average). Chromosomal DNA was then treated with proteinase K, purified, and amplified in three independent reactions (Simková et al., 2008). The amplification products were pooled to reduce amplification bias and at least 7 µg DNA was obtained from each line. To check the chromosome identity and the level of contamination in individual flow-sorted fractions, 2,000 chromosomes were sorted onto a microscopic slide, air-dried, and used for fluorescence in situ hybridization with a probe for GAA microsatellites (Kubaláková et al., 2003).
Chromosome 6A Sequence Analysis
The MutChromSeq protocol was used to identify the new mutations in the overgrowth mutants (Sánchez-Martín et al., 2016). All raw Illumina reads from chromosome 6A DNA of Icaro, Anhinga, and the overgrowth mutants M12, M17, M22, M24, and M54 were quality trimmed using Trimmomatic V0.32 with the default settings for paired end reads (Bolger et al., 2014). The quality trimmed reads from Icaro were de novo assembled using CLC Assembly Cell v 4.3.0 (https://www.qiagenbioinformatics.com/). Assembly quality control was assessed and assembly statistics generated using QUAST (quality assessment tool for genome assemblies; Gurevich et al., 2013). The de novo assembly produced an assembly with a total length of 444 Mb (∼70% of the total length of 6A) and an N50 contig length of 1851 bp. The de novo Icaro assembly was masked for highly repetitive repeats regions with RepeatMasker v4.0.6 (http://repeatmasker.org) using the Triticeae Repeat Database (trep-db_complete_Rel-16.fasta) as the external library (http://wheat.pw.usda.gov/ITMI/Repeats/). Quality trimmed reads from Icaro, M12, M17, M22, M24, and M54 were mapped to the de novo Icaro assembly using BWA v0.7.15 (bwa aln) with default settings (Li and Durbin, 2009). SAM files were converted to BAM format (samtools view), duplicates removed (samtools rmdup), and mapped files converted to the mpileup format (samtools mpileup –BQ0) using samtools v1.3.1 (Li et al., 2009). Two custom java scripts Pileup2XML.jar and MutChromSeq.jar (https://github.com/steuernb/MutChromSeq) were used to integrate the Icaro, M12, M17, M22, M24, and M54 mpileup data sets and to identify contigs where all five mutants had an independent SNV (Sánchez-Martín et al., 2016). Potential SNVs were discarded if the allele frequency was less than 99% or total coverage was <10 (Sánchez-Martín et al., 2016).
Comparison of Nucleotide and Amino Acid Sequences
The nucleotide sequences of GA2oxA9 from Icaro (GenBank accession no. KX163068; Vikhe et al., 2017), M12, M17, M22, M24, and M54 were aligned using MEGA v6.06 (Tamura et al., 2013). Predicted ORFs of GA2oxA9 from Icaro (Vikhe et al., 2017), M12, M17, M22, M24, and M54 were translated and aligned with the wheat and rice (Oryza sativa) GA 2-oxidases, GA2oxA6, GA2oxA9 (Pearce et al., 2015), OsGA2ox5, and OsGA2ox6 (Lo et al., 2008) using MEGA v6.06 (Tamura et al., 2013). The GA2oxA9 gene was amplified by PCR from genomic DNA from Castelporziano, Capelli, Halberd, Halberd-Rht18, and the Halberd overgrowth mutants M1, M10, M13, M26, M30, M46, M47, and M75. PCR primer sequences are listed in Supplemental Table S4. The products of the PCR reaction were sequenced and the nucleotide, and predicted amino acid sequences were aligned using MEGA v6.06 (Tamura et al., 2013). The sequence 100 kb upstream of the transcriptional start site of GA2oxA9 from Icaro was compared to Anhinga using the dotmatcher tool (http://www.bioinformatics.nl/cgi-bin/emboss/dotmatcher) with threshold and window size set to 50.
Expression Analysis
For gene expression in peduncles, the basal 25% of elongating peduncles from the main stem were harvested at 50% final length and immediately frozen in liquid nitrogen. For gene expression in the first leaf, the entire leaf was harvested 3 d after germination. RNA was extracted and DNase treated using the MaxwellRSC plant RNA kit (Promega) as per the manufacturer’s instructions. First-strand cDNA synthesis primed with oligo dT (18) was performed using Maxima H Minus Reverse Transcriptase (Thermo Scientific) as per the manufacturer’s instructions. qPCR was performed using the Bio-Rad384CFX real-time system. qPCR was performed using Platinum Taq DNA polymerase (Life Technologies) and SYBR green. TaACTIN was used as the reference gene because its expression was stable across genotypes. Relative transcript levels were calculated using the ΔΔCt method allowing for primer amplification efficiencies as previously described (Deng et al., 2015; Ford et al., 2016). The qPCR primer sequences used in this study are listed in Supplemental Table S4. The products of the GA2oxA9 and GA2oxB9 qPCRs were sequenced to confirm their specificity. The qPCR results are the mean values of four biological and two technical replicates, and appropriate no template controls were included and melt curve analysis conducted for all experiments.
GA Content Analysis
For GA content analyses the basal 25% of elongating peduncles including the node from the main stem were harvested at 50% final length and immediately frozen in liquid nitrogen. Each biological replicate consisted of ∼2 g fresh weight (FW) of pooled peduncles from at least six individual plants. GA was extracted as previously detailed (Boden et al., 2014). Briefly, tissue was homogenized, covered in 80% methanol overnight, internal standards ([2H2 ]GA1, [2H2 ]GA8, [2H2 ]GA12, [2H2 ]GA19, [2H2 ]GA20, and [2H2 ]GA53) were added, and solids were removed by centrifugation. Samples were extracted by passing though C18 Sep-Pak (Waters) followed by SAX column (Maxi-Clean; Grace Davison Discovery Sciences) as previously described. Samples were then derivatized as described (Li et al., 2017a) except that samples were incubated in 200 mm EDC (N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride; Sigma-Aldrich) in ethanol in 50 µL overnight at 40°C, then dried and resuspended in 30 μL Milli-Q water. Samples were then analyzed using a Waters Acquity H-Class UPLC instrument coupled to a Waters Xevo triple quadrupole mass spectrometer. A Waters Acquity UPLC BEH C18 column (2.1 mm × 100 mm × 1.7 µm) was used. The instrument was operated and multiple reaction monitoring (MRM) transitions were monitored (Li et al., 2017a). For all samples, peak areas for endogenous and labeled GAs were compared and combined with FW of sample to calculate ng/g FW.
Functional Assay
The coding sequences of Icaro and M24 were synthesized by Genscript with NdeI and SalI restriction sites at the 5′ and 3′ends, respectively. The coding sequence was inserted into pMCSG7 and transformed into BL21 (DE3) cells. Cells were grown overnight at 37°C and used as inoculum for LB cultures containing 200 μg/mL−1 ampicillin. At an OD600 of 0.6 cultures were placed at 23°C and induced with 0.5 mm IPTG overnight. Escherichia coli proteins were extracted by French press in extraction buffer according to Macmillan et al. (2005). A GA metabolic assay was performed as previously described (Macmillan et al., 2005). In brief, E. coli lysate was incubated with radiolabeled GA precursor in a buffered reaction mixture containing cofactors at 30°C for 1 h before the reaction was stopped with 0.25 volumes of acetic acid. Samples were resuspended in 0.4% acetic acid, and 20 ng of [2H2 ]GA29 catabolite was added as an internal standard for metabolite recovery. Samples were loaded onto preconditioned C18 Sep-Pak cartridges and eluted with 100% methanol. Samples were dried and resuspended in 1% acetic acid before running as free acids on a Waters Acquity H-Class UPLC instrument coupled to a Waters Xevo triple quadrupole mass spectrometer. A Waters Acquity UPLC BEH C18 column (2.1 mm × 100 mm × 1.7 µm) was used. The instrument was operated as described above. MRM transitions for labeled GA metabolites were determined experimentally from the analysis of standard compounds or analogous [2H2 ] labeled compounds. Samples were compared to various external standards, including genuine [2H2 ]GA110, kindly supplied by Prof. Shinjiro Yamaguchi and Dr. Kiyoshi Mashiguchi (Tohoku University, Japan). Peak areas for diagnostic MRM transitions for [14C1] GA12, [14C1] GA110, and [2H2 ]GA29 catabolite were recorded, and peak areas for [14C1] GA12 and [14C1] GA110 were adjusted using the [2H2 ]GA29 catabolite surrogate standard peak areas.
Statistical Analysis for Plant Height, Expression Analysis, and GA Content
All data presented for plant height, relative expression of GA2oxA9, and GA content are mean values ± se. Differences between mean values of two values were tested by a Student’s t test assuming a two-tailed distribution and equal variance. Differences between mean values of greater than two genotypes were determined by one-way ANOVA with Tukey’s honestly significant difference post-hoc test in the R statistical package.
Accession Numbers
Accession number are as follows: GA2oxA9, KX163068;
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Comparison of DNA sequence from Icaro and Anhinga 100Kb upstream of the GA2oxA9 transcription start site.
Supplemental Table S2. Identification of deletions in Icaro compared to Anhinga 5Mb upstream of the GA2oxA9 transcription start site.
Supplemental Table S3. Sequences of SNP based KASP markers.
Supplemental Table S4. Primer sequences.
Supplemental Figure S1. Nucleotide alignment of GA2oxA9 from Icaro and overgrowth mutants.
Supplemental Figure S2. Protein alignment of Icaro, overgrowth mutants, and rice and wheat C20-GA 2-oxidases.
Supplemental Figure S3. Nucleotide alignment of GA2oxA9 from Halberd Isolines.
Supplemental Figure S4. Protein alignment of GA2oxA9 from Hal-Rht18 and Halberd overgrowth mutants.
Supplemental Figure 5. Leaf elongation rate and GA2oxA9 expression in rapidly elongating first leaves.
Supplemental Figure S6. Crude and soluble protein extracts from E. coli expressing GA2oxA9 coding sequences of Icaro and M24 separated by SDS_PAGE.
Supplemental Figure 7. The Anhinga and Icaro sequence 100 kb upstream of GA2oxA9 is highly conserved.
Acknowledgments
We thank Sergio Gálvez Rojas for assisting with early steps in the bioinformatic analysis and Tanya Phongkham for excellent technical assistance. We also thank Greg Rebetzke for providing Halberd-Rht18 germplasm and A/Prof. John Ross from University of Tasmania and Philippa Borrill from the John Innes Centre for critical discussions and suggestions. We thank Zdeňka Dubská and Romana Šperková for technical assistance with chromosome sorting. We thank Brande Wulff for providing access to CLC license and computing facilities at John Innes Centre. Carl Davies has contributed photographic skills and generated the graphics.
Footnotes
M.K., J.V., and J.D. were supported by the Czech Science Foundation (award P501-12-G090) and by the Ministry of Education, Youth, and Sports of the Czech Republic (award LO1204 from the National Program of Sustainability I).
Articles can be viewed without a subscription.
References
- Appleford NE, Wilkinson MD, Ma Q, Evans DJ, Stone MC, Pearce SP, Powers SJ, Thomas SG, Jones HD, Phillips AL, Hedden P, Lenton JR (2007) Decreased shoot stature and grain α-amylase activity following ectopic expression of a gibberellin 2-oxidase gene in transgenic wheat. J Exp Bot 58: 3213–3226 [DOI] [PubMed] [Google Scholar]
- Boden SA, Weiss D, Ross JJ, Davies NW, Trevaskis B, Chandler PM, Swain SM (2014) EARLY FLOWERING 3 regulates flowering in spring barley by mediating gibberellin production and FLOWERING LOCUS T expression. Plant Cell 26: 1557–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrill P, Adamski N, Uauy C (2015) Genomics as the key to unlocking the polyploid potential of wheat. New Phytol 208: 1008–1022 [DOI] [PubMed] [Google Scholar]
- Busov VB, Meilan R, Pearce DW, Ma C, Rood SB, Strauss SH (2003) Activation tagging of a dominant gibberellin catabolism gene (GA 2-oxidase) from poplar that regulates tree stature. Plant Physiol 132: 1283–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler PM, Robertson M (1999) Gibberellin dose-response curves and the characterization of dwarf mutants of barley. Plant Physiol 120: 623–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler PM, Harding CA (2013) ‘Overgrowth’ mutants in barley and wheat: new alleles and phenotypes of the ‘Green Revolution’ DELLA gene. J Exp Bot 64: 1603–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng WW, Clausen J, Boden S, Oliver SN, Casao MC, Ford B, Anderssen RS, Trevaskis B (2015) Dawn and dusk set states of the circadian oscillator in sprouting Barley (Hordeum vulgare) seedlings. PLoS One 10: e0129781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias AA, Busov VB, Kosola KR, Ma C, Etherington E, Shevchenko O, Gandhi H, Pearce DW, Rood SB, Strauss SH (2012) Green revolution trees: semidwarfism transgenes modify gibberellins, promote root growth, enhance morphological diversity, and reduce competitiveness in hybrid poplar. Plant Physiol 160: 1130–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MH, Rebetzke GJ, Chandler P, Bonnett D, Spielmeyer W, Richards RA (2004) The effect of different height reducing genes on the early growth of wheat. Funct Plant Biol 31: 583–589 [DOI] [PubMed] [Google Scholar]
- Ford B, Deng W, Clausen J, Oliver S, Boden S, Hemming M, Trevaskis B (2016) Barley (Hordeum vulgare) circadian clock genes can respond rapidly to temperature in an EARLY FLOWERING 3-dependent manner. J Exp Bot 67: 5517–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale MD, Youssefian S, Russell GE (1985) Dwarfing genes in wheat. In GE Russell, ed, Progress in Plant Breeding 1. Butterworth-Heinemann, London, pp 1–35 [Google Scholar]
- Giorgi D, Farina A, Grosso V, Gennaro A, Ceoloni C, Lucretti S (2013) FISHIS: fluorescence in situ hybridization in suspension and chromosome flow sorting made easy. PLoS One 8: e57994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G (2013) QUAST: quality assessment tool for genome assemblies. Bioinformatics 29: 1072–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque MA, Martinek P, Watanabe N, Kuboyama T (2011) Genetic mapping of gibberellic acid-sensitive genes for semi-dwarfism in durum wheat. Cereal Res Commun 39: 171–178 [Google Scholar]
- Hedden P. (2003) The genes of the Green Revolution. Trends Genet 19: 5–9 [DOI] [PubMed] [Google Scholar]
- Hedden P, Thomas SG (2012) Gibberellin biosynthesis and its regulation. Biochem J 444: 11–25 [DOI] [PubMed] [Google Scholar]
- Huang J, Tang D, Shen Y, Qin B, Hong L, You A, Li M, Wang X, Yu H, Gu M, Cheng Z (2010) Activation of gibberellin 2-oxidase 6 decreases active gibberellin levels and creates a dominant semi-dwarf phenotype in rice (Oryza sativa L.). J Genet Genomics 37: 23–36 [DOI] [PubMed] [Google Scholar]
- Jia Q, Li C, Shang Y, Zhu J, Hua W, Wang J, Yang J, Zhang G (2015) Molecular characterization and functional analysis of barley semi-dwarf mutant Riso no. 9265. BMC Genomics 16: 927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konzak CF. (1988) Evaluation and genetic analysis of semi-dwarf mutants of wheat. In Semi-Dwarf Cereal Mutants and Their Use in Cross-Breeding: Research Coordination Meeting 1981. International Atomic Energy Agency, Vienna, Austria, pp 25–37 [Google Scholar]
- Koornneef M, Dellaert LWM, van der Veen JH (1982) EMS- and radiation-induced mutation frequencies at individual loci in Arabidopsis thaliana (L.) Heynh. Mutat Res 93: 109–123 [DOI] [PubMed] [Google Scholar]
- Kubaláková M, Valárik M, Barto J, Vrána J, Cíhalíková J, Molnár-Láng M, Dolezel J (2003) Analysis and sorting of rye (Secale cereale L.) chromosomes using flow cytometry. Genome 46: 893–905 [DOI] [PubMed] [Google Scholar]
- Li D, Guo Z, Liu C, Li J, Xu W, Chen Y (2017a) Quantification of near-attomole gibberellins in floral organs dissected from a single Arabidopsis thaliana flower. Plant J 91: 547–557 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Torres-Garcia J, Latrasse D, Benhamed M, Schilderink S, Zhou W, Kulikova O, Hirt H, Bisseling T (2017b) Plant-specific histone deacetylases HDT1/2 regulate GIBBERELLIN 2-OXIDASE 2 expression to control Arabidopsis root meristem cell number. Plant Cell 29: 2183–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo SF, Yang SY, Chen KT, Hsing YI, Zeevaart JA, Chen LJ, Yu SM (2008) A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20: 2603–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo SF, Ho TD, Liu YL, Jiang MJ, Hsieh KT, Chen KT, Yu LC, Lee MH, Chen CY, Huang TP, et al. (2017) Ectopic expression of specific GA2 oxidase mutants promotes yield and stress tolerance in rice. Plant Biotechnol J 15: 850–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macmillan CP, Blundell CA, King RW (2005) Flowering of the grass Lolium perenne: effects of vernalization and long days on gibberellin biosynthesis and signaling. Plant Physiol 138: 1794–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce S, Huttly AK, Prosser IM, Li YD, Vaughan SP, Gallova B, Patil A, Coghill JA, Dubcovsky J, Hedden P, Phillips AL (2015) Heterologous expression and transcript analysis of gibberellin biosynthetic genes of grasses reveals novel functionality in the GA3ox family. BMC Plant Biol 15: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, et al. (1999) ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400: 256–261 [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai M, Sakamoto T, Saito T, Matsuoka M, Tanaka H, Kobayashi M (2003) Expression of novel rice gibberellin 2-oxidase gene is under homeostatic regulation by biologically active gibberellins. J Plant Res 116: 161–164 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Kobayashi M, Itoh H, Tagiri A, Kayano T, Tanaka H, Iwahori S, Matsuoka M (2001) Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiol 125: 1508–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Morinaka Y, Ishiyama K, Kobayashi M, Itoh H, Kayano T, Iwahori S, Matsuoka M, Tanaka H (2003) Genetic manipulation of gibberellin metabolism in transgenic rice. Nat Biotechnol 21: 909–913 [DOI] [PubMed] [Google Scholar]
- Sánchez-Martín J, Steuernagel B, Ghosh S, Herren G, Hurni S, Adamski N, Vrána J, Kubaláková M, Krattinger SG, Wicker T, et al. (2016) Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol 17: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, Kitano H, Matsuoka M (2002) Green revolution: a mutant gibberellin-synthesis gene in rice. Nature 416: 701–702 [DOI] [PubMed] [Google Scholar]
- Schomburg FM, Bizzell CM, Lee DJ, Zeevaart JA, Amasino RM (2003) Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 15: 151–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simková H, Svensson JT, Condamine P, Hribová E, Suchánková P, Bhat PR, Bartos J, Safár J, Close TJ, Dolezel J (2008) Coupling amplified DNA from flow-sorted chromosomes to high-density SNP mapping in barley. BMC Genomics 9: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmeyer W, Ellis MH, Chandler PM (2002) Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc Natl Acad Sci USA 99: 9043–9048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spray CR, Kobayashi M, Suzuki Y, Phinney BO, Gaskin P, MacMillan J (1996) The dwarf-1 (d1) mutant of Zea mays blocks three steps in the gibberellin-biosynthetic pathway. Proc Natl Acad Sci USA 93: 10515–10518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T. (2015) Physiological and genetic studies of an alternative semi-dwarfing gene Rht18 in wheat. PhD dissertation. University of Tasmania, Australia [Google Scholar]
- Trick M, Adamski NM, Mugford SG, Jiang CC, Febrer M, Uauy C (2012) Combining SNP discovery from next-generation sequencing data with bulked segregant analysis (BSA) to fine-map genes in polyploid wheat. BMC Plant Biol 12: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikhe P, Patil R, Chavan A, Oak M, Tamhankar S (2017) Mapping gibberellin-sensitive dwarfing locus Rht18 in durum wheat and development of SSR and SNP markers for selection in breeding. Mol Breed 37: 28 [Google Scholar]
- Vrána J, Kubaláková M, Simková H, Cíhalíková J, Lysák MA, Dolezel J (2000) Flow sorting of mitotic chromosomes in common wheat (Triticum aestivum L.). Genetics 156: 2033–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrána J, Cápal P, Číhalíková J, Kubaláková M, Doležel J (2016) Flow sorting plant chromosomes – In SF Kianian PMA, Kianian, eds, Plant Cytogenetics: Methods and Protocols. Springer, New York, pp 119–134 [DOI] [PubMed] [Google Scholar]
- Wuddineh WA, Mazarei M, Zhang J, Poovaiah CR, Mann DG, Ziebell A, Sykes RW, Davis MF, Udvardi MK, Stewart CN Jr (2015) Identification and overexpression of gibberellin 2-oxidase (GA2ox) in switchgrass (Panicum virgatum L.) for improved plant architecture and reduced biomass recalcitrance. Plant Biotechnol J 13: 636–647 [DOI] [PubMed] [Google Scholar]
- Würschum T, Langer SM, Longin CFH, Tucker MR, Leiser WL (2017) A modern Green Revolution gene for reduced height in wheat. Plant J 92: 892–903 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S. (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59: 225–251 [DOI] [PubMed] [Google Scholar]
- Yang ZY, Zheng JC, Liu CY, Wang YS, Condon AG, Chen YF, Hu YG (2015) Effects of the GA-responsive dwarfing gene Rht18 from tetraploid wheat on agronomic traits of common wheat. Field Crops Res 183: 92–101 [Google Scholar]
- Zimin AV, Puiu D, Hall R, Kingan S, Clavijo BJ, Salzberg SL (2017) The first near-complete assembly of the hexaploid bread wheat genome, Triticum aestivum. Gigascience 6: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]