Abstract
Spontaneous preterm birth (PTB; birth prior to 37 weeks of gestation) is a complex phenotype with multiple risk factors that complicate our understanding of its etiology. A number of recent studies have supported the hypothesis that epigenetic modifications such as DNA methylation induced by pregnancy-related risk factors may influence the risk of PTB or result in changes that predispose a neonate to adult-onset diseases. The critical role of timing of gene expression in the etiology of PTB makes it a highly relevant disorder in which to examine the potential role of epigenetic changes. Because changes in DNA methylation patterns can result in long-term consequences, it is of critical interest to identify the epigenetic patterns associated with adverse pregnancy outcomes. This review examines the potential role of DNA methylation as a risk factor for PTB and discusses several issues and limitations that should be considered when planning DNA methylation studies.
Keywords: prematurity, preterm labor, epigenetics, genetics, DNA
Preterm Birth is a Global Health Challenge
The preterm birth (PTB) rate continues to rise in the United States, increasing by as much as 30% during the last 25 years. Despite advances in medical care, the World Health Organization recently estimated the global PTB rate for singleton gestation as 9.6%, which corresponds to approximately 13 million preterm babies born every year worldwide; this represents a substantial problem for overtaxed health, education, and social services.1
Of neonatal deaths between birth and 7 days not attributable to congenital conditions, 28% result from PTB.1 ,2 Children born preterm are more likely to have cerebral palsy, sensory deficits, learning disabilities, and respiratory illnesses.1 ,2 Lower gestational age increases the risk of asthma at 6 years of age,3 and PTB increased the risk of very low birth weight children to be diagnosed with asthma at age 12.4 Preterm birth also increases the risk of being hospitalized with infections during childhood.5 Among school aged children, those born preterm show diminished cognitive performance, increased externalizing and internalizing behaviors, and are more likely to develop attention-deficit hypertension disorder.6 Girls who were delivered preterm have a higher risk of developing anorexia nervosa,7 and boys who were born small for gestational age (SGA) and preterm are more likely to develop personality and psychotic disorders or be hospitalized for mental illness compared to boys born SGA at term.8
Preterm birth and reduced fetal growth have also been linked to a number of important chronic diseases of adulthood. Preterm birth is associated with the development of type 2 diabetes,9 independent of fetal growth rates.10 It is often difficult to delineate the effects of PTB from fetal growth restriction but such adverse intrauterine conditions associate with hypertension, coronary heart disease, and stroke.11–17 Taken as a whole, it is clear that PTB not only imparts a difficult start to life but may also impart a considerable risk of a disease-burdened life and tremendous economic costs.1,2
The 2 major classes of PTB are spontaneous (mostly unknown etiology) and indicated (known risk factors including preeclampsia, multiple gestations, gestational diabetes, and maternal or fetal anomalies).18 Studies have identified multiple risk factors for PTB including race, socioeconomic factors, psychosocial stressors, behavioral factors, maternal infection, prior history, and genetic variants.18 Despite the tremendous knowledge gained over the past decade, this information has not been translated into screening or effective intervention and has not decreased the rate of PTB.19–21 Understanding the specific initiators and effectors of the labor process will provide valuable insight into the pathophysiologic pathways that result in preterm labor and may result in novel interventions.
The etiology of PTB remains unclear, and the identification of biomarkers to predict high-risk pregnancies resulting in PTB would represent a significant advancement. In this respect, epigenetics is a particularly attractive area of investigation because of the increasing prevalence of PTB in recent years and the association of PTB with development of adult-onset diseases.
Epigenetics and Fetal Development
Epigenetics refers to structural modification of DNA sequence that produces stable, but potentially reversible, alterations in the transcriptional potential of a cell, without changes in the DNA sequence. Cytosine methylation is one such modification that occurs at position 5′ of the cytosine pyrimidine ring in CpG dinucleotides. Methylation of CpG sites in the promoter regions of genes may disrupt transcription factor binding or attract methyl-binding proteins. These changes to the promoter structure correlate with chromatin remodeling and transcriptional silencing.22
Epigenetics play an important role in fetal development. The literature on epigenetic regulation during embryogenesis has been reviewed by Shi and Wu.23 Following fertilization, areas that are not protected by imprinting undergo both active and passive demethylation.24,25 DNA methylation patterns then begin to differentiate by developmental stage and by tissue.26 Because fetal development is a period of extensive cellular replication and growth, environmentally induced epigenetic changes may result in stable patterns of gene expression and phenotypic differences among exposed individuals.27–29 In contrast, the rate of DNA synthesis is much lower during adulthood, and the epigenome may be more resilient to environmental insults.
DNA Methylation as a Risk Factor for PTB
Studies suggest that nutritional deficiencies and imbalances, psychosocial stressors, behavioral factors such as cigarette smoking, maternal infection (bacterial vaginosis or intra-amniotic infection), and race all increase the risk of PTB,18 and epigenetic mechanisms such as DNA methylation may partially underlie many of these risk factors. Therefore, a comprehensive risk model of PTB should incorporate the factors outlined in Figure. 1. Indeed, many ongoing studies are investigating their numerous interactions.
Figure 1.
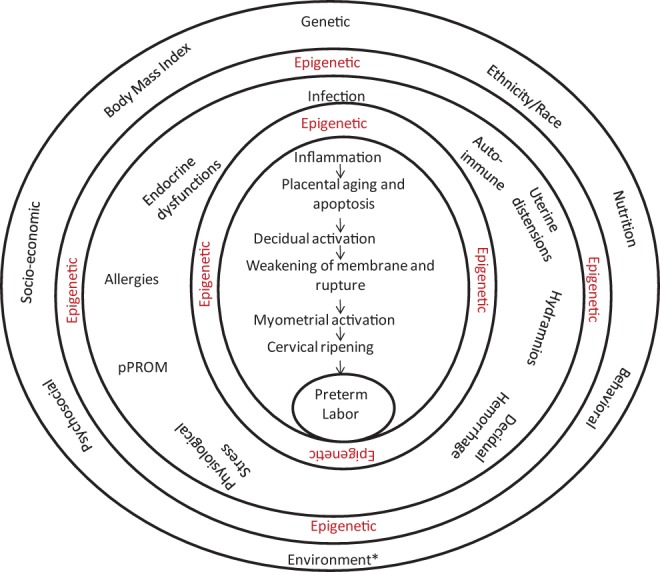
This figure provides a comprehensive schema of the processes that impact preterm birth (PTB) from a general level (outer circle) to the final steps (inner circle). These factors can potentially function independently or can interact to affect PTB. Each risk factor operates via multiple related clinical manifestations (middle circle), encompassing overlapping and often redundant biomolecules, culminating in a final effector cascade (innermost circle). Preterm birth is effectively a syndrome of multiple disease processes. These risk factors and disease processes are mediated by immune and inflammatory processes that produce uterotonins (Prostaglandins - PGs) and stimulate decidual activation, myometrial contractility, fetal membrane weakening and rupture, and cervical ripening and dilatation, culminating in premature labor and delivery.5 These pathologic processes are initiated by well-characterized risk factors (outer circle). *The environment alone may consist of multitude of exogenous factors not limited to a single factor. Environmental risk factors may lead to epigenetic changes, resulting in long-term alterations in gene expression that increase the risk of adult-onset diseases. Epigenetic changes can also occur as a result of risk associated clinical manifestations and also due to interventions, if any, at this stage. Unlike static genetic risk factors, epigenetic changes can happen at multiple levels as indicated in the figure.
Environmental factors that alter the intrauterine environment may induce epigenetic-mediated changes in gene expression that result in increased susceptibility to adverse pregnancy outcomes.27,30 In humans, differential methylation of storkhead box 1 gene (STOX1) has been associated with preeclampsia.31,32 In rats, intrauterine growth restriction (IUGR) has been associated with decreased p53 methylation, decreased CpG island and genome-wide methylation, and decreased expression of DNA methyltransferase 1 (DNMT1).33,34 DNMT1 is a maintenance methyltransferase that plays a role in placental development and can disrupt this process in the first trimester of pregnancy.
A review by Suter and Aagaard-Tillery12 discussed the environmental factors that can influence the epigenome during pregnancy, and indeed several studies have supported that environmental risk factors influence DNA methylation patterns during pregnancy. For example, animal studies show that insulin growth factor 2 (IGF2) imprinting in the placenta is altered by Campylobacter rectus infection during pregnancy,35 and it is likely that methylation patterns may differ depending on the type of infectious agent.
Diet can also modify the epigenome, and deficiency of methyl donors such as folate is directly correlated with changes in DNA methylation.36–40 Several studies have demonstrated the impact of maternal diet on offspring outcomes via epigenetic mechanisms. In a mouse model, a maternal high-fat diet increased the risk of offspring obesity by altering offspring preference for palatable foods via changes in DNA methylation and messenger RNA (mRNA) expression of dopamine and opioid-related genes.41 Using a protein restriction model in rats, Sohi et al42 recently reported that epigenetic changes mediate the association between maternal undernutrition and offspring cholesterol dysregulation. Consistent with this finding, IUGR, induced by a low-protein maternal diet through pregnancy and lactation, decreases the ratio of offspring liver to body weight and results in higher levels of hepatic cholesterol; the authors concluded that this change in circulating cholesterol associated with histone modifications in the promoter region of Cyp7a1 associated with chromatin silencing and reduced Cyp7a1 expression.42 Modifications of gene expression at imprinted loci and global and local placental DNA methylation patterns are also reported to result from a high-fat diet.43 These reports underscore the importance of a balanced diet during pregnancy to avoid epigenetic modifications that can contribute not only to pregnancy complications such as IUGR or PTB but also to long lasting effects in the offspring.44
Smoking has also been linked to PTB, and 1 study found suggestive evidence that genome-wide methylation is decreased in adults who were exposed prenatally to cigarette smoke.45 In placental samples obtained from smokers and nonsmokers, prenatal tobacco exposure was associated with hypomethylation of the CYP1A1 promoter region and increased CYP1A1 expression.46 Recent data suggest a potential mechanism for these changes as both markers of oxidative stress and apoptosis are increased in normal human fetal membrane cultures that were exposed to cigarette smoke extracts.47
The burden of PTB is disproportionately concentrated in Africa and Asia and is especially high for those of African descent in developed countries1 Pregnancies in which either parent is of African descent are at increased risk of PTB,48 and this effect appears to be largely independent of measured environmental risk factors.49 Consistent with the hypothesis that racial disparity in PTB may result from genetic or epigenetic changes, differences in genetic polymorphisms have been associated with PTB as well as biomarkers of infection and inflammation associated with PTB (tumor necrosis factor-α and interleukin 6 concentration) among African American and Caucasian mothers.50–53 Further, DNA methylation patterns vary in the umbilical cord blood of African American newborns.54
Although epidemiologic studies support the association of several environmental risk factors with adverse pregnancy outcome, no studies to date have directly examined DNA methylation as a molecular mediator between the environmental risk factors and PTB. Such studies will be required to determine whether epigenetic changes play a role in the relationship between environmental risk factors and PTB.
Developmental Origin of Adult Health and Diseases
The work of Barker and colleagues,15,16 who observed that impaired placental growth was associated with chronic heart failure in later life, stimulated interest in the hypotheses known as the developmental origins of adult health and diseases hypothesis. Environmental insults during prenatal development, such as restricted maternal nutrition, can result in growth restrictions and long-term consequences for affected offspring, and epigenetic changes may serve as mediators between environmental insults and the development of adult-onset diseases. Support has been provided by plant and animal studies that document epigenetic inheritance in the context of their potential contribution to adult health and diseases.55–57
Studies in rats have shown that nutritional deprivation during pregnancy results in intrauterine and postnatal growth restriction.58,59 Notably, IUGR pups born to food-restricted mothers but cross-fostered by normally fed mothers demonstrated accelerated growth that resulted in increased weight and body fat, suggesting that restricted maternal nutrition may contribute to obesity in later life.58,59
Similarly, among people born during the Dutch famine, exposure to famine during early gestation was associated with coronary heart disease, a more atherogenic plasma lipid profile, disturbed blood coagulation, increased stress responsiveness and higher rates of obesity. Those exposed to famine in midway through gestation had increased rates of microalbuminuria and obstructive airways disease, and those exposed to famine at any stage of gestation were more likely to have impaired glucose tolerance.60 These findings show that maternal undernutrition during gestation affects health in later life, but these effects are dependent on the timing of the exposure. While timing of the exposure is clearly important, these studies collectively suggest that epigenetic modifications resulting from such intense stressors may result in long-term consequences for an individual.60,61
Evaluating the impact of early environmental exposures on the fetal epigenome may offer insight into both regulatory processes inherent in development and adult-onset diseases. Exposure to environmental chemicals can alter the fetal epigenome, thereby changing gene expression patterns.29 Neonatal exposure to both estradiol and bisphenol A, an additive for many plastics including food containers and baby bottles, may cause multiple gene-specific changes in DNA methylation in the rat prostate, including hypomethylation of the phosphodiesterase type 4 variant 4 (PDE4D4), which has been associated with prostate cancer risk.62 Also, prenatal exposure to diethylstilbestrol (DES) results in hypomethylation in 2 critical DNA control regions.63 Individuals treated with DES during pregnancy had increased rates of reproductive disorders and clear cell adenocarcinoma of the vagina,63 and both DES-exposed mothers and their children were more likely to report lupus, arthritis, asthma, and respiratory tract infections in the 1985 National Health Interview Survey.64
Direct evidence implicating environmentally induced DNA methylation changes as a causal factor in PTB has yet to emerge. While the purpose for this review is to promote this line of research, future research will be required to delineate whether DNA methylation changes represent a cause or consequence of PTB.
A Widening Perspective on DNA Methylation
While many studies to date have focused on the examinations of 5-methylcytosine (5mC), few studies have been able to distinguish 5mC from its modification, 5hmC. 5-Hydroxymethylcytosine plays an intermediate role in oxidative demethylation pathways65 and is integral in the self-renewal and maintenance of embryonic stem cells66 from several mammalian species.67 5hmC is associated across the genome with increased transcription, and variations in expression due to 5hmC may influence the processes of pluripotency and lineage commitment.68
Similarly, while most conventional studies of methylation focus on CG methylation, almost 25% of DNA methylation in embryonic stem cells occurs at non-CG dinucleotides. This non-CG methylation is enriched in gene bodies and depleted in protein-binding sites and enhancers. It is also enriched near genes involved in pluripotency and differentiation. However, non-CG methylation is not evident after cellular differentiation occurs.69
Although 5hmC and methylation of non-CG dinuclotides have yet to be extensively evaluated in the context of human disease, the enrichment and likely regulatory roles of 5hmC and non-CG methylation in embryonic stem cells suggest that they could be relevant to early-life and developmental disorders. Hence, both 5hmC and non-CG methylation represent a previously unexplored aspect of epigenetic regulation that may contribute to PTB.
Genome-Wide Epigenetic Studies
Methods to conduct DNA methylation studies have been reviewed extensively,70,71 so here we will focus on the evolving field and the issues to consider before undertaking such studies. As with studies of sequence variants, the initial foray into examining epigenetic changes associated with PTB has focused on specific candidate genes. These studies are dependent on the choice of candidate genes and may lead to findings that have small effect sizes and do not replicate consistently. However, the recent availability of inexpensive commercial methylation microarrays has facilitated genome-wide methylation studies, which have already met with some success.72 -75 Although genome-wide studies of DNA methylation have much in common with both genetic association studies and gene expression studies, at this early stage a consensus has not been reached about the optimal study design or analysis methods for methylation studies.
The appropriate analytical approaches for methylation studies will depend on the technology used. For array-based platforms that rely on bisulfite treatment of DNA such as the Illumina GoldenGate and Infinium chips that provide estimates of the methylation proportion with single-CpG resolution, it is common to perform simple tests of association between the methylation proportion and the phenotype, adjusting for multiple testing via Bonferroni or false discovery rate procedures. Platforms that enrich methylated DNA through the use of methylation-specific restriction enzymes (McrBC) or methylated DNA immunoprecipitation (MeDIP) require more complex statistical approaches to estimate the absolute methylation level while accounting for sequence-dependent biases due to CpG density and other factors. To address this, strategies that model the dependence of enrichment on CpG density have been proposed for MeDIP76 ,77 and McrBC approaches.78
Although the basic quality control criteria leading to the removal of specific data points, samples, or CpG sites are standard for commercially available methylation arrays, there is currently no consistently used approach to normalization of methylation data. Methods to normalize signals abound in the gene expression literature,79 ,80 but these methods are based on the assumption of no global differences between samples, which is often inappropriate for methylation data.78 The absence of differences in the overall signal is generally thought to be an appropriate assumption for expression data, but global differences may be observed in methylation data when comparing cancer versus healthy tissue,81 different tissue types,82 or even individuals of different ages.83,84 If global methylation patterns vary with respect to the phenotype studied, the application of standard normalization methods such as Loess and quantile normalization could drastically reduce power by removing true differences between samples and thus are likely inappropriate for analysis of methylation data. The use of subset quantile normalization,85 which equalizes the quantiles of negative control probes instead of the signal probes, has been proposed for methylation microarray data.78 However, subset quantile normalization requires a large number of negative control features whose signals cover the entire range of the signal probes; hence, for experiments with only a small or moderate number of negative control probes, the development of further approaches would be valuable.
Population stratification, a well-documented issue in studies of sequence variants,86 is also likely to impact the studies of DNA methylation. Recent studies have reported differential methylation of genes between African American and Caucasian umbilical cord blood samples,54 and evaluation of participants of varying ancestry suggest that genetic and epigenetic variation may be somewhat correlated.87 It is well known that the frequency of sequence-based polymorphisms varies across different ethnic backgrounds, and the similarity of methylation patterns in families83 and within twin pairs88 suggests that sequence variation may impact methylation patterns. In fact, evidence of allele-specific methylation has been reported89 -91 and may be partially responsible for differences in methylation patterns associated with race or disease state. These results emphasize the importance of controlling for ancestry in epigenetic studies as well as in studies of sequence variants, ideally by careful ancestry-matching of cases and controls as part of the study design and inclusion of covariates representing self-reported or genetic ancestry.
Cell type heterogeneity between samples is an issue analogous to population stratification that may affect both gene expression and methylation studies but not studies of sequence variants. The cell type composition of blood is known to be altered in a variety of disease states92 -97 and can vary with other health-related phenotypes such as exercise level98 and obesity.99 Differences in methylation between cell types have been reported for specific genes 100,101 and genome-wide.102 Because cell type composition is correlated with both individual health and methylation, the association between DNA methylation and an outcome of interest may be confounded by variations in the proportion of cell types within a sample or tissue. Hence, it is important to control for cell type differences within a study; however, attempts to sort tissues into subpopulations may limit the amount of DNA available for the study. As an alternative, the development of statistical approaches to account for differences in cell type proportions across samples would be valuable.
Because DNA methylation varies with cell type, it is important to select a relevant tissue in which to study PTB. Placental tissue, fetal membranes, and umbilical cord blood have obvious biological relevance and can be obtained through noninvasive methods. Umbilical cord blood has the added advantage of being amenable to longitudinal studies in which blood sampling may occur at birth and then regularly over development. However DNA samples obtained from saliva are highly heterogeneous with the proportions of leukocytes and epithelial cells ranging from 20% to 80% among individuals. The sample volumes, especially from infants, may be too limited for cell-sorting methods so this may not be a feasible approach until statistical approaches are available to adjust for cell type proportions in different samples. As both maternal and fetal factors can contribute to PTB, one should also consider sampling maternal blood samples, cervical or myometrial tissues.
Another important confounding variable in DNA methylation studies is age. Recent studies have reported widespread age-related methylation differences at CpG sites across the genome.72,83,84,103 These results underscore the importance of matching for age in case–control studies of DNA methylation. To avoid confounding due to differences in the age distribution, cases and controls should be matched by age whenever possible. Adjustment for age as a covariate is also useful but may not be sufficient if the age distributions differ substantially between cases and controls. Moreover, if age is correlated with case–control status, its inclusion as a covariate may reduce power. Conversely, in well-matched samples, age will be independent of the phenotype of interest, so including age as a covariate can lead to increased power since it will explain the methylation differences that would otherwise be attributed to noise.
Summary
Preterm birth is a complex disease with many known epidemiologic and genetic risk factors, and awareness of the impact of prenatal conditions on an individual’s long-term health has focused more attention on PTB. Epigenetic changes induced by various risk factors for pregnancy may influence the risk of PTB or induce changes in the fetal epigenome that predispose a neonate to adult-onset diseases. Although epigenetic changes, such as DNA methylation, have been associated with many complex disease outcomes, their potential influence on PTB remains unexplored. Although many epidemiologic factors as listed in Figure 1 are associated with PTB, a link between these factor-induced DNA methylation changes and its association with adverse pregnancy outcome or long-term consequences during the life course are still lacking. Animal model studies clearly support the rationale for such phenomena and motivate future research in this area. Future PTB studies should focus on genome-wide epigenetic changes in human pregnancies, ideally in the context of longitudinal designs to assess the role played by DNA methylation in both short- and long-term outcomes related to PTB.
This review is meant to provide an overview of the potential role of DNA methylation in adverse pregnancy outcomes as well as to discuss the issues and limitations that should be considered when planning DNA methylation studies. Evaluating the potential role of epigenetic changes in PTB is a timely goal, and it is hoped that this review will aid researchers who are contemplating or designing these studies.
Footnotes
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
The authors received no financial support for the research, authorship, and/or publication of this article.
References
- 1. Beck S, W, ojdyla D, S, ay L, et al. The worldwide incidence of preterm birth: a systematic review of maternal mortality and morbidity. Bull World Health Organ. 2010;88(1):31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Behrman REB, AS Preterm birth: causes, consequences, and prevention In: Coupbaah, ed. Outcomes. Washington, DC: The National Academies Press; 2007. [PubMed] [Google Scholar]
- 3. Bernsen RM, de Jongste JC, Koes BW, Aardoom HA, van der Wouden JC. Perinatal characteristics and obstetric complications as risk factors for asthma, allergy and eczema at the age of 6 years. Clin Exp Allergy. 2005;35(9):1135–1140. [DOI] [PubMed] [Google Scholar]
- 4. Mai XM, Gaddlin PO, Nilsson L, et al. Asthma, lung function and allergy in 12-year-old children with very low birth weight: a prospective study. Pediatr Allergy Immunol. 2003;14(3):184–192. [DOI] [PubMed] [Google Scholar]
- 5. Yuan W, Basso O, Sorensen HT, Olsen J. Indicators of fetal growth and infectious disease in childhood–a birth cohort with hospitalization as outcome. Eur J Epidemiol. 2001;17(9):829–834. [DOI] [PubMed] [Google Scholar]
- 6. Bhutta AT, Cleves MA, Casey PH, Cradock MM, Anand KJ. Cognitive and behavioral outcomes of school-aged children who were born preterm: a meta-analysis. JAMA. 2002;288(6):728–737. [DOI] [PubMed] [Google Scholar]
- 7. Cnattingius S, Hultman CM, Dahl M, Sparen P. Very preterm birth, birth trauma, and the risk of anorexia nervosa among girls. Arch Gen Psychiatry. 1999;56(7):634–638. [DOI] [PubMed] [Google Scholar]
- 8. Monfils Gustafsson W, Josefsson A, Ekholm Selling K, Sydsjo G. Preterm birth or foetal growth impairment and psychiatric hospitalization in adolescence and early adulthood in a Swedish population-based birth cohort. Acta Psychiatr Scand. 2009;119(1):54–61. [DOI] [PubMed] [Google Scholar]
- 9. Burdge GC, Hanson MA, Slater-Jefferies JL, Lillycrop KA. Epigenetic regulation of transcription: a mechanism for inducing variations in phenotype (fetal programming) by differences in nutrition during early life? Br J Nutr. 2007;97(6):1036–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kajantie E, Osmond C, Barker DJ, Eriksson JG. Preterm birth–a risk factor for type 2 diabetes? The Helsinki birth cohort study. Diabetes Care. 2010;33(12):2623–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koupil I, Leon DA, Lithell HO. Length of gestation is associated with mortality from cerebrovascular disease. J Epidemiol Community Health 2005;59(6):473–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Suter MA, Aagaard-Tillery KM. Environmental influences on epigenetic profiles. Semin Reprod Med. 2009;27(5):380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Szyf M. The early life environment and the epigenome. Biochim Biophys Acta. 2009;1790(9):878–885. [DOI] [PubMed] [Google Scholar]
- 14. Chmurzynska A. Fetal programming: link between early nutrition, DNA methylation, and complex diseases. Nutr Rev. 2010;68(2):87–98. [DOI] [PubMed] [Google Scholar]
- 15. Barker DJ. The fetal and infant origins of adult disease. BMJ. 1990;301(6761):1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barker DJ, Gelow J, Thornburg K, Osmond C, Kajantie E, Eriksson JG. The early origins of chronic heart failure: impaired placental growth and initiation of insulin resistance in childhood. Eur J Heart Fail. 2010;12(8):819–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Champagne FA, Curley JP. Epigenetic mechanisms mediating the long-term effects of maternal care on development. Neurosci Biobehav Rev. 2009;33(4):593–600. [DOI] [PubMed] [Google Scholar]
- 18. Menon R. Spontaneous preterm birth, a clinical dilemma: etiologic, pathophysiologic and genetic heterogeneities and racial disparity. Acta Obstet Gynecol Scand. 2008;87(6):590–600. [DOI] [PubMed] [Google Scholar]
- 19. Raju TN, Higgins RD, Stark AR, Leveno KJ. Optimizing care and outcome for late-preterm (near-term) infants: a summary of the workshop sponsored by the National Institute of Child Health and Human Development. Pediatrics. 2006;118(3):1207–1214. [DOI] [PubMed] [Google Scholar]
- 20. Ananth CV, Vintzileos AM. Epidemiology of preterm birth and its clinical subtypes. J Matern Fetal Neonatal Med. 2006;19(12):773–782. [DOI] [PubMed] [Google Scholar]
- 21. Green NS, Damus K, Simpson JL, et al. Research agenda for preterm birth: recommendations from the March of Dimes. Am J Obstet Gynecol 2005;193(3 pt 1):626–635. [DOI] [PubMed] [Google Scholar]
- 22. Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330(6004):612–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shi L, Wu J. Epigenetic regulation in mammalian preimplantation embryo development. Reprod Biol Endocrinol. 2009;7:59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kafri T, Gao X, Razin A. Mechanistic aspects of genome-wide demethylation in the preimplantation mouse embryo. Proc Natl Acad Sci U S A. 1993;90(22):10558–10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rougier N, Bourc'his D, Gomes DM, et al. Chromosome methylation patterns during mammalian preimplantation development. Genes Dev. 1998;12(14):2108–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liang P, Song F, Ghosh S, et al. Genome-wide survey reveals dynamic widespread tissue-specific changes in DNA methylation during development. BMC Genomics. 2011;12(1):231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr. 2007;27:363–388. [DOI] [PubMed] [Google Scholar]
- 28. Waterland RA, Jirtle RL. Early nutrition, epigenetic changes at transposons and imprinted genes, and enhanced susceptibility to adult chronic diseases. Nutrition. 2004;20(1):63–68. [DOI] [PubMed] [Google Scholar]
- 29. Dolinoy DC, Weidman JR, Jirtle RL. Epigenetic gene regulation: linking early developmental environment to adult disease. Reprod Toxicol. 2007;23(3):297–307. [DOI] [PubMed] [Google Scholar]
- 30. Gordon L, Joo JH, Andronikos R, et al. Expression discordance of monozygotic twins at birth: effect of intrauterine environment and a possible mechanism for fetal programming. Epigenetics. 2011;6(5):579–592. [DOI] [PubMed] [Google Scholar]
- 31. van Dijk M, Drewlo S, Oudejans CB. Differential methylation of STOX1 in human placenta. Epigenetics. 2010;5(8):736–742. [DOI] [PubMed] [Google Scholar]
- 32. van Dijk M, Oudejans CB. STOX1: key player in trophoblast dysfunction underlying early onset preeclampsia with growth retardation. J Pregnancy. 2011;2011:521826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ke X, Lei Q, James SJ, et al. Uteroplacental insufficiency affects epigenetic determinants of chromatin structure in brains of neonatal and juvenile IUGR rats. Physiol Genomics. 2006;25(1):16–28. [DOI] [PubMed] [Google Scholar]
- 34. Pham TD, MacLennan NK, Chiu CT, Laksana GS, Hsu JL, Lane RH. Uteroplacental insufficiency increases apoptosis and alters p53 gene methylation in the full-term IUGR rat kidney. Am J Physiol Regul Integr Comp Physiol. 2003;285(5):R962–R970. [DOI] [PubMed] [Google Scholar]
- 35. Bobetsis YA, Barros SP, Lin DM, et al. Bacterial infection promotes DNA hypermethylation. J Dent Res. 2007;86(2):169–174. [DOI] [PubMed] [Google Scholar]
- 36. McKay JA, Wong YK, Relton CL, Ford D, Mathers JC. Maternal folate supply and sex influence gene-specific DNA methylation in the fetal gut. Mol Nutr Food Res. 2011;55:1717–1723. [DOI] [PubMed] [Google Scholar]
- 37. Hoyo C, Murtha AP, Schildkraut JM, et al. Methylation variation at IGF2 differentially methylated regions and maternal folic acid use before and during pregnancy. Epigenetics. 2011;6(7):928–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McKay JA, Waltham KJ, Williams EA, Mathers JC. Folate depletion during pregnancy and lactation reduces genomic DNA methylation in murine adult offspring. Genes Nutr. 2011;6(2):189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maloney CA, Hay SM, Rees WD. Folate deficiency during pregnancy impacts on methyl metabolism without affecting global DNA methylation in the rat fetus. Br J Nutr. 2007;97(6):1090–1098. [DOI] [PubMed] [Google Scholar]
- 40. McKay JA, Xie L, Harris S, Wong YK, Ford D, Mathers JC. Blood as a surrogate marker for tissue-specific DNA methylation and changes due to folate depletion in post-partum female mice. Mol Nutr Food Res. 2011;55(7):1026–1035. [DOI] [PubMed] [Google Scholar]
- 41. Vucetic Z, Kimmel J, Totoki K, Hollenbeck E, Reyes TM. Maternal high-fat diet alters methylation and gene expression of dopamine and opioid-related genes. Endocrinology. 2010;151(10):4756–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sohi G, Marchand K, Revesz A, Arany E, Hardy DB. Maternal protein restriction elevates cholesterol in adult rat offspring due to repressive changes in histone modifications at the cholesterol 7{alpha}-hydroxylase promoter. Mol Endocrinol. 2011;25(5):785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gallou-Kabani C, Gabory A, Tost J, et al. Sex-and diet-specific changes of imprinted gene expression and DNA methylation in mouse placenta under a high-fat diet. PLoS One. 2010;5(12):e14398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Attig L, Gabory A, Junien C. Nutritional developmental epigenomics: immediate and long-lasting effects. Proc Nutr Soc. 2010;69(2):221–231. [DOI] [PubMed] [Google Scholar]
- 45. Flom J, Ferris J, Gonzalez K, Santella R, Terry M. Prenatal tobacco smoke exposure and genomewide methylation in adulthood. Cancer Epidemiol Biomarkers Prev. 2011;20:720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Suter M, Abramovici A, Showalter L, et al. In utero tobacco exposure epigenetically modifies placental CYP1A1 expression. Metabolism. 2010;59(10):1481–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Menon R, Fortunato SJ, Yu J, et al. Cigarette smoke induces oxidative stress and apoptosis in normal term fetal membranes. Placenta. 2011;32(4):317–322. [DOI] [PubMed] [Google Scholar]
- 48. Adams MM, Elam-Evans LD, Wilson HG, Gilbertz DA. Rates of and factors associated with recurrence of preterm delivery. JAMA. 2000;283(12):1591–1596. [DOI] [PubMed] [Google Scholar]
- 49. Goldenberg RL, Cliver SP, Mulvihill FX, et al. Medical, psychosocial, and behavioral risk factors do not explain the increased risk for low birth weight among black women. Am J Obstet Gynecol. 1996;175(5):1317–1324. [DOI] [PubMed] [Google Scholar]
- 50. Menon R, Velez DR, Morgan N, Lombardi SJ, Fortunato SJ, Williams SM. Genetic regulation of amniotic fluid TNF-alpha and soluble TNF receptor concentrations affected by race and preterm birth. Hum Genet. 2008;124(3):243–253. [DOI] [PubMed] [Google Scholar]
- 51. Velez DR, Fortunato SJ, Williams SM, Menon R. Interleukin-6 (IL-6) and receptor (IL6-R) gene haplotypes associate with amniotic fluid protein concentrations in preterm birth. Hum Mol Genet. 2008;17(11):1619–1630. [DOI] [PubMed] [Google Scholar]
- 52. Menon R, Fortunato SJ, Edwards DR, Williams SM. Association of genetic variants, ethnicity and preterm birth with amniotic fluid cytokine concentrations. Ann Hum Genet 2010; 74:165–83. [DOI] [PubMed] [Google Scholar]
- 53. Williams SM, Velez DR, Menon R. Geographic ancestry and markers of preterm birth. Expert Rev Mol Diagn. 2010;10(1):27–32. [DOI] [PubMed] [Google Scholar]
- 54. Adkins RM, Krushkal J, Tylavsky FA, Thomas F. Racial differences in gene-specific DNA methylation levels are present at birth. Birth Defects Res A Clin Mol Teratol. 2011;91(8):728–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Johannes F, Porcher E, Teixeira FK, et al. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 2009;5(6):e1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dunn GA, Bale TL. Maternal high-fat diet promotes body length increases and insulin insensitivity in second-generation mice. Endocrinology. 2009;150(11):4999–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bocock PN, Aagaard-Tillery KM. Animal models of epigenetic inheritance. Semin Reprod Med. 2009;27(5):369–379. [DOI] [PubMed] [Google Scholar]
- 58. Woodall SM, Breier BH, Johnston BM, Gluckman PD. A model of intrauterine growth retardation caused by chronic maternal undernutrition in the rat: effects on the somatotrophic axis and postnatal growth. J Endocrinol. 1996;150(2):231–242. [DOI] [PubMed] [Google Scholar]
- 59. Desai M, Gayle D, Babu J, Ross MG. Programmed obesity in intrauterine growth-restricted newborns: modulation by newborn nutrition. Am J Physiol Regul Integr Comp Physiol. 2005;288(1):R91–R96. [DOI] [PubMed] [Google Scholar]
- 60. Roseboom T, de Rooij S, Painter R. The Dutch famine and its long-term consequences for adult health. Early Hum Dev. 2006;82(8):485–491. [DOI] [PubMed] [Google Scholar]
- 61. Susser M, Stein Z. Timing in prenatal nutrition: a reprise of the Dutch Famine Study. Nutr Rev. 1994;52(3):84–94. [DOI] [PubMed] [Google Scholar]
- 62. Ho SM, Tang WY, Belmonte de Frausto J, Prins GS. Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res. 2006;66(11):5624–5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A. 2007;104(32):13056–13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wingard DL, Turiel J. Long-term effects of exposure to diethylstilbestrol. West J Med. 1988;149(5):551–554. [PMC free article] [PubMed] [Google Scholar]
- 65. Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-Methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145(3):423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466(7310):1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wossidlo M, Nakamura T, Lepikhov K, et al. 5-hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2:241. [DOI] [PubMed] [Google Scholar]
- 68. Ficz G, Branco MR, Seisenberger S, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473(7347):398–402. [DOI] [PubMed] [Google Scholar]
- 69. Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Laird PW. Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet. 2010;11(3):191–203. [DOI] [PubMed] [Google Scholar]
- 71. Stolzenberg DS, Grant PA, Bekiranov S. Epigenetic methodologies for behavioral scientists. Horm Behav. 2011;59(3):407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Teschendorff AE, Menon U, Gentry-Maharaj A, et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 2010;20(4):440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang L, Aakre JA, Jiang R, et al. Methylation markers for small cell lung cancer in peripheral blood leukocyte DNA. J Thorac Oncol. 2010;5(6):778–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Javierre BM, Fernandez AF, Richter J, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20(2):170–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kim M, Long TI, Arakawa K, Wang R, Yu MC, Laird PW. DNA methylation as a biomarker for cardiovascular disease risk. PLoS One. 2010;5(3):e9692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Down TA, Rakyan VK, Turner DJ, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26(7):779–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Pelizzola M, Koga Y, Urban AE, et al. MEDME: an experimental and analytical methodology for the estimation of DNA methylation levels based on microarray derived MeDIP-enrichment. Genome Res. 2008;18(10):1652–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Aryee MJ, Wu Z, Ladd-Acosta C, et al. Accurate genome-scale percentage DNA methylation estimates from microarray data. Biostatistics. 2010;12(2):197–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002;32(Suppl):496–501. [DOI] [PubMed] [Google Scholar]
- 80. Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. [DOI] [PubMed] [Google Scholar]
- 81. Calvanese V, Lara E, Kahn A, Fraga MF. The role of epigenetics in aging and age-related diseases. Ageing Res Rev. 2009;8(4):268–276. [DOI] [PubMed] [Google Scholar]
- 82. Yang HH, Hu N, Wang C, et al. Influence of genetic background and tissue types on global DNA methylation patterns. PLoS One. 2010;5(2):e9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bjornsson HT, Sigurdsson MI, Fallin MD, et al. Intra-individual change over time in DNA methylation with familial clustering. JAMA. 2008;299(24):2877–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bollati V, Schwartz J, Wright R, et al. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev. 2009;130(4):234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wu Z, Aryee MJ. Subset quantile normalization using negative control features. J Comput Biol. 2010;17(10):1385–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nat Rev Genet. 2010;11(7):459–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Liu J, Hutchison K, Perrone-Bizzozero N, Morgan M, Sui J, Calhoun V. Identification of genetic and epigenetic marks involved in population structure. PLoS One. 2010;5(10):e13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kaminsky ZA, Tang T, Wang SC, et al. DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet. 2009;41(2):240–245. [DOI] [PubMed] [Google Scholar]
- 89. Kerkel K, Spadola A, Yuan E, et al. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet. 2008;40(7):904–908. [DOI] [PubMed] [Google Scholar]
- 90. Schalkwyk LC, Meaburn EL, Smith R, et al. Allelic skewing of DNA methylation is widespread across the genome. Am J Hum Genet. 2010;86(2):196–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhang D, Cheng L, Badner JA, et al. Genetic control of individual differences in gene-specific methylation in human brain. Am J Hum Genet 2010;86(3):411–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Bian C, Wu Y, Shi Y, et al. Predictive value of the relative lymphocyte count in coronary heart disease. Heart Vessels. 2010;25(6):469–473. [DOI] [PubMed] [Google Scholar]
- 93. Jafarzadeh A, Poorgholami M, Izadi N, Nemati M, Rezayati M. Immunological and hematological changes in patients with hyperthyroidism or hypothyroidism. Clin Invest Med. 2010;33(5):E271–E279. [DOI] [PubMed] [Google Scholar]
- 94. Dilli D, Oguz SS, Dilmen U, Koker MY, Kizilgun M. Predictive values of neutrophil CD64 expression compared with interleukin-6 and C-reactive protein in early diagnosis of neonatal sepsis. J Clin Lab Anal. 2010;24(6):363–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Gkrania-Klotsas E, Ye Z, Cooper AJ, et al. Differential white blood cell count and type 2 diabetes: systematic review and meta-analysis of cross-sectional and prospective studies. PLoS One. 2010;5(10):e13405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hinz T, Zaccaro D, Byron M, et al. Atopic dermo-respiratory syndrome is a correlate of eczema herpeticum. Allergy. 2011;66(7):925–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Singh K. Leucocyte counts in anaemia. Indian J Physiol Pharmacol. 2010;54(1):85–88. [PubMed] [Google Scholar]
- 98. Santos-Silva A, Rebelo MI, Castro EM, et al. Leukocyte activation, erythrocyte damage, lipid profile and oxidative stress imposed by high competition physical exercise in adolescents. Clin Chim Acta. 2001;306(1-2):119–126. [DOI] [PubMed] [Google Scholar]
- 99. Nascimento H, Rocha S, Rego C, et al. Leukocyte count versus C-reactive protein levels in obese portuguese patients aged 6-12 years old. Open Biochem J. 2010;4:72–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Moverare-Skrtic S, Mellstrom D, Vandenput L, Ehrich M, Ohlsson C. Peripheral blood leukocyte distribution and body mass index are associated with the methylation pattern of the androgen receptor promoter. Endocrine. 2009;35(2):204–210. [DOI] [PubMed] [Google Scholar]
- 101. Sakamoto S, Ortaldo JR, Young HA. Methylation patterns of the T cell receptor beta-chain gene in T cells, large granular lymphocytes, B cells, and monocytes. J Immunol. 1988;140(2):654–660. [PubMed] [Google Scholar]
- 102. Sun YV, Turner ST, Smith JA, et al. Comparison of the DNA methylation profiles of human peripheral blood cells and transformed B-lymphocytes. Hum Genet. 2010;127(6):651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Rakyan VK, Down TA, Maslau S, et al. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 2010;20(4):434–439. [DOI] [PMC free article] [PubMed] [Google Scholar]


