Abstract
In the central nervous system, the D1-alpha subtype receptor (Drd1α) is the most abundant dopamine (DA) receptor, which plays a vital role in regulating neuronal growth and development. However, the mechanisms underlying Drd1α receptor abnormalities mediating behavioral responses and modulating working memory function are still unclear. Using a novel RNA in situ hybridization assay, the current study identified dopamine Drd1α receptor and tyrosine hydroxylase (TH) RNA expression from DA-related circuitry in the nucleus accumbens (NAc) area and substantia nigra region (SNR), respectively. Drd1α expression in the NAc shows a "discrete dot" staining pattern. Clear sex differences in Drd1α expression were observed. In contrast, TH shows a "clustered" staining pattern. Regarding TH expression, female rats displayed a higher signal expression per cell relative to male animals. The methods presented here provide a novel in situ hybridization technique for investigating changes in dopamine system dysfunction during the progression of central nervous system diseases.
Keywords: Neuroscience, Issue 133, RNA hybridization, Dopamine D1-alpha receptor (Drd1α), in situ, Neuroscience, Biological Sex, Rat
Introduction
Dysfunction of the striatal dopamine system is involved in the progression of clinical symptoms observed in multiple neurocognitive diseases. Dopamine D1 receptors are present in the prefrontal cortex (PFC) and striatal regions of the brain and heavily influence cognitive processes1, including working memory, temporal processing, and locomotive behavior2,3,4,5,6,7. Previous studies elucidated that changes of dopamine D1 receptors were associated with the progression of attention deficit-hyperactivity disorder (ADHD)8, the neurocognitive symptoms in schizophrenia9,10 and stress susceptibility11. Specifically, in schizophrenia, positron emission tomography (PET) studies indicated that the binding ability of dopamine D1 receptors in the prefrontal cortices was highly related to cognitive deficits and the presence of negative symptoms11. The dendritic growth of excitatory neurons in the prefrontal cortex regulated by the dopamine D1 receptor alleviates stress susceptibility. Furthermore, the knockdown of D1 receptor in medial prefrontal cortical (mPFC) neurons could enhance the social defeat stress-induced social avoidance12.
Here, we introduce a novel technique of RNA in situ hybridization to visualize single RNA molecules in a cell with fresh-frozen tissue samples. The present technique has multiple advantages over methods that exist within the current literature. First, the current procedure preserves the spatial and morphological context of the tissue and was performed on fresh-frozen tissue samples so that other procedures requiring fresh, non-embedded tissues may be combined with the current methods. Similar procedures in formalin-fixed and paraffin-embedded tissues have illustrated that single transcription resolution can be achieved using an RNA in situ hybridization technique13. Detection of RNA at the single transcription level provides superior sensitivity to low copy number expression as well as the opportunity to compare gene expression at the level of individual cells that cannot be achieved by other nucleic acid detection methods, such as polymerase chain reaction (PCR) techniques. Additionally, the current method maintains images with a high signal-to-noise ratio through highly specific RNA probes that are hybridized to single target RNA transcripts, and sequentially bound with a cascade of signal amplification molecules in the detection system. Finally, the present technology provides the opportunity to evaluate multiple biological systems with its target-specific proprietary probes, rather than limiting our investigation to only one class of system-related markers such as protein detection by immunohistochemistry methods.
In our study, we used this novel RNA in situ hybridization to evaluate Drd1α receptor expression in the nucleus accumbens (NAc) and tyrosine hydroxylase (TH) expression in the substantia nigra (SNR) of both male and female F344/N rats. The innovative RNA in situ hybridization enabled us to investigate mechanisms influencing both DA uptake and DA release simultaneously, improving our understanding of the striatal DA system's complexities. Here, we describe the procedure for fresh-frozen brain slices and provide methods of data analysis for different staining patterns: "discrete dot" or "clusters".
Protocol
The experimental protocol was approved by the Animal Care and Use Committee (IACUC) at the University of South Carolina (federal assurance number: A3049-01).
1. Preparation of Fresh Frozen Brain Sections
Use the F344/N rat strain: three rats of each sex, 13 months of age, body weight approximately 320 g.
Adjust the sevoflurane concentration to 5% (overdose of sevoflurane). Continue sevoflurane exposure after breathing stops for an additional minute.
Decapitate the rat and remove the brain.
Submerge the rat brain in liquid nitrogen for 15 s within 5 min of tissue harvest.
Equilibrate the brain to -20 °C in a cryostat for 1 h.
Cut 30 µm sections and transfer onto slides (see Table of Materials).
Choose sections from the nucleus accumbens region, approximately 2.76 mm to 2.28 mm anterior to Bregma14.
Continuously slice the rat brain from the olfactory bulb through the nucleus accumbens region according to the stereotaxic brain structure of the rat.
Mount samples onto the slides. Keep the sections at -20 °C for 10 min to dry.
Immediately immerse slides in the pre-chilled 4% paraformaldehyde for 1 h at 4 °C.
Place the slides in an increasing ethanol gradient at room temperature (RT): 50% EtOH for 5 min; 70% EtOH for 5 min; and 100% EtOH for 5 min.
Repeat with fresh 100% EtOH.
Place slides on absorbent paper, and air dry.
Draw a barrier around each section with a barrier pen (see Table of Materials). Let the barrier dry completely for 1 min.
2. Pretreatment of Brain Sections
Turn on the oven and set the temperature to 40 °C.
Add 3 drops (90 µL) of Pretreatment 4 reagent (see Table of Materials) on each brain section (one section per slide).
Incubate the sections for 30 min at RT.
Submerge the slides in 1x PBS for 1 min at RT. Repeat with fresh 1x PBS. NOTE: Slides should not stay in 1x PBS for longer than 15 min.
3. RNA In Situ Hybridization Fluorescent Multiplex Assay
Warm the target probe for 10 min at 40 °C in a water bath, and then cool to RT.
Place the RNA reagent (e.g., Amp 1-4 FL) at RT.
Remove excess liquid from slides with absorbent paper and place back on the slide rack (see Table of Materials).
Add 3 drops (90 µL) of the Drd1α probe (C1) on the sample slice. Incubate for 2 h at 40 °C.
Submerge slides in 1x wash buffer for 2 minutes at RT. Repeat with fresh 1x wash buffer.
Remove excess liquid from the slides with absorbent paper and place back on the slide rack (see Table of Materials).
Add 3 drops (90 µL) of Amp 1-FL on the sample slice. Incubate for 30 min at 40 °C.
Submerge slides in 1 wash buffer for 2 min at RT. Repeat with fresh 1wash buffer.
Remove excess liquid from the slides with absorbent paper and place back on the slide rack (see Table of Materials).
Add 3 drops (90 µL) of Amp 2-FL on the sample slice. Incubate for 15 min at 40 °C.
Submerge slides in 1x wash buffer for 2 minutes at RT. Repeat with fresh 1x wash buffer.
Remove excess liquid from the slides with absorbent paper and place back on the slide rack (see Table of Materials).
Add 3 drops (90 µL) of Amp 3-FL on the sample slice. Incubate for 30 min at 40 °C.
Submerge slides in 1x wash buffer for 2 min at RT. Repeat with fresh 1x wash buffer.
Remove excess liquid from the slides with absorbent paper and place back on the slide rack (see Table of Materials).
Add 3 drops (90 µL) of Amp 4-FL-Alt A on the sample slice. Incubate for 15 min at 40 °C.
Submerge slides in 1x wash buffer for 2 minutes at RT. Repeat with fresh 1x wash buffer.
Remove excess liquid from slides and immediately place 2 drops of mounting reagent (see Table of Materials) onto each section.
Place a 22 mm 22 mm coverslip (see Table of Materials) over each brain section.
Store slides in the dark at 2-8 °C until dry.
Turn on the confocal microscope and switch to a 60X objective.
Obtain Z-stack images with a confocal microscope. See the supplemental video for a detail procedure of confocal imaging.
Acquire images of Drd1α labeled cells in the nucleus accumbens region (Excitation: 488 nm; Emission: 515/30 nm). NOTE: Do not let brain sections dry out between incubation steps.
4. Data Analysis
- Semi-quantitatively analyze the staining pattern: "discrete dots".
- To identify the individual cell from the image, perform DAPI staining as a nuclear marker. However, it is still difficult to analyze certain parts of brain tissue since it has multiple types of cells with irregular shape of nuclei and/or cells. Here, two experienced researchers separately assign each cell from images to define cell region.
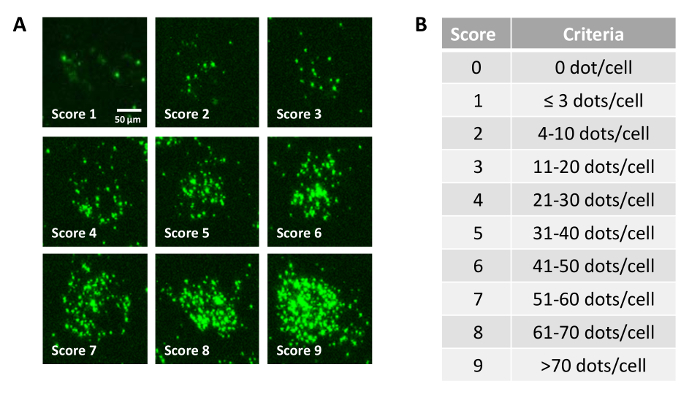
- Visually score each cell within the representative confocal images based on the number of dots per cell using the specific criteria (Figure 1B).
- Statistically analyze the "discrete dots" staining pattern (Optional).
- Examine the number of dots per cell to provide a categorical variable that is ordinal in nature (i.e., the number of cells which fall into each rank category from lowest cell expression (1) to highest cell expression (9)) for further statistical analyses.
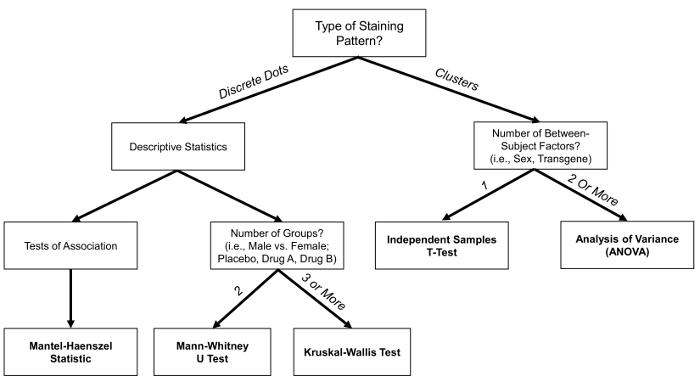
- After examination of descriptive statistics, use three main approaches: a Mantel-Haenszel statistic, a Mann-Whitney U test, and a Kruskal-Wallis test. Although the precise statistical approach will be dependent upon the experimental design and research question of interest, a statistical decision tree (Figure 4) may aid in making decisions regarding one's analytical approach.
- Quantify the signal intensity in the region of interest (ROI) for staining pattern: "clusters".
- Calculate the signal intensity of each image using the following equation: Signal intensity = Total intensity of ROI image - (Average background intensity × Total image area)
- Count the total number of cells within ROI image to calculate the approximate signal expression per cell. Group differences in signal intensity/image as well as the number of cells/image may be of interest to consider mechanisms that may contribute to group differences seen in signal expression/cell (Figure 3B-3D).
- Statistically analyze the "clusters" staining pattern (Optional).
- Examine the signal expression/cell to provide a continuous variable for further statistical analysis. Use two approaches, including an independent sample t-test and analysis of variance (ANOVA), based on the number of between-subjects factors included in the design. A statistical decision tree (Figure 4) may aid in making decisions regarding the most appropriate statistical approach.
Representative Results
The current study observed a "discrete dots" staining pattern for RNA expression in the dopamine D1-alpha receptors (Drd1α) of the NAc in F344/N rats (Figure 1A). Individual fluorescence signals were easily identified and can be seen as single "dots," each of which represents a single RNA transcript within the cell. For images from the NAc that display the "discrete dots" staining pattern, we assessed the dynamic range of expression using a semi-quantitative analysis (Figure 1A, Figure 2A&2B). Each cell was scored from "0-9" based on the number of dots per cell. This "hybridized signal"-score (H-score) method can thus assess the extent and variability for Drd1α expression across individual cells.
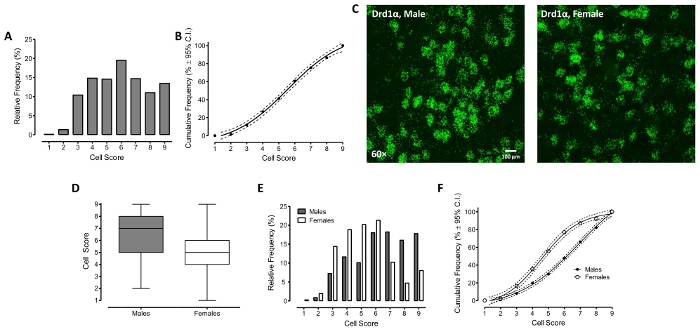
The semi-quantitative approach for the "discrete dot" staining pattern provides a categorical variable that is ordinal in nature (i.e., the number of dots per cell ranked from lowest (1) to highest (9)) for further statistical analysis. In the present study, we were interested in examining the effect of one between-subjects factor (i.e., sex) on Drd1α expression in the NAc. To account for the nested data structure (i.e., two Z-stack images of Drd1α labeled cells in NAc were obtained for each animal), average count values were calculated for each category and used for statistical analysis (Male: n=3, Female: n=3). Given our interest in examining differences in one between-subjects factor (i.e., sex) with two groups (i.e., Male, Female) a Mann-Whitney U test was conducted to assess differences in median cell count (Figure 2D). A Mann-Whitney U test revealed a significant main effect of sex [z=-5.8, p≤0.001], with female animals displaying a decreased median cell score relative to male animals. Figure 2E and 2F illustrate the frequency of cells in each category using two methods: relative frequency and cumulative frequency. Female animals displayed a significant shift in the distribution, with a greater frequency of cells in lower ordered categories relative to male animals. A Mantel-Haenszel test of association was conducted to statistically examine the distribution, revealing a significant effect of sex [χ2MH(1)=67.3, p≤0.001]. Thus, overall, results suggest clear sex differences in Drd1α expression in the NAc.
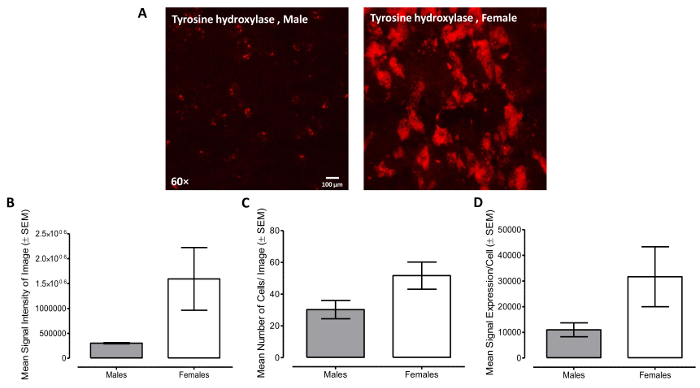
In cases where the copy number of the transcript is very high, the signals can appear as clusters. Here, tyrosine hydroxylase expression (TH) in SNR region showed the "clusters" staining pattern (Figure 3A). To quantify this high-expressing marker, we first averaged the background intensity from eight different background areas. After setting the minimum intensity threshold above the average background intensity, the adjusted image intensity was analyzed as signal per cell of TH.
Given our interest in examining one between-subjects factor (i.e., sex), an independent samples t-test is an appropriate statistical approach. For the analysis, data were log-transformed. To account for the nested data structure (i.e., two Z-stack images of TH labeled cells in SNR were obtained for each animal), average values were calculated and used for statistical analysis (Male: n=2, Female: n=3). Female animals displayed a significant increase in average signal intensity (Figure 3B), suggestive of a higher level of total TH expression, relative to male animals (t(3)=3.3, p≤0.05). However, no significant sex differences were observed in either the number of cells per image (t(3)=2.0, p>0.05; Figure 3C) or the mean signal expression per cell (t(3)=2.0, p>0.05; Figure 3D). A mixed-design ANOVA, considering the three measures as a within-subject factor, also revealed a significant main effect of sex [F(1,3)=10.6, p≤0.05].
Figure 1: Criteria of semi-quantitative analysis on the average number of dots per cell for staining pattern: "discrete dots".A. Representative confocal images (60X) of Drd1α expression at different scores (amplified 250% scale relative to original picture size) B. The specific "score" category for each criterion. Please click here to view a larger version of this figure.
Figure 2: Dopamine D1-alpha receptor (Drd1α) RNA level in Nucleus Accumbens region of F344/N rats.A. The relative frequency % of each "score" category. B. A sigmoidal dose-response curve provided a well-described fit (r2=0.99) with 95% confidence intervals (CI; indicated by the dotted lines) was fit to the cumulative frequency % of each "score" category. C. Representative confocal images (60X) of Drd1α expression in male and female rats, which have the "discrete dots" staining pattern. D. Representative graph of the mean cell score in male and female rats. The error bars stand for the standard error of mean (SEM). E. The relative frequency % of each "score" category, comparing expression in tissues from male and female rats. F. Sigmoidal dose-response curves provided a well-described fit (r2s=0.99) with 95% confidence intervals to the cumulative frequency % of each "score" category between male and female rats. Please click here to view a larger version of this figure.
Figure 3: Tyrosine hydroxylase RNA level (TH) in Substantia Nigra (SNR) region in F344/N rats.A. Representative confocal images (60X; Excitation: 543 nm; Emission: 590/50 nm) of TH expression in male and female rats, which have the "clusters" staining pattern. B-D. Representative analysis of signal intensity per cell. Signal expression/ cell can be derived from quantifying the signal intensity of the image (with intensity threshold set above background) and dividing it by the number of cells within the image. The error bars represent the standard error of the mean (SEM). Please click here to view a larger version of this figure.
Figure 4: Decision Tree for Recommended Statistical Analysis. The decision tree provides guidelines for determining which statistical analysis is most appropriate for the research question of interest. The decision tree is not exhaustive and in some cases, other analyses may be appropriate. Please click here to view a larger version of this figure.
Discussion
In this protocol, we describe a novel technique of in situ hybridization for fresh-frozen brain slices to evaluate Drd1α receptor expression in the nucleus accumbens (NAc) and tyrosine hydroxylase (TH) expression in the substantia nigra (SNR) region. We also provide methods of data analysis for different staining patterns: "discrete dot" or "clusters".
The critical steps for a successful multiplex fluorescence RNA in situ hybridization assay are included. Once decapitating the rat and removing the brain, freeze the brain in liquid nitrogen within 5 min. Keep the brain sections at -20 °C until 4% paraformaldehyde fixation. After the 30 min of incubation of pretreatment 4, slides should not stay in 1x PBS for longer than 15 min. Do not let brain sections dry out between amplification steps. Confocal imaging is very helpful for providing high quality images showing clear staining signals in tissue sample. Using the hybridization oven will rapidly and efficiently bring the samples to 40 °C.
The individual fluorescence signals of Drd1α receptor expression in the NAc were identified as single "dots," each of which represent a single RNA transcript within the cell. Importantly, "dot" size reflects differences in assay conditions, sample preparation, and other factors rather than Drd1α-RNA expression levels. Similarly, the signal intensity of individual "dots" is not relevant to Drd1α-RNA expression levels. We assessed the dynamic range of this expression as a semi-quantitative analysis scored with "0-9" based on the number of dots per cell, which can assess the extent and variability for Drd1α expression. Furthermore, examination of the number of dots per cell provides a categorical variable for further statistical analyses.
Following the examination of descriptive statistics, our protocol recommends three main approaches to statistically analyze signals that present the "discrete dot" staining pattern, which provides an ordinal variable for further analysis. Specifically, a nonparametric Mann-Whitney U test, a nonparametric Kruskal-Wallis test, and a Mantel-Haenszel test of association are recommended. A Mann-Whitney U test and a Kruskal-Wallis test are most appropriate when examining only one between-subjects factor (e.g., Sex). Additionally, a Mantel-Haenszel test of association may be appropriate for examining shifts in the distribution. More complex statistical analyses, including a generalized estimating equation or general linear mixed model may be appropriate when additional factors are included. The recommendations are not exhaustive, and the precise statistical approach will be dependent upon the experimental design and research question of interest.
However, in cases where the copy number of the transcript is very high, the signals can appear as clusters. Here, TH expression in the SNR region showed the "clusters" staining pattern. To quantify the high-expressing TH marker, we adjusted image intensity based on setting the minimum intensity threshold above the average background intensity, and analyzed as signal per cell of TH. Signals that present the "cluster" staining pattern, such as TH in the SNR, can be statistically analyzed by using an independent samples t-tests or ANOVA to compare groups.
There are limitations of this novel RNA in situ hybridization assay. For the "discrete dot" staining pattern, these include how to define an individual cell more accurately. In our study, to identify the individual cell from the image, we performed DAPI staining as a nuclear marker. However, it is still difficult to analyze certain parts of the brain tissue since it has multiple types of cells with irregular shapes of nuclei and/or cells. Here, two experienced researchers separately assign each cell from images to define the cell region. For the "clusters" staining pattern, specific software should be chosen to setup threshold or automatic cell recognization.
Thus, the innovative RNA in situ hybridization assay enabled us to investigate mechanisms influencing both DA uptake and DA release simultaneously, improving understanding of the complexities of the striatal DA system. Overall, researchers would benefit from the novel RNA in situ hybridization technique when investigating dysfunctional changes to dopaminergic markers during the emergence and progression of brain disorders.
Disclosures
There are no conflicts of interest to declare.
Acknowledgments
The present works were supported by National Institutes of Health (NIH) grants HD043680, MH106392, DA013137, and NS100624. Partial fund was provided by a NIH T32 training grant in Biomedical-Behavioral science.
References
- Goldman-Rakic PS, Castner SA, Svensson TH, Siever LJ, Williams GV. Targeting the dopamine D1 receptor in schizophrenia: insights for cognitive dysfunction. Psychopharmacology (Berl) 2004;174(1):3–16. doi: 10.1007/s00213-004-1793-y. [DOI] [PubMed] [Google Scholar]
- Beaulieu JM, Gainetdinov RR. The physiology, signaling, and pharmacology of dopamine receptors. Pharmacol Rev. 2011;63:182–217. doi: 10.1124/pr.110.002642. [DOI] [PubMed] [Google Scholar]
- Zahrt J, Taylor JR, Mathew RG, Arnsten AF. Supranormal stimulation of D1 dopamine receptors in the rodent prefrontal cortex impairs spatial working memory performance. J Neurosci. 1997;17:8528–8535. doi: 10.1523/JNEUROSCI.17-21-08528.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floresco SB, Magyar O, Ghods-Sharifi S, Vexelman C, Tse MT. Multiple dopamine receptor subtypes in the medial prefrontal cortex of the rat regulate set-shifting. Neuropsychopharmacology. 2006;31:297–309. doi: 10.1038/sj.npp.1300825. [DOI] [PubMed] [Google Scholar]
- Arnsten AF, Girgis RR, Gray DL, Mailman RB. Novel dopamine therapeutics for cognitive deficits in schizophrenia. Biol Psychiatry. 2017;81:67–77. doi: 10.1016/j.biopsych.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenbroek BA, Budde S, Cools AR. Prepulse inhibition and latent inhibition: the role of dopamine in the medial prefrontal cortex. Neuroscience. 1996;75(2):535–542. doi: 10.1016/0306-4522(96)00307-7. [DOI] [PubMed] [Google Scholar]
- Parker KL, Alberico SL, Miller AD, Narayanan NS. Prefrontal D1 dopamine signaling is necessary for temporal expectation during reaction time performance. Neuroscience. 2013;255:246–254. doi: 10.1016/j.neuroscience.2013.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manduca A, Servadio M, Damsteegt R, Campolongo P, Vanderschuren LJ, Trezza V. Dopaminergic Neurotransmission in the Nucleus Accumbens Modulates Social Play Behavior in Rats. Neuropsychopharmacology. 2016;41(9):2215–2223. doi: 10.1038/npp.2016.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubo Y, et al. Decreased prefrontal dopamine D1 receptors in schizophrenia revealed by PET. Nature. 1997;385(6617):634–636. doi: 10.1038/385634a0. [DOI] [PubMed] [Google Scholar]
- Abi-Dargham A, et al. Prefrontal dopamine D1 receptors and working memory in schizophrenia. J Neurosci. 2002;22(9):3708–3719. doi: 10.1523/JNEUROSCI.22-09-03708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara R, et al. Dopamine D1 receptor subtype mediates acute stress-induced dendritic growth in excitatory neurons of the medial prefrontal cortex and contributes to suppression of stress susceptibility in mice. Mol Psychiatry. 2017;19 doi: 10.1038/mp.2017.177. [DOI] [PubMed] [Google Scholar]
- Okubo Y, et al. Decreased prefrontal dopamine D1 receptors in schizophrenia revealed by PET. Nature. 1997;385:634–636. doi: 10.1038/385634a0. [DOI] [PubMed] [Google Scholar]
- Wang F, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14(1):22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The rat brain in stereotaxic coordinates. 7th ed. Burlington: Elsevier Academic Press; 2014. [Google Scholar]


