Abstract
Plant α-1,4 glucanotransferases (disproportionating enzymes, or D-enzymes) transfer glucan chains among oligosaccharides with the concomitant release of glucose (Glc). Analysis of Chlamydomonas reinhardtii sta11-1 mutants revealed a correlation between a D-enzyme deficiency and specific alterations in amylopectin structure and starch biosynthesis, thereby suggesting previously unknown biosynthetic functions. This study characterized the biochemical activities of the α-1,4 glucanotransferase that is deficient in sta11-1 mutants. The enzyme exhibited the glucan transfer and Glc production activities that define D-enzymes. D-enzyme also transferred glucans among the outer chains of amylopectin (using the polysaccharide chains as both donor and acceptor) and from malto-oligosaccharides into the outer chains of either amylopectin or glycogen. In contrast to transfer among oligosaccharides, which occurs readily with maltotriose, transfer into polysaccharide required longer donor molecules. All three enzymatic activities, evolution of Glc from oligosaccharides, glucan transfer from oligosaccharides into polysaccharides, and transfer among polysaccharide outer chains, were evident in a single 62-kD band. Absence of all three activities co-segregated with the sta11-1 mutation, which is known to cause abnormal accumulation of oligosaccharides at the expense of starch. To explain these data we propose that D-enzymes function directly in building the amylopectin structure.
In plants, the only α-1,4 glucanotransferases reported to be present at the time of starch synthesis are collectively called D-enzymes (Peat et al., 1956; Takaha et al., 1993). D-enzymes act on soluble oligosaccharides at least three Glc residues long (maltotriose) and disproportionate them into oligosaccharides of various lengths at the expense of Glc formation. In such a reaction an α-1,4 linkage is cleaved from a donor unbranched oligosaccharide of at least three Glc residues, and the resulting chain segment is transferred to another acceptor glucan, creating a novel α-1,4 linkage. Maltosyl residues are often transferred as a result of D-enzyme action, but maltose itself is not a product of the reaction. However, both Glc and maltose can be used as acceptors (Jones and Whelan, 1969). It is known that in Arabidopsis leaves, D-enzyme is the major maltotriose-metabolizing enzyme present (Lin and Preiss, 1988; Zeeman et al., 1998). The presence of this activity during potato tuber development has led investigators to suggest that D-enzyme might be required for some specific aspect of starch biosynthesis (Takaha et al., 1993). Like a few other glucanotransferases, such as branching enzyme (Takaha et al., 1996a), D-enzyme has been recently shown to lead to the formation of cyclic compounds after prolonged incubation of both amylose and amylopectin with high amounts of pure activity (Takaha et al., 1996b, 1998). It is not known if this property relates to the physiological function of this enzyme.
D-enzyme is believed to be part of the starch degradation pathway. The disproportionating of small malto-oligosaccharides into longer glucans facilitates their degradation through maltodextrin phosphorylases and glucosidases in a fashion similar to that described for amylomaltase, a similar α-1,4-glucanotransferase required for malto-oligosaccharide assimilation in Escherichia coli (for review, see Boos and Shuman, 1998).
We have previously shown that the absence of D-enzyme in sta11-1 mutants correlates with an accumulation of unbranched malto-oligosaccharides. However, we also found a large decrease in starch content in conditions of maximal starch synthesis, a result that does not fit the catabolic function envisioned for these α-1,4 glucanotransferases. We were surprised to find a modification of amylopectin chain-length distribution. The latter consisted of a relative increase of very small chains that could easily be distinguished from similar distributions witnessed in other mutant amylopectin structures (Fontaine et al., 1993; Libessart et al., 1995). This surprising change correlated with an alteration in granule size morphology and crystallinity. It also resulted in significant increases in the ratio of amylose to amylopectin. Because no other known enzyme of the starch metabolic pathway was altered in the mutant, all of these phenotypes have to be explained by the disappearance of D-enzyme. The most straightforward explanation would be to assume a direct function of D-enzyme in the building of the amylopectin structure; however, the biochemical evidence supporting such a function remains to be produced.
We demonstrate that the Chlamydomonas reinhardtii D-enzyme is active on the outer chains of amylopectin and glycogen. We propose a novel function for the plant α-1,4 glucanotransferase that explains all phenotypic traits observed simultaneously in the sta11-1 mutants of C. reinhardtii.
MATERIALS AND METHODS
Materials
[1,2-14C]Acetic acid (sodium salt) was purchased from Amersham. Maltotriose-forming amylase was purchased from Unipex (Reuil-Malmaison, France). Sweet potato β-amylase and Pseudomonas amyloderamosa isoamylase were from Megazyme International (Sydney, Australia). Glc and glucans were assayed through the amyloglucosidase using the starch determination kit from Boehringer Mannheim. Glc, maltose, maltotriose, maltotetraose, maltopentaose, maltohexaose, and maltoheptaose were from Sigma. Radioactive oligosaccharides were prepared by extracting in vivo-labeled amylopectin from the Chlamydomonas reinhardtii mutant strain BAFR1 defective for granule-bound starch synthase I and amylose synthesis (Delrue et al., 1992). The strain was grown in the presence of 500 μCi [1,2-14C] of sodium acetate at 702 μCi mg−1. The culture was starved of nitrogen in TP (Tris phosphate) medium for 5 d. Fifteen percent of the total label was successfully incorporated into amylopectin. The starch was purified through Percoll gradient centrifugation as detailed previously (Delrue et al., 1992), and stored at −20°C. Radioactive oligosaccharides were prepared freshly by debranching 10 mg of in vivo-labeled amylopectin to completion, as described previously (Dauvillée et al., 1999). The specific radioactivity of the debranched oligosaccharides was 1.8 μCi mg−1.
Radiolabeled maltotriose is not available commercially and was prepared as follows. Maltotriose-forming amylase (0.06 unit) from Microbacterium sp. was incubated with 500 μg of radioactive oligosaccharide prepared as described above in a 1-mL final volume of 10 mm CaCl2, 10 mm Tris/HCl, pH 7.0. The reaction was incubated for 15 min at 55°C and stopped by heating for 5 min in a boiling-water bath. The maltotriose produced was purified by high-performance anion-exchange chromatography with pulsed amperometric detection as described previously (Fontaine et al., 1993). The yield and purity were 46% and 95%, respectively. Maltotriose was lyophilized and stored at room temperature.
C. reinhardtii Strains, Growth Conditions, and Media
The wild-type reference C. reinhardtii strain used in this study was 137C (mt− nit1 nit2) and JV45J (mt− nit1 nit2 sta11-1). Wild-type and mutant segregants from a cross performed between JV45J and wild-type strain 37 (mt+ pab2 ac14) have been previously described and were used throughout this work. Starch from the granule-bound starch synthase I mutant BAFR1 (mt+ nit1 nit2 cw15 arg7-7 sta2-29::ARG7) was used as the source of pure amylopectin.
All experiments were carried out in continuous light (40 μE m−2 s−1) in the presence of acetate at 24°C in liquid cultures that were shaken vigorously without air or CO2 bubbling. Late-log-phase cultures were inoculated at 105 cells mL−1 and harvested at 2 × 106 cells mL−1. Standard TAP (Tris acetate phosphate) medium was used in this work and was fully detailed in Harris (1989).
Structural Analysis of Polysaccharides
1H-NMR of gel permeation chromatography-purified starch fractions were as described previously (Fontaine et al., 1993). The APTS-tagged chains produced by isoamylase-mediated debranching were separated by high-resolution slab gel electrophoresis on a DNA sequencer (O'Shea and Morell, 1996). Enzymatic debranching of polysaccharides was as previously described (Dauvillée et al., 1999).
Enzyme Purification
Frozen crude extract proteins (500 mg) were cleared by centrifugation and precipitated by 10% protamine sulfate for 20 min at 4°C. The supernatant was submitted to anion-exchange chromatography (Mono-Q columns, Pharmacia) in 50 mm sodium acetate and 2 mm DTT adjusted to pH 6.0 with acetic acid. The unretained fraction was subjected to (NH4)2SO4 precipitation at 30% saturation. The supernatant was further precipitated at 50% saturation. The pellet was ressuspended in the same buffer and subjected to gel permeation chromatography on a S100 column (Pharmacia). The pooled fractions containing D-enzyme activity were passed through a cation-exchange column (model UnoS12, Bio-Rad) in the same buffer. The enzyme activity was followed by either the previously described zymogram or by the quantitative D-enzyme assay. The latter consists of measuring the Glc produced from maltotriose by incubating up to 50 μL of sample with 75 μL of 50 mm sodium acetate adjusted to pH 6.0 with acetic acid and 25 μL of an 80 mg mL−1 maltotriose solution. The mixture was incubated at 30°C for 15 min and stopped by heating for 3 min in a boiling-water bath. The sample was cleared by centrifugation, and the Glc was assayed by measuring the production of NADPH during the standard hexokinase-Glc-6-P dehydrogenase reaction.
Zymogram Analyses
We used zymogram techniques adapted from the method developed by Lacks and Springhorn (1980), who used α-amylase as a model enzyme to study renaturation and detection. An in-depth discussion on renaturation-detection techniques can be found in Gabriel and Gersten (1992). Soluble crude extracts were always prepared from late-log-phase cells (2 × 106 cells mL−1) grown in high-salt acetate medium under continuous light (80 μE m−2 s−1). Algae were ruptured by passage in a French press (10,000 p.s.i.) at a density of 109 cells mL−1 and immediately stored at −80°C. After thawing, the lysate was cleared by centrifugation at 10,000g for 15 min at 4°C. The amount of protein was measured using a protein assay kit (Bio-Rad). Protein (500 μg) in 100 μL of 25 mm Tris-Gly, pH 8.3, 1% (w/v) SDS, 5% (v/v) β-mercaptoethanol was denatured by heating in a boiling-water bath for 5 min.
Our zymogram procedures were established to detect starch hydrolases in starch-containing gels in Mouille et al. (1996). The oligosaccharide-incorporation zymogram technique was modified from these procedures as follows. Denatured proteins were loaded on a 30:1 (acry:bis), 7.5% (v/v) acrylamide, 0.1% (v/v) SDS, 1.5-mm-thick denaturing polyacrylamide gel containing 0.3% rabbit liver glycogen (Sigma). Electrophoresis was performed at room temperature at 15 V cm−1 for 90 min using the Mini-Protean II cell (Bio-Rad) in 25 mm Tris-Gly, pH 8.3, 1 mm DTT, and 0.1% (v/v) SDS buffer. At the end of the run, the gel was washed twice with gentle shaking for 1 h in 100 mL of 40 mm Tris at room temperature to remove SDS and to renature the proteins. The gel was incubated overnight in 50 mm Tris, 5 mm EDTA, 10 mm DTT, and 2 mm maltoheptaose (Sigma) at 25°C. The reaction was stopped, and the gel was stained in an aqueous solution containing 0.25% KI and 0.025% I2.
Enzyme Treatment of Amylopectin
Five milligrams of amylopectin from the waxy cultivar of maize was dispersed in 100 μL of 90% DMSO. Seven-hundred microliters of 50 mm sodium acetate, adjusted to pH 6.0 with acetic acid and with an enzyme activity corresponding to 2 nmol of Glc produced from maltotriose per minute, was added subsequently and incubated overnight at 30°C; 500 μg of the sample was further subjected to a β-amylase (17 units) treatment in 50 μL of 50 mm sodium acetate adjusted to pH 4.0 with acetic acid. After 1 h of incubation, another dose of β-amylase was added and the incubation continued for an additional hour. The reaction was stopped and the enzyme inactivated by heating the sample for 5 min in a boiling-water bath. Both the β-amylase-treated and untreated samples were then debranched and analyzed.
Oligosaccharide Incorporation Assay
From 50 to 500 μg of radioactive oligosaccharides, prepared as described in “Materials and Methods,” were added to 50 μg to 5 mg of either nonradioactive amylopectin (purified from the same BAFR1 strain) or rabbit liver glycogen in a 1-mL final volume of 50 mm sodium acetate adjusted to pH 6.0 with acetic acid. An activity of D-enzyme corresponding to 106 nmol of Glc produced per minute from maltotriose was added to the sample and incubated for 150 min at 30°C. The activity in the presence of 500 μg of oligosaccharides was previously proven to be strictly proportional to time (up to 4 h) and activity amount (up to 400 nmol of Glc produced per minute from maltotriose). The labeled polysaccharide was then separated from the unincorporated oligosaccharides through gel permeation chromatography on either TSK-HW50 (Merck) for glycogen or on Sepharose CL2B (Pharmacia) for amylopectin. The reverse reaction, transfer of radioactive outer chains from amylopectin to cold oligosaccharides, was monitored at or above the physiological concentration of malto-oligosaccharides (200–500 μg malto-oligosaccharide mL−1) using gel permeation chromatography.
RESULTS
Enzyme Purification
D-enzyme was partially purified by a three-step chromatographic procedure involving anion exchange, selective ammonium sulfate precipitation, and gel permeation and cation exchange. The purified activity displayed a wide pH optimum over the range of 5.0 to 7.5. The activity decreased significantly above pH 9.0. It is worth noting that under our standard conditions we could not detect significant production of Glc from maltotriose in crude extracts from the sta11-1 mutant, suggesting that D-enzyme was the only enzyme present that was able to metabolize small oligosaccharides under physiological conditions.
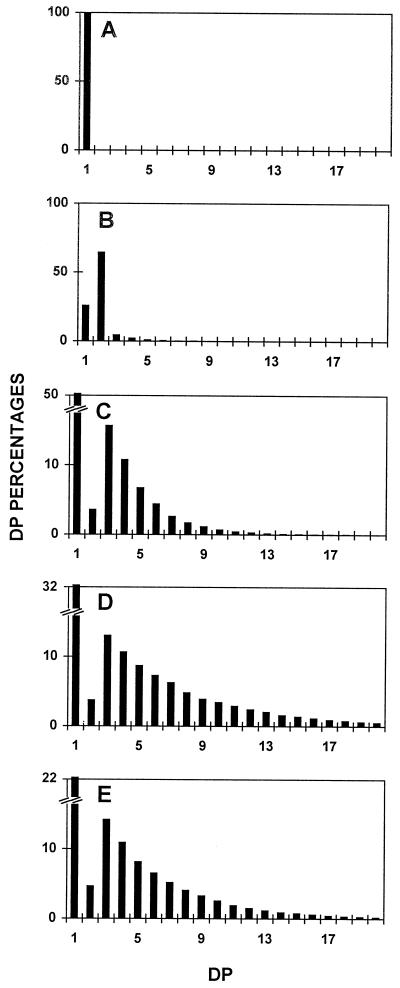
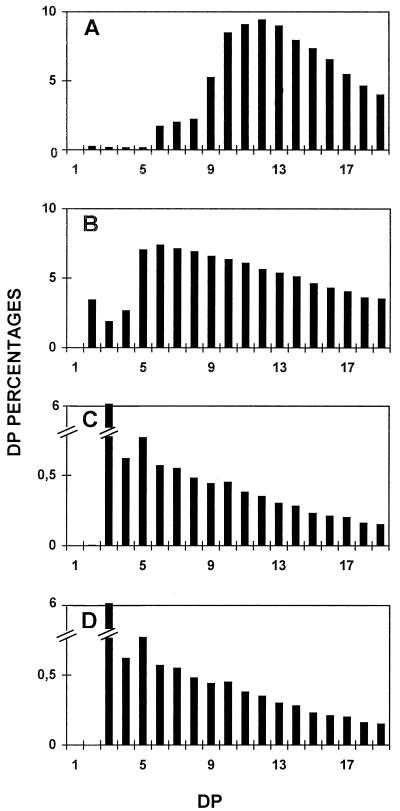
After these chromatographic steps, yields of approximatively 2.4% of D-enzyme activity were obtained (Table I). If all fractions containing the activity were harvested, the purification factor measured was 32-fold. However, if low-protein-containing fractions were selected, we were able to bring this factor up to 135-fold, with a 0.5% yield (Table I). The enzyme was freed from all other contaminating starch hydrolases after the second chromatographic step. These contaminating activities were monitored by assaying the fractions on starch-containing zymogram gels (Mouille et al., 1996). The activity was stable when stored for over 6 months at −20°C in purification buffer. We tested the action of the purified 62-kD enzyme separately on Glc, maltose, maltotriose, maltotetraose, maltopentaose, and maltoheptaose (Fig. 1). While the 62-kD α-1,4 glucanotransferase left Glc untouched, it successfully disproportionated all oligosaccharides above DP 3, as shown in Figure 1. The reduced but significant disproportionating activity witnessed with DP 2 was due to the presence of trace amounts of maltotriose and maltotetraose in the commercial source of maltose. The very low amounts of maltose detected whenever oligosaccharides were successfully disproportionated further confirmed that the enzyme obeys the rules established years ago by Whelan to define D-enzymes (Jones and Whelan, 1969).
Table I.
D-enzyme purification
| Purification Step | Purification Factor | Yield |
|---|---|---|
| % | ||
| Crude extract | 1 | 100 |
| Protamine sulfate | 2.8 | 40 |
| Mono Q | 5 | 22 |
| S100 + UnoS | 32 | 2.4 |
| Fraction 24 | 135 | 0.5 |
D-enzyme was measured through the release of Glc from maltotriose (see Methods).
Figure 1.
Chain-length distributions of oligosaccharides after incubation with the 62-kD transferase. Undebranched oligosaccharides were separated according to length after APTS fluorescence labeling and separation on a DNA sequencer. Percentages of chains ranging between DP 1 and 20 (chains containing 1–20 Glc residues) are scaled on the y axis. A, Undebranched malto-oligosaccharides generated through incubation of Glc with semi-pure C. reinhardtii D-enzyme. B, Undebranched malto-oligosaccharides generated through incubation of maltose with semi-pure C. reinhardtii D-enzyme. C, Undebranched malto-oligosaccharides generated through incubation of maltotriose with semi-pure C. reinhardtii D-enzyme. D, Undebranched malto-oligosaccharides generated through incubation of maltopentaose with semi-pure C. reinhardtii D-enzyme. E, Undebranched malto-oligosaccharides generated through incubation of maltoheptaose with semi-pure C. reinhardtii D-enzyme.
Activity of D-Enzyme toward the Outer Chains of Amylopectin
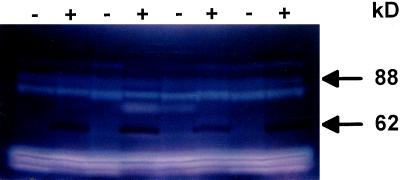
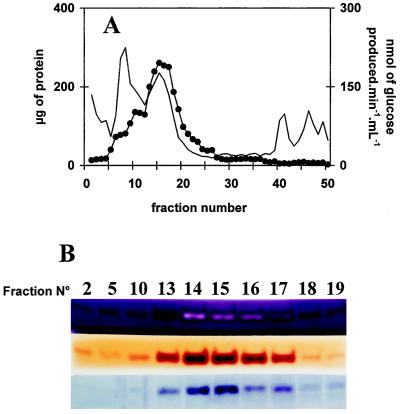
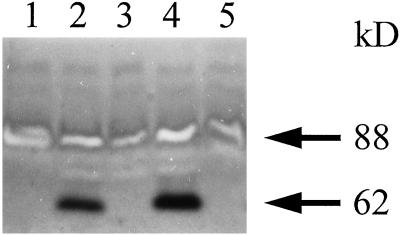
In the course of performing zymograms under denaturing conditions to detect the various starch hydrolases in sta11-1 and wild-type strains, iodine treatment of the starch-containing gels (Fig. 2) revealed the absence in the mutant of a band staining dark red. The 62-kD mass deduced from the zymogram fit that of the C. reinhardtii D-enzyme (Colleoni et al., 1999). It was absent in all meiotic progeny bearing the sta11-1 mutation (n = 75) and systematically present in wild-type strains. In addition, we noted co-elution throughout enzyme purification of this red-staining band and with both the quantitative D-enzyme assay and our zymogram procedure using maltotriose as a substrate (Colleoni et al., 1999). An example of this co-purification is displayed in Figure 3.
Figure 2.
Zymogram analysis. Denatured crude extracts (100 μg of protein) from eight wild-type (+) and sta11-1 (−) strains were loaded on denaturing polyacrylamide gels containing soluble potato starch (Mouille et al., 1996). The proteins were renatured after electrophoresis and incubated overnight. Starch hydrolases were revealed by iodine staining. The 88-kD blue-staining band was previously proven to be a debranching enzyme. The 62-kD dark-red-staining band can be distinguished from all other starch hydrolases by the absence of gel clearing within the staining region, which is consistent with the absence of oligosaccharide production during polysaccharide modification.
Figure 3.
Co-purification of starch structure modification, Glc production from maltotriose, and incorporation of maltoheptaose into the external chains of glycogen. A, Elution profile of FPLC chromatography. In this experiment, 60 mg of crude extract protein (after a protamine sulfate precipitation step) was loaded on an anion-exchange column linked directly to a cation-exchange column, which was only eluted with 5% (w/v) NaCl. Two-milliliter fractions were collected at a 1 mL min−1 flow rate. Proteins (thin line) were measured in each fraction according to the Bio-Rad assay. D-enzyme activity (•) was monitored through production of Glc from maltotriose (see Methods). B, Fractions from all cation exchanges were assayed through three distinct zymogram procedures involving denaturation of proteins and renaturation after migration (see Methods). Only the active fractions are shown in B. Three distinct zymogram procedures are displayed. These include modification of starch structure (top), incorporation of maltoheptaose in the outer chains of glycogen (middle), and Glc production from maltotriose (bottom). All active bands were shown to migrate as 62-kD proteins. Note that the dark-red band that characterizes D-enzyme in starch-containing zymograms turns to a white stain upon incubation with a vast excess of enzyme in the concentrated peak fractions.
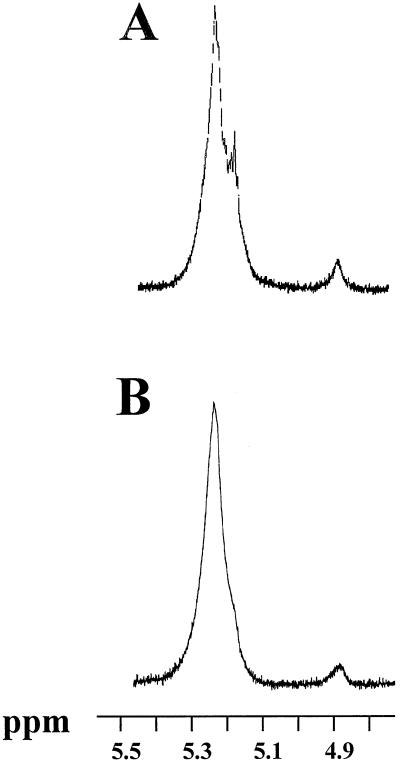
Because we have previously proven that the 62-kD zymogram band contained the D-enzyme activity, and because both D-enzyme and this activity co-purify and are clearly under the control of the same gene, we conclude that the modification of starch structure witnessed in these gels and the disproportionating activity are supported by the same 62-kD protein. Following the technique established in Mouille et al. (1996), polysaccharide was eluted from the bands present in the amylopectin-containing gels and subjected to 1H-NMR analysis. A similar analysis was performed after 135-fold purification of the enzyme activity (see above) and treatment of amylopectin in purification buffer.
The 1H-NMR signals of the incubated amylopectin displayed major changes. The latter consisted of the replacement of the bimodal proton signal at 5.3 to 5.2 ppm by a monomodal signal at the same 5.2 ppm position in our standard NMR conditions (Fig. 4). The relative heights of these two signals distinguishes the 1H-NMR signatures of glycogen and amylopectin (Mouille et al., 1996) and are therefore suspected to reflect differences in chain-length distribution. Enzymatic debranching does not affect the relative heights of these two signals, confirming that chain-length distribution modifications are present rather than increases in the number of branches (Mouille et al., 1996).
Figure 4.
1H-NMR analysis of amylopectin incubated with or without the 62-kD transferase. Ten milligrams of maize amylopectin was incubated overnight in 1 mL of 50 mm sodium acetate/acetic acid, pH 6.0, in the absence (A) or presence (B) of 50 μL of semi-pure enzyme fraction corresponding to 2 nmol of Glc produced from maltotriose per minute. Part of the 1H-NMR spectra revealing signals specific from the α-1,4-linked Glc residues (from 5.4–5.1 ppm) or for the α-1,6-linked Glc residues (around 4.9 ppm) is displayed. NMR analysis was as described in Mouille et al. (1996).
Upon incubation of amylopectin with the semi-pure D-enzyme, changes were witnessed in the chain-length distribution of the incubated amylopectin (Fig. 5, A and B). We found no evidence of oligosaccharide production by spotting the treated samples on TLC plates as described in Mouille et al. (1996). The amount of amyloglucosidase-resistant material (cyclic glucans) remained under 5% of the total polysaccharide amounts used in these experiments, suggesting that incubation times exceeding 12 h and higher enzyme activities are required to produce these structures. In addition, no release of radioactive material was witnessed when radioactive amylopectin was used as a substrate (see Methods).
Figure 5.
Chain-length distributions of amylopectin after incubation with the 62-kD transferase. Isoamylase-debranched chains were separated according to length after APTS fluorescence labeling and separation on DNA sequencer. Percentages of chains ranging between DP 1 and 19 (chains containing 1–19 Glc residues) are scaled on the y axis. A, Debranched chains from gel permeation chromatography-purified C. reinhardtii reference amylopectin (extracted from the amylose-free BAFR1 strain). B, Debranched chains from gel permeation chromatography-purified C. reinhardtii reference amylopectin (from strain BAFR1) after incubation with semi-pure C. reinhardtii D-enzyme. C, Debranched chains from gel permeation chromatography-purified C. reinhardtii reference amylopectin (from strain BAFR1) after incubation with semi-pure C. reinhardtii D-enzyme and subsequent digestion with β-amylase. D, Debranched chains from gel permeation chromatography-purified C. reinhardtii reference amylopectin (extracted from the amylose-free BAFR1 strain) after digestion with β-amylase. In C and D, the polysaccharide was not separated from the maltose generated by β-amylase. DP 2 is therefore not represented.
Because no oligosaccharides were liberated during amylopectin treatment with the 62-kD D-enzyme, and because the amount of α-1,6 linkages remained constant (5%), we conclude that amylopectin acts as an α-1,4 glucanotransferase, cleaving α-1,4 linkages present on donor amylopectin chains and transferring them to the nonreducing end of neighboring acceptor chains. To prove that the transfer reaction involved the outer chains of amylopectin, β-amylase digestions of the incubated polysaccharides were performed on the untreated and incubated amylopectin. The digested polysaccharides were then subjected to enzymatic debranching, yielding identical chain-length distributions for the treated and untreated samples (Fig. 5, C and D). Because β-amylase are processive enzymes that selectively digest the outer chains of the polysaccharides, this result proves that the major modifications seen in Figure 5B are confined to the polymer's external chains.
These dramatic changes are sufficient to explain a shift in color of the iodine polysaccharide complex in starch- or amylopectin-containing gels. Indeed, Banks et al. (1971) have shown that the λmax (the wavelength of the maximal absorbance of the iodine-polysaccharide complex) decreases with the average DP of linear α-1,4 glucan populations. Therefore, changes in exterior chain-length distributions of D-enzyme-treated amylopectin starches are likely to affect the λmax of the resulting polysaccharide. This could explain the typical dark-red stain detected in the zymograms.
D-Enzyme Favors the Incorporation of Oligosaccharides into Polysaccharide Outer Chains
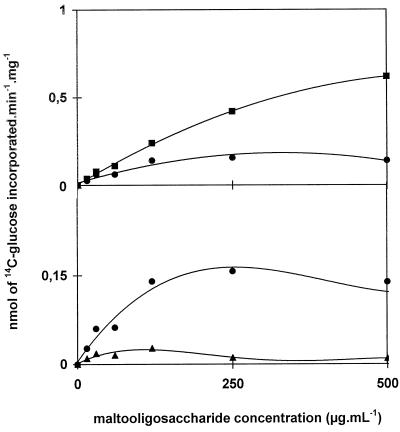
We characterized further the action of D-enzyme in the presence of both amylopectin and unbranched malto-oligosaccharides. We produced 14C labeled malto-oligosaccharides by debranching in vivo-labeled amylopectin. The chain-length distribution of the mixture of debranched chains was the same as that displayed in Figure 5A. Incorporation of the oligosaccharides into amylopectin (Fig. 6) or rabbit liver glycogen was monitored successfully. This incorporation was linear with time for incubation times ranging between 15 min and 6 h. This radioactivity disappeared upon subsequent treatment of the polysaccharide with β-amylase, proving that D-enzyme transfers efficiently segments of malto-oligosaccharides onto the outer chains of both glycogen and amylopectin. In contrast, incubation of radioactive amylopectin (1 mg mL−1) with up to 500 μg mL−1 of unbranched, cold malto-oligosaccharides prepared from the same source did not yield any measurable release of radioactive material into the malto-oligosaccharide fraction. The sensitivity of the assay was such that we would have detected this reverse reaction even if as little as 5% of the amount transferred in similar conditions onto the polysaccharide outer chains had occurred. The physiological concentration that we measured in C. reinhardtii for small malto-oligosaccharides ranged between 50 to at most 200 μg mL−1. We therefore conclude that at the starch granule surface (with amylopectin at over 10 mg mL−1 and malto-oligosaccharide concentrations well under 200 μg mL−1), incorporation of debranched oligosaccharides such as those generated through debranching of pre-amylopectin would be strongly favored.
Figure 6.
Malto-oligosaccharide incorporation assays. Amylopectin (2.5 mg, ▪; 1 mg, •; 0.25 mg, ▴) was incubated with increasing concentrations of radiolabeled malto-oligosaccharides produced by debranching radioactive C. reinhardtii amylopectin. The y axis represents two different scales giving net incorporation of oligosaccharides within the polysaccharide. An enzyme activity corresponding to 2 nmol of Glc produced from maltotriose per minute was used in these experiments. Incorporation was strictly proportional to enzyme activity for activities ranging between 0.5 and 20 nmol of Glc produced from maltotriose per minute. The label was confined to the outer chains of amylopectin, since all of the radioactive material was liberated upon treatment with β-amylase.
The Specific Requirements of D-Enzyme for the Incorporation of Oligosaccharides into Polysaccharide Outer Chains
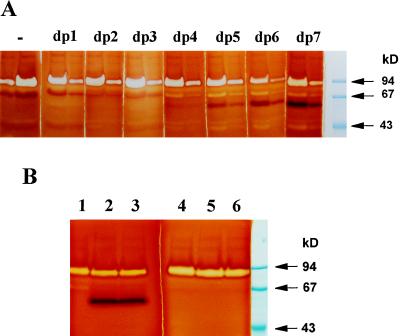
Incorporation of label from malto-oligosaccharides into glycogen gave us an additional reliable assay of D-enzyme activity. This enabled us to set-up a novel zymogram procedure for D-enzyme detection (Fig. 7) based on the incorporation of oligosaccharides into glycogen. Once again, our denaturing gels allowed us to estimate the mass of the enzyme at 62 kD. Moreover, co-elution of this activity stain was seen with those of our two other zymogram procedures and with the quantitative D-enzyme assay during purification of the protein (Fig. 3). We found co-segregation between all three types of activities (Glc production from maltotriose, amylopectin modification, and incorporation of unbranched glucans into glycogen) and the wild-type STA11 allele in the progeny of crosses involving wild-type and mutant C. reinhardtii strains. We then proceeded to monitor the incorporation of malto-oligosaccharides of controlled length into the external chains of glycogen using this novel zymogram procedure.
Figure 7.
Malto-oligosaccharide incorporation zymograms. Two-hundred micrograms of denatured crude extract protein from wild-type (lanes 2 and 4) and mutant (lanes 1, 3, and 5) recombinants obtained after crossing JV45J with the wild-type strain 37 were loaded in glycogen-containing gels in the presence of maltoheptaose. Note that the white-staining band at 88 kD represents the debranching enzyme missing in the glycogen-producing mutants of C. reinhardtii (Mouille et al., 1996).
It is evident from our results displayed in Figure 8A that efficiency of transfer onto glycogen external chains increases with the length of the donor chain. Particularly striking in Figure 8B is the absence of stain when using maltotriose as a donor substrate even at very high substrate concentrations and after prolonged incubation. To ensure that we were not dealing with some iodine staining artifact, we prepared radioactive maltotriose (see Methods) and compared the efficiency of transfer with that of debranched amylopectin. We found low incorporation of label from purified labeled maltotriose into the outer chains of amylopectin. Indeed, we observed 10% of the incorporation rates initially measured with the mix of debranched radioactive oligosaccharides that we displayed in Figure 6.
Figure 8.
Comparisons of malto-oligosaccharide incorporation into glycogen. A, Crude extract protein (100 and 300 μg) was loaded eight times in pairs on two identical gel systems and run simultaneously in glycogen-containing gels. The gels were then cut into eight different fragments and incubated separately with 2 mm DP1 (Glc) to DP 7 (maltoheptaose). One gel fragment was incubated without malto-oligosaccharides (−). The gels were incubated overnight at room temperature. B, Denatured crude extract protein (200 μg) from two wild-type reference strains (lanes 2 and 3; lanes 5 and 6) and mutant strain JV45J (lanes 1 and 4) were loaded in glycogen-containing gels and incubated for 48 h with 2 mm maltoheptaose (lanes 1–3) or 20 mm maltotriose (lanes 4–6).
Surprisingly, purified and labeled maltotetraose gave incorporation rates similar to those previously achieved by the mix of debranched and labeled oligosaccharides, while in zymograms maltotetraose always gave faint stains. It must be stressed that all malto-oligosaccharides added in the label incorporation experiments were immediately disproportionnated by D-enzyme into longer oligosaccharides, while in the zymogram experiments this modification was slowed because of the vast excess of substrate oligosaccharide contained in the large volume (30 mL) of incubation buffer. Therefore, we feel that glucans longer than maltotetraose will define the actual optimal substrates of the incorporation reaction. Such longer glucans are simply not produced fast enough by disproportionating maltotriose when a large excess of oligosaccharides is used on an immobilized D-enzyme such as in the zymogram procedures. We expect that in vivo glucans of the required lengths are produced by the processing of pre-amylopectin through isoamylases.
We would like to stress that the incorporation of labeled oligosaccharides into solubilized amylopectin, although more quantitatively reliable, is a poor reflection of the physiological situation. Both amylopectin and oligosaccharides readily diffuse in this system. However, in the zymogram procedure the oligosaccharides are allowed to diffuse in a large volume of buffer while the polysaccharide remains constrained in a small gel phase volume together with a high enzyme specific activity. This situation mimicks the synthesis of polysaccharide at the granule surface.
The Enhancement by D-Enzyme of the Phosphorylase-Mediated Malto-Oligosaccharide Degradation
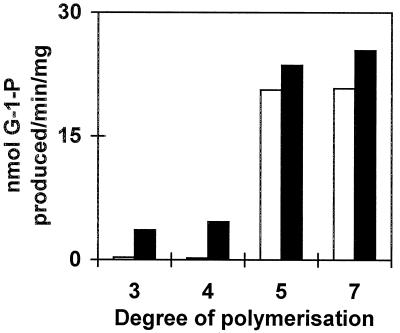
In bacteria it was suggested that amylomaltase, an α-1,4 glucanotransferase similar to D-enzyme, enhanced the production of Glc-1-P by maltodextrin phosphorylase through the generation of glucans long enough (DP 5) to be used by maltodextrin phosphorylase (for review, see Boos and Shuman, 1998). We tested this suggestion by measuring Glc-1-P production from maltotriose, maltotetraose, maltopentaose, and maltoheptaose in crude extracts of C. reinhardtii wild-type and mutant sta11-1 strains. As shown in Figure 9, it is evident that the presence of D-enzyme stimulates the degradation of both maltotetraose and maltotriose through phosphorylase by at least a factor of 5. Interestingly, the same treshold length (DP 5) of the glucan allowing Glc-1-P production is found both in bacteria and plants.
Figure 9.
Stimulation of malto-oligosacharide phosphorolysis by D-enzyme. Crude extract protein (100 μg) from wild-type strain 137C (black bars) and mutant strain JV45J (white bars) were incubated for 1 h at 30°C in the presence of 2.5 mm maltotriose, maltotetraose, maltopentaose, and maltoheptaose.
DISCUSSION
D-Enzyme Readily Transfers Segments of Chains onto Polysaccharide External Chains
D-enzyme was initially described as an enzyme using and producing soluble oligosaccharides (Peat et al., 1956; Jones and Whelan, 1969). It was assumed to be an integral part of the malto-oligosaccharide assimilation pathway during starch breakdown. Recent papers using high amounts of pure potato D-enzyme activity on both amylose and amylopectin showed that the enzyme was able to produce cyclic branched or unbranched glucans of various lengths after prolonged incubation (Takaha et al., 1996b, 1998). Similar results were obtained with branching enzymes (Takaha et al., 1996a). These experiments, however, did not investigate the effect of D-enzyme on the amylopectin chain-length distribution at lower concentrations of enzyme activities and for restricted time periods. It occurred to us that the reaction that we had first observed on amylopectin alone failed to generate oligosaccharides because it uses the polysaccharide outer chains both as donor and acceptor glucans.
The reaction is driven on an unbranched soluble malto-oligosaccharide at the expense of Glc formation from the reducing end of the donor chain. However, the analogous residue from the donor amylopectin outer chain is expected to remain bound to the polymer through an α-1,6 branch. Indeed, the concentration of external glucans within starch can be estimated at 450 mm (van de Wal et al., 1998). Such high local concentrations would outcompete as a recipient any soluble oligosaccharide that might be present within the plastid. We further reasoned that if unbranched oligosaccharides and preamylopectin co-exist in the plant cell, then D-enzyme would presumably act as a polymerase, using oligosaccharide chains as donors and yielding net incorporation into insoluble mature amylopectin ultimately at the expense of Glc formation.
Because of the extremely high local concentration of potential acceptor chains in amylopectin or its precursors (pre-amylopectin), the glucan once incorporated will only very slowly be freed from the polysaccharide. The energy cost of such a polymerization reaction would be exceedingly low and would consist only of two ATP high-energy bonds required to regenerate one ADP-Glc for each glucan chain incorporated into the polysaccharide. In the present study, we have demonstrated that the D-enzyme from C. reinhardtii (and probably from vascular plants, too) is able to incorporate segments of unbranched malto-oligosaccharides quickly and efficiently on the outer chains of amylopectin. We were, however, unable to observe the reverse reaction (transfer from amylopectin outer chains to malto-oligosaccharides) at physiological substrate concentrations. This is not that surprising, since fructosyl transferases, enzymes of analogous properties, have been proven to function in fructan polymerization (van der Meer et al., 1998). The rates of incorporation that we measured are physiologically relevant, especially if one assumes that the reaction takes place at the growing surface of the starch granule. Indeed, we have been unable to saturate D-enzyme with amylopectin in our in vitro incorporation experiments.
The Function of D-Enzyme in Amylopectin Synthesis
D-enzyme-defective mutants accumulate large amounts of oligosaccharides with no detectable α-1,6 branches. A likely source of such oligosaccharides can be sought in the action of debranching enzymes. We and others have recently proposed that amylopectin synthesis occurs through cycles of glucan trimming by selective debranching of a precursor (pre-amylopectin) into a mature semi-crystalline amylopectin molecule (Ball et al., 1996). During this process of pruning α-1,6 branches, the plant debranching enzymes will release unbranched malto-oligosaccharides. Because such molecules accumulate in the D-enzyme-deficient mutant, we believe that the normal function of plant α-1,4 glucanotransferases is to process these chains.
There are two possible types of processing. These involve distinct mechanisms aimed at retrieving more or less of the energy that would otherwise be lost with the spliced glucans. The first involves disproportionating the small linear oligosaccharides into Glc and longer oligosaccharides, rendering the latter accessible to phosphorolytic degradation. This fits better with the postulated function of D-enzyme in the normal process of starch degradation. We did observe stimulation of degradation by phosphorylase in the presence of D-enzyme. The second would consist of direct transfer of glucans back onto the outer chains of amylopectin. Such a mechanism would be more efficient, since most of the energy contained in the α-1,4 linkages of the spliced glucans would be recovered on the maturing polysaccharide.
In this study, we provide biochemical support for the physiological relevance of this reaction. Most importantly, the second mechanism offers a straightforward explanation for all of the phenotypic traits observed in the D-enzyme-defective mutants. These consist not only of malto-oligosaccharide accumulation, but also of a structural modification of the remaining amylopectin and a downfall in starch synthesis. It is not easy to understand the basis for the modification in starch structure if the first mechanism was operating on its own. It must be stressed, however, that changes in polysaccharide structure could also arise from competition between malto-oligosaccharides and the normal substrates of soluble starch synthases and branching enzymes. In addition, it remains possible that small modifications in structure originate from the activity of D-enzyme toward the external chains of the polysaccharide without implying that oligosaccharides are necessarily reinserted into the structure. It is also possible that both postulated mechanisms occur simultaneously, and certainly does not rule out an additional function of D-enzyme in the normal process of starch degradation.
While in C. reinhardtii the presence of debranching enzymes remains mandatory to obtain starch synthesis, the requirement for D-enzyme becomes acute only in conditions of maximal starch synthesis. Losses of metabolic energy generated by the action of debranching enzymes become critical only when the cell devotes its activity to carbohydrate storage and the energy supply remains limited. This is certainly the case in nitrogen-starved C. reinhardtii cells, but could vary according to the plant species or tissue. However, we do not expect to see differences in amylopectin structure and maltoligosaccharide accumulation vanish under any circumstances. This will probably enable the breeding of plants with modified starch structure and high yield.
We believe our data prove that D-enzyme is required for normal amylopectin synthesis. We expect not only to open new avenues for breeders to improve starch quality, but also to give new insights into our understanding of starch biosynthesis in plants. These insights stem from the functional tie that we establish between polysaccharide and malto-oligosaccharide metabolisms in building amylopectin structure. In return, the discovery of such ties in plants may encourage further investigations performed on amylomaltase, the product of the E. coli MALQ gene, which is the bacterial analog of D-enzyme. We have observed high rates of incorporation of maltoheptaose into the external chains of glycogen through the MALQ gene product, an observation that is relevant to our understanding of bacterial glycogen metabolism.
ACKNOWLEDGMENTS
We thank Alan Myers and Jens Kossman for helpful discussions.
Abbreviations:
- APTS
8-amino-1,3,6-pyrenetrisulfonic acid
- D-enzyme
disproportionating enzyme
- DP
degree of polymerization
Footnotes
This work was supported by grants from the Ministère de l'Education Nationale, by the Centre National de la Recherche Scientifique (Unité Mixte de Recherche du Centre National de la Recherche Scientifique no. 8576, Director André Verbert), by the University of Lille, and by a grant from Biogemma (Cambridge, UK).
LITERATURE CITED
- Ball S, Guan H-P, James M, Myers A, Keeling P, Mouille G, Buléon A, Colonna P, Preiss J. From glycogen to amylopectin: a model explaining the biogenesis of the plant starch granule. Cell. 1996;86:349–352. doi: 10.1016/s0092-8674(00)80107-5. [DOI] [PubMed] [Google Scholar]
- Banks W, Greenwood CT, Khan KM. The interaction of linear amylose oligomers with iodine. Carbohydr Res. 1971;17:25–33. [Google Scholar]
- Boos W, Shuman H. Maltose/maltodextrin system of Escherichia coli: transport, metabolism and regulation. Microbiol Mol Biol Rev. 1998;62:204–229. doi: 10.1128/mmbr.62.1.204-229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleoni C, Dauvillée D, Mouille G, Buléon A, Gallant D, Bouchet B, Morell M, Samuel M, Delrue B, d'Hulst C, et al. (1999) Genetic and biochemical Evidence for the involvement of α-1,4 glucanotransferases in amylopectin synthesis. Plant Physiol 120: 993–1003 [DOI] [PMC free article] [PubMed]
- Dauvillée D, Colleoni C, Shaw E, Mouille G, D'Hulst C, Morell M, Samuel MS, Bouchet B, Gallant DJ, Sinskey A. Plant Physiol. 1999;119:321–330. doi: 10.1104/pp.119.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delrue B, Fontaine T, Routier F, Decq A, Wieruszeski JM, Van Den Koornhuyse N, Maddelein M-L, Faunet B, Ball SG. Waxy Chlamydomonas reinhardtii: monocellular algal mutants defective in amylose biosynthesis and granule-bound starch synthase activity accumulate a structurally modified amylopectin. J Bacteriol. 1992;174:3612–3620. doi: 10.1128/jb.174.11.3612-3620.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine T, D'Hulst C, Maddelein M-L, Routier F, Marianne-Pepin T, Decq A, Wieruszeski JM, Delrue B, Van Den Koornhuyse N, Bossu JP. Toward an understanding of the biogenesis of the starch granule: evidence that Chlamydomonas soluble starch starch synthase II controls the synthesis of intermediate size glucans of amylopectin. J Biol Chem. 1993;268:16223–16230. [PubMed] [Google Scholar]
- Gabriel O, Gersten DM. Staining for enzymatic activity after gel electrophoresis, I. Anal Biochem. 1992;203:1–21. doi: 10.1016/0003-2697(92)90036-7. [DOI] [PubMed] [Google Scholar]
- Harris EH. Culture and storage methods. In: Harris E, editor. The Chlamydomonas Sourcebook. A Comprehensive Guide to Biology and Laboratory Use. San Diego: Academic Press; 1989. pp. 25–63. [DOI] [PubMed] [Google Scholar]
- Jones G, Whelan WJ. The action pattern of D-enzyme, a transmaltodextrinylase from potato. Carbohydr Res. 1969;9:483–490. [Google Scholar]
- Lacks SA, Springhorn SS. Renaturation of enzymes after polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate. J Biol Chem. 1980;255:7467–7473. [PubMed] [Google Scholar]
- Libessart N, Maddelein M-L, Van Den Koornhuyse N, Decq A, Delrue B, Ball SG. Storage, photosynthesis and growth: the conditional nature of mutations affecting starch synthesis and structure in Chlamydomonas reinhardtii. Plant Cell. 1995;7:1117–1127. doi: 10.1105/tpc.7.8.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TP, Preiss J. Characterization of D-enzyme (4-α-glucanotransferase) in Arabidopsis leaf. Plant Physiol. 1988;86:260–265. doi: 10.1104/pp.86.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouille G, Maddelein M-L, Libessart N, Talaga P, Decq A, Delrue B, Ball S. Phytoglycogen processing: a mandatory step for starch biosynthesis in plants. Plant Cell. 1996;8:1353–1366. doi: 10.1105/tpc.8.8.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea MG, Morell MK. High resolution slab gel electrophoresis of 8-amino-1,3,6-pyrenetrisulfonic acid (APTS) tagged oligosaccharides using a DNA sequencer. Electrophoresis. 1996;17:681–688. doi: 10.1002/elps.1150170410. [DOI] [PubMed] [Google Scholar]
- O'Shea MG, Samuel MS, Konik CM, Morell MK. Fluorophore-assisted carbohydrate electrophoresis (FACE) of oligosaccharides: efficiency of labelling and high-resolution separation. Carbohydr Res. 1998;307:1–12. [Google Scholar]
- Peat S, Whelan WJ, Rees WR. The enzymic synthesis and degradation of starch: the disproportionating enzyme of potato. J Chem Soc. 1956;1956:44–53. [Google Scholar]
- Takaha H, Takaha T, Okada S, Takagi M, Imanaka T. Cyclic reaction catalyzed by branching enzyme. J Bacteriol. 1996a;178:1600–1606. doi: 10.1128/jb.178.6.1600-1606.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaha T, Yanase M, Okada S, Smith SM. Disproportionating enzyme (4-α-glucanotransferase; EC 2.4.1.25) of potato: purification, molecular cloning, and potential role in starch metabolism. J Biol Chem. 1993;268:1391–1396. [PubMed] [Google Scholar]
- Takaha T, Yanase M, Takaha H, Okada S, Smith SM. Potato D-enzyme catalyzes the cyclization of amylose to produce cycloamylose, a novel cyclic glucan. J Biol Chem. 1996b;271:2902–2908. doi: 10.1074/jbc.271.6.2902. [DOI] [PubMed] [Google Scholar]
- Takaha T, Yanase M, Takaha H, Okada S, Smith SM. Cyclic glucans produced by the intramolecular transglycosylation activity of potato D-enzyme on amylopectin. Biochem Biophys Res Commun. 1998;247:493–497. doi: 10.1006/bbrc.1998.8817. [DOI] [PubMed] [Google Scholar]
- van de Wal M, D'Hulst C, Vincken J-P, Buléon A, Visser R, Ball S. Amylose is synthesized in vitro by extension of and cleavage from amylopectin. J Biol Chem. 1998;273:22232–22240. doi: 10.1074/jbc.273.35.22232. [DOI] [PubMed] [Google Scholar]
- van der Meer I, Koops AJ, Hakkert JC, van Tunen AJ. Cloning of the fructan biosynthesis pathway of Jerusalem artichoke. Plant J. 1998;15:489–500. doi: 10.1046/j.1365-313x.1998.00230.x. [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Northrop F, Smith AM, Rees T. A starch-accumulating mutant of Arabidopsis thaliana deficient in a chloroplastic starch hydrolyzing enzyme. Plant J. 1998;15:357–365. doi: 10.1046/j.1365-313x.1998.00213.x. [DOI] [PubMed] [Google Scholar]