Abstract
The article shows how to implement the LD index assay, which is a sensitive microplate assay to determine the accumulation of triacylglycerols (TAGs) in lipid droplets (LDs). LD index is obtained without lipid extraction. It allows measuring the LDs content in high-throughput experiments under different conditions such as growth in rich or nitrogen depleted media. Albeit the method was described for the first time to study the lipid droplet metabolism in Saccharomyces cerevisiae, it was successfully applied to the basidiomycete Ustilago maydis. Interestingly, and because LDs are organelles phylogenetically conserved in eukaryotic cells, the method can be applied to a large variety of cells, from yeast to mammalian cells. The LD index is based on the liquid fluorescence recovery assay (LFR) of the BODIPY 493/503 under quenching conditions, by the addition of cells fixed with formaldehyde. Potassium iodine is used as a fluorescence quencher. The ratio between the fluorescence and the optical density slopes is named LD index. Slopes are calculated from the straight lines obtained when BODIPY fluorescence and optical density at 600 nm (OD600) are plotted against sample addition. Optimal data quality is reflected by correlation coefficients equal or above 0.9 (r ≥ 0.9). Multiple samples can be read simultaneously as it can be implemented in a microplate. Since BODIPY 493/503 is a lipophilic fluorescent dye that partitions into the lipid droplets, it can be used in many types of cells that accumulate LDs.
Keywords: Biochemistry, Issue 134, Lipid index, lipid droplets, basidiomycete, fluorescence recovery, BODIPY 493/503, Ustilago maydis
Introduction
Lipid droplets (LDs) are ubiquitous intracellular fat bodies composed of a core of neutral lipids, mainly TAGs and sterol esters (SE). The core is surrounded by a monolayer of phospholipids that interacts with proteins like perilipin and enzymes involved in the synthesis of neutral lipids, such as the diacylglycerol acyl-transferase, acetyl-CoA carboxylase and acyl-CoA synthase1. Due to its dynamic behavior, the phospholipid monolayer also contains triacylglycerol lipases that hydrolyze TAGs and SE2. Depending on the cell type and organism, stored neutral lipids can be used to generate energy, or synthesize phospholipids and signaling molecules. In yeast and other fungi, the content of LDs changes in response to variations in the nitrogen/carbon ratio in the culture media, indicating that decreased nitrogen availability might be the key to increase neutral lipid production3,4,5. The production of high amounts of TAGs in yeast has a potential use in biotechnology as source of biofuels and in the food industry. Some stored lipids contain high proportions of polyunsaturated fatty acids, like omega 3 and omega 6, with nutritional and dietary importance 6,7,8,9,10,11. LDs of mammalian cells, including human cells, also contain TAGs and cholesterol esters. The phospholipid monolayer interacts with the mammalian homologous of the proteins described in the yeast LDs, and with an additional protein, perilipin, which is absent in S. cerevisiae12. One proposed role for the phospholipid monolayer and its associated proteins is to stabilize the LDs structure and to allow the interaction of LDs with organelles as mitochondria, endoplasmic reticulum, peroxisomes, and vacuoles, mainly for lipid exchange 13,14. Interestingly, in humans, LDs appear to be involved in pathologies like type 2 diabetes, atherosclerosis, steatohepatitis, and coronary heart disease, in which there is an increase in their number 15,16,17. Some types of viruses use LDs as platforms to assemble the virions 2,18,19.
Due to the implications of the LDs in human pathologies and their potential biotechnological use, the exact experimental determination of LDs formation is an important task. This article describes a reliable assay based on the recovery of the fluorescence (LFR) of BODIPY 493/503 (4, 4-difluoro-1, 3, 5, 7, 8-pentamethyl-4-bora-3a, 4a-diaza-s-indacene), to get a relative value of the content of neutral lipids in cells. With this assay, it is possible to follow the dynamics of the accumulation of neutral lipids in fungi like U. maydis and S. cerevisiae, and also in mammalian cells, without the need of lipid extraction 20. The method was applied for the first time in S. cerevisiae to identify the protein phosphatases and kinases involved in the regulation of lipid metabolism. This was possible without lipid extraction or protein purification21,22. LFR also has been used to establish the dynamics of LDs formation in peritoneal macrophages23. The use of BODIPY 493/503 has some advantages over other neutral lipid dyes as Nile Red. BODIPY 493/503 is highly specific for neutral lipids and has a narrow emission spectrum, facilitating the simultaneous detection of signals from dyes like red fluorescent protein or Mito-tracker, when the samples are analyzed by confocal microscopy. Unfortunately, BODIPY 493/503 is sensitive to photobleaching, but this process can be avoided by using an antiquenching reagent during exposure to light24.
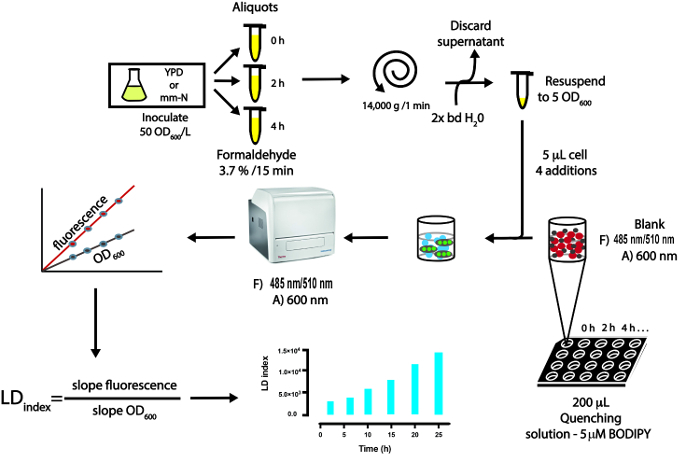
To carry out the LFR assay in yeast cells, they are cultured under the desired nutritional conditions, and aliquots are withdrawn at different times. Next, cells are fixed with formaldehyde, which preserves the integrity of LDs for months when cells are stored at 4 °C. Other fixation techniques should be avoided, especially those using methanol or cold acetone, since they lead to the degradation of LDs within the cells24. To measure the LD index, formaldehyde-fixed cells are suspended in water to obtain a defined concentration. Then, they are added to a solution containing the fluorophore BODIPY 493/503 quenched by KI, and when the fluorophore enters the cell and associates with the LDs, there is a recovery of its fluorescence. Concomitant to the fluorescence measurement (485 nm/510 nm), the concentration of cells is quantified by measuring the optical density at 600 nm. Each sample is read four times by adding subsequent 5 µL aliquots of a formaldehyde-fixed cell suspension to the same well. Blanks of fluorescence and absorbance are acquired before the addition of cells. The quality of the fluorescence and the absorbance data are evaluated by determining the linearity of the measurements: if r <0.9, data are discarded. The r value is important because basically the intensity of fluorescence should produce a linear response to increasing cell concentrations. If the cell concentration is too high, the linearity is lost. The LFR assay provides a fast, simple and economic method to select the desired LD phenotype in high-throughput experiments. After selecting the desired conditions, the LD content of individual cells can be studied by confocal microscopy, using the same formaldehyde-fixed cells stained with BODIPY, providing an image of the LDs in the cell. Their TAGs and SE content can now be further analyzed by thin layer chromatography.
Protocol
1. Preparation of Buffers and Solutions
To prepare 1 L of phosphate buffered saline (PBS, pH 7), dissolve 8 g of NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4, 0.24 g of KH2PO4 in 800 mL of distillated water. Adjust the pH to 7.0 with HCl, and then add water for a final volume of 1 L. PBS can be made as a 10x stock solution and stored at room temperature.
To prepare 10 mL of fixing solution, add 1 mL of 37% formaldehyde to 9 mL of PBS to reach a final concentration of 3.7%. Prepare this solution just before use. Caution: Use gloves to handle the formaldehyde solution.
To prepare the quenching-solution (500 mM), dissolve 8.3 g of KI in 100 mL of distilled water. Prepare this solution before use and store it at room temperature.
To prepare BODIPY 10 mM stock solution, dissolve 10 mg of BODIPY 493/503 in 3.8 mL of dimethyl sulfoxide (DMSO). Keep 100 µL aliquots in the dark at -70 °C.
To prepare BODIPY 5 µM-quenching solution, add 1 µL of 10 mM BODIPY 493/503 to 2 mL of 500 mM quenching solution. For the LFR assay, a final concentration of 5 µM of BODIPY is required.
To prepare the mineral solution, add 0.06 g of H3BO3, 0.14 g of MnCl2-4H2O, 0.4 g of ZnCl2, 0.04 g of Na2MoO4-2H2O, 0.1 g of FeCl3-6H2O and 0.4 g of CuSO4-5H2O to 1 L of distilled water.
To prepare the salt solution, add 16 g of KH2PO4, 4 g of Na2SO4, 8 g of KCl, 2 g of MgSO4, 1 g of CaCl2, and 8 mL of mineral solution to 1 L of distilled water.
To prepare the minimal medium without a nitrogen source for U. maydis FB2 (a2b2), add 10 g of glucose and 62.5 mL of salt solution to 1 L of distilled water, according to Holliday, et al.31. Adjust the pH to 7.0 and then dispense 100 mL of the media in 250 mL flasks and sterilize them. Autoclave for 20 min at 15 psi (1.05 kg/cm2) on liquid cycle or filter sterilize.
To prepare the yeast peptone dextrose medium (YPD), add 5 g of yeast extract, 2.5 g of peptone, and 5 g of glucose to 1 L of distilled water. Dispense 100 mL of the solution in 250 mL flasks and sterilize them. Autoclave for 20 min at 15 psi (1.05 kg/cm2) on liquid cycle or filter sterilize.
2. Culture Condition and Cell Fixation
NOTE: Preserve the structure of the LDs by fixing the cells as soon as possible. The fixation can be done in microcentrifuge tubes. Fixed cells can be kept at 4 °C for up to one month if dehydration is avoided leaving a thin layer of water.
Grow U. maydis cells under culture conditions relevant for the study. Start the culture with an initial OD600 of 0.05 (1.15 × 106 cells/ mL). Incubate the cells at 28 °C, 180 rpm for 24 h. An OD600 of 1 corresponds to 2.3 × 107 cells/ mL.
At the selected times, withdraw an aliquot of the cells. For U. maydis, withdraw 5 mL every 2 h during the first 8 h. After 8 h of growth, aliquots of 2 mL are enough.
Measure the optical density at 600 nm of each aliquot.
Centrifuge the cell suspension at 14,000 x g for 1 min at 4 °C using a tabletop centrifuge. Discard the supernatant.
Suspend the pellet of the cells in 1 mL of PBS-formaldehyde 3.7% buffer. Incubate the cells for 15 min at room temperature.
After the incubation, centrifuge the cell suspension at 14,000 x g for 1 min at 4 °C. Discard the supernatant.
Wash the cells pellet twice with a similar volume of distilled water (1 mL). Centrifuge the cell suspension at 14,000 x g for 1 min at 4 °C. Discard the supernatant after each washing step.
Adjust the cell pellet to 5 OD600 (1.15 x 108 cell/ mL) with distilled water (Equation 1).
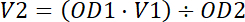 Equation 1.
Equation 1.Keep the sample at 4 °C until its use.
3. Liquid Fluorescence Recovery Assay (LFR)
Turn on the spectrophotometer and open the Skanlt software. Click on NEW SESSION, and choose START and then Varioskan Lux. Select PROTOCOL from the session tree, enter the settings for the fluorescence wavelengths (excitation 485 nm/emission 510 nm; optical density 600 nm) and choose automatic photomultiplier gain. For excitation bandwidth, select 12 nm. In the optics, select top (excitation of the sample is from the top of the well).
Enter a gentle and continuous agitation (300 rpm). Select RUN PLATE OUT, and PAUSE UNTIL THE USER ACTION (time used for the addition of the sample to the wells). Repeat the protocol five times. Click SAVE and write a name for the session on the SESSION NAME FIELD. The protocol is now ready for the samples analyses.
Add 200 µL of the BODIPY 5 µM- quenched solution to each well of a 96 well black-clear bottom plate.
Place the plate inside the spectrophotometer chamber and incubate 5 min at 30 °C. From this point, protect the plate from light as much as possible.
Click the START button and read the fluorescence (excitation 485/emission 510) and the OD600 corresponding to the blanks.
Add 5 µL of the formaldehyde-fixed cell suspension to the wells during the pause time. Mix the samples carefully with the pipette. Put the plate inside the spectrophotometer and click CONTINUE. Then, the samples will be processed according to the protocol designed above.
Repeat three successive additions of 5 µL of cell suspension. Make sure that the cells do not precipitate. (Figure 1).
4. Calculations of The LD Index
For each sample in the microplate, plot a graph of fluorescence and absorbance against the volume of each successive addition (5 points including the blank). In this study, use Microsoft Excel software for the calculations. To plot the data, enter the volume, fluorescence, and absorbance data in the first, second and third column of the datasheet, respectively. Then, select the whole data with the cursor, click INSERT and select (XY) SCATTER.
Select the points for each line in the graph and insert the respective TREND LINE with the SHOW EQUATION box selected. Write down the values of the slopes of the two straight lines, according to equation 2:
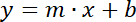 Equation 2 Where y= fluorescence or optical density, x = volume of sample (0, 5, 10, 15, 20 µL), m = slope of the fluorescence or optical density line, and b = y-intercept.
Equation 2 Where y= fluorescence or optical density, x = volume of sample (0, 5, 10, 15, 20 µL), m = slope of the fluorescence or optical density line, and b = y-intercept.Divide the slope of the fluorescence line by the slope of the optical density line to get the LD index, equation 3.
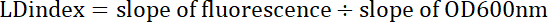 Equation 3 The LD index corresponds to the quotient of fluorescence and optical density slopes.
Equation 3 The LD index corresponds to the quotient of fluorescence and optical density slopes.
5. Data Quality Analysis
Calculate the correlation coefficient of each straight line: click on the FUNCTION WIZARD (fx) and choose CORREL. If r <0.9, discard the data and repeat the readings. If r ≥ 0.9, readings are reliable.
Representative Results
LFR, with BODIPY 493/503 as the fluorescent probe is a reliable and easy method to study the dynamic accumulation of LDs in U. maydis, regardless the growth conditions. When cells were cultured in YPD-medium, there was an increase in the LD index during the exponential phase, followed by a decline in the stationary phase (Figure 2A). In contrast, in cells grown under nitrogen starvation, there was a steady increase in LD index in both the exponential and stationary phases (Figure 2B). Because both cultures were started with the same optical density (similar number of cells), the results show the high capacity of the LFR assay to detect small changes in the lipid content (Figure 2). The sensitivity of the method was corroborated by confocal microscopy: YPD-cells contain many small LDs throughout the whole cell, while under nitrogen starvation there was an important production of large LDs, typically 4-5 LDs per cells, (Figure 2, right panels A and B, respectively). Since the LD index is not an absolute value of the neutral lipid content, a standard curve relating the LD index with the amount of triacylglycerols should be constructed. This approach was used to study the dynamics of TAGs synthesis in S. cerevisiae20. Based on each standard curve, the LFR assay can be used to determinate the rate of lipid accumulation in cells.
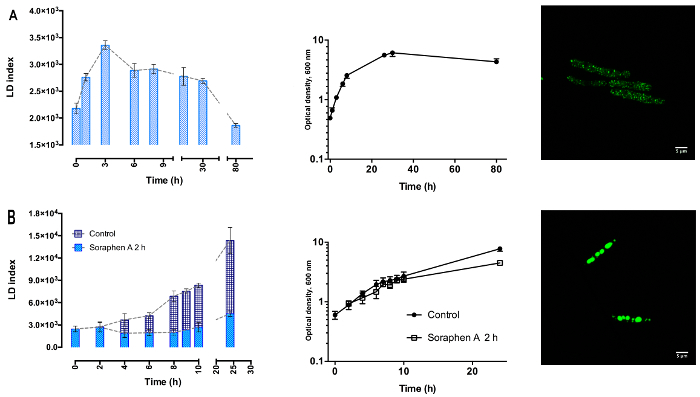
The LD index can be followed in the presence of soraphen A, a specific inhibitor of the acetyl-CoA carboxylase, an enzyme that is essential for the synthesis of fatty acids contained in the TAGs stored in LDs25. The inhibitor was added to the culture medium after 2 h of growth in the absence of a nitrogen source, a condition in which cells showed the higher rate of lipid accumulation (Figure 2B). In accordance with previous reports in S. cerevisiae, the addition of soraphen-A resulted in the full inhibition of LD accumulation (Figure 2B, light blue) 20,21.
To further investigate the mobilization of LDs in U. maydis, cells grown under nitrogen starvation for 72 h were transferred to YPD-medium (Figure 3). LD index decreased following an exponential function. After 24 h, the LD index reached similar values to that observed in cells cultured in YPD-medium without the stress of nitrogen absence (Figure 2A). Since the cells were able to mobilize the LDs accumulated during nitrogen starvation, the result suggests a reduction in the TAGs content. A similar response was reported for S. cerevisiae during the study of phosphatases involved in the regulation of lipid metabolism21.
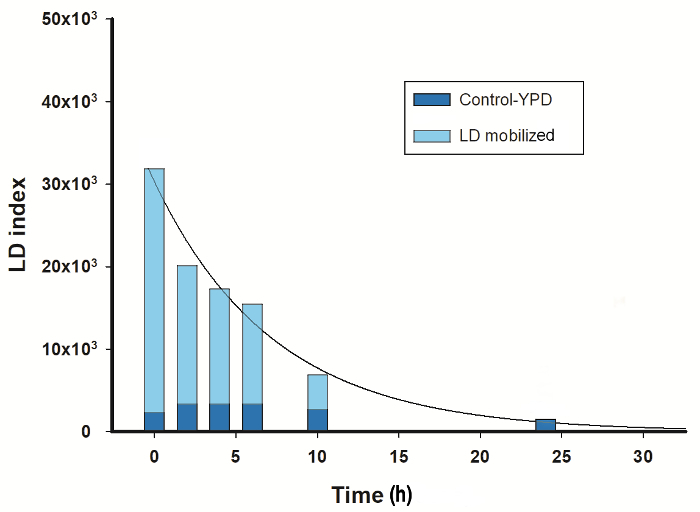
Figure 1: Schematic representation of the steps involved in the LFR. The steps of culture conditions are representative for U. maydis yeast but can be adapted to other microorganism or type of cells. URF, Units of Recovery Fluorescence; bd, bidistilled water. Please click here to view a larger version of this figure.
Figure 2: Dynamics of lipid droplets accumulation in U. maydis. Yeast cells were grown for 24 h in YPD-medium, harvested and suspended in (A) fresh YPD-medium (Top left panel) or in (B) minimal medium without a nitrogen source (bottom left panel-dark blue). Under nitrogen starvation, the increase in LD index was inhibited by adding 192 nM soraphen A in the culture media after 2 h of growth (B, light blue). n=3, data are the mean ± SEM. Right panel: confocal microscopy of LDs in cell harvested at 24 h. (A) Top panel: YPD- cells. (B) Bottom panel: cells without a nitrogen source. Please click here to view a larger version of this figure.
Figure 3: LDs mobilization. Yeast cells were grown for 24 h in YPD-medium, harvested and grown in fresh YPD for 24 h (control) or in minimal medium without a nitrogen source for 72 h, then transferred to fresh-YPD medium and incubated for additional 24 h. Aliquots were withdrawn at the indicated times for both cultures to measure the mobilization of LDs by LFR. Data was fitted by an exponential decay equation (y = A·e-kt). Please click here to view a larger version of this figure.
Discussion
LFR is a novel method that was applied for the first time to study the lipid metabolism in S. cerevisiae and later it was successfully implemented in U. maydis20,21. Although the LD index does not give an absolute value of the TAGs accumulated in cells, it provides a fast idea of the lipid content of cells, and the interpretation of the data is straightforward when LD index from two or more experimental conditions are compared. BODIPY 493/503 is highly specific for neutral lipids, the main component of the LDs, and it has a narrow emission spectrum facilitating the simultaneous detection of other signals coming from dyes like Mito-tracker or CMAC blue when cells are studied by confocal microscopy26. Like many fluorescent molecules, BODIPY 493/503 is sensitive to photobleaching during fluorescence microscopy experiments, but this process can be decreased by adding anti photobleaching reagents during the exposure to light27. In summary, BODIPY 493/503 can be used in a wide range of cells to study the accumulation and mobilization of neutral lipids21,22,23,28,29.
For the method to work properly, some point must be taken into consideration. Since the OD600 is used for the calculation of the LD index, the morphology of the cells should not have radical changes during the experiment. The preservation of the intracellular components during the fixation of cells also is a critical step, and this preservation should include the LDs, which are the main objective of this protocol. Fixing the cells with an appropriate compound is very important for the application of this method to many types of cells without any problem. Here, we used formaldehyde to fix the structures of the cells; the aldehyde reacts with the amino groups of proteins. It has been reported that other aldehydes also preserve the structures of a wide range of cells and it seems that there are not significant changes in the protein structure24,26. A disadvantage of formaldehyde is that it induces small membrane damage and vacuole formation near the mitochondrial and nuclear membranes, but, these negative effects are common to all aldehyde fixation methods26. Organic solvents like methanol or acetone should be avoided because they degrade the LDs in the cells. Fixation with organic solvents is based on the dehydration and precipitation of proteins, which can be considered a negative effect. In addition, the use of organic solvent is associated with non-specific staining.
As any other technique, the LD index assay might have limitations when implemented to other systems. Problems in the diffusion of BODIPY across the fungal cell wall, the dependence of the fluorescence on fatty acid composition, the type of lipids and the protein content within intracellular lipid bodies30, preclude the comparison of the amount of TAGs between different species or even with the same cells grown under different nutritional conditions. Also, large morphological changes such as the transition to mycelium or flocculation of cells are some of the problems that can be found.
LD index assay is a high-throughput method that requires short times and small amounts of biomass and reactants. The assay is reliable, and the data obtained are accurate and reproducible. In addition, it provides guidelines for the research on the dynamics of lipid metabolism in medicine, and it might become a tool in biotechnology for the high-throughput screening of oleaginous organism for the production of biofuels or feed oils.
Disclosures
The authors have nothing to disclosure
Acknowledgments
This work was supported by grants Instituto Politécnico Nacional-Secretaria de Investigación y Posgrado (IPN-SIP-20170864), Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica (PAPIIT IN222117)- Universidad Nacional Autónoma de México (UNAM), and Consejo Nacional de Ciencia y Tecnología (CONACyT 254904-JPP and 256520-GGS). Fundação de Amparo a Pesquisa do Rio de Janeiro (FAPERJ-Cientistas do Nosso Estado: E 26/103.353/2011). We thanks to QFB. Oscar Iván Luqueño Bocardo for the schematic overview. We thank Dr. Miguel Tapia Rodríguez for the valuable help in the confocal microscopy of LDs. We wish to thank Dr. Bruno Bozaquel Morais for his pioneering work in developing the technique in S. cerevisiae that we adapted for U. maydis.
References
- Guo Y, Cordes KR, Farese RV, Walther TC. Lipid droplets at a glance. Journal of Cell Science. 2009;122(6):749–752. doi: 10.1242/jcs.037630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte MA. Expanding roles for lipid droplets. Current Biology. 2015;25(11):R470–R481. doi: 10.1016/j.cub.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sestric R, Munch G, Cicek N, Sparling R, Levin DB. Growth and neutral lipid synthesis by Yarrowia lipolytica on various carbon substrates under nutrient-sufficient and nutrient-limited conditions. Bioresource Technology. 2014;164:41–46. doi: 10.1016/j.biortech.2014.04.016. [DOI] [PubMed] [Google Scholar]
- Raimondi S, et al. Getting lipids from glycerol: new perspectives on biotechnological exploitation of Candida freyschussii. Microbial Cell Factories. 2014;13:83. doi: 10.1186/1475-2859-13-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cescut J, Fillaudeau L, Molina-Jouve C, Uribelarrea JL. Carbon accumulation in Rhodotorula glutinis induced by nitrogen limitation. Biotechnology for Biofuels. 2014;7:164. doi: 10.1186/s13068-014-0164-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galafassi S, et al. Lipid production for second generation biodiesel by the oleaginous yeast Rhodotorula graminis. Bioresource Technology. 2012;111:398–403. doi: 10.1016/j.biortech.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Kot AM, Blazejak S, Kurcz A, Gientka I, Kieliszek M. Rhodotorula glutinis-potential source of lipids, carotenoids, and enzymes for use in industries. Applied Microbiology and Biotechnology. 2016;100(14):6103–6117. doi: 10.1007/s00253-016-7611-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakicka M, Lazar Z, Dulermo T, Fickers P, Nicaud JM. Lipid production by the oleaginous yeast Yarrowia lipolytica using industrial by-products under different culture conditions. Biotechnology for Biofuels. 2015;8 doi: 10.1186/s13068-015-0286-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu ZW, et al. Dynamics of the Lipid Droplet Proteome of the Oleaginous Yeast Rhodosporidium toruloides. Eukaryotic Cell. 2015;14(3):252–264. doi: 10.1128/EC.00141-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolouchova I, Mat'atkova O, Sigler K, Masak J, Rezanka T. Production of Palmitoleic and Linoleic Acid in Oleaginous and Nonoleaginous Yeast Biomass. International Journal of Analytical Chemistry. 2016. [DOI] [PMC free article] [PubMed]
- Radulovic M, et al. The emergence of lipid droplets in yeast: current status and experimental approaches. Current Genetics. 2013;59(4):231–242. doi: 10.1007/s00294-013-0407-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, et al. Perilipin-Mediated Lipid Droplet Formation in Adipocytes Promotes Sterol Regulatory Element-Binding Protein-1 Processing and Triacylglyceride Accumulation. Plos One. 2013;8(5):e64605. doi: 10.1371/journal.pone.0064605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam AR, Farese Jr RV, Walther TC. The biophysics and cell biology of lipid droplets. Nature Reviews Molecular Cell Biology. 2013;14(12):775–786. doi: 10.1038/nrm3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penno A, Hackenbroich G, Thiele C. Phospholipids and lipid droplets. Biochimica et Biophysica Acta. 2013;1831(3):589–594. doi: 10.1016/j.bbalip.2012.12.001. [DOI] [PubMed] [Google Scholar]
- Ageitos JM, Vallejo JA, Veiga-Crespo P, Villa TG. Oily yeasts as oleaginous cell factories. Applied Microbiology and Biotechnology. 2011;90(4):1219–1227. doi: 10.1007/s00253-011-3200-z. [DOI] [PubMed] [Google Scholar]
- Athenstaedt K, Daum G. The life cycle of neutral lipids: synthesis, storage and degradation. Cellular and Molecular Life Sciences. 2006;63(12):1355–1369. doi: 10.1007/s00018-006-6016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall LL, Stimpson SE, Hyland R, Coorssen JR, Myers SJ. Increased lipid droplet accumulation associated with a peripheral sensory neuropathy. Journal of Biological Chemistry. 2014;7(2):67–76. doi: 10.1007/s12154-014-0108-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipe A, McLauchlan J. Hepatitis C virus and lipid droplets: finding a niche. Trends in Molecular Medicine. 2015;21(1):34–42. doi: 10.1016/j.molmed.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Greenberg AS, et al. The role of lipid droplets in metabolic disease in rodents and humans. Journal of Clinical Investigation. 2011;121(6):2102–2110. doi: 10.1172/JCI46069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar LR, et al. Lipid droplets accumulation and other biochemical changes induced in the fungal pathogen Ustilago maydis under nitrogen-starvation. Archives of Microbiology. 2017. [DOI] [PubMed]
- Bozaquel-Morais BL, Madeira JB, Maya-Monteiro CM, Masuda CA, Montero-Lomeli M. A new fluorescence-based method identifies protein phosphatases regulating lipid droplet metabolism. PLoS One. 2010;5(10):e13692. doi: 10.1371/journal.pone.0013692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira JB, et al. TORC1 Inhibition Induces Lipid Droplet Replenishment in Yeast. Molecular and Cellular Biology. 2015;35(4):737–746. doi: 10.1128/MCB.01314-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maya-Monteiro CM, et al. Leptin induces macrophage lipid body formation by a phosphatidylinositol 3-kinase- and mammalian target of rapamycin-dependent mechanism. Journal of Biological Chemistry. 2008;283(4):2203–2210. doi: 10.1074/jbc.M706706200. [DOI] [PubMed] [Google Scholar]
- Listenberger LL, Brown DA. Current Protocols in Cell Biology. John Wiley & Sons, Inc; 2001. [Google Scholar]
- Jump DB, Torres-Gonzalez M, Olson LK. Soraphen A, an inhibitor of acetyl CoA carboxylase activity, interferes with fatty acid elongation. Biochemical Pharmacology. 2011;81(5):649–660. doi: 10.1016/j.bcp.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobro AJ, Smith NI. An evaluation of fixation methods: Spatial and compositional cellular changes observed by Raman imaging. Vibrational Spectroscopy. 2017;91:31–45. [Google Scholar]
- Chen YT, Wan L, Zhang DP, Bian YZ, Jiang JZ. Modulation of the spectroscopic property of Bodipy derivates through tuning the molecular configuration. Photochemical & Photobiological Sciences. 2011;10(6):1030–1038. doi: 10.1039/c1pp00001b. [DOI] [PubMed] [Google Scholar]
- Pagac M, et al. SEIPIN Regulates Lipid Droplet Expansion and Adipocyte Development by Modulating the Activity of Glycerol-3-phosphate Acyltransferase. Cell Reports. 2016;17(6):1546–1559. doi: 10.1016/j.celrep.2016.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu B, Simon MC. BODIPY 493/503 Staining of Neutral Lipid Droplets for Microscopy and Quantification by Flow Cytometry. Bio-protocol. 2016;6(17) doi: 10.21769/BioProtoc.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumin J, et al. The use of fluorescent Nile red and BODIPY for lipid measurement in microalgae. Biotechnology for Biofuels. 2015;8:42. doi: 10.1186/s13068-015-0220-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. Ustilago maydis. In: King R, editor. Handbook of genetics. New York: Plenum; 1974. pp. 575–595. [Google Scholar]


