Key Points
Question
Can tumor messenger RNA (mRNA) expression of HER3 predict response to anti–epidermal growth factor receptor (anti-EGFR) agents in RAS wild type advanced colorectal cancer?
Findings
In this analysis of patients in a large phase 3 trial of irinotecan with and without panitumumab, patients with RAS wild type whose tumors had high HER3 mRNA expression benefitted markedly from panitumumab, but patients with RAS wild type whose tumors had low HER3 expression gained no benefit. Interaction between the biomarker and the treatment was significant.
Meaning
Tumor HER3 mRNA expression may be a useful predictive biomarker for anti-EGFR therapy in RAS wild-type colorectal cancer; further study is warranted.
This biomarker study examines the efficacy of irinotecan plus panitumumab in patients with advanced colorectal cancer whose tumors had high HER3 messenger RNA expression.
Abstract
Importance
Epidermal growth factor receptor (EGFR) (HER1) signaling depends on ligand binding and dimerization with itself or other HER receptors. We previously showed in a randomized trial that high EGFR ligand expression is predictive of panitumumab benefit in advanced colorectal cancer. Tumor expression of HER3 may further refine the RAS wild-type (wt) population benefitting from anti-EGFR agents.
Objective
To examine HER3 messenger RNA expression as a prognostic and predictive biomarker for anti-EGFR therapy in a randomized clinical trial of panitumumab.
Design, Setting, and Participants
The study was a prospectively planned retrospective biomarker study of pretreatment samples from the PICCOLO trial that tested the addition of panitumumab to irinotecan therapy in patients with KRAS wt advanced colorectal cancer who experienced failure with prior fluoropyrimidine treatment. HER3 was assessed as a prognostic marker, then as a predictive biomarker in patients with RAS wt, first as a continuous variable and then as a binary (high vs low) variable. Relationship with MEK-AKT pathway mutations and EGFR ligands epiregulin and amphiregulin (EREG/AREG) were also assessed.
Main Outcomes and Measures
Primary end point was progression-free survival (PFS); secondary end points were response rate and overall survival (OS).
Results
In 308 patients (mean age at randomization, 61.6 years; 193 men) higher HER3 was weakly prognostic for OS (hazard ratio [HR] per 2-fold change, 0.91; 95% CI, 0.83-0.99; P = .04) but not PFS (HR, 0.93; 95% CI, 0.83-1.05; P = .25). Higher HER3 was predictive, being associated with prolonged PFS on irinotecan plus panitumumab (IrPan) (HR, 0.71; 95% CI, 0.61-0.82; P < .001), but not irinotecan (HR, 0.96; 95% CI, 0.82-1.13; P = .65) in patients with RAS wt, with significant interaction between biomarker and treatment (P = .001). Similar interaction was seen for OS (P = .004). In an exploratory binary model, dividing the population at the 66th percentile, HER3 was predictive of panitumumab benefit: in patients with high HER3 expression, median PFS was 8.2 months (IrPan) vs 4.4 months (irinotecan) (HR, 0.33; 95% CI, 0.19-0.58; P < .001). Patients with low HER3 expression gained no benefit in PFS: 3.3 months (IrPan) vs 4.3 months (irinotecan) (HR, 0.96; 95% CI, 0.67-1.38; P = .84), with significant interaction (P = .002). The binary model was also predictive for OS, with significant interaction (P = .01). Combining HER3 and ligand data, patients with HER3-high, AREG/EREG-high tumors gained markedly from panitumumab (PFS HR, 0.24; 95% CI, 0.11-0.51; P < .005 and OS HR, 0.36; 95% CI, 0.18-0.73; P = .004). Conversely, patients with HER3-low, AREG/EREG-low tumors did not benefit (PFS HR, 1.14; 95% CI, 0.73-1.79; P = .57 and OS HR, 1.44; 95% CI, 0.92-2.26; P = .11).
Conclusions and Relevance
High HER3 expression identified patients with RAS wt who gained markedly from panitumumab, and those who did not, with statistically significant biomarker-treatment interactions for PFS and OS. This finding provides insight into the mechanism of anti-EGFR agents and is of potential clinical utility.
Introduction
While RAS mutations identify patients with advanced colorectal cancer (aCRC) who will not benefit from anti–epidermal growth factor receptor (anti-EGFR) agents, RAS wild type (wt) status does not reliably predict who will. We recently published clinical validation of combined tumor messenger RNA (mRNA) overexpression of the EGFR ligands amphiregulin (AREG) and epiregulin (EREG) as a predictive biomarker for panitumumab in RAS-wt aCRC in a large randomized clinical trial (PICCOLO [Panitumumab, Irinotecan, and Ciclosporin in COLOrectal cancer], a study of irinotecan-panitumumab [IrPan] vs irinotecan alone for second-line treatment of KRAS-wt aCRC).
The activity of agents targeting EGFR (HER1) may also depend on interactions with other HER receptors. HER family interdependence is well documented; HER3, unlike other HER receptors, cannot generate signaling through homodimerization but is an obligate heterodimer, predominantly with EGFR or HER2. The equilibrium between HER3 heterodimers may be a key factor in downstream EGFR family signaling.
We investigate the role of HER3 tumor mRNA expression in aCRC in pretreatment samples from the PICCOLO trial, first as a prognostic biomarker, then as a predictive biomarker for panitumumab efficacy. We further investigate its relationship with AREG and EREG, with a view to future development as a clinically applicable selection tool.
Methods
Following United Kingdom national ethics committee approval, written consent was obtained for the retrieval of stored tumor blocks.
The results of the PICCOLO trial have been reported previously. This study includes all patients with adequate stored tumor material. Ribonucleic acid was extracted from formalin-fixed, paraffin-embedded tissue sections and HER3 expression measured by reverse transcription polymerase chain reaction (primer sequences are shown in eTable 1 in the Supplement).
The primary end point was progression-free survival (PFS); secondary end points were overall survival (OS) and 12-week response rate. The primary analysis assessed HER3 expression as a continuous variable (log-transformed to base 2). In a planned exploratory analysis, HER3 expression was assessed as a dichotomous variable, using a range of cut points to achieve optimum discrimination for interaction between biomarker and treatment. Further details are provided in the Supplement.
Results
Three hundred and thirty-one patients had adequate tumor material for RNA extraction; HER3 expression measurement was successful in 308. Baseline characteristics by treatment arm were well balanced (eTable 2 in the Supplement), with no significant differences from the overall trial population.
RAS/RAF genotype data (KRAS c.12-13,61,146; NRAS c.12-13,61; and BRAF V600E) were available for all cases. Of 308 patients, 209 (67.8%) were wt for all KRAS/NRAS codons and form the primary analysis population for this study (eFigure 1 in the Supplement). HER2 expression data were available for 210 of 308 (68%) patients; however, overexpression was found in only 3 of 210 (1.4%), precluding meaningful analysis.
HER3 was not significantly associated with RAS mutation status, but was lower in BRAF-mutant and PIK3CA-mutated groups compared with wt. HER3 levels were weakly positively correlated with both AREG and EREG levels (Spearman ρ correlation, 0.26 for each; P < .001). In patients with RAS wt, HER3 levels were lower in right-sided compared with left-sided tumors, but there was no association in the whole population.
Higher log2 HER3 was prognostic for improved OS (hazard ratio [HR] per 2-fold change, 0.91; 95% CI, 0.83-0.99; P = .04) but not for PFS (HR, 0.93; 95% CI, 0.83-1.05; P = .25) in the whole population (eTable 3 in the Supplement).
In patients with RAS wt, increasing HER3 expression was significantly predictive for panitumumab benefit to PFS (IrPan: HR, 0.71; 95% CI, 0.61-0.82 per 2-fold increase; P < .001 vs irinotecan: HR, 0.96; 95% CI, 0.82-1.13; P = .65). Interactions were significant (unadjusted P = .001) (Table 1). Estimates were similar after adjusting for performance status and previous response, with significant interaction (P = .003). HER3 was also significantly predictive for panitumumab benefit on OS in patients with RAS wt (IrPan: HR, 0.73; 95% CI, 0.64-0.83; P < .001 vs irinotecan: HR, 0.93; 95% CI, 0.83-1.05; P = .25). Interactions were significant (unadjusted P = .004; adjusted P = .01) (Table 1). This effect was independent of BRAF mutation status and primary tumor location (eTable 4 in the Supplement).
Table 1. Effect of Log2 Transformed HER3 Expression on PFS and OS .
| Survival | Mutation Subgroup | All Patients | Irinotecan | IrPan | P Value for Interactionb | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Events/Patients, No. | Unadjusted HR for log2 HER3a (95% CI) | P Value | Events/Patients, No. | Unadjusted HR for log2 HER3 (95% CI) | P Value | Events/Patients, No. | Unadjusted HR for log2 HER3 (95% CI) | P Value | |||
| PFS | RAS wt | 190/208 | 0.82 (0.74-0.90) |
<.001 | 106/114 | 0.96 (0.82-1.13) |
.65 | 84/94 | 0.71 (0.61-0.82) |
<.001 | .001 |
| RAS mut | 95/99 | 0.95 (0.87-1.05) |
.34 | 48/49 | 0.88 (0.72-1.07) |
.19 | 47/50 | 0.99 (0.88-1.12) |
.91 | .37 | |
| BRAF mut | 44/47 | 0.79 (0.66-0.94) |
.01 | 18/19 | 1.03 (0.70-1.53) |
.88 | 26/28 | 0.77 (0.63-0.93) |
.01 | .11 | |
| PIK mut | 32/35 | 0.87 (0.69-1.10) |
.26 | 22/23 | 0.95 (0.72-1.27) |
.74 | 10/12 | 0.53 (0.28-1.02) |
.06 | .01 | |
| All patients | 285/307 | 0.88 (0.83-0.94) |
<.001 | 154/163 | 0.93 (0.83-1.05) |
.25 | 131/144 | 0.87 (0.81-0.94) |
<.001 | .21 | |
| OS | RAS wt | 192/209 | 0.86 (0.80-0.94) |
<.001 | 106/115 | 0.93 (0.83-1.05) |
.25 | 86/94 | 0.73 (0.64-0.83) |
<.001 | .004 |
| RAS mut | 97/99 | 0.94 (0.86-1.03) |
.17 | 47/49 | 0.83 (0.70-0.98) |
.03 | 50/50 | 1.01 (0.89-1.14) |
.93 | .07 | |
| BRAF mut | 45/47 | 0.81 (0.70-0.94) |
.01 | 18/19 | 0.89 (0.61-1.31) |
.56 | 27/28 | 0.80 (0.68-0.95) |
.01 | .67 | |
| PIK mut | 33/35 | 0.96 (0.79-1.16) |
.67 | 21/23 | 0.93 (0.74-1.17) |
.54 | 12/12 | 0.65 (0.37-1.15) |
.14 | .31 | |
| All patients | 289/308 | 0.90 (0.85-0.95) |
<.001 | 153/164 | 0.91 (0.83-0.99) |
.04 | 136/144 | 0.89 (0.83-0.96) |
.001 | .74 | |
Abbreviations: HR, hazard ratios; IrPan, irinotecan with panitumumab; mut, mutation; OS, overall survival, PFS, progression-free survival; wt, wild type.
HR per 2-fold increase in HER3.
P value is from a likelihood ratio test comparing a model including the main effects for log2 HER3 and treatment (IrPan vs irinotecan) plus the log2 HER3 treatment interaction term with a model including only the main effects. HER3 is the independent variable in this equation.
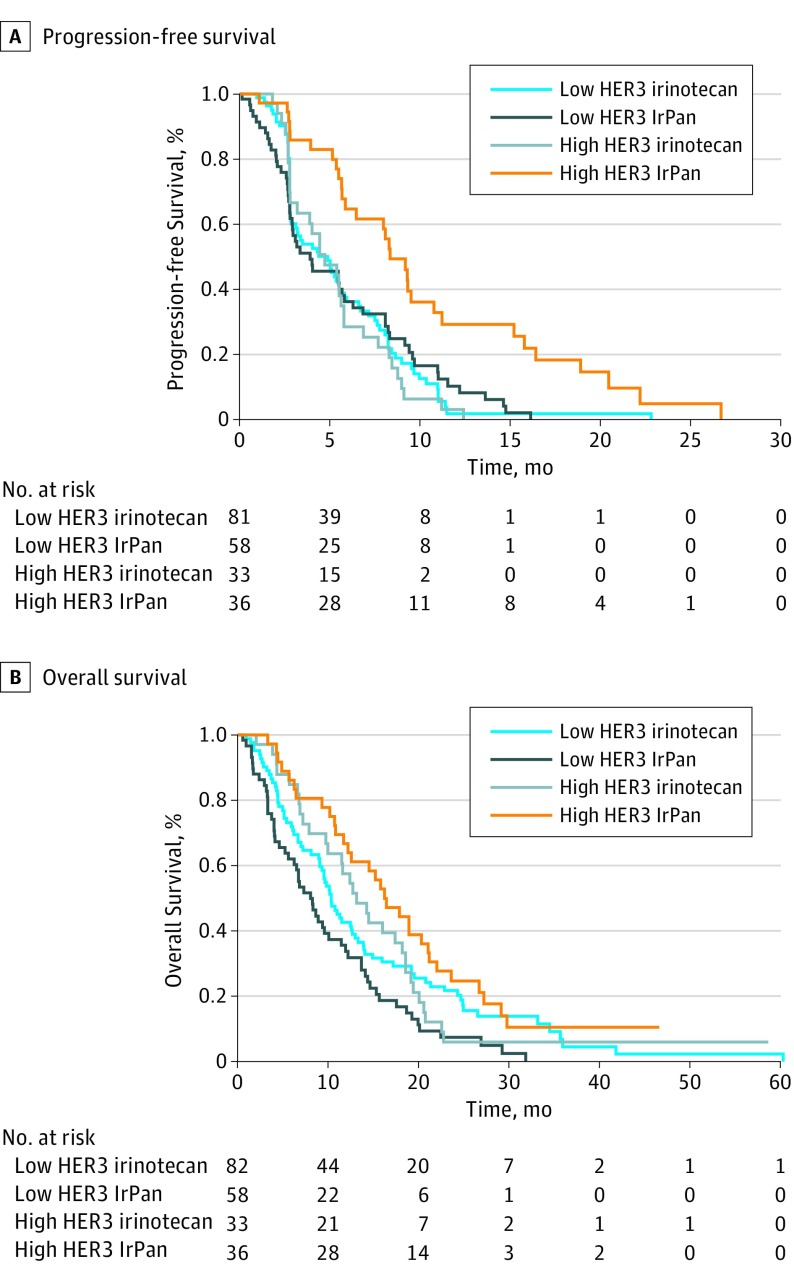
Preplanned predictive analyses of HER3 expression dichotomized at the 50th, 66th, 80th, and 90th percentiles in patients with RAS wt (eTable 5 in the Supplement) showed that at the 66th percentile, there was no PFS benefit from panitumumab in low expressers (median PFS, 3.3 months [IrPan] vs 4.3 months [irinotecan]; HR, 0.96; 95% CI, 0.67-1.38; P = .84), clear benefit in high expressers (HR, 0.33; 95% CI, 0.19-0.58; P < .001) (Figure), and strong evidence for interaction. Secondary analyses adjusting for performance status and previous response showed similar estimates. This cut point was therefore selected for all subsequent analyses to define “HER3-high” and “HER3-low” populations. Dichotomized HER3 was also predictive for OS in patients with RAS wt, though here the interaction was driven in part by a negative impact of panitumumab in patients with low HER3 expression. In patients with RAS wt and high HER3 expression, median OS was 14.6 months (IrPan) vs 13.2 months (irinotecan) (HR, 0.66; 95% CI, 0.40-1.10; P = .11), while in patients with RAS wt and low HER3 expression, median OS was 8.3 months (IrPan) vs 10.3 months (irinotecan) (HR, 1.56; 95% CI, 1.09-2.23; P = .02), with significant interaction (P = .01) (Table 2 and Figure). Secondary analyses adjusting for performance status and previous response showed similar estimates for interaction (P = .02). The dichotomized HER3 model was also independent of BRAF mutation status and primary tumor location (eTable 4 in the Supplement).
Figure. Survival of Patients With High and Low HER3 Expression Treated With IrPan vs Irinotecan.
A, Progression-free survival in RAS wild type patients for high HER3 expressers and low HER3 expressers treated with IrPan vs irinotecan. B, Overall survival in high HER3 expressers and low HER3 treated with IrPan vs irinotecan. IrPan indicates irinotecan with panitumumab.
Table 2. Effect of Treatment (IrPan vs Irinotecan) on PFS and OS in Low and High HER3 Expression.
| Survival | Mutation Subgroupa | All Patients | Low HER3 Expression (<66th Percentile) | High HER3 Expression (>66th Percentile) | P Value for Interaction | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Events/Patients, No. | Unadjusted HR for IrPan vs Irinotecan (95% CI) | P Value | Events/Patients, No. | Unadjusted HR for IrPan vs Irinotecan (95% CI) | P Value | Events/Patients, No. | Unadjusted HR for IrPan vs Irinotecan (95% CI) | P Value | |||
| PFS | RAS wt | 190/208 | 0.67 (0.50-0.90) |
.01 | 128/139 | 0.96 (0.67-1.38) |
.84 | 62/69 | 0.33 (0.19-0.58) |
<.001 | .002 |
| RAS mut | 95/99 | 1.18 (0.78-1.77) |
.43 | 60/63 | 1.17 (0.70-1.97) |
.54 | 35/36 | 1.26 (0.63-2.52) |
.51 | .89 | |
| BRAF mut | 44/47 | 1.07 (0.57-1.98) |
.84 | 33/35 | 1.29 (0.60-2.74) |
.51 | 11/12 | 0.29 (0.06-1.40) |
.12 | .06 | |
| All patients | 285/307 | 0.81 (0.64-1.03) |
.09 | 188/202 | 1.05 (0.78-1.40) |
.76 | 97/105 | 0.55 (0.36-0.84) |
.006 | .01 | |
| OS | RAS wt | 192/209 | 1.10 (0.82-1.46) |
.52 | 130/140 | 1.56 (1.09-2.23) |
.02 | 62/69 | 0.66 (0.40-1.10) |
.11 | .01 |
| RAS mut | 97/99 | 1.36 (0.91-2.04) |
.14 | 63/63 | 1.02 (0.62-1.69) |
.92 | 34/36 | 2.09 (1.0-4.37) |
.05 | .09 | |
| BRAF mut | 45/47 | 1.33 (0.73-2.43) |
.35 | 33/35 | 1.41 (0.67-2.98) |
.36 | 12/12 | 0.41 (0.11-1.57) |
.19 | .11 | |
| All patients | 289/308 | 1.17 (0.93-1.48) |
.18 | 193/203 | 1.36 (1.02-1.81) |
.03 | 96/105 | 0.95 (0.64-1.42) |
.80 | .15 | |
Abbreviations: HR, hazard ratios; IrPan, irinotecan with panitumumab; mut, mutation; OS, overall survival; PFS, progression-free survival; wt, wild type.
PIK3CA mutant cases excluded owing to small patient numbers.
Dichotomized HER3 was not significantly prognostic for OS (HR, 0.85; 95% CI, 0.60-1.20; P = .35) or PFS (HR, 1.03; 95% CI, 0.73-1.45; P = .86) (eTable 3 and eFigure 2 in the Supplement).
In patients with RAS wt and high HER3 expression, 12-week response rate was 48.6% (IrPan) vs 12.1% (irinotecan) (relative risk, 4.01; 95% CI, 1.50-10.68), while in patients with RAS wt and low HER3 expression response rate was 24.6% (IrPan) vs 11.1% (irinotecan) (relative risk, 2.21; 95% CI, 1.03-4.75; interaction P = .34) (eTable 6 in the Supplement).
We previously reported that in a dichotomous ligand mRNA expression model, “EREG-high and/or AREG-high” was strongly predictive of panitumumab PFS benefit in patients with RAS wt, and that as a continuous variable AREG was the more predictive of the 2 ligands.
As continuous variables in a joint model, HER3 and AREG were independent predictors of IrPan PFS benefit, while for OS the evidence was equivocal (eTable 7 in the Supplement).
In an exploratory dichotomous model, the RAS wt population was divided into 4 groups: high HER3, high AREG/EREG (n = 42); high HER3, low AREG/EREG (n = 27); low HER3/high AREG/EREG (n = 53); and low HER3, low AREG/EREG (n = 87). Marked panitumumab benefit was observed in the high-HER3, high-AREG/EREG group (PFS HR, 0.24; 95% CI, 0.11-0.51; P < .001 and OS HR, 0.36; 95% CI, 0.18-0.73; P = .004). Conversely, the low-HER3, low-AREG/EREG group showed no evidence of benefit (PFS HR, 1.14; 95% CI, 0.73-1.79; P = .57 and OS HR, 1.44; 95% CI, 0.92-2.26; P = .11). Patients in the 2 intermediate groups had intermediate effects for PFS (eTable 8 in the Supplement).
Discussion
To our knowledge, the work described herein represents the largest dedicated analysis of HER3 expression in aCRC to date. We found no evidence for HER3 as a prognostic marker but found HER3 overexpression to be significantly associated with benefit from panitumumab.
Overall in PICCOLO panitumumab produced a significant improvement in PFS but not OS. It is therefore encouraging that HER3 was significantly predictive for OS as well as PFS. Furthermore, in our combined HER3-and-ligands model, patients with high-HER3, high-AREG/EREG tumors achieved marked and significant OS benefit with panitumumab (HR, 0.36; 95% CI, 0.18-0.73; P = .004). These findings are interesting but must be treated with caution, especially given that the HER3 cut point for dichotomization was derived internally from this data set.
The interaction between HER3 expression and anti-EGFR agent efficacy in aCRC has been studied previously in 2 smaller series. In these studies HER3 overexpression was associated with inferior clinical outcomes with anti-EGFR agents. The present study is a dedicated comprehensive analysis in a mature, large, randomized trial, allowing for adjustments for likely confounders. Preclinical work supports both hypotheses: heregulin activation of HER3 might trigger an alternate signaling pathway circumventing EGFR blockade, supporting HER3 overexpression as a negative predictive marker. Alternatively, given its role as an obligate heterodimer, HER3 expression could identify those tumors most reliant on EGFR signaling through autocrine feedback loops, and more likely to respond to EGFR-targeted agents.
Limitations
These findings are interesting, but results should be treated with caution, especially given that the HER3 cut point for dichotomization was derived internally from this dataset. Additionally, tissue was available for only 331 (47.6%) of the 696 patients in the IrPan vs irinotecan randomization in PICCOLO.
Conclusions
This study suggests that high HER3 expression is related to anti-EGFR agent activity in aCRC and may offer a clinically useful selection biomarker. Prior to clinical application, revalidation of the findings and refining of the cut point is required, in well-designed hypothesis-based studies using other randomized data sets.
eFigure 1. Consort diagram of study population
eFigure 2. OS KM curves for high vs low HER3 expression levels
eTable 1. Synthetic standards for RT-PCR
eTable 2. Baseline patient characteristics by treatment arm, including mutation status
eTable 3. Prognostic analysis for the effect of the HER3 dichotomous HER3 classifier and log2 HER3 on PFS and OS
eTable 4. BRAF and primary tumour location adjusted HRs and 95% CIs for the effect of log2 HER3 and the dichotomous classifier on PFS and OS by treatment arm in RAS-wt patients
eTable 5. Estimated crude HRs and 95% CIs for the effect of treatment arm (IrPan vs irinotecan) on PFS and OS in RAS wt patients stratified by HER3 expression 4 ways to explore different cut-points
eTable 6. Estimated crude relative risk and 95% CIs for 12-week response rate (RR) by HER3 expression status
eTable 7. AREG and EREG adjusted HRs and 95% CIs for the effect of log2 HER3 on PFS and OS by treatment arm in RAS wt patients
eTable 8. Estimated crude HRs and 95% CIs for the effect of treatment on PFS and OS in 4 biomarker groups depending upon dichotomous HER3 and ligand classification
References
- 1.Douillard JY, Oliner KS, Siena S, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369(11):1023-1034. [DOI] [PubMed] [Google Scholar]
- 2.Seymour MT, Brown SR, Middleton G, et al. Panitumumab and irinotecan versus irinotecan alone for patients with KRAS wild-type, fluorouracil-resistant advanced colorectal cancer (PICCOLO): a prospectively stratified randomised trial. Lancet Oncol. 2013;14(8):749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seligmann JF, Elliott F, Richman SD, et al. Combined epiregulin and amphiregulin expression levels as a predictive biomarker for panitumumab therapy benefit or lack of benefit in patients with RAS wild-type advanced colorectal cancer. JAMA Oncol. 2016;2(5):633-642. [DOI] [PubMed] [Google Scholar]
- 4.Tzahar E, Waterman H, Chen X, et al. A hierarchical network of interreceptor interactions determines signal transduction by Neu differentiation factor/neuregulin and epidermal growth factor. Mol Cell Biol. 1996;16(10):5276-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sierke SL, Cheng K, Kim HH, Koland JG. Biochemical characterization of the protein tyrosine kinase homology domain of the ErbB3 (HER3) receptor protein. Biochem J. 1997;322(Pt 3):757-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi BK, Fan X, Deng H, Zhang N, An Z. ERBB3 (HER3) is a key sensor in the regulation of ERBB-mediated signaling in both low and high ERBB2 (HER2) expressing cancer cells. Cancer Med. 2012;1(1):28-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engelman JA, Jänne PA, Mermel C, et al. ErbB-3 mediates phosphoinositide 3-kinase activity in gefitinib-sensitive non-small cell lung cancer cell lines. Proc Natl Acad Sci U S A. 2005;102(10):3788-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sergina NV, Rausch M, Wang D, et al. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature. 2007;445(7126):437-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richman SD, Southward K, Chambers P, et al. HER2 overexpression and amplification as a potential therapeutic target in colorectal cancer: analysis of 3256 patients enrolled in the QUASAR, FOCUS and PICCOLO colorectal cancer trials. J Pathol. 2016;238(4):562-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scartozzi M, Mandolesi A, Giampieri R, et al. The role of HER-3 expression in the prediction of clinical outcome for advanced colorectal cancer patients receiving irinotecan and cetuximab. Oncologist. 2011;16(1):53-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cushman SM, Jiang C, Hatch AJ, et al. Gene expression markers of efficacy and resistance to cetuximab treatment in metastatic colorectal cancer: results from CALGB 80203 (Alliance). Clin Cancer Res. 2015;21(5):1078-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eFigure 1. Consort diagram of study population
eFigure 2. OS KM curves for high vs low HER3 expression levels
eTable 1. Synthetic standards for RT-PCR
eTable 2. Baseline patient characteristics by treatment arm, including mutation status
eTable 3. Prognostic analysis for the effect of the HER3 dichotomous HER3 classifier and log2 HER3 on PFS and OS
eTable 4. BRAF and primary tumour location adjusted HRs and 95% CIs for the effect of log2 HER3 and the dichotomous classifier on PFS and OS by treatment arm in RAS-wt patients
eTable 5. Estimated crude HRs and 95% CIs for the effect of treatment arm (IrPan vs irinotecan) on PFS and OS in RAS wt patients stratified by HER3 expression 4 ways to explore different cut-points
eTable 6. Estimated crude relative risk and 95% CIs for 12-week response rate (RR) by HER3 expression status
eTable 7. AREG and EREG adjusted HRs and 95% CIs for the effect of log2 HER3 on PFS and OS by treatment arm in RAS wt patients
eTable 8. Estimated crude HRs and 95% CIs for the effect of treatment on PFS and OS in 4 biomarker groups depending upon dichotomous HER3 and ligand classification