Key Points
Question
Is treatment with the protein farnesyltransferase inhibitor lonafarnib associated with a lower mortality rate among children with the rare, fatal premature aging disease Hutchinson-Gilford progeria syndrome?
Findings
In this cohort study of 27 treated patients with Hutchinson-Gilford progeria syndrome compared with a pool of 103 contemporaneous untreated patients, treatment with lonafarnib monotherapy compared with no treatment was associated with a significantly lower mortality rate (3.7% vs 33.3%) after a median of 2.2 years of follow-up.
Meaning
This preliminary study suggests that treatment with lonafarnib may have therapeutic benefit for children with Hutchinson-Gilford progeria syndrome, but the findings are limited by its observational design.
Abstract
Importance
Hutchinson-Gilford progeria syndrome (HGPS) is an extremely rare fatal premature aging disease. There is no approved treatment.
Objective
To evaluate the association of monotherapy using the protein farnesyltransferase inhibitor lonafarnib with mortality rate in children with HGPS.
Design, Setting, and Participants
Cohort study comparing contemporaneous (birth date ≥1991) untreated patients with HGPS matched with treated patients by age, sex, and continent of residency using conditional Cox proportional hazards regression. Treatment cohorts included patients from 2 single-group, single-site clinical trials (ProLon1 [n = 27; completed] and ProLon2 [n = 36; ongoing]). Untreated patients originated from a separate natural history study (n = 103). The cutoff date for patient follow-up was January 1, 2018.
Exposure
Treated patients received oral lonafarnib (150 mg/m2) twice daily. Untreated patients received no clinical trial medications.
Main Outcomes and Measures
The primary outcome was mortality. The primary analysis compared treated patients from the first lonafarnib trial with matched untreated patients. A secondary analysis compared the combined cohorts from both lonafarnib trials with matched untreated patients.
Results
Among untreated and treated patients (n = 258) from 6 continents, 123 (47.7%) were female; 141 (54.7%) had a known genotype, of which 125 (88.7%) were classic (c.1824C>T in LMNA). When identified (n = 73), the primary cause of death was heart failure (79.4%). The median treatment duration was 2.2 years. Median age at start of follow-up was 8.4 (interquartile range [IQR], 4.8-9.5) years in the first trial cohort and 6.5 (IQR, 3.7-9.0) years in the combined cohort. There was 1 death (3.7%) among 27 patients in the first trial group and there were 9 deaths (33.3%) among 27 patients in the matched untreated group. Treatment was associated with a lower mortality rate (hazard ratio, 0.12; 95% CI, 0.01-0.93; P = .04). In the combined cohort, there were 4 deaths (6.3%) among 63 patients in the treated group and 17 deaths (27.0%) among 63 patients in the matched untreated group (hazard ratio, 0.23; 95% CI, 0.06-0.90; P = .04).
Conclusions and Relevance
Among patients with HGPS, lonafarnib monotherapy, compared with no treatment, was associated with a lower mortality rate after 2.2 years of follow-up. Study interpretation is limited by its observational design.
This cohort study compares mortality rates among patients with Hutchinson-Gilford progeria syndrome who were treated with the protein farnesyltransferase inhibitor lonafarnib vs contemporaneous matched untreated patients.
Introduction
Hutchinson-Gilford progeria syndrome (HGPS) is an extremely rare, fatal, autosomal dominant segmental premature aging disease, with an estimated incidence of 1 per 4 million births and a prevalence of 1 in 20 million living individuals. It has no sex, ethnic, or regional predisposition. Morbidity includes failure to thrive, generalized lipodystrophy, alopecia, bone dysplasia, and progressive atherosclerosis leading to cardiac disease and stroke. There are no established biochemical biomarkers that predict clinical benefit in HGPS. Neither serum cholesterol nor high-sensitivity C-reactive protein levels are elevated in this disease. Mortality is caused primarily by heart failure at a mean age of 14.6 years.
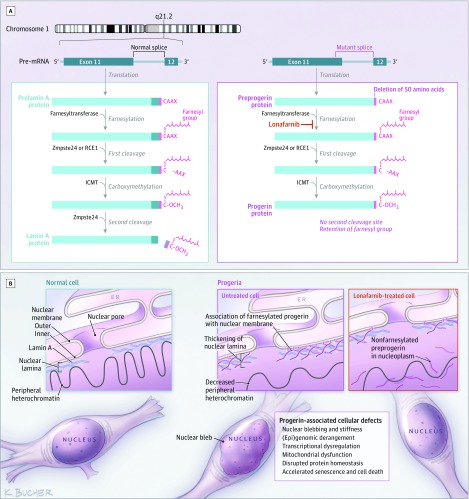
Hutchinson-Gilford progeria syndrome is caused by single base pathogenic variants in the LMNA gene that activate a cryptic splice site and result in the production of a farnesylated mutant lamin A protein called progerin (Figure 1). Lamin A, an inner nuclear membrane protein, is crucial to many cellular functions. Persistent farnesylation of the mutant protein causes it to intercalate into the inner nuclear membrane, where it accumulates and exerts damage to cells as they age. Preclinical studies with protein farnesyltransferase inhibitors have yielded improved disease phenotypes.
Figure 1. Posttranslational Processing Pathways Producing Lamin A and Progerin, Including the Target Site for Lonafarnib.
Panel A: A prelamin polypeptide chain with its C-terminal −CAAX box, representing cysteine (C), aliphatic amino acids (AA), and any amino acid (X). The α-helical rod domain is divided into segments to assist in displaying the progerin defect. Posttranslational processing consists of 4 steps: (1) A farnesyl group is attached to the cysteine residue of the −CAAX box by farnesyltransferase; (2) the last 3 residues are proteolytically cleaved by the zinc metalloprotease Zmpste24 or by Ras-converting enzyme (RCE1); (3) carboxymethylation by isoprenylcysteine carboxyl methyltransferase (ICMT); and (4) the terminal 15 C-terminal residues, including the farnesylated and carboxymethylated cysteine, are cleaved off by Zmpste24. Panel B: Representative progerin-expressing cell type (fibroblasts) demonstrating (left) lamin A associated with the inner nuclear membrane in a normal cell, (center) reduced lamin A and presence of farnesylated progerin in a Hutchinson-Gilford progeria syndrome (HGPS) cell, and (right) decreased progerin with appearance of nonfarnesylated preprogerin in a lonafarnib-treated HGPS cell. Progerin affects every level of cellular function; major progerin-associated cellular effects are listed in the box.
No drugs are approved for the treatment of HGPS. Two phase 2 single-group treatment trials have evaluated monotherapy with the farnesyltransferase inhibitor lonafarnib. In treatment trial 1 (ProLon1), lonafarnib was well tolerated. Rate of weight gain, arterial pulse wave velocity, carotid artery echodensity, skeletal rigidity, and sensorineural hearing were improved. Preliminary evidence of decreased rates of strokes, headaches, and seizures was also reported. Lipodystrophy, skin features, alopecia, and joint contractures were unaffected, underscoring that lonafarnib treats some aspects of disease but is not a cure for HGPS. Treatment trial 2 (ProLon2) has completed accrual and is ongoing (https://clinicaltrials.gov/show/NCT000916747). Neither trial has evaluated mortality as an outcome measure. The current study assessed the association between lonafarnib monotherapy and mortality rate compared with no treatment.
Methods
General Study Design and Approvals
This observational cohort study compared treated patients with contemporaneous untreated participants. The study was approved by the institutional review board of Rhode Island Hospital, Providence. Data were compiled at the Brown University Center for Gerontology and Healthcare Research, Providence, Rhode Island (L.B.G., J.B., and S.E.C.), and data analysis was performed at Boston University (H.S., J.M., and R.B.D.). Some data were obtained through a Data Use Agreement among The Progeria Research Foundation, Rhode Island Hospital, and Brown University, for which patient consent was not required, as approved by the institutional review board. Patients in the full natural history cohort were born between 1876 and 2015. The earliest patient observation for both the treated cohorts and the untreated contemporaneous controls used in treatment mortality analyses was in 1991. The study data inclusion cutoff date was January 1, 2018.
Participants
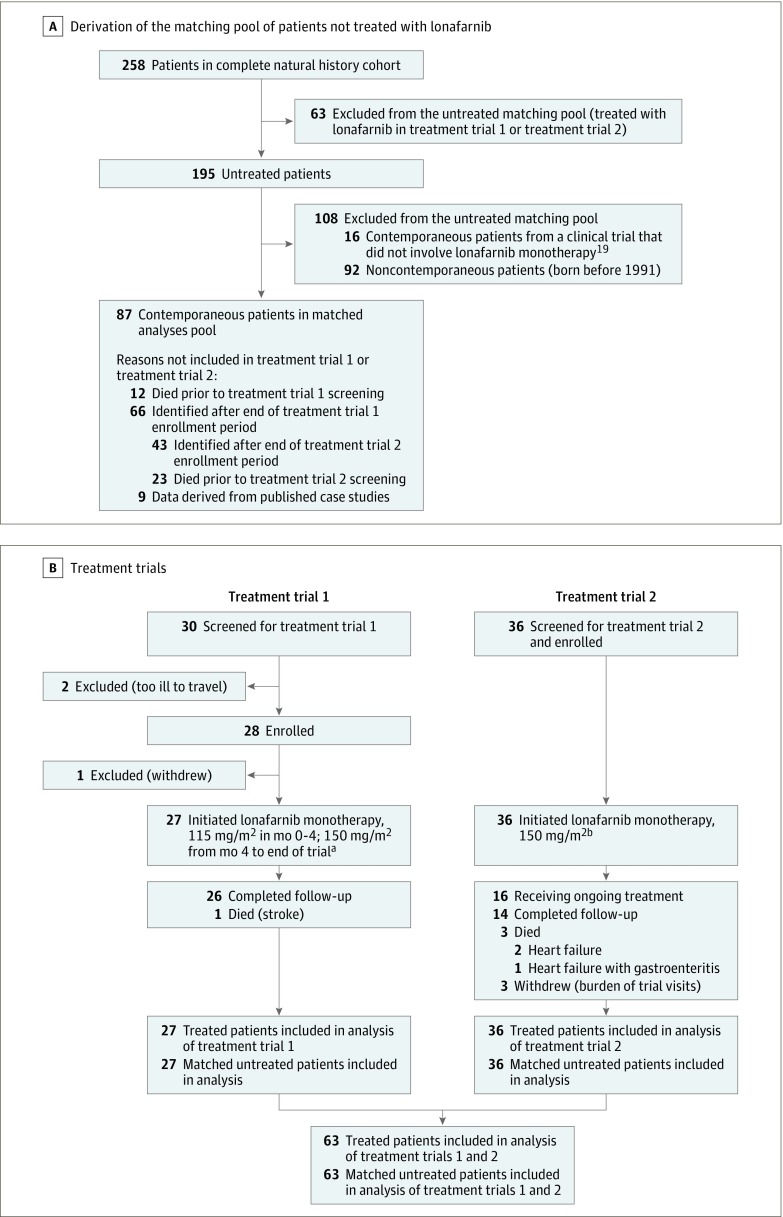
Study patients and their associated data were identified using The Progeria Research Foundation International Registry, Diagnostics Program, and Medical and Research Database as well as published scientific and news articles and publicly available databases (Figure 2). Of 258 total patients in the complete natural history cohort, 211 were identified from the Progeria Research Foundation International Registry. Among these 211 patients, some inclusion data from other Progeria Research Foundation programs were also used in 42 patients and some inclusion data from public data sources were also used in 37 patients. Forty-seven patients’ study inclusion information was derived purely from scientific publications. When applicable, consent was obtained in the language of the participant. Minimum inclusion criteria were HGPS phenotype confirmation by study investigators and information on living age or age at death. Hutchinson-Gilford progeria syndrome has a highly consistent phenotype that can be reliably differentiated from non–progerin-producing progeroid laminopathies using pathognomonic physical findings. Although exclusion of non–progerin-producing laminopathies is reliably accomplished using phenotype in the absence of genotype, instances in which the splicing mutation yields very low levels of progerin result in a clinically different phenotype, which is not categorized as HGPS. Such patients were excluded from the analysis. Additional data collected included sex, country of origin, cause of death, and genotype, when available.
Figure 2. Flow of Patients Through the Study.
Two hundred fifty-eight patients were used to construct the untreated natural history analysis (panel A). Of these, 87 patients were both contemporaneous with the treatment trial patients and not included in a treatment trial at any time. To compare mortality rates among treated vs untreated patients, random matching was performed whereby (panel B) 27 patients from treatment trial 1, 63 patients from treatment trial 1 plus treatment trial 2, and 36 patients from treatment trial 2 were randomly matched on age, sex, and continent of residency in 3 separate analyses.
aThese are the treated patients used in treatment trial 1–treated vs matched untreated comparison of mortality rates.
bThese are the treated patients used in the treatment trial 2–treated vs matched untreated comparison of mortality rates.
Untreated patients received no clinical trial medications. The treated cohort was derived from the 2 treatment trials, conducted at Boston Children’s Hospital, where lonafarnib (Merck) monotherapy was administered to children with HGPS (Figure 2). Trial participants had not previously been enrolled in any clinical treatment trial and had not received HGPS-specific medications. Treated and matched untreated patient dates of birth were 1991-2004 in treatment trial 1 and 1997-2014 in treatment trial 2.
Treatment
Selection bias for trial inclusion was minimized because inclusion was facilitated for all identified patients, including those who eventually enrolled and those who did not. Facilitation included access to logistical assistance, no-cost availability of genetic testing, interpreters, and coverage of trial-related expenses for travel, lodging, research testing, and medications.
Treatment trial 1 was initiated in 2007 and completed in 2010. Lonafarnib was administered every 12 ( ± 2) hours orally at 115 mg/m2 for 4 months and then escalated to 150 mg/m2 for the trial duration. Toxic effects included mild diarrhea, fatigue, nausea, vomiting, anorexia, and liver function and hemoglobin abnormalities, all of which generally improved with time. Treatment trial 2 was initiated in 2013 as a lonafarnib monotherapy extension of a completed 3-drug combination therapy trial. Oral lonafarnib dosing was 150 mg/m2 throughout.
Outcomes
The primary study outcome was all-cause mortality. First, in a natural history study, all-cause mortality was assessed in untreated children. Second, mortality was compared between untreated and lonafarnib-treated children enrolled in treatment trial 1. A secondary analysis compared mortality between untreated children and children enrolled in both trials combined. A post hoc secondary analysis compared mortality between untreated children and participants in the ongoing second treatment trial. Cause of death was also assessed.
Statistical Analysis
Demographic characteristics are reported using counts with percentages for dichotomous outcomes and using means with standard deviations and medians with interquartile ranges (IQRs) for continuous outcomes.
To estimate survival age for untreated children with HGPS, a Kaplan-Meier survival curve was generated (follow-up started at birth) with the full untreated cohort plus treatment trial participants censored at the time of treatment initiation. Untreated patients who were living at the start of data analysis and patients from a published report who were living at the time of the report but then lost to follow-up were censored at the time of last known age.
Comparisons of Kaplan-Meier survival curves between subgroups were performed for (1) male vs female patients; (2) all patients with dates of birth before 1991 vs during or after 1991; and (3) patients with genetic diagnoses vs unknown progerin-producing LMNA mutation information. The first 2 subgroup analyses were performed with the full untreated cohort plus treatment trial participants censored at the time of treatment initiation. For the subgroup analysis of known vs unknown genetic diagnoses, treatment trial patients were excluded to avoid inherent bias because 100% of trial patients had known genetic information (a trial inclusion criterion) compared with 40.5% of untreated patients.
To compare mortality rates among children treated in treatment trial 1 vs among untreated children, treated children were first matched to untreated children by age, sex, and continent of residency to control for potential confounding. For every treated patient, all untreated patients of the same sex and from the same continent of residency who were alive at the age when the treated patient began lonafarnib were identified. From this group of untreated patients, 1 was randomly selected and used as the matched untreated patient in the analysis; that patient was then no longer available for matching. Follow-up began at the age at treatment initiation for the treated patient in the matched pair. This is similar to the matched design approach discussed by Li et al.
Kaplan-Meier mortality estimates for treated and untreated matched groups are presented. Unadjusted Cox proportional hazards regression, conditioned on the matched pair, was used to compare treated and untreated matched groups on mortality rate after matching and was used to calculate unadjusted hazard ratios (HRs) and their 2-sided 95% confidence intervals for mortality in treated vs untreated patients. Treated patients who were living at completion of the first treatment trial were censored at that time (2-2.5 years following treatment initiation). When the treated patient in a matched pair was censored, his/her untreated match was censored at the same length of follow-up. If any patient in the matched pair was lost to follow-up, the corresponding match was censored at the same follow-up time, and logistic regression multiple imputation was used to impute death status at the end of follow-up for both patients in the matched pair prior to carrying out Cox regression. The logistic regression imputation model included sex, continent of residency, age at start of follow-up, and treatment status (treated or untreated) as predictors. Ten data sets were imputed, and the conditional unadjusted Cox regression was used to compare treated vs untreated patients on mortality within each imputed data set. The 10 treatment β coefficients and their standard errors were combined across the 10 imputed data sets using the method described by Rubin to create 1 overall treatment β coefficient and standard errors. These were used to generate the overall P value assessing treatment effect and the confidence interval of the β coefficient. The confidence interval and β coefficient were then exponentiated to create the HR and 2-sided 95% confidence interval for treated vs untreated patients. The proportional hazards assumption was assessed within each imputed data set using the Kolmogorov-type supremum test (and the proportional hazards assumption was always met with a nonsignificant P≥.20).
Two secondary analyses were performed. The first was prespecified and compared survival among participants from both trials combined with a 1-to-1 matched set of untreated children. Once matching was complete, multiply imputed unadjusted conditional Cox proportional hazards regressions were performed for each study separately, and the final imputed treatment β coefficient and standard error from each study were combined using a random-effects meta-analysis. A post hoc secondary analysis was carried out in participants from treatment trial 2 in the same manner as participants in treatment trial 1; follow-up of patients in treatment trial 2 is not complete and these results are preliminary. In all cases, the proportional hazards assumption was always met (P > .20).
Sensitivity analyses were conducted by repeating the above described analyses but removing or censoring patients with confounding factors. Specifically, 2 patients who intended to enroll in treatment trial 1 but could not because of health issues were omitted from the untreated group; and 1 patient in the treated group in treatment trial 1 who received clinical care at the trial hospital site was censored at age 18.4 years when care was administered.
Statistical analyses were carried out using SAS software, version 9.4 (SAS Institute Inc). P values were 2-sided and deemed significant at P<.05.
Results
Patient Characteristics
Characteristics of the patient groups are presented in the Table. All participants were positive for the HGPS phenotype. All known mutations were progerin producing. The untreated group (n = 195) consisted of 179 patients who never received treatment in a clinical trial plus pretreatment follow-up data from 16 patients who received treatment in a clinical trial not involving lonafarnib monotherapy. The untreated group was 47.7% female, and continent of origin in descending frequency was North America, Asia, Europe, South America, Africa, and Australia; the genotype was known for 79 untreated patients (40.5%). The lonafarnib-treated group (100% of clinical trial patients; n = 63) was born in 1991 or later and was 47.6% female (n = 30). The median age at start of treatment was 6.5 (IQR, 3.7-9.0) years; continent of origin in descending frequency was North America, Asia, South America, Europe, Africa, and Australia; the genotype was known for all 63 treated patients (100%).
Table. Patient Characteristics.
| Characteristics | No. (%)a | |||||||
|---|---|---|---|---|---|---|---|---|
| Full Natural History Cohort (n = 258) | Untreated (n = 195)b | Treatment Trial 1 (n = 27) | Matched Untreated for Treatment Trial 1 (n = 27) | Treatment Trial 2 (n = 36) | Matched Untreated for Treatment Trial 2 (n = 36) | Treatment Trial 1 + Treatment Trial 2 (n = 63) | Matched Untreated for Treatment Trial 1 + Treatment Trial 2 (n = 63) | |
| Female | 123 (47.7) | 93 (47.7) | 16 (59.3) | 16 (59.3) | 14 (38.9) | 14 (38.9) | 30 (47.6) | 30 (47.6) |
| Male | 135 (52.3) | 102 (52.3) | 11 (40.7) | 11 (40.7) | 22 (61.1) | 22 (61.1) | 33 (52.4) | 33 (52.4) |
| Age at start of follow-up in treated vs untreated comparisons, y | ||||||||
| Mean (SD) | NAc | NAc | 7.6 (3.2) | 7.6 (3.2) | 6.4 (4.0) | 6.4 (4.0) | 6.9 (3.7) | 6.9 (3.7) |
| Median (IQR) | NAc | NAc | 8.4 (4.8-9.5) | 8.4 (4.8-9.5) | 5.2 (3.6-8.4) | 5.2 (3.6-8.4) | 6.5 (3.7-9.0) | 6.5 (3.7-9.0) |
| Born ≥1991d | 166 (64.3) | 103 (52.8) | 27 (100.0) | 27 (100.0) | 36 (100.0) | 36 (100.0) | 63 (100.0) | 63 (100.0) |
| Known genotype | 141 (54.7) | 79 (40.5) | 27 (100.0) | 13 (48.1) | 36 (100) | 19 (52.8) | 62 (98.4) | 34 (54.0) |
| Mutationd | ||||||||
| c.1824C>T; p.G608G | 125 (88.6) | 66 (83.5) | 26 (96.3) | 13 (100.0) | 33 (94.3) | 17 (89.5) | 59 (95.2) | 28 (82.4) |
| c.1822G>A, p.G608S | 5 (3.5) | 4 (5.1) | 1 (3.7) | 0 | 0 | 0 | 1 (1.6) | 1 (2.9) |
| Intron 11, c.1968+1G>C | 2 (1.4) | 2 (2.5) | 0 | 0 | 0 | 0 | 0 | 0 |
| Intron 11, c.1968+1G>A | 5 (3.5) | 4 (5.1) | 0 | 0 | 1 (2.9) | 1 (5.3) | 1 (1.6) | 3 (8.8) |
| Intron 11, c.1968+2T>A | 2 (1.4) | 1 (1.3) | 0 | 0 | 1 (2.9) | 0 | 1 (1.6) | 1 (2.9) |
| Intron 11, c.1968+2T>C | 1 (0.7) | 1 (1.3) | 0 | 0 | 0 | 1 (5.3) | 0 | 1 (2.9) |
| Intron 11, c.1968+5G>C | 1 (0.7) | 1 (0.7) | 0 | 0 | 0 | 0 | 0 | 0 |
| Continente | ||||||||
| Africa | 12 (4.7) | 10 (5.1) | 0 | 0 | 2 (5.6) | 1 (2.8) | 2 (3.2) | 1 (1.6) |
| Asia | 67 (26.0) | 50 (25.6) | 4 (14.8) | 4 (14.8) | 13 (36.1) | 12 (33.3) | 17 (27.0) | 19 (30.2) |
| Australia | 3 (1.2) | 2 (1.0) | 0 | 0 | 1 (2.8) | 0 | 1 (1.6) | 0 |
| Europe | 48 (18.6) | 38 (19.5) | 8 (29.6) | 8 (29.6) | 2 (5.6) | 4 (11.1) | 10 (15.9) | 15 (23.8) |
| North America | 85 (33.0) | 63 (32.3) | 11 (40.7) | 8 (29.6) | 11 (30.6) | 11 (30.6) | 22 (34.9) | 13 (20.6) |
| South America | 43 (16.7) | 32 (16.4) | 4 (14.8) | 7 (25.9) | 7 (19.4) | 8 (22.2) | 11 (17.5) | 15 (23.8) |
Abbreviation: IQR, interquartile range.
Unless otherwise indicated.
Of the 195 patients, 16 were treated in a separate combination therapy trial and were included in the overall assessment of untreated survival (Figure 3, A and B), in which they were censored at the time of combination trial treatment initiation; these 16 patients were not included in any analyses comparing lonafarnib-treated with completely untreated patients in this investigation.
Data not applicable (NA) because not all untreated patients were included in the matched treated vs untreated analysis (born before 1991 or not matched to an untreated patient); hence, age at start of follow-up in treated vs untreated comparisons is not applicable for the populations that contain all untreated patients. In Figure 3A (overall untreated survival), these patients, as with all patients described in Figure 3, were followed up from birth.
Patients with known genotype.
For 5 of 27 treated patients in the treatment trial 1 analysis, 16 of 63 treated patients in the treatment trial 1 + treatment trial 2 analysis, and 4 of 36 treated patients in the treatment trial 2 analysis, an exact match on continent of residency could not be found, so the requirement for matching on continent was relaxed for these patients; the overall distribution of continents between the treated and untreated groups was still similar (P ≥ .20).
Mortality Rate Among Untreated Patients
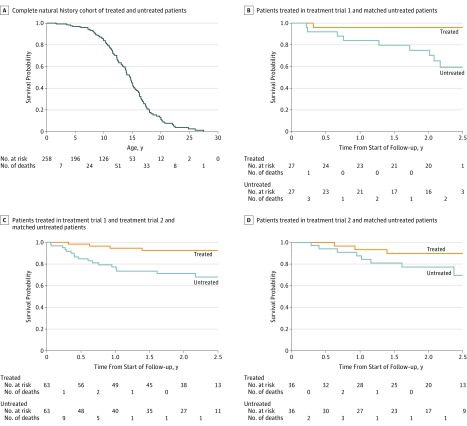
Figure 3A presents the Kaplan-Meier survival curve for untreated HGPS patients plus treated patients censored at treatment initiation, with follow-up starting at birth (n = 258). Of these, 124 (48.1%) died. Mean and median survival ages were 14.5 years and 14.6 years, respectively.
Figure 3. Kaplan-Meier Survival Curves for Untreated and Treated Hutchinson-Gilford Progeria Syndrome Cohorts.
Number of patients at risk are presented below the x-axis. If any patient in a matched pair was censored, the corresponding match was censored at the same follow-up time and the number at risk decreased. Also shown are numbers of deaths that occurred before or at that time point and after the previous time point on the x-axis. For panel A, time 0 on the x-axis (ie, beginning of patient risk) is date of birth. For panels B-D, time 0 on the x-axis is defined for each matched pair as the age at treatment initiation for the treated patient in the matched pair. Panel A: The untreated cohort includes all patients never treated (median follow-up time, 11.8 [interquartile range {IQR}, 7.8-15.2] years); treated patients were also included but were censored at age of treatment initiation (median follow-up time, 6.0 [IQR, 3.5-8.9] years). There were 124 deaths among 258 patients. Mean and median survival times were 14.5 years and 14.6 years, respectively. Median follow-up time was 9.9 (IQR, 5.7-14.0) years. Panel B: For treatment trial 1, there was 1 death in 27 treated patients and there were 9 deaths in 27 matched untreated patients. P = .04 based on Cox proportional hazards regression conditioned on the matched pair. Median follow-up time was 2.2 (IQR, 1.9-2.2) years for treated and 2.1 (IQR, 1.0-2.2) years for untreated patients. Panel C: For treatment trial 1 + treatment trial 2, there were 4 deaths in 63 treated patients and 17 deaths in 63 matched untreated patients. P = .03 based on Cox proportional hazards regression conditioned on the matched pair. The median follow-up time was 2.2 (IQR, 1.4-2.3) years for treated and 1.7 (IQR, 0.6-2.2) years for untreated patients. Panel D: For treatment trial 2, there were 3 deaths in 36 treated patients and 8 deaths in 36 matched untreated patients. P = .18 based on Cox proportional hazards regression conditioned on the matched pair. The median follow-up time was 2.0 (IQR, 1.3-2.5) years for treated patients and 1.9 (IQR, 1.0-2.4) years for untreated patients.
There was no significant difference in untreated patient mortality rates for (1) female vs male patients (HR, 1.42; 95% CI, 0.98-2.04; P = .06) (eFigure, A, in the Supplement); (2) patients with dates of birth before 1991 vs during or after 1991 (HR, 0.78; 95% CI, 0.53-1.14; P = .20) (eFigure, B, in the Supplement); and (3) completely untreated patients with known genetic diagnoses vs with unknown progerin-producing LMNA mutation information (HR, 1.33; 95% CI, 0.91-1.93; P = .14) (eFigure, C, in the Supplement).
Association Between Lonafarnib Treatment and Mortality Rate
There were 103 contemporaneous completely untreated patients. Data for 94 (91.3%) of the 103 untreated patients and 100% of the treated patients were derived from The Progeria Research Foundation International Registry; data for 9 (8.7%) of the 103 completely untreated patients were derived from scientific publications.
In treatment trial 1, the median age at start of follow-up was 8.4 (IQR, 4.8-9.5) years. Treatment was associated with a lower mortality rate among the treated cohort vs the untreated cohort (Figure 3B). There was 1 death (3.7%) among 27 patients in the treated group and there were 9 deaths (33.3%) among 27 patients in the matched untreated group. The conditional unadjusted HR for mortality rates of treated vs untreated patients was 0.12 (95% CI, 0.01-0.93; P = .04). One untreated patient did not have complete follow-up, so that patient and the corresponding matched patient were included in the analysis via multiple imputation. Median follow-up time was 2.2 (IQR, 1.9-2.2) years for treated patients and 2.1 (IQR, 1.0-2.2) years for untreated patients.
For the secondary outcome analysis of the combined trials (median age, 6.5 [IQR, 3.7-9.0] years at start of follow-up), mortality was statistically significantly lower in treated patients vs matched untreated patients (Figure 3C). There were 4 deaths (6.3%) among 63 patients in the treated group and 17 deaths (27.0%) among 63 patients in the matched untreated group. The random-effects meta-analytical conditional HR for treated vs matched untreated patients across the 2 studies was 0.23 (95% CI, 0.06-0.90; P = .04). Two untreated patients did not have complete follow-up and were included in the analysis via multiple imputation. Median follow-up time was 2.2 (IQR, 1.4-2.3) years for treated patients and 1.7 (IQR, 0.6-2.2) years for untreated patients.
For the post hoc analysis of treatment trial 2 (median age, 5.2 [IQR, 3.6-8.4] years at start of follow-up), there were 3 deaths (8.3%) among 36 patients in the treated group, and 8 deaths (22.2%) among 36 patients in the matched untreated group (Figure 3D). There was no significant difference in mortality between treated and untreated patients (HR, 0.33; 95% CI, 0.07-1.59; P = .17). Two untreated and 3 treated patients did not have complete follow-up, and these patients and their matches were included in the analysis via multiple imputation. Median follow-up time was 2.0 (IQR, 1.3-2.5) years for treated patients and 1.9 (IQR, 1.0-2.4) years for untreated patients.
Results of prespecified sensitivity analyses omitting 2 patients from the untreated group who intended to enroll in treatment trial 1 but could not because of health issues, plus censoring 1 patient in the treated group at age 18.4 years when clinical care was administered, yielded similar HRs and P values: HR, 0.09 (95% CI, 0.01-0.70; P = .04) for treatment trial 1; HR, 0.11 (95% CI, 0.03-0.47; P = .01) for the combined trials; and HR, 0.33 (95% CI, 0.07-1.59; P = .17) for treatment trial 2.
Causes of Death
Cause of death was identified in 69 (55%) of 124 deceased untreated patients. Among these 69 deaths, 55 (80%) were due to heart failure; 5 of these were additionally precipitated by superimposed respiratory infection, 1 by complications of surgery, and 1 by a concurrent stroke. Six deaths (9%) were due to head injury. Three deaths (4%) were due to complications of surgery, 2 by cardiac failure possibly precipitated by general anesthesia and 1 by respiratory arrest. Two deaths (3%) were due to stroke, 2 (3%) to trauma from motor vehicle crashes, and 1 (1%) to complications of gastroenteritis and pneumonia.
Cause of death was identified in 4 (100%) of 4 treated patients: 3 (75%) due to heart failure, 1 of which was additionally precipitated by superimposed infectious gastroenteritis, and 1 (25%) due to stroke.
Discussion
Among patients with HGPS, lonafarnib monotherapy compared with no treatment was associated with lower mortality after 2.2 years of follow-up. Association with a lower mortality rate was observed both within a single trial cohort and when additional patients were added from a separate currently ongoing lonafarnib monotherapy trial. Several strategies were used to minimize sample bias. Cohorts were matched on age at treatment initiation, sex, and continent of residency, and analyses were conditioned on the matched pair. Matching was contemporaneous such that medical care was similarly available for both the treated and untreated groups. In addition, untreated matching for each of the 2 main analyses (treatment trial 1 and the combined trials) was performed independently such that the matching process was repeated separately for each analysis. A series of sensitivity and subgroup analyses in both the untreated and treated cohorts was also performed; none showed significance.
A previous evaluation of the drug combination of lonafarnib, pravastatin, and zoledronic acid showed an association with a lower mortality rate in patients with HGPS. However, the relative influences of these 3 drugs were not evaluated and are crucial to decisions regarding which medication(s) should be administered to potentially lower mortality in this fatal disease.
Because children with HGPS die of premature atherosclerosis, the lower mortality rate may have been attributable to cardiovascular and possibly cerebrovascular benefit. This premise is supported by treatment trial 1 showing evidence of decreased carotid-femoral pulse wave velocity, carotid artery wall echodensity, stroke incidence, headache, and seizures.
Limitations
This study had several limitations. First, because of the extreme rarity of the disease, the sample sizes were small, resulting in wide confidence intervals. However, in the primary and larger cohort secondary analyses, the lower mortality rate was statistically significant. Although the raw number of deaths was lower in the treated matched group, sensitivity analysis of the ongoing treatment trial alone was not significant. Second, this was a cohort study with contemporaneous controls, in which data were gathered from 2 separate clinical trials and an external untreated control group. For this fatal pediatric disease with no known treatments, only single-group clinical trials have been conducted to date because of ethical considerations and are therefore the sole source of data to demonstrate safety and efficacy of any potential new treatment. Although the current study design was prespecified, mortality rate was not an end point within either clinical trial because of the lack of an untreated control group within the trials. Nonetheless, the combination of a well-defined end point (survival) and a well-matched contemporaneous historical control group has become acceptable for drug evaluation in other pediatric rare diseases that lack controlled trials. Third, because of the rarity of this disease, the contemporaneous control and treated groups spanned similar birth years rather than each case being matched to a control by birth year. Given that a case-by-case match was achieved for sex and continent of residency, it is unlikely that this type of matching would have altered the results. Fourth, because this was not a randomized study, there is likely to be residual confounding. Fifth, monotherapy was conducted for a maximum of 2.5 years. Longer-term effects on mortality rate would be valuable to study given the potential for lifelong treatment with lonafarnib.
Conclusions
Among patients with HGPS, lonafarnib monotherapy, compared with no treatment, was associated with a lower mortality rate after 2.2 years of follow-up. Study interpretation is limited by its observational design.
eFigure. Natural History Kaplan-Meier Sensitivity Analyses for Untreated Cohort by Baseline Characteristics
References
- 1.Merideth MA, Gordon LB, Clauss S, et al. Phenotype and course of Hutchinson-Gilford progeria syndrome. N Engl J Med. 2008;358(6):592-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hennekam RC. Hutchinson-Gilford progeria syndrome: review of the phenotype. Am J Med Genet A. 2006;140(23):2603-2624. [DOI] [PubMed] [Google Scholar]
- 3.Gordon LB. PRF by the numbers. https://www.progeriaresearch.org/prf-by-the-numbers/. Accessed February 27, 2018.
- 4.Gordon LB, Harten IA, Patti ME, Lichtenstein AH. Reduced adiponectin and HDL cholesterol without elevated C-reactive protein: clues to the biology of premature atherosclerosis in Hutchinson-Gilford progeria syndrome. J Pediatr. 2005;146(3):336-341. [DOI] [PubMed] [Google Scholar]
- 5.Gordon LB, Massaro J, D’Agostino RB Sr, et al. ; Progeria Clinical Trials Collaborative . Impact of farnesylation inhibitors on survival in Hutchinson-Gilford progeria syndrome. Circulation. 2014;130(1):27-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Sandre-Giovannoli A, Bernard R, Cau P, et al. Lamin A truncation in Hutchinson-Gilford progeria. Science. 2003;300(5628):2055. [DOI] [PubMed] [Google Scholar]
- 7.Eriksson M, Brown WT, Gordon LB, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423(6937):293-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gruenbaum Y, Foisner R. Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu Rev Biochem. 2015;84(1):131-164. [DOI] [PubMed] [Google Scholar]
- 9.Casasola A, Scalzo D, Nandakumar V, et al. Prelamin A processing, accumulation and distribution in normal cells and laminopathy disorders. Nucleus. 2016;7(1):84-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strandgren C, Revêchon G, Sola-Carvajal A, Eriksson M. Emerging candidate treatment strategies for Hutchinson-Gilford progeria syndrome. Biochem Soc Trans. 2017;45(6):1279-1293. [DOI] [PubMed] [Google Scholar]
- 11.Gordon LB, Kleinman ME, Miller DT, et al. Clinical trial of a farnesyltransferase inhibitor in children with Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A. 2012;109(41):16666-16671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ullrich NJ, Kieran MW, Miller DT, et al. Neurologic features of Hutchinson-Gilford progeria syndrome after lonafarnib treatment. Neurology. 2013;81(5):427-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Progeria Research Foundation International Progeria Registry. https://www.progeriaresearch.org/international-progeria-registry/. Accessed January 1, 2018.
- 14.Progeria Research Foundation Diagnostics Program. https://progeriaresearch.org/diagnostic_testing.html. Accessed January 1, 2018.
- 15.Progeria Research Foundation Medical and Research Database. https://progeriaresearch.org/medical-database/. Accessed January 1, 2018.
- 16.Gordon LB, Brown WT, Collins FC Hutchinson-Gilford progeria syndrome. In: Adam MP, Ardinger HH, Pagon RA, et al, eds. GeneReviews. December 12, 2003. (last update: January 8, 2015). https://www.ncbi.nlm.nih.gov/books/NBK1121/.
- 17.Shalev SA, De Sandre-Giovannoli A, Shani AA, Levy N. An association of Hutchinson-Gilford progeria and malignancy. Am J Med Genet A. 2007;143A(16):1821-1826. [DOI] [PubMed] [Google Scholar]
- 18.Hisama FM, Lessel D, Leistritz D, et al. Coronary artery disease in a Werner syndrome-like form of progeria characterized by low levels of progerin, a splice variant of lamin A. Am J Med Genet A. 2011;155A(12):3002-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon LB, Kleinman ME, Massaro J, et al. Clinical trial of the protein farnesylation inhibitors lonafarnib, pravastatin, and zoledronic acid in children with Hutchinson-Gilford progeria syndrome. Circulation. 2016;134(2):114-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Schaubel DE, He K. Matching methods for obtaining survival functions to estimate the effect of a time-dependent treatment. Stat Biosci. 2014;6(1):105-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubin D. Multiple Imputation for Nonresponse in Surveys. Vol 81 Hoboken, NJ: John Wiley & Sons; 2004. [Google Scholar]
- 22.Moscicki RA, Tandon PK. Challenges for small biopharmaceutical companies. N Engl J Med. 2017;376(17):1698. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eFigure. Natural History Kaplan-Meier Sensitivity Analyses for Untreated Cohort by Baseline Characteristics