Abstract
The expression patterns of senescence-related genes were determined during ozone (O3) exposure in Arabidopsis. Rosettes were treated with 0.15 μL L−1 O3 for 6 h d−1 for 14 d. O3-treated leaves began to yellow after 10 d of exposure, whereas yellowing was not apparent in control leaves until d 14. Transcript levels for eight of 12 senescence related genes characterized showed induction by O3. SAG13 (senescence-associated gene), SAG21, ERD1 (early responsive to dehydration), and BCB (blue copper-binding protein) were induced within 2 to 4 d of O3 treatment; SAG18, SAG20, and ACS6 (ACC synthase) were induced within 4 to 6 d; and CCH (copper chaperone) was induced within 6 to 8 d. In contrast, levels of photosynthetic gene transcripts, rbcS (small subunit of Rubisco) and cab (chlorophyll a/b-binding protein), declined after 6 d. Other markers of natural senescence, SAG12, SAG19, MT1 (metallothionein), and Atgsr2 (glutamine synthetase), did not show enhanced transcript accumulation. When SAG12 promoter-GUS (β-glucuronidase) and SAG13 promoter-GUS transgenic plants were treated with O3, GUS activity was induced in SAG13-GUS plants after 2 d but was not detected in SAG12-GUS plants. SAG13 promoter-driven GUS activity was located throughout O3-treated leaves, whereas control leaves generally showed activity along the margins. The acceleration of leaf senescence induced by O3 is a regulated event involving many genes associated with natural senescence.
Leaf senescence is the sequence of degradative processes leading to the remobilization of nutrients and eventual leaf death. The senescence process is highly regulated, involving photosynthetic decline, protein degradation, lipid peroxidation, and chlorophyll degradation (Smart, 1994). Total RNA levels decline during senescence as RNase activity increases (Blank and McKeon, 1991). Chloroplasts are one of the earliest sites of catabolism, while mitochondria remain intact until late in the senescence process in order for respiration to continue (Smart, 1994). Plant hormones are involved in regulating the senescence process, with cytokinins delaying senescence, ethylene modulating the timing of senescence, and the other hormones playing less prominent roles (Smart, 1994). Leaf senescence, like other developmental processes, is actively regulated by differential gene expression. Transcript levels for photosynthetic genes such as rbcS (small subunit of Rubisco) and cab (chlorophyll a/b-binding protein) decline (Bate et al., 1991), while other genes become activated (Buchanan-Wollaston, 1997; Weaver et al., 1997).
Using differential screening and subtractive hybridization techniques, researchers have identified genes with increased expression during senescence. These genes have been identified in Arabidopsis, oilseed rape, tomato, barley, potato, cucumber, rice, wheat, and maize (for reviews, see Buchanan-Wollaston, 1997; Weaver et al., 1997). Such genes are often referred to as SAGs or senescence-up-regulated genes. Among the identified senescence-induced genes are genes encoding proteases, RNases, Gln synthetase, metallothioneins, protease regulators, ACC oxidase, lipases, glyoxylate cycle enzymes, catalase, endoxyloglucan transferase, pathogenesis-related proteins, ATP sulfurylase, glutathione S-transferase, Cyt P450, and polyubiquitin (Buchanan-Wollaston, 1997; Weaver et al., 1997). Some identified cDNA clones have no obvious senescence-related function and other senescence-induced clones remain unidentified.
While the initiation of leaf senescence depends upon the age of the leaf and the reproductive phase of the plant, external factors such as nutrient deficiency, pathogenic attack, drought, light limitation, and temperature can induce premature senescence (Smart, 1994). Researchers have begun to examine the similarities and differences in gene expression during natural senescence, hormone treatment, and stress by measuring the induction of senescence-related genes (Becker and Apel, 1993; Oh et al., 1996; Chung et al., 1997; Park et al., 1998; Weaver et al., 1998). Studies with ABA, ethylene, cytokinin, methyl jasmonate, wounding, dehydration, and dark treatment have shown that these genes are differentially regulated, suggesting that there are multiple signaling pathways leading to their induction (Gan and Amasino, 1997; Park et al., 1998; Weaver et al., 1998). Expression of some senescence-related genes appears to be quite specific to natural senescence, whereas other transcripts are induced by treatments in addition to natural senescence (Weaver et al., 1998).
Ozone (O3) is a stress known to induce accelerated foliar senescence in many plant species including potato, radish, alfalfa, wheat, and hybrid poplar (Pell and Pearson, 1983; Reich, 1983; Held et al., 1991; Nie et al., 1993; Pell et al., 1997). O3 exposure accelerates chlorophyll and protein loss and reduces photosynthetic capacity and efficiency in older leaves (Reich, 1983; Held et al., 1991; Nie et al., 1993). Accelerated loss of Rubisco protein is also closely associated with O3-induced senescence (Pell and Pearson, 1983; Nie et al., 1993; Pell et al., 1997). O3 exposure reduced transcript levels for cab, rbcS, and rbcL (large subunit of Rubisco) in potato (Glick et al., 1995) and cab and rbcS in Arabidopsis (Conklin and Last, 1995) and tobacco (Bahl and Kahl, 1995). Accelerated yellowing of older leaves occurred in Arabidopsis plants following exposure to 0.10 to 0.15 μL L−1 O3 given continuously for 2 d (Kubo et al., 1995). Exposure to 0.15 μL L−1 O3 for 8 d reduced Arabidopsis rosette dry weight by 44% and reduced total chlorophyll, carotenoids, Rubisco activity, and levels of Rubisco large and small subunits (Rao et al., 1995). These results demonstrate that O3 induces changes associated with natural senescence in many species including Arabidopsis. While a decline in message level for photosynthetic genes has been observed during O3-induced accelerated leaf senescence, other molecular changes known to occur during natural senescence have not, to our knowledge, been reported.
The main objective of this study was to determine whether O3 exposure regulates the expression of SAGs. The expression pattern of SAG12 and SAG13 was determined by fluorometric quantification of GUS activity in transgenic Arabidopsis carrying either the SAG12 promoter-GUS or the SAG13 promoter-GUS fusion. Expression levels for SAG12 and SAG13 were also characterized by northern analysis. The spatial distribution of SAG13 expression was determined by staining for GUS activity in O3-treated and control transgenic SAG13-GUS plants. The expression patterns of 10 additional senescence-related genes were characterized by northern analysis in relation to the decline in PAG expression. SAG transcript levels were also analyzed following removal of the O3 treatment to determine whether transcript levels remained elevated or returned to control levels.
MATERIALS AND METHODS
Plant Growth and O3 Exposure Experiments
Seeds of Arabidopsis ecotype Lansberg erecta transformed with the SAG12 promoter-GUS fusion or SAG13 promoter-GUS fusion, were provided by S. Gan and Richard Amasino (University of Wisconsin, Madison). SAG12-GUS, SAG13-GUS, and wild-type Lansberg erecta seeds were planted on a commercial soil mix (Redi-earth Plug and Seedling Mix, Scotts-Sierra, Marysville, OH) supplemented with 20:20:20 fertilizer (Peters Professional, Scotts-Sierra) and imbibed overnight at room temperature. Seeds were placed in 4°C for four nights and then transferred to growth chambers to ensure uniform timing of germination. The plants were grown in growth chambers (Environmental Growth Chambers, Chagrin Falls, OH) at 23°C and 60% RH under a 12-h light/dark cycle at 200 μmol m−2 s−1. Seedlings germinated within 2 d and were thinned to a single plant per cell pack.
Plants were treated with O3 at 15 d post germination, when the fifth leaf, as counted by order of emergence from the meristem (cotyledons were not counted), was 3 to 4 d old. Half of the plants were exposed to 0.15 μL L−1 O3 for 6 h d−1 and the other half remained nontreated in another growth chamber. O3 was generated by passing oxygen through an ozonator (OREC V1-0, Ozone Research and Equipment, Phoenix), and O3 concentrations in the growth chamber were monitored continuously with a UV photometric O3 analyzer (model 49, Thermo Environmental Instruments, Franklin, MA). In experiments 1 and 2, plants were exposed to O3 for 8 and 14 consecutive d, respectively. GUS activity was analyzed every 2 d in the fifth and sixth leaves harvested from four replicate SAG12-GUS and SAG13-GUS transgenic plants per treatment. For staining of GUS activity, the fifth and sixth leaves were collected from three replicate SAG13-GUS transgenic plants per treatment per sampling time. Leaves for GUS staining were harvested on d 3, 6, and 8 in experiment 1, and on d 4, 8, 12, and 14 in experiment 2. For northern analysis, three replicate samples of wild-type plants were collected per treatment every 2 d; each sample consisted of the fifth and sixth leaves pooled from six plants. In addition, one wild-type rosette was collected at each sampling time per treatment in experiment 2.
In a third experiment, wild-type plants were exposed to O3 for 10 consecutive d. Two replicate samples of the fifth and sixth leaves pooled from six plants were collected per treatment at the end of the 6-h exposure and 18 h later, on d 6, 8, and 10 of the O3 exposure. The samples were analyzed for SAG transcript levels.
GUS Activity Assays
For fluorometric quantification of GUS activity, samples were ground in microcentrifuge tubes under liquid nitrogen. Leaf tissue was lysed in 150 to 200 μL of extraction buffer (50 mm sodium phosphate buffer, pH 7.0, 10 mm EDTA, 0.1% [v/v] Triton X-100, 0.1% [w/v] N-lauroylsarcosine, and 10 mm β-mercaptoethanol) and stored at −80°C for later analysis (Jefferson et al., 1987). Following centrifugation of the crude extract, 50 μL was incubated at 37°C in 500 μL of assay buffer (2 mm 4-methylumbelliferyl β-d-glucuronide in extraction buffer). At 1-h intervals, 100-μL aliquots were removed and the reaction was stopped with 900 μL of 0.2 m Na2CO3. Fluorescence of the methyl umbelliferone product was quantified with a fluorometer (CytoFluor II multi-well plate reader, PE Biosystems). Protein concentrations were measured with the protein-dye-binding assay (Bradford, 1976) using Coomassie Plus protein assay reagent (Pierce) with BSA as a standard.
For staining of GUS activity, leaves were vacuum infiltrated with 50 mm sodium phosphate buffer, pH 7.0, 1 mm EDTA, 0.01% (v/v) Triton X-100, and 1 mm 5-bromo-4-chloro-3-indolyl β-d-glucuronide (Gold BioTechnology, St. Louis) (Jefferson et al., 1987; Thoma et al., 1996). Leaves were incubated at 37°C until blue staining became evident, 72 h after infiltration. Following staining, leaves were cleared of chlorophyll with 70% (v/v) ethanol.
RNA Extraction and Analysis
Northern analysis was conducted with the probes listed in Table I. Leaf tissue was ground under liquid nitrogen and total RNA was extracted from 100 mg of tissue (RNeasy, Qiagen, Chatsworth, CA). Total RNA was fractionated in a 1% (w/v) agarose-formaldehyde gel, transferred to a membrane (Hybond-N, Amersham), and fixed to the membrane by baking for 2 h at 80°C. The membranes were prehybridized in 0.5 m sodium phosphate buffer and 7% (w/v) SDS at 65°C for 1 h (Church and Gilbert, 1984). Probes were random-primed labeled with [α-32P]dCTP and unincorporated nucleotides were removed with spin columns (Quick Spin, Boehringer Mannheim). The membranes were hybridized overnight at 65°C. Following hybridization, the membranes were washed at 65°C twice in 40 mm sodium phosphate buffer, 5% SDS, and 1 mm EDTA for 20 min, and twice in 40 mm sodium phosphate buffer, 1% SDS, and 1 mm EDTA for 20 min. Membranes were exposed to film (X-Omat, Kodak) at −80°C with two intensifying screens. Membranes were stripped with boiling 0.1% SDS for rehybridizing with other probes. The final hybridization on each membrane was performed with cDNA for pea rRNA as a loading check (Jorgenson et al., 1982).
Table I.
SAGs used in the study of O3-induced accelerated leaf senescence
| Gene | Identity/Similarity | Reference | Time of Induction |
|---|---|---|---|
| d of O3 exposure | |||
| SAG12 | Cys protease | Lohman et al. (1994) | NIa |
| Gan and Amasino (1997) | |||
| SAG13 | Short-chain alcohol dehydrogenase | Lohman et al. (1994) | 2 –4 |
| Weaver et al. (1997) | |||
| BCB (SAG14) | Blue copper-binding protein (membrane) | Van Gysel et al. (1993) | 2 –4 |
| Lohman et al. (1994) | |||
| Weaver et al. (1997) | |||
| ERD1 (SAG15) | ClpC-like gene (chloroplast) | Kiyosue et al. (1993) | 2 –4 |
| Lohman et al. (1994) | |||
| Weaver et al. (1998) | |||
| MT1 (SAG17) | Metallothionein | Zhou and Goldsbrough (1994) | NI |
| Lohman et al. (1994) | |||
| Weaver et al. (1997) | |||
| SAG18 | Novel gene | Weaver et al. (1998) | 4 –6 |
| SAG19 | Unidentified | L.M. Weaver and R.M. Amasino (personal communication) | NI |
| SAG20 | Novel gene | Weaver et al. (1998) | 4 –6 |
| SAG21 | Late embryogenesis-abundant gene | Weaver et al. (1998) | 2 –4 |
| CCH | Copper chaperone | Himelblau et al. (1998) | 6 –8 |
| Atgsr2 | Glutamine synthetase (cytosol) | Peterman and Goodman (1991) | NI |
| Bernhard and Matile (1994) | |||
| ACS6 | ACC synthase | Vahala et al. (1998) | 4 –6 |
| Arteca and Arteca (1999) |
Selected references include information on clone identification and expression patterns.
NI, Not induced by 14-d O3 exposure.
RESULTS
Arabidopsis plants exhibited downward leaf rolling after 4 d of treatment with 0.15 μL L−1 O3. O3 treatment reduced rosette leaf growth and accelerated the yellowing of older leaves. The fifth leaf began to show signs of senescence after 10 d of O3 exposure, whereas control leaves did not begin to show signs of senescence until d 14, the last day of the experiment. In an independent experiment, chlorophyll levels per unit area declined more rapidly in O3-treated leaves (data not shown). These changes in growth and development occurred without any visible signs of hypersensitive-response-like necrosis.
Effects of O3 Exposure on SAG12 and SAG13 Expression
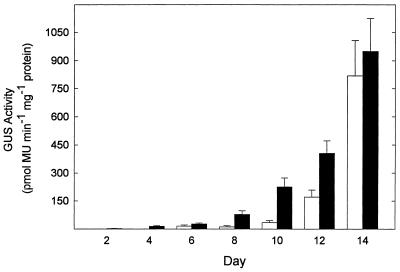
In experiment 1, the O3 exposure was 8 d in duration, and in experiment 2 the exposure was for 14 d. As the results in experiment 1 were supported in experiment 2, only the more extensive data of the latter experiment are presented here. SAG12 promoter-driven GUS activity was not detected in control or O3-treated plants on any sampling day throughout the 14 d of the experiment (data not shown), while O3 exposure did accelerate the onset of SAG13 promoter-driven GUS activity (Fig. 1). O3-induced, SAG13 promoter-driven GUS activity was first detected on d 2, whereas GUS activity was not detected until d 6 in control leaves. GUS activity gradually increased in O3-treated and control leaves through the remainder of the experiment. SAG13 promoter-driven GUS activity in O3-treated leaves always exceeded the level found in control leaves, except on d 14, when the difference between treatments was no longer detected (Fig. 1). SAG13 promoter-driven GUS activity appeared after 2 d in O3-treated leaves, while yellowing did not occur on the fifth leaf until d 10. No SAG12 or SAG13 promoter-driven GUS activity was detected in treated or control nontransformed plants.
Figure 1.
SAG13 promoter-driven GUS activity was induced by O3 treatment. Fifteen-day-old Arabidopsis ecotype Landsberg erecta plants transformed with the SAG13 promoter-GUS fusion were exposed to 0.15 μL L−1 O3 for 6 h d−1 for 14 d. The fifth and sixth leaves were harvested from a single plant and GUS activity was measured by fluorometric quantification of 4-methyl umbelliferone (MU). Black bars, O3-Treated leaves; white bars, nontreated leaves. Each bar represents the mean of four samples ± se, except control bars on d 2 and 10, where the mean of three samples was taken. No GUS activity was detected in nontransformed plants (data not shown).
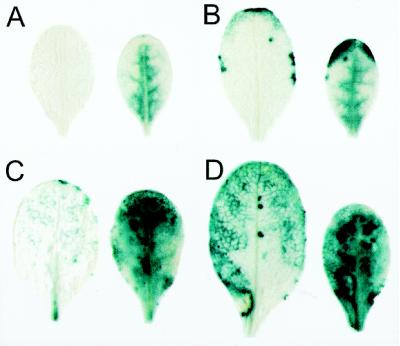
The localization of SAG13 expression was determined by staining for GUS activity (Fig. 2). The staining pattern was altered spatially and temporally by O3 treatment. GUS staining was diffusely distributed in the interior of O3-treated leaves on d 4, while no staining could be detected in control leaves. By d 8 of the experiment, intense GUS staining was present at the leaf margin and interior of O3-treated leaves. In control leaves, staining was localized to discrete areas along the margins, with some faint and variable staining at the leaf tip. Following d 12 and 14, O3-treated leaves showed intense blue staining throughout the entire leaf, and control leaves began to show stronger staining in the leaf interior as senescence progressed from the margins inward.
Figure 2.
Photographs showing O3-induced GUS staining in the fifth leaf of transgenic SAG13-GUS plants. Fifteen-day-old SAG13-GUS plants were exposed to 0.15 μL L−1 O3 for 6 h d−1 for 14 d. Leaves were vacuum infiltrated with 1 mm 5-bromo-4-chloro-3-indolyl β-d-glucuronide, incubated at 37°C for 72 h, and cleared of chlorophyll with 70% ethanol. Nontreated leaves are shown on the left and O3-treated leaves on the right from samples harvested 4, 8, 12, and 14 d after exposure in A through D, respectively. A similar pattern of expression was found in the sixth leaf (data not shown). The leaves shown are representative of three leaves per treatment per day.
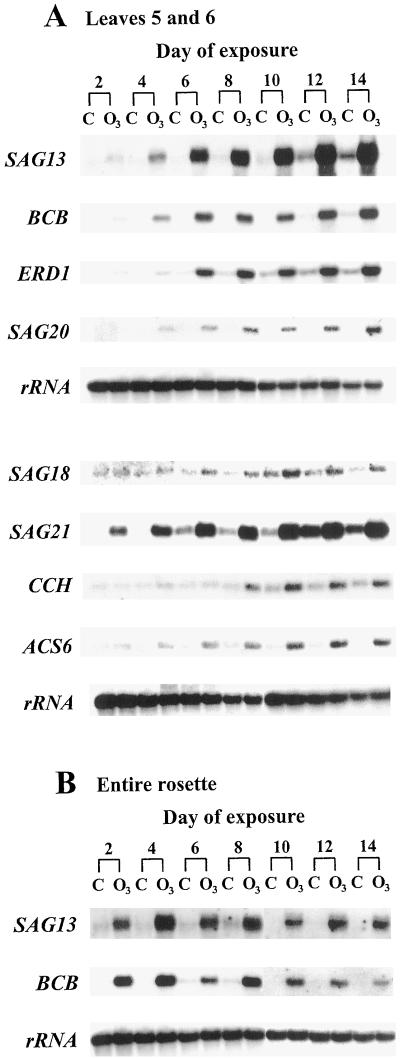
The effect of O3 exposure on SAG12 and SAG13 expression was also determined by northern analysis. Increased abundance of the SAG13 transcript was detected after 2 to 4 d of O3 exposure (Fig. 3), whereas the SAG12 transcript remained undetectable in O3-treated and nontreated leaves and rosettes on all sampling days (Fig. 4). These results support the GUS activity data obtained from SAG12-GUS and SAG13-GUS transgenic leaves (Fig. 1). SAG13 transcript levels gradually increased in O3-treated leaves five and six at later time points and did not appear in control leaves until d 10 to 12 (Fig. 3A). SAG13 transcript levels in entire rosettes did not show this gradual increase in abundance, yet levels did remain elevated in O3-treated rosettes compared with nontreated rosettes (Fig. 3B). The SAG13 transcript was always more abundant in O3-treated leaves than in control leaves (Fig. 3). In contrast, SAG13 promoter-driven GUS activity was similar in O3-treated and control leaves on d 14 (Fig. 1). This discrepancy may be due to the long half-life of GUS, which is approximately 50 h in living mesophyll protoplasts (Jefferson et al., 1987).
Figure 3.
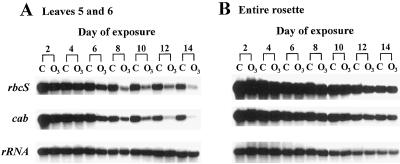
Induction of senescence-related transcripts in O3-treated Arabidopsis plants. Fifteen-day-old plants were exposed to 0.15 μL L−1 O3 for 6 h d−1 or remained nontreated. Total RNA was extracted and 3 μg of RNA was separated on 1% formaldehyde-agarose gels, transferred to membranes, and hybridized with the radiolabeled probes indicated. A, Each lane contains RNA extracted from the fifth and sixth leaves pooled from six plants. The samples shown are one representative replicate from a total of three. B, Each lane contains RNA extracted from one rosette and only one replicate was analyzed. C, Control, nontreated plants; O3, O3-treated plants.
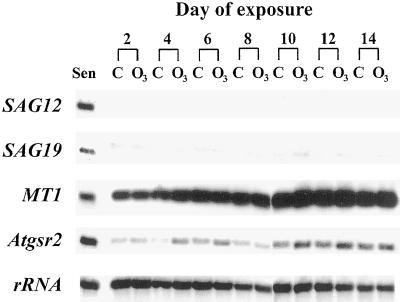
Figure 4.
SAG12, SAG19, MT1, and Atgsr2 transcript levels were not altered by O3 treatment. Fifteen-day-old Arabidopsis plants were exposed to 0.15 μL L−1 O3 for 6 h d−1 or remained nontreated. Samples were prepared as in Figure 3. Each lane contains RNA extracted from the fifth and sixth leaves pooled from six plants. The samples shown are one representative replicate from a total of three. Sen, RNA sample extracted from yellowing (senescent) leaves older than 30 d; C, control, nontreated plants; O3, O3-treated plants.
Effects of O3 Exposure on PAG and SAG Expression
Transcript levels for the PAGs rbcS and cab showed a strong reduction in the fifth and sixth leaves after 6 d of O3 exposure (Fig. 5A). PAG transcript levels continued to decline gradually throughout the remainder of the experiment. Only a slight decline in PAG mRNA levels was found in control leaves (Fig. 5A). The O3-induced decline in PAG expression, as found in the fifth and sixth leaves, was not readily detectable in RNA samples extracted from entire rosettes (Fig. 5B). PAG transcript levels declined with age in both O3-treated and nontreated rosettes.
Figure 5.
PAG transcript levels declined after treatment with O3. Fifteen-day-old Arabidopsis plants were exposed to 0.15 μL L−1 O3 for 6 h d−1 or remained nontreated. Samples were prepared as in Figure 3. A, Each lane contains RNA extracted from the fifth and sixth leaves pooled from six plants. The samples shown are one representative replicate from a total of three. B, Each lane contains RNA extracted from one rosette and only one replicate was analyzed. C, Control, nontreated plants; O3, O3-treated plants.
SAG expression levels were determined in three replicate samples, and the range of days given for the time of induction represents the variability within these samples. SAG13, SAG21, BCB (blue copper-binding protein), and ERD1 (early responsive to dehydration) were induced in the fifth and sixth leaves between d 2 and 4 of O3 treatment (Fig. 3A), prior to any detectable decline in PAG transcript levels. SAG18, SAG20, and ACS6 (ACC synthase) were induced between d 4 and 6 and CCH (copper chaperone) was induced between d 6 and 8 of the O3 treatment in the fifth and sixth leaves (Fig. 3A). Transcripts for all of these genes continued to accumulate throughout the 14 d of exposure. Transcripts for most of these genes were detected in control leaves, but did not appear until later and levels remained below those found in O3-treated samples. The SAG21 transcript was detected in the fifth and sixth control leaves on d 6; ERD1 between d 8 and 10; SAG13, SAG18, and CCH between d 10 and 12; and BCB between d 12 and 14 (Fig. 3A). SAG20 and ACS6 did not show any appreciable accumulation in the fifth and sixth control leaves during the experimental period (Fig. 3A). Transcript levels for SAG13 and BCB were greater in O3-treated rosettes compared with nontreated rosettes; however, transcript accumulation throughout the 14 d of exposure, as found for leaves five and six, was not detected in rosettes (Fig. 3B). Similar results were obtained for SAG21 and ERD1 transcript levels in O3-treated rosettes and SAG18, SAG20, CCH, and ACS6 transcript levels were more abundant in O3-treated rosettes on only some of the harvest days (data not shown).
Not all of the characterized SAGs were induced by O3 treatment. SAG12, SAG19, MT1 (metallothionein), and Atgsr2 (glutamine synthetase) were not induced by O3 treatment during the 8-d exposure in experiment 1 (data not shown). The O3 exposure period in experiment 2 was extended for a total of 14 d to determine if the expression of these SAGs could be induced with a longer O3 treatment. SAG12, SAG19, MT1, and Atgsr2 were not induced to any measurable degree during the 14-d exposure (Fig. 4). MT1 and Atgsr2 transcripts were present in all samples and transcript levels gradually increased in abundance equally in O3-treated and control leaves. Slightly greater signals for the MT1 and Atgsr2 transcripts were found in a few of the O3-treated samples, but this response was not consistent. SAG12 transcript was not detected in any sample and SAG19 transcript remained nearly undetectable (Fig. 4). RNA was extracted from partially yellow leaves harvested from nontreated plants older than 30 d post germination and was included on membranes to demonstrate that SAG12 and SAG19 transcripts could be detected (Fig. 4).
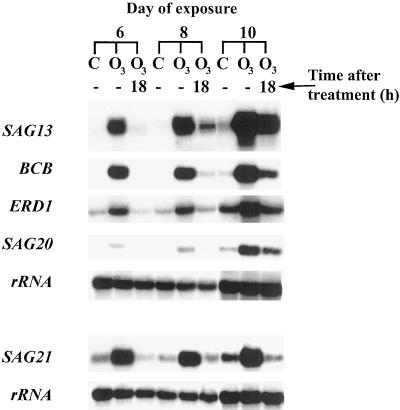
The ability of O3-treated leaves to recover from the accelerated induction of SAGs was investigated by analyzing transcript levels following removal of O3 on d 6, 8, and 10 of the exposure. The fifth and sixth leaves were harvested from plants immediately following the daily 6-h O3 treatment and from another set of plants allowed to recover from the treatment for an additional 18 h in O3-free air. Transcript levels for SAG13, BCB, ERD1, SAG20, and SAG21 were greater in leaves treated with 6 h of O3 on d 6, 8, and 10 than in nontreated leaves (Fig. 6). Transcript levels for these genes declined following 18 h in O3-free air. On d 6, SAG transcripts were nearly undetectable in control leaves, but were induced in O3-treated leaves. Following 18 h in O3-free air, transcript levels were undetectable in O3-treated samples. On d 8, transcript levels remained nearly undetectable in control leaves, but were induced in O3-treated samples and once again declined in O3-treated samples following the 18-h period. By d 10, SAG transcripts were detected in controls and O3-treated samples. The decline in transcript level 18 h after the removal of O3 was still apparent.
Figure 6.
SAG13, BCB, ERD1, SAG20, and SAG21 transcript levels declined following a recovery period in O3-free air. Fifteen-day-old Arabidopsis plants were exposed to 0.15 μL L−1 O3 for 6 h d−1 or remained nontreated. The fifth and sixth leaves were harvested from six plants immediately following 6 h of exposure to O3 or 18 h after removal of O3. Samples were prepared as in Figure 3. The samples shown are one of two replicates. C, Control, nontreated plants; O3, O3-treated plants.
DISCUSSION
In the present study, chronic O3 treatment accelerated the normal rate of foliar senescence in Arabidopsis plants. This response occurred in the absence of the necrosis observed in response to higher O3 concentrations reported previously for other species (Pell et al., 1997). O3-induced leaf yellowing in Arabidopsis was previously observed in older leaves exposed to O3 continuously for 2 d (Kubo et al., 1995). Rosette growth was reduced and downward leaf curling was evident within 4 d of O3 exposure, similar to results obtained by Sharma and Davis (1994) and Rao et al. (1995). Leaf curling appeared to be an altered growth response and was not the result of dehydration, since the percent dry matter did not vary between treatments in an independent experiment (data not shown). A suite of O3-induced changes in transcript levels were observed, including reductions in levels of PAGs and increased levels of many but not all SAGs measured (Table I; Figs. 3–5). These changes were only expressed in leaves of a discrete developmental age. Hence, observations of O3-induced decline in PAG transcript levels, for example, were observed in the fifth and sixth leaves but were not detected when whole rosettes were analyzed (Fig. 5).
Similarities and Contrasts to Natural Senescence
O3 induces many changes common to natural senescence, but at an accelerated rate: for example, loss of total protein, Rubisco, chlorophyll, and increased leaf abscission (Pell and Pearson, 1983; Reich, 1983; Held et al., 1991; Nie et al., 1993). Diminishing rbcS and cab transcript levels are indicators of declining photosynthetic activity during natural senescence; the observation in this experiment that O3 treatment reduced the level of these transcripts was supported by previous investigations (Bahl and Kahl, 1995; Conklin and Last, 1995; Glick et al., 1995). Transcript levels for two other genes, SDG1 (senescence-down-regulated gene) and SDG2, declined during the O3 exposure with an expression pattern similar to rbcS and cab (data not shown). SDG1 and SDG2 showed reduced transcript abundance in a differential screen of nonsenescent versus senescent leaves (Lohman et al., 1994).
O3 treatment induced the early expression of many molecular markers of senescence, providing additional evidence that changes in gene expression during chronic O3 treatment are similar to natural senescence. Two metal-binding proteins, CCH (copper chaperone) and BCB (blue copper-binding protein), are among the genes induced by O3. These genes were previously shown to be induced by acute O3 exposure; BCB was induced within a 3-h exposure to 0.30 μL L−1 O3 (Richards et al., 1998) and CCH transcript levels increased by 30% after a 30-min exposure to 0.80 μL L−1 O3 (Himelblau et al., 1998). Metal-binding proteins may play an important role in metal remobilization during senescence. O3 treatment also induced transcript accumulation of a protease regulator, ERD1; proteases are involved in protein degradation during natural senescence and may be further required for degradation of oxidized proteins during O3-induced accelerated senescence. Transcript accumulation of other genes, including SAG13, SAG18, SAG20, and SAG21, was also induced by O3 treatment; the function of these genes in senescence remains unclear. While O3 induced the buildup of SAG transcripts, it is not known how this translates into accumulation of the protein products.
Transcripts for SAG12, SAG19, Atgsr2, and MT1 accumulate during natural senescence, but were not induced by chronic O3 treatment. These genes may lack responsive elements able to recognize O3-induced signaling compounds. Proteases other than the Cys protease SAG12 may have been available for proteolysis and adequate quantities of Gln synthetase, Atgsr2, and metallothionein, MT1, may have been present due to high basal transcript levels. If all senescence-related genes play critical roles in cellular degradation and nutrient remobilization during natural senescence, the lack of these gene products during O3-induced accelerated senescence may reduce the efficiency of nutrient recovery.
Specific and perhaps premature induction of gene expression in response to O3 is reminiscent of molecular changes in response to other stresses (Weaver et al., 1997). Genes induced during chronic O3 exposure have also been shown to be induced by darkness, dehydration, and treatment with ethylene or ABA. Dark treatment induced the O3-responsive genes, ERD1, BCB, and SAG20, dehydration induced ERD1, BCB, SAG20, and SAG21, ethylene treatment induced ERD1, BCB, SAG13, SAG20, and SAG21, and ABA treatment induced ERD1 and SAG13 (Kiyosue et al., 1993; Nakashima et al., 1997; Weaver et al., 1998). The overlap in gene expression suggests that O3 treatment, darkness, and dehydration may induce similar signaling molecules. Ethylene and ABA may play a role as signals during O3-induced accelerated leaf senescence, as discussed below.
In addition to affecting the timing of induction of some SAGs, O3 also seems to influence the spatial distribution of that induction. SAG13-promoter driven GUS activity first appeared at the leaf margin in control leaves, which resembles the pattern of yellowing found in naturally senescing leaves (Weaver et al., 1998). In contrast, O3 treatment induced SAG13 expression throughout the leaves. This distribution of SAG13 expression probably coincided with regions where O3 entered through open stomata.
Potential Signals of Molecular Events
Elevated SAG13, SAG20, SAG21, BCB, and ERD1 transcript levels in O3-treated leaves were not sustained following the removal of O3. Daily O3 exposures were required to provide a signal to maintain enhanced SAG transcript levels, suggesting that the leaves may retain some ability to recover from exposure to O3. A similar recovery was shown for rbcS and cab transcripts in Arabidopsis following a 24-h O3-free period after treatment with 0.175 μL L−1 O3 for 8 h d−1 for 4 d (Conklin and Last, 1995).
Since O3 treatment induced premature changes in transcript levels of genes associated with natural senescence, O3 may elicit some of the same signals involved in natural senescence. The common mechanism regulating O3-induced accelerated leaf senescence and natural leaf senescence may involve reactive oxygen species. Oxidative stress has long been associated with senescence (Thompson et al., 1987), and recently this link was shown in the late-flowering (or extended longevity) Arabidopsis mutant gigantea (gi-3), which exhibited enhanced tolerance to methyl viologen-induced oxidative stress (Kurepa et al., 1998). Following stomatal uptake of O3, internal O3 concentrations rapidly drop (Laisk et al., 1989) as decomposition products, including reactive oxygen species, are formed. These reactive oxygen species can react with membrane lipids to produce more reactive oxygen intermediates. A second sustained peak of reactive oxygen species was found in the O3-sensitive tobacco cv Bel W3 following O3 exposure, and was not found in the O3-tolerant cv Bel B (Schraudner et al., 1998). An O3-responsive region in the stilbene synthase promoter has been identified (Schubert et al., 1997), and a comparison of this 150-bp region with the SAG13 promoter (S. Gan and R.M. Amasino, personal communication) did not reveal any strong sequence similarity (data not shown). The presence of O3 or reactive oxygen species responsive elements in SAGs is worthy of future investigation.
Alternatively, the O3-induced changes in gene expression could have been induced through a secondary signal. Ethylene treatment induces many of the O3-responsive SAGs (Weaver et al., 1998), and plants exposed to high doses of O3 produce large quantities of ethylene (Pell et al., 1997). Ethylene has been shown to regulate the timing of leaf senescence in Arabidopsis (Grbic and Bleecker, 1995). In our experiments, ACS6, one member of the gene family encoding ACC synthase in Arabidopsis, was detected within 4 to 6 d of O3 exposure. This gene is induced by many stresses, including O3, while ACS1, ACS2, ACS4, and ACS5 are not induced by O3 treatment (Vahala et al., 1998; Arteca and Arteca, 1999). At high O3 concentrations, ethylene emission is one of the first responses and is correlated with the degree of lesion formation (Tuomainen et al., 1997). The importance of ethylene in regulating the response to low O3 concentrations in the induction of accelerated leaf senescence remains to be determined. We are currently investigating the need for ethylene perception in the induction of this suite of SAGs.
Other potential signaling molecules include ABA, salicylic acid, and calcium. ABA is another senescence-promoting hormone, and some of the O3-responsive SAGs are inducible by ABA treatment (Weaver et al., 1998). Salicylic acid and calcium increase during exposure to high O3 concentrations and are involved in the induction of antioxidant gene expression (Yalpani et al., 1994; Sharma and Davis, 1997; Clayton et al., 1999); however, it is not known whether they are involved in the response to chronic O3.
In conclusion, chronic O3 treatment induced SAG expression while suppressing PAG expression. An initial pattern of senescence-related gene induction by O3 has emerged. Future experiments should focus on determining which genes are essential for the induction of O3-induced accelerated leaf senescence and what, if any, interdependency exists between these genes. Further investigation will determine the identity of signals required for O3-induced accelerated leaf senescence and elucidate the role of oxidative stress in the progression of natural leaf senescence.
ACKNOWLEDGMENTS
The authors thank Richard Amasino for the generous gift of the SAG clones and the Arabidopsis Biological Resource Center (Columbus, OH) for the Atgsr2 clone, stock no. CD3-195, donated by T.K. Peterman. We are also grateful to Ed Himelblau and Michael Weaver for helpful discussions and Nan Eckardt and Judy Sinn for critical reading of the manuscript.
Abbreviations:
- PAG
photosynthesis-associated gene
- SAG
senescence-associated gene
Footnotes
External funding for this research was provided by the Environmental Protection Agency (grant no. U915212–01–1) and by the U.S. Department of Agriculture (grant no. 93–38420–8742). This research was also supported in part by the Pennsylvania Agricultural Experiment Station and the Environmental Resources Research Institute. It is contribution no. 2064 from the Department of Plant Pathology, The Pennsylvania State University.
LITERATURE CITED
- Arteca JM, Arteca RN. A multi-responsive gene encoding 1-aminocyclopropane-1-carboxylate synthase (ACS6) in mature Arabidopsis leaves. Plant Mol Biol. 1999;39:209–219. doi: 10.1023/a:1006177902093. [DOI] [PubMed] [Google Scholar]
- Bahl A, Kahl G. Air pollutant stress changes the steady-state transcript levels of three photosynthesis genes. Environ Pollut. 1995;88:57–65. doi: 10.1016/0269-7491(95)91048-p. [DOI] [PubMed] [Google Scholar]
- Bate NJ, Rothstein SJ, Thompson JE. Expression of nuclear and chloroplast photosynthesis-specific genes during leaf senescence. J Exp Bot. 1991;42:801–811. [Google Scholar]
- Becker W, Apel K. Differences in gene expression between natural and artificially induced leaf senescence. Planta. 1993;189:74–79. [Google Scholar]
- Bernhard WR, Matile P. Differential expression of glutamine synthetase genes during the senescence of Arabidopsis thaliana rosette leaves. Plant Sci. 1994;98:7–14. [Google Scholar]
- Blank A, McKeon TA. Expression of three RNase activities during natural and dark-induced senescence of wheat leaves. Plant Physiol. 1991;97:1409–1413. doi: 10.1104/pp.97.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V. The molecular biology of leaf senescence. J Exp Bot. 1997;48:181–199. [Google Scholar]
- Chung BC, Lee SY, Oh SA, Rhew TH, Nam HG, Lee CH. The promoter activity of sen1, a senescence-associated gene of Arabidopsis, is repressed by sugars. J Plant Physiol. 1997;151:339–345. [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton H, Knight MR, Knight H, McAinsh MR, Hetherington AM. Dissection of the ozone-induced calcium signature. Plant J. 1999;17:575–579. doi: 10.1046/j.1365-313x.1999.00411.x. [DOI] [PubMed] [Google Scholar]
- Conklin PL, Last RL. Differential accumulation of antioxidant mRNAs in Arabidopsis thaliana exposed to ozone. Plant Physiol. 1995;109:203–212. doi: 10.1104/pp.109.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan S, Amasino RM. Making sense of senescence: molecular genetic regulation and manipulation of leaf senescence. Plant Physiol. 1997;113:313–319. doi: 10.1104/pp.113.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick RE, Schlagnhaufer CD, Arteca RN, Pell EJ. Ozone-induced ethylene emission accelerates the loss of ribulose1,5-bisphosphate carboxylase/oxygenase and nuclear-encoded mRNAs in senescing potato leaves. Plant Physiol. 1995;109:891–898. doi: 10.1104/pp.109.3.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbic V, Bleecker AB. Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 1995;8:595–602. [Google Scholar]
- Held AA, Mooney HA, Gorham JN. Acclimation to ozone stress in radish: leaf demography and photosynthesis. New Phytol. 1991;118:417–423. [Google Scholar]
- Himelblau E, Mira H, Lin SJ, Culotta VC, Peñarrubia L, Amasino RM. Identification of a functional homolog of the yeast copper homeostasis gene ATX1 from Arabidopsis. Plant Physiol. 1998;117:1227–1234. doi: 10.1104/pp.117.4.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgenson RA, Cuellar RE, Thompson WF. Modes and tempos in the evolution of nuclear encoded ribosomal RNA genes in legumes. Carnegie Inst Wash Year Book. 1982;81:98–101. [Google Scholar]
- Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. Characterization of cDNA for a dehydration-inducible gene that encodes a CLP A, B-like protein in Arabidopsis thaliana L. Biochem Biophys Res Commun. 1993;196:1214–1220. doi: 10.1006/bbrc.1993.2381. [DOI] [PubMed] [Google Scholar]
- Kubo A, Saji H, Tanaka K, Kondo N. Expression of Arabidopsis cytosolic ascorbate peroxidase gene in response to ozone or sulfur dioxide. Plant Mol Biol. 1995;29:479–489. doi: 10.1007/BF00020979. [DOI] [PubMed] [Google Scholar]
- Kurepa J, Smalle J, Van Montagu M, Inzé D. Oxidative stress tolerance and longevity in Arabidopsis: the late-flowering mutant gigantea is tolerant to paraquat. Plant J. 1998;14:759–764. doi: 10.1046/j.1365-313x.1998.00168.x. [DOI] [PubMed] [Google Scholar]
- Laisk A, Kull O, Moldau H. Ozone concentration in leaf intercellular air spaces is close to zero. Plant Physiol. 1989;90:1163–1167. doi: 10.1104/pp.90.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman KN, Gan S, John MC, Amasino RM. Molecular analysis of natural leaf senescence in Arabidopsis thaliana. Physiol Plant. 1994;92:322–328. [Google Scholar]
- Nakashima K, Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. A nuclear gene, erd1, encoding a chloroplast-targeted Clp protease regulatory subunit homolog is not only induced by water stress but also developmentally up-regulated during senescence in Arabidopsis thaliana. Plant J. 1997;12:851–861. doi: 10.1046/j.1365-313x.1997.12040851.x. [DOI] [PubMed] [Google Scholar]
- Nie GY, Tomasevic M, Baker NR. Effects of ozone on the photosynthetic apparatus and leaf proteins during leaf development in wheat. Plant Cell Environ. 1993;16:643–651. [Google Scholar]
- Oh SA, Lee SY, Chung IK, Lee CH, Nam HG. A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol Biol. 1996;30:739–754. doi: 10.1007/BF00019008. [DOI] [PubMed] [Google Scholar]
- Park JH, Oh SA, Kim YH, Woo HR, Nam HG. Differential expression of senescence-associated mRNAs during leaf senescence induced by different senescence-inducing factors in Arabidopsis. Plant Mol Biol. 1998;37:445–454. doi: 10.1023/a:1005958300951. [DOI] [PubMed] [Google Scholar]
- Pell EJ, Pearson NS. Ozone-induced reduction in quantity of ribulose-1,5-bisphosphate carboxylase in alfalfa foliage. Plant Physiol. 1983;73:185–187. doi: 10.1104/pp.73.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pell EJ, Schlagnhaufer CD, Arteca RN. Ozone-induced oxidative stress: mechanisms of action and reaction. Physiol Plant. 1997;100:264–273. [Google Scholar]
- Peterman TK, Goodman HM. The glutamine synthetase gene family of Arabidopsis thaliana: light-regulation and differential expression in leaves, roots and seeds. Mol Gen Genet. 1991;230:145–154. [Google Scholar]
- Rao MV, Paliyath G, Ormrod DP. Differential response of photosynthetic pigments, Rubisco activity and Rubisco protein of Arabidopsis thaliana exposed to UVB and ozone. Photochem Photobiol. 1995;62:727–735. [Google Scholar]
- Reich PB. Effects of low concentrations of O3 on net photosynthesis, dark respiration, and chlorophyll contents in aging hybrid poplar leaves. Plant Physiol. 1983;73:291–296. doi: 10.1104/pp.73.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards KD, Schott EJ, Sharma YK, Davis KR, Gardner RC. Aluminum induces oxidative stress genes in Arabidopsis thaliana. Plant Physiol. 1998;116:409–418. doi: 10.1104/pp.116.1.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraudner M, Moeder W, Wiese C, Van Camp W, Inzé D, Langebartels C, Sandermann H. Ozone-induced oxidative burst in the ozone biomonitor plant, tobacco Bel W3. Plant J. 1998;16:235–245. doi: 10.1046/j.1365-313x.1998.00294.x. [DOI] [PubMed] [Google Scholar]
- Schubert R, Fischer R, Hain R, Schreier PH, Bahnweg G, Ernst D, Sandermann H. An ozone-responsive region of the grapevine resveratrol synthase promoter differs from the basal pathogen-responsive sequence. Plant Mol Biol. 1997;34:417–426. doi: 10.1023/a:1005830714852. [DOI] [PubMed] [Google Scholar]
- Sharma YK, Davis KR. Ozone-induced expression of stress-related genes in Arabidopsis thaliana. Plant Physiol. 1994;105:1089–1096. doi: 10.1104/pp.105.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma YK, Davis KR. The effects of ozone on antioxidant responses in plants. Free Radic Biol Med. 1997;23:470–488. doi: 10.1016/s0891-5849(97)00108-1. [DOI] [PubMed] [Google Scholar]
- Smart CM. Gene expression during leaf senescence. New Phytol. 1994;126:419–448. doi: 10.1111/j.1469-8137.1994.tb04243.x. [DOI] [PubMed] [Google Scholar]
- Thoma S, Sullivan ML, Vierstra RD. Members of two gene families encoding ubiquitin-conjugating enzymes, AtUBC1–3 and AtUBC4–6, from Arabidopsis thaliana are differentially expressed. Plant Mol Biol. 1996;31:493–505. doi: 10.1007/BF00042223. [DOI] [PubMed] [Google Scholar]
- Thompson JE, Legge RL, Barber RF. The role of free radicals in senescence and wounding. New Phytol. 1987;105:317–344. doi: 10.1111/j.1469-8137.1987.tb00871.x. [DOI] [PubMed] [Google Scholar]
- Tuomainen J, Betz C, Kangasjärvi J, Ernst D, Yin ZH, Langebartels C, Sandermann H. Ozone induction of ethylene emission in tomato plants: regulation by differential accumulation of transcripts for the biosynthetic enzymes. Plant J. 1997;12:1151–1162. [Google Scholar]
- Vahala J, Schlagnhaufer CD, Pell EJ. Induction of an ACC synthase cDNA by ozone in light-grown Arabidopsis thaliana leaves. Physiol Plant. 1998;103:45–50. [Google Scholar]
- Van Gysel A, Van Montagu M, Inze D. A negatively light-regulated gene from Arabidopsis thaliana encodes a protein showing high similarity to blue copper-binding proteins. Gene. 1993;136:79–85. doi: 10.1016/0378-1119(93)90450-h. [DOI] [PubMed] [Google Scholar]
- Weaver LM, Gan S, Quirino B, Amasino RM. A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol Biol. 1998;37:455–469. doi: 10.1023/a:1005934428906. [DOI] [PubMed] [Google Scholar]
- Weaver LM, Himelblau E, Amasino RM (1997) Leaf senescence: gene expression and regulation. In JK Setlow, ed, Genetic Engineering, Vol 19. Plenum Press, New York, pp 215–234
- Yalpani N, Enyedi AJ, León J, Raskin I. Ultraviolet light and ozone stimulate accumulation of salicylic acid, pathogenesis-related proteins and virus resistance in tobacco. Planta. 1994;193:372–376. [Google Scholar]
- Zhou J, Goldsbrough PB. Functional homologs of fungal metallothionein genes from Arabidopsis. Plant Cell. 1994;6:875–884. doi: 10.1105/tpc.6.6.875. [DOI] [PMC free article] [PubMed] [Google Scholar]