Abstract
Beneficial plant-microbe interactions offer a sustainable biological solution with the potential to boost low-input food and bioenergy production. A better mechanistic understanding of these complex plant-microbe interactions will be crucial to improving plant production as well as performing basic ecological studies investigating plant-soil-microbe interactions. Here, a detailed description for ecosystem fabrication is presented, using widely available 3D printing technologies, to create controlled laboratory habitats (EcoFABs) for mechanistic studies of plant-microbe interactions within specific environmental conditions. Two sizes of EcoFABs are described that are suited for the investigation of microbial interactions with various plant species, including Arabidopsis thaliana, Brachypodium distachyon, and Panicum virgatum. These flow-through devices allow for controlled manipulation and sampling of root microbiomes, root chemistry as well as imaging of root morphology and microbial localization. This protocol includes the details for maintaining sterile conditions inside EcoFABs and mounting independent LED light systems onto EcoFABs. Detailed methods for addition of different forms of media, including soils, sand, and liquid growth media coupled to the characterization of these systems using imaging and metabolomics are described. Together, these systems enable dynamic and detailed investigation of plant and plant-microbial consortia including the manipulation of microbiome composition (including mutants), the monitoring of plant growth, root morphology, exudate composition, and microbial localization under controlled environmental conditions. We anticipate that these detailed protocols will serve as an important starting point for other researchers, ideally helping create standardized experimental systems for investigating plant-microbe interactions.
Keywords: Environmental Sciences, Issue 134, Laboratory ecosystems, EcoFAB, plant-microbe interactions, microbiome, root morphology, root exudates, LC-MS, NIMS, metabolomics, microscopic imaging
Introduction
The application of beneficial plant microbes in agriculture offers great potential to increase sustainable food and biofuel production to provide for a growing population1,2,3,4. A significant amount of work supports the importance of plant microbiomes in plant nutrient uptake, tolerance to stresses, and resistance to disease5,6,7,8. However, it is difficult to investigate these mechanisms of plant-microbe interactions in field ecosystems due to the complexity and associated irreproducibility and inability to precisely control microbiome composition and genetics (e.g., using microbial mutants)4,9,10.
One strategy is to construct simplified model ecosystems to enable controlled, replicated laboratory experiments investigating plant-microbe interactions to generate insights that can be further tested in the field10,11,12. This concept builds on traditional approaches using plants grown in soil-filled pots or on agar slabs within greenhouses or incubators13. Although these will likely remain the most widely used approaches, they lack the ability to precisely monitor and manipulate plant growth environments. To these ends, rhizoboxes and rhizotrons represent a major improvement in the ability to study below-ground processes14,15, and, first protocols were published for analyzing rhizosphere metabolites in soil16. More recently, to enable high throughput analysis, advanced microfluidic devices13,17 such as Plant Chip18,19, RootArray20, and RootChip21, have been developed as efficient tools for plant phenotyping with micrometer-scale spatial resolution to monitor the early growth stages of the small model plant Arabidopsis thaliana in liquid flow medium. Recently, a two-layer imaging platform was described that enables root hair imaging of Arabidopsis thaliana at seedling stage with a microfluidic platform22.
Here, detailed protocols for constructing controlled laboratory devices (EcoFABs) are provided, for studying plant microbe interactions and show that they can be used to study diverse plants including Arabidopsis thaliana, Brachypodium distachyon23, the ecologically important wild oat Avena barbata, and the bioenergy crop Panicum virgatum (switchgrass). EcoFAB is a sterile plant growth platform that includes two primary components: the EcoFAB device and sterile plant-sized transparent container. The EcoFAB device is made from a polydimethylsiloxane (PDMS) manufacturing process that involves casting PDMS layers from a 3D printed plastic mold and bonding PDMS layers onto microscope slides using methods previously reported24,25. The detailed procedures of EcoFAB workflow, such as device fabrication, sterilization, seed germination, seedling transplantation, microbe inoculation/cocultivation, sample preparation, and analysis, are described in this protocol (Figure 1). Further modifications of the basic workflow are described, including the installation of computer controlled LED grow lights and the utilization of solid substrates. The utilization of imaging techniques to investigate root morphology changes, microbial colonization of roots, and mass spectroscopic imaging of root exudates are described. We anticipate that the simple, inexpensive design based on readily available materials, as well as the detailed protocols presented here, will turn the EcoFAB platform into a community resource, standardizing laboratory plant-microbiome studies.
Protocol
Caution: This protocol includes the use of hazardous chemicals, sharp objects, electrical devices, hot objects, and other hazards that may result in injury. Appropriate personal protective equipment (PPE, e.g., chemically resistant gloves, safety glasses, lab coat, long clothes, closed-toed shoes, etc.) should be worn, and the appropriate safety procedures (safety training, use of a fume hood, etc.) should be followed.
1. EcoFAB Device Fabrication: Casting PDMS Layers (Figure 2 & Figure 3)

Construct the EcoFAB molds using 3D printing techniques (the design files are available at . Each mold includes three parts: a casting frame, a featured mold base, and an insert, as shown in Figure 2. Print the mold base and insert out of rigid opaque photopolymers using a 3D plastic printer. Utilize a minimum of 100 µm resolution, and print the casting frame with acrylonitrile butadiene styrene (ABS).
Mix 40 mL of siloxane elastomer base with curing agent in a disposable 1 L container. Depending on the desired experiment (steps 2.1 and 2.2), use different ratios (v/v) of elastomer base/curing agent (5:1, 15:1, or 30:1). Proceed to steps 1.3 to 1.8 for all kinds of mixtures. Caution: Wear chemically resistant gloves, safety glasses, and other PPEs.
Place the container in a vacuum chamber for at least 30 min to remove air bubbles from the elastomer mixture.
Pour the mixture into the assembled 3D printed plastic mold (Figure 3A) and keep the mold on a heating block at 85 °C for 4 h (Figure 3B). Caution: Wear PPE to avoid burns.
Let the mold cool down for 5 min. Then pull out the insert from the mold gently (Figure 3C), and then slowly insert a utility knife between the casting frame and PDMS (the solidified elastomer mixture) to separate them (Figure 2D).
Press the mold base with PDMS up out of the casting frame (Figure 3E). Use a knife or other tools to gently separate the PDMS layer from the mold base at the edges (Figure 3F), and then slowly peel it off from the mold surface (Figure 3G).
Create inlet and outlet channels on the PDMS layers by making ~1.6 mm holes for the inlet and outlet ports with a 15 gauge blunt needle (Figure 3H, I). NOTE: The standard mold has an inlet and outlet port, while the wide-outlet mold only needs the inlet port (Figure 3H, I).
Use scissors to trim the edges of PDMS layers. NOTE: The trimmed PDMS layers should be ≥76 mm x 51 mm rectangles for small EcoFAB devices and ≥102 mm x 83 mm rectangles for large EcoFAB devices.
2. EcoFAB Device Fabrication: Chemically Attaching PDMS Layers onto Microscope Slides (Figure 3 & Figure 4)
- Permanently bonding the PDMS layers to microscope slides
- Rinse the bonding side of the PDMS layer (made of a 15:1 elastomer base to curing agent mixture) and 7.6 × 5 cm microscope slide with methanol and then blow dry with compressed air or an ultra-pure nitrogen gun. Caution: Methanol is toxic. Work in a fume hood and wear protective eye covering, gloves and other PPEs.
- Place the microscope slide and PDMS layer into a plasma cleaner with their bonding sides facing up (Figure 3J). If a plasma cleaner is not available, skip to step 2.2.
- Close the chamber and the gas vent valve of the plasma cleaner, and turn on the vacuum and pump down the chamber for 1 min.
- Turn on the power of the plasma generator, and switch the radio frequency (RF) level to "HI" for 1 min.
- Turn off both the vacuum pump and the plasma power, and vent the chamber to atmosphere.
- Take out the microscope slide and PDMS layer from the plasma chamber, and quickly press all four edges of the PDMS layer onto the slide with even pressure (Figure 3L). Make sure the center oval region of the PDMS layer (the root chamber) does not touch the slide.
- Place the sealed EcoFAB device on a 120 °C heating block for 20 min to further secure the permanent bonding between the PDMS layer and the microscope slide.
- Let the device cool down for 5 min. Trim off the extra edges of the PDMS layer with a knife.
- Reversible physical sealing of the PDMS layers to microscope slides
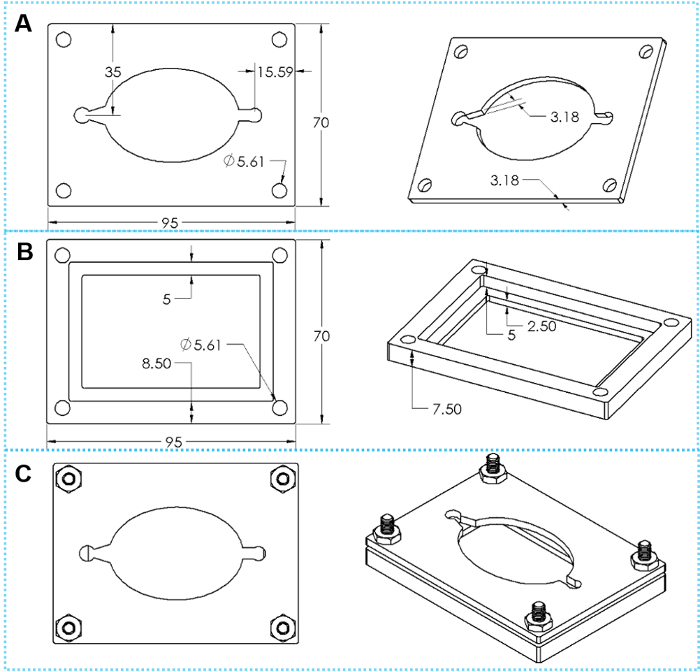
- The reversible sealing technique utilizes a set of custom clamps (either printed by a 3D plastic printer or machined in metal, the drawings are shown in Figure 4).
- Place the microscope slide into the cutout on the bottom clamp plate, and then align the PDMS layer (made of a 5:1 elastomer base to curing agent mixture) on top of the slide.
- Place the top clamp plate over the PDMS layer. Secure the top and bottom plates together using four hex cap screws, orienting the screws so that the nuts are threaded on from the top of the clamp.
- Adhering PDMS directly to microscope slides
- Position the PDMS layer (made of a 30:1 elastomer base to curing agent mixture) on top of a microscope slide.
- Press the PDMS layer to the slide. The soft, very adhesive PDMS layer (30:1) should stick to the slide creating a waterproof seal without the permanent chemical bonding or physical press from a clamp (Figure 3L).
3. EcoFABs Sterilization
Rinse EcoFAB devices with ultrapure water.
Place one EcoFAB device in an EcoFAB container, and add 70% ethanol until the device is submerged. Close the container lid and shake gently to wet all surfaces inside with ethanol. Make sure the root growth chamber of the EcoFAB device is filled with ethanol, with very few or no air bubbles.
After 30 min incubation at room temperature, pour off 70% ethanol, and repeat the incubation with 100% ethanol for 5 min.
Drain out ethanol, and incubate the sterilized EcoFAB for 16 h in a laminar flow hood to dry it completely. If available, sterilize the system by turning on the UV light within hood for 1 h. Caution: Wear appropriate PPE when working with UV lights.
Store the sterilized EcoFABs in a sterile hood or autoclaved bags for future use.
4. EcoFABs with LED Grow Lights (Figure 5)
- Attaching LED strips onto EcoFAB containers
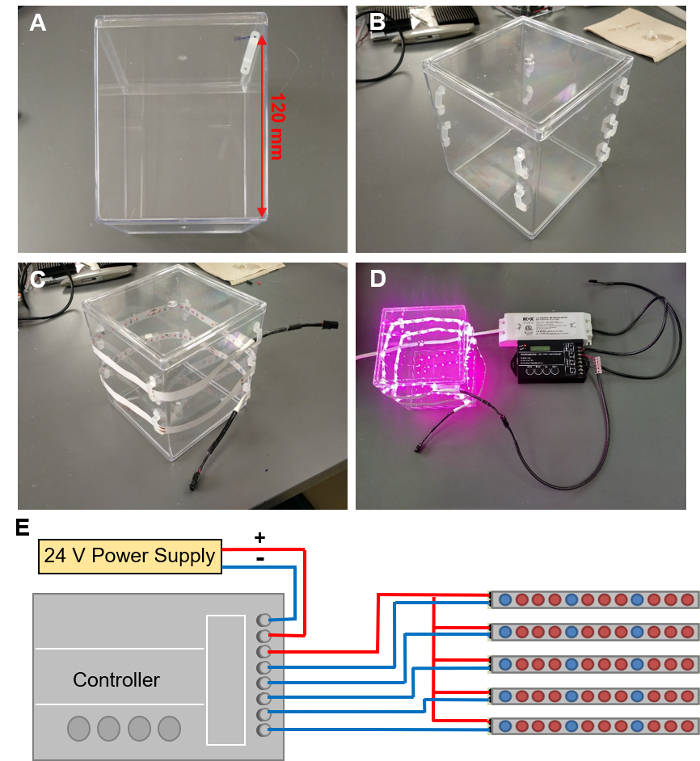
- Mark out the locations on the EcoFAB container for 9 LED clips. Start with the first clip 120 mm up from the bottom of the container along an edge (Figure 5A), and proceed to mark out clip locations in a spiral around the container, with each next clip dropping 10 mm. A spiral of 9 clips that allows a 1 m LED strip to wrap around the container twice.
- Hot glue an LED clip to each marked position by adding two dabs of hot glue onto the container, aligned with the position of the clips' mounting holes. Press the clip holes into these two dabs of glue, then add another dab of glue on top of the holes. Repeat the procedure for all clips until 9 clips form a spiral (Figure 5B). Caution: Wear gloves and other PPE when working with hot glue to avoid burns.
- Thread the LED strip through the clips in a spiral shape, with LEDs facing into the container. The strip should circle around twice (Figure 5C).
- Connecting LED strips to the power supply with a controller (Figure 5D displays an EcoFAB chamber with illuminated lights, the programming of the controller is described in step 4.3). Caution: Electrical shock hazard: make sure the power supply is unplugged when connecting wires.
- Connect the positive and negative terminals of the power supply to the "INPUT: V+" and "INPUT: V-" terminals of the controller using 2-wire cable (Figure 5E displays a schematic drawing of the controller setup).
- Connect the negative lead from the bare end of a female-to-bare cable to one "OUTPUT" channel on the controller. NOTE: There are five channels on the controller that is utilized in this protocol, so it can support up to five 1 m LED strips.
- Connect all positive leads of the cables to a compact splicing connector (if more than one channels are needed), and then link this connector to the "OUTPUT V+" terminal of the controller.
- Plug each LED strip into the female end of the cables, so each LED has its own channel to be controlled. If desired, use female-to-male cables to extend the reach.
Program the controller for a desired light cycle according to manufacturer's instructions,
5. Growing Plants in EcoFABs
- Seed sterilization and germination NOTE: The seed sterilization and all following steps with seedlings must be performed in sterile conditions. The sterilization process discussed below is suitable to the seeds of Arabidopsis thaliana, Avena barbata, Brachypodium distachyon, and Panicum virgatum. Panicum virgatum seeds should be suspended in 60% sulfuric acid for 1 h before the sterilization process. It is advised to prepare 1 - 2 seeds per EcoFAB device, considering the germination rate and the homogeneity of germination.
- Soak the seeds in 70% ethanol for 2 min.
- Remove ethanol with a pipette, and rinse the seeds with sterile water three times.
- Leave the seeds in 10% bleach solution for 5 min.
- Remove the bleach solution, and thoroughly wash the seeds using sterile water three times.
- Add sterile water to the seeds, and incubate the microcentrifuge tube in a 4 °C refrigerator for 7 days.
- Evenly spread the seeds on 0.5 Murashige & Skoog (MS) medium with 0.6% phytagel and seal the plates with micropore tape.
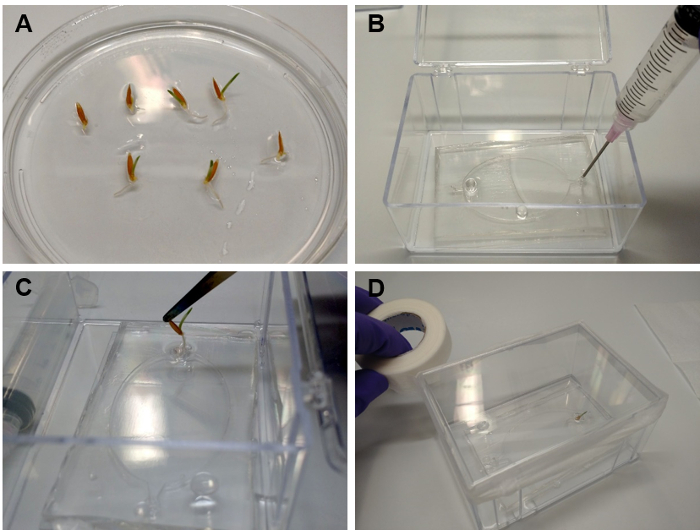
- Grow the plants to a root length of about 5 mm for transfer to EcoFABs (Figure 6A). For the experiments presented here, apply a 16 h light/8 h dark illumination regime in a 22 °C growth chamber, and incubate the plants 2 - 7 d before transfer to EcoFAB (2 days for Avena barbata and Brachypodium distachyon, 7 days for Arabidopsis thaliana and Panicum virgatum).
- Transferring seedlings into EcoFABs with liquid medium (Figure 6)
- Using a sterile syringe or micropipette, flush the root chamber of an EcoFAB device with sterile water for three times, and then fill the root chamber with the growth medium of interest, for example 0.5 MS medium (Figure 6B, step 5.1.6).
- Carefully insert a single seedling into the plant reservoir of the EcoFAB device (Figure 6C). NOTE: The root should be fully submerged inside the root chamber, with the shoot sticking out of the reservoir.
- Add 3 mL of sterile water into the container, avoiding the EcoFAB device. This will increase the humidity and reduce the evaporation of the medium from the root chamber.
- Close the container, and seal the lid with micropore tape (Figure 6D).
- Place the EcoFAB into a plant incubator, or utilize the EcoFAB illumination system in a controlled temperature environment suitable for the growth of the respective plant (step 4). For this study, set the chamber to 24 °C.
- Periodically check the EcoFAB to refill growth media inside the root growth chamber and add water to the container. Perform all steps in sterile conditions. NOTE: For early plant growth stages, refilling of the root growth chamber is necessary every 5 to 7 days. For later growth stages, a refilling is necessary every 2 to 3 days. If desired, use a syringe or pipette to collect root exudate solution from the root growth chambers into an microcentrifuge tube and store it in a -80 °C freezer; also, image the root morphology with a gel imager or microscope.
- Transferring seedlings into EcoFABs with solid substrates
- Use the root chambers fabricated with a 5:1 mixture of elastomer base to curing agent if using a set of custom clamps to attach it to a microscope slide (Figure 3K, Figure 4); or choose a PDMS layer made of 30:1 base to curing agent mixture if adhering PDMS layers to slides directly (as described in step 2.2).
- Sterilize the EcoFAB chambers, as described in step 3.
- Carefully add sterilized soil/sand into the root chamber, turn the PDMS layer upside down, and add the substrate to the root chamber. Avoid any particulate fall on the area that will be in contact with the microscope slide, since this will reduce adhesion.
- Place the microscope slide on top of the PDMS layer, and press all the edges firmly. Carefully flip the EcoFAB device so that no soil/sand falls out of the reservoir opening. NOTE: For EcoFAB devices made of a 5:1 base to curing agent mixture, use a custom clamp to secure the seal.
- Flow liquid medium or water through the inlet or outlet channel of the EcoFAB device, and transfer a seedling into its plant reservoir, as described in step 3.3.
- Adding microbes into EcoFABs
- Transfer a microbial colony to an incubation tube with 8 mL of LB broth, and grow it to OD 0.5 (approximately 12 h).
- Transfer the culture solution into a 15 mL centrifuge tube, and centrifuge it at room temperature for 5 min at 3000 x g to pellet microbes.
- Remove the supernatant, and add 8 mL of plant growth medium used in the target EcoFAB. Suspend the pellet of microbes, and centrifuge the tube at room temperature for 5 min at 3000 x g.
- Repeat step 5.4.3. twice to completely remove any LB nutrient traces.
- Add plant growth medium to the washed microbe pellet until its optical density is about 0.5 at 600 nm.
- Add 20 µL of the microbe solution into the root chamber through the EcoFAB outlet. The strains used in this publication traveled to plant roots within 2 - 3 days, and started colonizing root surfaces.
- For engineered chemiluminescent, make sure to include the inducer (1 mM IPTG) in plant growth medium to induce luciferase expression.
6. Metabolite Profiling of Root Exudates from EcoFABs
- Sample preparation for LC/MS based metabolomics analysis
- Put the microcentrifuge tubes with root exudates collected from EcoFABs in a lyophilizer, and turn on the lyophilizer to remove all the water from the tubes.
- Add 300 µL of LC-MS grade methanol into each tube, and sonicate for 30 min. Caution: Wear PPE when working with methanol.
- Place the tubes in a centrifuge, and centrifuge them at 3000 x g for 5 min.
- Transfer the supernatant solutions into new microcentrifuge tubes, and evaporate methanol in a vacuum concentrator.
- Add 150 µL of methanol with 1 mM LC-MS internal standards into each tube, and incubate the tubes in a 4 °C refrigerator for 12 h.
- Centrifuge the tubes at 3000 x g for 5 min, and transfer the supernatant into 0.22 µm filter tubes.
- Centrifuge the filter tubes, and transfer the filtered solutions into 2.0 mL LC/MS vials with 200 µL of inserts.
- Place the vials inside a LC/MS rack, and load the rack inside the LC/MS auto-sampler.
7. Mass Spectroscopic Imaging of Plant Roots in EcoFABs (Figure 7)
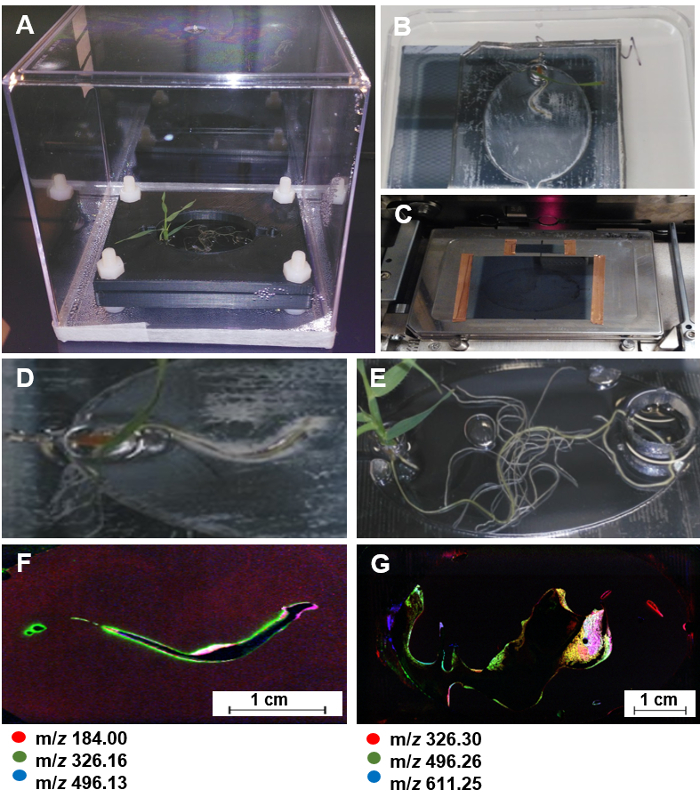
NOTE: EcoFAB devices made of a 5:1 elastomer base to curing agent mixture with custom clamps (Figure 7A) are used for root stamping onto nanostructure-initiator mass spectrometry (NIMS) chips,28,29,30 since PDMS layers can be reversely bonded to the surfaces of NIMS chips.
Sterilize a NIMS chip surface with UV light for 1 h.
Pick an EcoFAB with a growing plant from the incubator, and place it in a sterile hood.
Open the EcoFAB container, and remove the top plate of the clamp.
Lift up the PDMS layer together with the plant inside, and carefully attach the PDMS layer with the plant onto a NIMS chip (Figure 7B, D, E). NOTE: Once the root touches the NIMS chip surface, it should not be moved. This prevents "smearing" of the root metabolites.
Gently press down on the roots through the PDMS layer until the roots fully contact the NIMS surface. Leave the roots on the NIMS surface for 20 min.
Lift off the PDMS layer including the plant from the NIMS chip, again avoiding moving the root across the NIMS surface. Return the plant to the clamp if desired.
Attach the NIMS chip onto a custom MALDI plate and load the plate into a MALDI spectrometer for mass imaging (Figure 7C).
Use OpenMSI program to generate the NIMS image of root metabolites (Figure 7D-G)31.
Representative Results
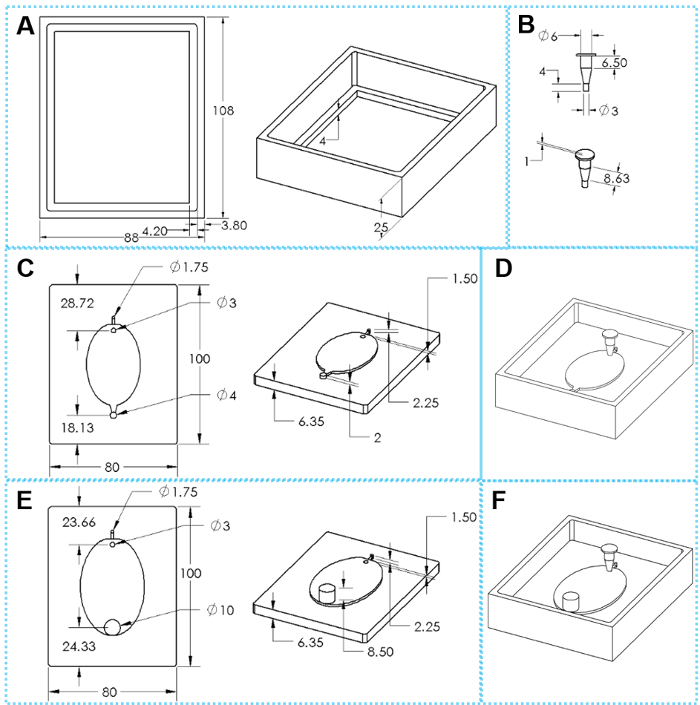
Each EcoFAB system includes an EcoFAB device and a plant sized transparent plastic container. One EcoFAB device has a plant reservoir, a root growth chamber, a 1.6 mm flow inlet, and a 1.6 mm outlet for standard EcoFAB device (Figure 2D & Figure 3H) or a 10 mm outlet for wide-outlet EcoFAB device (Figure 2F & Figure 3I). The plant reservoir is designed in a trapezoid shape that has a 6 mm top opening and 3 mm bottom opening, and this design reduces the chance of flow leakage during liquid injection and also ensures enough space for plant growth. The root growth chamber adopts an oval shape with 2 mm depth to fit many model plants' root systems, as shown in Figure 2C and E. Both inlet and outlet channels of a standard EcoFAB device can be connected with PTFE tubing so nutrient solutions can flow into the root growth chamber without opening the EcoFAB container. The wide-outlet EcoFAB device greatly reduces the flow resistance of the outlet, and is preferably used when growing plants with thick root systems or periodically collecting root exudates after complex root systems are derived from plants.
The casting molds for fabricating PDMS layers of EcoFAB devices are created in a design software, and then 3D printed in rigid opaque photopolymers, as shown in Figure 2 and Figure 3. Plants inside EcoFABs can be directly observed with a microscope using a long work distance, ensuring the sterile grow environment (Figure 8A, Supplementary File 1). EcoFAB devices with plants can also fit onto a high-resolution microscope stage, which enables higher resolution imaging of plant-microbe interactions (Figure 8B, Supplementary File 2). Sterility is not maintained in this environment, and high-resolution imaging is therefore only suitable for endpoint measurements.
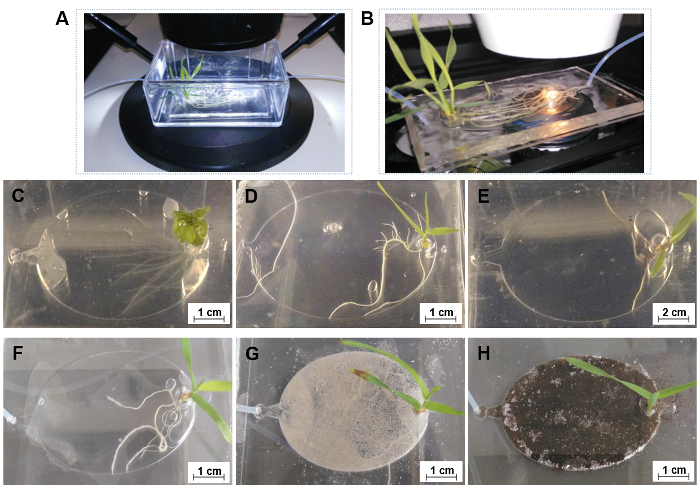
EcoFABs are designed to enable systematic studies of plants, such as their morphology, metabolisms, and microbial communities at their different growth stages across their life cycles. Here, EcoFABs were examined as a general platform to study a variety of plant species. Figure 8C-E show 7-day old Arabidopsis thaliana, Brachypodium distachyon, and Panicum virgatum growing in EcoFABs. All three plants were found to grow well in the EcoFAB for over one month. Both the dicot, Arabidopsis thaliana and the monocot, Brachypodium distachyon were found to live up to their reproduction stages in the EcoFABs.
The reversible sealing system allows the use of solid substrates (e.g., soil) within the EcoFABs (step 2.2). This reversible sealing approach enables loading of solid substrates into root growth chambers, and also enables sample collection from specific regions of root rhizospheres. Figure 8F-H show a group of 14-day old Brachypodium distachyon growing in hydroponic medium, as well as sand and soil supplemented with hydroponic medium (sand) and water (soil). The thin solid substrate layer in root growth chambers allows light to penetrate through for microscopic imaging of root systems.
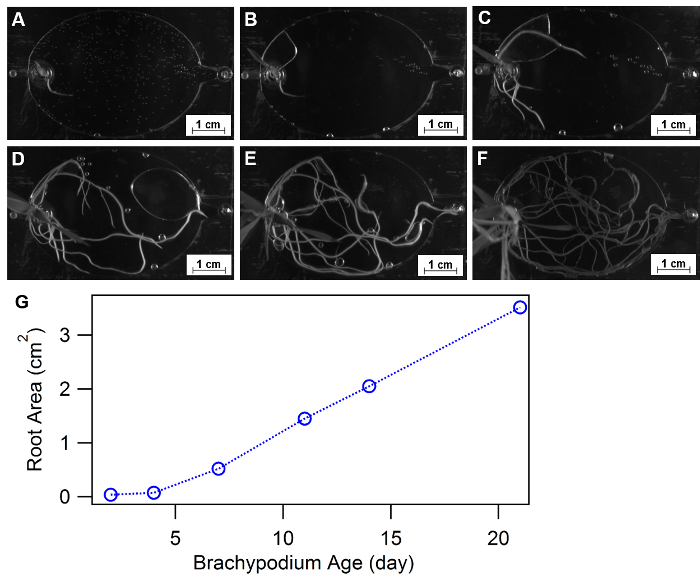
Root morphology is defined as spatial configuration and distribution of a plant root system and has been approved as an essential physiology response to diverse growth environments, such as nutrient or water availability32,33,34. EcoFABs provide a convenient approach of studying plant morphology over time or under different nutrient conditions. Figure 9A-F show an example of using EcoFABs to track root morphologies of Brachypodium distachyon in the first three weeks. A Brachypodium distachyon seedling was transferred into the EcoFAB device, and its root structure was recorded by a camera inside a BIO-RAD gel imager. Image processing program, such as Image J, python and matlab, can be further applied to quantify the changes of root morphologies over time or at different medium environments. The quantification of total root area over the course of three weeks showed a gradual increase at the early stage (<1 week) followed by a linear growth trend to the end of three weeks, as shown in Figure 9G.
A primary motivation for constructing the EcoFAB is to investigate plant-microbe interactions. As described in step 5.4, microorganisms are transferred into root growth chambers of EcoFAB devices through the inlet channel. Figure 10 shows, an EcoFAB containing Pseudomonas simea (formerly, fluorescens) WCS417 (WCS417), a plant growth promoting rhizobacteria with chemiluminescent labels, was added into the plant root systems with a concentration of 106 cells per plant. The WCS417 signal was detected with a gel imager, which indicated a distinct spatial distribution of WCS417 microbes in root growth chambers. In both MS liquid medium with and without the sand solid substrate, WCS417 microbes colonized the surfaces of the entire root systems with microbes concentrated around the root tip areas, possibly due to the active nutrient production of root tips (Figure 10G&H)35. On the other hand, the WCS417 microbes in soil substrate accumulated around the plant reservoir region instead of root tips (Figure 10I). As the microbes were added through the outlet channel, the microbes were also able to move in the soil substrate, but did not accumulate on the root, as observed in liquid medium with or without sand. This could indicate that the soil is a sufficient nutrient source, and the microbes migrated to the plant reservoir for optimal respiration conditions.

To study metabolite profiling of plant root exudates as well as metabolite uptake and release from plant-microbe interactions, the exudate solutions from the root growth chambers was collected across various growth stages of plants in EcoFABs. As described in step 6, exudate samples are then extracted for LC-MS analysis. Using this method, a range of metabolites exuded by the plant and consumed by the microbes was detected, and the related metabolite profiling of root exudates with and without microbes colonization is currently under investigation.
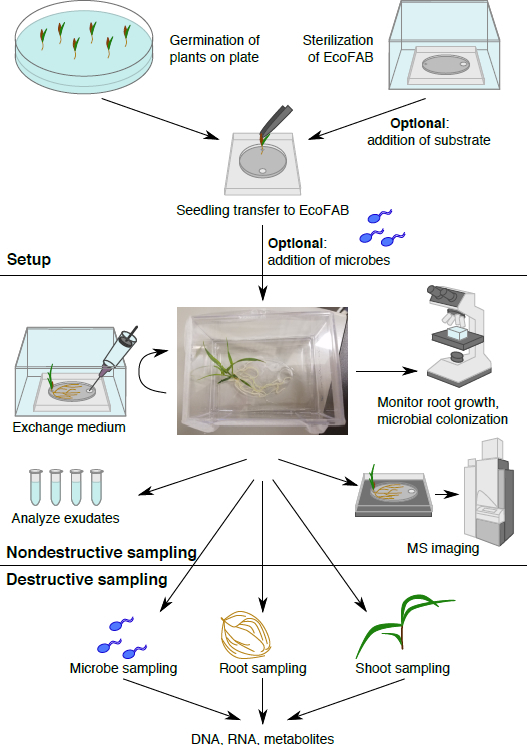
Figure 1: The EcoFAB workflow. Plants are germinated on plate, and transferred to the sterilized EcoFAB, microbes can be added. Nondestructive sampling: root exudates can be sampled and imaged, and root morphology can be visualized. Destructive sampling allows analysis of microbe, root, and shoot parameters in detail.
Figure 2: The components of 3D printed molds for EcoFAB device fabrication. (A) Top and tilted views of a casting frame. (B) Top and tilted views of an insert. (C) Top and tilted views of a standard mold base. (E) Top and tilted views of a wide-outlet mold base. (D, F) Assembled molds for fabricating standard and wide-outlet EcoFAB devices, respectively. The oval dimensions are 51 mm x 34 mm for small EcoFAB mold and 76 mm x 62 mm for large EcoFAB mold. Please click here to view a larger version of this figure.
Figure 3: EcoFAB device fabrication. (A) Pouring the mixture of elastomer base and curing agent into the mold. (B) Heating the mold with mixture at 85 °C for 4 h. (C) Removing the insert from the mold. (D) Separating the PDMS from the casting frame. (E) Pushing the mold base out of the casting frame. (F) Using a knife to separate the PDMS from the mold along the edges. (G) Peeling the PDMS layer slowly off the mold base. (H) Poking holes for both inlet and outlet channels of the standard PDMS layer. (I) Poking a hole for the inlet channel of the wide-outlet PDMS layer. (J) The PDMS layer (made of a 15:1 elastomer base to curing agent mixture) and a microscope slide are rinsed, and transferred into a plasma cleaner for bonding. (K) Using clamps to hold the PDMS layer (made of a 5:1 elastomer base to curing agent mixture) onto a microscope slide. (L) Pressing the PDMS layer (made of a 30:1 elastomer base to curing agent mixture) directly onto a microscope slide. Please click here to view a larger version of this figure.
Figure 4: The design of custom clamps. (A) Top and tilted views of a top clamp plate. (B) Top and tilted views of a bottom clamp plate. (C) Top and tilted views of assembled clamp with four sets of hex cap screws. Please click here to view a larger version of this figure.
Figure 5: Installing LED grow lights. (A) Marking out the locations for 9 LED clips in a spiral around the EcoFAB container. (B) LED clips attached to the EcoFAB container. (C) Threading a LED strip through these clips. (D) Connecting the LED strip to a controller wired with a 24V power supply. (E) The schematic of wire connections to the controller. Please click here to view a larger version of this figure.
Figure 6: Transferring seedlings into EcoFABs. (A) Brachypodium distachyon plants grown for 2 days on a 0.5 MS plate. (B) Filling the root chamber with plant growth medium. (C) Using a tweezer to carefully insert the root into the plant reservoir. (D) Sealing the EcoFAB container with micropore tape, after adding 3 mL of water to the bottom of the container. Please click here to view a larger version of this figure.
Figure 7: NIMS imaging of plant roots in EcoFABs. (A) A Brachypodium distachyon growing in a sterile EcoFAB. (B) Attaching the PDMS layer with the plant onto a NIMS chip for 20 min. (C) Using copper tape to attach the NIMS chip onto a custom MALDI plate, and loading it into a MALDI mass spectrometer. (D-G) one 7-day old and one 20-day old Brachypodium distachyon plant used for NIMS imaging (D, E) and the corresponding NIMS images (F, G). The predominant ions were highlighted in red, green, and blue. Please click here to view a larger version of this figure.
Figure 8: The general applications of EcoFABs. (A) Directly capturing root growth of Brachypodium distachyon in an EcoFAB with a long work distance microscope setup. (B) Directly observing root-microbe interactions with a high-resolution microscope setup. (C-E) 7-day old Arabidopsis thaliana (C), Brachypodium distachyon (D), and Panicum virgatum (E) in 0.5 MS hydroponic medium, (F-H) 14-day old Brachypodium distachyon grown in 0.5 MS hydroponics (F), in sand (G) and soil (H) substrate supplied with 0.5 MS medium and water, respectively. Please click here to view a larger version of this figure.
Figure 9: Using EcoFABs to study root morphology. (A-F) Root development of Brachypodium distachyon growing in EcoFABs filled with 0.5 MS medium during first three weeks: (A) 2 days, (B) 4 days, (C) 7 days, (D) 11 days, (E) 14 days, (F) 21 days of growth. (G) Averaged root surface areas were estimated by ImageJ software. Please click here to view a larger version of this figure.
Figure 10: Using EcoFABs to study root-microbe interactions. (A, B, C) A group of 15-day old Brachypodium distachyon colonizing with Pseudomonas fluorescens WCS417 in different forms of media-MS liquid solution, sand and soil substrates. (D, E, F) Bright field pictures of their root systems. (G, H, I) The corresponding chemiluminescent images of these root systems after 14 days co-cultivation. Please click here to view a larger version of this figure.
Supplementary File 1. Using EcoFAB to capture root growth. Please click here to download this file.
Supplementary File 2. Using EcoFAB to capture root-microbes interactions. Please click here to download this file.
Discussion
The protocols reported here for using ecosystem fabrication to create EcoFABs provides community resources for systematic plant biology studies in highly controlled laboratory conditions. Advances in 3D printing provide widely accessible technologies for constructing and iteratively refining EcoFAB designs. The root chamber presented here is found to be well suited for imaging microscopy and maintaining sterility, enabling controlled addition of microbes to investigate plant-microbe interactions.The EcoFAB platform is compatible with various plant species. It is important to recognize physiological effects of growing the plants within the narrow root chamber such that additional experiments will be required to generalize findings to plants growing in natural environments.
The use of sterile chambers and LED grow light enables the investigation of the effects of various light conditions, including wavelength, intensity, and duration, on plant growth and related physiological parameters in parallel. Reversible bonding root chambers allow the use of solid substrates as well as to spatially collect solid samples for biochemical and genetic analysis. The applications of solid substrates, such as soils, sand, and quartz beads, offer the possibilities of using EcoFABs to construct more ecologically relevant laboratory ecosystems. However, all the systems presented here use saturated liquid (hydroponic cultures) which are not an accurate reflection of most soils and it will be important to further refine these designs to maintain air pockets within the soil such that they better represent natural soils.
The use of both simple cameras and microscopes is described to image root system morphology development at both bulk to cellular levels. This suitability for monitoring root morphology imaging and quantification will likely be helpful for understanding the regulatory mechanisms of plant physiological and molecular signals triggered by plant genotypic adaptions to growth conditions. However, a limitation for studying physiological root development is the current horizontal placement of the EcoFAB device. In natural environments, the roots gravitropic response leads to a predominantly vertical development of the root system. Thus, the horizontal system presented here likely differs in some factors from a natural environment, and the fabrication of EcoFAB systems with vertical placement of the root chamber is a desirable goal for future EcoFAB versions. Although the current EcoFAB devices are placed horizontally, the analysis of root morphology parameters in various conditions, or in response to microbes, is possible. High resolution imaging can be applied to capture root colonization dynamics of single isolates or communities, providing information about which plant parts are colonized in various nutrient sufficient and deficient conditions. It is anticipated that such studies will provide important new insights into how plant microbiomes are assembled, and how these dynamics change over time, for example as the roots develop.
Microfluidic devices enable imaging of very young plants, and usually the amount of metabolites collected is not sufficient for LCMS analysis. Soil-based systems, such as rhizotrons, allow the imaging of root morphology when either the plants are transformed with chemiluminescent construct (Glo-root) or with NMR-based methods33,34. Metabolite extractions from these systems are time consuming because of large volume of samples. EcoFABs are a combination of both: the fabrication is similar to microfluidic devices. EcoFABs were designed to be simple and inexpensive to reproduce, but the size of the chamber can be adjusted to grow plants with small or large root systems, up to their reproductive stages. Simultaneous observations of root morphology changes and root exudation are possible. The system is sterile, enabling controlled addition of specific microbes.
EcoFABs are designed to enable controlled introduction and sampling of microbes and metabolites. Specifically, samples collected from root growth chambers are found to be sufficient for mass spectroscopic metabolite profiling. The integration of mass spectrometry imaging (e.g., NIMS technique presented here) provides a non-destructive approach of studying metabolite spatial distributions of root systems. This technique will likely be helpful in future stable isotope tracing experiments and mapping microbial localization to specific metabolites36. While this protocol has focused on single isolates, the same design can certainly be used for more complex communities. The sample volumes and biomass within the EcoFABs are likely more than sufficient for further integration with DNA sequencing technologies, which will be important to characterizing and monitoring microbial community structure and gene expression.
In conclusion, this protocol details the fabrication of laboratory ecosystems designed for the investigation of plant-microbe interactions, with emphasis on simple and accessible methods that can easily be implemented and extended by researchers around the world. Current efforts are aimed at demonstrating the reproducibility between labs and the integration of a temperature control system such that each EcoFAB will have independently controlled light and temperature. A further advancement of the system will be the integration of automated sampling and refilling of the EcoFAB root chambers and the development of reproducible protocols for establishing relevant plant microbiomes within EcoFABs.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This work was supported by Laboratory Directed Research and Development (LDRD) program of Lawrence Berkeley National Laboratory supported by the Office of Science, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231 and an award DE-SC0014079 from the U.S. Department of Energy Office of Science to UC Berkeley. Work at the Molecular Foundry was supported under U.S. Department of Energy Contract No. DE-AC02-05CH11231. We also thank Suzanne M. Kosina, Katherine Louie, Benjamin P. Bowen, and Benjamin J. Cole at Lawrence Berkeley National Laboratory for all their help.
References
- Morrissey JP, Dow JM, Mark GL, O'Gara F. Are microbes at the root of a solution to world food production. EMBO Rep. 2004;5(10):922–926. doi: 10.1038/sj.embor.7400263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar K, Bryant D, Cope-Selby N. Understanding and engineering beneficial plant-microbe interactions: plant growth promotion in energy crops. Plant Biotechnol J. 2014;12(9):1193–1206. doi: 10.1111/pbi.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh JS, Abhilash PC, Gupta VK. Agriculturally Important Microbes in Sustainable Food Production. Trends Biotechnol. 2016;34(10):773–775. doi: 10.1016/j.tibtech.2016.06.010. [DOI] [PubMed] [Google Scholar]
- Dubey RK, Tripathi V, Dubey PK, Singh HB, Abhilash PC. Exploring rhizospheric interactions for agricultural sustainability: the need of integrative research on multi-trophic interactions. J Clean Prod. 2016;115:362–365. [Google Scholar]
- Hunter P. Plant microbiomes and sustainable agriculture. EMBO Rep. 2016;17(12):1696–1699. doi: 10.15252/embr.201643476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden MGA, Hartmann M. Networking in the Plant Microbiome. PLoS Biol. 2016;14(2):e1002378. doi: 10.1371/journal.pbio.1002378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vessey JK. Plant growth promoting rhizobacteria as biofertilizers. Plant Soil. 2003;255(2):571–586. [Google Scholar]
- Yang J, Kloepper JW, Ryu C-M. Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci. 2009;14(1):1–4. doi: 10.1016/j.tplants.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Reynolds HL, Packer A, Bever JD, Clay K. GRASSROOTS ECOLOGY: PLANT-MICROBE-SOIL INTERACTIONS AS DRIVERS OF PLANT COMMUNITY STRUCTURE AND DYNAMICS. Ecology. 2003;84(9):2281–2291. [Google Scholar]
- Finkel OM, Castrillo G, Herrera Paredes S, Salas González I, Dangl JL. Understanding and exploiting plant beneficial microbes. Curr Opin Plant Biol. 2017;38:155–163. doi: 10.1016/j.pbi.2017.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northen TR, Zhang Z, Gao J, Swenson T, Yoshikuni Y. National Academies of Sciences, Engineering, and Medicine. 2017. The Chemistry of Microbiomes: Proceedings of a Seminar Series. Washington, DC: The National Academies Press; 2017. Advancing Our Understanding of the Chemistry of Soil Microbiomes. [Google Scholar]
- Busby PE, et al. Research priorities for harnessing plant microbiomes in sustainable agriculture. PLOS Biology. 2017;15(3):e2001793. doi: 10.1371/journal.pbio.2001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanati Nezhad A. Microfluidic platforms for plant cells studies. Lab Chip. 2014;14(17):3262–3274. doi: 10.1039/c4lc00495g. [DOI] [PubMed] [Google Scholar]
- Oburger E, et al. Evaluation of a novel tool for sampling root exudates from soil-grown plants compared to conventional techniques. Environ Exp Bot. 2013;87:235–247. [Google Scholar]
- Van Der Krift TAJ, Berendse F. Root life spans of four grass species from habitats differing in nutrient availability. Funct Ecol. 2002;16(2):198–203. [Google Scholar]
- Massalha H, Korenblum E, Malitsky S, Shapiro OH, Aharoni A. Live imaging of root-bacteria interactions in a microfluidics setup. P Natl. Acad. Sci. USA. 2017;114(17):4549–4554. doi: 10.1073/pnas.1618584114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanati Nezhad A, Naghavi M, Packirisamy M, Bhat R, Geitmann A. Quantification of cellular penetrative forces using lab-on-a-chip technology and finite element modeling. P Natl. Acad. Sci. USA. 2013;110(20):8093–8098. doi: 10.1073/pnas.1221677110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Xu Z, Aluru MR, Dong L. Plant chip for high-throughput phenotyping of Arabidopsis. Lab Chip. 2014;14(7):1281–1293. doi: 10.1039/c3lc51326b. [DOI] [PubMed] [Google Scholar]
- Parashar A, Pandey S. Plant-in-chip: Microfluidic system for studying root growth and pathogenic interactions in Arabidopsis. Appl. Phys. Lett. 2011;98(26):263703. [Google Scholar]
- Busch W, et al. A microfluidic device and computational platform for high-throughput live imaging of gene expression. Nat Methods. 2012;9(11):1101–1106. doi: 10.1038/nmeth.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann G, et al. The RootChip: An Integrated Microfluidic Chip for Plant Science. Plant Cell. 2011;23(12):4234–4240. doi: 10.1105/tpc.111.092577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufrecht JA, Ryan JM, Hasim S, Allison DP, Nebenführ A, Doktycz MJ, Retterer ST. Imaging the Root Hair Morphology of Arabidopsis Seedlings in a Two-layer Microfluidic Platform. J. Vis. Exp. 2017. [DOI] [PMC free article] [PubMed]
- Garvin DF, et al. Development of Genetic and Genomic Research Resources for Brachypodium distachyon, a New Model System for Grass Crop Research. Crop Sci. 2008;48(Supplement_1):S69–S84. [Google Scholar]
- Lisensky GC, et al. Replication and Compression of Surface Structures with Polydimethylsiloxane Elastomer. J. Chem. Educ. 1999;76(4):537. [Google Scholar]
- Friend J, Yeo L. Fabrication of microfluidic devices using polydimethylsiloxane. Biomicrofluidics. 2010;4(2):026502. doi: 10.1063/1.3259624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, et al. Analysis of Metabolomics Datasets with High-Performance Computing and Metabolite Atlases. Metabolites. 2015;5(3):431–442. doi: 10.3390/metabo5030431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner LW, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3(3):211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, de Raad M, Bowen BP, Zuckermann RN, Northen TR. Application of Black Silicon for Nanostructure-Initiator Mass Spectrometry. Anal. Chem. 2016;88(3):1625–1630. doi: 10.1021/acs.analchem.5b03452. [DOI] [PubMed] [Google Scholar]
- Gao J, et al. Morphology-Driven Control of Metabolite Selectivity Using Nanostructure-Initiator Mass Spectrometry. Anal. Chem. 2017;89(12):6521–6526. doi: 10.1021/acs.analchem.7b00599. [DOI] [PubMed] [Google Scholar]
- Woo H-K, Northen TR, Yanes O, Siuzdak G. Nanostructure-initiator mass spectrometry: a protocol for preparing and applying NIMS surfaces for high-sensitivity mass analysis. Nat. Protoc. 2008;3(8):1341–1349. doi: 10.1038/nprot.2008.110. [DOI] [PubMed] [Google Scholar]
- Rübel O, et al. OpenMSI: A High-Performance Web-Based Platform for Mass Spectrometry Imaging. Anal. Chem. 2013;85(21):10354–10361. doi: 10.1021/ac402540a. [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L. The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol. 2003;6(3):280–287. doi: 10.1016/s1369-5266(03)00035-9. [DOI] [PubMed] [Google Scholar]
- Lynch JP. Steep, cheap and deep: an ideotype to optimize water and N acquisition by maize root systems. Ann. Bot. 2013;112(2):347–357. doi: 10.1093/aob/mcs293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rellán-Álvarez R, et al. GLO-Roots: an imaging platform enabling multidimensional characterization of soil-grown root systems. eLife. 2015;4:e07597. doi: 10.7554/eLife.07597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamilova F, Validov S, Azarova T, Mulders I, Lugtenberg B. Enrichment for enhanced competitive plant root tip colonizers selects for a new class of biocontrol bacteria. Environ. Microbiol. 2005;7(11):1809–1817. doi: 10.1111/j.1462-2920.2005.00889.x. [DOI] [PubMed] [Google Scholar]
- Klitgaard A, Nielsen JB, Frandsen RJN, Andersen MR, Nielsen KF. Combining Stable Isotope Labeling and Molecular Networking for Biosynthetic Pathway Characterization. Anal. Chem. 2015;87(13):6520–6526. doi: 10.1021/acs.analchem.5b01934. [DOI] [PubMed] [Google Scholar]


