Abstract
Chromosome mis-segregation leads to aneuploidy, a condition in which cells harbor an imbalanced chromosome number. Several lines of evidence strongly indicate that aneuploidy triggers genome instability, ultimately generating cells with complex karyotypes that arrest their proliferation. Isolation and characterization of cells harboring complex karyotypes are crucial to study the impact of an imbalanced chromosome number on cell physiology. To date, no methods have been established to reliably isolate such aneuploid cells. This paper provides a protocol for the enrichment and analysis of aneuploid cells with complex karyotypes utilizing standard, inexpensive tissue culture techniques. This protocol can be used to analyze several features of aneuploid cells with complex karyotypes including their induced senescence-associated secretory phenotype, pro-inflammatory properties, and ability to interact with immune cells. Because cancer cells often harbor imbalances in chromosome number, it is crucial to decipher how aneuploidy impacts cell physiology in normal cells, with the ultimate goal of uncovering both its pro- and anti-tumorigenic effects.
Keywords: Cancer Research, Issue 134, aneuploidy, chromosome mis-segregation, genome instability, complex karyotype, cell cycle arrest, senescence
Introduction
Errors in the process of chromosome segregation lead to aneuploidy, a condition characterized by a chromosome number that is not a multiple of the haploid complement1,2,3,4. Aneuploid karyotypes trigger replication stress that generates further genomic instability5,6,7,8,9,10, increases karyotype complexity, and ultimately leads to cell cycle arrest of a subpopulation of cells10. The purpose of the method presented here is to generate and separate such a subpopulation from cycling cells. By employing inexpensive tissue culture techniques, this protocol facilitates the isolation and characterization of cell cycle-arrested aneuploid cells with complex karyotypes. These cells are referred to as ArCK (Arrested with Complex Karyotype) cells and their euploid cycling counterparts as controls.
This protocol is the first one to be established for this purpose and allows for the isolation and further study of ArCK lines including, but not limited to, their induced senescence and the senescence-associated secretory phenotype (SASP), their pro-inflammatory features, and their ability to engage with immune cells. The method presented here has been developed in untransformed, immortalized human cells but has not yet been tested in cancer lines. Some transformed cells may be insensitive to cell cycle arrest due to suppression of one or multiple pathways; therefore, further validation should be performed in other cell lines.
Protocol
Culture condition
RPE-1 hTERT cell line was cultured in Dulbecco's Modified Eagle Medium (Table of Materials) supplemented with 10% Fetal Bovine Serum, 2 mM L-glutamine, and 100 U/ml penicillin/streptomycin. Cells were incubated at 37 °C with 5% CO2 in a humidified environment. The protocol described below has been developed by using 10-cm dishes and all technical details reported refer to those dishes. If using different dishes, scale up or down accordingly.
1. Synchronization of RPE-1 cells
Use freshly thawed cells that have not undergone more than 15 passages for all experiments. On day 1, thaw and dispense ~2.5 - 5.0 x 105 RPE-1 hTERT cells into a 10-cm tissue culture dish. Add 8 mL of standard growth medium and incubate overnight. Determine cell number using a standard cell counter (such as a hemocytometer). NOTE: Cell density refers to growth conditions of RPE-1 cells. Growth conditions might vary among different cell lines; consider adjusting them accordingly.
On day 2, synchronize cells at the G1/S border by aspirating regular growth medium and adding 8 mL medium containing 5 mM thymidine. NOTE: Thymidine concentration and length of treatment might vary across different cell lines. It is advisable to perform pilot experiments to determine the best conditions for the cell line(s) to be tested.
On day 3, 24 h after adding thymidine medium (step 1.2), release from thymidine block by aspirating medium and washing cells three times with 1x PBS. Add 8 mL of regular growth medium and return plates to the incubator.
2. Generation of Aneuploid Cells by Interference with the Activity of the Mitotic Kinase Mps1
On day 3, 6 h after release from thymidine block (step 1.3; this is about 3 - 6 h before cells enter mitosis), aspirate medium, wash once with 1x PBS, and replace with medium containing 500 nM reversine (the Mps1 inhibitor). NOTE: RPE-1 cells enter mitosis 9 - 12 h after thymidine wash-out10,11. However, cell cycle kinetics vary among different cell lines. Therefore, it is crucial to determine those kinetics in pilot experiments by performing cell cycle analysis by established methods (e.g., by measuring DNA content via FACS analysis). Additionally, the optimal working concentration of Mps1 inhibitor may vary between cell lines. Previous experiments using RPE1 cells have been successful using reversine at a working concentration of 500 nM10,11,12. It is advisable to perform pilot experiments to determine the optimal concentration for the cell line to be tested.
On day 4, 12 h after reversine treatment (about 6 h after cells were in mitosis), aspirate reversine medium and wash cells three times with 1x PBS. Add 8 mL regular growth medium to the plate and return cells to the incubator.
3. Removal of Aneuploid Cycling Cells and Enrichment of ArCK Population
On day 6, about 72 h after cells enter the first mitosis (step 2.2; this is about 66 h after reversine wash-out, which corresponds to about 2 - 3 cell cycles), aspirate medium, wash once with 1x PBS and replace with medium containing 300 nM nocodazole. NOTE: Recent work has shown that RPE-1 cells harboring aneuploid karyotypes are genomically unstable and arrest their proliferation about 2 - 3 cell cycles after the first faulty mitosis10. However, this kinetics might vary among different cell lines. Therefore, it is crucial to perform pilot experiments to determine the proper timing for the removal of aneuploid cycling cells.
On day 7, 12 h after nocodazole treatment, aspirate medium and add 3 mL of 1x PBS to the plate. Shake off the mitotic cells by tapping the side of the plate. Rotate the plate between each tap to allow even removal of mitotic cells.
To ensure complete removal of mitotic cells, repeat the shake-off process for multiple rounds (3 - 5 times recommended), aspirating and adding 3 mL of 1x PBS between each round.
After shake-off, gently aspirate any liquid that adhered to the lid or the edge of the plate. Briefly, check the plate under a microscope at 10X magnification. Ensure that few mitotic cells are observed. If more than five mitotic cells are seen under 10X objective, repeat another round of shake-off. NOTE: Mitotic cells would appear rounded and may be partially detached after shake-off. It is crucial to ensure the complete removal of mitotic cells before exposing the cells to the next round of nocodazole treatment. Failure to do so may lead to mitotic cell death and/or result in the generation of tetraploid cells.
After final shake-off, wash cells once with 5 mL of 1x PBS. Add 8 mL medium containing 330 nM nocodazole into the plate and incubate for 12 h.
Repeat nocodazole treatment/shake-off (steps 3.1 - 3.5) every 12 h until no mitotic cells are observed after incubation. Usually, 4-5 rounds of treatment/shake-off are recommended. NOTE: The information provided here refer to RPE-1 cells. The number of rounds of treatment/shake-off might vary among different cell lines. To avoid prolonged and unnecessary exposure to nocodazole it is crucial to carefully determine, in a pilot experiment, the number of rounds required for efficient and complete removal of aneuploid cycling cells.
After 4 - 5 rounds of nocodazole treatment/shake-off (steps 3.1 - 3.6), add 10 mL of regular growth medium and incubate. Allow cells to rest for 12 - 24 h before future manipulation or assay use. NOTE: After final shake-off, cells remaining on the plate are cell cycle arrested and, as shown recently10, harbor complex karyotypes. These cells are referred to as ArCK (Arrested with Complex Karyotype) cells. It is advisable to perform assays on ArCK populations within a week of isolation.
Optionally, to assess the percentage of arrested cells with complex karyotypes, collect and count cells from each shake-off using a standard cell counter.
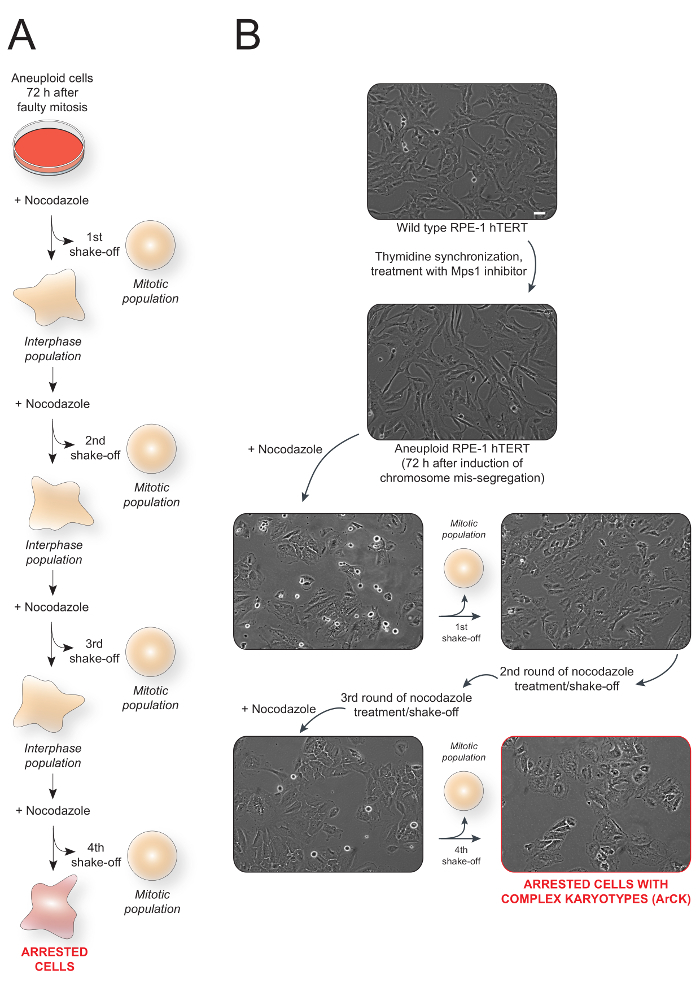
Figure 1: Overall schematic of the method used to generate and isolate aneuploid cells with complex karyotype (ArCK) and representative images.(A) Schematic of ArCK cells generation and isolation protocol. (B) Representative images of RPE-1 hTERT cells at each stage of the treatment. The last panel (outlined in red) represents the isolated ArCK cells. Scale bar 100 µm. Please click here to view a larger version of this figure.
4. Characterization of ArCK Population
- Confirm successful isolation of ArCK population by performing the following assays.
- Asses beta-galactosidase staining (the marker of senescence) using a commercially available kit (see Table of Materials).
- Measure secretion of pro-inflammatory cytokines, such as CCL2, by employing commercially available kits (see Table of Materials).
- Confirm increased levels of the cell cycle inhibitors p53, p21, p16 by Western Blot. NOTE: To properly control these assays, ArCK populations should be compared to both euploid and aneuploid cycling cells. The former is low passage, wild-type RPE-1 hTERT cells exponentially growing. The latter can be obtained by inducing chromosome mis-segregation in RPE-1 hTERT and harvesting cells 72 h after the first aberrant mitosis. To do so, follow the protocol described above from step 1.1 and harvest cells right before the first nocodazole treatment (step 3.1).
Perform single-cell sequencing to assess the karyotype of the arrested cells. NOTE: Single-cell sequencing is a well-established method to determine chromosomal and sub-chromosomal changes across a cell population at the single cell level13. Detailed procedures for how to perform single-cell sequencing can be found elsewhere10.
Representative Results
This method utilizes an in vitro tissue culture system to isolate aneuploid cells with complex karyotypes that arrest their proliferation. This population is referred to as ArCK (Arrested with Complex Karyotypes) cells. Figure 1A shows the scheme of the experiment. Wild-type cycling RPE-1 hTERT cells are synchronized at the G1/S border with thymidine and treated with a Mps1 inhibitor to induce chromosome mis-segregation. The spindle poison nocodazole is added to the cells 72 h after induction of chromosome mis-segregation. 12 h after nocodazole treatment, the aneuploid cells that are still able to proliferate enter mitosis and are trapped in this cell cycle stage due to interference with microtubule polymerization. These trapped cells are then easily removed by gentle shake-off. The shake-off process is repeated 3 - 5 times to ensure complete removal of cycling cells. Figure 1B shows the representative images of RPE-1 cells at each stage of the treatment.
ArCK cells display increased levels of cell cycle inhibitors, such as p53, p21, and p16 compared to euploid and aneuploid cycling cells, as shown in Figure 2A. Additionally, Figure 2B shows positive β-galactosidase staining in ArCK cells in comparison to euploid control cells, a well-established marker for cellular senescence.
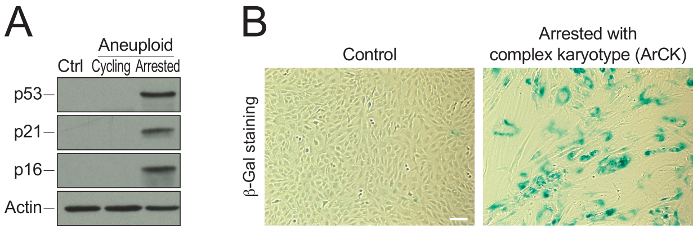
Figure 2: Representative results utilizing ArCK cells.(A) Western blot assessing cell cycle inhibitor levels in euploid, aneuploid cycling, and ArCK cells. The levels of p53, p21, and p16 were determined by western blot analysis. Actin served as a loading control. (B) β-galactosidase staining assessing the level of senescence in ArCK cells. Senescence-associated β-galactosidase (β-Gal) activity was determined in euploid cells and ArCK cells. Scale bar 100 µm. Please click here to view a larger version of this figure.
This cell cycle arrest is a prominent feature of aneuploid cells with complex karyotypes, as shown by representative karyotype analysis from single-cell sequencing in Figure 3, in which ArCK cells display multiple, random chromosome gains and losses. This finding is in full agreement with previous reports10.
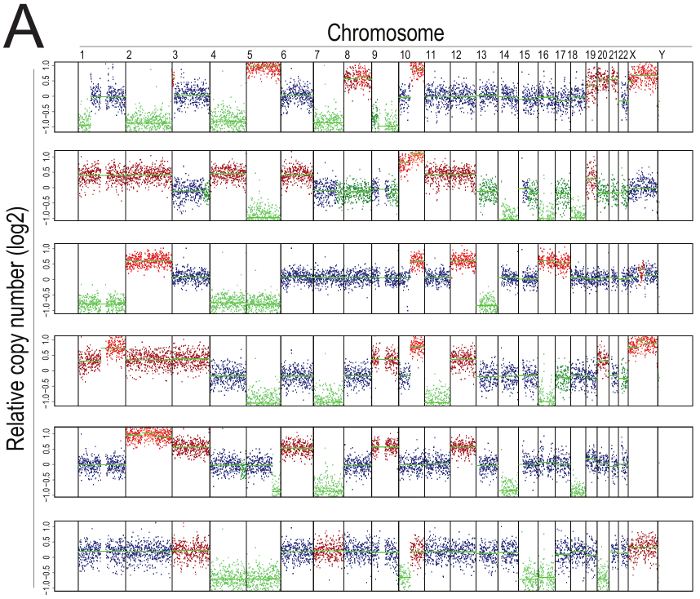
Figure 3: Representative single-cell sequencing of ArCK cells. Segmentation plots showing the karyotype of six representative ArCK cells. Segmentation plots show the copy number of all chromosomes from 1 to X relative to a euploid reference on a log2 scale (wild type RPE-1 is a diploid cell line of female origin). Chromosome gains are highlighted in red, chromosome losses in green. Please click here to view a larger version of this figure.
Discussion
This novel method to generate and enrich for arrested cells with complex karyotypes (ArCK) allows for the study of cells that have multiple chromosome gains or losses and cease to divide. The method setup has been designed to facilitate the isolation of ArCK cells in a quick and reliable way.
The most critical step in this assay is rigorously controlling the timeline of drug treatment and, most importantly, the removal of aneuploid cycling cells. For optimal results, the timing of nocodazole treatment and shake-off is of particular importance to ensure that cycling cells are removed from the plate while they are still rounded and mitotic, preventing the possibility of mitotic cell death or slippage into G1, potentially creating a tetraploid population. It is not recommended that the timing of shake-off deviates more than two hours from the recommended 12-h nocodazole incubation.
Future studies on ArCK cells have the potential to facilitate a deeper understanding of how complex karyotypes affect cell physiology. In particular, a recent study demonstrated that cells of the immune system are able to interact with and trigger immune clearance of ArCK cells10. The method described here provides an excellent starting point for further characterization of ArCK cells, including the clarification of the molecular mechanisms underlying immune clearance in untransformed cells and the study of how oncogenic transformation may bypass this surveillance mechanism, a crucial question in the field14,15.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This work was supported in part by the Koch Institute Support (core) Grant P30-CA 14051 from the National Cancer Institute, by the National Institutes of Health Grant (CA206157-22 and GM118066) and Curt Marble Cancer Research Fund to Angelika Amon. Angelika Amon is also an investigator of the Howard Hughes Medical Institute and the Glenn Foundation for Biomedical Research. S. S. was supported by the American Italian Cancer Foundation (AICF), by a Fellowship in Cancer Research from Marie Curie Actions and the Italian Association for Cancer Research (AIRC), and by a KI Quinquennial Cancer Research Fellowship. E.M. was supported by a fellowship from the Howard Hughes Medical Institute.
References
- Santaguida S, Amon A. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nature Reviews Molecular Cell Biology. 2015;16(8):473–485. doi: 10.1038/nrm4025. [DOI] [PubMed] [Google Scholar]
- Pfau SJ, Amon A. Chromosomal instability and aneuploidy in cancer: from yeast to man. EMBO reports. 2012;13(6):515–527. doi: 10.1038/embor.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nature Reviews Genetics. 2012;13(3):189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nature Reviews Molecular Cell Biology. 2009;10(7):478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meena JK, Cerutti A, et al. Telomerase abrogates aneuploidy-induced telomere replication stress, senescence and cell depletion. The EMBO Journal. 2015. pp. 1–14. [DOI] [PMC free article] [PubMed]
- Ohashi A, Ohori M, et al. Aneuploidy generates proteotoxic stress and DNA damage concurrently with p53-mediated post-mitotic apoptosis in SAC-impaired cells. Nature Communications. 2015;6:7668. doi: 10.1038/ncomms8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passerini V, Ozeri-Galai E, et al. The presence of extra chromosomes leads to genomic instability. Nature Communications. 2016;7:10754. doi: 10.1038/ncomms10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Blank HM, et al. Aneuploidy drives genomic instability in yeast. Science. 2011;333(6045):1026–1030. doi: 10.1126/science.1206412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Pavelka N, Bradford WD, Rancati G, Li R. Karyotypic determinants of chromosome instability in aneuploid budding yeast. PLoS Genetics. 2012;8(5):e1002719. doi: 10.1371/journal.pgen.1002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Richardson A, et al. Chromosome Mis-segregation Generates Cell-Cycle-Arrested Cells with Complex Karyotypes that Are Eliminated by the Immune System. Developmental Cell. 2017;41(6):638.e5–651.e5. doi: 10.1016/j.devcel.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Vasile E, White E, Amon A. Aneuploidy-induced cellular stresses limit autophagic degradation. Genes & Development. 2015;29(19):2010–2021. doi: 10.1101/gad.269118.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Tighe A, D'Alise AM, Taylor SS, Musacchio A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. The Journal of Cell Biology. 2010;190(1):73–87. doi: 10.1083/jcb.201001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nature Reviews Genetics. 2016;17(3):175–188. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- López-Soto A, Gonzalez S, López-Larrea C, Kroemer G. Immunosurveillance of Malignant Cells with Complex Karyotypes. Trends in cell biology. 2017. pp. 1–4. [DOI] [PubMed]
- Watson EV, Elledge SJ. Aneuploidy Police Detect Chromosomal Imbalance Triggering Immune Crackdown! Trends in genetics: TIG. 2017. pp. 1–3. [DOI] [PubMed]


