Abstract
Previous screening of a cDNA library of leaf poly(A+) RNA from Urochloa panicoides, a phosphoenolpyruvate carboxykinase (PCK)-type C4 monocot, led to the characterization of cDNAs encoding the U. panicoides PCK subunit PCK1. A second PCK sequence, designated PCK2, has now been found by rescreening the library. The deduced PCK2 polypeptide is 626 residues in length, has a predicted molecular mass of 68,686 D, and is 96% identical to the deduced PCK1 sequence. Isolation and characterization of genomic DNA fragments revealed that the PCK1 and PCK2 genes are each closely linked to another PCK gene. These additional genes have been designated PCK3 and PCK4, respectively. In each case, the second gene is located upstream and in the same transcriptional orientation as the gene characterized through cDNA analysis. A reverse transcription-polymerase chain reaction assay was used to demonstrate that PCK1 and PCK2 transcripts predominate in leaves, whereas PCK3 and PCK4 transcripts predominate in roots. Moreover, accumulation of PCK1 and PCK2 transcripts is light dependent. Direct N-terminal sequencing of PCK polypeptides purified from leaves demonstrated that PCK2 is produced. These results strongly suggest that PCK1 and PCK2 are involved in the photosynthetic CO2-concentrating mechanism active in U. panicoides.
PCK (ATP [GTP]: oxaloacetate carboxylase [transphosphorylase], EC 4.1.1.32 [GTP dependent] or EC 4.1.1.49 [ATP dependent]) is widespread in nature and catalyzes the reversible decarboxylation of oxaloacetate to PEP. The GTP-dependent enzyme found in animals catalyzes the first committed step in gluconeogenesis (Utter and Kolenbrander, 1972). The ATP-dependent enzyme of plants performs this function in the cotyledons of species with fat-storing seeds, mobilizing reduced carbon from lipids for use in other tissues of the seedling (Leegood and ap Rees, 1979). The plant enzyme also has a key role in photosynthetic carbon assimilation in one group of C4 (Hatch, 1987) and CAM (Dittrich et al., 1973) plants, and in some species of algae (Reiskind and Bowes, 1991), and may be involved in the response of Brassica napus to chilling (Sáez-Vásquez et al., 1995).
In PCK-type C4 grasses such as Urochloa panicoides, PCK is involved in the carbon-concentrating mechanism inherent in photosynthetic tissues (Hatch, 1987). Located in the cytosol (Ku et al., 1980; Chapman and Hatch, 1983), PCK is the major decarboxylating enzyme found in the bundle sheath cells (Hatch, 1987). Through the decarboxylation of oxaloacetate to PEP, PCK helps raise the CO2 concentration in the bundle sheath cells to levels much higher than that found in mesophyll cells or the atmosphere. The increased level of CO2 then suppresses photorespiration in these plants (Hatch, 1987).
The plant PCK is a multimeric enzyme of identical subunits (Burnell, 1986; Walker et al., 1995). In PCK-type C4 plants, the leaf enzyme involved in photosynthesis is hexameric (Burnell, 1986). In contrast, the enzyme found in gluconeogenic cucumber (Cucumis sativus) cotyledons is tetrameric (Walker and Leegood, 1995; Walker et al., 1995). The full-length U. panicoides PCK subunit is 68 kD (Finnegan and Burnell, 1995), but the subunit size varies from 67 to 78 kD in other species (Walker et al., 1995; Walker and Leegood, 1996). The N terminus of the plant PCK subunit may contain regions important for enzyme regulation, because it contains a target site for dark-dependent, light-reversible phosphorylation in most species examined (Walker and Leegood, 1996; Walker et al., 1997). The enzyme from the leaves of U. panicoides and several other C4 grasses is not susceptible to phosphorylation (Walker and Leegood, 1996). However, the N terminus of the leaf subunit is extremely labile (Finnegan and Burnell, 1995; Walker et al., 1995), which is true for the enzyme subunit from all species examined (Walker and Leegood, 1996; Walker et al., 1997).
A single complete cDNA sequence for a PCK subunit has been reported for cucumber (Cucumis sativus) (Kim and Smith, 1994) and the PCK-type C4 grasses U. panicoides (Finnegan and Burnell, 1995), Spartina anglica (accession no. E12730), and Zoysia japonica (accession no. E12731). However, genome sequencing of Arabidopsis has revealed three possible PCK gene sequences (accession nos. AC004705, CAA16690, and CAB38935). Moreover, analysis of U. panicoides cDNAs (Finnegan and Burnell, 1995) indicated that the enzyme subunit is also encoded by a multigene family in this species. Northern analysis using the U. panicoides PCK1 cDNA as a probe showed that the accumulation of PCK transcripts in dark-grown U. panicoides seedlings is induced by light (Finnegan and Burnell, 1995), which is in keeping with the role of the enzyme in photosynthesis. We report the cDNA sequence corresponding to a second U. panicoides PCK gene, PCK2, as well as the partial genomic characterization of two other PCK genes, PCK3 and PCK4. Results of a RT-PCR assay indicated that PCK1 and PCK2 are expressed in a leaf-predominant manner, and are therefore likely to be the subunits involved in photosynthesis. In contrast, transcripts from PCK3 and PCK4 accumulate predominantly in roots.
MATERIALS AND METHODS
Plant Material
All plant material used for nucleic acid extractions was from Urochloa panicoides (accession no. CQ2798) supplied by Commonwealth Scientific and Industrial Research Organization, Division of Tropical Crops and Pastures (Brisbane, Queensland, Australia). Plant tissue was usually stored at −70°C prior to use. Light-grown plants were grown in full sunlight during the summer months. Dark-grown plants were grown at 28°C from seeds sown on wet tissue in aluminum foil-wrapped 60- × 60- × 95-mm polystyrene boxes. For light induction experiments, 7-d-old dark-grown plants were subjected to 100 μmol quanta m−2 s−1 light.
Library Construction and Screening
A U. panicoides cDNA expression library, constructed using poly(A+) RNA isolated from leaves of a light-grown plant, was screened with an anti-PCK antiserum and cDNA probes as previously described (Finnegan and Burnell, 1995). For construction of a genomic library, total U. panicoides DNA was isolated from leaves (Komari et al., 1989), partially digested with Sau3A, and size-fractionated on a linear NaCl gradient (Sambrook et al., 1989). The DNA between 15 and 23 kb was ligated (Sambrook et al., 1989) into Lambda DASH II (Stratagene) digested with BamHI. The library was packaged using a packaging extract (Gigapack II, Stratagene), and contained 1.8 × 106 independent clones when grown on Escherichia coli strain XL-1 Blue (Stratagene) plating bacteria (Sambrook et al., 1989). Approximately 1.5 × 106 phage were plated, transferred to Hybond-N+ membranes (Amersham), and probed with a heat-denatured, radiolabeled restriction fragment probe as described previously (Finnegan and Burnell, 1995). The probe was an 873-bp EcoRI/HindIII restriction fragment from λPCK170204 (Fig. 1) lying entirely within the PCK2 ORF.
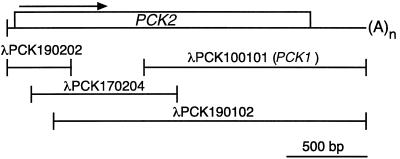
Figure 1.
Cloning of cDNAs encoding U. panicoides PCK2. A cDNA library derived from U. panicoides leaf poly(A+) RNA was screened by hybridization with progressively more 5′ fragments of U. panicoides PCK cDNA until a full-length PCK2 sequence was obtained. A schematic diagram of the composite PCK2 cDNA is shown above the constituent cDNA fragments. The ORF (box) and the direction of transcription (arrow) for PCK2 are indicated. The length and position of the PCK1 cDNA used as the hybridization probe in Southern analysis is also shown.
Southern-Blot Analysis
Purified U. panicoides genomic DNA (Komari et al., 1989) was digested with restriction enzymes under conditions recommended by the manufacturer (AMRAD-Pharmacia) and concentrated by precipitation with isopropanol (Sambrook et al., 1989). The fragments were separated on a 0.7% (w/v) agarose gel (Sambrook et al., 1989) and transferred to Hybond-N+ membranes by capillary blotting with 0.4 m NaOH. The membrane was prehybridized at 65°C for 1 h in 5× SSC (1× SSC = 0.15 m NaCl and 15 mm trisodium citrate), 1% (w/v) SDS, 50 mm sodium phosphate (pH 7.0), 0.1% (w/v) Ficoll (AMRAD-Pharmacia), 0.1% (w/v) PVP, 0.1% (w/v) BSA, and 0.5 mg mL−1 heat-denatured herring sperm DNA. A heat-denatured, radiolabeled restriction fragment probe was added and hybridization was allowed to proceed for 16 h at 65°C. The membrane was washed twice at 65°C for 15 min in 2× SSC, 0.1% (w/v) SDS, and twice at 65°C for 30 min in 0.1× SSC, 0.1% (w/v) SDS before autoradiography. The probe was the partial PCK1 cDNA insert from clone λPCK100101 (Finnegan and Burnell, 1995) and extended 1.4 kb upstream from the poly(A+) addition site (Fig. 1).
Oligonucleotide Primers
The primers used in this study were synthesized by the Macquarie University Centre for Analytical Biotechnology (Sydney, Australia), the Queensland University of Technology Centre for Molecular Biotechnology (Brisbane, Australia) or Gene Works (Adelaide, Australia). The sequences of the universal U. panicoides PCK gene primers are 5′-GCGCGCGCGGCCGCAAGATGCAAAGCACGCCC-3′ (UP1) and 5′-CGTCAACACCTGGACGGACA-3′ (UP2). The sequences for the PCK gene-specific primers are 5′-AGCATACAGAGCTGGTCTACTC-3′ (PCK1), 5′-AGCATACAGAGCTGGTCTACTG-3′ (PCK2), 5′-ATCATCGTAACACACGCACCAG-3′ (PCK3), and 5′-GTTGG-CACATCGATCCAACACA-3′ (PCK4).
RT-PCR Assays
Total RNA was purified from various U. panicoides tissues according to the method of Chomczynski and Sacchi (1987). Reverse transcriptase reactions were performed in 20-μL reactions containing 1 μg of total RNA, 10 mm Tris-HCl (pH 8.4), 50 mm KCl, 5 mm MgCl2, 2.5 μm (50 pmol) each primer, 1 mm each dATP, dGTP, dCTP and dTTP, 5 mm DTT, 20 units of RNA Guard (AMRAD-Pharmacia), and 200 units of Moloney murine leukemia virus reverse transcriptase (Promega). Stock solutions of RNA, buffer, salts, and primers were combined, heated at 95°C for 2 min, and chilled on ice. Appropriate volumes of ice-cold dNTP and DTT stocks were added, followed by the RNA Guard and reverse transcriptase. Reactions were incubated at 42°C for 60 min. For RT-PCR assays, 5 μL of the reverse transcriptase reaction was added to 45 μL of ice-cold PCR cocktail containing 10 mm Tris-HCl (pH 8.4), 50 mm KCl, 4.5 mm MgCl2, 200 μm each dATP, dGTP, dCTP and dTTP, 10 pmol of each primer, and 1 unit of DNA polymerase (AmpliTaq, Cetus-Perkin-Elmer). Complete PCR mixtures were moved from ice to a thermal cycler (Cetus-Perkin-Elmer) prewarmed to 95°C, and incubated for 2 min. Templates were amplified by incubating the reactions at 95°C for 45 s and 70°C for 2 min for 30 cycles. The products were analyzed by electrophoresis on agarose gels (Sambrook et al., 1989).
Miscellaneous Methods
Standard procedures (Sambrook et al., 1989) were used to prepare phage DNA from plate lysates and to subclone inserts into pBluescript II KS− (Stratagene). Deletion mutants were generated by exonuclease III treatment and sequenced as described previously (Finnegan and Burnell, 1995). Restriction fragments to be used as probes were purified from agarose gels (Sambrook et al., 1989) and radiolabeled with [α-32P]dCTP (NEN-DuPont) by random priming using a DNA-labeling kit (Gigaprime, Gene Works, Adelaide, Australia).
RESULTS
Multiple Genes Encode PCK in U. panicoides
After analysis of cDNAs, we previously reported that the PCK1 gene of U. panicoides encodes a PCK subunit with a molecular mass of 68.5 kD (Finnegan and Burnell, 1995). During the isolation of PCK1 cDNAs, it became apparent that there was another abundant cDNA sequence in the library that was similar but not identical to the PCK1 cDNA sequence. Using probes derived from progressively more 5′ sequences to screen the library (Finnegan and Burnell, 1995), a group of cDNAs (Fig. 1) was isolated that formed a sequence encoding this second PCK subunit. The gene specifying the new U. panicoides subunit was thus designated PCK2. Both strands of each clone were sequenced entirely and the overlapping regions of the clones found to be identical in sequence. A total of 2,268 bp of PCK2 cDNA sequence was determined, comprised of an ORF of 1,878 bp flanked by 5′ and 3′ UTRs of 86 and 304 bp, respectively (Fig. 1). Although there were no in-frame stop codons in the 5′ UTR, 5′-RACE experiments indicated that the ORF shown in Figure 1 was complete (results not shown). The 3′ UTR in clone λPCK190102 was followed by 30 adenosine residues, presumably representing the poly(A+) tail of the PCK2 cDNA.
When compared over their entire lengths, the PCK1 and PCK2 cDNAs were 94.4% identical at the nucleotide level. Much of the variation occurred in the 3′ UTRs of the cDNAs, which were 82.5% identical (Fig. 2). The variation between the 3′ UTRs was mainly due to a 26-bp insertion in PCK1 that includes one copy of a 23-bp direct repeat. The PCK2 cDNA has an additional 44-bp 3′ extension compared with PCK1.
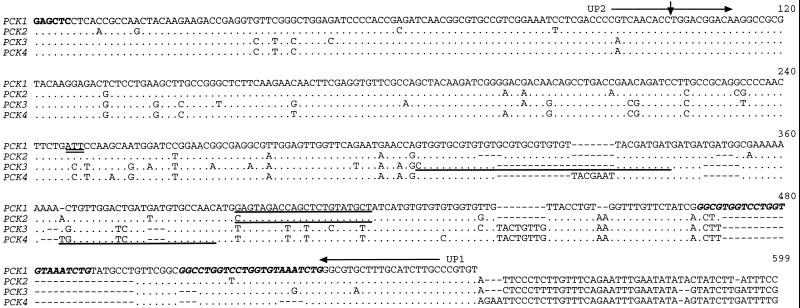
Figure 2.
Comparison of the nucleotide sequences at the 3′ ends of four putative U. panicoides PCK mRNAs. The sequence of the PCK1 cDNA (accession no. U09241) from the SacI site (bold) located 234 bp upstream of the stop codon (double underline) to the poly(A+) addition site is shown on the top line. The homologous sequences of the PCK2 cDNA (accession no. AF136161) and the PCK3 (accession no. AF136162) and PCK4 (accession no. AF136163) genes are shown below. Dots represent residues identical to PCK1 and dashes denote gaps introduced to optimize the alignment. The position of the single intron covered by this comparison and conserved in all four U. panicoides PCK genes is indicated by the vertical arrowhead. The intron sequences have been omitted from this comparison. The position and direction of the “universal” PCK primers UP1 and UP2 (horizontal arrows) and the sequences to which primers specific for each PCK gene anneal (underlines) are shown. The 23-bp repeat sequences in the PCK1 3′ UTR are shown in bold italics.
The deduced PCK2 polypeptide had 626 amino acid residues (Fig. 3), two more than the PCK1 polypeptide, and a calculated molecular mass of 68,686 D. The latter corresponds exactly to the estimated size of the U. panicoides protein obtained from immunoblots (68–69 kD; Finnegan and Burnell, 1995; Walker et al., 1995). The PCK2 ORF had 74 nucleotide differences compared with that of PCK1. This variation gave rise to 26 amino acid differences (Fig. 3), of which 21 (81%) were nonconservative. The differences between the U. panicoides PCK1 and PCK2 polypeptides were mainly localized to the N- and C-terminal regions of the proteins. Only one difference, the conservative substitution of Thr-236 in PCK1 for a Ser in PCK2, occurred within any of the subsequences previously identified as having above-average similarity among ATP-dependent PCKs (Linss et al., 1993; Finnegan and Burnell, 1995). Moreover, none of the differences involved residues thought to be present in the PCK active site as defined by analysis of the crystal structure of the E. coli enzyme (Fig. 3; Matte et al., 1996).
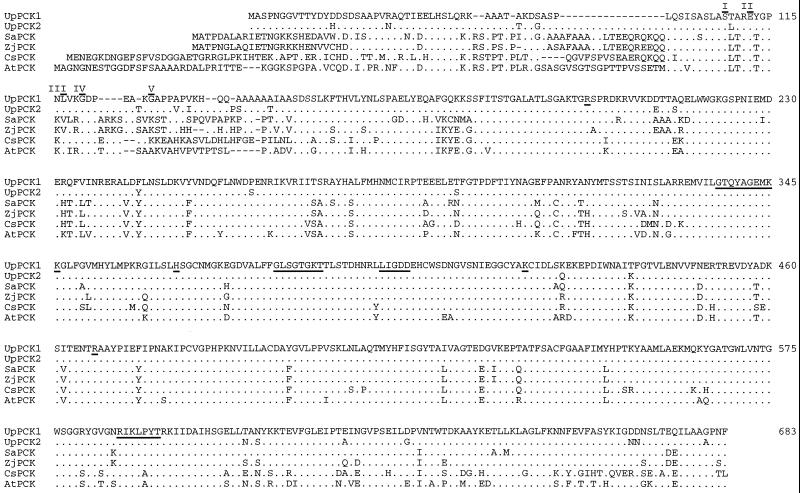
Figure 3.
Comparison of plant PCK subunit sequences. The entire U. panicoides PCK1 amino acid sequence is shown on the top line in the single-letter code. Residues present in the PCK active site (Matte et al., 1996) are underlined. N-terminal residues determined directly from PCK polypeptides are overlined and are assigned Roman numerals corresponding to the polypeptides in Figure 8. Dots in the U. panicoides (Up) PCK2, S. anglica (Sa), Z. japonica (Zj), cucumber (Cs), and Arabidopsis (At) subunit sequences indicate residues found in the U. panicoides PCK1 sequence. Gaps (dashes) were introduced to maximize identity. The numbering refers to the positions within the comparison.
The complete primary structure of a single PCK subunit has been determined for cucumber (Kim and Smith, 1994) and the PCK-type C4 monocots S. anglica (accession no. E12730) and Z. japonica (accession no. E12731), whereas genes for two complete subunits have been characterized for Arabidopsis (accession nos. CAA16690 and CAB38935). All of these proteins had N-terminal extensions compared with the PCKs of U. panicoides, ranging from 29 residues for the Z. japonica subunit to up to 48 residues for one of the Arabidopsis subunits (Fig. 3). Within the overlapping sequences (Fig. 3), the S. anglica and Z. japonica PCKs were 80% identical to U. panicoides PCK1, while the cucumber and Arabidopsis subunits were 75% identical to this subunit. Many of the differences among the subunits were located in the N-terminal portion of the comparison (Fig. 3). When the comparison was restricted to the sequences following Ser-95 of U. panicoides PCK1, the identity with U. panicoides PCK1 increased to 90% for the S. anglica and Z. japonica subunits and to nearly 85% for the cucumber and Arabidopsis sequences.
Genomic Organization of PCK Genes
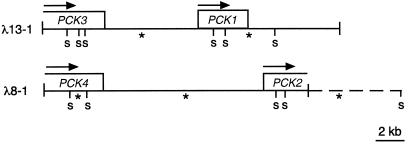
A U. panicoides genomic DNA library was screened for PCK genes by hybridization of membrane-bound plaques with the 873-bp cDNA insert from λPCK170204 (Fig. 1). This probe covered the N-terminal portion of the PCK2 ORF. The inserts of several hybridizing clones were characterized. Restriction mapping, Southern analysis, and partial sequencing of clones λ8-1 and λ13-1 indicated that each possessed two unique PCK-related sequences (Fig. 4). The approximately 21-kb insert of λ13-1 encompassed the entire PCK1 gene and an estimated 80% of a presumed PCK gene designated PCK3. PCK1 and PCK3 had the same transcriptional sense, with the initiation codon of PCK1 located about 7.5 kb downstream of the PCK3 termination codon. The 18-kb insert of λ8-1 contained the 5′ 90% of PCK2 and the 3′ 85% of another presumed PCK gene, PCK4. Again, PCK2 and PCK4 were transcribed in the same direction, with the stop codon of PCK4 being about 11.5 kb upstream of the PCK2 start codon. The length of the PCK2 gene is assumed to be similar to that of PCK1, which is about 3.2 kb. The PCK3 and PCK4 genes appear to be somewhat longer, but the 5′ ends of these genes have not yet been fully characterized.
Figure 4.
Organization of PCK genes in U. panicoides. Schematic representations of the SalI restriction fragment maps of two genomic bacteriophage lambda clones encoding the four identified PCK genes in U. panicoides are shown and include the ORF (boxes) and the direction of transcription (arrows) for each gene. The SalI fragments that hybridize to a 1.4-kb partial PCK1 cDNA (see Fig. 1) are indicated with asterisks. The dashed line extends to a SalI site mapped by genomic Southern analysis.
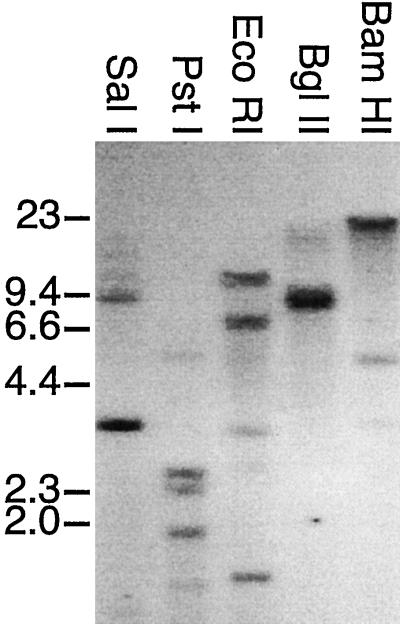
The possibility that U. panicoides may have more than four PCK subunit genes was examined by genomic Southern hybridization analysis (Fig. 5). Restriction digests of U. panicoides total cellular DNA were separated on an agarose gel, transferred to a membrane, and probed with the 1.4-kb λPCK100101 cDNA insert covering the C-terminal 60% of the ORF and the entire 3′ UTR of the PCK1 mRNA (Fig. 1). This probe detected five fragments in SalI digests of U. panicoides genomic DNA (Fig. 5, lane 1). Examination of the maps of clones λ8-1 or λ13-1 (Fig. 4) indicated that each of these labeled fragments corresponded to a SalI fragment predicted to hybridize to the probe. The 10-kb hybridizing fragment corresponded in size to the fragment extending from the C-terminal end of PCK3 into the N-terminal portion of PCK1 on the map of λ13-1, whereas the 3.3-kb hybridizing fragment spanned the C terminus of PCK1. The 1.2- and 14-kb hybridizing fragments were located on λ8-1; the 1.2-kb fragment was entirely within PCK4. The 14-kb fragment extended from within the C-terminal portion of PCK4, across the PCK4-PCK2 intergenic region, and into the N-terminal part of PCK2. From the maps of the λ8-1 and λ13-1 inserts, the only other fragment larger than 500 bp expected to hybridize to the probe would encode the C terminus of PCK2 and would be greater than 1.2 kb. It is likely, then, that the 8-kb hybridizing fragment corresponded to this fragment. Due to the high-stringency wash conditions used, the variation in hybridization signals among the bands probably reflects sequence divergence among the PCK genes. In this regard, the 3.3-kb SalI fragment, which contains sequences identical to the probe, produced the strongest signal.
Figure 5.
Genomic Southern analysis of PCK genes. Approximately 10 μg of U. panicoides total genomic DNA was digested with the indicated restriction endonucleases. The fragments were separated on a 0.7% (w/v) agarose gel, transferred to a nylon membrane, and probed with a 1.4-kb U. panicoides PCK1 partial cDNA (Fig. 1). The positions and sizes (in kb) of the HindIII fragments of bacteriophage lambda are shown on the left as markers.
When the genomic DNA digested with PstI, EcoRI, BglII or BamHI (Fig. 5, lanes 2–5) was probed with the 1.4-kb PCK1 cDNA fragment, relatively simple hybridization patterns were obtained (Fig. 5). In both the BglII and BamHI digests, only three hybridizing fragments were observed. Given the length of the PCK3-PCK1 (7.5 kb) and PCK4-PCK2 (11 kb) intergenic regions, and the fact that the coding regions for PCK1 and PCK2 were each about 3.5 kb (Fig. 4), it is unlikely that the hybridizing 20-kb BamHI fragment would have segments of more than two PCK genes. The 4.9- and 3.4-kb BamHI fragments were each too small to carry more than one PCK gene. Similarly, the 17-kb BglII fragment is unlikely to span more than two PCK genes, whereas the 8.5- and 7.9-kb fragments were not large enough to span the distance between regions known to hybridize to the probe. Taken together, these observations indicate that the PCK multigene family contains only four closely related members, but the possibility of other more divergent members cannot be eliminated.
3′-End Sequence Analysis of the PCK Genes
Northern analysis has previously demonstrated that the expression of a 2.7-kb PCK mRNA is light inducible in U. panicoides leaves but undetectable in roots (Finnegan and Burnell, 1995). As a first step in determining the relative contributions of the four U. panicoides PCK genes to the PCK transcript pool, the feasibility of designing PCK-specific probes was examined by sequencing the 3′ UTR of the four genes. Figure 2 shows a comparison of the four PCK sequences from the conserved SacI site 244 bp upstream of the stop codon to the poly(A+) addition site for the PCK1 and PCK2 cDNAs, or to the position analogous to the PCK2 poly(A+) addition site for the PCK3 and PCK4 genomic sequences. Each PCK gene had a conserved intron (results not shown) located after position 102 in Figure 2. The C termini of the four PCK genes and their 3′ UTRs were very similar but not identical. There were a number of single nucleotide differences among the four 3′ UTRs and several small insertions/deletions ranging in length from one to 26 bp, with many of the insertions/deletions involving repeat sequences. These characteristics prevented the differentiation of gene-specific transcripts through hybridization (results not shown).
Tissue-Dependent Expression of PCK Genes
A RT-PCR assay was used to examine the tissue-dependent expression of the four U. panicoides PCK genes. Two convergent oligonucleotide primers, UP1 and UP2 (Fig. 2), were designed to detect all PCK transcripts present in preparations of total RNA. In the assay (Fig. 6A), the downstream universal PCK gene primer UP1 was used to prime cDNA synthesis. The resulting cDNA was then amplified using UP1 and the upstream universal PCK gene primer UP2. Because primer UP2 spans the intron excision site in the 3′ region of the PCK genes (Fig. 2), the UP1/UP2 primer pair should not amplify PCK genomic sequences that may contaminate preparations of total RNA.
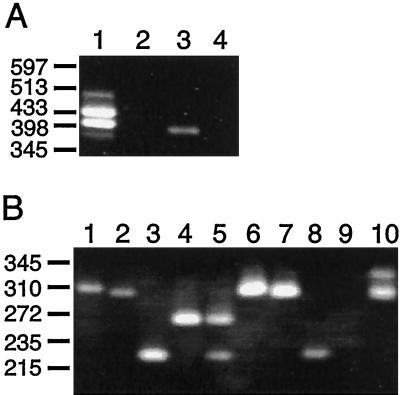
Figure 6.
Tissue-specific expression of U. panicoides PCK genes. A, RT-PCR assay using universal PCK primers. The universal PCK primer UP1 (see Fig. 2) was used to prime reverse transcription of 1 μg of total RNA isolated from leaves (lanes 1 and 2) or roots (lanes 3 and 4) of a light-grown plant. The reactions either contained (lanes 1 and 3) or lacked (lanes 2 and 4) reverse transcriptase. The products were amplified by PCR using the universal PCK primers UP1 and UP2 (see Fig. 2) before separation on an agarose gel. B, Semi-nested RT-PCR using PCK-specific primers (see Fig. 2). The RT-PCR products from A were purified, diluted 105-fold, and reamplified by standard PCR (see Methods). The products obtained from the reamplification of the root (lanes 1–5) or leaf (lanes 6–10) RT-PCR products are shown. The semi-nested PCR step was primed with universal primer UP2 and the primers specific for PCK1 (lanes 1 and 6), PCK2 (lanes 2 and 7), PCK3 (lanes 3 and 8), or PCK4 (lanes 4 and 9). Competitive PCRs containing primer UP2 and all four gene-specific primers are also shown (lanes 5 and 10). The positions and sizes (in kb) of the AvaII fragments of bacteriophage lambda are shown on the left as markers.
When total RNA from the green leaves of U. panicoides was subjected to RT-PCR, two predominant products of 435 and 405 bp were detected (Fig. 6A, lane 1) corresponding in size to the products expected from PCK1 and PCK2 transcripts, respectively. Two additional products of about 490 and 380 bp were also obtained. The origins of these products are not known, but neither hybridized to PCK1 cDNA probes (not shown). Total RNA isolated from root tissue gave rise to a single product band of about 400 bp (Fig. 6A, lane 3) that was indistinguishable in size from the 393- and 396-bp products expected from PCK3 and PCK4 transcripts, respectively. None of the RT-PCR products observed in this experiment were due to genomic DNA contamination of the leaf and root RNA preparations, because no products were detected when reverse transcriptase was omitted from the assay (Fig. 6A, lanes 2 and 4). The results of this experiment indicate that PCK1 and PCK2 are expressed in a leaf-predominant manner, whereas some combination of PCK3 and PCK4 is expressed in a root-predominant manner.
As the RT-PCR products from PCK3 and PCK4 transcripts could not be distinguished from one another by length or restriction site polymorphisms due to the high similarity of the 3′ regions of these genes (Fig. 2), a semi-nested PCR assay was employed to examine their expression. The UP1/UP2 RT-PCR products from leaf and root RNA were purified and re-amplified using the universal primer UP2 together with individual primers specific for each PCK gene (Fig. 2). Each primer pair yielded only the single predicted product when used to amplify the RT-PCR product from leaf and root RNA (Fig. 6B). These products were 314 bp for PCK1 (lanes 1 and 6), 308 bp for PCK2 (lanes 2 and 7), 231 bp for PCK3 (lanes 3 and 8), and 273 bp for PCK4 (lane 4). The only variation in this pattern was that the UP2/PCK4-specific primer pair did not always produce a product when leaf RNA was the starting material (Fig. 6B, lane 9).
The relative abundance of the products from these four reactions was dependant on the tissue source of the RNA template. When leaf RNA was tested, similar amounts of the PCK1 and PCK2 products were obtained. These amounts were consistently greater than those of the PCK3 and PCK4 products produced (Fig. 6B, compare lanes 6 and 7 with lanes 8 and 9). This result is similar to that seen for the direct RT-PCR experiment described above, and suggests that transcripts from the former two genes are more abundant in leaf tissue than transcripts from the latter two genes. However, when root RNA was examined, PCK1- and PCK2-specific products were always obtained in much lower amounts than the PCK3 and PCK4 products (Fig. 6B, compare lanes 1 and 2 with 3 and 4), implying that the transcripts from the latter two genes are more abundant in root tissue than PCK1 and PCK2 transcripts.
The qualitative differences observed when the RT-PCR products were amplified with the gene-specific primers in separate reactions was verified by semicompetitive PCR. Root RNA was amplified in a semi-nested RT-PCR assay in which primer UP2 and all four gene-specific primers were combined in a single reaction. This produced only the PCK3- and PCK4-specific products in similar amounts (Fig. 6B, lane 5). These products were not detected when leaf RNA was the template (Fig. 6B, lane 10); instead, two different products, neither of which was evident when root RNA was tested, were obtained. The 310-bp species is a doublet of the PCK1- and PCK2-specific products. The other product, with slightly lower mobility, was of unknown origin and only arose when the four gene-specific primers were combined. Therefore, this product probably arose from amplification of an unknown template through priming by two of these primers. Although the PCR assays described here are not quantitative, the results strongly indicate that the PCK1 and PCK2 transcripts are the most abundant in leaf tissue and that PCK3 and PCK4 transcripts are the most abundant in root tissue.
Light Induction of Individual PCK Genes
We previously reported that U. panicoides PCK transcript accumulation was induced by exposure to light (Finnegan and Burnell, 1995). The results of the present study indicated that PCK1 and PCK2 transcripts are the most abundant PCK transcripts in leaf tissue. To determine if the accumulation of transcripts from either of these genes is induced by light, the RT-PCR assay was used to examine PCK transcripts in greening shoots. U. panicoides seeds were germinated and seedlings were grown in the dark for 7 d before being exposed to continuous light. Cotyledons were harvested after various exposure times, and RNA was isolated from each individual. The RNA was then subjected to the RT-PCR assay using the UP1/UP2 primer pair. As demonstrated above, RNA isolated from roots yielded a product of about 400 bp (Fig. 7, lane 1), corresponding to a mixture of PCK3- and PCK4-specific products, whereas total leaf RNA from a light-grown plant gave two prominent products of 435 and 405 bp (Fig. 7, lane 8), corresponding to PCK1 and PCK2 transcripts, respectively.
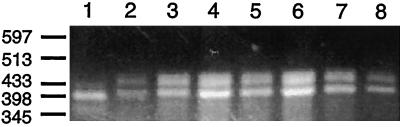
Figure 7.
Light induction of U. panicoides PCK genes. Total RNA (1 μg) isolated from the roots (lane 1) or leaves (lane 8) of a light-grown plant or from the cotyledons of 7-d-old dark-grown plants illuminated with 100 μmol quanta m−2 s−1 light for 0 (lane 2), 6 (lane 3), 12 (lane 4), 24 (lane 5), 30 (lane 6), or 36 (lane 7) h just prior to harvesting was subjected to RT-PCR using the universal PCK primers UP1 and UP2, as described in the legend to Figure 6A. The positions and sizes (in kb) of the AvaII fragments of bacteriophage lambda are shown on the left as markers.
When RNA from dark-grown shoots was tested for the presence of PCK transcripts, products were only obtained for one of six individuals. These products were identical to those from RNA from green leaves (Fig. 7, lane 2), but the band intensities were lower than normally observed using 1 μg of leaf total RNA (Fig. 7, lane 8). After 6 h of illumination, the PCK RT-PCR pattern in all six dark-grown cotyledons tested was exactly the same as in a green leaf (Fig. 7, compare lane 3 with lane 8). This pattern did not change with continuous illumination for 12 to 36 h (Fig. 7, lanes 4–7). The lack of detectable RT-PCR products from five of six dark-grown shoots was probably not due to failure of the assay, because RT-PCR products were obtained from all 23 light-exposed cotyledons examined; rather, it suggests a low abundance of PCK transcripts in dark-grown shoots.
Presence of PCK1 and PCK2 in Leaves
To determine if both PCK1 and PCK2 proteins are produced within the leaves of U. panicoides, N-terminal sequence information previously obtained for PCK (Finnegan and Burnell, 1995) was re-evaluated (Fig. 8). In an earlier study (Finnegan and Burnell, 1995), the first attempt to obtain a N-terminal sequence was made by subjecting a mixture of the 60- to 63-kD PCK polypeptides to 12 cycles of automated Edman degradation. Each cycle released three or four different amino acids, allowing the identification of five distinct sequences with high identity to the deduced PCK1 sequence (marked I–V in Fig. 8). Examination of the differences between the predicted PCK1 and PCK2 sequences over these short regions allowed the origin of all the sequences except sequence II to be identified (Fig. 8). The Leu residue released at cycle 2 suggested that sequence I was derived from PCK2, whereas the Lys residue released in cycle 9 indicated that sequence III arose from PCK1. These two N termini were the most abundant in the mixture, together accounting for the most abundant amino acids released from 11 of the 12 cycles (Fig. 8). The minor N termini represented by sequences IV and V arose from PCK1, as each contained residues specific for this subunit.
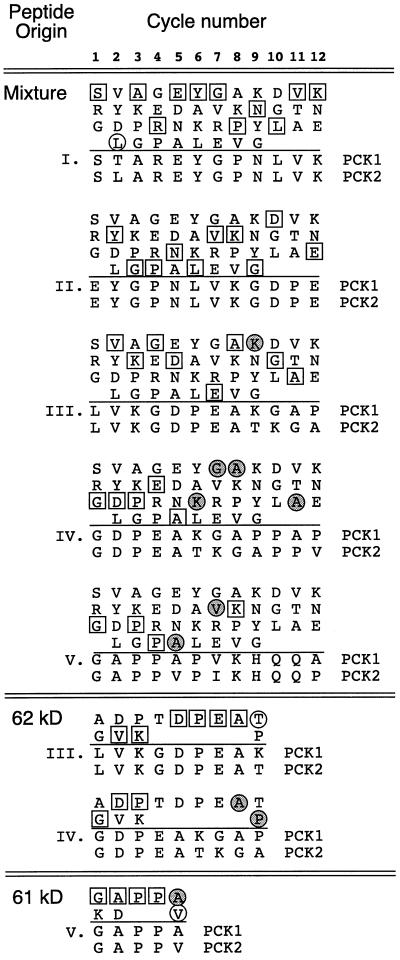
Figure 8.
N-terminal amino acid analysis of U. panicoides PCK polypeptides. PCK was purified from U. panicoides leaves using a method that results in the stepwise cleavage of up to 8 kD from the N terminus of a high proportion of the polypeptide chains (Finnegan and Burnell, 1995). The mixture of cleavage products was subjected to Edman degradation. The amino acid residues released in each cycle are shown in the matrices and are arranged in order of decreasing abundance (at the top of each column is the most abundant residue released). The sequences below the line in each matrix were deduced from the PCK1 and PCK2 cDNAs. The Roman numerals correspond to the sequences previously identified (Finnegan and Burnell, 1995). In the body of each matrix, residues found in both PCK1 and PCK2 sequences are indicated by white boxes, whereas residues found in only the PCK1 or the PCK2 sequence are indicated by shaded and white circles, respectively. This analysis was also performed with the SDS-PAGE-purified 62- and 61-kD PCK degradation products (Finnegan and Burnell, 1995).
In the previous study (Finnegan and Burnell, 1995), the N-terminal sequences present in the isolated 62- and 61-kD PCK fragments were also determined. Re-analysis of these results supported the conclusion that both PCK1 and PCK2 are produced within leaves (Fig. 8). The 62-kD band clearly contained N-terminal sequences III and IV derived from PCK2 and PCK1, respectively. The 61-kD band contained sequence V and was derived from both PCK1 and PCK2. The differences in the N termini present in the PCK mixture versus the isolated 62- and 61-kD bands may have been due to the increased ability to detect the amino acids released from low-abundance polypeptides in the purified bands compared with the mixture containing several different N termini. Alternatively, there may have been variations between the two enzyme preparations used in the analyses.
DISCUSSION
The U. panicoides PCK Multigene Family
In the C4 grass U. panicoides, PCK is encoded by a multigene family with at least four members. Multigene families encoding the subunits of this enzyme may be widespread in plants as genome sequencing has revealed three possible PCK genes in Arabidopsis. Moreover, genomic Southern hybridization analysis indicated that B. napus, as well as its progenitor species Brassica campestris and Brassica oleracea, also contain PCK multigene families (Sáez-Vásquez et al., 1995). In contrast, similar analysis detected only a single gene in cucumber (Kim and Smith, 1994).
The sequencing of cDNAs spanning the entire PCK2 ORF has shown that the PCK2 subunit is 96% identical to PCK1. None of the differences are likely to affect the active site of the ATP-dependent PCK (Matte et al., 1996). The concentration of sequence differences at the N and C termini has allowed us to show that both proteins are expressed in U. panicoides leaves. In fact, all four PCK genes examined here may yield a protein product. RT-PCR analysis indicated that both PCK3 and PCK4 are also transcriptionally active, but the possibility that one or both is translationally silent has not been ruled out.
The finding of two members of a gene family in close proximity to one another and in the same transcriptional orientation is not unknown in plants. Similar gene arrangements have been observed for the alternative oxidase genes AOX1a and AOX1b (Saisho et al., 1997) and the drought-induced genes rd29A/rd29B (Yamaguchi-Shinozaki and Shinozaki, 1993) in Arabidopsis, and the catalase genes cat1 and cat2 in castor bean (Suzuki et al., 1994).
Control of PCK Gene Expression
Expression of PCK subunit genes in plants apparently follows specific developmental programs. Transcripts from the U. panicoides genes accumulate in a tissue-dependent manner, with PCK1 and PCK2 transcripts predominating in leaves and PCK3 and PCK4 transcripts predominating in roots. Moreover, PCK1 and PCK2 transcripts accumulate in a light-dependent manner. In gluconeogenic cucumber cotyledons (Kim and Smith, 1994), PCK transcripts and protein are at maximal levels a few days after seed imbibition, and thereafter decline to undetectable levels until low levels of both transcripts and protein reappear during cotyledon senescence. The regulation of PCK gene expression may be triggered in part by metabolic cues. Transcripts accumulate during cold acclimation in B. napus, an adaptive response that alters the metabolic status of the affected tissue (Sáez-Vásquez et al., 1995).
Role of PCK Subunits
The demonstration that PCK1 and PCK2 transcripts accumulate in a light-inducible manner and are the most predominant PCK transcripts in leaves indicates that the corresponding proteins are likely to be those involved in the C4 photosynthetic pathway in U. panicoides. This conclusion is supported by the identification of N-terminal sequences corresponding to PCK1 and PCK2 in leaf extracts. The accumulation of PCK3 and PCK4 transcripts in a root-predominant manner indicates that the proteins encoded by these genes are involved in some other unknown, possibly anaplerotic, function. The requirement of PCK in roots may have an important physiological role. It has been proposed that the PCK enzyme detected in cucumber roots may perform a gluconeogenic function, converting storage lipids to sugar (Walker and Leegood, 1995). Our experiments indicate that there is likely to be low-level PCK3 and PCK4 expression in photosynthetically active leaves as well. Whether this level of expression is physiologically relevant remains to be elucidated, but low levels of PCK expression in photosynthetic organs has also been documented in C3 plants (Kim and Smith, 1994; Walker et al., 1995).
Regulation of PCK in Plants
In addition to the coarse regulation of PCK abundance possibly afforded by the regulation of gene expression, fine regulation of enzyme activity may also occur in most plants at the level of protein phosphorylation. Examination of PCK from a number of species (Walker and Leegood, 1996; Walker et al., 1997) revealed that there are two distinguishable types of enzymes. One type is found in gluconeogenic seedlings, including cucumber, and in the leaves of CAM plants and some C4 grasses, including S. anglica. This enzyme has a molecular mass of 71 to 74 kD and is subject to phosphorylation in vivo. The phosphorylation is dark dependent and light reversible, indicating that it may have a regulatory role (Walker and Leegood, 1996; Walker et al., 1997). Interestingly, the site of phosphorylation is located in the N-terminal extension found in plant PCKs (Walker and Leegood, 1996). This extension is rapidly cleaved from the enzyme during purification, suggesting that it is at the surface of the enzyme.
The second type of plant PCK is slightly smaller, 67 to 70 kD, and is not subject to phosphorylation. This enzyme type has only been found in the leaves of some PCK-type C4 grasses, including U. panicoides (Walker and Leegood, 1996; Walker et al., 1997). So far, only the predominant PCK found in leaf tissues, and therefore involved in photosynthetic carbon assimilation, has been examined in these species. It will be interesting to see whether the enzyme composed of PCK3 and/or PCK4 subunits is regulated differently than the photosynthetic enzyme.
Abbreviations:
- PCK
PEP carboxykinase
- RT-PCR
reverse transcription-PCR
Footnotes
This work was supported by a Joint Research and Development Grant from Japan Tobacco to J.N.B.
LITERATURE CITED
- Burnell JN. Purification and properties of phosphoenolpyruvate carboxykinase from C4 plants. Aust J Plant Physiol. 1986;13:577–587. [Google Scholar]
- Chapman KSR, Hatch MD. Intracellular location of phosphoenolpyruvate carboxykinase and other C4 photosynthetic enzymes in mesophyll and bundle sheath protoplasts of Panicum maximum. Plant Sci Lett. 1983;29:145–154. [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Dittrich P, Campbell WH, Black C. Phosphoenolpyruvate carboxykinase in plants exhibiting Crassulacean acid metabolism. Plant Physiol. 1973;52:357–361. doi: 10.1104/pp.52.4.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan PM, Burnell JN. Isolation and sequence analysis of cDNAs encoding phosphoenolpyruvate carboxykinase from the PCK-type C4 grass Urochloa panicoides. Plant Mol Biol. 1995;27:365–376. doi: 10.1007/BF00020190. [DOI] [PubMed] [Google Scholar]
- Hatch MD. C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochim Biophys Acta. 1987;895:81–106. [Google Scholar]
- Kim D-J, Smith SM. Molecular cloning of cucumber phosphoenolpyruvate carboxykinase and developmental regulation of gene expression. Plant Mol Biol. 1994;26:423–434. doi: 10.1007/BF00039551. [DOI] [PubMed] [Google Scholar]
- Komari T, Saito Y, Nakakido F, Kumasiro T. Efficient selection of somatic hybrids in Nicotiana tabacum L. using a combination of drug-resistance markers introduced by transformation. Theor Appl Genet. 1989;77:547–552. doi: 10.1007/BF00274277. [DOI] [PubMed] [Google Scholar]
- Ku MSB, Spalding MH, Edwards GE. Intracellular localization of phosphoenolpyruvate carboxykinase in leaves of C4 and CAM plants. Plant Sci Lett. 1980;19:1–9. [Google Scholar]
- Leegood RC, ap Rees T. Phosphoenolpyruvate carboxykinase and gluconeogenesis in cotyledons of Cucurbita pepo. Biochim Biophys Acta. 1979;524:207–218. doi: 10.1016/0005-2744(78)90119-5. [DOI] [PubMed] [Google Scholar]
- Linss H, Goldenberg S, Urbina JA, Amzel LM. Cloning and characterization of the gene encoding ATP-dependent phosphoenolpyruvate carboxykinase in Trypanosoma cruzi: comparison of primary and predicted secondary structure with the host GTP-dependent enzyme. Gene. 1993;136:69–77. doi: 10.1016/0378-1119(93)90449-d. [DOI] [PubMed] [Google Scholar]
- Matte A, Goldie H, Sweet RM, Delbaere TJ. Crystal structure of Escherichia coli phosphoenolpyruvate carboxykinase: a new structural family with the P-loop nucleoside triphosphate hydrolase fold. J Mol Biol. 1996;256:126–143. doi: 10.1006/jmbi.1996.0072. [DOI] [PubMed] [Google Scholar]
- Reiskind JB, Bowes G. The role of phosphoenolpyruvate carboxykinase in a marine macroalga with C-4-like photosynthetic characteristics. Proc Natl Acad Sci USA. 1991;88:2883–2887. doi: 10.1073/pnas.88.7.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáez-Vásquez J, Raynal M, Delseny M. A rapeseed cold-inducible transcript encodes a phoshoenolpyruvate carboxykinase. Plant Physiol. 1995;109:611–618. doi: 10.1104/pp.109.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saisho D, Nambara E, Naito S, Tsutsumi N, Hirai A, Nakazono M. Plant Mol Biol. 1997;35:585–596. doi: 10.1023/a:1005818507743. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Suzuki M, Ario T, Hattori T, Nakamura K, Asahi T. Isolation and characterization of two tightly linked catalase genes from castor bean that are differentially regulated. Plant Mol Biol. 1994;25:507–516. doi: 10.1007/BF00043878. [DOI] [PubMed] [Google Scholar]
- Utter MF, Kolenbrander HM (1972) Formation of oxaloacetate by CO2 fixation on phosphoenolpyruvate. In PD Boyer, ed, The Enzymes, Ed 3, Vol 6. Academic Press, New York, pp 117–168
- Walker RP, Acheson RM, Técsi LI, Leegood RC. Phosphoenolpyruvate carboxykinase in C4 plants: its role and regulation. Aust J Plant Physiol. 1997;24:459–468. [Google Scholar]
- Walker RP, Leegood RC. Purification, and phosphorylation in vivo and in vitro, of phosphoenolpyruvate carboxykinase from cucumber cotyledons. FEBS Lett. 1995;362:70–74. doi: 10.1016/0014-5793(95)00212-r. [DOI] [PubMed] [Google Scholar]
- Walker RP, Leegood RC. Phosphorylation of phosphoenolpyruvate carboxykinase in plants: studies in plants with C4 photosynthesis and Crassulacean acid metabolism and in germinating seeds. Biochem J. 1996;317:653–658. doi: 10.1042/bj3170653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker RP, Trevanion SJ, Leegood RC. Phosphoenolpyruvate carboxykinase from higher plants: purification from cucumber and evidence of rapid proteolytic cleavage in extracts from a range of plant tissues. Planta. 1995;196:58–63. [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol Gen Genet. 1993;236:331–340. doi: 10.1007/BF00277130. [DOI] [PubMed] [Google Scholar]