Abstract
The rickettsiae are obligate intracellular alphaproteobacteria that exhibit a complex infectious life cycle in both arthropod and mammalian hosts. As obligate intracellular bacteria, rickettsiae are highly adapted to living inside a variety of host cells, including vascular endothelial cells during mammalian infection. Although it is assumed that the rickettsiae produce numerous virulence factors that usurp or disrupt various host cell pathways, they have been challenging to genetically manipulate to identify the key bacterial factors that contribute to infection. Motivated to overcome this challenge, we sought to expand the repertoire of available rickettsial loss-of-function mutants, using an improved mariner-based transposon mutagenesis scheme. Here, we present the isolation of over 100 transposon mutants in the spotted fever group species Rickettsia parkeri. Transposon insertions disrupted genes whose products are implicated in a variety of pathways, including bacterial replication and metabolism, the type IV secretion system, factors with previously established roles in host cell interactions and pathogenesis, or are of unknown function. Given the need to identify critical virulence factors, forward genetic screens such as this will provide an excellent platform to more directly investigate rickettsial biology and pathogenesis.
Introduction
Bacteria in the genus Rickettsia are obligate intracellular alphaproteobacteria that are divided into four groups—the spotted fever group (SFG), typhus group (TG), ancestral group (AG), and transitional group (TRG) [1]. They inhabit arthropods (ticks, fleas, and mites), and many can be transmitted to humans and other mammals. Pathogenic species primarily target endothelial cells in the vasculature, causing a variety of vascular diseases such as typhus and Rocky Mountain spotted fever [2]. Despite the prevalence of rickettsial diseases throughout the world, we know little about the bacterial factors required for infection and pathogenesis.
The SFG species Rickettsia parkeri, a tick-borne pathogen that causes a mild form of spotted fever in humans [3,4], is emerging as a model organism to study SFG rickettsial pathogenesis. R. parkeri can be studied under BL2 conditions and has animal models of pathogenesis that mimic aspects of human infection [5,6]. Furthermore, the R. parkeri life cycle closely matches that of the more virulent SFG species Rickettsia rickettsii [7,8], the causative agent of Rocky Mountain spotted fever. Like its more virulent relative, R. parkeri invades non-phagocytic cells and is taken into a primary phagocytic vacuole [9]. They then break out of this vacuole and enter the cytosol to replicate and grow [10]. R. parkeri and many other Rickettsia species also hijack the host cell actin cytoskeleton to polymerize actin tails and undergo actin-based motility [11–13]. During spread, motile R. parkeri move to a host cell-cell junction but then lose their actin tails before entering into a short (~1 μm) plasma membrane protrusion that is subsequently engulfed into the neighboring cell. The bacterium then lyses the double-membrane secondary vacuole to enter the neighboring cell cytosol to begin the process of spread again [14]. Because of its experimental tractability and the fact that its lifecycle is indistinguishable from more virulent species, R. parkeri provides an attractive system for investigating rickettsial host-pathogen interactions.
As an obligate intracellular pathogen, R. parkeri must produce virulence factors that usurp or disrupt various host cell pathways. However, due to their obligate growth requirement, it has been challenging to genetically manipulate rickettsiae to identify the key bacterial factors that contribute to infection [15]. Fortunately, recent advances have expanded the genetic toolkit that can be used in the rickettsiae, allowing us to peer more directly into the molecular mechanisms that drive rickettsial biology. Chief among these advances was the development of a Himar1 mariner-based transposon system for random mutagenesis of rickettsial genomes [16]. To date, smaller-scale mutagenesis studies have been completed in the TG species R. prowazekii [16–18] and the SFG species R. rickettsii [18,19].
Despite these advances, we still do not know all of the critical bacterial factors that mediate interactions with the host. Moreover, many of the genes in R. parkeri are annotated to encode hypothetical proteins, which limits our ability to rationally explore their functions. Therefore, we set out to expand the repertoire of available R. parkeri mutants using a forward genetic screen. We used the mariner-based transposon system [16] and developed a more streamlined protocol to rapidly isolate R. parkeri mutants that alter plaque size [14]. To date, we have isolated over 100 mutants that disrupt genes predicted to function in a variety of pathways. We have previously published our detailed analysis of three mutants–in sca2, rickA, and sca4 [14,20]. Here, we present the full panel of mutants to demonstrate the potential and ease of developing rickettsial transposon libraries.
Materials and methods
Cell lines
Vero cells (monkey, kidney epithelial) were obtained from the University of California, Berkeley tissue culture facility and grown in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen) containing 5% fetal bovine serum (FBS) at 37°C in 5% CO2.
Transposon mutagenesis in R. parkeri
R. parkeri Portsmouth strain was a gift from Dr. Chris Paddock (Centers for Disease Control and Prevention). Wild-type R. parkeri were expanded and purified by centrifugation through a 30% MD-76R solution, as previously described [14]. The pMW1650 plasmid carrying the Himar1 mariner-based transposon [16] (a gift from Dr. David Wood, University of South Alabama) was used to generate R. parkeri strains carrying transposon insertions, as previously described [14] and reintroduced here. To isolate small plaque mutants, we implemented a small-scale electroporation protocol. A T75 cm2 flask of confluent Vero cells was infected with approximately 107 WT R. parkeri. When Vero cells were at least 90% rounded at 3 d post infection, they were scraped from the flask. Infected cells were spun down for 5 min at 1800 x g at 4°C and resuspended in 3–6 ml of K-36 buffer. To release bacteria, infected cells were mechanically disrupted either by passing them through a 27.5 gauge syringe needle 10 times, or by vortexing at ~2900 rpm using a Vortex Genie 2 (Scientific Industries Inc.) in a 15 ml conical tube containing 2 g of 1 mm glass beads, with two 30 s pulses and 30 s incubations in ice after each pulse. This bead disruption procedure was adopted for a majority of the screen, because it was faster and reduced the possibility of a needle stick. Host cell debris was pelleted by centrifugation for 5 min at 200 x g at 4°C. The supernatant containing R. parkeri was transferred to 1.5 ml microcentrifuge tubes and spun for 2 min at 9000 x g at 4°C. Bacterial pellets were then washed three times in cold 250 mM sucrose, resuspended in 50 μl cold 250 mM sucrose, mixed with 1 μg of pMW1650 plasmid, placed in a 0.1 cm cuvette, and electroporated at 1.8 kV, 200 ohms, 25 μF, 5 ms using a Gene Pulser Xcell (Bio-Rad). Bacteria were immediately recovered in 1.2 ml brain heart infusion (BHI) medium. For infections of confluent Vero cells in 6-well plates, medium was removed from each well, and cells were washed with phosphate-buffered saline (PBS). Electroporated bacteria (100 μl) was added to each well, and plates were placed in a humidified chamber and rocked for 30 min at 37°C. An overlay of DMEM with 5% FBS and 0.5% agarose was then added to each well. Infected cells were incubated at 33°C, 5% CO2 for 24 h, and then to select for transformants, a second overlay was added containing rifampicin (Sigma) to a final concentration 200 ng/ml to select for transformants. Stock solutions of rifampicin were prepared in DMSO at 25 mg/ml and stored at -20°C. After at least 3 or 4 d, plaques were visible by eye in the cell monolayer, and plaques smaller or bigger relative to neighboring plaques were harvested and re-plated for further analysis, as described below.
To isolate and expand mutant strains before mapping the sites of transposon insertion, plaques were picked and resuspended in 200 μl of BHI. Medium was aspirated from confluent Vero cells in 6-well plates, and the isolated plaque resuspension was used to infect the cells at 37°C for 30 min with rocking. Then 3 ml DMEM with 2% FBS and 200 ng/ml rifampicin was added to each well, and infections were allowed to progress until monolayers were fully infected. Infected cells were isolated using mechanical disruption as described above, except that bacteria were immediately resuspended in BHI without a sucrose wash and stored at -80°C. These plaque-purified strains were then used as described below to map the transposon insertion sites.
Semi-random nested PCR
To map the transposon insertion sites, plaque-purified R. parkeri strains were boiled for 10 min and used as templates for PCR reactions. Genomic DNA at insertion sites was amplified for sequencing using semi-random nested PCR. The first “external” PCR reaction used transposon-specific primers (ExTn1 5’-CACCAATTGCTAAATTAGCTTTAGTTCC-3’; or ExTn2 5’-GTGAGCTATGAGAAAGCGCCACGC-3’) and a universal primer (Univ1 5’-GCTAGCGGCCGCACTAGTCGANNNNNNNNNNCTTCT-3’). Univ1 has a specific sequence at the 5’ end and a random sequence near the 3’ end to allow for random annealing throughout the chromosome. The first PCR reaction yielded the “external” product that served as a template in the subsequent “internal” PCR reaction using transposon-specific primers (InTn1 5’-GCTAGCGGCCGCGGTCCTTGTACTTGTTTATAATTATCATGAG-3’; or InTn2 5’-GCTAGCGGCCGCCCTGGTATCTTTATAGTCCTGTCGG-3’) and a different universal primer (Univ2 5’-GCTAGCGGCCGCACTAGTCGA-3’). Univ2 contains the same specific sequence as Univ1, allowing for specific amplification of the external PCR product. PCR products were cleaned using ExoSAP-IT PCR Product Cleanup Reagent (Affymetrix) and sequenced using transposon-specific primers SR095 5’-CGCCACCTCTGACTTGAGCGTCG-3’ and SR096 5’-CCATATGAAAACACTCCAAAAAAC-3’. Genomic locations were determined using BLAST against the R. parkeri strain Portsmouth genome (GenBank/NCBI accession NC_017044.1).
Results
Design of an improved transposon mutagenesis scheme
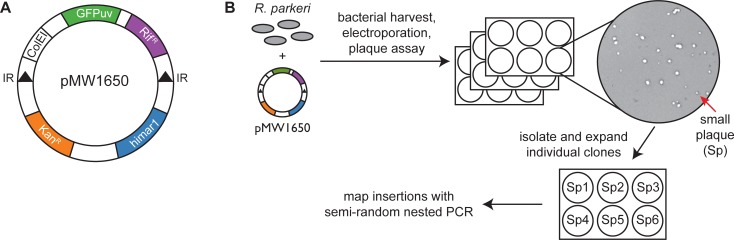
We used the pMW1650 plasmid, which carries a Himar1 mariner-based transposon [16], to randomly mutate the R. parkeri genome. pMW1650 encodes the Himar1 transposase, a transposon cassette that contains the R. prowazekii arr-2 rifampin resistance gene, and a variant of green fluorescent protein (GFPuv) [16] (Fig 1A). The first reported application of this system in R. prowazekii [16] and R. rickettsii [18] yielded some transposon mutants, but we sought to improve the mutagenesis scheme to increase the chances of identifying genes important for infection. Therefore, we developed a simple and rapid procedure to extract bacteria from infected host cells [14]. In the past, we had purified R. parkeri from infected host cells using an hours-long process involving mechanical disruption and density gradient centrifugation prior to electroporation [21]. In recent work, we optimized this procedure to more quickly isolate and electroporate bacteria and re-infect host cells in under an hour [14]. To mechanically disrupt infected cells, we either passed infected cells through a syringe needle or vortexed cells in the presence of 1 mm glass beads. Samples were then spun at low speed for 5 min to pellet host cell debris, followed by a 2 min high-speed spin to pellet bacteria. Bacteria were then quickly washed 2–3 times in cold sucrose prior to electroporation.
Fig 1. Transposon mutagenesis of R. parkeri.
(A) Map of the pMW1650 plasmid used in this study for transposon mutagenesis (IR, inverted repeats). (B) Experimental scheme for transposon mutagenesis and isolation of individual mutants.
To identify genes involved in infection, we screened for transformants that showed altered plaque size and/or morphology (Fig 1B). We predicted that plaque size changes would result from defects at different stages of the rickettsial life cycle, including in intracellular growth, replication, motility, and/or spread. To screen for such mutants, pMW1650-electroporated bacteria were immediately used to setup plaque assays in the presence of rifampicin to select for transformants. Plaque size was monitored visually over the course of 3–9 days, and those displaying a small plaque (Sp) or big plaque (Bp) phenotype relative to their neighbors were selected for expansion. After independently repeating this process 7 times, 120 Sp mutant and 2 Bp mutants were selected for further analysis, as detailed below.
Expansion and mapping of the transposon mutants
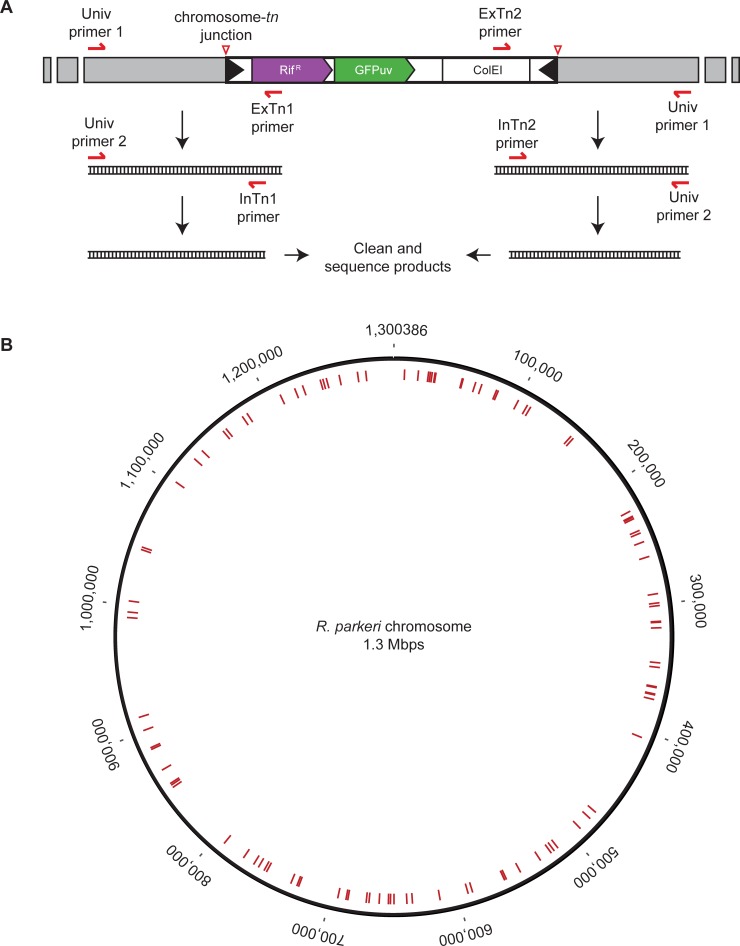
To expand the isolated transformants, plaques were picked and transferred to uninfected host cells to propagate each bacterial strain. Once the host cells were >75% infected, bacteria were purified using the rapid isolation procedure outlined above. Nine isolates did not grow in this expansion procedure, possibly due to a lack of live bacteria in the original plaque or poor isolation of the infected cells. The remaining transformants could be expanded, and for these the transposon insertion site was mapped using a semi-random nested PCR protocol. In short, the junctions between the transposon and the flanking genomic regions were amplified via two nested PCR reactions, using transposon-specific and universal primers (Fig 2A). PCR products were sent directly for sequencing. Mapping of the transposon insertion sites to the R. parkeri chromosome (accession number CP003341) revealed no preference for specific regions (Fig 2B), similar to what was observed in R. rickettsii [18,19] and R. prowazekii [16,18]. Using this procedure, we identified the transposon insertion sites for 106 mutants. For 6 isolates the transposon insertion site could not be mapped, and the strains did not express GFPuv (data not shown), suggesting these were spontaneous rifampicin-resistant strains. Of the 106 transposon mutations mapped, 81 were within the coding regions of 75 distinct genes and 25 were in intergenic regions (Table 1). These results lay the groundwork for critical follow-up studies (e.g. purification of clonal populations, complementation, phenotypic analyses, etc.) required for revealing gene function. They also highlight that transposon mutagenesis can be readily adapted for large-scale forward genetic screening in R. parkeri.
Fig 2. Mapping the transposon insertion sites.
(A) Diagram showing the insertion of the transposon cassette into a chromosomal region (in grey). Primers specific to the transposon ends were paired with universal primers to amplify the chromosome- transposon junctions (red triangles), using semi-random nested PCR. Two nested PCR reactions were done to improve amplification of the chromosome-transposon junction directly from boiled bacteria. (B) R. parkeri chromosomal map showing all transposon insertion sites (see red lines) identified in this screen.
Table 1. Transposon insertion sites in R. parkeri.
| Name | Genome position | Gene symbol | Gene product description |
|---|---|---|---|
| Sp1 | 101427 | MC1_00610 | Putative cytoplasmic protein |
| Sp2 | 112315 | MC1_00650 ** | Surface cell antigen 2 (Sca2) |
| Sp3 | 681322–681323 | MC1_03895 | Single-stranded-DNA-specific exonuclease RecJ |
| Sp6 | 365840 | MC1_02010 | Cytochrome c1, heme protein |
| Sp7 | 670632 | MC1_03810 | Folylpolyglutamate synthase |
| Sp8 | 1047805–1047806 | Intergenic | n/a |
| Sp9 | 151196–151197 | MC1_00820 | VirB6 Type IV secretory pathway (rvhB6e) |
| Sp10 | 491813 | Intergenic | n/a |
| Sp11 | 1136147 | MC1_06660 | DNA polymerase I |
| Sp13 | 563189–563190 | MC1_03195 | RND efflux transporter |
| Sp14 | 518698–518699 | MC1_02960 | CTP synthetase |
| Sp15 | 520939 | MC1_02980 | Hypothetical protein |
| Sp17 | 1248850–1248851 | MC1_07220 | Transcriptional regulator |
| Sp18 | 70364 | MC1_00450 | Hypothetical protein |
| Sp19 | 654506–654507 | MC1_03740 | Antigenic heat-stable 120 kDa protein (Sca4) |
| Sp20 | 531536 | MC1_03025 ** | ampG protein |
| Sp21 | 20179 | Intergenic | n/a |
| Sp22 | 29609 | MC1_00175 | F0F1 ATP synthase subunit B |
| Sp23 | 474265 | MC1_02665 | Outer membrane assembly protein |
| Sp24 | 753916 | Intergenic | n/a |
| Sp25 | 728290 | MC1_04100 | Isopentenyl pyrophosphate isomerase |
| Sp26 | 33338 | MC1_00210 | Transcriptional regulator |
| Sp27 | 30722 | Intergenic | n/a |
| Sp28 | 301811 | MC1_01650 | Protease |
| Sp29 | 225255 | MC1_01180 | Acriflavin resistance protein D |
| Sp30 | 886852 | Intergenic | n/a |
| Sp31 | 262955–262956 | MC1_01410 | Hypothetical protein |
| Sp33 | 299510 | MC1_01640 | Putative toxin of toxin-antitoxin (TA) system |
| Sp34 | 888003 | MC1_05085 ** | Actin polymerization protein RickA |
| Sp35 | 589425 | MC1_03370 | Thiol:disulfide interchange protein dsbA |
| Sp36 | 230327 | MC1_01215 | Prolyl endopeptidase |
| Sp37 | 637085 | MC1_03670 | Hypothetical protein |
| Sp38 | 292360–292361 | MC1_01595 | S-adenosylmethionine:tRNA ribosyltransferase-isomerase |
| Sp39 | 912985 | MC1_05235 | Hypothetical protein (RARP-2) |
| Sp40 | 1279632 | Intergenic | n/a |
| Sp41 | 995818 | Intergenic | n/a |
| Sp42 | 372020 | MC1_02055 ** | GTP-binding protein LepA |
| Sp43 | 651603–651604 | MC1_03735 | ADP, ATP carrier protein |
| Sp44 | 868641 | MC1_04970 | HAD-superfamily hydrolase |
| Sp45 | 761156 | MC1_04295 | Microcin C7 resistance protein |
| Sp46 | 852817 | MC1_04870 | Methylated-DNA-protein-cysteine methyltransferase |
| Sp47 | 856486 | MC1_04920 | Hypothetical protein |
| Sp48 | 243782–243780 | MC1_01300 | DNA repair protein RecN |
| Sp49 | 687339 | MC1_03930 | Hypothetical protein |
| Sp50 | 1158028 | MC1_06730 | Hypothetical protein |
| Sp51 | 888088 | MC1_05085 ** | Actin polymerization protein RickA |
| Sp52 | 346470 | Intergenic | n/a |
| Sp53 | 793762 | MC1_04525 | Hypothetical protein |
| Sp54 | 1258878 | MC1_07285 | Hypothetical protein |
| Sp55 | 1245896 | MC1_07200 | tig Trigger factor |
| Sp56 | 1243741–1243742 | Intergenic | n/a |
| Sp57 | 1004249–1004250 | Intergenic | n/a |
| Sp58 | 1273429 | MC1_07360 | NAD(P) transhydrogenase subunit alpha |
| Sp59 | 1181227 | Intergenic | n/a |
| Sp60 | 1158028 | MC1_01115 | Hypothetical protein |
| Sp62 | 350030–350031 | MC1_01915 | Cytochrome c oxidase assembly protein |
| Sp63 | 109070 | MC1_00650 ** | Surface cell antigen (Sca2) |
| Sp64 | 314932 | MC1_01745 ** | Ankyrin repeat-containing protein (RARP-1) |
| Sp65 | 726967 | Intergenic | n/a |
| Sp66 | 615509–615510 | MC1_03545 | Hypothetical protein |
| Sp71 | 371351 | MC1_02055 ** | GTP-binding protein LepA |
| Sp72 | 83786 | MC1_00525 | Stage 0 sporulation protein J |
| Sp73 | 655844 | MC1_03745 | Putative transcriptional regulator |
| Sp74 | 991759–991760 | MC1_05745 | Hypothetical protein |
| Sp75 | 251100 | MC1_01335 | Ankyrin repeat-containing protein |
| Sp76 | 9674 | Intergenic | n/a |
| Sp78 | 672659 | Intergenic | n/a |
| Sp79 | 65481 | Intergenic | n/a |
| Sp80 | 1210788–1210789 | MC1_07040 | Outer membrane protein OmpA |
| Sp81 | 365135 | MC1_02000 | Cytochrome b |
| Sp82 | 549578–549579 | MC1_03115 | Cytochrome c oxidase polypeptide |
| Sp83 | 480829 | MC1_02715 | Hypothetical protein |
| Sp84 | 689140 | Intergenic | n/a |
| Sp85 | 514488 | Intergenic | n/a |
| Sp88 | 241435 | MC1_01295 | Thermostable carboxypeptidase |
| Sp90 | 1127301 | MC1_06610 | Hypothetical protein |
| Sp91 | 82796 | MC1_00515 | 16S rRNA methyltransferase GidB |
| Sp92 | 1229489–1229490 | MC1_07110 | 17 kDa surface antigen |
| Sp93 | 1223170 | MC1_07070 | Undecaprenyl-phosphate alpha-N-acetylglucosaminyltransferase |
| Sp94 | 774831 | Intergenic | n/a |
| Sp95 | 902617 | MC1_05150 | Patatin b1 |
| Sp96 | 561640–561641 | MC1_03180 | Hypothetical protein |
| Sp97 | 641129–641130 | MC1_03685 | miaA tRNA delta(2)-isopentenylpyrophosphate transferase |
| Sp98 | 34100–34101 | MC1_00220 | Putative methyltransferase |
| Sp99 | 1104365 | Intergenic | n/a |
| Sp100 | 375061 | Intergenic | n/a |
| Sp101 | 152889–152890 | Intergenic | n/a |
| Sp102 | 406474–406475 | MC1_02260 | DNA mismatch repair protein MutS |
| Sp103 | 662735 | MC1_03780 | Hypothetical protein |
| Sp104 | 1161553 | MC1_06745 | Hypothetical protein |
| Sp105 | 593543–593544 | MC1_03405 | Acylamino acid-releasing protein |
| Sp106 | 1045462–1045463 | MC1_06065 | Outer membrane protein OmpB |
| Sp107 | 531709 | MC1_03025 ** | ampG protein |
| Sp108 | 1177263 | MC1_06810 | F0F1 ATP synthase subunit beta |
| Sp109 | 54288–54289 | MC1_00370 ** | Chaperone ClpB |
| Sp111 | 854916 | Intergenic | n/a |
| Sp112 | 55657 | MC1_00370 ** | Chaperone ClpB |
| Sp113 | 319455 | MC1_01760 | Histidine kinase sensor protein |
| Sp114 | 765596 | MC1_04335 | Ribonuclease D |
| Sp115 | 229548 | Intergenic | n/a |
| Sp116 | 314408 | MC1_01745 ** | Ankyrin repeat-containing protein (RARP-1) |
| Sp117 | 733347 | MC1_04135 | Hypothetical protein |
| Sp118 | 695571 | Intergenic | n/a |
| Sp119 | 231259 | MC1_01235 | Prolyl endopeptidase |
| Sp120 | 27839 | MC1_00155 | Hypothetical protein |
| Bp2 | 756531 | MC1_04275 | Hypothetical protein |
Spontaneous rifampicin resistant mutants: Sp4-5, 69–70, 86, Bp1. Clones that did not expand: Sp16, 32, 61, 67–68, 77, 87, 89, 110. Mapping for Sp12 revealed two different insertion sites and was not included in the list above.
** Indicates more than one isolated transposon mutant/gene. n/a, not applicable.
Discussion
A critical barrier to identifying and characterizing virulence factors in obligate intracellular bacterial pathogens has been the inability to easily manipulate their genomes. In this study, we sought to overcome this barrier and harness recent advances in rickettsial genetics to build a library of transposon mutants of the SFG Rickettsia species, R. parkeri. We streamlined previous protocols to introduce a mariner-based transposon into the R. parkeri genome and isolated 106 transposon insertion mutations. Our study represents the first such transposon mutant library in this species, and the most extensive reported library in the rickettsiae.
In our study, we selected for mutants that showed an altered plaque size phenotype in infected host cell monolayers. Transposon mutations may cause a small plaque phenotype due to any number of defects, including: poor bacterial replication, reduced access to or survival within the cytosol, impaired cytosolic actin-based motility, and defective cell-to-cell spread. It was thus not surprising that we identified genes with a diverse set of predicted functions. Many genes with products predicted to perform bacterial-intrinsic functions (e.g. DNA replication) were identified and are expected to indirectly influence host-pathogen interactions through their role in bacterial growth and division. Other genes had more direct connections to the infectious life cycle and were further characterized in our recent studies to reveal their specific functions in intracellular infection [14,20]. For example, we previously described transposon mutations that disrupt the rickA (Sp34) and sca2 (Sp2) genes and showed that these gene products are required for two independent phases of R. parkeri actin-based motility [20]. We also identified a transposon insertion (Sp19) in sca4 gene and showed this encodes a secreted effector that promotes cell-to-cell spread [14].
Other genes mutated in this screen have been suggested to play critical roles during the infectious life cycle of other Rickettsia species but have yet to be characterized in R. parkeri. For example, we isolated transposon insertion mutants in the ompA (Sp80) and ompB (Sp106) genes, encoding the outer membrane proteins OmpA and OmpB. Work with SFG species R. conorii and R. rickettsii showed that OmpA and OmpB may regulate adhesion to and/or invasion of host cells [22–25]. However, some of this work relied on expression of these proteins in other bacterial species because mutants were not available. Interestingly, targeted knockout of ompA in R. rickettsii via an LtrA group II intron retrohoming system revealed no clear requirement for OmpA in invasion [26], suggesting it alone is not necessary. This highlights the importance of studying loss-of-function mutants to reveal gene function. The fact that ompA and ompB mutants exhibit a small plaque phenotype suggest additional functions of these proteins, putative indirect effects of the truncated products, or simply a need for efficient invasion of neighboring cells after host cells lyse during plaque development. Future work on these mutants should help reveal the relative importance of these proteins during invasion and/or other stages of the R. parkeri life cycle.
Our screen also revealed genes for which no specific role has been ascribed during the infectious life cycle, although the sequence of their protein products suggests a role in interaction with host cells. These proteins include some with eukaryotic-like motifs such as ankyrin repeats, which often mediate protein-protein interactions [27], and are a common motif in secreted bacterial effector proteins or virulence factors that target host pathways [27,28]. In particular, mutations in genes encoding R. parkeri orthologs of RARP-1 and RARP-2 from R. typhi (accession numbers MC1_01745 and MC1_05235, respectively) were identified in our screen (Sp64, Sp116, and Sp39). Work in R. typhi has revealed that RARP-1 and RARP-2 are secreted into the host cell, but their precise functions remain unknown [29,30].
Another mystery in rickettsial biology relates to the functional importance of the putative type IV secretion system (T4SS) encoded in their genomes [31], which in other species is involved in translocating DNA, nucleoproteins, and effector proteins into host cells [32]. Strikingly, the Rickettsia T4SS has an unusual expansion of the VirB6-like genes (i.e. Rickettsiales vir homolog, rvhB6), which are predicted to encode inner membrane protein components at the base of the T4SS [30,31,33]. Interestingly, we isolated a strain with a transposon insertion mutation in the fifth VirB6-like gene, rvhB6e (Sp9). This mutant will prove useful to explore the function of the T4SS in rickettsial infection.
Finally, we identified 20 strains, each carrying a transposon insertion disrupting a gene encoding a hypothetical protein. One of these caused a big plaque phenotype, suggesting enhanced growth or cell-to-cell spread. Further study of these mutants has the potential to reveal the function of these uncharacterized gene products during rickettsial infection. We also identified 25 small plaque mutants with insertions in intergenic regions. In these cases, the mutant phenotype could be caused by transposon insertion into a promoter region that alters gene expression. These mutants may represent tools for exploring gene regulation during the R. parkeri life cycle.
Overall, our mutant collection provides an important resource that can be used to uncover key bacterial players that regulate rickettsial interactions with their host cells. This will also allow for more direct analysis of gene function in the rickettsiae without the reliance on introducing genes into heterologous organisms. This forward genetics approach has the potential to reveal new insights into rickettsial biology and pathogenesis; however, limitations remain. For example, because the rickettsiae are obligate intracellular pathogens, screens such as these are unlikely to reveal genes that are essential for invasion or intracellular growth. Therefore, we cannot necessarily assess the relative importance of genes not identified in forward genetic screens. Additionally, the reported protocol still has limits with regard to electroporation efficiency and recovery on host cells, which makes it harder to adapt for large-scale mutagenesis work. Continued optimization of the protocols we report—by further characterizing and enhancing transformation efficiency and bacterial viability—will help investigators expand the available toolkit to generate more Rickettsia mutants. Additional advancements in rickettsial genetic methods will also be necessary to complement our work and more effectively probe the molecular mechanisms of all genes whose products may control critical host-pathogen interactions.
Acknowledgments
We thank David Wood and Chris Paddock for reagents and Shawna Reed for technical help. We also thank the staff of UC Berkeley’s Cell Culture Facility and DNA Sequencing Facility for critical experimental support.
Data Availability
All relevant data are within the paper.
Funding Statement
R.L.L. was supported by a Helen Hay Whitney Foundation postdoctoral fellowship and National Institutes of Health grant K99 GM115765. This work in the lab of M.D.W. is supported by National Institutes of Health grant R01 AI109044. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gillespie JJ, Williams K, Shukla M, Snyder EE, Nordberg EK, Ceraul SM, et al. Rickettsia phylogenomics: unwinding the intricacies of obligate intracellular life. PLoS ONE. 2008;3: e2018 doi: 10.1371/journal.pone.0002018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walker DH, Ismail N. Emerging and re-emerging rickettsioses: endothelial cell infection and early disease events. Nat Rev Microbiol. 2008;6: 375–386. doi: 10.1038/nrmicro1866 [DOI] [PubMed] [Google Scholar]
- 3.Paddock CD, Sumner JW, Comer JA, Zaki SR, Goldsmith CS, Goddard J, et al. Rickettsia parkeri: a newly recognized cause of spotted fever rickettsiosis in the United States. Clin Infect Dis. 2004;38: 805–811. doi: 10.1086/381894 [DOI] [PubMed] [Google Scholar]
- 4.Paddock CD, Finley RW, Wright CS, Robinson HN, Schrodt BJ, Lane CC, et al. Rickettsia parkeri rickettsiosis and its clinical distinction from Rocky Mountain spotted fever. Clin Infect Dis. 2008;47: 1188–1196. doi: 10.1086/592254 [DOI] [PubMed] [Google Scholar]
- 5.Grasperge BJ, Reif KE, Morgan TD, Sunyakumthorn P, Bynog J, Paddock CD, et al. Susceptibility of inbred mice to Rickettsia parkeri. Infect Immun. 2012;80: 1846–1852. doi: 10.1128/IAI.00109-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banajee KH, Embers ME, Langohr IM, Doyle LA, Hasenkampf NR, Macaluso KR. Amblyomma maculatum feeding augments Rickettsia parkeri infection in a rhesus macaque model: a pilot study. PLoS ONE. 2015;10: e0135175 doi: 10.1371/journal.pone.0135175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ray K, Marteyn B, Sansonetti PJ, Tang CM. Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat Rev Microbiol. 2009;7: 333–340. doi: 10.1038/nrmicro2112 [DOI] [PubMed] [Google Scholar]
- 8.Lamason RL, Welch MD. Actin-based motility and cell-to-cell spread of bacterial pathogens. Curr Opin Microbiol. 2017;35: 48–57. doi: 10.1016/j.mib.2016.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reed SCO, Serio AW, Welch MD. Rickettsia parkeri invasion of diverse host cells involves an Arp2/3 complex, WAVE complex and Rho-family GTPase-dependent pathway. Cell Microbiol. 2012;14: 529–545. doi: 10.1111/j.1462-5822.2011.01739.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welch MD, Reed SCO, Haglund CM. Establishing intracellular infection: escape from the phagosome and intracellular colonization (rickettsiaceae) In: Palmer GH, Azad AF, editors. Washington DC: Intracellular Pathogens II: Rickettsiales; 2012. pp. 154–174. doi: 10.1128/9781555817336.ch5 [Google Scholar]
- 11.Heinzen RA, Hayes SF, Peacock MG, Hackstadt T. Directional actin polymerization associated with spotted fever group Rickettsia infection of Vero cells. Infect Immun. 1993;61: 1926–1935. iai.asm.org/content/61/5/1926.full.pdf+html [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serio AW, Jeng RL, Haglund CM, Reed SC, Welch MD. Defining a core set of actin cytoskeletal proteins critical for actin-based motility of Rickettsia. Cell Host Microbe. 2010;7: 388–398. doi: 10.1016/j.chom.2010.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choe JE, Welch MD. Actin-based motility of bacterial pathogens: mechanistic diversity and its impact on virulence. Pathog Dis. 2016;74: ftw099 doi: 10.1093/femspd/ftw099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamason RL, Bastounis E, Kafai NM, Serrano R, Del Álamo JC, Theriot JA, et al. Rickettsia Sca4 reduces vinculin-mediated intercellular tension to promote spread. Cell. 2016;167: 670–683.e10. doi: 10.1016/j.cell.2016.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McClure EE, Chávez ASO, Shaw DK, Carlyon JA, Ganta RR, Noh SM, et al. Engineering of obligate intracellular bacteria: progress, challenges and paradigms. Nat Rev Microbiol. 2017;15: 544–558. doi: 10.1038/nrmicro.2017.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Z-M, Tucker AM, Driskell LO, Wood DO. Mariner-based transposon mutagenesis of Rickettsia prowazekii. Appl Environ Microbiol. 2007;73: 6644–6649. doi: 10.1128/AEM.01727-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin A, Tucker AM, Hines A, Wood DO. Transposon mutagenesis of the obligate intracellular pathogen Rickettsia prowazekii. Appl Environ Microbiol. 2004;70: 2816–2822. doi: 10.1128/AEM.70.5.2816-2822.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark TR, Lackey AM, Kleba B, Driskell LO, Lutter EI, Martens C, et al. Transformation frequency of a mariner-based transposon in Rickettsia rickettsii. 2011;193: 4993–4995. doi: 10.1128/JB.05279-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleba B, Clark TR, Lutter EI, Ellison DW, Hackstadt T. Disruption of the Rickettsia rickettsii Sca2 autotransporter inhibits actin-based motility. Infect Immun. 2010;78: 2240–2247. doi: 10.1128/IAI.00100-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reed SCO, Lamason RL, Risca VI, Abernathy E, Welch MD. Rickettsia actin-based motility occurs in distinct phases mediated by different actin nucleators. Curr Biol. 2014;24: 98–103. doi: 10.1016/j.cub.2013.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welch MD, Reed SCO, Lamason RL, Serio AW. Expression of an epitope-tagged virulence protein in Rickettsia parkeri using transposon insertion. PLoS ONE. 2012;7: e37310 doi: 10.1371/journal.pone.0037310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uchiyama T. Adherence to and invasion of Vero cells by recombinant Escherichia coli expressing the outer membrane protein rOmpB of Rickettsia japonica. Ann N Y Acad Sci. 2003;990: 585–590. doi: 10.1111/j.1749-6632.2003.tb07431.x [DOI] [PubMed] [Google Scholar]
- 23.Li H, Walker DH. rOmpA is a critical protein for the adhesion of Rickettsia rickettsii to host cells. Microb Pathog. 1998;24: 289–298. doi: 10.1006/mpat.1997.0197 [DOI] [PubMed] [Google Scholar]
- 24.Hillman RD, Baktash YM, Martinez JJ. OmpA-mediated rickettsial adherence to and invasion of human endothelial cells is dependent upon interaction with α2β1 integrin. Cell Microbiol. 2013;15: 727–741. doi: 10.1111/cmi.12068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan YGY, Cardwell MM, Hermanas TM, Uchiyama T, Martinez JJ. Rickettsial outer-membrane protein B (rOmpB) mediates bacterial invasion through Ku70 in an actin, c-Cbl, clathrin and caveolin 2-dependent manner. Cell Microbiol. 2009;11: 629–644. doi: 10.1111/j.1462-5822.2008.01279.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noriea NF, Clark TR, Hackstadt T. Targeted knockout of the Rickettsia rickettsii OmpA surface antigen does not diminish virulence in a mammalian model system. MBio. 2015;6 doi: 10.1128/mBio.00323-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al-Khodor S, Price CT, Kalia A, Abu Kwaik Y. Functional diversity of ankyrin repeats in microbial proteins. Trends Microbiol. 2010;18: 132–139. doi: 10.1016/j.tim.2009.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan X, Lührmann A, Satoh A, Laskowski-Arce MA, Roy CR. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science. 2008;320: 1651–1654. doi: 10.1126/science.1158160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaur SJ, Rahman MS, Ammerman NC, Beier-Sexton M, Ceraul SM, Gillespie JJ, et al. TolC-dependent secretion of an ankyrin repeat-containing protein of Rickettsia typhi. J Bacteriol. 2012;194: 4920–4932. doi: 10.1128/JB.00793-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gillespie JJ, Kaur SJ, Rahman MS, Rennoll-Bankert K, Sears KT, Beier-Sexton M, et al. Secretome of obligate intracellular Rickettsia. FEMS Microbiol Rev. 2015;39: 47–80. doi: 10.1111/1574-6976.12084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gillespie JJ, Ammerman NC, Dreher-Lesnick SM, Rahman MS, Worley MJ, Setubal JC, et al. An anomalous type IV secretion system in Rickettsia is evolutionarily conserved. PLoS ONE. 2009;4: e4833 doi: 10.1371/journal.pone.0004833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grohmann E, Christie PJ, Waksman G, Backert S. Type IV secretion in Gram-negative and Gram-positive bacteria. Mol Microbiol. 2018;107: 455–471. doi: 10.1111/mmi.13896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gillespie JJ, Phan IQH, Driscoll TP, Guillotte ML, Lehman SS, Rennoll-Bankert KE, et al. The Rickettsia type IV secretion system: unrealized complexity mired by gene family expansion. Pathog Dis. 2016;74: ftw058 doi: 10.1093/femspd/ftw058 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.