Abstract
We aimed to understand the molecular changes in host cells that accompany infection by the seasonal influenza A H1N1 virus because the initial response rapidly changes owing to the fact that the virus has a robust initial propagation phase. Human epithelial alveolar A549 cells were infected and total RNA was extracted at 30 min, 1 h, 2 h, 4 h, 8 h, 24 h, and 48 h post infection (h.p.i.). The differentially expressed host genes were clustered into two distinct sets of genes as the infection progressed over time. The patterns of expression were significantly different at the early stages of infection. One of the responses showed roles similar to those associated with the enrichment gene sets to known ‘gp120 pathway in HIV.’ This gene set contains genes known to play roles in preventing the progress of apoptosis, which infected cells undergo as a response to viral infection. The other gene set showed enrichment of ‘Drug Metabolism Enzymes (DMEs).’ The identification of two distinct gene sets indicates that the virus regulates the cell’s mechanisms to create a favorable environment for its stable replication and protection of gene metabolites within 8 h.
Keywords: A549 Cells, Influenza A virus H1N1 subtype, High-throughput nucleotide sequencing, Sequence analysis RNA, Apoptosis, Gene expression regulation
INTRODUCTION
Influenza A H1N1 Virus (INV) is a RNA virus belonging to the family Orthomyxoviridae. Commonly referred to as the flu virus, H1N1 is specific to birds and mammals. The latest worldwide flu pandemic occurred in April 2009 and accounted for an estimated 284,500 global deaths (Dawood et al., 2012).
In general, a variety of host responses during viral infection have been identified, including activation of numerous cell death and survival pathways. These pathways include: 1) programmed cell death I (apoptosis), 2) programmed cell death II (autophagy), and 3) endoplasmic reticulum stress with subsequent unfolded protein response (UPR). There has been extensive research on the regulatory roles of these pathways during the influenza virus life cycle (Yeganeh et al., 2013). Pathogen sensory pathways (e.g., RIG-I) displayed long-lasting associations with cytokine/chemokine signaling through day 8 (Dimitrakopoulou et al., 2014). Transcriptome research for H1N1 infection revealed that host cell innate immunity is induced at 3 to 5 days post infection (Zou et al., 2013). Another report described that the influenza induced an early immune response 3 to 7 days after infection (Park et al., 2015). However, influenza symptoms typically begin two days after exposure to the virus (Nair et al., 2011). In order to understand host cell pathway hijacking in H1N1 infection within two days, transcriptome changes during the first 48 h post infection could provide important insights into the mechanisms underlying the pathogenicity of influenza A viruses. The host cell transcriptome profile was rapidly changed in the early stages of infection (within 8 h of infection). Several mediators of apoptosis were activated in this stage. After 8 h, the most significant change was found to be that in expression of genes encoding drug-metabolizing enzymes.
MATERIALS AND METHODS
Host cell preparation
A549 cells are adenocarcinoma human alveolar basal epithelial cells. The A549 cell line is widely used as an in vitro model as a type II pulmonary epithelial cell model for studies of drug metabolism and as a transfection host. A549 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM). All media contained 10% fetal bovine serum and antibiotics (penicillin streptomycin).
Viral infection and total RNA extraction
A549 cells were infected with influenza A virus at a multiplicity of infection (MOI) of 3 for 30 min at 37°C. The virus was propagated at 37°C in 11-day-old chicken embryos. After being washed with phosphate-buffered saline (PBS), cells were infected with influenza virus (IV) at an MOI of 3 for 30 min at 37°C. Infection media consisted of 0.1% glucose, 0.05% vitamin solution, and 0.5 mg/ml L-(tosylamido-2-phenyl) ethyl chloromethyl ketone (TPCK)-treated trypsin in DMEM. Infected cells were then washed and incubated in medium without serum for varying lengths of time. All infected cells were inoculated at less than 37 CT (cycle of threshold) for RT-PCR validation experiments. Total RNA was extracted from A549 cells with the RNeasy Mini Kit (74104, QIAGEN, MD, USA).
Sample QC & RNA quantification
RNA integrity was assessed using an Agilent Technologies 2100 Bioanalyzer with an RNA Integrity Number (RIN) value greater than or equal to 7. Cell-to-cell variability of influenza viral infection was quantified by subjecting the cell supernatant to a plaque assay to determine the load of infectious viral particles and then performing real-time reverse transcription quantitative PCR (RT–qPCR) on cell lysates to quantify the level intracellular viral RNA (vRNA) of individual genome segments. The size of PCR-enriched fragments was verified by assessing the template size distributions on an Agilent Technologies 2100 Bioanalyzer using a DNA 1000 chip (Agilent, CA, USA).
Next generation sequencing
The sequencing library was prepared by random fragmentation of the DNA or cDNA sample, followed by 5’ and 3’ adapter ligation. The Illumina Hiseq 4000 generated raw images utilizing HiSeq Control Software (HCS, Illumina, CA, USA) v3.3 for system control and base calling through the integrated primary analysis software Real Time Analysis (RTA, Illumina) v2.5.2. The binary BCL (base calls) value was converted into FASTQ utilizing Illumina package bcl2fastq (V2.16.0.10, Illumina).
Sequence alignment
The alignment software application “New Tuxedo Protocol” was used for transcript assembly (Pertea et al., 2016). In the alignment step, HISAT2 was used to generate the alignment data (Kim et al., 2015a), which is a fast alignment program for mapping next-generation sequencing to read segments of the human genome (Siren et al., 2014).
Transcript assembly
In the transcript assembly, StringTie (Pertea et al., 2015) is known a fast and highly efficient assembler of RNA-Seq alignments into potential transcripts. It uses a novel network flow algorithm and an optional de novo assembly step to assemble and quantitate full-length transcripts representing multiple splice variants for each gene locus. StringTie estimates gene-level measurements by appropriately combining fragments per kilobase of exon per million reads (FPKMs) from the transcripts of each gene.
Time-series expression analysis
Ballgown (Frazee et al., 2015) is a bioconductor-based suite that hosts a comprehensive set of bioconductor communities. The “stattest” function of Ballgown is available with a gene feature that prevents data skewing in the time series and was used for statistical tests of time-series expression. Gene-level measurements were performed by appropriately combining FPKMs from the transcripts comprising the gene (Li and Dewey, 2011) and it proposed a directed graph model combined with an expectation-maximization algorithm to estimate abundance. Ballgown generates a reference index by preparing reference transcript data and calculated RSEM values by inputting RNA-Seq data. A natural splines model with default settings was used to fit time profiles and six degrees of freedom (df) were used to fit the splines. “Timecourse=TRUE” parameters were assigned for time profiling using the natural splines function (Supplementary Table 1).
Functional annotation
DAVID bioinformatics resources consist of an integrated biological knowledgebase and analytic tools aimed at systematically extracting biological meaning from large gene/protein lists (Huang et al., 2009).
Gene set variation analysis (GSVA)
GSVA is a gene set enrichment method which estimates variation of pathway activity over a sample population in an unsupervised manner. Program operation was employed using basic parameters in ‘RNA-seq’ mode and ‘kernel=TRUE’ using ‘gsva’ function (release version 3.5). The known-gene sets database was updated with the ‘Hallmark class’ and ‘c7 class’ by extending the default class (Hänzelmann et al., 2013).
Weighted correlation network analysis (WGCNA)
Time-series differentially expressed genes (DEGs) were categorized by WGCNA. Categorized data were divided into 300–1000 gene categories with respect to their biological annotation-interpretation (Supplemetal Table 2). Constructing a weighted gene network entails calculation of the soft thresholding power β, to which co-expression similarity is increased to calculate adjacency. We did this with a power of 16, which is the lowest power for which the scale-free topology fit index curve flattens upon reaching a high value (in this case, approximately 0.95), a relatively large minimum module size of 30, and a moderate sensitivity (Supplementary Table 3).
Biological metabolic network in-silico validation
Elsevier’s Pathway Studio Mammalian, ChemEffect, DiseaseFX analysis: The set of biological networks in this study was created using Pathway Studio 11.0 (Ariadne) software (https://mammalcedfx.pathwaystudio.com/). Elsevier’s Pathway Studio® enables users to explore molecular interactions and cause-and-effect relationships associated with biological processes by integrating a vast knowledge base of biological relationships with analytical and visualization tools.
Ingenuity Pathway Analysis (IPA) metabolomics analysis: DEGs were analyzed using QIAGEN’s IPA software. The canonical pathways and functional processes with the most significant biological importance were identified using the list of DEGs identified with RNA-seq and the Ingenuity Pathways Knowledge Base. Pathway enrichment p-values (Fisher’s exact test) and activation z-scores were calculated using IPA.
RESULTS
The biological functions are divided into two groups approximately 8 h after infection
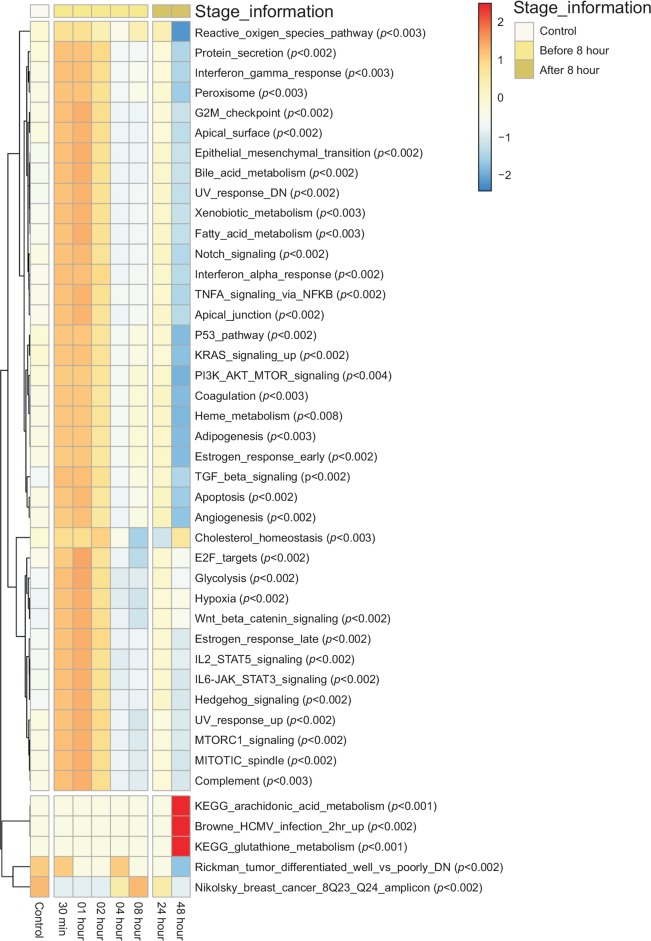
In hierarchical clustering, all samples were divided two groups (Supplementary Fig. 1). Early infection and late infection state samples were clustered into separate groups (Supplementary Fig. 2). Known biological function pathways were calculated using GSVA and all samples were divided into two groups by pathway activity score (Fig. 1). GSVA provides increased power to detect subtle pathway activity changes with the ‘Hallmark class’ and ‘c7 class’: ‘Apoptosis (p<0.002)′, ‘G2M checkpoint (p<0.002)′, ‘Interferon alpha response (p<0.002)′ and other biological functions were highly enriched in samples collected from cells infected for less than 8 h. ‘Arachidonic acid metabolism (p<0.001)′, ‘glutathione metabolism (p<0.001)′ and others were highly enriched in samples collected from cells infected for more than 8 h. The clustering result for whole transcriptome profile and pathway activity showed the same pattern, indicating that 8 h is a critical point for functional changes in host cells.
Fig. 1.
Unsupervised hierarchical clustering of gene set variation analysis. Gene set variation analysis (GSVA) is a non-parametric, unsupervised method for estimating variation of gene set enrichment through the expression data of infected cell. GSVA transformed each line gene-based expression matrix of gene by sample to matrix of a gene-set by sample. The matrix shows clear separation between the groups.
Changes in gene expression were observed in the two groups at 8 h
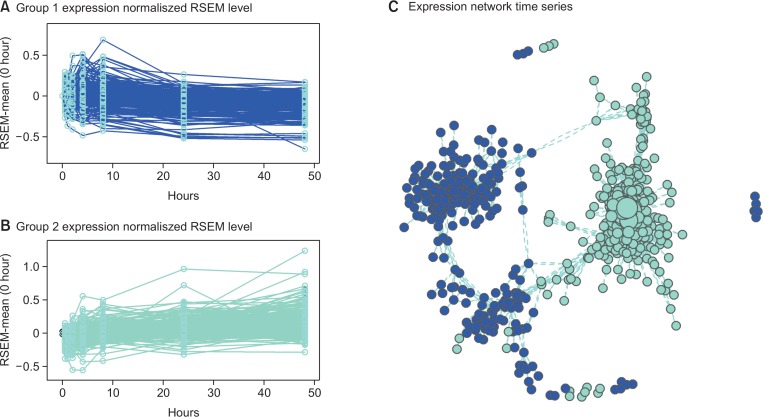
To identify host cell pathway hijacking, DEGs were selected by statistical testing of time-series expression data (Fig. 2). Gene expression changes were divided into two major groups. Fig. 2 shows 1,031 genes of the time-series DEGs, which were filtered by a threshold Q-value<0.05 (Supplementary Table 1). To find gene expression patterns in a functional module, WGCNA was used to predict groups of highly correlated genes. The results of the WGCNA analysis package analysis divided the DEGs into two groups. The biological functions are divided into two groups at approximately 8 h after infection (Supplementary Table 2). The blue module (blue line) shows increases expression after 4 h and decreases after 8 h, while the turquoise module (turquoise line) decreases at 4 h and then increases after 24 h.
Fig. 2.
Visualization of differential expression gene sets (DEGs) in the two groups. (A) early stage expression from 30 min to 8 h and (B) the late stage expression pattern, which progressively increased after 8 h of infection. (C) A network graph showing correlated genes. The graph shows clear separation between the groups.
Gene ontology (GO) enrichment analysis (Dennis et al., 2003) results of these two modules are displayed in Table 1. The envelop surface glycoprotein gp120 (p<0.007) was enriched in group 1. The neuroactive ligand-receptor interaction (p<1.97E−04) and metabolism of xenobiotics by cytochrome P450 (p<2.60E−04) were enriched in group 2.
Table 1.
Time series DEGs: GO annotation and pathway menrichment analysis for all timepoints and early- and late-specific timepoints
| Gene groups | Category | Term | Count | % | p-value | Fold enrichment | Bonferroni | Benjamini | FDR | |
|---|---|---|---|---|---|---|---|---|---|---|
| Early stage changed gene sets (<8 hour) | HIV_Interaction | env | env:Envelope surface glycoprotein gp120 | 20 | 5.68 | 0.007 | 1.65 | 0.05 | 0.05 | 3.54 |
| Molecular function | GO:0015267 | Channel activity | 28 | 5.53 | 2.24E-05 | 2.49 | 0.01 | 0.01 | 0.03 | |
| GO:0022803 | Passive transmembrane transporter activity | 28 | 5.53 | 2.34E-05 | 2.49 | 0.01 | 0.01 | 0.03 | ||
| GO:0030955 | Potassium ion binding | 14 | 2.77 | 4.50E-05 | 4.01 | 0.03 | 0.01 | 0.07 | ||
| GO:0022838 | Substrate specific channel activity | 26 | 5.14 | 9.05E-05 | 2.40 | 0.05 | 0.01 | 0.13 | ||
| KEGG Pathway | hsa04080 | Neuroactive ligand-receptor interaction | 19 | 3.75 | 1.97E-04 | 2.68 | 0.02 | 0.02 | 0.23 | |
| hsa00980 | Metabolism of xenobiotics by cytochrome 450 | 9 | 1.78 | 2.06E-04 | 5.41 | 0.02 | 0.01 | 0.24 | ||
| hsa00982 | Drug metabolism | 9 | 1.78 | 2.60E-04 | 5.24 | 0.03 | 0.01 | 0.30 |
Category refers to the GO functional category; Count refers to the number of enrichment DEGs.
Biological features of host cell before 8 h after infection: apoptosis
The protein gp120 is essential for viral entry into cells as it plays a vital role in attachment to specific cell surface receptors (Gram and Hansen, 1998). Viruses, such as HIV, with gp120 in their genome induce cellular apoptosis (Kapasi et al., 2002). Apoptosis is important in an infection response and is activated at the early infection stage (Fig. 1). Supplementary Fig. 3 shows that apoptosis marker genes were expressed in early infection stage (Subramanian et al., 2005). The viral gene gp120 is known to be associated with BCL-2 expression (Gaubin et al., 1999). BCL-2 and other apoptosis-related genes are down-regulated in the early stages of influenza A infection. At timepoints before 8 h post-infection, expression of apoptosis marker genes, including BCL2, was decreased (Supplementary Fig. 4A).
Specific biological features after 8 h: DME-centric pathways
DMEs are the set of metabolic pathways that modify the chemical structure of xenobiotics, which are compound of foreign materials such as replicated viral RNA and other metabolites (Mackenzie et al., 2017). Suppression of DMEs is necessary for viruses to maintain their presence in a cell and replicate themselves. In general, ‘endoplasmic reticulum (ER) stress response’ and ‘cytochrome P450’ are typical examples of DMEs. Both were found to be DEGs in the experimental results (Supplementary Fig. 4B, 4C).
In the Supplementary Fig. 4B, the endoplasmic reticulum (ER) stress response, also known as the unfolded protein response (UPR), is a primitive, evolutionary conserved molecular signaling cascade which has been implicated in multiple biological processes, including innate immunity and the pathogenesis of certain viral infections. Influenza A virus induces ER stress in a pathway-specific manner (Hassan et al., 2012). In the Supplementary Fig. 4C, various cytochrome P450 (p< 2.06E−04) genes were differentially expressed 8 h post infection. The major role of cytochrome P450s is in steroid and bile acid syntheses (Nelson, 2009; Lorbek et al., 2012). They are essential for the solubilization and absorption of lipids and fat-soluble vitamins in the intestine and they contribute to the major DME pathways by which cholesterol is excreted from the body (Lu et al., 2000).
DISCUSSION
The timepoint of 8 h post infection is an important inflection point for gene expression and biological functions
Kim et al. (2015b) reported that relatively few host cell genes were differentially expressed at 8 h post infection. In this study, our data are in agreement with the report by Kim et al. (2015b). This study indicates that 8 h post infection is an inflection point for host cell function and viral transition from the early to late infection stage.
In the early infection stage ‘Apoptosis’, ‘G2M checkpoint’, ‘Interferon alpha response’, and other biological functions are enriched. One study reported that apoptosis of virus-infected cells is one important host strategy used to limit viral infection (Scott and Norris, 2008). Cell cycle arrest may inhibit early cell death of infected cells, allow the cells to evade immune defenses, or help promote virus assembly (Bagga and Bouchard, 2014). Innate cytokine responses, such as alpha interferon (IFN-alpha) have roles in determining the rate of virus replication in the initial stages of infection (Price et al., 2000). These results indicate that influenza virus naturally establishes an environment for self-reproduction in early infection stage and hijacks host cell pathways, termed ‘cytopathic viral effects’ (Heaton, 2017) (Fig. 1).
In the late infection stage, ‘Arachidonic acid metabolism’ and ‘glutathione metabolism’ were found to be enriched. Lu et al. (2012) reported results from metabolic profiling and biochemical detection which indicated significant metabolic changes in the arachidonic acid metabolic pathway. Glutathione is partially depleted in influenza A virus infection (Alsuwaidi et al., 2013). These result suggest that late expressed genes create circumstances necessary to hijack host cell pathways by inhibiting the inflammatory response and the oxidation required for metabolic breakdown of xenobiotic materials by living organisms, termed the ‘Drug metabolism pathway’ (Commandeur et al., 1995).
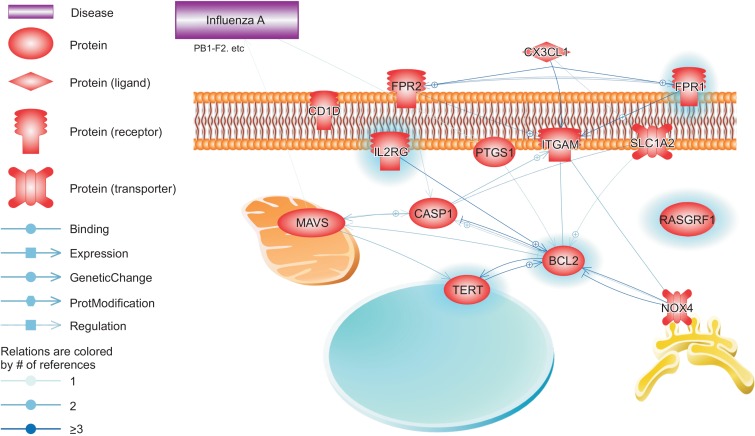
However, driver modules in host cell function hijacking was still unclear. To identify driver function in the early infection stage, WGCNA used co-expression to predict functional modules. We predicted an early infection module (blue module) and found enriched ‘GP120 pathway in HIV’ (Fig. 3). The MAVS gene (IPS-1, VISA or Cardif) is critical for host defenses to viral infection by inducing type-1 interferons (IFN-I), though its role in virus-induced apoptotic responses has not been elucidated (Lei et al., 2009). Like gp120 in HIV, the PB1-F2 gene has been shown to induce BCL2 by changing the level of MAVS gene expression (Varga et al., 2011). Unlike HIV, influenza A requires a mediator such as gp120. The PB1-F2 viral gene in the influenza genome is a candidate to be a mediator of gene sets regulated by gp120 (Fig. 3). The PB1-F2 viral gene is known to induce cell death (Chen et al., 2001) and apoptosis (Nicholson et al., 1998), similar to gp120.
Fig. 3.
The reconstruction signaling and regulatory network of gp120 at apoptosis pathway at the early stage. Schema of the gp120 protein regulated pathway in influenza-infected cells. The genes highlighted in blue are the genes related to “glycoprotein gp120” from the enrichment gene analysis. The remaining genes were identified through network analysis (PATHWAY studio).
In the WGCNA results for late stages of infection, we predicted a late module (turquoise module) and found the ‘Drug metabolism enzyme pathway (DMEs)’ to be enriched (Supplementary Fig. 5). DMEs were additionally found to be enriched in the late stage of infection by DAVID: ‘metabolism of xenobiotics by cytochrome 450 (p<0.004)′ and ‘drug metabolism (p<0.0003)′ (Supplementary Table 3). P450 converts exotic materials to chemically reactive metabolites, which, if not detoxified, may lead to various forms of hepatic and extrahepatic toxicity, including cellular necrosis (Park et al., 1995). In Supplementary Table 4, we show that CYP2C9 (P-value=7.46E−08), CYP2C18 (p-value=9.41E-06), CYP2F1 (p-value=0.0012), and CYP3A7 (p-value=9.95E−05) are involved in drug and steroid metabolism (Nelson, 2009).
The hub gene of the transcriptome-driven reconstruction network is a candidate drug targeting for influenza A
A therapeutic strategy that might escape viral resistance is to target host cellular mechanisms involved in viral replication and pathogenesis. The DGIdb integrates drug-gene interactions from 15 different sources (Wagner et al., 2016) (Table 2). Reconstruction of biological networks has been vital to understanding complex biological activities (Yu et al., 2013). In the gp120-regulated pathway of the early stages of infection, two genes (BCL-2, TERT) that surpassed the score thresholds (Sum score of DGIdb >2) were found. BCL2 was enriched in Paclitaxel, Oblimersen, Docetaxel, and Obatoclax treatments, while the TERT gene was only enriched in GV1001-treated cells.
Table 2.
Drug-gene interaction database for candidate genes
| Gene | Drug | CFB | CFCT | DB | GPI | PG | TALC | TE | TT | TC | #of DB | # of PMID | Sum score (DB+PMID) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BCL2 | PACLITAXEL | - | TRUE | TRUE | - | - | - | TRUE | - | TRUE | 4 | 5 | 9 |
| BCL2 | DOCETAXEL | - | - | TRUE | - | - | - | - | - | TRUE | 2 | 6 | 8 |
| BCL2 | RASAGILINE | - | - | TRUE | - | - | - | - | - | - | 1 | 5 | 6 |
| BCL2 | OBLIMERSEN | - | - | - | - | - | TRUE | - | TRUE | TRUE | 3 | 0 | 3 |
| FPR1 | NEDOCROMIL | - | - | TRUE | - | - | - | - | - | - | 1 | 3 | 4 |
| NR1H4 | CHENODEOXYCHOLIC ACID | - | - | TRUE | TRUE | - | - | - | - | - | 2 | 2 | 4 |
| NR1H4 | FEXARAMINE | - | - | TRUE | TRUE | - | - | - | - | - | 2 | 2 | 4 |
| TERT | GV1001 | - | - | TRUE | - | - | - | - | - | TRUE | 2 | 1 | 3 |
| TNF | THALIDOMIDE | - | - | TRUE | - | - | - | TRUE | TRUE | TRUE | 4 | 7 | 11 |
| TNF | AMRINONE | - | - | TRUE | - | - | - | - | - | - | 1 | 5 | 6 |
| TNF | CHLOROQUINE | - | - | TRUE | - | - | - | - | - | - | 1 | 5 | 6 |
| TNF | CLENBUTEROL | - | - | TRUE | - | - | - | - | - | - | 1 | 5 | 6 |
| TNF | PSEUDOEPHEDRINE | - | - | TRUE | - | - | - | - | - | - | 1 | 2 | 3 |
CFB: Clearity Foundation Biomarkers, CFCT: Clearity Foundation Clinical Trial, DB: Drugbank, GPI: Guide to Pharmacology Interactions Version: 04-March-2015, PG: PharmGKB - The Pharmacogenomics Knowledgebase, TALC: Targeted Agents in Lung Cancer, TE: Trends in the exploitation of novel drug targets, TT: Therapeutic Target Database, Version 4.3.02, TC: The Druggable Genome: Evaluation of Drug Targets in Clinical Trials Suggests Major Shifts in Molecular Class and Indication.
Brown et al. reported that Docetaxel should be further studied for its use in influenza vaccine production (Chen et al., 2012; Brown et al., 2015). Obatoclax is a novel inhibitor of endosomal acidification which prevents viral fusion and could be pursued as a potential broad-spectrum antiviral candidate (Varghese et al., 2017). The TERT gene decreased rapidly at 8 h post infection (ENSG00000164362, p-value=3.34E−05). TERT is the major catalytic subunit of telomerase. TERT has additionally reported to be associated with apoptosis (Fu et al., 1999).
The final goal of studying host-microbe interactions is to inform in silico models in order to identify crucial pathways, hubs, and bottlenecks, which will lead to development of new targets and strategies for prevention, diagnosis, risk assessment, and treatment of severe influenza infection in humans (Kollmus et al., 2014). This study adopted functional transcriptome analysis with transcriptome sequencing and identification of genes after focusing on key time-series processes, providing several gene set enrichment analysis methods to identify genes related to seasonal influenza strains. In the future, a series of functional validation experiments need to be performed to compare more virulent influenza strains.
Acknowledgments
We thank Dr. Hueeman Kim for his advice regarding experimental design. This research was funded by the post-genome multi-ministerial genome project (NRF-2014M3C9A3064815) of the Ministry of Science, ICT, and Future Planning, Republic of Korea. This research was supported by National Cancer Center Grant (NCC-1711290).
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
REFERENCES
- Alsuwaidi AR, Almarzooqi S, Albawardi A, Benedict S, Kochiyil J, Mustafa F, Hartwig SM, Varga SM, Souid AK. Cellular bioenergetics, caspase activity and glutathione in murine lungs infected with influenza A virus. Virology. 2013;446:180–188. doi: 10.1016/j.virol.2013.07.034. [DOI] [PubMed] [Google Scholar]
- Bagga S, Bouchard MJ. Cell cycle regulation during viral infection. Methods Mol Biol. 2014;1170:165–227. doi: 10.1007/978-1-4939-0888-2_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SG, Knowell AE, Hunt A, Patel D, Bhosle S, Chaudhary J. Interferon inducible antiviral MxA is inversely associated with prostate cancer and regulates cell cycle, invasion and Docetaxel induced apoptosis. Prostate. 2015;75:266–279. doi: 10.1002/pros.22912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Yuan L, Fan Q, Su F, Chen Y, Hu S. Adjuvant effect of docetaxel on the immune responses to influenza A H1N1 vaccine in mice. BMC Immunol. 2012;13:36. doi: 10.1186/1471-2172-13-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, Basta S, O’Neill R, Schickli J, Palese P, Henklein P, Bennink JR, Yewdell JW. A novel influenza A virus mitochondrial protein that induces cell death. Nat Med. 2001;7:1306–1312. doi: 10.1038/nm1201-1306. [DOI] [PubMed] [Google Scholar]
- Commandeur JN, Stijntjes GJ, Vermeulen NP. Enzymes and transport systems involved in the formation and disposition of glutathione S-conjugates. Role in bioactivation and detoxication mechanisms of xenobiotics. Pharmacol Rev. 1995;47:271–330. [PubMed] [Google Scholar]
- Dawood FS, Iuliano AD, Reed C, Meltzer MI, Shay DK, Cheng PY, et al. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect Dis. 2012;12:687–695. doi: 10.1016/S1473-3099(12)70121-4. [DOI] [PubMed] [Google Scholar]
- Dennis G, Sherman B, Hosack D, Yang J, Gao W, Lane H, Lempicki R. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:R60. doi: 10.1186/gb-2003-4-9-r60. [DOI] [PubMed] [Google Scholar]
- Dimitrakopoulou K, Dimitrakopoulos GN, Wilk E, Tsimpouris C, Sgarbas KN, Schughart K, Bezerianos A. Influenza A immunomics and public health omics: the dynamic pathway interplay in host response to H1N1 infection. OMICS. 2014;18:167–183. doi: 10.1089/omi.2013.0062. [DOI] [PubMed] [Google Scholar]
- Frazee AC, Pertea G, Jaffe AE, Langmead B, Salzberg SL, Leek JT. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat Biotechnol. 2015;33:243–246. doi: 10.1038/nbt.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Begley JG, Killen MW, Mattson MP. Anti-apoptotic role of telomerase in pheochromocytoma cells. J Biol Chem. 1999;274:7264–7271. doi: 10.1074/jbc.274.11.7264. [DOI] [PubMed] [Google Scholar]
- Gaubin M, Autiero M, Basmaciogullari S, Metivier D, Mishal Z, Culerrier R, Oudin A, Guardiola J, Piatier-Tonneau D. Potent inhibition of CD4/TCR-mediated T cell apoptosis by a CD4-binding glycoprotein secreted from breast tumor and seminal vesicle cells. J Immunol. 1999;162:2631–2638. [PubMed] [Google Scholar]
- Gram GJ, Hansen JE. Characterization of HIV gp120 envelope glycoprotein by lectin analysis. Methods Mol Med. 1998;9:167–174. doi: 10.1385/0-89603-396-1:167. [DOI] [PubMed] [Google Scholar]
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan IH, Zhang MS, Powers LS, Shao JQ, Baltrusaitis J, Rutkowski DT, Legge K, Monick MM. Influenza A viral replication is blocked by inhibition of the inositol-requiring enzyme 1 (IRE1) stress pathway. J Biol Chem. 2012;287:4679–4689. doi: 10.1074/jbc.M111.284695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton NS. Revisiting the concept of a cytopathic viral infection. PLoS Pathog. 2017;13:e1006409. doi: 10.1371/journal.ppat.1006409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Kapasi AA, Patel G, Franki N, Singhal PC. HIV-1 gp120-induced tubular epithelial cell apoptosis is mediated through p38-MAPK phosphorylation. Mol Med. 2002;8:676–685. [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015a;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Bheda-Malge A, Lin Y, Sreekrishna K, Adams R, Robinson MK, Bascom CC, Tiesman JP, Isfort RJ, Gelinas R. A systems approach to understanding human rhinovirus and influenza virus infection. Virology. 2015b;486:146–157. doi: 10.1016/j.virol.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmus H, Wilk E, Schughart K. Systems biology and systems genetics - novel innovative approaches to study host–pathogen interactions during influenza infection. Curr Opin Virol. 2014;6:47–54. doi: 10.1016/j.coviro.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Lei Y, Moore CB, Liesman RM, O’Connor BP, Bergstralh DT, Chen ZJ, Pickles RJ, Ting JP. MAVS-mediated apoptosis and its inhibition by viral proteins. PLoS ONE. 2009;4:e5466. doi: 10.1371/journal.pone.0005466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorbek G, Lewinska M, Rozman D. Cytochrome P450s in the synthesis of cholesterol and bile acids--from mouse models to human diseases. FEBS J. 2012;279:1516–1533. doi: 10.1111/j.1742-4658.2011.08432.x. [DOI] [PubMed] [Google Scholar]
- Lu C, Jiang Z, Fan X, Liao G, Li S, He C, Han L, Luo S, Liu Y, Lin H, Li L, Li X, Liang Q, Wang Y, Luo G. A metabonomic approach to the effect evaluation of treatment in patients infected with influenza A (H1N1). Talanta. 2012;100:51–56. doi: 10.1016/j.talanta.2012.07.076. [DOI] [PubMed] [Google Scholar]
- Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6:507–515. doi: 10.1016/S1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- Mackenzie PI, Somogyi AA, Miners JO. Advances in drug metabolism and pharmacogenetics research in Australia. Pharmacol Res. 2017;116:7–19. doi: 10.1016/j.phrs.2016.12.008. [DOI] [PubMed] [Google Scholar]
- Nair H, Brooks WA, Katz M, Roca A, Berkley JA, Madhi SA, et al. Global burden of respiratory infections due to seasonal influenza in young children: a systematic review and meta-analysis. Lancet. 2011;378:1917–1930. doi: 10.1016/S0140-6736(11)61051-9. [DOI] [PubMed] [Google Scholar]
- Nelson DR. The cytochrome p450 homepage. Hum. Genomics. 2009;4:59–65. doi: 10.1186/1479-7364-4-1-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson KG, Webster RG, Hay AJ. Textbook of Influenza. Blackwell Science; 1998. [Google Scholar]
- Park BK, Pirmohamed M, Kitteringham NR. The role of cytochrome P450 enzymes in hepatic and extrahepatic human drug toxicity. Pharmacol Ther. 1995;68:385–424. doi: 10.1016/0163-7258(95)02013-6. [DOI] [PubMed] [Google Scholar]
- Park SJ, Kumar M, Kwon HI, Seong RK, Han K, Song JM, Kim CJ, Choi YK, Shin OS. Dynamic changes in host gene expression associated with H5N8 avian influenza virus infection in mice. Sci Rep. 2015;5:16512. doi: 10.1038/srep16512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GE, Gaszewska-Mastarlarz A, Moskophidis D. The role of alpha/beta and gamma interferons in development of immunity to influenza A virus in mice. J Virol. 2000;74:3996–4003. doi: 10.1128/JVI.74.9.3996-4003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott I, Norris KL. The mitochondrial antiviral signaling protein, MAVS, is cleaved during apoptosis. Biochem Biophys Res Commun. 2008;375:101–106. doi: 10.1016/j.bbrc.2008.07.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siren J, Valimaki N, Makinen V. Indexing graphs for path queries with applications in genome research. EEE/ACM Trans. Comput Biol Bioinform. 2014;11:375–388. doi: 10.1109/TCBB.2013.2297101. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga ZT, Ramos I, Hai R, Schmolke M, García-Sastre A, Fernandez-Sesma A, Palese P. The influenza virus protein PB1-F2inhibits the induction of type I interferon at the level of the MAVS adaptor protein. PLoS Pathog. 2011;7:e1002067. doi: 10.1371/journal.ppat.1002067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese FS, Rausalu K, Hakanen M, Saul S, Kummerer BM, Susi P, Merits A, Ahola T. Obatoclax inhibits alphavirus membrane fusion by neutralizing the acidic environment of endocytic compartments. Antimicrob Agents Chemother. 2017;61:02227–16. doi: 10.1128/AAC.02227-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner AH, Coffman AC, Ainscough BJ, Spies NC, Skidmore ZL, Campbell KM, Krysiak K, Pan D, McMichael JF, Eldred JM, Walker JR, Wilson RK, Mardis ER, Griffith M, Griffith OL. DGIdb 2.0: mining clinically relevant druggene interactions. Nucleic Acids Res. 2016;44:D1036–D1044. doi: 10.1093/nar/gkv1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeganeh B, Rezaei Moghadam A, Tran AT, Rahim MN, Ande SR, Hashemi M, Coombs KM, Ghavami S. Asthma and influenza virus infection: focusing on cell death and stress pathways in influenza virus replication. Iran J Allergy Asthma Immunol. 2013;12:1–17. [PubMed] [Google Scholar]
- Yu D, Kim M, Xiao G, Hwang TH. Review of biological network data and its applications. Genomics Inform. 2013;11:200–210. doi: 10.5808/GI.2013.11.4.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W, Chen D, Xiong M, Zhu J, Lin X, Wang L, Zhang J, Chen L, Zhang H, Chen H, Chen M, Jin M. Insights into the increasing virulence of the swine-origin pandemic H1N1/2009 influenza virus. Sci Rep. 2013;3:1601. doi: 10.1038/srep01601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.