Abstract
Taste buds contain multiple cell types with each type expressing receptors and transduction components for a subset of taste qualities. The sour sensing cells, Type III cells, release serotonin (5-HT) in response to the presence of sour (acidic) tastants and this released 5-HT activates 5-HT3 receptors on the gustatory nerves. We show here, using Htr3a-EGFP mice, that 5-HT3-expressing nerve fibers preferentially contact and synapse with Type III taste cells. Further, these 5-HT3-expressing nerve fibers terminate in a restricted central-lateral portion of the nucleus of the solitary tract (nTS) – the same area that shows increased c-Fos expression upon presentation of a sour tastant (30 mM citric acid). This acid stimulation also evokes c-Fos in the laterally adjacent mediodorsal spinal trigeminal nucleus (DMSp5), but this trigeminal activation is not associated with the presence of 5-HT3-expressing nerve fibers as it is in the nTS. Rather, the neuronal activation in the trigeminal complex likely is attributable to direct depolarization of acid-sensitive trigeminal nerve fibers, e.g. polymodal nociceptors, rather than through taste buds. Taken together, these findings suggest that transmission of sour taste information involves communication between Type III taste cells and 5-HT3-expressing afferent nerve fibers that project to a restricted portion of the nTS consistent with the mapping of taste quality information in the primary gustatory nucleus.
Keywords: citric acid, serotonin receptor, taste Bud, solitary tract, trigeminal, chemesthesis
INTRODUCTION
The primary function of the sense of taste is to promote the consumption of palatable nutrients (salty, sweet, and umami) and rejection of aversive toxins and poisons (sour and bitter). Of the five primary tastes, sour is perhaps the most complex in terms of both perception and underlying neurobiology. In particular, acidic substances stimulate not only taste cells that comprise taste buds, but also free nerve endings of acid-sensitive nerve fibers (Damann et al., 2008; Roper, 2014). Together these systems are responsible for the multimodal experiences of “sour” taste and pain-like sensations (e.g. “tart”, “tangy”) called ‘chemesthesis.’ Given the perceptual and anatomical complexity of intraoral acidification, it is not surprising that the precise neural underpinnings of sour taste perception remain unresolved.
In this regard, taste buds contain 3 types of elongate cells, classified into one of three distinct categories: Type I cells are glia-like in that they do not respond directly to canonical taste stimuli and are considered to be support cells. Type II cells, also called “receptor cells,” express the canonical G-protein coupled taste receptors for sweet, umami and bitter tastes (Nelson et al., 2001; DeFazio et al., 2006; Tomchik et al., 2007). Type III taste cells are the most likely candidate for sour taste transduction as acidic, “sour” stimuli activate Type III cells, resulting in the release of serotonin (5-HT) and possibly other neurotransmitters (Chan et al., 2004; Dvoryanchikov et al., 2007; Huang et al., 2008; Huang et al., 2011). Moreover, genetric deletion of or toxin induced blockade of synaptic transmission from Type III cells expressing PKD-2L1 receptors eliminates electrophysiological responses to sour stimuli (Huang et al., 2006; Kataoka et al., 2008; Oka et al., 2013). Taken together, these studies strongly imply that Type III taste cells are the transducers for sour taste and are necessary for transmission of sour information to the nerves.
As for other taste qualities, transmission of sour taste information to taste nerves requires purinergic signalling through P2X-containing neural receptors (Finger et al., 2005; Vandenbeuch et al., 2015) but 5-HT signaling also contributes to transmission from Type III cells. Genetic deletion or pharmacological blockade of 5-HT3 receptors on gustatory afferent nerve fibers decreases neural responses to sour taste stimuli (Larson et al., 2015). Therefore, 5-HT3-expression may serve as a marker of sour-responsive nerve fibers within taste buds as well as a means to mapping projections of sour-responsive nerve fibers into the brainstem.
The gustatory nerves terminate centrally within the rostral and intermediate two-thirds nucleus of the solitary tract (nTS), a complex nucleus extending throughout the rostral medulla (Whitehead and Frank, 1983; Corson et al., 2012; Corson and Erisir, 2013). Non-taste oral afferents from the trigeminal nerve also terminate within the lateral portions of the rostral-intermediate nTS (Corson et al., 2012) as well as in the laterally adjacent descending trigeminal complex including especially the mediodorsal spinal trigeminal nucleus (DMSp5; Stratford et al., 2016) which receives input predominantly from the lingual branch of the trigeminal nerve (Corson et al., 2012). Taste-related activity due to intraoral infusion of acidic solutions is restricted to the dorsocentral subnucleus of the nTS and lateral portions of nTS at intermediate AP levels, while non-taste activation by intraoral acidification is pronounced within the rostrolateral subnucleus of the nTS and in the DMSp5 (Stratford et al., 2016).
In the present study, we used quantitative confocal microscopy and immuno-electron microscopy to clarify the peripheral anatomical relationship between Type III taste cells (which contain 5-HT) and the subset of gustatory nerve fibers expressing 5-HT3A receptors. Then, we examined which areas of the brainstem (nTS vs. DMSp5) receive input from 5-HT3A-labeled taste fibers and tested whether these areas are responsive to sour-taste stimulation measured by citric acid-evoked c-Fos activity. Finally, we compared 5-HT3A fiber distribution to calcitonin gene-related peptide (CGRP), a marker for polymodal nociceptors that we previously showed is involved in the non-taste component of perception of intraoral acids (Stratford et al., 2016). Our results provide support for the role of serotonergic neurotransmission in peripheral sour-sensing taste systems and the taste quality-specific mapping of this system into the primary gustatory nucleus in the brainstem.
MATERIALS AND METHODS
Animals
All animal experiments were conducted in accordance with the University of Colorado Animal Care and Use Committee. Mice were housed in ventilated cages on a 12–12 h light cycle and fed standard chow ad libitum. 3 – 7 month old male and female 5-HT3AGFP mice (n = 5 of each sex), originally on FVB/N-Swiss Webster background (JAX stock # 000273-UNC; Tg(Htr3a-EGFP)DH30Gsat/Mmnc; RRID:IMSR_MMRC:000273) but crossed for 2 – 4 generations with C57BL/6J mice (RRID:IMSR_JAX:000664; Jackson Laboratories, Bar Harbor, ME), or age-matched C57BL6/J (control) mice (n = 3) were used for all experiments (Table 1). Previous studies on the taste system in the Htr3a-EGFP line have shown the exact correlation between GFP-expressing gustatory ganglion cells and those showing physiological responses to 5-HT and 5-HT3 agonists (Larson et al., 2015).
Table 1.
Age and Sex of All Animals Used
| Mouse | Age (days) | Sex | Animal Identification |
|---|---|---|---|
| 5-HT3A GFP | 171 | M | 140724 |
| 5-HT3A GFP | 176 | M | 140729 |
| 5-HT3A GFP | 104 | F | 141009 |
| 5-HT3A GFP | 99 | M | 141215 |
| 5-HT3A GFP | 193 | F | 150214 |
| 5-HT3A GFP | 184 | F | 150130c |
| 5-HT3A GFP | 181 | F | 150130b |
| 5-HT3A GFP | 182 | M | 150130a |
| 5-HT3A GFP | 177 | M | 150125b |
| 5-HT3A GFP | 153 | F | 150105 |
| C57Bl6/J | 156 | F | 120306a |
| C57Bl6/J | 156 | F | 120309a |
| C57Bl6/J | 203 | F | 120310a |
Peripheral Taste Field Tissue Preparation
Animals were deeply anesthetized with Fatal-Plus® (50 mg/kg intraperitoneally; MWI, Boise, ID) or pentobarbital (i.p. injection 100mg/kg in saline), then perfused transcardially with saline (0.9% sodium chloride) followed by 4% paraformaldehyde (PFA) in 0.1M phosphate buffer, pH 7.2–7.4 (PB). For 5-HT/5-HT3A GFP colocalization analysis, animals were injected i.p. with 5-hydroxy-L-tryptophan (5-HTP, 50 mg/kg; Sigma, St. Louis, MO) 1 hour before perfusion. After perfusion, tongues were removed, post-fixed for three hours at room temperature in 4% PFA in PB, and then cryoprotected overnight in 20% sucrose in PB at 4˚C. After cryoprotection, the circumvallate (CV), foliate (Fol), fungiform (FF), and soft palatal (SP) taste fields were blocked. Tissues were then embedded in OCT (Optimal Cutting Temperature compound; Fisher Scientific, Pittsburgh, PA), frozen, cryosectioned at 12 – 16 μm and directly mounted onto Superfrost Plus slides (Fisher Scientific, Pittsburgh, PA). Slides were then processed using standard immunohistochemical methods (see General Immunohistochemistry Methods).
Immuno-Electron Microscopy
5-HT3A GFP mice were anesthetized with pentobarbital (20 μg/g, i.p.) and perfused for 10 seconds through the left ventricle with 0.1% sodium nitrite, 0.9% sodium chloride and 100 units sodium heparin in 100 ml 0.1M PB (pH 7.3). This was followed by perfusion fixation for 10 minutes with 4% PFA in 0.1M PB. All perfusates were warmed to 42˚C before use. After perfusion the excised circumvallate papillae were fixed in fresh fixative for 3 h at 4˚C. Some tissues were sectioned on a vibratome at 50–100μm tickness, other tissue was cryo-protected by passage through 3 solutions according to the method of Eldred (1983): Solution 1, 5% glycerin and 10% sucrose in 0.1 M PB (plus 0.1 ml of 2.2% calcium chloride per 100 ml) for 1 h; Solution 2, 10% glycerin and 15% sucrose in 0.1 M PB (plus 0.1 ml of 2.2% calcium chloride per 100 ml) for 1 h; Solution 3, 15% glycerin and 20% sucrose in 0.1 M PB (plus 0.1 ml of 2.2% calcium chloride per 100 ml) overnight.
Frozen sections (50 μm thick) were cut into 0.1 M phosphate buffered saline (PBS) on a freezing stage sliding microtome and then blocked with 5% normal donkey serum, 0.3% BSA in 0.1M PBS (pH 7.3) for 2 h at 4˚C. This was followed by incubation for 24 h at 4˚C with chicken anti- GFP polyclonal antibody (Table 2) in 0.1M PBS. After rinsing in 0.1M PBS, the sections were incubated in affinity purified biotinylated donkey anti-chicken IgG secondary antibody (Jackson ImmunoResearch Labs; Cat# 703-065-155; RRID: AB_2313596; diluted to 1:2000) in PBS for 2 h at room temperature. The sections then were incubated with avidin-biotin complex (Elite ABC-Peroxidase Vectastain Kit; Vector Laboratories; Cat# PK-6100; RRID: AB_2336819) in PBS for 1 h at room temperature. After rinsing, sections were treated for 10 min in 0.05M Tris buffer (pH 7.3) containing 0.05% DAB then transferred for 2–4 min into a fresh aliquot of the DAB mixture containing H2O2 at a final concentration of 0.002%. Vibratome sections were prepared similarly.
Table 2.
Primary Antibodies
| Antibody (Antibody Registry ID) | Antigen (sequence; if applicable) | Source (catalog #) | Working Dilution |
|---|---|---|---|
| Chicken polyclonal anti-GFP (AB_90890) (for immuno- electron microscopy) | Purified recombinant full length green fluorescent protein (GFP) | Millipore Billerica, MA (AB16901) | 1:2,000 |
| Chicken polyclonal anti-GFP (AB_2307313) (for immunohistochemistry) | Purified recombinant full length green fluorescent protein (GFP) | Aves Labs, Inc. Tigard, Oregon (GFP-1010) | 1:2,000 |
| Goat polyclonal anti-GNAT3 (AB_10882823) | synthetic peptide to the internal region of the protein human protein (C-KNQFLDLNLKKEDKE) | Aviva System Biology San Diego, CA (OAEB00418) | 1:500 |
| Rabbit polyclonal anti-5-HT (AB_572263) | serotonin coupled to BSA with paraformaldehyde | ImmunoStar, Inc. Hudson, WI (20080) | 1:5,000 |
| Rabbit polyclonal anti-αCGRP (AB_518147) | synthetic peptide to mouse/rat N′ terminus (HSCNTATCVTHRLAGLLSRSGGVVKDNFVPTNVGSEAF) | Peninsula Laboratories, LLC San Carlos, CA (T-4032) | 1:2,500 |
| Rabbit polyclonal anti-c-Fos (AB_2314042) | synthetic peptide to human 4–17 residues (SGFNADYEASSSRC) | EMD Millipore Billerica, MA (PC38) | 1:3,000 |
After the DAB reaction, the sections were washed 3 times for 5 min each in 0.1 M phosphate buffer (pH 7.3) and post-fixed in 1% OsO4 in 0.1M PB for 15 min. After rinsing in 0.05 M sodium maleate buffer (pH 5.2), the sections were stained en bloc in 1% uranyl acetate in 0.025 M sodium maleate buffer (pH 6.0) overnight at 4˚C, followed by dehydration in an alcohol series, processed through propylene oxide, and embedded with Eponate 12 (Ted Pella, Inc.). Thin sections (90–120 nm) were cut with a diamond knife on a Reichert Ultracut E ultramicrotome and then examined with a HITACHI H-7000 transmission electron microscope at 75 kV. Ultrastructural images were obtained with a FEI Tecnai G2 Transmission Electron Microscope with a XR80 AMT digital camera (Advanced Microscopy Techniques, Woburn, MA). Controls consisted of omitting the primary antiserum. No reactivity was observed under these conditions.
Citric Acid Stimulation via Intraoral Cannula and Brainstem Tissue Preparation
Bilateral intraoral cannulae were implanted into 5-HT3AGFP animals as described previously (Stratford and Thompson, 2016; Stratford et al., 2016). Briefly, mice were anesthetized with an intramuscular (IM) injection of a combination of medetomidine hydrochloride (Domitor®; 0.4 mg/kg; Pfizer, Helsinki, Finland) and ketamine hydrocholoride (40 mg/kg; Bioniche Pharma, Lake Forest, IL). Intraoral cannulae were inserted via midline incision caudal to the pinnae and a sterile, 1.5 inch, stainless steel hypodermic needle (19 gauge; Hamilton, Reno, NV) was inserted in the back of the neck and guided subcutaneously into the oral cavity, lateral to the first maxillary molar.
A polyethylene tube (50 gauge; Becton Dickinson and Company, Sparks, MD), flared with a cautery iron, was first fed though a small, flat # 0 nylon washer (Product Components Corporation; Martinez, CA) and then through the tip of the hypodermic needle. The needle was withdrawn, leaving the cannula and washer in place. The wound was closed with Dexon II 6.0 mm suture (Syneture, Norwalk, CT), and a second nylon washer secured the loose end of the tubing. The remaining tubing was flared by a cautery and a blunt 1” hypodermic needle (25 gauge; McKesson, San Francisco, CA) was secured to the loose end of the tubing. Animals were allowed to recover for 5 days prior to training.
Fluids were delivered into intraoral cannulae via a 5 cc syringe (Becton Dickinson and Company, Sparks, MD) connected to a syringe pump (Model R99-E, Razel Scientific Instruments, St. Albans, VT). Three days prior to stimulation, all animals were placed on 23 h/day water restriction, and received 3 ml of dH2O infused over 30 min each day into one of two intraoral cannulae at a flow rate of 0.10 ml/ min. On stimulation day, 3 ml of either deionized water (control; n = 3) or 30 mM citric acid (n = 5) was infused into a single intraoral cannula at a constant flow rate (0.10 ml/ min) over the course of 30 min. Then, animals were left undisturbed for forty-five min prior to perfusion using the same methods as described above (see Peripheral Taste Field Tissue Preparation). After 3 h post fixation and overnight cryopreservation, brainstems were blocked just anterior to the cerebellum, embedded in OCT, frozen and cryosectioned at 40 μm. The olfactory bulbs were also blocked, sectioned, and used as a positive control for c-Fos immunohistochemistry. Free-floating sections were collected in 0.9 % NaCl in PB (PBS).
General Immunohistochemistry Methods
Staining was carried out using standard immunohistochemical procedures. Briefly, tissue (slides or free-floating sections) were washed three times in PBS, and then transferred to a blocking medium consisting of 2% normal donkey serum (NDS, Jackson ImmunoResearch, West Grove, PA; RRID: AB_2337258) diluted in antibody medium (AB medium; 0.3% triton, 0.15 M sodium chloride, and 1% bovine serum albumin in PB) for one hour at room temperature. Primary antisera (Table 2) were diluted in AB medium and incubated for 1 night at 4˚C, with the exception of c-Fos antisera, which was incubated for 2 nights at 4˚C. Then, tissue was rinsed 3 times in 0.1 M PBS, followed by a 2 h incubation at room temperature in secondary antibodies each diluted 1:400 in AB media.
For taste tissue, an Alexa Fluor 488 (donkey anti-chicken, Jackson ImmunoResearch Labs; Cat# 703-546-155; RRID: AB_141708), an AlexaFluor 568 (donkey anti-rabbit, Invitrogen; Cat # A10042; RRID: AB_11180183), and an AlexaFluor 647 (donkey anti-goat, Invitrogen; Cat # A21447; RRID: AB_141844 were incubated concurrently. For brainstem tissue, an Alexa Fluor 488 (donkey anti-chicken, Jackson ImmunoResearch Labs; Cat# 703-546-155; RRID: AB_141708) was incubated concurrently with a Rhodamine Red-X Fab Fragment (donkey anti-rabbit, Jackson ImmunoResearch Labs; Cat# 711-297-003; RRID: AB_2340615).
Peripheral tissue was cover-slipped using Fluoromount – G (Southern Biotech, Birmingham, Alabama). Brainstem sections were counterstained with NeuroTrace Nissl stain 640/680 (1:100; Thermo Fisher Scientific; Cat# N21483; RRID: AB_2572212), which was included in the AB medium during incubation in secondary antisera. Following an additional three 0.1M PBS washes, free floating brainstem sections were mounted onto Superfrost Plus slides and then cover-slipped using Fluoromount – G.
Image Acquisition
Taste field images were collected using a Leica TCS SP5 II (Buffalo Grove, IL) laser scanning confocal microscope with a 63x oil-immersion objective (n.A. 1.4). Images were collected at 1024 x 1024 pixels with 2.5x digital zoom using the Leica Application Suite, Advanced Fluorescence (version 2.7.3.9723). Whole slide images of brainstem sections were photographed using Surveyor by Objective Imaging software (RRID: SCR_014433; Cambridge, UK; http://www.objectiveimaging.com/Surveyor/OI_Turboscan.htm) with a monochrome Leica DFC 365FX camera on a Leica DM6000B microscope. For each fluorophore, a series of 10X grid images were obtained using the Multiscan setting. Images were then stitched together in real time using the best focus algorithm in the Surveyor software, which yielded a mosaic image of the whole microscope slide. Then images of whole brain sections were exported as single images using the Region of Interest selection tool in Surveyor.
Post Image Acquisition Processing
All images were unaltered prior to being analyzed. Brightness and contrast of all photographs shown as representative sections in all figures were optimized in Adobe Photoshop CS5 (RRID: SCR_014199; San Jose, CA) using the levels and hue tools. For use in Figs. 4 and 6, spatial distribution plots generated by the PixelIntensity program (see details below) were first saved as .tiff images within MATLAB. Then these images were converted into binary images within Adobe Photoshop CS6 using the ‘mode’ and ‘levels’ tools. Finally, each binary density image was then false colored (magenta or green) and combined into a single image.
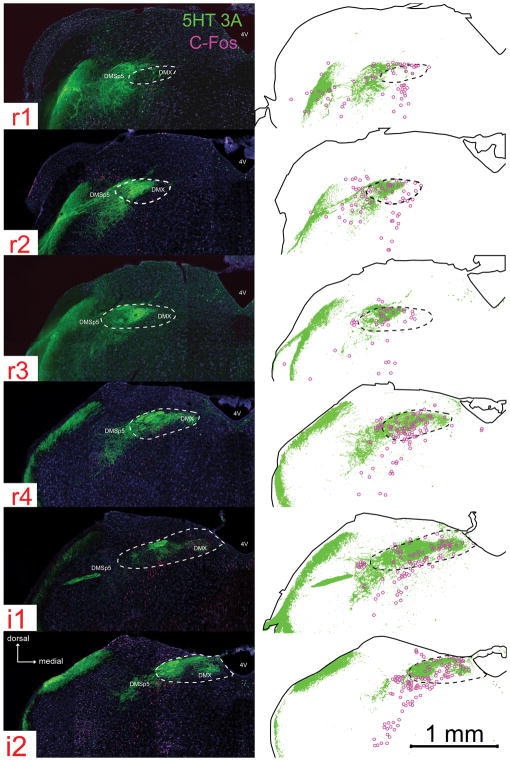
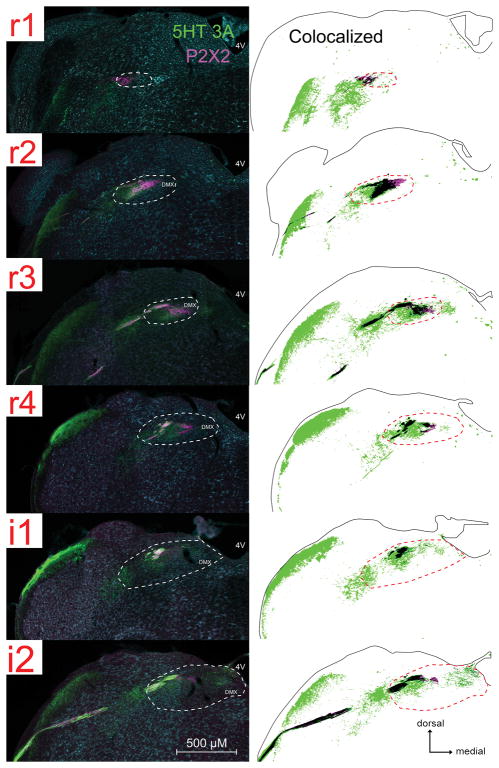
Fig. 4. Neuronal activation in response to citric acid intraoral taste stimulation is found in areas of the nTS and adjacent DMSp5 that are densely innervated by 5-HT3A-positive fibers.
For each image-drawing pair: Left: photomicrographs of 5-HT3A GFP (green) and c-Fos (magenta) immunoreactivity. The white dotted line demarcates the boundary of the nTS. 4V: 4th ventricle; DMSP5: dorsomedial nucleus of the descending trigeminal complex; DMX: dorsal motor nucleus of the vagus. Right: Spatial distribution of 5-HT3A labeled pixels that are 2X standard deviation of background (identified using a customized MATLAB program). The magenta circles are chartings of citric acid- evoked c-Fos activation in the brainstem. The solid black outline indicates the boundaries of the tissue; whereas the black dotted line demarcates the boundary of the nTS.
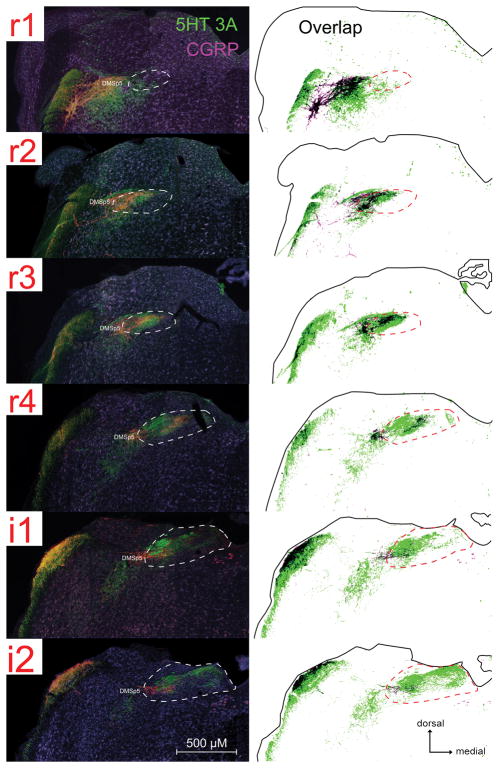
Fig. 6. 5-HT3A-positive fibers are found in some brainstem regions where CGRP immunoreactivity is also found within the nTS and adjacent DMSp5.
For each image-drawing pair: Left: photomicrographs of 5-HT3A GFP (green) and CGRP (magenta) immunoreactivity. The white dotted line demarcates the boundary of the nTS. DMSP5: dorsomedial nucleus of the descending trigeminal complex. Right: Spatial distribution of CGRP and 5-HT3A labeled pixels that are 2X standard deviation of background (identified using a customized MATLAB program). Labeled pixels that overlap are shown as black. The solid black outline indicates the boundaries of the tissue; whereas the black dotted line demarcates the boundary of the nTS.
MATLAB Image Analyses
Peripheral taste tissue 5-HT3a GFP/ immunofluroscence colocalization analysis
The theory underlying the method for this analysis are described in Corson and Erisir, 2013). In brief, if two differently labeled objects are in physical contact, then because of limits on resolution of light microscopy, one or more lines of pixels along the apposition will exhibit dual labeling, whereas if objects are close, but not in contact, then pixels will not exhibit dual label if the object separation is wider than the limits of resolution.
For colocalization analysis, z-stack images (0.7 um spacing) of the entire tissue section were analyzed using imstack in the 2014a (serial update 2) of the Matlab software (The MathWorks, Natick, MA; RRID: SCR_001622; program available on Mathworks Exchange: http://www.mathworks.com/matlabcentral/fileexchange/55546-imstack). First, planes lacking immunofluorescence for Type II cells, Type III cells, or GFP positive nerve fibers were removed from the analysis. Then, to eliminate background staining, a threshold mask was applied to images using Otsu’s method (Otsu, 1979) modified for 3D images stacks, which resulted in binary, post-threshold images. Then the total number of labeled pixels for each channel (GFP, 5-HT, GNAT3) as well as total number of colocalized pixels (GFP/5-HT or GFP/GNAT3) were measured from these binarized images.
Representative levels of the nucleus of the solitary tract
In order to thoroughly quantify immunohistochemical staining, as well as citric-acid evoked c-Fos activity within the rostral and intermediate parts of the nTS (and adjacent DMSp5), we divided the nTS into 6 rostral-caudal levels as described in detail previously (Stratford et al., 2016). The rostrocaudal levels are designated as rostral (r1 – r4) and intermediate (i1 – i2); situated respectively at −6.36, −6.48, −6.72, −6.96, −7.08, and −7.20 from bregma). For each level, we further divided the region of the nTS into 6 subfields: lateral, mid, and medial for dorsal and ventral tiers.
Citric-acid evoked c-Fos positive brainstem cell quantification
To quantify the number of c-Fos positive cells in an unbiased fashion, we used ImageBWconvertGUI, a custom-made program running in the 2013a Matlab Computing Environment as described previously (Stratford et al., 2016) and available at https://github.com/neuropil/ImageBWconvert/. In brief, the red (c-Fos) color channel in each image was filtered based on a stringent threshold (mean + 2 standard deviations (SD) of background pixel intensity level), and then the red color channel was converted to a binary channel. Then, the number of c-Fos positive cells was quantified using the cell counter plug-in in ImageJ (version 1.47, Bethesda, MD; RRID: SCR_003070). Cases were counted only when substantial c-Fos immunoreactivity (Fos-LI) was observed in the olfactory bulb, as c-Fos expression is robust in the olfactory bulb in all animals (Guthrie et al., 1993).
Spatial mapping of immunofluorescence in brainstem regions
Visualization of the spatial distribution within each nTS section of 5-HT3A GFP and CGRP was done using PixelIntensity as described previously (Stratford et al., 2016) and available at https://github.com/neuropil/HeatMappingProgram. Briefly, background pixel intensity was calculated for each channel separately using a (150 x 150 pixel) square polygon averaged across all 6 representative nTS sections. To quantify pixel intensity in regions of interest, a polygon was drawn around the region of interest and immunofluorescence label was defined based on a threshold (mean + 2 SD of background pixel intensity level). Then the pixels that met this 2 + SD criteria were used to create a 2-D spatial distribution map for each immunohistochemical stain.
Percent of 5-HT3a GFP labeled pixels in brainstem areas
In order to quantify the amount of 5-HT3a GFP labeled pixels within each nTS subregion, as well as with in the DMSp5, the percent of GFP positive label within each region was measured using ImageJ. To do this, spatial distribution maps of 5HT3a GFP label pixels obtained from the PixelIntensity program described above (see Spatial mapping of immunofluorescence in brainstem regions) were loaded into ImageJ. Then, standard-size boxes (for the nTS) or circles (for the DMSp5) were drawn with the ‘region of interest’ tool to demarcate each nTS subregion or DMSp5 and the percent of labeled pixels within each region of interest was calculated using the ‘area fraction’ measurement within the ‘measure’ toolbar.
Primary antibody Characterization
Green Fluorescent Protein (GFP) Millipore (for immuno-electron microscopy)
The antiserum, directed against recombinant green fluorescent protein (GFP) containing a 6-his tag, recognizes the full length recombinant green fluorescent protein, as determined by immunoblot analysis. Specificity controls show immunoreaction product when applied to GFP-transfected cells, but not in cells lacking GFP (Millipore data sheet). Further, no immunoreactivity is observed when this antibody is applied to tissue from wildtype mice.
Green Fluorescent Protein (GFP) Aves (for immunohistochemistry)
The antiserum recognizes the entire recombinant full length green fluorescent protein as determined by western blot analysis, and colabels the endogenously expressed GFP in transgenic GFP-expressing mice (Aves data sheet). This antibody does not label tissue from wildtype mice lacking GFP expression.
Guanine nucleotide-binding protein G(t) subunit α-3 (GNAT3)
The polyclonal antibody recognizes sequence C-KNQFLDLNLKKEDKE from the internal region of the human GNAT3 protein and is predicted to recognize the cow, dog, mouse, pig, rabbit and rat GNAT3 based on sequence homology. Moreover, a Western blot analysis found a 27 kDa band in the human colon, which was successfully blocked by incubation with the immunizing peptide (Aviva Systems Biology data sheet). This antiserum stains identically to another well-characterized anti-GNAT3 (gustducin) antiserum (Santa Cruz Gα gust Antibody (I-20): sc-395; described in Jang et al., 2007).
Serotonin (5-HT)
The antiserum does not cross-react to 5-hydroxy-tryptophan, 5-hydroxyindole, 5-hydroxy-indole-3-aceticacid, or dopamine in Bn-SA/HRP labeling assays (Immunostar data sheet). In mouse tissue, this antibody shows reaction with known serotonergic groups in the brainstem (raphe nucleus) and co-localizes with other markers of Type III taste cells (Yee et al., 2001).
Calcitonin Gene-Related Peptide (α-CGRP)
The antiserum recognizes canine, rat, and mouse α-CGRP as determined by radioimmunoassay (Peninsula data sheet). A Western blot revealed a 4 kDa band in the mouse trigeminal ganglion and preabsorption with CGRP eliminated staining on the mouse trigeminal ganglion (Kosaras et al., 2009) The antibody staining was in agreement with that shown for the mouse trigeminal ganglion (Kosaras et al., 2009).
c-Fos
The polyclonal antibody recognizes residues 4–17 of the human c-Fos protein (Calbiochem data sheet). This antiserum stains a single band at ~50 – 55 kDa as observed by Western Blot analysis of Fibroblast-like BHK 21 C13 cells (Archer et al., 1999). Moreover, omission of rabbit anti-c-Fos antibody resulted in no labeled cells (data not shown; Ingham et al., 2009; Kelsch et al., 2012).
Statistical Analyses
Data in Tables 3– 5 and Fig. 5 are presented as group means ± SD. Data shown in Fig. 2 are presented as group means ± SEM. Data were analyzed using Pearson’s R correlations and appropriate one- and two- way analysis of variance (ANOVA)s (Statistica; RRID:SCR_014213; StatSoft, Tulsa, OK). Tukey’s honest significant difference tests were used to assess statistically significant (p < 0.05) main effects or interactions (see Results for details).
Table 3. Water- Evoked c-Fos (raw count).
Data are shown as means ± SD.
| nTS | DMSp5 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L | I | M | |||||||||||
| r1 | D | 2.0 | ± | 0 | 1.0 | ± | 1.0 | 1.3 | ± | 1.5 | 0.3 | ± | 0.6 |
| V | 1.3 | ± | 1.5 | 0.3 | ± | 0.6 | 0.3 | ± | 0.6 | ||||
| r2 | D | 0.7 | ± | 1.2 | 1.3 | ± | 0.6 | 0.7 | ± | 1.2 | 0.3 | ± | 0.6 |
| V | 1.3 | ± | 1.5 | 0 | ± | 0 | 0.3 | ± | 0.6 | ||||
| r3 | D | 3.3 | ± | 0.6 | 1.7 | ± | 1.5 | 0 | ± | 0 | 1.0 | ± | 0 |
| V | 5.0 | ± | 2.0 | 1.7 | ± | 1.5 | 0.3 | ± | 0.6 | ||||
| r4 | D | 1.7 | ± | 0.6 | 1.3 | ± | 1.5 | 1.0 | ± | 1.7 | 0 | ± | 0 |
| V | 7.7 | ± | 2.1 | 1.0 | ± | 1.7 | 0 | ± | 0 | ||||
| i1 | D | 3.0 | ± | 1.0 | 2.0 | ± | 1.7 | 1.3 | ± | 1.2 | 0 | ± | 0 |
| V | 4.3 | ± | 1.2 | 3.0 | ± | 1.7 | 1.0 | ± | 1.0 | ||||
| i2 | D | 3.3 | ± | 2.5 | 4.7 | ± | 2.3 | 2.7 | ± | 2.5 | 0.7 | ± | 0.6 |
| V | 7.0 | ± | 1.0 | 4.0 | ± | 4.0 | 1.0 | ± | 1.7 | ||||
Table 5. 5-HT3A GFP Pixel Counts (number of GFP-positive pixels > 2 SD of background).
Data are shown as means ± SD.
| DMSp5 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L | I | M | |||||||||||
| r1 | D | 60.8 | ± | 16.3 | 15.1 | ± | 3.9 | 4.9 | ± | 2.7 | 96.5 | ± | 2.3 |
| V | 90.7 | ± | 15.7 | 3.6 | ± | 1.5 | 5.0 | ± | 2.0 | ||||
| r2 | D | 41.1 | ± | 9.0 | 74.3 | ± | 7.3 | 4.4 | ± | 2.3 | 96.1 | ± | 2.2 |
| V | 88.0 | ± | 2.9 | 38.1 | ± | 6.1 | 2.8 | ± | 1.1 | ||||
| r3 | D | 63.2 | ± | 11.2 | 69.3 | ± | 6.0 | 1.8 | ± | 0.6 | 72.7 | ± | 3.8 |
| V | 60.5 | ± | 6.8 | 44.6 | ± | 6.0 | 1.9 | ± | 0.3 | ||||
| r4 | D | 62.3 | ± | 6.1 | 93.7 | ± | 4.2 | 75.0 | ± | 5.0 | 75.0 | ± | 4.1 |
| V | 81.6 | ± | 5.3 | 90.1 | ± | 3.9 | 46.0 | ± | 3.7 | ||||
| i1 | D | 82.5 | ± | 5.6 | 92.8 | ± | 2.5 | 43.8 | ± | 5.7 | 48.1 | ± | 3.7 |
| V | 92.9 | ± | 2.4 | 76.6 | ± | 4.0 | 48.1 | ± | 5.4 | ||||
| i2 | D | 90.5 | ± | 4.1 | 74.6 | ± | 4.6 | 45.1 | ± | 4.3 | 34.8 | ± | 4.2 |
| V | 92.9 | ± | 4.3 | 75.1 | ± | 4.1 | 39.0 | ± | 6.9 | ||||
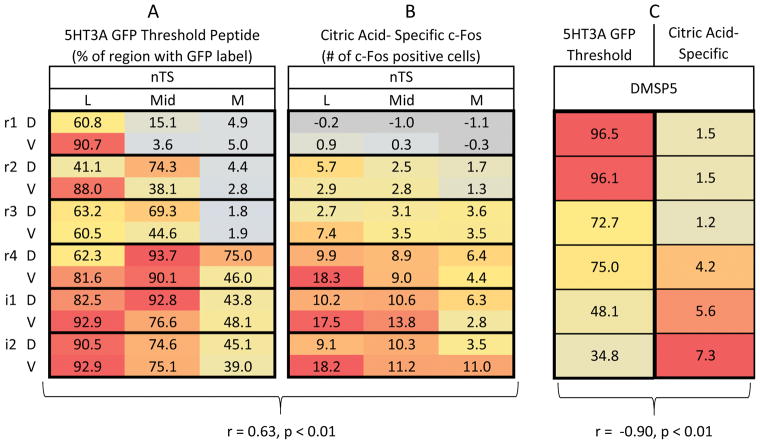
Fig. 5. Citric acid evoked Fos-LI is positively correlated with 5-HT3AGFP fiber distribution in the nTS, but negatively correlated within the DMSp5.
A and B: Spatial pattern ‘heat maps’ of: the number of 5-HT3A GFP labeled pixels (A) or citric acid- specific (raw count – avg. water) Fos-LI (B) with the nTS. Each 3×2 box within each column represents one level of the nTS subdivided into subfields: lateral (L), Mid, medial (M) in dorsal (D) & ventral (V) tiers. Each heat map is color coded so that blue = minimal; red = maximal. In B, for clarity, negative numbers (i.e. where citric acid counts were less than water counts) are shown in grey. Citric acid-specific Fos-LI is positively correlated with 5-HT3A GFP labeled pixels within the nTS (r = 0.65, p < 0.01). C: Apatial pixel density of 5-HT3A GFP pixels (left) vs. citric acid- specific Fos-LI (right). Each box represents quantifications within the DMSp5 at one level of the nTS and each heat map is color coded so that blue = minimal; red = maximal. The number of 5-HT3A GFP labeled pixels within the nTS is negatively (inversely) correlated with citric acid-specific Fos-LI (r = -090, p < 0.01). All numbers represent the average value for each anatomical area (nTS subregions or DMSp5).
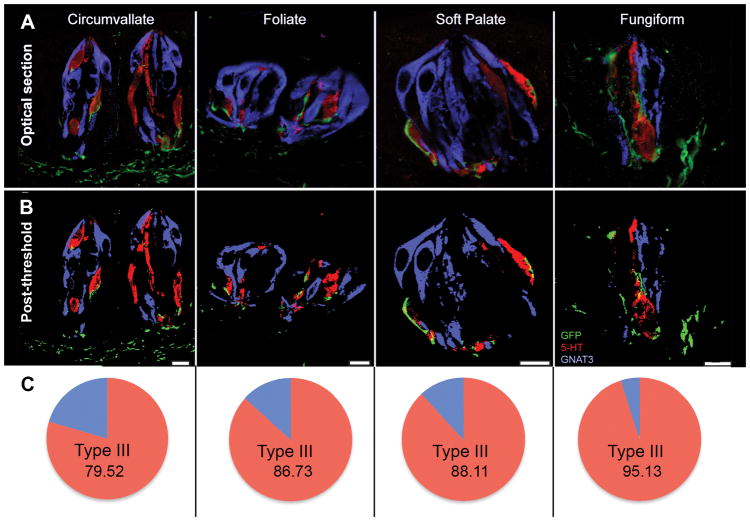
Fig. 2. 5-HT3A-positive nerve fibers colocalize with Type III taste cells to a greater extent than with Type II taste cells in all taste fields.
The percentage of 5-HT3A GFP positive fibers that colocalize with Type II cells (5-HT; yellow) is significantly greater than the percentage of 5-HT3A GFP positive fibers that colocalize with Type III cells (GNAT3; teal). Data represent the average percent of dual-labeled pixels in every optical section of multiple z-stacks ± SEM. CV: circumvallate; FF: fungiform; Fol: foliate; SP: soft palate. * = yellow bar significantly greater than teal bar.
Peripheral taste tissue 5HT3a GFP/ immunofluroscence colocalization
The proportion of 5HT3A GFP labeled pixels that colocalize with Type II and Type III labeled cells (identified by GNAT3 or PLCβ2, and 5-HT, respectively) was analyzed with a two-way (cell type x taste field) ANOVA.
c-Fos Statistical Analyses
The number of c-Fos positive cells in response to water and citric acid stimulation within the nTS and adjacent DMSp5 were compared with individual one-way (tastant) ANOVAs. To quantify citric acid-specific Fos-LI, for each subdivision of the nTS or the dorsomedial nucleus of the descending trigeminal complex (DMSp5), counts of c-Fos positive cells following water stimulation were subtracted from counts following acid stimulation. This yielded an index of the number of cells specifically activated by oral exposure to acid. The relationship between citric acid-specific Fos-LI and percent of 5HT3A GFP pixel label was compared using a Pearson’s r correlation for the nTS and DMSp5 individually.
RESULTS
5HT3A-positive nerve fibers are preferentially associated with Type III, but not Type II, taste cells
As shown in Figs. 1 and 2, 5HT3A GFP-positive nerve fibers (green) predominately neighbor Type III cells (as identified by the Type III cell type marker, 5HT; red), but not Type II cells (as indicated by the Type II cell marker, GNAT3; blue or PLCβ2) across all taste fields (CV, Fol, FF, SP). Moreover, rigorous co-label analysis of Type II, Type III, and 5HT3A GFP labeled pixels further support this association. As shown by Corson and Erisir (2013), when two objects are in contact, resolution limits of optical microscopy dictate that one or more pixels will show colocalization of labels, although the objects are anatomically separate. In taste buds labeled for 5HT3A GFP nerve fibers, significantly more pixels colocalize label with Type III-5HT (red) pixels, than with Type II (blue) pixels [F (3, 68) = 3.62, p < 0.05], across all taste fields (all p’s < 0.01; Fig. 2). This relationship was similar whether the Type II marker was GNAT3 or PLCβ2. This indicates that 5-HT3A GFP fibers preferentially contact Type III taste cells.
Fig. 1. 5-HT3A-positive nerve fibers innervate Type III, but not Type II, taste cells in all taste fields.
A: Single optical sections of taste buds with 5-HT3A GFP (green), 5-HT (red; Type III cell marker) and GNAT3 (blue; Type II cell marker) immunoreactivity. B: The same image from A after threshold and binarization.
Type III taste cells synapse on to 5HT3A-positive nerve fibers
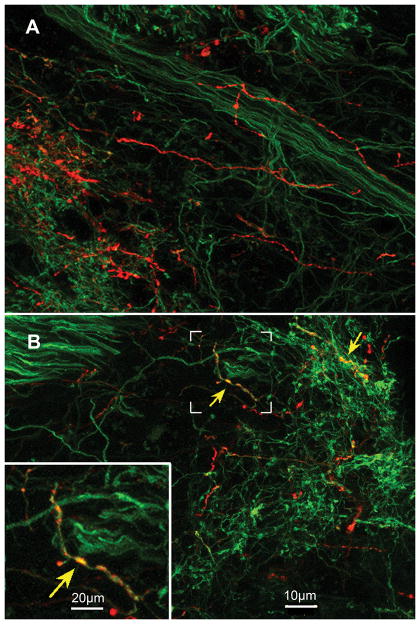
Although fluorescence microscopic analysis indicates a strong relationship between 5HT3A-GFP fibers and Type III cells, this technique lacks the resolution to visualize this association at an ultrastructural level. Thus, to determine whether 5HT3A-GFP fibers synapse onto Type III cells, we performed immuno-electron microscopy on Htr3a-EGFP mouse CV.
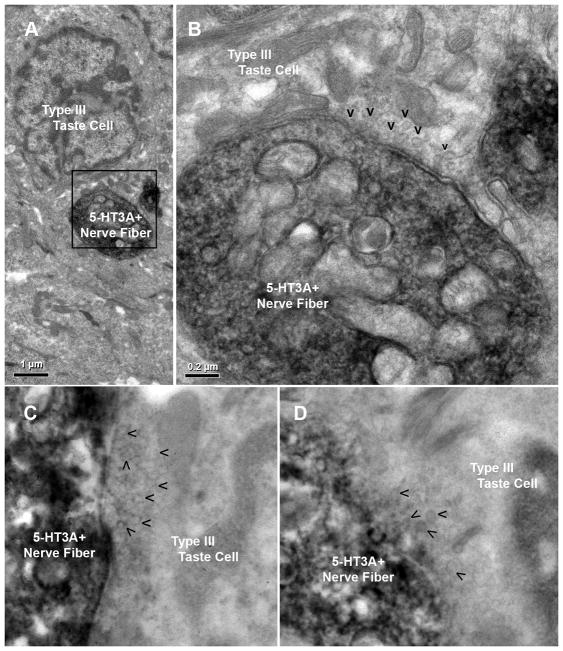
Synapses are observed at points of contact between 5-HT3A GFP positive nerve fibers and Type III cells in both circumvallate and fungiform papillae, as identified by small clear vesicles in proximity to the plasma membrane of Type III cells (Fig. 3). In contrast, we found no synapses between 5-HT3A GFP positive nerve fibers and Type II cells. These results suggest that information from Type III taste cells, which respond to acidic (sour) stimuli, is preferentially communicated to 5-HT3A-expressing nerve fibers.
Fig. 3. 5-HT3A GFP-positive nerve fibers synapse onto Type III cells.
Left: Immuno-electron microscopy of 5-HT3A GFP mouse CV taste buds with 5-HT3A GFP-positive nerve fibers clearly identified as dark, labeled nerve fibers (DAB reaction for GFP immunolabel). Right: An enlarged field of view from left that highlights a synapse between a 5-HT3A GFP-positive nerve fiber and a Type III cell, as evident by small clear vesicles (V’s) in close association with the plasma membrane of the Type III cell. Identification of the presynaptic cell as Type III is based primarily on nuclear morphology which shows substantial heterochromatin and several indentations. No synapses are seen with Type II cells (not shown).
5-HT3A GFP fibers are distributed heterogeneously within the nTS
Since information about acidic (sour) stimuli is preferentially communicated to 5-HT3A GFP expressing nerve fibers, we tested whether GFP-labeled fibers are distributed heterogeneously in particular subdomains of the nTS. Significant 5-HT3A GFP label (as identified by pixel density) is present through the entire nTS (Fig. 4), with the greatest percentage of GFP label occurring in the lateral regions of r1- r2, i1 – i2 and the mid r4 – i1 region of the nTS including the rostrocentral and rostrolateral subnuclei (Fig. 5A). In addition, substantial 5-HT3A GFP label is present in the adjacent DMSp5, but based on projection patterns of gustatory nerves, this label is not attributable to primary gustatory fibers, but likely arises from the general mucosal innervation from the lingual trigeminal (Corson et al., 2012) and perhaps glossopharyngeal nerves (Ciriello et al., 1981). Similarly, the GFP label in the rostralmost part of the rostrolateral subnucleus likely is attributable to fibers of the trigeminal nerve rather than gustatory afferents (Corson et al., 2012).
Citric acid evoked c-Fos activity is correlated with 5-HT3A GFP fiber distribution in the nTS, but not in the DMSp5
Within the nTS and DMSp5, few c-Fos positive cells were observed after stimulation of the oral cavity with water (Table 3), in keeping with previous studies (Stratford et al., 2016). This water-evoked Fos-LI was significantly less than that evoked by citric acid for both the nTS (water: 72.7 ± 21; citric acid: 253.6 ± 65; [F (1, 6) = 20.7, p < 0.01]) and DMSp5 (water: 2.3 ± 1.2; citric acid: 19.6 ± 4.9; [F (1, 6) = 33.6, p < 0.01]. Citric acid-specific Fos-LI (raw count – avg. water count) occurs throughout the entire rostral-intermediate nTS, with Fos-LI highest in the lateral and mid nTS of r4 – i4 (Fig. 5B and Table 4). Substantial Fos-LI also occurs in the adjacent DMSp5, with Fos-LI greatest in the i1 and i2 regions of this area.
Table 4. Citric Acid- Specific c-Fos (raw count – avg. water).
Data are shown as means ± SD.
| DMSp5 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L | I | M | |||||||||||
| r1 | D | −0.2 | ± | 0.6 | −1.0 | ± | 0 | −1.1 | ± | 0.4 | 1.5 | ± | 0.6 |
| V | 0.9 | ± | 0.7 | 0.3 | ± | 0.4 | −0.3 | ± | 0 | ||||
| r2 | D | 5.7 | ± | 1.1 | 2.5 | ± | 1.0 | 1.7 | ± | 1.3 | 1.5 | ± | 0.4 |
| V | 2.9 | ± | 0.5 | 2.8 | ± | 0.8 | 1.3 | ± | 0.5 | ||||
| r3 | D | 2.7 | ± | 1.2 | 3.1 | ± | 1.2 | 3.6 | ± | 1.3 | 1.2 | ± | 0.2 |
| V | 7.4 | ± | 0.5 | 3.5 | ± | 1.0 | 3.5 | ± | 0.8 | ||||
| r4 | D | 9.9 | ± | 0.8 | 8.9 | ± | 0.4 | 6.4 | ± | 1.7 | 4 | ± | 0.2 |
| V | 18.3 | ± | 0.6 | 9.0 | ± | 1.6 | 4.4 | ± | 0.5 | ||||
| i1 | D | 10.2 | ± | 1.6 | 10.6 | ± | 1.4 | 6.3 | ± | 2.5 | 6 | ± | 0.2 |
| V | 17.5 | ± | 0.8 | 13.8 | ± | 0.9 | 2.8 | ± | 0.9 | ||||
| i2 | D | 9.1 | ± | 1.9 | 10.3 | ± | 1.8 | 3.5 | ± | 2.6 | 7.3 | ± | 0.5 |
| V | 18.2 | ± | 1.1 | 11.2 | ± | 1.3 | 11.0 | ± | 0.4 | ||||
We tested whether the citric acid- specific Fos-LI preferentially was located within regions of the nTS and DMSp5 that were rich in 5-HT3A GFP. 5-HT3A GFP regional pixel density is positively correlated with citric acid-specific Fos-LI (raw count – avg. water) across the entire nTS (r = 0.63, n = 36, p < 0.01; Fig. 5A and B). In contrast, although 5-HT3A GFP label is present within the DMSp5, 5-HT3A GFP pixel density is negatively correlated with citric acid-specific Fos-LI in this region, with citric acid Fos-LI greatest in the intermediate (i1 and i2) levels, but 5-HT3A GFP pixel density greatest in the rostral (r1 and r2) levels (r = −0.90, n = 6, p < 0.01; Fig. 5C). These findings show that, for the gustatory nTS, sour-evoked Fos-LI correlates with the presence of 5-HT3A GFP fibers; whereas within the trigeminal complex, no such association exists.
Relationship of 5-HT3A GFP to CGRP-expressing polynociceptors
Because our previous work suggested the possible involvement of CGRP-expressing polymodal nociceptors in the detection and behavioral avoidance of acidic taste stimuli (Stratford et al., 2016), we also compared the spatial distribution of 5-HT3A GFP in relation to CGRP within the brainstem. As shown in Figs. 6 and 7, although CGRP labeled pixels are found within 5-HT3A GFP -rich areas within the nTS and DMSp5 (Fig. 6), the two labels do not co-localize in nerve fibers within the nTS although some colocalization occurs within the trigeminal complex (Fig. 7).
Fig. 7. 5-HT3A GFP and CGRP mostly do not colocalize within the lateral nTS or DMSp5 or anterior taste fields.
LSCM images of (A) the rostral nTS (R4), and (B) the DMSp5, and (C) palatal taste bud showing little colocalization of CGRP (red) and 5-HT3A-driven GFP (green). No fibers show co-localization in the image of the rostral nTS (A) whereas a few fibers (yellow arrow) show co-localization in the DMSp5. Inset at lower left of panel B shows a higher magnification of the bracketed region in the main figure showing a double-labeled varicose axon.
DISCUSSION
Despite advances in neuroscience methodologies that have helped delineate the receptor cells and neural pathways that underlie the perception of many taste qualities, including sweet, salty and umami (Chen et al., 2011; Barretto et al., 2015; Tokita and Boughter, 2016; Carleton et al., 2010), comparatively less is known about the transduction and subsequent encoding of acidic, sour stimuli. In particular, Type III cells within the taste bud are known be crucial for sour taste transduction (Huang et al., 2006; Huang et al., 2008; Kataoka et al., 2008; Chang et al., 2010; Ye et al., 2016), although the exact mechanism for transduction is unclear. Upon taste stimulation, Type III cells release several neurotransmitters, including 5-HT (Huang et al., 2008). The released 5-HT then plays a role in activation of gustatory afferent fibers, a portion of which express 5-HT3A mRNA and functional 5-HT3 receptors (Larson et al., 2015). The current study tested whether the subset of gustatory afferents that express the 5-HT3A receptors are preferentially associated with and are post-synaptic to Type III taste cells. We found that 5-HT3A-containing nerve fibers preferentially innervate and receive synapses from Type III cells, but seldom contact Type II cells, suggesting a preferential involvement of serotonergic transmission with sour taste. Somewhat confounding the situation is the likelihood that some Type III cells respond to NaCl and therefore are implicated in transmission of salty taste information (Lewandowski et al., 2016). Thus, we cannot conclude that 5-HT3-expressing gustatory nerve fibers constitute a strict “labeled line” for sour taste information. Nonetheless, electrophysiological analyses of single fibers in gustatory nerves show that distinct, response-specific classes of nerve fibers exist including a category of “H-fibers,” which show preferential responsiveness to acidic stimuli (e.g. Ninomiya et al., 1982; Frank et al., 1983; Spector and Travers, 2005) with weaker activation by salts. This specificity approximately corresponds to the response spectrum of Type III taste cells and so may represent a line of information transmission dedicated to Type III cell activity.
The preferential contact of 5-HT3A-expressing nerve fibers with Type III taste cells that we report is at odds with a recent study showing that glossopharyngeal nerve ganglion cells that receive synapses from Type III cells do not preferentially express 5-HT3 receptors (Maeda et al., 2016). That study, however, relied on specific transcellular transfer of WGA from PDK1L3-expressing cells to identify which ganglion cells receive synapses from Type III taste cells. Although PKD1L3 may be a reliable marker for sour-responsive taste cells, the fidelity of transfer of WGA in this system is questionable. When barley lectin is expressed from Type II cells within taste buds, the lectin tends to fill extracellular space of the bud, labeling non-selectively other taste cells and the afferent nerve fibers (Voigt et al., 2015). If the same leakage of lectin from the site of release occurs from Type III cells, then this would explain the lack of specificity reported by Maeda and co-workers. It should be noted, however, that whereas Type II cells utilize a non-vesicular release of neurotransmitter (Kinnamon et al., 1985; Vandenbeuch et al., 2010); Type III cells do show classical synapses. Hence, release and transcellular transfer from Type III cells may be different and more selective than the similar process involving Type II cells.
The association of 5-HT3A-expressing nerve fibers with Type III taste cells enabled us to utilize 5-HT3A -driven expression of GFP as a marker for nerve fibers likely associated with sour-transducing taste cells. We found that these GFP-labeled fibers do not end diffusely throughout the gustatory primary afferent zone of the nTS, but rather are restricted to the lateral and central subnuclei. This finding suggests that if 5-HT3-expressing fibers preferentially convey sour-related information, then these subregions of the nTS should preferentially show responses to sour.
Regional mapping of taste quality within the nTS in rodents was shown previously in studies by relying on expression of c-Fos and other immediate-early genes (Travers, 2002). In particular, bitter tastants activate medial nTS neurons, MSG and NaCl activate intermediate nTS neurons, and sucrose stimulates neurons more diffusely in the nucleus (Harrer and Travers, 1996; King et al., 1999; Travers et al., 1999; Travers, 2002; Chan et al., 2004; Travers et al., 2007; Travers and Travers, 2007; Stratford and Thompson, 2016). Consistent with our previous findings, citric acid infusion into the oral cavity of mice activates neurons in lateral and intermediate sectors of the nTS (Stratford et al., 2016). Significant citric acid-induced activity in the lateral nTS is seemingly at odds with data previous study in rats, which reports the highest citric acid-induced Fos-LI in the middle parts of the rostral nTS (Travers, 2002). This apparent discrepancy may be attributable to several factors. Most obvious is a potential species difference between rats vs. mice, but we consider this to be unlikely as the source of the difference. Another possibly is that differences in stimulus strength between the two studies (i.e. 100 mM citric acid used by Travers vs. 30 mM used in the present study) may account for the differences observed. Higher concentrations of tastants tend to activate a wider, less-selective population of primary afferent fibers (Wu et al., 2015), which may subsequently produce different spatial patterns of c-Fos activity. Finally, in the Travers (2002) study, the main focus was on the rostralmost levels of the nTS whereas our analysis extended more caudally to include intermediate levels, and it is in these intermediate levels that we see the highest levels of acid-induced activation in the lateral part of the nucleus. Similarly, this lateral portion of the intermediate nTS (named “caudal gustatory” nTS in Gaillard et al., 2008) showed the greatest activation in response to orally applied linoleic acid. Moreover, we show that nerve fibers from 5-HT3A-expressing ganglion cells preferentially innervate this acid-responsive region of nTS, in keeping with their role in sour-related taste transmission.
Interestingly, we also identified 5-HT3-expressing nerve fibers within the adjacent DMSp5 (Fig. 6)- a region implicated in the chemesthetic component of sour taste perception (Stratford and Finger, 2011). Only trigeminal fibers, and not gustatory afferents, terminate within the DMSp5, so cell activation in this nucleus likely reflects activation of trigeminal rather than gustatory afferents. In the DMSp5, citric acid-induced Fos-LI is inversely related to 5-HT3A GFP fiber density, suggesting that 5-HT3-expressing trigeminal afferents are not responsive to peripheral acidification. Many peripheral sensory fibers, including the class of peptidergic polymodal nociceptors, possess acid-sensitive ion channels, e.g. TrpV1, TrpA1 and ASICs (Bae et al., 2004; Dusenkova et al., 2014; Usoskin et al., 2015) and would thus respond to the presence of citric acid in the oral cavity. The 5-HT3A GFP fibers within the DMSp5 do not, however, colocalize with CGRP, one of the hallmarks of many polymodal nociceptors. Similarly, molecular analysis of somatosensory ganglion cells shows that 5-HT3 is expressed highly only in the PEP2 class of ganglion cells which do not highly express TrpV1 or TrpA1, two of the possible acid-responsive ion channels, which are mostly expressed in NP1 (for TrpA1), NP2, NP3 and PEP1 ganglion cells (Usoskin et al., 2015). Further evidence that DMSp5 is involved in trigeminal rather than taste-related activity is the recent electrophysiological analysis by Breza and Travers (2016) showing that taste responses occur only in the P2X2-rich terminal field in the nTS and that lateral to that, i.e. in lateralmost parts of nTS and in DMSp5, neurons show responses to mechanical stimuli.
In summary, our results are consistent with a tendency for regional segregation of taste quality information within nTS, the primary sensory relay nucleus for taste. Taken together with the reports of quality-segregation of responses within other levels of the taste neuroaxis (Chen et al., 2011; Tokita and Boughter, 2016), our results are consonant with the “labeled line” hypothesis of taste coding, i.e. that dedicated chains of neurons convey information about a specific taste quality: sour, sweet, bitter, etc. Nonetheless, many other studies report that quality representation of taste is less specific depending on stimulus concentration (Wu et al., 2015), or plastic depending on behavioral or past experience of the animal (e.g. Accolla et al., 2007; Accolla and Carleton, 2008). Timing of activity in relation to lick status also appears to be a key factor in encoding of taste information (Lemon and Katz, 2007; Roussin et al., 2012). Since mapping of c-Fos activity is but a snapshot of averaged activity over a long time scale, all temporal considerations are lost. Thus, although our results are consistent with a mapped representation of gustatory quality, many other factors should be considered before concluding that taste quality follows a strict labeled line topographic organization.
Table 6.
List of Anatomical Abbreviations
| Abbreviation | Anatomical Structure |
|---|---|
| 4V | 4th ventricle |
| CV | circumvallate papillae |
| DMSp5 | dorsomedial nucleus of the descending trigeminal complex |
| DMX | dorsal motor nucleus of the vagus |
| FF | fungiform papillae |
| Fol | foliate papillae |
| nTS | nucleus of the solitary tract |
| SP | soft palate |
| V | synaptic vesicle |
Acknowledgments
The authors thank Nicole Shultz and Mei Li for immunohistochemistry and other technical support. This work was supported by an NIH grants to JMS (5F32DC012025-03), to Dr. Thomas E. Finger (5R01DC012931-03), Dr. Sue Kinnamon (R01 DC012555) and the Rocky Mountain Taste and Smell Center (P30 DC04657).
References
- Accolla R, Carleton A. Internal body state influences topographical plasticity of sensory representations in the rat gustatory cortex. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:4010–4015. doi: 10.1073/pnas.0708927105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Accolla R, Bathellier B, Petersen CC, Carleton A. Differential spatial representation of taste modalities in the rat gustatory cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:1396–1404. doi: 10.1523/JNEUROSCI.5188-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer S, Li TT, Evans AT, Britland ST, Morgan H. Cell reactions to dielectrophoretic manipulation. Biochem Biophys Res Commun. 1999;257:687–698. doi: 10.1006/bbrc.1999.0445. [DOI] [PubMed] [Google Scholar]
- Bae YC, Oh JM, Hwang SJ, Shigenaga Y, Valtschanoff JG. Expression of vanilloid receptor TRPV1 in the rat trigeminal sensory nuclei. J Comp Neurol. 2004;478:62–71. doi: 10.1002/cne.20272. [DOI] [PubMed] [Google Scholar]
- Barretto RP, Gillis-Smith S, Chandrashekar J, Yarmolinsky DA, Schnitzer MJ, Ryba NJ, Zuker CS. The neural representation of taste quality at the periphery. Nature. 2015;517:373–376. doi: 10.1038/nature13873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breza JM, Travers SP. P2X2 Receptor Terminal Field Demarcates a “Transition Zone” for Gustatory and Mechanosensory Processing in the Mouse Nucleus Tractus Solitarius. Chemical senses. 2016 doi: 10.1093/chemse/bjw055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carleton A, Accolla R, Simon SA. Coding in the mammalian gustatory system. Trends Neurosci. 2010;33:326–334. doi: 10.1016/j.tins.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CY, Yoo JE, Travers SP. Diverse bitter stimuli elicit highly similar patterns of Fos-like immunoreactivity in the nucleus of the solitary tract. Chem Senses. 2004;29:573–581. doi: 10.1093/chemse/bjh062. [DOI] [PubMed] [Google Scholar]
- Chang RB, Waters H, Liman ER. A proton current drives action potentials in genetically identified sour taste cells. Proc Natl Acad Sci U S A. 2010;107:22320–22325. doi: 10.1073/pnas.1013664107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Gabitto M, Peng Y, Ryba NJ, Zuker CS. A gustotopic map of taste qualities in the mammalian brain. Science. 2011;333:1262–1266. doi: 10.1126/science.1204076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello J, Hrycyshyn AW, Calaresu FR. Glossopharyngeal and vagal afferent projections to the brain stem of the cat: a horseradish peroxidase study. Journal of the autonomic nervous system. 1981;4:63–79. doi: 10.1016/0165-1838(81)90007-2. [DOI] [PubMed] [Google Scholar]
- Corson J, Aldridge A, Wilmoth K, Erisir A. A survey of oral cavity afferents to the rat nucleus tractus solitarii. J Comp Neurol. 2012;520:495–527. doi: 10.1002/cne.22715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corson JA, Erisir A. Monosynaptic convergence of chorda tympani and glossopharyngeal afferents onto ascending relay neurons in the nucleus of the solitary tract: a high-resolution confocal and correlative electron microscopy approach. J Comp Neurol. 2013;521:2907–2926. doi: 10.1002/cne.23357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damann N, Voets T, Nilius B. TRPs in our senses. Curr Biol. 2008;18:R880–889. doi: 10.1016/j.cub.2008.07.063. [DOI] [PubMed] [Google Scholar]
- DeFazio RA, Dvoryanchikov G, Maruyama Y, Kim JW, Pereira E, Roper SD, Chaudhari N. Separate populations of receptor cells and presynaptic cells in mouse taste buds. J Neurosci. 2006;26:3971–3980. doi: 10.1523/JNEUROSCI.0515-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusenkova S, Ru F, Surdenikova L, Nassenstein C, Hatok J, Dusenka R, Banovcin P, Jr, Kliment J, Tatar M, Kollarik M. The expression profile of acid-sensing ion channel (ASIC) subunits ASIC1a, ASIC1b, ASIC2a, ASIC2b, and ASIC3 in the esophageal vagal afferent nerve subtypes. American journal of physiology Gastrointestinal and liver physiology. 2014;307:G922–930. doi: 10.1152/ajpgi.00129.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvoryanchikov G, Tomchik SM, Chaudhari N. Biogenic amine synthesis and uptake in rodent taste buds. J Comp Neurol. 2007;505:302–313. doi: 10.1002/cne.21494. [DOI] [PubMed] [Google Scholar]
- Eldred WD, Zucker C, Karten HJ, Yazulla S. Comparison of fixation and penetration enhancement techniques for use in ultrastructural immunocytochemistry. J Histochem Cytochem. 1983;31:285–292. doi: 10.1177/31.2.6339606. [DOI] [PubMed] [Google Scholar]
- Finger TE, Danilova V, Barrows J, Bartel DL, Vigers AJ, Stone L, Hellekant G, Kinnamon SC. ATP signaling is crucial for communication from taste buds to gustatory nerves. Science. 2005;310:1495–1499. doi: 10.1126/science.1118435. [DOI] [PubMed] [Google Scholar]
- Frank ME, Contreras RJ, Hettinger TP. Nerve fibers sensitive to ionic taste stimuli in chorda tympani of the rat. J Neurophysiol. 1983;50:941–960. doi: 10.1152/jn.1983.50.4.941. [DOI] [PubMed] [Google Scholar]
- Gaillard D, Laugerette F, Darcel N, El-Yassimi A, Passilly-Degrace P, Hichami A, Khan NA, Montmayeur JP, Besnard P. The gustatory pathway is involved in CD36-mediated orosensory perception of long-chain fatty acids in the mouse. FASEB J. 2008;22:1458–1468. doi: 10.1096/fj.07-8415com. [DOI] [PubMed] [Google Scholar]
- Guthrie KM, Anderson AJ, Leon M, Gall C. Odor-induced increases in c-fos mRNA expression reveal an anatomical “unit” for odor processing in olfactory bulb. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:3329–3333. doi: 10.1073/pnas.90.8.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrer MI, Travers SP. Topographic organization of Fos-like immunoreactivity in the rostral nucleus of the solitary tract evoked by gustatory stimulation with sucrose and quinine. Brain Res. 1996;711:125–137. doi: 10.1016/0006-8993(95)01410-1. [DOI] [PubMed] [Google Scholar]
- Huang AL, Chen X, Hoon MA, Chandrashekar J, Guo W, Trankner D, Ryba NJ, Zuker CS. The cells and logic for mammalian sour taste detection. Nature. 2006;442:934–938. doi: 10.1038/nature05084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YA, Pereira E, Roper SD. Acid stimulation (sour taste) elicits GABA and serotonin release from mouse taste cells. PLoS One. 2011;6:e25471. doi: 10.1371/journal.pone.0025471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YA, Maruyama Y, Stimac R, Roper SD. Presynaptic (Type III) cells in mouse taste buds sense sour (acid) taste. The Journal of physiology. 2008;586:2903–2912. doi: 10.1113/jphysiol.2008.151233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham ES, Gunhan E, Fuller PM, Fuller CA. Immunotoxin-induced ablation of melanopsin retinal ganglion cells in a non-murine mammalian model. The Journal of comparative neurology. 2009;516:125–140. doi: 10.1002/cne.22103. [DOI] [PubMed] [Google Scholar]
- Jang HJ, Kokrashvili Z, Theodorakis MJ, Carlson OD, Kim BJ, Zhou J, Kim HH, Xu X, Chan SL, Juhaszova M, Bernier M, Mosinger B, Margolskee RF, Egan JM. Gut-expressed gustducin and taste receptors regulate secretion of glucagon-like peptide-1. Proc Natl Acad Sci U S A. 2007;104:15069–15074. doi: 10.1073/pnas.0706890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka S, Yang R, Ishimaru Y, Matsunami H, Sevigny J, Kinnamon JC, Finger TE. The candidate sour taste receptor, PKD2L1, is expressed by type III taste cells in the mouse. Chem Senses. 2008;33:243–254. doi: 10.1093/chemse/bjm083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsch W, Sim S, Lois C. Increasing heterogeneity in the organization of synaptic inputs of mature olfactory bulb neurons generated in newborn rats. The Journal of comparative neurology. 2012;520:1327–1338. doi: 10.1002/cne.22799. [DOI] [PubMed] [Google Scholar]
- King CT, Travers SP, Rowland NE, Garcea M, Spector AC. Glossopharyngeal nerve transection eliminates quinine-stimulated fos-like immunoreactivity in the nucleus of the solitary tract: implications for a functional topography of gustatory nerve input in rats. J Neurosci. 1999;19:3107–3121. doi: 10.1523/JNEUROSCI.19-08-03107.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnamon JC, Taylor BJ, Delay RJ, Roper SD. Ultrastructure of mouse vallate taste buds. I. Taste cells and their associated synapses. J Comp Neurol. 1985;235:48–60. doi: 10.1002/cne.902350105. [DOI] [PubMed] [Google Scholar]
- Kosaras B, Jakubowski M, Kainz V, Burstein R. Sensory innervation of the calvarial bones of the mouse. The Journal of comparative neurology. 2009;515:331–348. doi: 10.1002/cne.22049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson ED, Vandenbeuch A, Voigt A, Meyerhof W, Kinnamon SC, Finger TE. The Role of 5-HT3 Receptors in Signaling from Taste Buds to Nerves. J Neurosci. 2015;35:15984–15995. doi: 10.1523/JNEUROSCI.1868-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon CH, Katz DB. The neural processing of taste. BMC neuroscience. 2007;8(Suppl 3):S5. doi: 10.1186/1471-2202-8-S3-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowski BC, Sukumaran SK, Margolskee RF, Bachmanov AA. Amiloride-Insensitive Salt Taste Is Mediated by Two Populations of Type III Taste Cells with Distinct Transduction Mechanisms. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2016;36:1942–1953. doi: 10.1523/JNEUROSCI.2947-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda N, Ohmoto M, Yamamoto K, Kurokawa A, Narukawa M, Ishimaru Y, Misaka T, Matsumoto I, Abe K. Expression of serotonin receptor genes in cranial ganglia. Neuroscience letters. 2016;617:46–51. doi: 10.1016/j.neulet.2016.01.050. [DOI] [PubMed] [Google Scholar]
- Nelson G, Hoon MA, Chandrashekar J, Zhang Y, Ryba NJ, Zuker CS. Mammalian sweet taste receptors. Cell. 2001;106:381–390. doi: 10.1016/s0092-8674(01)00451-2. [DOI] [PubMed] [Google Scholar]
- Ninomiya Y, Tonosaki K, Funakoshi M. Gustatory neural response in the mouse. Brain Res. 1982;244:370–373. doi: 10.1016/0006-8993(82)90100-7. [DOI] [PubMed] [Google Scholar]
- Oka Y, Butnaru M, von Buchholtz L, Ryba NJ, Zuker CS. High salt recruits aversive taste pathways. Nature. 2013;494:472–475. doi: 10.1038/nature11905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsu N. A Threshold Selection Method from Gray-Level Histograms. IEEE Transactions on Systems, Man and Cybernetics. 1979;9:62–66. [Google Scholar]
- Roper SD. TRPs in taste and chemesthesis. Handb Exp Pharmacol. 2014;223:827–871. doi: 10.1007/978-3-319-05161-1_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussin AT, D’Agostino AE, Fooden AM, Victor JD, Di Lorenzo PM. Taste coding in the nucleus of the solitary tract of the awake, freely licking rat. J Neurosci. 2012;32:10494–10506. doi: 10.1523/JNEUROSCI.1856-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector AC, Travers SP. The representation of taste quality in the mammalian nervous system. Behav Cogn Neurosci Rev. 2005;4:143–191. doi: 10.1177/1534582305280031. [DOI] [PubMed] [Google Scholar]
- Stratford JM, Finger TE. Central representation of postingestive chemosensory cues in mice that lack the ability to taste. J Neurosci. 2011;31:9101–9110. doi: 10.1523/JNEUROSCI.0404-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratford JM, Thompson JA. MSG-Evoked c-Fos Activity in the Nucleus of the Solitary Tract Is Dependent upon Fluid Delivery and Stimulation Parameters. Chemical senses. 2016;41:211–220. doi: 10.1093/chemse/bjv082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratford JM, Thompson JA, Finger TE. Immunocytochemical Organization and Sour Taste Activation in the Rostral Nuc. Solitary Tract of Mice. The Journal of comparative neurology. 2016 doi: 10.1002/cne.24059. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokita K, Boughter JD., Jr Topographic organizations of taste-responsive neurons in the parabrachial nucleus of C57BL/6J mice: An electrophysiological mapping study. Neuroscience. 2016;316:151–166. doi: 10.1016/j.neuroscience.2015.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomchik SM, Berg S, Kim JW, Chaudhari N, Roper SD. Breadth of tuning and taste coding in mammalian taste buds. J Neurosci. 2007;27:10840–10848. doi: 10.1523/JNEUROSCI.1863-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers JB, Urbanek K, Grill HJ. Fos-like immunoreactivity in the brain stem following oral quinine stimulation in decerebrate rats. Am J Physiol. 1999;277:R384–394. doi: 10.1152/ajpregu.1999.277.2.R384. [DOI] [PubMed] [Google Scholar]
- Travers JB, Herman K, Yoo J, Travers SP. Taste reactivity and Fos expression in GAD1-EGFP transgenic mice. Chem Senses. 2007;32:129–137. doi: 10.1093/chemse/bjl038. [DOI] [PubMed] [Google Scholar]
- Travers SP. Quinine and citric acid elicit distinctive Fos-like immunoreactivity in the rat nucleus of the solitary tract. Am J Physiol Regul Integr Comp Physiol. 2002;282:R1798–1810. doi: 10.1152/ajpregu.00590.2001. [DOI] [PubMed] [Google Scholar]
- Travers SP, Travers JB. Taste-evoked Fos expression in nitrergic neurons in the nucleus of the solitary tract and reticular formation of the rat. J Comp Neurol. 2007;500:746–760. doi: 10.1002/cne.21213. [DOI] [PubMed] [Google Scholar]
- Usoskin D, Furlan A, Islam S, Abdo H, Lonnerberg P, Lou D, Hjerling-Leffler J, Haeggstrom J, Kharchenko O, Kharchenko PV, Linnarsson S, Ernfors P. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci. 2015;18:145–153. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- Vandenbeuch A, Tizzano M, Anderson CB, Stone LM, Goldberg D, Kinnamon SC. Evidence for a role of glutamate as an efferent transmitter in taste buds. BMC Neurosci. 2010;11:77. doi: 10.1186/1471-2202-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbeuch A, Larson ED, Anderson CB, Smith SA, Ford AP, Finger TE, Kinnamon SC. Postsynaptic P2X3-containing receptors in gustatory nerve fibres mediate responses to all taste qualities in mice. J Physiol. 2015;593:1113–1125. doi: 10.1113/jphysiol.2014.281014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt A, Bojahr J, Narukawa M, Hubner S, Boehm U, Meyerhof W. Transsynaptic Tracing from Taste Receptor Cells Reveals Local Taste Receptor Gene Expression in Gustatory Ganglia and Brain. J Neurosci. 2015;35:9717–9729. doi: 10.1523/JNEUROSCI.0381-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead MC, Frank ME. Anatomy of the gustatory system in the hamster: central projections of the chorda tympani and the lingual nerve. The Journal of comparative neurology. 1983;220:378–395. doi: 10.1002/cne.902200403. [DOI] [PubMed] [Google Scholar]
- Wu A, Dvoryanchikov G, Pereira E, Chaudhari N, Roper SD. Breadth of tuning in taste afferent neurons varies with stimulus strength. Nature communications. 2015;6:8171. doi: 10.1038/ncomms9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W, Chang RB, Bushman JD, Tu YH, Mulhall EM, Wilson CE, Cooper AJ, Chick WS, Hill-Eubanks DC, Nelson MT, Kinnamon SC, Liman ER. The K+ channel KIR2.1 functions in tandem with proton influx to mediate sour taste transduction. Proc Natl Acad Sci U S A. 2016;113:E229–238. doi: 10.1073/pnas.1514282112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee CL, Yang R, Bottger B, Finger TE, Kinnamon JC. “Type III” cells of rat taste buds: immunohistochemical and ultrastructural studies of neuron-specific enolase, protein gene product 9.5, and serotonin. J Comp Neurol. 2001;440:97–108. doi: 10.1002/cne.1372. [DOI] [PubMed] [Google Scholar]