Abstract
Acquired drug resistance prevents cancer therapies from achieving stable and complete responses.1 Emerging evidence implicates a key role for nonmutational drug resistance mechanisms underlying the survival of residual cancer “persister” cells.2-4 The persister cell pool constitutes a reservoir from which drug-resistant tumours may emerge. Targeting persister cells therefore presents a therapeutic opportunity to impede tumour relapse.5 In an earlier report, we found that cancer cells in a high mesenchymal therapy-resistant cell state are dependent on the lipid hydroperoxidase GPX4 for survival.6 Here, we describe the discovery that a similar therapy-resistant cell state underlies the behavior of persister cells derived from a wide range of cancers and drug treatments. Consequently, we show that persister cells acquire a dependency on GPX4. We demonstrate that loss of GPX4 function results in selective persister cell ferroptotic death in vitro and prevents tumour relapse in vivo. These findings support targeting GPX4 as a therapeutic strategy to prevent acquired drug resistance.
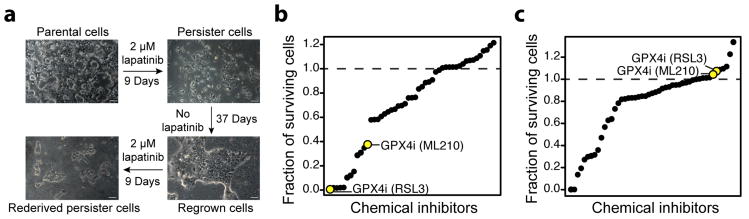
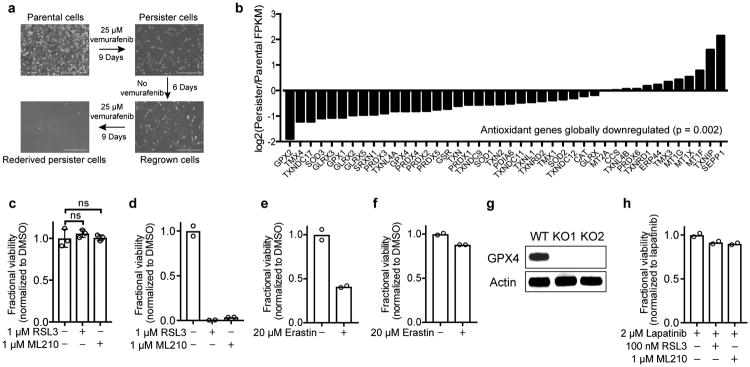
We sought to identify therapeutically exploitable vulnerabilities in an experimental model of minimal residual disease, drug-tolerant persister cells. Persister cells, which are present across a wide range of tumour types, survive cytotoxic exposure to targeted therapy or chemotherapy through poorly understood reversible, nonmutational mechanisms.2 We focused our initial work on the HER2-amplified breast cancer line BT474 which, upon treatment for nine or more days with cytotoxic concentrations of lapatinib, reveals a small population of quiescent surviving persister cells (Fig. 1a). Removal of lapatinib allows the persister cells to regrow and re-acquire sensitivity to lapatinib. Subsequent lapatinib treatment re-derives persister cells. The reversibility of BT474 persister cell drug resistance, which is also observed in other persister cell models2 including A375 melanoma (Extended Data Fig. 1a), is indicative of a nonmutational resistance mechanism.
Figure 1. RNAseq and small-molecule screen in drug-tolerant persister cells.
a, A small fraction of BT474 cells enter into a reversible, quiescent drug-tolerant persister state in response to nine or more days of treatment with 2 μM lapatinib. Scale bars indicate 100 μm. b, Small-molecule inhibitor screen in which prederived BT474 persister cells were treated with 1 μM small-molecule inhibitors while maintained in 2 μM lapatinib. c, Small-molecule inhibitor counter-screen in parental BT474 cells. a is representative of two independent experiments. b, c, n = 1 biologically independent sample from a single screening experiment.
To identify cellular processes that may represent selective dependencies in persister cells, we performed RNAseq on BT474 persister cells and drug naïve parental cells (Extended Data Tables 1, 2 and Supplementary Tables 1-4). In persister cells, we observed upregulation of stemness markers CD133 and CD44 which corroborated earlier observations in other persister cell models2 (Extended Data Table 1). We also observed upregulation of mesenchymal markers and downregulation of epithelial markers (Extended Data Table 1), and this encouraged us to further explore the role that mesenchymal state biology may play in persister cell therapy resistance. Furthermore, a broad downregulation of antioxidant gene-expression in persister cells, including Nrf2 target genes, directed our attention toward possible persister cell vulnerabilities in oxidative stress defense mechanisms (Extended Data Fig. 1b and Extended Data Table 2).
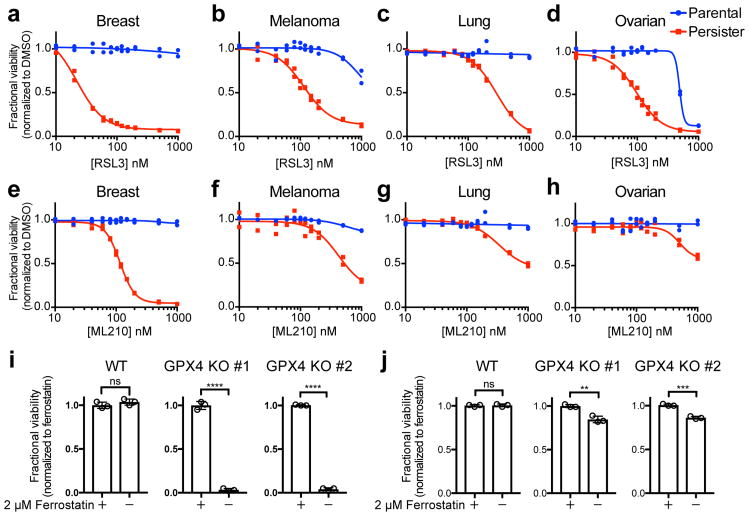
We recently reported that cancer cells in a high mesenchymal therapy-resistant state are selectively sensitive to ferroptosis, an oxidative form of cell death which can be induced by inhibition of the lipid hydroperoxidase GPX4.6 We included GPX4 inhibitors within a diverse collection of compounds screened against BT474 parental and persister cells and found GPX4 inhibitors RSL3 and ML210 to be among the compounds most selectively lethal to persister cells, with minimal effect on parental cells or nontransformed MCF10A cells (Fig. 1b, c, and Extended Data Fig. 1c). Given the observed phenotypic and gene-expression similarities between persister cells from different cancer types,2-4,7 we wondered whether sensitivity to GPX4 inhibition might be a general persister cell vulnerability. Remarkably, we found that sensitivity to GPX4 inhibition extended beyond BT474 persister cells to a variety of other persister cell models (Fig. 2a-h and Extended Data Fig. 1d). In each case, we found the parental cells to be far less sensitive to GPX4 inhibitors (Fig. 2a-h). We also observed a differential sensitivity of persister cells, although to a lesser extent, to the ferroptosis inducer erastin which acts upstream of GPX4 by depleting GPX4 cofactor glutathione (Extended Data Fig. 1e, f).8
Figure 2. Persister cells are vulnerable to GPX4 inhibition.
Breast (BT474), melanoma (A375), lung (PC9) and ovarian (Kuramochi) cancer parental or persister cells (see Methods) were treated with GPX4 inhibitor RSL3 (a-d) or ML210 (e-h) for three days. i, A375 GPX4 WT or KO persister cells and j, parental cells with ferrostatin-1 withdrawn for three days. Data are plotted as means and error bars represent standard deviation. a-h, n = 2, and i, j, n = 3 biologically independent samples. From two-tailed t tests,*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, P > 0.05. All data are representative of two separate experiments.
We further confirmed the GPX4 dependency of persister cells using GPX4 knockout (KO) A375 melanoma cells (Fig. 2i, j, Extended Data Fig. 1g and Supplementary Data). A hallmark of ferroptosis is the accumulation of lipid hydroperoxides, and lipophilic, small-molecule antioxidants have been shown to rescue cells from ferroptosis.9-11 Upon withdrawal of the ferroptosis-rescuing lipophilic antioxidant ferrostatin-1, we observed that A375 GPX4 KO, but not GPX4 wild type (WT), persister cells undergo ferroptotic death (Fig. 2i).6 In contrast, GPX4 loss has minimal effect on parental A375 cell viability (Fig. 2j). Furthermore, we found that GPX4 inhibitors are not synergistic with lapatinib treatment of parental BT474 cells, demonstrating that GPX4 dependence is specific to the persister cell state (Extended Data Fig. 1h). In addition, though p53 can regulate ferroptosis sensitivity,12 we found that persister cells are GPX4 dependent regardless of p53 status (Fig. 2). These findings support the compelling hypothesis that GPX4 is a selective dependency of a drug-resistant persister cell state that is shared by cancer cells from many lineages subjected to diverse therapeutic regimens.
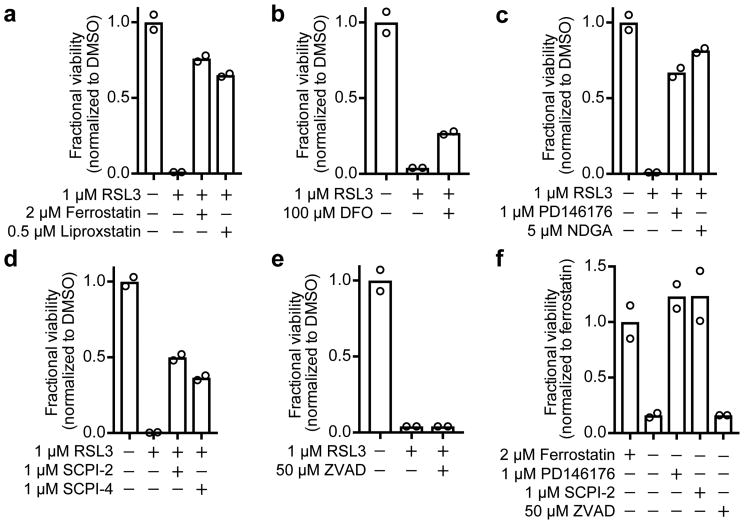
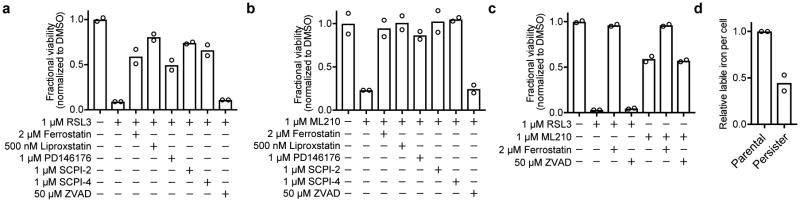
To test whether diverse persister cell types undergo canonical ferroptosis10 upon GPX4 inhibition, we treated persister cells with a series of ferroptosis rescue compounds. We found that co-treatment with the lipophilic antioxidants ferrostatin-1 or liproxstatin-1 rescued various persister cell types from GPX4 inhibition (Fig. 3a and Extended Data Fig. 2a-c). Ferroptosis is an iron-dependent process, and although persister cells have less labile iron than parental cells (Extended Data Fig. 2d), we observed that iron is nonetheless essential for GPX4 inhibitor-induced persister cell death because iron chelation with deferoxamine (DFO) partially rescues persister cells (Fig. 3b). Furthermore, consistent with the established role for lipoxygenase-catalyzed lipid hydroperoxidation for ferroptosis, we observed that treatment with lipoxygenase inhibitors rescued persister cells from GPX4 inhibition (Fig. 3c and Extended Data Fig. 2a, b). Lipid hydroperoxides must be transported by the lipid transporter SCP2 for cell death to occur.13,14 Accordingly, we found that persister cells were protected from GPX4 inhibition by chemical inhibition of SCP2 (Fig. 3d and Extended Data Fig. 2a, b). Also, in agreement with the non-apoptotic, caspase-independent nature of ferroptosis, we found that persister cell death from GPX4 inhibition is not rescued by the pan-caspase inhibitor Z-VAD-FMK (Fig. 3e and Extended Data Fig. 2a-c). These ferroptosis characteristics were also observed in A375 GPX4 KO persister cells upon removal of ferrostatin-1 (Fig. 3f). Taken together, these observations indicate that GPX4 inhibition induces canonical ferroptosis in persister cells.
Figure 3. GPX4 inhibition causes ferroptosis in persister cells.
BT474 persister cells co-treated with RSL3 and a, lipophilic antioxidants ferrostatin-1 and liproxstatin-1, b, iron chelator deferoxamine (DFO), c, lipoxygenase inhibitors PD146176 and NDGA, d, lipid transporter SCP2 inhibitors SCPI-2 and SCPI-4, and e, pan-caspase inhibitor Z-VAD-FMK (ZVAD). f, A375 GPX4 KO cells (clone 1) persister cells with ferrostatin-1 replaced by ferroptosis rescue compounds or ZVAD. Data are plotted as means. a-f, n = 2 biologically independent samples. All data are representative of two separate experiments.
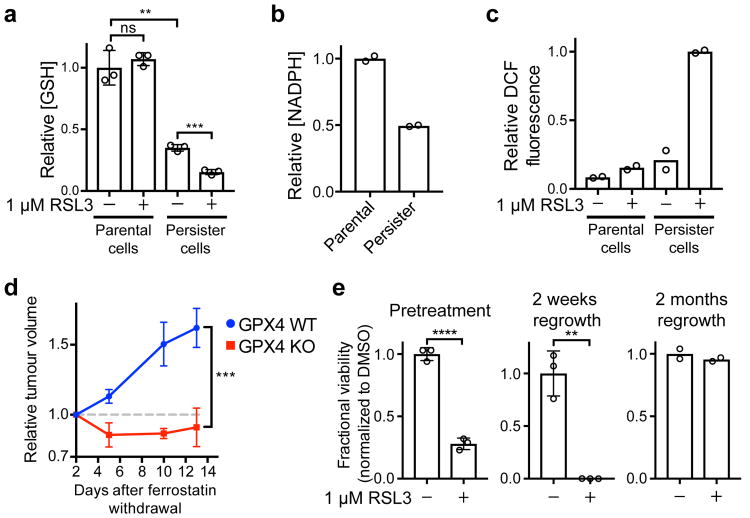
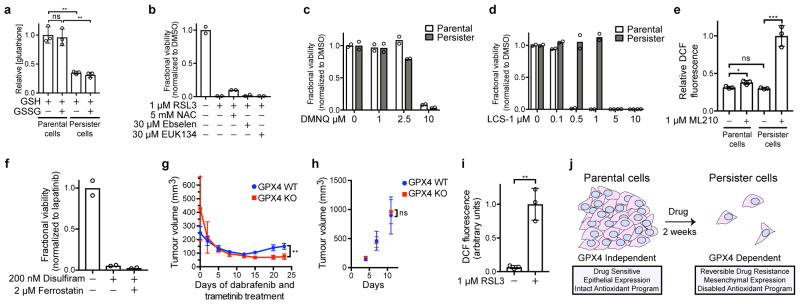
Based on our observation that antioxidant genes are downregulated in persister cells (Extended Data Fig. 1b and Extended Data Table 2), we hypothesized that there may be a disabled antioxidant program which contributes to the sensitivity to ferroptosis. Glutathione and NADPH are two reducing cofactors that have been demonstrated to protect cells from the toxic effects of lipid peroxidation and prevent ferroptosis.10,15,16 We found that persister cells have markedly decreased levels of both reducing cofactors (Fig. 4a, b and Extended Data Fig. 3a). This is consistent with our observation that persister cells have downregulated expression of glutathione and NADPH biosynthesis genes (Extended Data Table 2). Also, replacement of glutathione via treatment with glutathione metabolic precursor N-acetyl cysteine (NAC) partially rescues persister cells from ferroptosis (Extended Data Fig. 3b). Persister cells have a specific sensitivity to lipid peroxidation, rather than general sensitivity to oxidative stress, because persister cells are not rescued from GPX4 inhibition by antioxidants ebselen and EUK134 (Extended Data Fig. 3b), exhibit only modestly increased sensitivity to endogenous reactive oxygen species (ROS) induced by DMNQ treatment (Extended Data Fig. 3c), and are relatively insensitive to inhibition of an unrelated antioxidant gene, SOD1 (Extended Data Fig. 3d). These findings indicate that global downregulation of Nrf2 targets and other antioxidant genes, and decreased glutathione and NADPH act to impair persister cell lipid peroxidation defense and contribute to a dependence on GPX4 for survival.
Figure 4. A disabled antioxidant program underlies persister cell sensitivity to ferroptosis.
a, Glutathione levels in BT474 cells treated with RSL3 for one hour. b, NADPH levels in BT474 cells. c, ROS levels (DCF stain) in BT474 cells treated with RSL3 for one hour. d, Relapse of A375 GPX4 WT or KO (clone 1) tumours. Mice bearing GPX4 WT and KO tumours on opposing flanks dosed with ferrostatin-1 were treated with dabrafenib and trametinib to shrink tumours to their minimal size. Ferrostatin-1 was then withdrawn and tumour relapse was monitored. See source data for individual data points. e, left, twenty-four hour RSL3 pretreatment of BT474 cells prior to derivation of persister cells, middle, RSL3 treatment of cells regrown from persister cells upon lapatinib removal for fifteen days, or right, two months. Data are plotted as means and error bars represent standard deviation. a, e left and middle, n = 3, and b, c, e right, n = 2 biologically independent samples, and d, n = 4 animals. From two-tailed t tests, *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, P > 0.05. All data are representative of two separate experiments.
Consistent with a disabled lipid peroxidation defense, we observed that after one hour of GPX4 inhibition and prior to any cell death, there is a drastic elevation in 2′,7′-dichlorofluorescein (DCF) staining of ROS which indicates lipid peroxidation in persister cells but not parental cells (Fig. 4c, and Extended Data Fig. 3e). Furthermore, upon lipid hydroperoxide accumulation, free reduced glutathione levels are expected to decrease because reduced glutathione covalently ligates, via glutathione-S-transferase catalyzed or non-enzymatic glutathionylation, to oxidized products of lipid peroxidation such as aldehydes.17 Consistent with this, we observed a decrease in reduced glutathione levels specifically in persister cells immediately after GPX4 inhibition that mirrored the increase in DCF staining (Fig. 4a and Extended Data Fig. 3a). Interestingly, a recent study found that persister cells are sensitive to the aldehyde dehydrogenase inhibitor disulfiram.3 While disulfiram was shown to cause apoptosis rather than ferroptosis, and is not rescued by ferrostatin-1 (Extended Data Fig. 3f), the persister cell sensitivity to disulfiram may be reflective of the broader disabled antioxidant program.
We reasoned that an effective GPX4 inhibitor could potentially be used in a clinical setting to prevent tumour relapse. To test this possibility in a preclinical model, we developed an A375 melanoma xenograft model wherein tumours respond and relapse upon treatment with dabrafenib and trametinib. Because neither RSL3 nor ML210 are systemically bioavailable, we instead adopted a recently developed genetic strategy6 to test GPX4 dependence of residual tumours in vivo. Using GPX4 KO or GPX4 WT A375 cells (Extended Data Fig. 1g and Supplementary Data), we formed xenograft tumours in nude mice. We then treated the mice with dabrafenib and trametinib to shrink the tumours while concurrently dosing the mice with ferrostatin-1 to mask any effects of GPX4 deletion before or during the initial response to drug treatment. Once tumours had been reduced to their minimal volume, ferrostatin-1 was withdrawn, unmasking the GPX4 KO effect in these residual tumours. Upon further dosing of mice with dabrafenib and trametinib, without ferrostatin-1, the GPX4 WT tumours relapsed and the GPX4 KO tumours did not (Fig. 4d and Extended Data Fig. 3g). In contrast, parental A375 GPX4 KO and WT cells both formed tumours without ferrostatin-1 equally well (Extended Data Fig. 3h). Taken together, these A375 melanoma xenograft data are consistent with an essential role for GPX4 in residual A375 tumours in vivo.
Our data reveal that co-treatment of cancer cells with a targeted therapy or chemotherapy agent together with a GPX4 inhibitor effectively reduces the residual persister cell pool. However, co-treatment with multiple drugs often results in increased toxic side effects that can compromise patient safety and treatment efficacy. Interestingly, we found that pretreatment of BT474 parental cells with RSL3 alone for twenty-four hours decreases the number of persister cells remaining after subsequent lapatinib treatment (Fig. 4e). This finding is consistent with recent reports demonstrating that persister cells may preexist prior to drug treatment.3,18 We also observed that BT474 persister cells allowed to regrow after washout of lapatinib retain their full sensitivity to RSL3 for at least two weeks but for less than two months (Fig. 4e and Extended Data Fig. 3i), indicating that the disabled antioxidant program in persister cells is not immediately repaired upon drug removal and regrowth. These findings demonstrate that pre- or post-treatment with GPX4 inhibitors, rather than co-treatment, may be adequate to deplete the pool of persister cells that survive targeted therapy or chemotherapy. This staggered treatment strategy may be clinically beneficial in maximizing efficacy while minimizing the potential increase in toxicity that might be anticipated from simultaneous co-treatment with two agents.
Nonmutationally drug-tolerant persister cells have recently gained attention for their potential role in acquired drug resistance where they may constitute a reservoir of surviving cells from which fully drug-resistant cells eventually emerge to cause tumour relapse.2,4,5 Here we have shown that GPX4 inhibition is a promising strategy to selectively eradicate this residual cell reservoir(Extended Data Fig. 3j). While existing GPX4 inhibitors, including RSL3 and ML210, are valuable tool compounds in cell culture settings, their poor pharmacokinetic properties preclude their systemic use in vivo. Given the therapeutic promise for inducing ferroptosis in persister cells and drug-resistant cancers,6,19 the development of a potent bioavailable GPX4 inhibitor is an urgent priority. Because GPX4 genetic deletion is lethal in adult mice,13,20 further study will be needed to determine whether a suitable therapeutic window exists for treatment with GPX4 inhibitors. In summary, we have found that GPX4 inhibition results in ferroptotic death in cancer persister cells across many lineages, presenting a novel strategy to prevent tumour relapse.
Methods
Cell culture
HER2 amplified breast cancer BT474 (ATCC), EGFR mutant non-small cell lung cancer PC9 (Sigma-Aldrich), and Kuramochi ovarian cancer (JCRB) cells were cultured in RPMI-1640 medium, and BRAF mutant A375 melanoma (ATCC) cells were cultured in Dulbecco's Modified Eagle Medium, each supplemented with penicillin, streptomycin and 10% FBS. A375 GPX4 KO cells were cultured in the presence of 2 μM ferrostatin-1 unless otherwise noted. MCF10A (ATCC) cells were cultured in Mammary Epithelial Cell Growth Medium supplemented with bovine pituitary extract, gentamycin, amphotericin B (Lonza MEGM BulletKit), and 100 ng/mL cholera toxin (Sigma). Each cell line was maintained in a 5% CO2 atmosphere at 37°C. Cell line identities were confirmed by STR fingerprinting and all were found to be negative for mycoplasma using the MycoAlert kit (Lonza).
Persister cell derivation and treatments
Persister cells were derived from treatment of HER2 amplified breast cancer BT474, EGFR mutant non-small cell lung cancer PC9 or Kuramochi ovarian cancer cells with 2 μM lapatinib, 2 μM erlotinib, or co-treatment with 10 μM carboplatin and 100 nM paclitaxel, respectively, for at least nine days with fresh drug added every three days. A375 persister cells were derived from nine or more days of 2 μM vemurafenib (Fig. 2i), 25 μM vemurafenib (Extended Data Fig. 1a), 1 μM dabrafenib and 100 nM trametinib (Fig. 3f and Extended Data Fig. 2a, b), or 2 μM dabrafenib and 200 nM trametinib (Fig. 2b, f). A375 GPX4 KO cells were maintained in media with 2 μM ferrostatin-1 during persister cell derivation with drug. Chemotherapy-derived BT474 persister cells were similarly derived from co-treatment with 25 μM carboplatin and 1.25 nM paclitaxel (Extended Data Fig. 1d). Unless otherwise noted, small-molecule inhibitors were added to prederived persister cells in twelve well tissue culture plates that were maintained under constant drug exposure throughout subsequent small-molecule inhibitor treatment. Cell viability was assessed using CellTiter Glo (Promega) after three days of small-molecule treatment, or three days after ferrostatin-1 removal in the case of A375 GPX4 KO cell experiments, unless otherwise noted.
For testing RSL3 pretreatment prior to persister cell derivation and RSL3 treatment on cells regrown from persister cells (Fig. 4e), the following experimental setup was used. Fig. 4e, left, BT474 parental cells were pretreated for 24 hours with 1 μM RSL3 alone, then RSL3 was removed and persister cells were subsequently derived from two weeks of treatment with 2 μM lapatinib alone followed by cell viability assay. For Fig. 4e, middle and right, prederived BT474 persister cells from 2 μM lapatinib were allowed to regrow upon removal of lapatinib for 15 days (middle) or 63 days (right) and then were treated for three days with 1 μM RSL3 alone followed by cell viability assay.
Chemicals
(Z)-4-Hydroxytamoxifen, altretamine, baicalein, BWA4C, deferoxamine mesylate salt (DFO), erlotinib hydrochloride, ferrostatin-1, gossypol, HA14-1, hydroxychloroquine, liproxstatin-1, ML210, PBIT, salinomycin, trigonelline hydrochloride, tunicamycin, zileuton and Z-VAD-FMK were purchased from Sigma-Aldrich. ABT-263, ABT-737, dabrafenib, paclitaxel, canertinib, fluvastatin, GSK2110183, GX15-070, LBH589, 1S,3R-RSL3 (RSL3), romidepsin, saracatinib, trametinib, trichostatin A, TW-37 and vorinostat were purchased from Selleck Chemical. Mifepristone, pladienolide B, SB-431542, vemurafenib and XAV939 were purchased from Santa Cruz Biotechnology. Compound A (Mirk inhibitor), erastin, MIM1 and SP600125 were purchased from EMD Millipore. SCPI-2 and SCPI-4 were purchased from ChemBridge. Metformin and PD146176 were purchased from Enzo Life Sciences. AEW541 and JQ1 were purchased from Cayman Chemical. WEHI-539 was purchased from MedChem Express. Everolimus was purchased from Cell Signaling Technology. 17-DMAG was purchased from Biotang USA. Herboxidiene was purchased from Apollo Scientific. Sorafenib tosylate was purchased from AK Scientific. Spliceostatin A was purchased from AdooQ Bioscience. Lapatinib ditosylate was purchased from LC Laboratories. Nordihydroguaiaretic acid (NDGA) was purchased from Acros Organics. Carboplatin was purchased from Hospira or Sigma. All chemicals were stored as stock solutions in DMSO (Sigma-Aldrich) or nuclease-free water (Life Technologies).
RNAseq library preparation
To generate BT474 persister cells for RNAseq, BT474 cells were plated in five 15 cm tissue culture plates per replicate, for three replicates, and were allowed to reach 75% confluency before being treated with 2 μM lapatinib for nine days. Simultaneously, parental BT474 cells were cultured for nine days in lapatinib-free media in a 15 cm tissue culture plate for each of two replicates. 2 million persister or parental cells were used per replicate for RNA isolation. RNA was isolated using Trizol (Life Technologies), following the manufacturer's instructions. RNA samples were quantified using a NanoDrop (ThermoFisher), and RNA integrity was determined using a RNA 6000 Nano Kit (Agilent) on an Agilent 2100 Bioanalyzer. All RNA samples had a measured RNA Integrity Number (RIN) > 9.3. RNA samples were sent to the UC Davis Genome Center for non-strand specific polyA-enrichment, Illumina library preparation including multiplex barcodes and RNAseq across six lanes of an Illumina HiSeq 2000 instrument. 145 million to 160 million single end, 50 base pair reads were obtained per replicate.
RNAseq data analysis
Fastq sequence files were aligned to hg18 using TopHat v2.0.6, allowing only uniquely mapped reads with the -g 1 option. Differential expression testing and assignment of FPKM and fold change values to genes was performed with CuffDiff v2.0.2, comparing the two BT474 parental cell replicates to the three BT474 persister cell replicates. For CuffDiff, quartile normalization and -compatible-hits-norm options were used to assign reads to a GTF file containing all human RefSeq NM and NR genes which was downloaded from the UCSC Genome Browser and converted into a CuffDiff readable format using Cuffcompare as described in the Cufflinks manual.
The set of genes with minimum FPKM > 1 in either parental cells or persister cells, greater than two-fold upregulation or downregulation in persister cells relative to parental cells, and significant FDR corrected p value (q < 0.05) as calculated by CuffDiff were used for Core Analysis with Ingenuity Pathway Analysis (IPA) software to generate IPA results for Canonical Pathways, Upstream Regulators, and Functions (Supplementary Tables 2-4). Analysis of differential expression among genes within previously established panels of mesenchymal and epithelial marker genes21 (Extended Data Table 1) and human antioxidant genes22 (Extended Data Fig. 1b) was also performed using FPKMs and fold changes calculated by CuffDiff.
Chemical inhibitor screens
BT474 cells were seeded in duplicate twelve well tissue culture plates and were allowed to reach 75% confluency. To derive persister cells, each well was treated with 2 μM lapatinib for nine days, with fresh media containing 2 μM lapatinib added every three days. Small-molecule inhibitors at 1 μM concentration were then added in lapatinib-containing media and, following five days of incubation, each well was measured using CellTiter Glo reagent. CellTiter Glo measurements were made by transferring 200 μL of the CellTiter Glo solution per well into Costar 96 well clear flat bottom plates for readout using a Promega GloMax Explorer luminometer. CellTiter Glo luminescent signal was normalized to negative control wells by calculating the ratio of luminescence signal in the test versus negative control wells. Negative control wells contained media with 2 μM lapatinib but no additional chemical inhibitors. Each small-molecule inhibitor was also tested at 1 μM concentration in parental BT474 cells in 12 well plates, where cells were seeded in duplicate wells at 25% confluency and treated with chemical inhibitors. Following incubation for five days, cell viability was measured with CellTiter Glo.
CRISPR-mediated GPX4 deletion
Guide RNAs targeting the first exon of human GPX4 were designed using the CRISPR Design tool (Zhang lab, MIT). pX330 (Zhang lab, MIT) expression plasmids encoding the single guide RNAs 5′-CACGCCCGATACGCTGAGTG-3′ (for GPX4 KO clone 1) or 5′-CTTGGCGGAAAACTCGTGCA-3′ (for GPX4 KO clone 2) and spCas9 were generated and transfected into A375 cells in the presence of 2 μM ferrostatin-1 using Lipofectamine 3000 (Life Technologies). The cells were subjected to two rounds of transfection. In the first round, the CRISPR plasmids were co-transfected with a vector encoding mCherry and puromycin resistance. 24 hours after the transfection, the cells were selected with 1 μg/mL puromycin for three days. The selected cells were expanded and transfected again with the pX330 CRISPR constructs and a pMax-GFP reporter. 24 hours after the transfection, the cells were sorted and the GFP-positive, mCherry-negative cells were retained. Cells were then transferred into 96 well plates at ∼1 cell/well and expanded as clones. Genomic DNA was extracted from all clones with the NucleoSpin Blood kit (Macherey-Nagel). The GPX4 target sites were amplified from 100 ng of genomic DNA with Phusion DNA Polymerase (NEB) with primers For (5′-GTAAAACCGGACCAGAAGTACAAG-3′) and Rev (5′-CCCACCTGCTTCCCGAACTG-3′). Purified PCR products were TOPO cloned (Life Technologies) and sequenced with primer M13F (Quintara). 5 TOPO colonies from each clone were sequenced to confirm frameshifts in GPX4.
A375 GPX4 KO clones 1 and 2 were confirmed to lack GPX4 protein by western blot. For this, 3 million cells were seeded in 10 cm tissue culture plates and grown to 75% confluence, and the following day, cells were trypsinized, washed with PBS, and lysed using RIPA buffer (Thermo Scientific) supplemented with phosphatase inhibitor (Sigma) and protease inhibitor (Sigma). Lysates were centrifuged at 13,000 rpm at 4°C for twelve minutes, and the protein concentration of the supernatant was determined using the Bio-Rad Protein Assay. 40 μg of protein per sample was mixed with sample buffer (Bio-Rad) and denatured at 95°C for five minutes. Samples were separated by SDS-PAGE (NuPage 4–12% Bis-Tris Gel, Life Technologies), run with BioRad Precision Plus Protein Kaleidoscope ladder, and transferred to a nitrocellulose membrane using an iBlot system (Life Technologies). Membranes were blocked with 5% BSA for one hour at room temperature, and then incubated with either a 1:1000 dilution of anti-GPX4 (rabbit) polyclonal Ab (Abcam, ab41787) or 1:5000 dilution of anti-β-Actin (mouse) mAb (Sigma, A5441) at 4°C overnight. LICOR secondary antibodies were then incubated with the membrane for one hour at room temperature, and the membrane was imaged using the LICOR Odyssey Imaging System.
Glutathione and NADPH measurements
NADP/NADPH-Glo and GSH-Glo assay kits (Promega) were used to measure NADPH levels in 20,000 cells and glutathione levels in 50,000 cells per replicate following manufacturer's instructions. To measure total glutathione (GSH + GSSG), oxidized glutathione (GSSG) was reduced by incubating test samples with 500 μM tris(2-carboxyethyl)phosphine (TCEP) for 10 minutes prior to luminescence measurement, following the manufacturer's instructions. Each assay was read out using a Promega GloMax Explorer luminometer.
Intracellular reactive oxygen species measurements
BT474 cells plated in replicate 10 cm tissue culture plates were treated with 2 μM lapatinib for at least nine days, with fresh media added every three days, to derive persister cells. For cells re-grown from persister cells (Extended Data Fig. 3i), lapatinib was subsequently removed from persister cells and fresh lapatinib-free media was replaced every three days for fifteen days. Then, for both the persister cell experiments (Fig. 4c and Extended Data Fig. 3e) and regrown cell experiments (Extended Data Fig. 3i), cells were treated with 1 μM RSL3 or ML210 for one hour. RSL3 or ML210 was removed and cells were trypsinized. 100,000 cells were pelleted per replicate, and cell pellets were resuspended in full media containing 25 μM 6-carboxy-2′,7′-dichlorodihydrofluorescein diacetate, di(acetoxymethyl ester) (carboxy-DCFDA, acetoxymethyl ester) (Thermo Fisher Scientific) in HBSS with 1% FBS. After thirty minutes of incubation with DCF dye, cells were pelleted again, resuspended in HBSS with 1% FBS and green fluorescence was analyzed by with a SONY SH800S Cell Sorter. Because HER2-bound lapatinib is green fluorescent,23 data for persister cells were normalized to unstained persister cells, and parental cell data were normalized to unstained parental cells. A minimum of 3,000 cells were analyzed for each persister and parental replicate. Data were processed with FloJo software.
Iron measurements
Total labile iron (Fe II and Fe III combined) was measured using the colourimetric Iron Assay Kit (Abcam) following the manufacturer's instructions. 1.4 million BT474 persister or parental cells were tested per replicate for two biological replicate samples. Colourimetric signal was read out using a BioTek Synergy 2 Microplate Reader.
Xenograft experiments
The University of California San Francisco Institutional Animal Care and Use Committee (IACUC) approved the xenograft studies in Protocol #AN142193. Tumour size criteria permitted a maximum volume of 2000 mm3, however, careful observation of the animals and BCS score of 2 or less superseded any laboratory measurement. These limits were not exceeded in any experiments. For the formation of xenograft tumours without ferrostatin-1 (Extended Data Fig. 3h), 10 million A375 GPX4 WT or GPX4 KO (clone 1) cells were mixed in a 1:1 PBS:matrigel mixture and injected into the left and right flanks of female athymic mice aged 10-12 weeks with a mean body weight of 24 grams (NCR Nude, Taconic), respectively. Tumour volume measurements were made twice a week. All mice were included in the analysis. For the tumour relapse experiments (Fig. 4d), 10 million A375 GPX4 WT or KO (clone 1) cells were mixed in a 1:1 PBS:matrigel mixture containing 2.5 μM ferrostatin-1 and injected into left and right flanks, respectively, of male athymic nude mice aged 10-12 weeks with a mean body weight of 25 grams. Mice were dosed daily i.p. with 0.2 mg/kg ferrostatin-1 while tumours were initially established and during subsequent treatment with 100 mg/kg dabrafenib and 1 mg/kg trametinib twice daily by oral gavage. Once tumours had shrunk and tumour volumes stabilized, ferrostatin-1 treatment was ceased while dabrafenib and trametinib treatment was continued and tumours relapsed. All mice for which any tumour relapsed were included in the analysis.
Statistical analyses
No statistical methods were used to predetermine sample size. For mouse experiments, the mice were not randomized. The investigators performing tumour volume measurements were blinded. Statistical tests were performed with GraphPad Prism 7.0a. Unless otherwise noted, p values were calculated using unpaired, two-tailed t tests assuming unequal variance. Graphing of data was performed with GraphPad Prism 7.0a and R v3.2.1.
Data availability
All data are available from the authors upon reasonable request. This manuscript is accompanied by four Supplementary tables and data are deposited at the NCBI GEO accession GSE84896.
Extended Data
Extended Data Figure 1. Additional data demonstrating persister cell ferroptosis sensitivity.
a, A375 melanoma persister cells are reversibly drug-resistant. Scale bars indicate 400 μm. b, Global antioxidant gene-expression is downregulated in BT474 persister cells. P value calculated using a two-tailed Wilcoxon signed rank test. c, MCF10A parental cells and d, BT474 persister cells derived from carboplatin and paclitaxel were treated with RSL3 or ML210 for three days. e, BT474 persister cells or f, parental cells were treated with erastin for five days. g, Western blot demonstrating GPX4 knockout in two distinct A375 clones (clone 1, KO1; clone 2, KO2). For gel source data, see Supplementary Figure 1. h, BT474 parental cells co-treated with 2 μM lapatinib and RSL3 or ML210 for three days. Data are plotted as means and error bars represent standard deviation. c, n = 3 and d-h, n = 2 biologically independent samples. c, P value calculated from a two-tailed t test where ns represents P > 0.05. All data are representative of two separate experiments.
Extended Data Figure 2. Additional data demonstrating that GPX4 inhibition causes ferroptosis in persister cells.
A375 persister cells were treated with a, RSL3 or b, ML210 and ferroptosis rescue compounds for three days. c, PC9 persister cells were treated with RSL3 or ML210 and ferroptosis rescue compounds for three days. d, Relative concentration of total labile iron in BT474 parental and persister cells. Data are plotted as means and error bars represent standard deviation. a-d, n = 2 biologically independent samples. From two-tailed t tests, *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, P > 0.05. All data are representative of two separate experiments.
Extended Data Figure 3. Additional data demonstrating that persister cells have a disabled antioxidant program and depend on GPX4 in vivo.
a, Reduced glutathione (GSH) and reduced plus oxidized glutathione (GSH + GSSG) levels in BT474 cells. BT474 persister cells were b, treated with RSL3 and antioxidant compounds, c, treated with endogenous ROS generating compound DMNQ, or d, treated with SOD1 inhibitor LCS-1, for three days. e, ROS levels (DCF stain) in BT474 cells treated with ML210 for one hour. f, BT474 persister cells were co-treated with ALDH inhibitor disulfiram and ferrostatin-1 for three days. g, Tumour volume measurements for the full time course of the experiment presented in Fig. 4d. Ferrostatin-1 was withdrawn on Day 10. See source data for individual data points. h, Untreated A375 GPX4 WT or GPX4 KO (clone 1) tumour formation without ferrostatin-1 dosing. See source data for individual data points. i, Reactive oxygen species levels (DCF stain) in cells regrown without lapatinib for fifteen days from BT474 persister cells and then treated with 1 μM RSL3 for one hour. j, Persister cell GPX4 dependence model. Data are plotted as means and error bars represent standard deviation. a, e, i, n = 3 and b-d, f, n = 2 biologically independent samples, g, n = 4 and h, n = 5 animals. From two-tailed t tests, *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, P > 0.05. All data are representative of two separate experiments.
Extended Data Table 1. Expression of EMT and stem markers.
RNAseq data for mesenchymal, epithelial and stemness marker genes in BT474 parental and persister cells.
| Parental Cell FPKM | Persister Cell FPKM | Fold Change | |
|---|---|---|---|
| Mesenchymal | |||
| VIM | 2.2 | 37.1 | 16.9 |
| FN1 | 0.3 | 0.6 | 2.0 |
| ZEB1 | 0.0 | 0.0 | N/A |
| ZEB2 | 0.0 | 0.0 | N/A |
| TWIST1 | 2.6 | 19.9 | 7.7 |
| TWIST2 | 0.0 | 0.0 | N/A |
| SNAI1 | 1.4 | 0.4 | 0.3 |
| SNAI2 | 0.009 | 0.04 | 4.4 |
| CDH2 | 0.3 | 1.0 | 3.0 |
| Epithelial | |||
| CDH1 | 142.8 | 90.0 | 0.6 |
| CLDN4 | 179.4 | 128.2 | 0.7 |
| CLDN7 | 93.2 | 49.2 | 0.5 |
| TJP3 | 26.3 | 16.1 | 0.6 |
| MUC1 | 55.3 | 42.8 | 0.8 |
| Sternness | |||
| CD133 | 0.4 | 1.7 | 4.3 |
| CD44 | 2.1 | 4.6 | 2.2 |
Extended Data Table 2. Expression of Nrf2 target genes.
RNAseq data for Nrf2 target genes in BT474 parental and persister cells.
| Parental Cell FPKM | Persister Cell FPKM | Fold Change | |
|---|---|---|---|
| NRF2 | 26.1 | 28.7 | 1.1 |
| Iron sequestration | |||
| FTL | 387.3 | 451.7 | 1.2 |
| FTH1 | 736.4 | 530.0 | 0.7 |
| HMOX1 | 2.2 | 3.9 | 1.8 |
| HMOX2 | 23.4 | 15.8 | 0.7 |
| Glutathione utilization | |||
| GPX2 | 4.3 | 1.1 | 0.3 |
| GSTM1 | 0.8 | 1.2 | 1.4 |
| GSTM2 | 0.4 | 2.5 | 5.6 |
| GSTM3 | 2.3 | 3.0 | 1.3 |
| Quinone detoxification | |||
| NQ01 | 96.7 | 55.7 | 0.6 |
| NADPH production | |||
| G6PD | 50.2 | 19.4 | 0.4 |
| PGD | 63.3 | 31.2 | 0.5 |
| ME1 | 16.8 | 5.3 | 0.3 |
| IDH1 | 96.4 | 35.9 | 0.4 |
| Glutathione production and regeneration | |||
| GCLM | 4.8 | 3.4 | 0.7 |
| GCLC | 14.3 | 16.5 | 1.2 |
| GSR | 70.5 | 42.8 | 0.6 |
| SLC7A11 | 1.6 | 1.2 | 0.7 |
| Thioredoxin production, regeneration and utilization | |||
| TXN | 159.7 | 105.2 | 0.7 |
| PRDX1 | 431.5 | 293.2 | 0.7 |
| TXNRD1 | 25.2 | 28.4 | 1.1 |
Supplementary Material
Acknowledgments
We acknowledge technical support from the UCSF ES Cell Targeting Core and UCSF Preclinical Therapeutics Core. This work was supported by grants from the National Cancer Institute (NCI) of the National Institutes of Health (NIH) (Cancer Target Discovery and Development Network grant U01CA168370 to M.T.M. and F.M., U01CA217882 to M.T.M., U01CA176152 to S.L.S., and U01CA168397 to M.E.B., and R01CA212767 to M.T.M.), Susan G Komen for the Cure Postdoctoral Fellowship KG1101214 to M.J.H., and the Howard Hughes Medical Institute (S.L.S.).
Footnotes
Supplementary Information is available in the online version of the paper.
Author contributions: M.T.M. and F.M. directed the project; M.J.H., V.S.V., S.L.S., F.M. and M.T.M. wrote the manuscript; M.J.H. performed chemical screens, RNAseq analysis, and all cell culture experiments except V.S.V. and M.J.R. performed Kuramochi cell GPX4 inhibitor assays, D.B. assisted with ROS, glutathione and NADPH assays, and J.G. performed western blots; M.J.H. directed CRISPR editing of A375 cells and in vivo experiments which were performed at UCSF core facilities; V.S.V. and J.K.E. contributed reagents; A.M., H.D.D., and M.B. provided advice and project support.
Author information: The authors declare no competing financial interests.
Contributor Information
Matthew J. Hangauer, Department of Microbiology and Immunology and UCSF Diabetes Center, University of California San Francisco, 513 Parnassus Avenue, San Francisco, California 94143, USA; UCSF Helen Diller Family Comprehensive Cancer Center, University of California San Francisco, 1450 3rd Street, San Francisco, California 94143, USA
Vasanthi S. Viswanathan, Broad Institute, 415 Main Street, Cambridge, Massachusetts 02142, USA
Matthew J. Ryan, Broad Institute, 415 Main Street, Cambridge, Massachusetts 02142, USA
Dhruv Bole, UCSF Helen Diller Family Comprehensive Cancer Center, University of California San Francisco, 1450 3rd Street, San Francisco, California 94143, USA.
John K. Eaton, Broad Institute, 415 Main Street, Cambridge, Massachusetts 02142, USA
Alexandre Matov, Data Set Analysis LLC, 155 Jackson Street, San Francisco, California 94111, USA.
Jacqueline Galeas, UCSF Helen Diller Family Comprehensive Cancer Center, University of California San Francisco, 1450 3rd Street, San Francisco, California 94143, USA.
Harshil D. Dhruv, Cancer and Cell Biology Division, The Translational Genomics Research Institute, 445 N 5th Street, Phoenix, Arizona 85004, USA
Michael E. Berens, Cancer and Cell Biology Division, The Translational Genomics Research Institute, 445 N 5th Street, Phoenix, Arizona 85004, USA
Stuart L. Schreiber, Broad Institute, 415 Main Street, Cambridge, Massachusetts 02142, USA; Howard Hughes Medical Institute, Chevy Chase, Maryland 20815, USA
Frank McCormick, UCSF Helen Diller Family Comprehensive Cancer Center, University of California San Francisco, 1450 3rd Street, San Francisco, California 94143, USA.
Michael T. McManus, Department of Microbiology and Immunology and UCSF Diabetes Center, University of California San Francisco, 513 Parnassus Avenue, San Francisco, California 94143, USA
References
- 1.Groenendijk FH, Bernards R. Drug resistance to targeted therapies: deja vu all over again. Mol Oncol. 2014;8:1067–1083. doi: 10.1016/j.molonc.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raha D, et al. The cancer stem cell marker aldehyde dehydrogenase is required to maintain a drug-tolerant tumor cell subpopulation. Cancer Res. 2014;74:3579–3590. doi: 10.1158/0008-5472.CAN-13-3456. [DOI] [PubMed] [Google Scholar]
- 4.Hata AN, et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med. 2016;22:262–269. doi: 10.1038/nm.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oxnard GR. The cellular origins of drug resistance in cancer. Nat Med. 2016;22:232–234. doi: 10.1038/nm.4058. [DOI] [PubMed] [Google Scholar]
- 6.Viswanathan VS, et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature. 2017;547:453–457. doi: 10.1038/nature23007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan W, et al. MET-independent lung cancer cells evading EGFR kinase inhibitors are therapeutically susceptible to BH3 mimetic agents. Cancer Res. 2011;71:4494–4505. doi: 10.1158/0008-5472.CAN-10-2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yagoda N, et al. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature. 2007;447:864–868. doi: 10.1038/nature05859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang WS, Stockwell BR. Ferroptosis: Death by Lipid Peroxidation. Trends Cell Biol. 2016;26:165–176. doi: 10.1016/j.tcb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dixon SJ, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skouta R, et al. Ferrostatins inhibit oxidative lipid damage and cell death in diverse disease models. J Am Chem Soc. 2014;136:4551–4556. doi: 10.1021/ja411006a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang L, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedmann Angeli JP, et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol. 2014;16:1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kriska T, Pilat A, Schmitt JC, Girotti AW. Sterol protein-2 (SCP-2) involvement in cholesterol hydroperoxide cytotoxicity as revealed by SCP-2 inhibitor effects. J Lipid Res. 2010;51:3174–3184. doi: 10.1194/jlr.M008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimada K, Hayano M, Pagano NC, Stockwell BR. Cell-Line Selectivity the Predictive Power of Pharmacogenomic Analyses and Helps Identify NADPH as Biomarker for Ferroptosis Sensitivity. Cell Chem Biol. 2016;23:225–235. doi: 10.1016/j.chembiol.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang WS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gueraud F, et al. Chemistry and biochemistry of lipid peroxidation products. Free Radic Res. 2010;44:1098–1124. doi: 10.3109/10715762.2010.498477. [DOI] [PubMed] [Google Scholar]
- 18.Shaffer SM, et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature. 2017;546:431–435. doi: 10.1038/nature22794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roh JL, Kim EH, Jang HJ, Park JY, Shin D. Induction of ferroptotic cell death for overcoming cisplatin resistance of head and neck cancer. Cancer Lett. 2016;381:96–103. doi: 10.1016/j.canlet.2016.07.035. [DOI] [PubMed] [Google Scholar]
- 20.Yoo SE, et al. Gpx4 ablation in adult mice results in a lethal phenotype accompanied by neuronal loss in brain. Free Radic Biol Med. 2012;52:1820–1827. doi: 10.1016/j.freeradbiomed.2012.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Additional references
- 21.Salt MB, Bandyopadhyay S, McCormick F. Epithelial--to-mesenchymal transition rewires the molecular path to PI3K-dependent proliferation. Cancer Discov. 2014;4:186–199. doi: 10.1158/2159-8290.CD-13-0520. [DOI] [PubMed] [Google Scholar]
- 22.Gelain DP, et al. A systematic review of human antioxidant genes. Front Biosci (Landmark Ed) 2009;14:4457–4463. doi: 10.2741/3541. [DOI] [PubMed] [Google Scholar]
- 23.Wilson JN, Liu W, Brown AS, Landgraf R. Binding-induced, turn-on fluorescence of the EGFR/ERBB kinase inhibitor, lapatinib. Org Biomol Chem. 2015;13:5006–5011. doi: 10.1039/c5ob00239g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available from the authors upon reasonable request. This manuscript is accompanied by four Supplementary tables and data are deposited at the NCBI GEO accession GSE84896.