Abstract
Mammalian target of rapamycin (mTOR) activity is required for memory and is dysregulated in disease. Activation of mTOR promotes protein synthesis; however, new studies are demonstrating that mTOR activity also represses the translation of mRNAs. Almost three decades ago, Kandel and colleagues hypothesised that memory was due to the induction of positive regulators and removal of negative constraints. Are these negative constraints repressed mRNAs that code for proteins that block memory formation? Herein, we will discuss the mRNAs coded by putative memory suppressors, how activation/inactivation of mTOR repress protein expression at the synapse, how mTOR activity regulates RNA binding proteins, mRNA stability, and translation, and what the possible implications of mRNA repression are to memory and neurodegenerative disorders.
Keywords: local translation, mammalian target of rapamycin, mRNA
Without losers, where would the winners be?
– Casey Stengel (Major League baseball player and manager of the ‘loveable losers,’ the New York Mets, 1962)
A referee is ‘an official whose job is to make sure that a game is played fairly and that the rules are not broken [1]’. Researchers since the 1970s have been deciphering the strict rules that govern synaptic plasticity, the long-lasting changes that strengthen or weaken synapses. Adherence to these rules mediates precision in synaptic communication, which is required for interconnected networks of synapses to form a memory. Breaking these rules leads to neurological disorders such as Alzheimer’s disease (AD), autism spectrum disorders (ASD), and epilepsy, where activity may end up being excitotoxic [2]. Determining the molecular referee for memory is a critical task as it may lead to more targeted pharmacological therapies for neurological disorders that express impaired memory.
In the ‘game’ of memory, two teams compete. Kan-del and colleagues in the late 1990s referred to the two teams as positive regulators and negative constraints of memory. In this visionary review, they likened negative constraints to tumour suppressor genes [3]. The presence of negative constraint proteins in the synapse suppresses memories. Just as the disruption of a single tumour repressor protein is probably not enough to promote cancer, aberrant expression of a single memory suppressor protein will probably not affect cognition [3]. Dysregulation of proteins that serve as hubs of translational regulation (i.e. single factors that coordinate the expression of many mRNAs), however, could compromise memory and lead to disease [4]. Examples of translational regulation hubs include signalling pathways that control protein synthesis, microRNAs – small RNAs known to repress mRNA translation – or RNA-binding proteins that repress or promote translation. Fundamental studies on molecular mechanisms that ensure that memory suppressor proteins are removed from the synapse during cellular conditions that model learning and memory are beginning to emerge.
Evidence for memory suppressor proteins
The quest to determine the cellular and molecular correlates of memory can be traced back to Santiago Ramon y Cajal, who, over a century ago, envisioned that alterations in synapses might underlie learning [5]. Since then, different research teams have discovered pieces of evidence to support Cajal’s proposition on synaptic plasticity. The historic work of Timothy Bliss and Terje Lømo laid the physiological evidence that synaptic plasticity and efficacy can endure for days [6]. These lasting changes in synaptic behaviour – commonly called ‘long-term potentiation’ (LTP) – are deemed to be the physiological expressions of memory and have fuelled the ongoing search to identify the molecular basis of memory.
Subsequent studies on LTP revealed that it is expressed in two distinct phases or stages: early and late. The late-phase LTP is highly relevant to this review not only because it is thought to be a model of memory consolidation, but also because it relies on protein synthesis [3,7–9]. While the requirement for protein synthesis in late-phase LTP is well-known, the necessity of reducing protein expression in LTP has been largely neglected. A careful reconsideration of a study on LTP and protein synthesis from Bliss and colleagues interestingly shows that reduction of proteins accompanies the expression of persistent LTP [10]. Although it was unclear from this study whether reduced protein expression was due to protein degradation or reduced synthesis, many have taken an interest in activity-dependent protein degradation (for review see [11,12]). In contrast, repression of protein synthesis, as a means of limiting the expression of memory suppression proteins, remains underexplored.
Mammalian target of rapamycin – the referee for remembering or forgetting?
mTOR activity is required for memory
Mammalian target of rapamycin (mTOR) controls a signalling pathway whose activity and downstream regulation of protein synthesis is required for long-lasting forms of synaptic plasticity and memory (reviewed in [13–15]). mTOR is a serine/threonine kinase and has a prime function in promoting mRNA translation initiation [16]. Experiments that reduce mTOR signalling with rapamycin, an mTOR-specific inhibitor, prior to LTP induction, block late-LTP [17–19]. Conversely, genetic manipulations in mice that result in elevated basal mTOR activity enhance late-LTP [20]. In support of these findings, behavioural experiments that assess learning and memory in rodents indicate the requirement of mTOR activity in the formation, consolidation and reconsolidation of memories [20–25].
Herein, we present studies that establish mTOR as a referee of synaptic plasticity and memory, with an emphasis on mechanisms by which mTOR activation suppresses protein synthesis and mTOR inactivation promotes protein expression. We will refer to the active mTOR state as mTOR-On and inactive as mTOR-Off. We consider the role of mTOR-On and mTOR-Off in local protein synthesis, the expression of RNA-binding proteins, RNA competition, mRNA stability and neurological disorders. The role of mTOR in promoting protein synthesis has been extensively reviewed elsewhere (for review see [26]), thus we will limit our discussion of mTOR-On proteins and refer to them only in the context of how they may influence the expression of mTOR-Off proteins.
mTOR acts as a switch for long-lasting changes in synaptic efficacy
mTOR protein is localised to dendrites and can modify synapses in a site-specific manner by determining which proteins win (are translated) or lose (are repressed) [18,27]. Previous studies theorised that den-drites contained polyribosomes, mRNAs, translation factors, as well as components that resemble the secretory pathway (endoplasmic reticulum and golgi membranes) to explain local protein synthesis (for review [28]). However, the identification of a referee, a single protein that can turn mRNA translation on and off, such as mTOR, fulfils the requirement for temporal and site-specific protein expression at synapses.
How is a synapse coached to be potentiated or depontentiated? N-methyl-D-aspartate receptor (NMDAR) activation turns mTOR on in dendrites during LTP [29]. Thus, activated synapses have to be ready or primed to respond, accordingly. As in many sports, a referee signals during a game to preside over the players and enforce the rules. This analogy can be extended to a synaptic arena, in which mTOR, acting as a referee, presides over molecular players of plasticity and memory. While the referee typically enforces rules, there are times where he or she has to use his or her experience in the game to make critical judgement calls thus, influencing the outcome of the game. The same is true for synapses. For example, to prime a synapse, mTOR officiates over molecular players [e.g. mRNAs that encode plasticity-related proteins (PRPs)] during the plasticity and memory game using the following guidelines. Guideline 1: mRNAs made in the nucleus are transported to the dendrites where they reside until mTOR signals to promote their translation. mTOR makes a critical judgment call based on the amount of synaptic stimulation and previous history of activity. Guideline 2: Since mRNAs can be silenced, repression of mRNA translation ensures the availability of dendritic mRNAs for temporal expression. The mRNAs repressed at the synapse may code for mRNAs that will potentiate or depontentiate the synapse. Toggling the activity of mTOR may dictate the direction the synapse will respond. Guideline 3: mTOR decides which mRNAs are ejected from the game. This often determines the outcome of the game. Removal of mRNAs from dendrites through decay processes may shift the odds toward whether a memory is consolidated or forgotten.
With such tight regulation over protein synthesis at the synapse, one questions why synapses contain mRNAs that code for proteins that block memory. To answer this question, the mRNAs that are repressed by mTOR need to be identified.
mTOR activity represses protein synthesis – the mRNAs that lose in the competition for translation
Our lab has focused on identifying proteins that may serve as memory suppressor proteins. Based on the simplistic idea that influencing the referee (i.e. inhibiting mTOR activity with the drug rapamycin), we identified, by tandem mass spectrometry (MS/MS), proteins whose expressions increase when mTOR activity is low. We hypothesise that these proteins fall into two groups: (a) memory suppressor proteins and (b) homeostatic proteins that turn mTOR back on. Thus, when mTOR is on, the memory suppressor proteins are absent, and when mTOR is off, they are present. Moreover, homeostatic proteins are necessary to prime the synapse to turn mTOR back on, in order for new learning to occur [30].
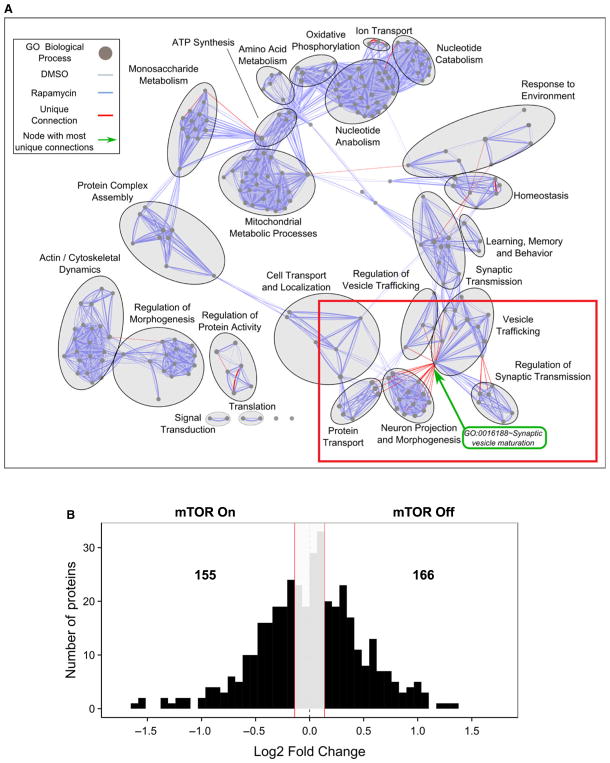
Interestingly, MS/MS analysis on cellular lysates of cortices isolated from rats that were briefly exposed to dimethyl sulfoxide (DMSO/control) or rapamycin via intraperitoneal injection, confirmed that mTOR activity greatly influences the synapse (Fig. 1). Analysis of the subcellular fraction that is enriched with postsynaptic density (PSD) determined that acutely perturbing mTOR activity changes ~75% of the proteins in PSD [31]. Moreover, roughly the same number of unique proteins were detected at the PSD when mTOR was active (mTOR-On proteins) and when mTOR was turned off (mTOR-Off proteins) (Fig. 1). Several mechanisms may account for increased expression in the PSD of mTOR-Off proteins, including local synthesis, translocation into the synapse, an increase in protein stability, or a combination of these. We will focus on those proteins whose mRNAs are repressed when mTOR is active and translated when mTOR is off and how they may fit the bill as memory suppressor proteins.
Fig. 1.
Mammalian target of rapamycin inhibition rapidly remodels the synapse. (A) Enrichment network obtained by mass spectrometry of total cortical lysates isolated from rats intraperitoneal injected with the mTORC1 inhibitor rapamycin or carrier. Rat corticies were isolated 1 h postinjection. Network analysis of gene ontology (GO) terms from mass spectrometry data reveals that mTOR has its biggest effect in synaptic processes as indicated by the red lines between GO terms and outlined in the red box. (B) Histogram of mass spectrometry-identified proteins from the PSD fractions of the same cortices as in (A) demonstrates that roughly equal number of unique proteins go up with mTOR inhibition (right of 0, expressed as fold change base 2 of rapamycin/control; 166 mTOR OFF proteins) as those that increase with mTOR activity (left of 0; 155 mTOR ON proteins). Figure reproduced from Niere et al. [31].
Potassium channels, the lovable losers, as a putative memory suppressor protein
What mTOR-Off proteins are good candidates to block memory? One would predict that proteins that increase the threshold for synaptic activation and, therefore, will reduce the probability that LTP will be sustained would be contenders for memory suppressor proteins. The field at large has focused on how blocking protein synthesis reduces proteins that increase excitatory drive and are required to maintain L-LTP. These are typically proteins such as glutamate receptors or signalling proteins that amplify synaptic signals like CaMKIIα (i.e. winners; for review see [32]). However, both synaptic and nonsynaptic plasticity underlie long-term potentiation and memory. Nonsynaptic plasticity refers to dynamic changes in heterogeneous population of ion channels including K+, Na+, and Ca2+, and is often referred to as plasticity of intrinsic excitability [33]. Notably, Schuman and colleagues have found that mRNAs coding for all three classes of voltage-gated ion channels are present in the dendritic fields of hippocampal CA1 [34]. While an increase in voltage-gated Na+ and Ca2+ channels is likely to reduce the threshold for synaptic activation, K+ channels will dampen the effect of synaptic activation. Thus, one would predict that mechanisms that block K+ channel activity and/or expression in dendrites are required for long-lasting synaptic efficacy (loveable loser). Some of the first studies that demonstrate the relationship between reduced K+ channel and long-lasting behaviours were done in invertebrates in the early 1980s (for review see [33]). Progress toward understanding how plasticity of intrinsic excitability contributes to information storage in mammals has been slower, likely due to the misconception that dendrites were passive and did not contain active ion channels [35]. However, manipulation of K+ channel activity in rodent dendrites, pharmacologically or using knockout mice, has been shown to have profound effects on LTP, LTD and memory [36–40] (for review see [35]). mRNAs coding for the voltage-gated potassium channel subfamilies Kv1, Kv4, Kvβ, the large conductance calcium sensitive potassium channel BK, and G-protein activated inwardly rectifying potassium channel (GIRK or Kir 3) have been localised to dendrites [27,34,41,42]. It’s still early days in our understanding of how intrinsic excitability contributes to memory, but local synthesis of ion channels provides a solution for site-specific titration of synaptic activation, which in turn will increase the capacity of memory storage [43].
Kcna1, 2, 3, 4, and 6
The Kcna gene family codes for the pore-forming subunits of the Kv1 or shaker subfamily (for review see [44]). Typically, this gene family codes for delayed rectifiers or A-type channels depending on its amino-terminus or subunit composition. In Drosophila, shaker mRNA is alternatively spliced with different amino-termini that dictate how fast the channel inactivates [45]. The amino-terminus has been termed the inactivation ball that, upon channel opening, occludes the pore, preventing further potassium movement across the membrane [46]. Channels with inactivation balls, such as Kv1.4, inactivate quickly leading to currents that have been described as A-type current (for review see [47]). In contrast, Kv1.1, 1.2, 1.3 and 1.6 are delayed rectifiers, which open slowly and do not inactivate. These channels help set the resting membrane potential, regulate transmitter release, and determine frequency of action potentials [48]. Notably, Kv1.1, 1.2 and 1.4 have been characterised as axonal channels in the hippocampus [49]. More recently, their mRNAs have been detected in dendrites [27,34]. Kv1.1 is locally synthesised in dendrites when NMDAR or mTOR activity is inhibited, making it tempting to speculate that translational repression of its mRNA is a positive feedback mechanism to sustain LTP and promote memory formation. However, due to Kv1.1 being localised both in axons and transiently expressed in dendrites, its role in the maintenance of LTP has been difficult to assess. For example, blocking Kv1.1 with its specific toxins dendrotoxin K or 4-aminopyrimidine (4AP) results in an increase in presynaptic transmitter release due to inhibition of presynaptic channels and, thus, confounds results. Tools that target Kv1 channels specifically in dendrites are necessary to assess its functional contribution to synaptic plasticity.
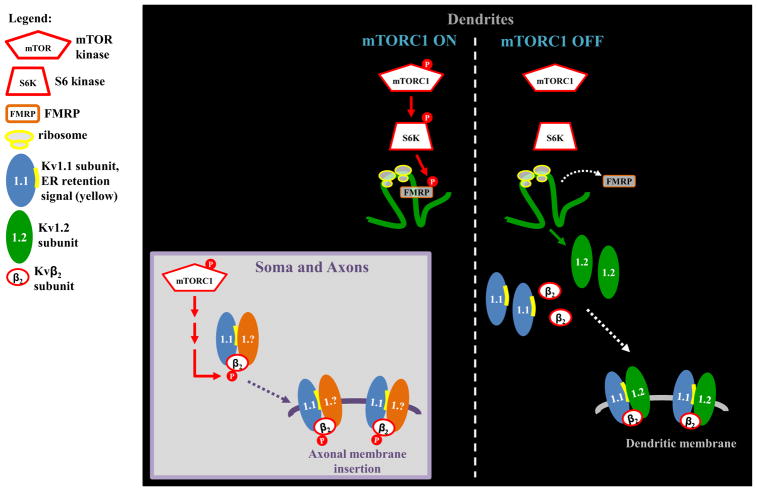
To date, it is unclear if the expression of other members of the Kv1 subfamily is regulated by NMDARs and mTOR in dendrites. Data from heterologous cells suggest that Kv1.1 is an obligate heteromultimer and assembles with other subfamily members to target the membrane [50]. Therefore, it is likely that other subfamily members are also expressed in an mTOR-regulated manner. Interestingly, mTOR activates S6 ribosomal subunit kinase (S6 kinase), which has been reported to phosphorylate the dendritic RNA binding protein (RBP) fragile X mental retardation protein (FMRP), promoting FMRP to bind and repress translation of its target mRNAs [51]. Since Kv1.2 mRNA is a verified target of FMRP, the model in Fig. 2 provides a reasonable mechanism by which Kv1.2 mRNA translation is repressed during increased mTOR activity (Fig. 2) [52].
Fig. 2.
Model of mTOR repression of Kv channel mRNAs. (Left) mTOR activity is ON. mTOR represses the mRNA translation of Kv1.1 and 1.2 via different RNA binding factors. Kv1.1 is repressed by the microRNA miR-129 (not shown) as reported in [88]. Kv1.2 (shown) and Kv4.2 mRNA (not shown but reported in [61]) may be repressed via mTOR dependent phosphorylation of S6 kinase which in turn phosphorylates FMRP. FMRP when phosphorylated binds to its target mRNAs, such as Kv1.2 and Kv4.2, to repress their translation. When mTOR is active, Kv1.1, and 1.2 are expressed in the soma where the axonal targeting domain in the amino terminus is exposed and binds to Kvβ2 targeting the channel complex to the axon [56,57]. When mTOR activity is reduced (right) FMRP is dephosphorylated and releases its target mRNAs. Kv1.1 mRNA is translated but must associate with Kv1.2 to mask its endoplasmic reticulum retention signal. Thus, the coordinated release of miR-129 from Kv1.1mRNA and FMRP from Kv1.2 mRNA allows for Kv1.1, Kv1.2 and Kvβ2 to form a functional channel and be inserted into the dendritic membrane.
While the model we propose provides a reasonable mechanism for local synthesis of Kv1.2 and perhaps other voltage-gated potassium channels (see Kv4), it should be noted that S6 kinase phosphorylation of FMRP is controversial [53]. Thus, future experiments are required to test this model.
Kcnb1 and 2
Kvβ subfamily is coded by the Kcnb1 and 2 genes. Kvβ1 and 2 differ in function [47]. Both Kvβ subunits assemble with the alpha pore-forming subunit at the amino-terminus. Kvβ1 acts like an inactivation ball and converts delayed rectifiers to A-type channels. To date, there is no evidence that Kvβ1 expression is regulated by mTOR activity. However, local synthesis of Kvβ1 would significantly impact the kinetics of the channel, which in turn would affect the membrane potential. It should be noted that neuronal excitability and synaptic plasticity are reduced in preclinical models of ageing, perhaps accounting for cognitive decline [54]. Aged Kvβ1 knockout mice have enhanced synaptic plasticity, suggesting that Kvβ1 may contribute to deficits in cellular models of learning and memory with ageing [55].
Kvβ2 promotes the surface expression of the Kv1 subfamily. Thus, its activity-dependent expression will increase the current density of the channel. Moreover, it traffics the alpha subunit to the axon [56,57]. Interestingly, Kvβ2 is differentially regulated by mTOR in a subcellular fashion. When mTOR is active, Kvβ2 expression increases in total lysate samples but is repressed in the PSD fraction [31]. The differential expression of Kvβ2 raises an interesting possibility. mTOR activity could increase somatic Kvβ2 to traffic Kv1 subunits made in the soma to the axon; when mTOR is inhibited, Kvβ2 could provide local insertion of Kv1 pore forming subunits, also locally synthesised into the PSD (Fig. 2). Interestingly, cyclin dependent kinase 5 (CDK5) phosphorylates Kvβ2, which facilitates axonal membrane insertion of the channel complex [58]. Our mass spectrometry data suggest that mTOR activity increases CDK5 expression in the lysate (somatic population), which is consistent with axonal expression and function of Kv1-containing channels when mTOR is active (Fig. 2, left, bottom schematic) [31].
Kcnd4
The Kv4 subfamily (Shal) is coded by the gene Kcnd4 and conducts A-type currents with fast inactivation when expressed. The Kv4 subfamily of ion channels is the best characterised in the context of synaptic plasticity, by far. Pharmacological inhibition of Kv4 channels in acute hippocampal slices increases back-propagating action potentials. Back-propagating action potentials facilitate associative synaptic plasticity by depolarising the membrane sufficiently to dislodge the magnesium block from the pore of NMDARs, which leads to increased synaptic potentiation [59]. Therefore, repression of Kv4 channel expression increases the likelihood that associative plasticity occurs and synaptic efficacy is increased. In support for this theory, Kv4.2 knockout mice show reduced LTP [36].
Kv4.2, like Kv1.2, is a known target of FMRP [52]; however, whether FMRP represses or promotes Kv4.2 mRNA translation is still a matter of debate [60–62]. In the hippocampus, physiological evidence supports both an increase and reduction of Kv4 channel activity with the induction of LTP [36,59]. Moreover, Brager and colleagues demonstrate a cell-specific increase in Kv4-mediated A-current in the prefrontal cortex of Fmr1 knockout mice [63], opposite to what they observed in CA1 hippocampal neurons [62]. As described in our model for Kv1.2 (Fig. 2), phosphorylation of FMRP by S6 kinase downstream of mTOR activity should affect Kv4 expression, one way or the other. Jan and colleagues demonstrated that Kv4.2 expression, like Kv1.1, increases with mTOR inhibition. Moreover, there was a corresponding reduction in FMRP phosphorylation, consistent with mTOR repressing the expression of Kv4.2 through FMRP [61]. Notably, we did not detect significant changes in Kv4 expression in our mass spectrometry data. Thus, the regulation of Kv4 mRNA translation and channel activity may be more complicated than simply being repressed or initiated by mTOR.
Kcma1
The Kcma1 gene codes for the large conductance calcium activated potassium channel, BK, and the mRNA is alternatively spliced in a cell specific manner. Interestingly, one splice variant retains an intron that serves as a dendritic targeting sequence, deeming this isoform dendrite-specific. The mRNA expression of this variant is dendritic. Knockdown of the intron-containing mRNA reduces BK expression in spines [41]. How local excitatory postsynaptic potentials (EPSPs) are affected by synthesis of BK near or in spines is not known. However, it is speculated that reduced BK would allow for a back-propagating action potential to repolarise the membrane potential more slowly, which would unblock NMDARs and increase synaptic efficacy [64]. Recently, Kato and colleagues suggested that fear conditioning suppresses BK expression in the amygdala [65]. Interestingly, our MS/ MS data did not detect BK protein at synapses; however, the channel was upregulated with mTOR inhibition in the total cellular lysate fraction, consistent with its repression with fear memory [31].
Kcnj2
The Kcnj2 gene codes for the G-protein activated inwardly rectifying potassium channel 2 (GIRK2 or Kir 3.2). At the physiological level, GIRK current is required for the depotentiation of EPSPs during LTP [37], and potentiation of the slow inhibitory postsynaptic current (sIPSC) during LTP [66]. While counterintuitive with GIRK’s role in depotentiation, the role of GIRK current has been shown to be required for contextual fear [39]. While these studies do not directly test the requirement for mTOR activity or local protein synthesis of synaptic expression of GIRK channels, Kcnj2 mRNA has been found to be trafficked to the dendrites by the RNA splicing factor protein Nova [42]. In contrast to Kcnma1, which requires splicing and the retention of a specific intron in order to target the dendrites, Nova binds to the 3′ untranslated region (UTR) of Kcnj2 mRNA, independent of its splicing function. Interestingly, slow inhibitory postsynaptic potential (sIPSP) of LTP is absent in the Nova2 knockout mouse, suggesting that dendritic targeting of Kcnj2 mRNA and local protein synthesis of GIRK2 are required [66].
In summary, even small changes in local ion channel expression in dendrites, perhaps undetectable by traditional biochemistry, may have a dramatic impact on EPSPs [67]. As the sensitivity of detection methods improves to visualise local synthesis of ion channels in dendrites and dendritic branches, and as tools are developed to specifically manipulate the expression of K+ channels in a site-specific manner, the role of local synthesis of K+ channels will help answer the question which channels increase information storage and which ones reduce it.
microRNAs – winners and losers
How does mTOR activity choose which transcripts are the winners and consequently synthesised? Multiple RNA binding factors repress or promote mRNA translation. Emerging data suggest that single RNA-binding factors, such as RBPs or microRNAs, may act as post-transcriptional regulons, regulating the synthesis of proteins with coordinated functions [68]. Recently, the model of this regulon has expanded to include regulation of noncoding mRNAs [69]. While this literature is quite extensive, below we focus on mechanisms by which mTOR regulates mRNA regulons to mediate memory. As mTOR activity is required for memory consolidation, we will consider the literature that explicitly studies memory and also requires mTOR activation.
MicroRNAs (miRs) are a group of small, noncoding RNAs that bind to regulatory sequences in the 3′UTR of mRNAs. miRs either lead to the degradation of the mRNAs they bind, or serve as translational repressors [70]. As translational repressors, miRs ensure proper temporal and spatial syntheses of proteins [71]. miRs have to be processed to their mature, binding-competent form by the enzyme Dicer. Interestingly, a study that examined the expression of miRs with fear conditioning demonstrated that the levels of the mature form change; however, the unprocessed or pre-miR levels remain the same [72]. Recently, miR processing was shown to occur at the synapse. These data suggest that dynamic regulation of mRNA repression and degradation can occur in a synapse-specific manner with active Dicer proteins dynamically binding to those mRNAs localised to synapses [73]. Mature (processed) miRs are loaded into the RNA induced silencing complex (RISC), which directs the miR to its target mRNA and facilitates base pairing between the miR and mRNA that will be silenced (for review see [74]).
miRs have been suggested to be memory suppressors. This stems from the theory that specific miRs release mRNAs that code for proteins that underlie L-LTP and consolidation. By the same logic, miRs may serve as memory enhancers by binding and repressing the translation of mRNAs that code for memory suppressor proteins, such as K+ channels. Evidence that miRs can serve as ‘memory suppressors’ is best characterised through olfactory learning in Drosophila. The neuronal circuits and cell types for olfactory learning in Drosophila are known and described in detail (for review see [75]). Some of the earliest data emerged from mutating RISC proteins, which disrupted miR function. These mutations lead to normal learning with deficits in long-term memory in Drosophila [76]. However, disruption of the RISC does not specifically indicate if de-repressing mRNA translation of memory enhancer or suppressor proteins is causative. To specifically address this question, manipulation of miRs in this tractable system has allowed for unbiased screens for memory-related miRs. Expression of sponges, specific complimentary oligonucleotides that sequester miRs from binding to their target mRNAs, were used to probe olfactory learning and memory against 134 individual miRs. 18–21% of the tested miR-sponges produced significant altered memory phenotypes; importantly, some increased memory and some led to memory deficits [77]. Many neuronal mRNAs have multiple miR binding sites, which are thought to provide a highly complex code for mRNA repression/ degradation, allowing for translation to be fine-tuned [78]. Therefore, it is striking that even roughly 20% of the individually expressed miRs screened had a phenotype. Collectively, these data argue that miRs play a role in repressing both memory enhancer and suppressor proteins, as predicted by the bidirectional protein expression regulated by mTOR activity.
What regulates miR binding to its target mRNAs? One might predict that mTOR may regulate the expression of memory-related miRs. Fear conditioning produced both up- and down-regulation of miRs at several time points after acquisition [72]. These data suggest that translation and repression of mRNAs that promote fear memory are regulated by miRs. Interestingly, fear conditioning upregulated several miRs that target mRNAs that code for proteins that inhibit the activation of mTOR, thus preventing the translation of these proteins [72]. Because these proteins would prevent the activation of mTOR during the consolidation phase, they fit the bill for memory suppressor proteins. Future work characterising other miRs that are upregulated during fear condition and their mRNA targets may lead to a better understanding of how NMDAR and mTOR regulate synaptic efficacy via mRNA translation.
HuD, an RNA-binding protein that bumps off its competitor (microRNAs)
Do the levels of miRs have to change with mTOR activity to repress translation of memory suppressor mRNAs? Dynamic changes in miR expression suggest a corresponding change in target mRNA translation. However, activity-dependent changes in the expression of RBPs that displace miRs may also be the trigger for the translation or repression of an mRNA. One of the best examples is the Hu family of RBPs (Elav, HuR, HuB, HuC, and HuD). Hu typically promotes the translation of its targets by binding to AU rich elements. HuD’s expression increases with learning, seizures and cocaine exposure [79,80]. Both seizure activity and cocaine exposure are reported to lead to hyperactive mTOR signalling [81–84].
How do Hu RBPs de-repress mRNA translation? The typical thought of action is through competitive binding with miRs for the same binding motif. However, through oligomerisation of the RBP, HuR proteins can displace bound miRs by binding many basepairs away from the miR seed sequence [85]. It should be noted, that the translation of HuD mRNA, itself, is highly regulated by machinery in the RISC complex and polyadenylation [86,87]; thus, activity-dependent local changes in Hu expression is another way to control site-specific protein expression of its target mRNAs.
mTOR activity level referees competition between high and low affinity HuD target mRNAs, favouring translation of memory enhancers
What happens when miR and RBP expression levels do not change? Are there other mechanisms for shifting the probability that an mRNA will be translated? One way to favour a winner is by removing a competitor. Hu proteins promote translation of their targets through stabilising the mRNAs that they bind to. We have shown that mTOR activity stabilises CaMKIIα mRNA through the binding of HuD. When mTOR activity is reduced, CaMKIIα mRNA degrades through deadenylation [82,88]. Moreover, we found that HuD disrupts miR-129 binding to Kv1.1 mRNA when mTOR is inactive. Surprisingly, mTOR activity does not alter HuD or miR-129 expression levels. These data suggest that perhaps the mRNAs that HuD stabilises – when mTOR is active – degrade when mTOR activity is reduced, allowing for free HuD to bind translationally repressed mRNAs, such as Kv1.1 [88]. CaMKIIα mRNA has 32 predicted HuD binding sites in its 3′UTR, and Kv1.1 mRNA has only three in its coding region. Based on the significant difference in the number of binding sites and the location of binding, we suggest that CaMKIIα mRNA represents a putative population of high affinity-HuD mRNAs that are stabilised when mTOR activity is high, and Kv1.1 mRNA represents low affinity mRNAs, with HuD levels being the limiting factor [88]. In agreement with this premise, HuD binding in the coding region only represents 8% of its target mRNAs, suggesting that it prefers binding to sites within the 3′UTR [89]. Thus, switching mTOR off removes the competition through mRNA decay (memory enhancers) and allows the underdog (memory suppressors) its opportunity to be expressed, for better or for worse.
How does mTOR referee other RBPs – is competition a general mechanism?
Is mRNA competition for RNA binding proteins a general mechanism that is employed to control the syntheses of select proteins and a means of shaping global and/or local function? mRNA competition, either due to differences in mRNA abundance or affinity to RBP, is just beginning to be explored [90]. The field of synthetic biology has embarked on pioneering studies on mRNA competition and considers mRNA competition to be a source of cross talk in protein synthesis [91]. An additional example beyond HuD is FMRP. FMRP and its mRNA targets are well-studied and characterised (for review see [92]). FMRP, encoded by the Fmr1 gene, is an RBP that is generally regarded as a suppressor of mRNA translation, such that excessive protein synthesis is deemed to underlie the pathological phenotypes in fragile X syndrome (FXS). Unlike HuD that typically promotes protein synthesis, cases have been reported where FMRP binding can repress or promote translation. For example, FMRP binds to superoxide dismutase 1 (SOD1) mRNA to stabilise it and promotes its translation [93]. In addition, FMRP has been reported to promote Kv4.2 mRNA translation. This unique finding exposes the dual role of FMRP as a repressor of protein synthesis for some mRNAs and as an activator for others. The exact mechanism of how FMRP switches from a repressor to an activator of translation is yet to be elucidated.
Could the decision of FMRP repression be dependent on differing affinities to its mRNA targets like HuD? The mRNAs of γ-aminobutyric acid B receptors 1 and 2 (GABABR1 and GABABR2) bind to and are translationally repressed by FMRP [52,94]. Basal levels of GABABR1 and 2 proteins are elevated in an Fmr1 knockout mouse model of FXS [94]. However, at 1 h after administration of a rapid antidepressant or ethanol, an NMDAR blocker, when FMRP level is reduced, only GABABR2 displays increased protein synthesis. This finding is curious because one would expect that the synthesis of proteins that serve a similar function and are translationally repressed by the same RBP would be identically affected upon perturbing the shared RBP. Is FMRP expression the limiting factor such that GABABR1 has a higher affinity for FMRP than R2? These findings intriguingly suggest that mRNAs that bind to translation suppressors may also compete for repression with the higher affinity mRNAs remaining repressed. Although studies will need to be conducted to test this idea, mRNA competition for translation de-repression may serve as a mechanism that dictates which mRNAs are translated in an activity-dependent fashion when bound to translation suppressors, like FMRP and miRs.
A caveat to the theory that FMRP target mRNAs compete for FMRP binding based on affinity of its binding site is that the FMRP binding motif is highly debated in the field. Some suggest that FMRP does not bind in a sequence specific manner [52]. Still others suggest that FMRP binds to certain motifs or structures [95,96]. Thus, if there is no sequence specificity how would mRNAs compete for binding? A recent bioinformatics analysis that compares different FMRP-mRNA binding motifs that were proposed by previous studies, have determined FMRP consensus binding sequence (FCBS) motifs: TGGA, GAC and TAY (for a comprehensive list of studies that examine the different RNA binding motifs of FMRP, see [97]). Thus, we propose that competition arises from how the different RNA binding motifs of FMRP determine which mRNAs are translated. For example, an isoleucine to asparagine mutation (I304N) in the hnRNPK homology 2 (KHs) domain of FMRP does not seem to affect the association of Kif1a and Lingo1 –established FMRP mRNA targets – to I304N-FMRP [52]. However, for another known FMRP target mRNA, Map1B, mRNA association with I304N-FMRP appears different.
As mentioned above, FMRP’s phosphorylation state has been shown to dictate its association with mRNAs. Phosphorylation of FMRP generally increases FMRP-mRNA association and suppresses protein synthesis. Because FMRP functions as a translation repressor and activator, it would be useful to examine which mRNAs associate with FMRP when it is phosphorylated or dephosphorylated as well as to the different RNA binding motifs of FMRP. This work will be invaluable to our understanding of how different symptoms arise in FXS.
Although we focus on mTOR’s role in regulating translation of local mRNAs, exciting findings from other groups demonstrate that the mitogen-activated protein kinase (MAPK)-interacting kinases (MNKs) may also referee the translation of select mRNAs that encode PRPs, independent of mTOR. Interestingly, these transcripts are repressed when bound to MNK and FMRP. Phosphorylation of MNK releases FMRP and allows translation initiation to occur. Thus, understanding how these two modes of translational repression fit together to remodel a synapse is an important future direction [98].
What happens when the ref favours one team over the other – is this the root of cognitive disorders?
For a game to be played fairly the ref has to be balanced in his calls. By the same analogy, it is essential that mTOR operates within its optimal operating range, remaining balanced for cells to remain healthy and to respond accordingly to extra- and intracellular cues [99]. But what happens when the ref favours one team over the other? Constitutively active or inactive mTOR states are strongly linked to disease. For example, epilepsy, ASD and AD generally display overactive mTOR, while Parkinson’s disease and Rett syndrome (RTT) exhibit underactive mTOR [83,100–106]. While excessive protein synthesis has been considered to be the root of hyperactive mTOR-related diseases, the discovery that mTOR is overactive in epilepsy argues otherwise (reviewed in [106–108]). Early studies on epilepsy have primarily focused on deficiencies in potassium channel function and expression [64]. Recent studies have started to focus on the relationship between mTOR activity and ion channel expression [27,82,109,110].
Is there a reciprocal relationship between memory suppressor protein expression and mTOR activity levels? Can tapping into memory suppressor proteins return mTOR to its normal operating range in disease states? Dendritic ion channels that help set the resting membrane potential have the ability to reduce synaptic activity and thus reduce activation of mTOR. The best example of this possible reciprocal relationship is through the overexpression of the putative memory suppressor protein Kv1.1 in epilepsy. Kv1.1 protein expression is tightly coupled to epileptogenesis. Although mTOR inhibition by rapamycin reduces seizures by increasing Kv1.1 protein level, its long-term use is not viable [111–113]. Notably, viral mediated expression of Kv1.1was reported to prevent seizures in a focal model of epilepsy [114]. Whether mTOR activity is altered in this model and Kv1.1 overexpression restores mTOR activity to baseline remains to be determined. Another example has been reported in the case of treating depression where mTOR is reported to be hypoactive. It has been demonstrated that knockdown of the hyperpolarising cyclic nucleotide channel (HCN), a nonselective ion channel that is critical in establishing the resting membrane potential, normalising synaptic inputs, and regulating membrane resistance [115], rescues depressive-like behaviours and increases mTOR activity [116]. Similarly, the increased expression of 14-3-3 eta, an adaptor protein that decouples GABABR signalling from dendritic K+ channels and thus removes slow inhibition, facilitates increased mTOR activity in the hippocampus and results in rapid antidepressant efficacy [117]. It is therefore important to determine disease-associated proteins whose expressions rely on mTOR activity level, with the idea that mTOR-On and mTOR-Off proteins work synergistically to preserve normal neuronal function.
Future questions and conclusions
Our understanding of how mTOR activity influences proteostasis is evolving. What is the next step? Most of these studies have been done examining global changes in mTOR activity. To this end, the mTOR inhibitor rapamycin has shown some promise in overactive mTOR related diseases such as TSC and AD (for review see [118–120]). However, long-term use of rapamycin is prohibited due to side effects that cause many patients to stop taking it (for review see [120]). In light of these studies, it’s been suggested that finding downstream targets of mTOR activity may provide better therapeutic options [121]. This point is driven home since we have recently shown that levels of mTOR activity may affect mRNA translation or repression of specific transcripts differently depending on its subcellular localisation [31]. Thus, targeting the mTOR-dependent differential expression of these proteins might provide a way to titrate site-specific changes in excitability without turning mTOR off throughout the cell. How would one do this? Perhaps the first step would be to know if competition for miRs or RBPs occurs in specific compartments (i.e. PSD, dendrite, axon and soma). Over 1000 RBPs have been identified to date, and many of the mRNA targets of these RBPs have also been catalogued [91]. Mapping RBPs, miRs, and mRNA populations isolated to single dendritic branches that may compete for binding will be important to understand how synapses or groups of synapses are uniquely modified and contribute to long-lasting changes in synaptic efficacy. Further studies that explore the mechanisms that lead to site-specific differential expression of proteins when mTOR activity is perturbed will yield insight into cognition, as well as provide new disease targets to correct mTOR-related disorders while avoiding systemic or cell-wide changes that occur with rapamycin. Together, these data strongly support the continued search for mTOR-regulated memory suppressor proteins as a means to enhance memory, normalise mTOR activity and treat disease.
Acknowledgments
This work was supported by NSF IOS-1355158 (KRG), NSF PRFB DBI-1306528 (FN), NIH-NIAAA pilot grant provided by the Integrated Neuroscience Initiative on Alcoholism AA013517 (KRG), Department of Defense USAMRMC Award W81XWH-14-10061 (KRG), NIH-NIA pilot grant provided by Wake Forest University Health Sciences Alzheimer’s Disease Core Center P30AG049638 (KRG), and NIH-NIDA pilot grant provided by Wake Forest University Health Sciences Center for the Neurobiology of Addiction Treatment (CNAT) P50DA06634 (KRG). We would like to thank West Graham for his helpful discussion regarding what makes a good referee. We would like to thank Dr. Chelcie Heaney for her critical review of our manuscript.
Abbreviations
- AD
Alzheimer’s disease
- ASD
autism spectrum disorders
- CDK5
cyclin dependent kinase 5
- DMSO
dimethyl sulfoxide
- EPSPs
excitatory postsynaptic potentials
- FCBS
FMRP consensus binding sequence
- FMRP
fragile X mental retardation protein
- FXS
fragile X syndrome
- LTP
long-term potentiation
- MAPK
mitogen-activated protein kinase
- miRs
MicroRNAs
- mTOR
mammalian target of rapamycin
- PSD
post-synaptic density
- RBP
RNA binding protein
- RISC
RNA induced silencing complex
- sIPSP
slow inhibitory postsynaptic potential
Footnotes
Author contributions
KRG and FN wrote the manuscript.
References
- 1.The American Heritage Dictionary of the English Language. 5. Houghton Mifflin Harcourt; Boston: 2011. [Google Scholar]
- 2.Crino PB. The mTOR signalling cascade: paving new roads to cure neurological disease. Nat Rev Neurol. 2016;12:379–392. doi: 10.1038/nrneurol.2016.81. [DOI] [PubMed] [Google Scholar]
- 3.Abel T, Martin KC, Bartsch D, Kandel ER. Memory suppressor genes: inhibitory constraints on the storage of long-term memory. Science. 1998;279:338– 341. doi: 10.1126/science.279.5349.338. [DOI] [PubMed] [Google Scholar]
- 4.Pernice HF, Schieweck R, Kiebler MA, Popper B. mTOR and MAPK: from localized translation control to epilepsy. BMC Neurosci. 2016;17:73. doi: 10.1186/s12868-016-0308-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cajal SRY. The croonian lecture: La fine structure des centres nerveux. Proc Royal Society London. 1894;55:444–468. [Google Scholar]
- 6.Bliss TV, Lomo T. Long-lasting potentiation of synaptic transmission in the dentate area of the anaesthetized rabbit following stimulation of the perforant path. J Physiol. 1973;232:331–356. doi: 10.1113/jphysiol.1973.sp010273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frey U, Huang YY, Kandel ER. Effects of cAMP simulate a late stage of LTP in hippocampal CA1 neurons. Science. 1993;260:1661–1664. doi: 10.1126/science.8389057. [DOI] [PubMed] [Google Scholar]
- 8.Krug M, Lossner B, Ott T. Anisomycin blocks the late phase of long-term potentiation in the dentate gyrus of freely moving rats. Brain Res Bull. 1984;13:39–42. doi: 10.1016/0361-9230(84)90005-4. [DOI] [PubMed] [Google Scholar]
- 9.Pittenger C, Kandel ER. In search of general mechanisms for long-lasting plasticity: Aplysia and the hippocampus. Philos Trans R Soc Lond B Biol Sci. 2003;358:757–763. doi: 10.1098/rstb.2002.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fazeli MS, Corbet J, Dunn MJ, Dolphin AC, Bliss TV. Changes in protein synthesis accompanying long-term potentiation in the dentate gyrus in vivo. J Neurosci. 1993;13:1346–1353. doi: 10.1523/JNEUROSCI.13-04-01346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanus C, Schuman EM. Proteostasis in complex dendrites. Nat Rev Neurosci. 2013;14:638–648. doi: 10.1038/nrn3546. [DOI] [PubMed] [Google Scholar]
- 12.Schieweck R, Popper B, Kiebler MA. Co-translational folding: a novel modulator of local protein expression in mammalian neurons? Trends Genet. 2016;32:788–800. doi: 10.1016/j.tig.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 13.Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61:10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graber TE, McCamphill PK, Sossin WS. A recollection of mTOR signaling in learning and memory. Learn Mem. 2013;20:518–530. doi: 10.1101/lm.027664.112. [DOI] [PubMed] [Google Scholar]
- 15.Hoeffer CA, Klann E. mTOR signaling: at the crossroads of plasticity, memory and disease. Trends Neurosci. 2010;33:67–75. doi: 10.1016/j.tins.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 17.Cammalleri M, Lutjens R, Berton F, King AR, Simpson C, Francesconi W, Sanna PP. Time-restricted role for dendritic activation of the mTOR-p70S6K pathway in the induction of late-phase long-term potentiation in the CA1. Proc Natl Acad Sci USA. 2003;100:14368–14373. doi: 10.1073/pnas.2336098100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang SJ, Reis G, Kang H, Gingras AC, Sonenberg N, Schuman EM. A rapamycin-sensitive signaling pathway contributes to long-term synaptic plasticity in the hippocampus. Proc Natl Acad Sci USA. 2002;99:467–472. doi: 10.1073/pnas.012605299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vickers CA, Dickson KS, Wyllie DJ. Induction and maintenance of late-phase long-term potentiation in isolated dendrites of rat hippocampal CA1 pyramidal neurones. J Physiol. 2005;568:803–813. doi: 10.1113/jphysiol.2005.092924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoeffer CA, Tang W, Wong H, Santillan A, Patterson RJ, Martinez LA, Tejada-Simon MV, Paylor R, Hamilton SL, Klann E. Removal of FKBP12 enhances mTOR-Raptor interactions, LTP, memory, and perseverative/repetitive behavior. Neuron. 2008;60:832–845. doi: 10.1016/j.neuron.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antion MD, Merhav M, Hoeffer CA, Reis G, Kozma SC, Thomas G, Schuman EM, Rosenblum K, Klann E. Removal of S6K1 and S6K2 leads to divergent alterations in learning, memory, and synaptic plasticity. Learn Mem. 2008;15:29–38. doi: 10.1101/lm.661908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bekinschtein P, Katche C, Slipczuk LN, Igaz LM, Cammarota M, Izquierdo I, Medina JH. mTOR signaling in the hippocampus is necessary for memory formation. Neurobiol Learn Mem. 2007;87:303–307. doi: 10.1016/j.nlm.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Blundell J, Kouser M, Powell CM. Systemic inhibition of mammalian target of rapamycin inhibits fear memory reconsolidation. Neurobiol Learn Mem. 2008;90:28–35. doi: 10.1016/j.nlm.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casadio A, Martin KC, Giustetto M, Zhu H, Chen M, Bartsch D, Bailey CH, Kandel ER. A transient, neuron-wide form of CREB-mediated long-term facilitation can be stabilized at specific synapses by local protein synthesis. Cell. 1999;99:221–237. doi: 10.1016/s0092-8674(00)81653-0. [DOI] [PubMed] [Google Scholar]
- 25.Parsons RG, Gafford GM, Helmstetter FJ. Translational control via the mammalian target of rapamycin pathway is critical for the formation and stability of long-term fear memory in amygdala neurons. J Neurosci. 2006;26:12977–12983. doi: 10.1523/JNEUROSCI.4209-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 27.Raab-Graham KF, Haddick PC, Jan YN, Jan LY. Activity- and mTOR-dependent suppression of Kv1.1 channel mRNA translation in dendrites. Science. 2006;314:144–148. doi: 10.1126/science.1131693. [DOI] [PubMed] [Google Scholar]
- 28.Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci. 2007;8:776–789. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- 29.Gong R, Park CS, Abbassi NR, Tang SJ. Roles of glutamate receptors and the mammalian target of rapamycin (mTOR) signaling pathway in activity-dependent dendritic protein synthesis in hippocampal neurons. J Biol Chem. 2006;281:18802–18815. doi: 10.1074/jbc.M512524200. [DOI] [PubMed] [Google Scholar]
- 30.Abraham WC, Bear MF. Metaplasticity: The plasticity of synaptic plasticity. Trends Neurosci. 1996;19:126–130. doi: 10.1016/s0166-2236(96)80018-x. [DOI] [PubMed] [Google Scholar]
- 31.Niere F, Namjoshi S, Song E, Dilly GA, Schoenhard G, Zemelman BV, Mechref Y, Raab-Graham KF. Analysis of proteins that rapidly change upon mechanistic/mammalian target of rapamycin complex 1 (mTORC1) repression identifies Parkinson protein 7 (PARK7) as a novel protein aberrantly expressed in tuberous sclerosis complex (TSC) Mol Cell Proteomics. 2016;15:426–444. doi: 10.1074/mcp.M115.055079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 33.Zhang W, Linden DJ. The other side of the engram: experience-driven changes in neuronal intrinsic excitability. Nat Rev Neurosci. 2003;4:885–900. doi: 10.1038/nrn1248. [DOI] [PubMed] [Google Scholar]
- 34.Cajigas IJ, Tushev G, Will TJ, Tom Dieck S, Fuerst N, Schuman EM. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012;74:453–466. doi: 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnston D, Christie BR, Frick A, Gray R, Hoffman DA, Schexnayder LK, Watanabe S, Yuan LL. Active dendrites, potassium channels and synaptic plasticity. Philos Trans R Soc Lond B Biol Sci. 2003;358:667–674. doi: 10.1098/rstb.2002.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X, Yuan LL, Zhao C, Birnbaum SG, Frick A, Jung WE, Schwarz TL, Sweatt JD, Johnston D. Deletion of Kv4.2 gene eliminates dendritic A-type K+ current and enhances induction of long-term potentiation in hippocampal CA1 pyramidal neurons. J Neurosci. 2006;26:12143–12151. doi: 10.1523/JNEUROSCI.2667-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chung HJ, Ge WP, Qian X, Wiser O, Jan YN, Jan LY. G protein-activated inwardly rectifying potassium channels mediate depotentiation of longterm potentiation. Proc Natl Acad Sci USA. 2009;106:635– 640. doi: 10.1073/pnas.0811685106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Truchet B, Manrique C, Sreng L, Chaillan FA, Roman FS, Mourre C. Kv4 potassium channels modulate hippocampal EPSP-spike potentiation and spatial memory in rats. Learn Mem. 2012;19:282–293. doi: 10.1101/lm.025411.111. [DOI] [PubMed] [Google Scholar]
- 39.Victoria NC, Marron Fernandez de Velasco E, Ostrovskaya O, Metzger S, Xia Z, Kotecki L, Benneyworth MA, Zink AN, Martemyanov KA, et al. G Protein-Gated K+ Channel ablation in forebrain pyramidal neurons selectively impairs fear learning. Biol Psychiatry. 2016;80:796–806. doi: 10.1016/j.biopsych.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wickens JR, McKenzie D, Costanzo E, Arbuthnott GW. Effects of potassium channel blockers on synaptic plasticity in the corticostriatal pathway. Neuropharmacology. 1998;37:523–533. doi: 10.1016/s0028-3908(98)00054-9. [DOI] [PubMed] [Google Scholar]
- 41.Bell TJ, Miyashiro KY, Sul JY, McCullough R, Buckley PT, Jochems J, Meaney DF, Haydon P, Cantor C, Parsons TD, et al. Cytoplasmic BK (Ca) channel intron-containing mRNAs contribute to the intrinsic excitability of hippocampal neurons. Proc Natl Acad Sci USA. 2008;105:1901–1906. doi: 10.1073/pnas.0711796105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Racca C, Gardiol A, Eom T, Ule J, Triller A, Darnell RB. The neuronal splicing factor nova co-localizes with target RNAs in the dendrite. Front Neural Circuits. 2010;4:5. doi: 10.3389/neuro.04.005.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kastellakis G, Silva AJ, Poirazi P. Linking memories across time via neuronal and dendritic overlaps in model neurons with active dendrites. Cell Rep. 2016;17:1491–1504. doi: 10.1016/j.celrep.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jan LY, Jan YN. Voltage-gated potassium channels and the diversity of electrical signalling. J Physiol. 2012;590:2591–2599. doi: 10.1113/jphysiol.2011.224212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Timpe LC, Jan YN, Jan LY. Four cDNA clones from the Shaker locus of Drosophila induce kinetically distinct A-type potassium currents in Xenopus oocytes. Neuron. 1988;1:659–667. doi: 10.1016/0896-6273(88)90165-1. [DOI] [PubMed] [Google Scholar]
- 46.Zagotta WN, Hoshi T, Aldrich RW. Restoration of inactivation in mutants of Shaker potassium channels by a peptide derived from ShB. Science. 1990;250:568–571. doi: 10.1126/science.2122520. [DOI] [PubMed] [Google Scholar]
- 47.Pongs O, Leicher T, Berger M, Roeper J, Bahring R, Wray D, Giese KP, Silva AJ, Storm JF. Functional and molecular aspects of voltage-gated K+ channel beta subunits. Ann N Y Acad Sci. 1999;868:344–355. doi: 10.1111/j.1749-6632.1999.tb11296.x. [DOI] [PubMed] [Google Scholar]
- 48.Brew HM, Hallows JL, Tempel BL. Hyperexcitability and reduced low threshold potassium currents in auditory neurons of mice lacking the channel subunit Kv1.1. J Physiol. 2003;548:1–20. doi: 10.1113/jphysiol.2002.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monaghan MM, Trimmer JS, Rhodes KJ. Experimental localization of Kv1 family voltage-gated K+ channel alpha and beta subunits in rat hippocampal formation. J Neurosci. 2001;21:5973–5983. doi: 10.1523/JNEUROSCI.21-16-05973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manganas LN, Trimmer JS. Subunit composition determines Kv1 potassium channel surface expression. J Biol Chem. 2000;275:29685–29693. doi: 10.1074/jbc.M005010200. [DOI] [PubMed] [Google Scholar]
- 51.Narayanan U, Nalavadi V, Nakamoto M, Thomas G, Ceman S, Bassell GJ, Warren ST. S6K1 phosphorylates and regulates fragile X mental retardation protein (FMRP) with the neuronal protein synthesis-dependent mammalian target of rapamycin (mTOR) signaling cascade. J Biol Chem. 2008;283:18478–18482. doi: 10.1074/jbc.C800055200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bartley CM, O’Keefe RA, Bordey A. FMRP S499 is phosphorylated independent of mTORC1- S6K1 activity. PLoS One. 2014;9:e96956. doi: 10.1371/journal.pone.0096956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosenzweig ES, Barnes CA. Impact of aging on hippocampal function: plasticity, network dynamics, and cognition. Prog Neurobiol. 2003;69:143–179. doi: 10.1016/s0301-0082(02)00126-0. [DOI] [PubMed] [Google Scholar]
- 55.Murphy GG, Fedorov NB, Giese KP, Ohno M, Friedman E, Chen R, Silva AJ. Increased neuronal excitability, synaptic plasticity, and learning in aged Kvbeta1.1 knockout mice. Curr Biol. 2004;14:1907–1915. doi: 10.1016/j.cub.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 56.Gu C, Zhou W, Puthenveedu MA, Xu M, Jan YN, Jan LY. The microtubule plus-end tracking protein EB1 is required for Kv1 voltage-gated K+ channel axonal targeting. Neuron. 2006;52:803–816. doi: 10.1016/j.neuron.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 57.Gu C, Jan YN, Jan LY. A conserved domain in axonal targeting of Kv1 (Shaker) voltagegated potassium channels. Science. 2003;301:646–649. doi: 10.1126/science.1086998. [DOI] [PubMed] [Google Scholar]
- 58.Vacher H, Trimmer JS. Diverse roles for auxiliary subunits in phosphorylation-dependent regulation of mammalian brain voltage-gated potassium channels. Pflugers Arch. 2011;462:631–643. doi: 10.1007/s00424-011-1004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoffman DA, Magee JC, Colbert CM, Johnston D. K+ channel regulation of signal propagation in dendrites of hippocampal pyramidal neurons. Nature. 1997;387:869–875. doi: 10.1038/43119. [DOI] [PubMed] [Google Scholar]
- 60.Gross C, Yao X, Pong DL, Jeromin A, Bassell GJ. Fragile X mental retardation protein regulates protein expression and mRNA translation of the potassium channel Kv4.2. J Neurosci. 2011;31:5693–5698. doi: 10.1523/JNEUROSCI.6661-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee HY, Ge WP, Huang W, He Y, Wang GX, Rowson-Baldwin A, Smith SJ, Jan YN, Jan LY. Bidirectional regulation of dendritic voltage-gated potassium channels by the fragile X mental retardation protein. Neuron. 2011;72:630–642. doi: 10.1016/j.neuron.2011.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Routh BN, Johnston D, Brager DH. Loss of functional A-type potassium channels in the dendrites of CA1 pyramidal neurons from a mouse model of fragile X syndrome. J Neurosci. 2013;33:19442–19450. doi: 10.1523/JNEUROSCI.3256-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kalmbach BE, Johnston D, Brager DH. Cell-type specific channelopathies in the prefrontal cortex of the fmr1-/y mouse model of fragile X syndrome. eNeuro. 2015 doi: 10.1523/ENEURO.0114-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Poolos NP, Johnston D. Dendritic ion channelopathy in acquired epilepsy. Epilepsia. 2012;53(Suppl 9):32–40. doi: 10.1111/epi.12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun P, Zhang Q, Zhang Y, Wang F, Wang L, Yamamoto R, Sugai T, Kato N. Fear conditioning suppresses large-conductance calcium-activated potassium channels in lateral amygdala neurons. Physiol Behav. 2015;138:279–284. doi: 10.1016/j.physbeh.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 66.Huang CS, Shi SH, Ule J, Ruggiu M, Barker LA, Darnell RB, Jan YN, Jan LY. Common molecular pathways mediate long-term potentiation of synaptic excitation and slow synaptic inhibition. Cell. 2005;123:105–118. doi: 10.1016/j.cell.2005.07.033. [DOI] [PubMed] [Google Scholar]
- 67.Burdakov D, Ashcroft FM. Shedding new light on brain metabolism and glial function. J Physiol. 2002;544:334. doi: 10.1113/jphysiol.2002.029090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Keene JD, Tenenbaum SA. Eukaryotic mRNPs may represent posttranscriptional operons. Mol Cell. 2002;9:1161–1167. doi: 10.1016/s1097-2765(02)00559-2. [DOI] [PubMed] [Google Scholar]
- 69.Kang MJ, Abdelmohsen K, Hutchison ER, Mitchell SJ, Grammatikakis I, Guo R, Noh JH, Martindale JL, Yang X, Lee EK, et al. HuD regulates coding and noncoding RNA to induce APP–>Abeta processing. Cell Rep. 2014;7:1401–1409. doi: 10.1016/j.celrep.2014.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 71.Schratt G. Fine-tuning neural gene expression with microRNAs. Curr Opin Neurobiol. 2009;19:213–219. doi: 10.1016/j.conb.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 72.Kye MJ, Neveu P, Lee YS, Zhou M, Steen JA, Sahin M, Kosik KS, Silva AJ. NMDA mediated contextual conditioning changes miRNA expression. PLoS One. 2011;6:e24682. doi: 10.1371/journal.pone.0024682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sambandan S, Akbalik G, Kochen L, Rinne J, Kahlstatt J, Glock C, Tushev G, Alvarez-Castelao B, Heckel A, Schuman EM. Activity-dependent spatially localized miRNA maturation in neuronal dendrites. Science. 2017;355:634–637. doi: 10.1126/science.aaf8995. [DOI] [PubMed] [Google Scholar]
- 74.Aksoy-Aksel A, Zampa F, Schratt G. MicroRNAs and synaptic plasticity–a mutual relationship. Philos Trans R Soc Lond B Biol Sci. 2014;369 doi: 10.1098/rstb.2013.0515. pii: 20130515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Busto GU, Guven-Ozkan T, Davis RL. MicroRNA function in Drosophila memory formation. Curr Opin Neurobiol. 2016;43:15–24. doi: 10.1016/j.conb.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, Koppetsch BS, Theurkauf WE, Zamore PD. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004;116:831–841. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- 77.Busto GU, Guven-Ozkan T, Fulga TA, Van Vactor D, Davis RL. microRNAs that promote or inhibit memory formation in Drosophila melanogaster. Genetics. 2015;200:569–580. doi: 10.1534/genetics.114.169623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bolognani F, Qiu S, Tanner DC, Paik J, Perrone-Bizzozero NI, Weeber EJ. Associative and spatial learning and memory deficits in transgenic mice overexpressing the RNA-binding protein HuD. Neurobiol Learn Mem. 2007;87:635–643. doi: 10.1016/j.nlm.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 80.Tiruchinapalli DM, Caron MG, Keene JD. Activity-dependent expression of ELAV/Hu RBPs and neuronal mRNAs in seizure and cocaine brain. J Neurochem. 2008;107:1529–1543. doi: 10.1111/j.1471-4159.2008.05718.x. [DOI] [PubMed] [Google Scholar]
- 81.Brewster AL, Lugo JN, Patil VV, Lee WL, Qian Y, Vanegas F, Anderson AE. Rapamycin reverses status epilepticus-induced memory deficits and dendritic damage. PLoS One. 2013;8:e57808. doi: 10.1371/journal.pone.0057808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sosanya NM, Brager DH, Wolfe S, Niere F, Raab-Graham KF. Rapamycin reveals an mTOR-independent repression of Kv1.1 expression during epileptogenesis. Neurobiol Dis. 2015;73:96–105. doi: 10.1016/j.nbd.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 83.Zeng LH, Rensing NR, Wong M. The mammalian target of rapamycin signaling pathway mediates epileptogenesis in a model of temporal lobe epilepsy. J Neurosci. 2009;29:6964–6972. doi: 10.1523/JNEUROSCI.0066-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sutton LP, Caron MG. Essential role of D1R in the regulation of mTOR complex1 signaling induced by cocaine. Neuropharmacology. 2015;99:610–619. doi: 10.1016/j.neuropharm.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kundu P, Fabian MR, Sonenberg N, Bhattacharyya SN, Filipowicz W. HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Nucleic Acids Res. 2012;40:5088–5100. doi: 10.1093/nar/gks148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Banerjee S, Neveu P, Kosik KS. A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron. 2009;64:871–884. doi: 10.1016/j.neuron.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 87.Udagawa T, Swanger SA, Takeuchi K, Kim JH, Nalavadi V, Shin J, Lorenz LJ, Zukin RS, Bassell GJ, Richter JD. Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex. Mol Cell. 2012;47:253–266. doi: 10.1016/j.molcel.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sosanya NM, Huang PP, Cacheaux LP, Chen CJ, Nguyen K, Perrone-Bizzozero NI, Raab-Graham KF. Degradation of high affinity HuD targets releases Kv1.1 mRNA from miR-129 repression by mTORC1. J Cell Biol. 2013;202:53–69. doi: 10.1083/jcb.201212089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bolognani F, Contente-Cuomo T, Perrone-Bizzozero NI. Novel recognition motifs and biological functions of the RNA-binding protein HuD revealed by genome-wide identification of its targets. Nucleic Acids Res. 2010;38:117–130. doi: 10.1093/nar/gkp863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lodish HF. Molecular Cell Biology. 7. W.H. Freeman and Co; New York: 2013. [Google Scholar]
- 91.Mather WH, Hasty J, Tsimring LS, Williams RJ. Translational cross talk in gene networks. Biophys J. 2013;104:2564–2572. doi: 10.1016/j.bpj.2013.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Darnell JC, Klann E. The translation of translational control by FMRP: Therapeutic targets for FXS. Nat Neurosci. 2013;16:1530–1536. doi: 10.1038/nn.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bechara EG, Didiot MC, Melko M, Davidovic L, Bensaid M, Martin P, Castets M, Pognonec P, Khandjian EW, Moine H, et al. A novel function for fragile X mental retardation protein in translational activation. PLoS Biol. 2009;7:e16. doi: 10.1371/journal.pbio.1000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wolfe SA, Workman ER, Heaney CF, Niere F, Namjoshi S, Cacheaux LP, Farris SP, Drew MR, Zemelman BV, Harris RA, et al. FMRP regulates an ethanol-dependent shift in GABABR function and expression with rapid antidepressant properties. Nat Commun. 2016;7:12867. doi: 10.1038/ncomms12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 96.Ascano M, Jr, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Anderson BR, Chopra P, Suhl JA, Warren ST, Bassell GJ. Identification of consensus binding sites clarifies FMRP binding determinants. Nucleic Acids Res. 2016;44:6649–6659. doi: 10.1093/nar/gkw593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bramham CR, Jensen KB, Proud CG. Tuning specific translation in cancer metastasis and synaptic memory: control at the MNK-eIF4E Axis. Trends Biochem Sci. 2016;41:847–858. doi: 10.1016/j.tibs.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 99.Raab-Graham KF, Workman ER, Namjoshi S, Niere F. Pushing the threshold: How NMDAR antagonists induce homeostasis through protein synthesis to remedy depression. Brain Res. 2016;1647:94–104. doi: 10.1016/j.brainres.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bove J, Martinez-Vicente M, Vila M. Fighting neurodegeneration with rapamycin: mechanistic insights. Nat Rev Neurosci. 2011;12:437–452. doi: 10.1038/nrn3068. [DOI] [PubMed] [Google Scholar]
- 101.Ma T, Hoeffer CA, Capetillo-Zarate E, Yu F, Wong H, Lin MT, Tampellini D, Klann E, Blitzer RD, Gouras GK. Dysregulation of the mTOR pathway mediates impairment of synaptic plasticity in a mouse model of Alzheimer’s disease. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012845. pii: e12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pei JJ, Hugon J. mTOR-dependent signalling in Alzheimer’s disease. J Cell Mol Med. 2008;12:2525–2532. doi: 10.1111/j.1582-4934.2008.00509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ricciardi S, Boggio EM, Grosso S, Lonetti G, Forlani G, Stefanelli G, Calcagno E, Morello N, Landsberger N, Biffo S, et al. Reduced AKT/mTOR signaling and protein synthesis dysregulation in a Rett syndrome animal model. Hum Mol Genet. 2011;20:1182–1196. doi: 10.1093/hmg/ddq563. [DOI] [PubMed] [Google Scholar]
- 104.Santini E, Heiman M, Greengard P, Valjent E, Fisone G. Inhibition of mTOR signaling in Parkinson’s disease prevents L-DOPA-induced dyskinesia. Sci Signal. 2009;2:ra36. doi: 10.1126/scisignal.2000308. [DOI] [PubMed] [Google Scholar]
- 105.Sharma A, Hoeffer CA, Takayasu Y, Miyawaki T, McBride SM, Klann E, Zukin RS. Dysregulation of mTOR signaling in fragile X syndrome. J Neurosci. 2010;30:694–702. doi: 10.1523/JNEUROSCI.3696-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Russo E, Citraro R, Constanti A, De Sarro G. The mTOR signaling pathway in the brain: focus on epilepsy and epileptogenesis. Mol Neurobiol. 2012;46:662–681. doi: 10.1007/s12035-012-8314-5. [DOI] [PubMed] [Google Scholar]
- 107.Ostendorf AP, Wong M. mTOR inhibition in epilepsy: rationale and clinical perspectives. CNS Drugs. 2015;29:91–99. doi: 10.1007/s40263-014-0223-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sadowski K, Kotulska-Jozwiak K, Jozwiak S. Role of mTOR inhibitors in epilepsy treatment. Pharmacol Rep: PR. 2015;67:636–646. doi: 10.1016/j.pharep.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 109.Dwyer BE, Wasterlain CG, Fujikawa DG, Yamada L. Brain protein metabolism in epilepsy. Adv Neurol. 1986;44:903–918. [PubMed] [Google Scholar]
- 110.Huang X, McMahon J, Yang J, Shin D, Huang Y. Rapamycin down-regulates KCC2 expression and increases seizure susceptibility to convulsants in immature rats. Neuroscience. 2012;219:33–47. doi: 10.1016/j.neuroscience.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ventura-Aguiar P, Campistol JM, Diekmann F. Safety of mTOR inhibitors in adult solid organ transplantation. Exp Opin Drug Safety. 2016;15:303–319. doi: 10.1517/14740338.2016.1132698. [DOI] [PubMed] [Google Scholar]
- 112.Eiden AM, Zhang S, Gary JM, Simmons JK, Mock BA. Molecular pathways: increased susceptibility to infection is a complication of mTOR inhibitor use in cancer therapy. Clin Cancer Res. 2016;22:277–283. doi: 10.1158/1078-0432.CCR-14-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Barlow AD, Nicholson ML, Herbert TP. Evidence for rapamycin toxicity in pancreatic beta-cells and a review of the underlying molecular mechanisms. Diabetes. 2013;62:2674–2682. doi: 10.2337/db13-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wykes RC, Heeroma JH, Mantoan L, Zheng K, MacDonald DC, Deisseroth K, Hashemi KS, Walker MC, Schorge S, Kullmann DM. Optogenetic and potassium channel gene therapy in a rodent model of focal neocortical epilepsy. Sci Transl Med. 2012;4:161ra152. doi: 10.1126/scitranslmed.3004190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Magee JC. Dendritic hyperpolarization-activated currents modify the integrative properties of hippocampal CA1 pyramidal neurons. J Neurosci. 1998;18:7613–7624. doi: 10.1523/JNEUROSCI.18-19-07613.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kim CS, Chang PY, Johnston D. Enhancement of dorsal hippocampal activity by knockdown of HCN1 channels leads to anxiolytic-and antidepressant-like behaviors. Neuron. 2012;75:503–516. doi: 10.1016/j.neuron.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Workman ER, Haddick PC, Bush K, Dilly GA, Niere F, Zemelman BV, Raab-Graham KF. Rapid antidepressants stimulate the decoupling of GABA(B) receptors from GIRK/Kir3 channels through increased protein stability of 14-3-3eta. Mol Psychiatry. 2015;20:298– 310. doi: 10.1038/mp.2014.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Richardson A, Galvan V, Lin AL, Oddo S. How longevity research can lead to therapies for alzheimer’s disease: the rapamycin story. Exp Gerontol. 2015;68:51–58. doi: 10.1016/j.exger.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Caban C, Khan N, Hasbani DM, Crino PB. Genetics of tuberous sclerosis complex: implications for clinical practice. Appl Clin Genet. 2017;10:1–8. doi: 10.2147/TACG.S90262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sahin M. Targeted treatment trials for tuberous sclerosis and autism: no longer a dream. Curr Opin Neurobiol. 2012;22:895–901. doi: 10.1016/j.conb.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Neasta J, Barak S, Hamida SB, Ron D. mTOR complex 1: a key player in neuroadaptations induced by drugs of abuse. J Neurochem. 2014;130:172–184. doi: 10.1111/jnc.12725. [DOI] [PMC free article] [PubMed] [Google Scholar]