Abstract
Saline-soluble glycinins and insoluble glutelins are the major storage proteins in soybean (Glycine max) and rice (Oryza sativa), respectively. In spite of their differences in solubility properties, both proteins are members of the 11S globulin gene family based on their similarities in primary sequences and processing of the coded protein. Wild-type and methionine-modified glycinin coding sequences were expressed in transgenic rice plants under the control of the rice glutelin GluB-1 promoter. Glycinins were specifically synthesized in the endosperm tissue and co-localized with glutelins in type II protein bodies. They assembled into 7S and 11S species, similar to what was observed in developing soybean seeds. This pattern was quite different from that displayed by the rice glutelins in untransformed plants, in which processed subunits sedimenting at 2S were apparent. In glycinin-expressing transgenic plants, however, glutelins were observed sedimenting at 7S and 11S with lesser amounts in the 2S region. A portion of the glycinins was also found associated in the insoluble glutelin fraction. Renaturation experiments suggested that the hybrid glycinin-glutelin oligomers were formed through specific interactions. Overall, these results indicate that despite significant differences in the assembly of soybean glycinin and rice glutelin, both proteins can assemble with each other to form soluble hexameric oligomers or insoluble aggregates.
Seed storage proteins were initially classified into albumins (water soluble), globulins (saline soluble), prolamins (alcohol soluble), and glutelins (residue) by Osborne (1924) according to their solubility properties. Based on more recent and extensive molecular and biochemical analysis of the storage protein genes and their coded products, the storage proteins fall into two major groups, the globulins and the prolamins (Shewry and Tatham, 1990). The rice (Oryza sativa) glutelins and soybean (Glycine max) glycinins are excellent examples of this reclassification of storage proteins. The rice glutelins, which comprise up to 70% to 80% of the total seed protein, are insoluble in a neutral saline solution but soluble in a diluted acid/alkaline solution. They exist as large macromolecular complexes formed by disulfide and hydrophobic interactions of acidic and basic polypeptides. The soybean glycinins, which account for 40% of the total proteins (Utsumi, 1992; Utsumi et al., 1997), are soluble in neutral saline solutions. These proteins accumulate as 11S oligomers comprised of six pairs (subunits) of acidic and basic polypeptides interlinked by a conserved disulfide bond. Although glutelin and glycinin have different properties (such as their solubility), they nevertheless are related and are both members of the 11S globulin family of storage proteins. These proteins share 32% to 37% identity in their primary sequences. Moreover, both proteins are synthesized as a larger precursor on the ER, are proteolytically processed into acidic and basic polypeptides, and are accumulated and stored in a vacuolar compartment.
The cellular events that lead from the synthesis of soybean glycinin to their assembly into a protein storage vacuole are well understood. When translocated in the ER lumen, the newly synthesized proglycinins (2S) assemble into trimers that sediment at 7S to 8S in Suc density gradients. These trimers are competent for transport to the protein storage vacuole via the Golgi complex. At this storage site, each proglycinin subunit of the trimer is proteolytically cleaved into acidic and basic polypeptides (Staswick et al., 1984) whereupon assembly into a hexamer (11S–12S) occurs. Soluble proteins destined for the vacuole must carry sorting determinants that allow them to be identified and sorted from the secretory pathway (Okita and Rogers, 1996). Horse bean 11S globulin, legumin, was demonstrated to have an internal signal determinant (Saalbach et al., 1991; Müntz, 1998) and a similar sorting signal likely occurs for the soybean glycinin. Recently, physical aggregation was proposed as a sorting mechanism of pea legumin: the higher hydrophobicity of prolegumin than mature legumin would be a driving force of aggregate formation and binding to the membrane of cargo vesicles that are targeted to the protein storage vacuole (Hinz et al., 1997; Müntz, 1998; Neuhaus and Rogers, 1998; Robinson et al., 1998).
The cellular processes that lead to the transport of rice glutelin to the type II protein body (equivalent to the protein storage vacuole) are not as well understood as the processes following the synthesis of soybean glycinin. Pulse-chase labeling studies readily demonstrate that the glutelin is initially synthesized as a larger precursor, which is then proteolytically processed into acidic and basic subunits. No evidence has been obtained showing that the rice glutelins are transported to the vacuole by an ordered process of 7S trimer formation, proteolysis, and 11S hexamer formation, as has been demonstrated for the soybean glycinins. Instead, the precursor form itself may be transported to the Golgi, where it is packaged into dense vesicles associated with the Golgi cisternae (Krishnan et al., 1986). The concentration of glutelins in dense vesicles and their hydrophobicity may result in protein aggregation, which would follow an aggregation mechanism for sorting (Okita and Rogers, 1996; Hinz et al., 1997; Müntz, 1998; Neuhaus and Rogers, 1998; Robinson et al., 1998).
Rice and soybean proteins are deficient in Lys and sulfur-containing amino acids, respectively. When viewed nutritionally, these proteins can compensate for one another and collectively provide a better balance of essential amino acids. In addition to their desired physicochemical properties, e.g. heat-induced gel forming and emulsifying abilities, soybean proteins such as glycinin lower the cholesterol levels in human serum (Kito et al., 1993), while rice glutelins have no significant physicochemical and physiological properties. Because of these functional properties, genetic engineering efforts have been directed at improving the nutritional value and physicochemical properties of soybean glycinins (Kim et al., 1990b; Utsumi et al., 1993a; Katsube et al., 1994, 1998a), with the aim of introducing these modified genes into rice (Katsube et al., 1998b).
We have previously demonstrated that glycinin can accumulate to significant levels in the endosperm tissue of tobacco seeds using a glutelin GluB-1 promoter (Takaiwa et al., 1995). In the present study, we expressed the soybean glycinin gene and a Met-rich glycinin gene in transgenic rice. We show that glycinin is assembled into trimeric and hexameric structures, is proteolytically processed into subunits, and is packaged into a protein storage vacuole by events in rice much like those observed in developing soybean seeds. In contrast, rice glutelins do not exhibit such trimeric and hexameric structures. However, in transgenic plants, glutelins can assemble with glycinin to form soluble hetero-trimers and -hexamers and insoluble complexes. Our results indicate that the secondary and tertiary structures of glycinin and glutelin are compatible to form higher order protein structures. Hence, assembly and packaging of glycinin should not be a limiting factor in expression in rice.
MATERIALS AND METHODS
Construction of Chimeric Genes and Transformation
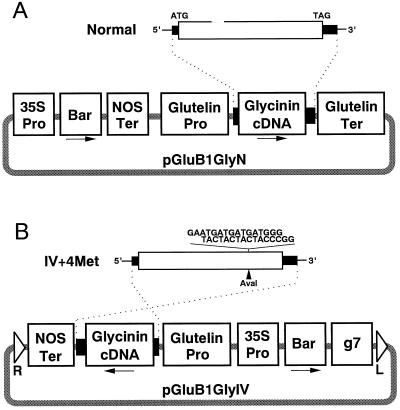
The chimeric gene consisting of the GluB-1 promoter (−1302 to +18) and the normal glycinin A1aB1b cDNA was recovered from pBGG1 (Takaiwa et al., 1995) after digestion with HindIII and SacI, and then inserted into the corresponding sites of pUC18 containing the 0.6-kb 3′ noncoding region of the GluB-1 gene to give pUGluB1Gly. A DNA fragment containing the CaMV 35S promoter-bialaphos resistance gene (Bar)-nopaline synthase gene terminator hybrid gene was then inserted into the filled-in HindIII site of pUGluB1Gly to give pGluB1Gly. A similar construct containing a modified glycinin cDNA (encoding the modified glycinin IV+4Met where four contiguous Met residues were inserted) was constructed from pBGG1IV (Takaiwa et al., 1995) and inserted in the binary plasmid pGPTV-bar/Fer (Goto et al., 1999) to give pGluB1GlyIV.
The plasmids containing the chimeric genes for the normal and modified glycinins were transferred into rice (Oryza sativa cv Matsuyama-mii) protoplasts by electroporation (Tada et al., 1990) and Agrobacterium tumefaciens-mediated transformation (Goto et al., 1999), respectively. Bialaphos-resistant calli were selected and plants were regenerated and rooted in the presence of 10 mg/L bialaphos as described previously (Caplan et al., 1992). A total of 68 plants containing pGluB1Gly and 34 plants containing pGluB1GlyIV were obtained and analyzed for the inserted glycinin gene by PCR amplification.
Glutelin and glycinin cDNAs encoding the mature primary sequences were amplified by PCR using the following pair of oligonucleotides; 5′-cagcagctattaggccagagcactag-3′ and 5′-gggaagctttatccgcaagccgacctaag-3′ for GluA-1, 5′-cagctatttaatcccagcacaaaccc-3′ and 5′-gggaagcttacattactctgaggtctcgc-3′ for GluB-1, 5′-ttcagttccagagagcagcc-3′ and 5′-cgcggatccatacaaaaagggctctaag-3′ for A1aB1b. The latter primer of each pair corresponds to the downstream primer and contained either a HindIII site for GluA-1 and GluB-1 or a BamHI site for A1aB1b. The ends of the PCR products were polished using a blunting kit (TaKaRa-Shuzo, Shiga, Japan) and then digested with HindIII or BamHI. The resultant DNA fragments were inserted into the filled-in NcoI and the HindIII sites or the filled-in NcoI and the BamHI sites of the pET-21d vector (Novagen) to yield the Escherichia coli expression plasmids pEGluA-1, pEGluB-1, and pEA1aB1b.
For protein induction, E. coli BL21(DE3) cells harboring individual expression plasmids were grown in Luria-Bertani medium. At an A600 of 0.6, isopropyl-β-d-thiogalactopyranoside (final concentration 1 mm) was added and the cells were allowed to grow for 24 h at 37°C for GluA-1 and GluB-1 and at 20°C for A1aB1b.
RNA Analysis
RNA was isolated from leaf and developing seeds as described by Takaiwa et al. (1987). Fifteen micrograms of total RNA was electrophoresed in a 1.2% (w/v) formaldehyde-containing agarose gel and then blotted to a charged nylon membrane (Amersham). The resulting nylon membrane was then hybridized in 50% (v/v) formamide, 5× Denhardt′s solution, 0.5% (w/v) SDS, and 250 μg/mL denatured salmon-sperm DNA at 45°C for 24 h with 32P-labeled glycinin A1aB1b and a glutelin GluB-1 cDNA inserts of pUG1 (Utsumi et al., 1993b) and pREEK1 (Takaiwa et al., 1989), respectively. The cDNA inserts were labeled with 32P by the random-priming method (Feinberg and Vogelstein, 1983). Membranes were washed under high-stringency conditions to exclude cross-hybridization four times in 2× SSC and 0.1% (w/v) SDS at room temperature and then twice in 0.1× SSC and 0.1% (w/v) SDS at 55°C.
Preparation of Anti-Proglutelin GluA-1 and GluB-1 Sera
E. coli cells were resuspended in 35 mm Na-Pi buffer (pH 8.0) containing 0.1 m NaCl, 1 mm EDTA, and 1.5 mm PMSF and disrupted by sonication. After centrifugation, the pellet was extracted with a 1% (v/v) lactic acid, 1 mm EDTA solution to solubilize the recombinant expressed proglutelins GluA-1 and GluB-1. The extracted proglutelins were dialyzed against buffer A composed of 50 mm Tris-HCl buffer (pH 8.0) containing 6 m urea, 0.1 m ME, 1 mm EDTA, and 1 mm PMSF, and then subjected to fast-flow chromatography (2.6- × 10-cm columns; SP Sepharose FF, Pharmacia). The proglutelins were eluted with a linear gradient of 0 to 0.2 m NaCl in buffer A. GluA-1 and GluB-1 were further purified by SDS-PAGE (11%, w/v) (Laemmli, 1970), yielding a single purified band. The purified proglutelins were electroeluted from the polyacrylamide gel and then thoroughly dialyzed against 10 mm K-Pi buffer (pH 7.6) containing 6 m urea to remove excess SDS. Rabbit antibodies were raised against the purified GluA-1 and GluB-1 as described previously (Utsumi et al., 1993b).
Screening of Transgenic Rice Plants Accumulating Glycinin at a High Level and Its in Situ Localization by Tissue Printing
Six seeds from independent transgenic plant were soaked overnight at 4°C in 10 mm phosphate buffer (pH 7.4) containing 0.8% (w/v) NaCl, 0.2% (w/v) KCl, 10 mm NaN3, and 0.4 m Suc, and then horizontally dissected into two pieces using a razor blade as described by Manteuffel and Panitz (1993). The pieces were soaked in acetone for 20 s to remove lipids and then pressed onto a nitrocellulose membrane for 5 to 10 s. After the prints had been made the membrane was baked for 30 min at 60°C. Glycinins were detected with anti-glycinin serum followed by goat anti-rabbit IgG-alkaline phosphatase conjugate (Promega).
For in situ localization of glycinin, a grain from a rice plant expressing glycinin at the highest level was vertically dissected into two pieces and treated as described above.
Electron Microscope Immunocytochemical Localization of Glycinin
Analysis of immunocytochemical localization was carried out essentially as described by Takeuchi and Nishimura (1993). Developing rice seeds were cut into 1.5- to 2.0-mm sections and fixed overnight at 4°C in 3% (v/v) glutaraldehyde solution. Tissues were washed three times (10 min for each wash) with phosphate buffer and then dehydrated through a 30%, 50%, 60%, 70%, 80%, 90% (v/v) series of ethanol washes (15 min for each wash). The specimens were then further dehydrated by three wash steps in 95% ethanol (15 min for each wash) followed by three wash steps in 100% ethanol (30 min per wash). The dehydrated tissues were infiltrated for 2 h in a 1:1 (v/v) London white resin:ethanol mixture, in 2:1 (v/v) overnight, followed by 100% London white resin over the next 24 h. Polymerization was in gelatin capsules for 2 to 7 d at 52°C under nitrogen gas.
Ultrathin sections (60–80 nm) were obtained with a glass knife and placed onto formal/carbon-coated grids. The sections were blocked with 5% (w/v) BSA-PBS and then incubated for 1 h at room temperature on a drop of anti-glycinin serum diluted 1:20 in PBS (10 mm Na-Pi buffer, pH 7.4, containing 150 mm NaCl) supplemented with 0.8% (w/v) BSA (Sigma), 0.1% (w/v) gelatin, and 2 mm NaN3 (BSA-PBS). The sections were washed four times for 5 min each on a drop of BSA-PBS and then incubated for 1 h at room temperature on a drop of goat antirabbit IgG (H+L) (Auro Probe EM, Amersham) diluted 1:40 in BSA-PBS. After washing twice with BSA-PBS, the sections were treated with 2.5% (w/v) glutaraldehyde solution for 10 min and washed twice with PBS and then once with distilled water (5 min per step). The sections were stained for 30 min with 2% (w/v) uranyl acetate followed by a 1-min incubation in 80 mm lead nitrate, 120 mm trisodium citrate dihydrate, and 0.16 n NaOH for 1 min. The grids were examined and photographed using an electron microscope (model H-700H, Hitachi, Tokyo).
Protein Extraction and Immunological Detection
For segregation analysis, 50 mature grains from each primary transgenic rice plant (T0), which accumulated significant amounts of normal or modified glycinins, were ground separately with a mortar and pestle in 62.5 mm Tris-HCl buffer (pH 6.8) containing 2% (w/v) SDS, 30% (v/v) glycerol, 0.1 m ME, 1.5 mm PMSF, and 1 mm EDTA at 4°C. Aliquots were then spotted on a nitrocellulose membrane and screened immunologically with anti-glycinin serum. The glycinin accumulation levels were estimated by comparing the densitometry signals obtained from extracts prepared from the transgenic plants against those of known amounts of purified glycinin.
Analysis of Self-Assembly by Suc Density Gradient Centrifugation
Soluble protein fractions were obtained by extracting 100 mg of mature rice seeds in 35 mm K-Pi buffer (pH 7.6) containing 0.4 m NaCl, 1.5 mm PMSF, and 1 mm EDTA (buffer B). A sample was then resolved by Suc density gradient centrifugation and the fractions analyzed by SDS-PAGE followed by immunoblotting as described previously (Utsumi et al., 1993b). The 2S, 7S, and 11S fractions purified from soybean (Glycine max) seeds according to the method of Thanh and Shibasaki (1976) were run in parallel as size markers.
Solubility Analysis of Glycinin Accumulated in Rice Seeds
Rice seeds (18 mg) were finely ground and proteins were sequentially extracted six times for 1 h each with 1.8 mL of saline solution (buffer B) containing 25 mm NEM with vigorous shaking at room temperature. The residual proteins were then extracted three times with 1.8 mL of 1% (v/v) lactic acid containing 1 mm EDTA and 25 mm NEM.
Renaturation Analysis of Reduced-Denatured Glutelin, Glycinin, and Proglycinin
Finely ground rice seeds (1.5 g) were sequentially extracted with 10 mL of buffer B containing 25 mm NEM followed by 10 mL of 55% (v/v) n-propyl alcohol to solubilize albumins, globulins, and prolamins. The residue was then extracted with 10 mL of 1% (v/v) lactic acid containing 1 mm EDTA and 25 mm NEM from the insoluble fraction to yield an enriched glutelin fraction. Glycinins were purified from soybean as described by Nagano et al. (1992). Recombinant proglycinin A1aB1b was purified as described previously (Kim et al., 1990a).
Glutelin, glycinin, and recombinant proglycinin A1aB1b (all at 100 μg/mL), and mixtures (100 μg each/mL) of glutelin with glycinin, recombinant proglycinin A1aB1b, ovalbumin (Sigma) or BSA (Sigma), and zein (Nacalai, Kyoto) with glycinin or proglycinin A1aB1b were denatured and reduced in 35 mm K-Pi buffer (pH7.6) containing 1 mm EDTA, 1 mm PMSF, 6 m urea, and 0.1 m ME. The denatured and reduced proteins were renaturated using a two-step dialysis method (Katsube et al., 1998a) based on the procedures of Utsumi et al. (1980) and Hirose et al. (1989). The denatured and reduced samples were dialyzed against 35 mm K-Pi buffer (pH 7.6) containing 0.1 m NaCl, 40% (w/v) glycerol, and 0.1 m ME for 12 h at 4°C, followed by dialysis for 24 h at 20°C, and then against the same buffer without ME for 24 h at 20°C. The renatured samples were dialyzed against 35 mm K-Pi buffer (pH 7.6) containing 0.1 m NaCl. The supernatant and the precipitate fractions of the dialysate were subjected to SDS-PAGE.
RESULTS
Transformation of Rice with Chimeric Normal and Modified Glycinin Genes
Normal and modified preproglycinin A1aB1b cDNAs composed of 27-bp 5′ noncoding, 1,488- or 1,506-bp coding, and 189-bp 3′ noncoding sequences were fused to the 5′-flanking region (−1,302 and +18 nucleotides from the transcriptional start site) of the rice storage protein glutelin GluB-1 gene (Fig. 1). These chimeric constructs were introduced into rice protoplasts by electroporation or into rice callus cells by A. tumefaciens-mediated transformation, and regenerated plants were obtained.
Figure 1.
Schematic representation of the normal (A) and Met-modified (B) glycinin chimeric genes. White and shaded boxes indicate the coding region and the 5′ and 3′ noncoding regions of glycinin cDNA, respectively. Thin and light-shaded lines depict vector DNA. Arrows indicate direction of transcription. 35S Pro, CaMV 35S promoter; Bar, the bialaphos resistance gene; Nos Ter, nopaline synthase gene terminator; g7, gene 7 terminator; Glutelin Pro, GluB-1 promoter; Glycinin cDNA, preproglycinin A1aB1b cDNA; Glutelin Ter, GluB-1 terminator; R, right T-DNA border; L, left T-DNA border.
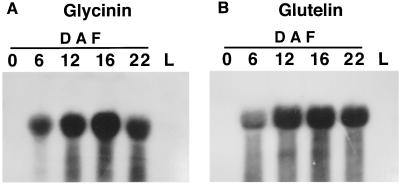
To investigate the temporal- and spatial-specific expression of the introduced chimeric genes, total RNA was prepared from developing seeds and leaves of T2 transgenic homozygous plants. As shown in Figure 2A, glycinin mRNA levels increased from 6 to 16 DAF, reached a plateau at 16 DAF, and then gradually decreased at 22 DAF. These profiles were identical to those of the glutelin GluB mRNA detected by GluB-1 cDNA (Fig. 2B), indicating that the glycinin gene was expressed specifically in the maturing seeds under the control of the GluB-1 promoter. Glycinin mRNAs were not detected in leaves or any nonseed tissues. There was no apparent difference in temporal and spatial patterns between the normal and modified glycinin constructs (data not shown).
Figure 2.
Northern analysis of developing seed and leaf RNAs isolated from transgenic rice plants. Total RNAs were isolated and fractionated by electrophoresis on a 1.2% agarose gel containing formaldehyde and then blotted onto nylon membranes. The RNAs were hybridized with the coding regions of preproglycinin A1aB1b cDNA (A) or proglutelin GluB-1 cDNA (B). L, Leaf.
Screening of Transgenic Rice Plants Accumulating Soybean Glycinin and Its Localization
The insertion of normal and modified glycinin genes into the rice genome of bialaphos-resistant plants was verified by PCR analysis. The accumulation levels of glycinin in individual plants (total of 70 plants) were roughly estimated immunologically by tissue printing. Most seeds were positively stained with anti-glycinin sera, although the antigen levels varied considerably among independent plants. Transgenic lines 11[pGluB1Gly] and 417[pGluB1GlyIV] exhibited the highest accumulation level among independent transgenic plants for each construct.
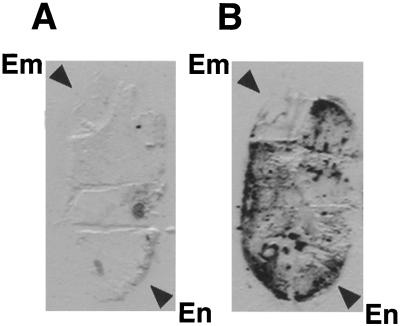
Immunological analysis of tissue prints prepared from longitudinally sectioned rice seeds revealed that glycinin was confined to the endosperm (Fig. 3). The same results were observed for seeds containing the modified glycinin. This accumulation pattern is consistent with the GUS staining pattern directed by the GluB-1 promoter (Wu et al., 1998a, 1998b), indicating that the accumulation pattern driven by the GluB-1 promoter was retained in the transgenic rice.
Figure 3.
Histochemical localization of glycinin in mature rice seeds. Seed section imprints from nontransgenic (A) and transgenic rice 11 [pGluB1Gly] (B) plants on nitrocellulose were visualized using anti-glycinin serum and a goat anti-rabbit IgG-alkaline phosphatase conjugate. Em, Embryo; En, endosperm.
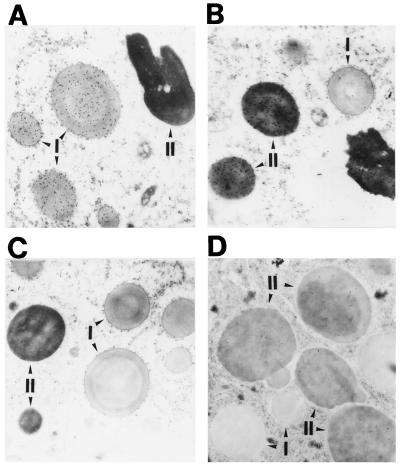
Glycinin is deposited into the protein storage vacuole in developing soybean seeds. Similarly, the rice glutelins are packaged in a vacuolar compartment called protein body II (Yamagata et al., 1982; Yamagata and Tanaka, 1986). Rice also contains a second storage site called protein body I, which contains the prolamins. Immunocytochemical studies at the electron microscope level were conducted to determine the intracellular location of glycinin in transgenic rice plants. Figure 4, A and B, shows the separate accumulation and packaging of prolamins and glutelins into protein bodies I and II, respectively. A similar immunocytochemical analysis for the soybean glycinin showed that the gold particles that denote reactivity with anti-glycinin were restricted to the matrix of the glutelin-containing protein body II. In addition to its absence in protein body I (Fig. 4D), immunogold particles were not evident on the ER or the Golgi apparatus, presumably due to the low concentration of glycinin in these organelles. These results indicate that glycinins are correctly targeted to the matrix of protein body II. Moreover, similar results were obtained for the modified glycinin, indicating that the modification did not cause mistargeting.
Figure 4.
Immunolocalization of accumulated glycinin in transgenic rice endosperm. Sections of embedded tissue were labeled with anti-prolamin (A), anti-glutelin (B), or anti-glycinin (C and D) sera, followed by gold-conjugated anti-rabbit IgG (A and B, 30 nm gold particles; C and D, 10 nm). A, B, and D, Transgenic rice 11[pGluB1Gly]; C, nontransgenic rice.
Progeny Segregation Analysis of the Expression Level
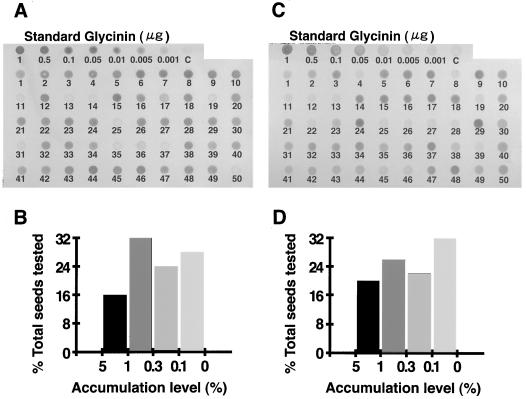
Total proteins extracted from 50 T1 seeds of the transgenic plants 11[pGluB1Gly] and 417[pGluB1GlyIV] were spotted onto nitrocellulose membranes and glycinin levels were estimated immunologically (Fig. 5, A and C). The highest levels of both normal and modified glycinins were around 5% of total proteins. Comparison of the relative accumulation levels of 50 grains within each plant indicated that the ratio of the grains with high (1%–5%), medium (0.3%–1.0%), low (0.1%–0.3%), and little (<0.1%) expression was approximately 1:1:1:1, as shown in Figure 5, B and D. The bialaphos resistance trait segregated into a 3:1 ratio of resistant to sensitive seeds. These segregation patterns are consistent with the view that the transgene was inserted in the rice genome at a single locus or closely linked loci. This was confirmed by Southern analysis of genomic DNA isolated from selected transgenic plants, which showed that one to a few copies of the intact fragment were integrated into the rice genome (data not shown).
Figure 5.
Comparison of the accumulation levels of normal and Met-modified glycinins in individual rice seeds by dot immunoblotting. Extracts (10 μg of proteins) of matured seeds from transgenic rice plants 11[pGluB1Gly] (A) and 417[pGluB1GlyIV] (C) were analyzed by immunoblotting. Between 0.001 and 1 μg of purified glycinin mixed with 10 μg of proteins of seed extract from nontransgenic rice plant was used as standard. The number below each dot indicates an individual seed. Frequencies of the accumulation levels of the expressed proteins in the individual seeds of 11[pGluB1Gly] (B) and 417[pGluB1GlyIV] (D) were calculated from dot blots.
Ten T2 generation seeds, each derived from T1 plants that gave the highest accumulation of normal and modified glycinins by half-seed analysis, were analyzed for the glycinin accumulation level, and the degree of glycinin accumulation was stably inherited in the third generation (data not shown).
Assembly of Glycinin in Rice Seeds
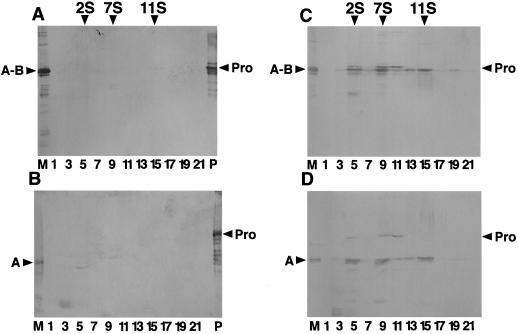
To investigate the ability of glycinin to self-assemble into hexamers in rice seeds, the salt-soluble proteins from the nontransgenic rice seeds and the T2 seeds of 11[pGluB1Gly] and 417[pGluB1GlyIV] were subjected to Suc density gradient centrifugation. After fractionation, proteins in each fraction were analyzed by SDS-PAGE in the absence of ME, followed by immunoblotting using anti-glycinin serum (Fig. 6, A and C). No immunoreactive bands were detected in the extract from nontransgenic rice seeds (Fig. 6A). The band with a molecular size similar to that of the mature disulfide-bonded subunit of glycinin was mainly observed in fractions 5, 9, and 15 of the extract from the transgenic rice seeds, corresponding to a size of 2S (monomer), 7S to 8S (trimer), and 11S to 12S (hexamer), respectively (Fig. 6C). In addition, the band with a molecular size similar to that of the proglycinin subunit expressed in E. coli (which is larger than that of mature glycinin) was detected in fractions 5 and 9 to 11. Soluble protein extracts from 417[pGluB1Gly IV] gave similar sedimentation patterns of 2S, 7S to 8S, and 11S to 12S, as shown in Figure 6C. These results indicate that newly synthesized glycinins assemble into trimeric and hexameric oligomers in a manner similar to that observed in developing soybean seeds.
Figure 6.
Analysis of the assembly of the normal glycinins accumulated in the seeds of the transgenic rice plant. Seed extracts were subjected to centrifugation on a 12-mL, 10% to 30% (w/v) linear Suc density gradient and analyzed by SDS-PAGE in the absence (A and C) or presence (B and D) of ME followed by immunoblotting. Sedimentation is from left to right. Sedimentation standards in Svedberg units are given. Arrowheads indicate the positions of recombinant proglycinin (Pro) synthesized in E. coli (lane P) and native mature glycinin (A-B; lane M). A and B, Nontransgenic rice; C and D, 11[pGluB1Gly].
To investigate whether the expressed glycinin was processed into mature form, protein fractions from the Suc density gradients were analyzed by SDS-PAGE in the presence of ME. Although immunoreactive bands were not detected in the extract from nontransgenic rice seeds (Fig. 6B), the band with a molecular size corresponding to the acidic polypeptide of mature glycinin was observed in fractions 5, 9, and 15 of the extract from the transgenic rice seeds, with the concomitant disappearance of the mature disulfide-bonded subunit (Fig. 6D). The band with a molecular size corresponding to proglycinin was also detected in fractions 5 and 9 to 11. These results indicate that processing from proglycinin to the mature form occurred in transgenic rice seeds in a manner similar to that in soybean seed, although a part of glycinin remained as proglycinin in the state of a monomer or a trimer. The anti-proglycinin antibody reacted very weakly with the basic polypeptide.
Glutelins Display a Distinct Sedimentation Pattern
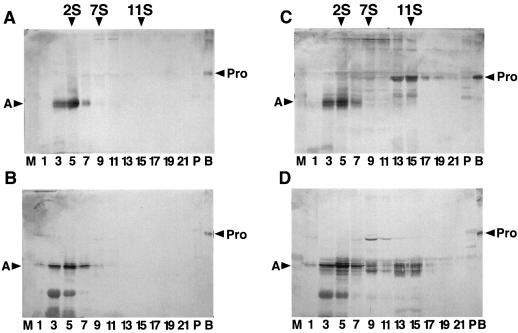
The processing and assembly of rice glutelins are poorly understood. To determine whether the rice glutelins are assembled into higher order oligomeric structures (as observed for the soybean glycinins), the same Suc density gradient fractions depicted in Figure 6, A and B, were analyzed by immunoblotting using anti-proglutelin GluA-1 serum. Trace amounts of the proglutelin were evident between the 2S and 7S regions of the Suc density gradient (Fig. 7A). The bulk of the soluble glutelin polypeptides was observed in the 2S region, with a molecular size comparable to the acidic subunit of 32 kD (Fig. 7A). Reactive glutelin polypeptides were essentially absent or at very low levels in the 7S and 11S regions of the Suc density gradients. When subjected to reducing conditions, bands corresponding to the acidic and basic polypeptides as well as a smaller reactive polypeptide band were evident (Fig. 7B). These results show that even though glutelin shares considerable homology with glycinin, the rice protein is processed and assembled in higher order structures by a different mechanism.
Figure 7.
Analysis of the formation of hybrid-11S storage protein complexes between glycinin and glutelin in seeds of transgenic rice plants. The same Suc density gradient fractions depicted in Figure 6 were analyzed by SDS-PAGE in the absence (A and C) and presence (B and D) of ME followed by immunoblotting using anti-proglutelin GluA-1 serum. Sedimentation is from left to right. Sedimentation standards are given in Svedberg units. Arrowheads indicate the positions of recombinant proglutelin (Pro) (lane B) and the native acidic polypeptide (A) of glutelin. One microgram each of recombinant proglycinin (lane P) and native mature glycinin (lane M) was electrophoresed as negative controls. A and B, Nontransgenic rice; C and D, 11[pGluB1Gly].
Formation of Hybrid Proteins between Glycinin and Glutelin
The sedimentation pattern of glutelin polypeptides was also investigated in the glycinin-expressing transgenic plants and yielded a much different pattern than that evident in normal plants. In addition to the polypeptide pattern at the 2S region observed earlier in nontransgenic plants, proglutelin polypeptides were evident at the 11S region with significantly smaller amounts at the 7S region of the Suc density gradient (Fig. 7C). In the presence of ME, the proglutelin polypeptides in 11S region dissociated into constituent polypeptides with a molecular size corresponding to the acidic polypeptides (Fig. 7D). Likewise, the bulk of the proglutelin in the 7S region dissociated into constituent polypeptides but a proportion remained undissociated, indicating intact unprocessed proglutelin. These results indicate that glutelin polypeptides assembled with glycinin proteins to form hetero-trimeric (7S) and hetero-hexameric (11S) structures. The amount of glutelin forming hybrid globulins with glycinin was estimated by densitometric measurements to be around 5% of the total glycinin extracted with a neutral saline solution.
Rice glutelins are composed of two classes, class A and class B. To examine whether polypeptides of both glutelin classes or only a single class can assemble with glycinin, proteins from the 11S fraction of the Suc density gradient (from 11[pGluB1Gly]) were examined by immunoblot analysis using anti-proglutelin GluA-1 and GluB-1 sera (Fig. 8). Anti-proglutelin GluA-1 serum reacts strongly with proglutelin GluA-1 and weakly with GluB-1 (Fig. 8A, lanes 1 and 2), whereas the reactivity of anti-proglutelin GluB-1 serum is just the opposite (Fig. 8B, lanes 1 and 2). Immunoblot analysis indicated that both antibodies reacted to glutelin polypeptides to about the same level irrespective of the absence or presence of ME (Fig. 8, lanes 4 and 6). These results indicate that glutelins A and B contributed equally to the formation of hybrid globulins with glycinin subunits.
Figure 8.
Analysis of the glutelin type involved in the hybrid globulin in seeds of transgenic rice plants. Proteins in fraction 15 in Figure 7 were analyzed immunologically using antibodies specific for GluA-1 (A, lanes 3–6) and GluB-1 (B, lanes 3–6) glutelin polypeptides. GluA-1 (100 ng, lane 1) and GluB-1 (100 ng, lane 2) were electrophoresed as positive controls. Lanes 1 to 4 were electrophoresed in the absence of ME; lanes 5 and 6 in the presence of ME. Lanes 3 and 5, Nontransgenic rice; lanes 4 and 6, 11[pGluB1Gly].
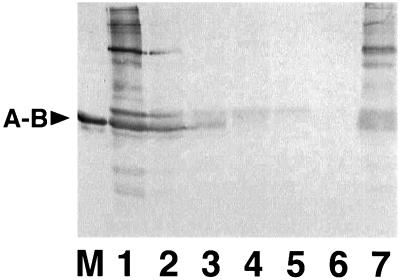
Glycinin was extracted repeatedly from the seeds of transgenic rice to examine whether glycinin is incorporated into the glutelin fraction. Glycinin was effectively solubilized by neutral saline solution after five extractions (Fig. 9, lane 6). When the residual protein fraction was extracted with dilute lactic acid solution, significant levels of glycinins were released (Fig. 9, lane 7), suggesting that glycinin also assembled with glutelins to form an insoluble complex. The amount of glycinin in the insoluble fraction was estimated to be around 30% of the total accumulated glycinin.
Figure 9.
Solubility analysis of glycinins accumulated in the seeds of transgenic rice plants. Seed proteins were extracted six successive times with saline solution followed by three successive times with lactic acid solution. Each saline extract (lanes 1–6) and a mixture of the lactic acid solution extract (lane 7) were electrophoresed in the absence of ME and then analyzed by immunoblotting using anti-glycinin serum. A-B denotes the position of the native mature glycinin (lane M).
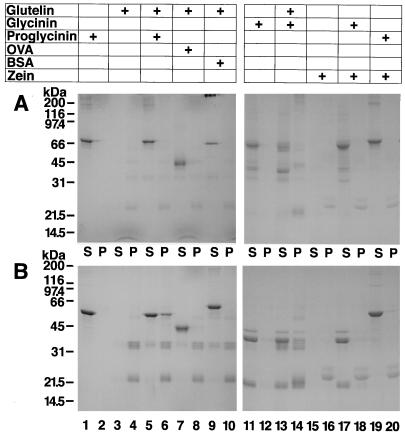
The results obtained here strongly suggest that glycinin and glutelin subunits form hybrid-oligomeric complexes in addition to their homologous assembly in the transgenic rice seeds. Alternatively, there is also the possibility that the formation of hybrid proteins is caused by nonspecific disulfide bonding and/or hydrophobic interaction. We analyzed the renaturation behavior of glutelin, glycinin, and proglycinin and mixtures of glutelin with either glycinin, proglycinin, ovalbumin or BSA, and glycinin or proglycinin with zein by SDS-PAGE in the absence (Fig. 10A) and presence (Fig. 10B) of ME. Ovalbumin contains four free Cys residues and one disulfide bond, and BSA contains one free Cys residue and 17 disulfide bonds. Both of these proteins are soluble in a neutral saline solution. The latter protein binds fatty acids and bile acids, indicating that the molecular surface has hydrophobic patches.
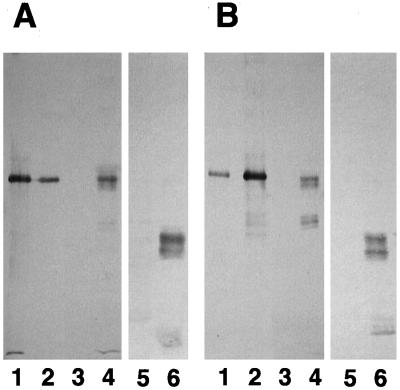
Figure 10.
Renaturation of glutelin, glycinin, and proglycinin either alone or in combination with other proteins. Purified glutelin, glycinin, or proglycinin was denatured and reduced in urea solution containing ME and then slowly renatured by dialysis either alone or in combination with other proteins. The dialysates were then centrifuged to obtain supernatant (S) and pellet (P) fractions, which were analyzed by SDS-PAGE in the absence (A) or presence (B) of ME. Lanes 1 and 2, Proglycinin; lanes 3 and 4, glutelin; lanes 5 and 6, mixture of glutelin and proglycinin; lanes 7 and 8, mixture of glutelin and ovalbumin (OVA); lanes 9 and 10, mixture of glutelin and BSA; lanes 11 and 12, glycinin; lanes 13 and 14, mixture of glutelin and glycinin; lanes 15 and 16, mixture of glycinin and zein; lanes 17 and 18, mixtures of proglycinin and zein. Protein molecular size markers were loaded onto lane M (200, 116, 97.4, 66, 45, 31, 21.5, and 14.5 kD).
Zein is not soluble in a neutral saline solution and contains a variable number of Cys residues (1-15) depending on the zein species (Shewry and Tatham, 1990). These proteins were denatured and reduced in urea containing ME and slowly renatured by dialysis as outlined in Methods. Upon renaturation, the bulk of the glutelin formed a precipitate with a small amount of acidic and basic polypeptides present in the supernatant (Fig. 10, lanes 3 and 4). This pattern was more readily apparent when the proteins were resolved by SDS-PAGE in the presence (Fig. 10B) of ME than in its absence (Fig. 10A). All of the glycinin (Fig. 10, lanes 11 and 12) and proglycinin A1aB1b (Fig. 10, lanes 1 and 2) renatured in a soluble form. However, when glutelin was renatured along with glycinin (Fig. 10, lanes 13 and 14) or proglycinin A1aB1b (Fig. 10, lanes 5 and 6), about one third of the total glycinin and proglycinin A1aB1b was recovered in the insoluble fraction.
These results suggest that glycinin and proglycinin A1aB1b were incorporated into the insoluble glutelin network through disulfide bonding. In contrast, much of the ovalbumin and BSA was recovered in soluble fraction when renatured in the presence of glutelin (Fig. 10, lanes 7–10), indicating no significant interaction among these proteins. Likewise, most of zein was recovered in the insoluble fraction and did not interact with glycinin (Fig. 10, lanes 15 and 16) or proglycinin (Fig. 10, lanes 17 and 18). These results suggest that the formation of hybrid proteins between glutelin and glycinin is through specific interactions, especially in the case of insoluble hybrid proteins through specific disulfide bonding of Cys residues.
DISCUSSION
In this study, we expressed the soybean glycinin genes in rice to determine whether they would be synthesized, processed, and assembled into higher order oligomeric structures, as was observed in developing soybean seeds. Our results clearly demonstrate that the majority (70%) of the newly synthesized glycinin molecules accumulate as salt-soluble proteins (globulins) and are assembled into trimeric (7S) and hexameric (11S) structures. Moreover, the glycinin polypeptides are properly processed into acidic and basic polypeptides interlinked by disulfide bonding in a manner similar to that in soybean embryonic tissue and cotyledons. In contrast, rice glutelins do not appear to be assembled into such oligomeric structures, as the only soluble form was proteolytically processed and sedimented at the 2S region of the Suc density gradient. Since proteolytic processing has been suggested to occur in the protein storage vacuole, these results indicate that glutelins are transported either as a monomer or as aggregates much larger than 11S. Alternatively, glutelins are assembled into trimers and hexamers similar to glycinin, but the transport from the ER to the protein storage vacuole and the processing step occurs at a much faster rate than for glycinins, resulting in low levels of these higher order oligomeric forms.
An important finding of this study was that a portion of the trimers and hexamers resolved by Suc density gradient centrifugation were hybrid globulins containing both glutelin and glycinin subunits. About 30% of newly synthesized glycinin molecules formed hybrid glutelins through specific interactions with glutelin subunits. Thus, glycinin and glutelin have the capacity to assemble with one another to form higher order protein structures. It is known that the formation of glycinin hexamers requires the processing of proglycinin to the mature form (Dickinson et al., 1989). Both proglycinin and proglutelin were observed in the trimer fraction but not in the hexamer fraction, indicating that glycinin and glutelin assemble into a trimeric and hexameric structural conformation very similar to the homoglycinin oligomers. These common structural features together with the similarities in their processing and sorting mechanisms further supports the hypothesis that these proteins are derived from a common ancestral gene (Takaiwa et al., 1987; Utsumi, 1992). Moreover, it may be concluded that transport of the rice glutelin occurs by an ordered process of protein folding at the secondary, tertiary, and quaternary levels, much like what occurs with soybean glycinin.
Assembly and processing of glycinins synthesized in rice were not complete, as shown in Figure 6. Glycinin monomers, trimers, and hexamers were observed in the proportion around 1:1:1, and unprocessed proglycinins were observed in the monomer and trimer fractions. A similar pattern of insufficient assembly and processing was observed for glycinins expressed under the control of the GluB-1 promoter in the endosperm tissue of tobacco seeds (Takaiwa et al., 1995). In contrast, the assembly and processing of glycinin into 11S species was much more efficient when the transgene was expressed under the control of CaMV 35S promoter in transgenic tobacco seeds. The CaMV 35S promoter drives maximum levels of glycinin expression of only approximately 0.1% of the total proteins (Utsumi et al., 1993b) compared with the stronger GluB-1 promoter, which yields up to 4% to 5% of the total proteins in rice or tobacco (Takaiwa et al., 1995). As the formation of the 11S hexamer is ultimately dependent on the transport of the 7S trimer to the protein storage vacuole, these observations suggest that the endomembrane system of rice and tobacco endosperm cells has a limiting capacity for protein transport to the vacuole.
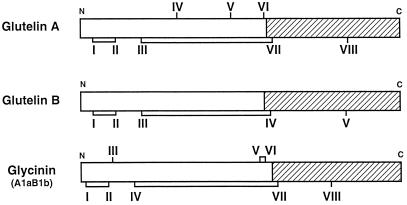
Glutelin subunits are classified into two types, GluA and GluB, on the basis of their amino acid sequences (Takaiwa et al., 1991). GluA and GluB contain eight and five Cys residues, respectively. Four of these Cys residues are conserved in the glutelins and in all of the 11S globulins found in legume and nonlegume seeds (Katsube et al., 1998b). These four conserved Cys residues form two disulfide bonds; one is an interchain bond between the acidic (Cys residue III of both types) and a basic polypeptide (VII of GluA and IV of GluB), while the other is an intrachain disulfide bond in the acidic polypeptide (I and II of both types) (Fig. 11) (Utsumi, 1992; Katsube et al., 1998b). Cys residues VIII of GluA and V of GluB are probably free, since most of glycinin-type proteins have a free Cys residue at the homologous site; e.g. cysteine residue VIII of glycinin A1aB1b (Katsube et al., 1998b).
Figure 11.
Schematic representation of the proposed positions of free Cys residues and disulfide bonds in glutelins A and B and glycinin A1aB1b. White and hatched areas are the acidic and the basic polypeptides, respectively. N and C represent the NH2 and COOH termini, respectively. The Cys residues are numbered by Roman numerals from the NH2 terminus. Disulfide bonds are represented by lines connecting two Cys residues.
We had speculated that some or all of the Cys residues IV, V, and VI of GluA contribute to the formation of multiple interchain disulfide bonds, resulting in the formation of insoluble glutelin aggregates, and that GluB is able to assemble into hexamers together with glycinin subunits (Katsube et al., 1998b). However, we observed both types of glutelins can assemble with glycinin to form hexamer structures (Fig. 8). This observation does not eliminate the possibility that Cys residues are responsible for the formation of glutelin aggregate, but indicates that the interaction of glutelin with glycinin is independent of the number of Cys residues in the glutelin primary sequences.
This glutelin-glycinin interaction not only results in soluble protein complexes but insoluble ones as well, since glycinin is observed in the glutelin fraction. In the highest glycinin-expressing transgenic rice seeds, glutelin is present at around 15-fold excess over glycinin. Therefore, we can speculate that: Interaction between glycinin subunits is stronger than that between glycinin and glutelin subunits, since homoglycinin is dominant (around 70% of the expressed glycinin), and once a glycinin subunit interacts with a glutelin subunit, the formation of hybrid globulin or glutelin proceeds depending on the population of glycinin and glutelin. Since glutelin levels are much larger than glycinin levels, the proportion of glycinin in the hybrid-glutelin complexes is much larger than in the hybrid globulin.
It has been demonstrated that soybean globulins exhibit a significant hypocholesterolemic effect if the dietary intake of these proteins is more than 6 g/d (Imura et al., 1996). Glycinin is likely to be primarily responsible for this effect on cholesterol levels (Makino et al., 1988; Sugano, 1996). Since glycinin accounts for around 40% of the soybean globulins, a dietary intake of 2.4 g/d of glycinin would be required. Seeds of 11[pGluB1Gly] contain 8% protein (Momma et al., 1999), including 400 mg of glycinin/100 g seeds. Since the average daily consumption of rice in Japan is 200 to 250 g, a transgenic rice plant containing 3 times higher glycinin than our present highest level would be necessary to fulfill this dietary requirement. By exploiting the processing and assembly of glycinin proteins into homo- and hetero-hexameric structures and into insoluble aggregates with glutelin, such heterologous expression levels in rice may be achieved in the near future.
Abbreviations:
- DAF
days after flowering
- ME
2-mercaptoethanol
- NEM
N-ethylmaleimide
Footnotes
This work was supported in part by grants from the Program for Promotion of Basic Research Activities for Innovative Biosciences (to S.U. and F.T.), the Ministry of Agriculture, Forestry, and Fisheries of Japan (to T.K., S.U., and F.T.), and Takano Life Science Research Foundation (to S.U.).
LITERATURE CITED
- Caplan A, Dekeyser R, Montagu MV. Selectable markers for rice transformation. Methods Enzymol. 1992;216:426–441. doi: 10.1016/0076-6879(92)16039-m. [DOI] [PubMed] [Google Scholar]
- Dickinson CD, Hussein EH, Nielsen NC. Role of postranslational cleavage in glycinin assembly. Plant Cell. 1989;1:459–469. doi: 10.1105/tpc.1.4.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Goto F, Yoshihara T, Shigemoto N, Toki S, Takaiwa F. Iron fortification of rice seed by the soybean ferritin gene. Nat Biotechnol. 1999;17:282–286. doi: 10.1038/7029. [DOI] [PubMed] [Google Scholar]
- Hinz G, Menze A, Hohl I, Vaux D. Isolation of prolegumin from developing pea seeds: its binding to endomembranes and assembly into prolegumin hexamers in the protein storage vacuole. J Exp Bot. 1997;48:139–149. [Google Scholar]
- Hirose M, Akuta T, Takahashi N. Renaturation of ovotransferrin under two-step conditions allowing primary folding of the fully reduced form and the subsequent regeneration of the intramolecular disulfides. J Biol Chem. 1989;264:16867–16872. [PubMed] [Google Scholar]
- Imura T, Tanaka M, Watanabe T, Kudo S, Uchida T, Kanazawa T. Effect of soy protein on serum lipid in adult volunteers (in Japanese) Ther Res. 1996;17:573–578. [Google Scholar]
- Katsube T, Gidamis AB, Kanamori J, Kang IJ, Utsumi S, Kito M. Modification tolerability of the hypervariable region of soybean proglycinin. J Agric Food Chem. 1994;42:2639–2645. [Google Scholar]
- Katsube T, Kang IJ, Takenaka Y, Adachi M, Maruyama N, Morisaki T, Utsumi S. N-Glycosylation does not affect assembly and tartgeting of proglycinin in yeast. Biochim Biophys Acta. 1998a;1379:107–117. doi: 10.1016/s0304-4165(97)00082-2. [DOI] [PubMed] [Google Scholar]
- Katsube T, Maruyama N, Takaiwa F, Utsumi S (1998b) Food protein engineering of soybean proteins and development of soy-rice. In PR Shewry, JA Napier, P Davis, eds, Engineering Crop Plants for Industrial End Uses. Portland Press, London, pp 65–76
- Kim C-S, Kamiya S, Kanamori J, Utsumi S, Kito M. High-level expression, purification and functional properties of soybean proglycinin from Escherichia coli. Agric Biol Chem. 1990a;54:1543–1550. [Google Scholar]
- Kim C-S, Kamiya S, Sato T, Utsumi S, Kito M. Improvement of nutritional value and functional properties of soybean glycinin by protein engineering. Protein Eng. 1990b;3:725–731. doi: 10.1093/protein/3.8.725. [DOI] [PubMed] [Google Scholar]
- Kito M, Moriyama T, Kimura Y, Kambara H. Changes in plasma lipid levels in young healthy volunteers by adding an extruder-cooked soy protein to conventional meals. Biosci Biotechnol Biochem. 1993;57:354–355. doi: 10.1271/bbb.57.354. [DOI] [PubMed] [Google Scholar]
- Krishnan HB, Franceschi VR, Okita TW. Immunochemical studies on the role of the Golgi complex in protein body formation in rice seeds. Planta. 1986;169:471–480. doi: 10.1007/BF00392095. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Makino S, Nakashima H, Minami K, Moriyama R, Takao S. Bile acid-binding protein from soybean seed: isolation, partial characterization and insulin-stimulating activity. Agric Biol Chem. 1998;52:803–809. [Google Scholar]
- Manteuffel RM, Panitz R. In situ localization of faba bean and oat legumin-type proteins in transgenic tobacco seeds by a highly sensitive immunological tissue print technique. Plant Mol Biol. 1993;22:1129–1134. doi: 10.1007/BF00028981. [DOI] [PubMed] [Google Scholar]
- Momma K, Hashimoto W, Ozawa S, Kawai S, Katsube T, Takaiwa F, Kito M, Utsumi S, Murata K. Quality and safety evaluation of genetically engineered rice with soybean glycinin: analyses of the grain composition and digestibility of glycinin in transgenic rice. Biosci Biotechnol Biochem. 1999;63:314–318. doi: 10.1271/bbb.63.314. [DOI] [PubMed] [Google Scholar]
- Müntz K. Deposition of storage proteins. Plant Mol Biol. 1998;38:77–99. [PubMed] [Google Scholar]
- Nagano T, Hirotsuka M, Mori H, Kohyama K, Nishinari K. Dynamic viscoelastic study on the gelation of 7S globulin from soybeans. J Agric Food Chem. 1992;40:941–944. [Google Scholar]
- Neuhaus J-M, Rogers JC. Sorting of proteins to vacuoles in plant cells. Plant Mol Biol. 1998;38:127–144. [PubMed] [Google Scholar]
- Okita TW, Rogers JC. Compartmentation of proteins in the endomembrane system of plant cells. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:327–350. doi: 10.1146/annurev.arplant.47.1.327. [DOI] [PubMed] [Google Scholar]
- Osborne TB (1924) The Vegetable Proteins. Longmans, London
- Robinson DG, Hinz G, Holstein SEH. The molecular characterization of transport vesicles. Plant Mol Biol. 1998;38:49–76. [PubMed] [Google Scholar]
- Saalbach G, Jung R, Kunze G, Saalbach I, Adler K, Müntz K. Different legumin protein domains act as vacuolar targeting signals. Plant Cell. 1991;3:695–708. doi: 10.1105/tpc.3.7.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry PR, Tatham AS. The prolamin storage proteins of careal seeds: structure and evolution. Biochem J. 1990;267:1–12. doi: 10.1042/bj2670001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Hermodson MA, Nielsen NC. Identification of the cystines which link the acidic and the basic components of the glycinin subunits. J Biol Chem. 1984;259:13431–13435. [PubMed] [Google Scholar]
- Sugano M. Soybean proteins and their hypocholesterolemic effect (in Japanese) Shokuhin-kogyo. 1996;39(18):59–68. [Google Scholar]
- Tada Y, Sakamoto M, Fujimura T. Efficient gene introduction into rice by electroporation and analysis of transgenic plants: use of electroporation buffer lacking chloride ions. Theor Appl Genet. 1990;80:475–480. doi: 10.1007/BF00226748. [DOI] [PubMed] [Google Scholar]
- Takaiwa F, Katsube T, Kitagawa S, Higasa T, Kito M, Utsumi S. High level accumulation of soybean glycinin in vacuole-derived protein bodies in the endosperm tissue of transgenic tobacco seed. Plant Sci. 1995;111:39–49. [Google Scholar]
- Takaiwa F, Kikuchi S, Oono K. A rice glutelin family-A major type of glutelin mRNAs can be divided into two classes. Mol Gen Genet. 1987;208:15–22. [Google Scholar]
- Takaiwa F, Kikuchi S, Oono K. The complete nucleotide sequence of new type cDNA coding for rice storage protein glutelin. Nucleic Acids Res. 1989;17:3289. doi: 10.1093/nar/17.8.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaiwa F, Oono K, Wing D, Kato A. Sequence of three members and expression of a new major subfamily of glutelin genes from rice. Plant Mol Biol. 1991;17:875–885. doi: 10.1007/BF00037068. [DOI] [PubMed] [Google Scholar]
- Takeuchi Y, Nishimura M. Studies on intracellular localization of proteins by immunoelectric microscopy using colloidal gold. Plant Cell Technol. 1993;5:115–123. [Google Scholar]
- Thanh VH, Shibasaki K. Major proteins of soybean seeds: a straightforward fractionation and their characterization. J Agric Food Chem. 1976;24:1117–1121. doi: 10.1021/jf60208a030. [DOI] [PubMed] [Google Scholar]
- Utsumi S. Plant food protein engineering. Adv Food Nutr Res. 1992;36:89–208. doi: 10.1016/s1043-4526(08)60105-9. [DOI] [PubMed] [Google Scholar]
- Utsumi S, Gidamis AB, Kanamori J, Kang IJ, Kito M. Effects of deletion of disulfide bonds by protein engineering on the conformation and functional properties of soybean proglycinin. J Agric Food Chem. 1993a;41:687–691. [Google Scholar]
- Utsumi S, Inaba H, Mori T. Formation of pseudo- and hybrid-11S globulins from subunits of soybean and broad bean 11S globulins. Agric Biol Chem. 1980;44:1891–1896. [Google Scholar]
- Utsumi S, Kitagawa S, Katsube T, Kang IJ, Gidamis AB, Takaiwa F, Kito M. Synthesis, processing and accumulation of modified glycinins of soybean in the seeds, leaves and stems of transgenic tobacco. Plant Sci. 1993b;92:191–202. [Google Scholar]
- Utsumi S, Matsumura Y, Mori T (1997) Structure-function relationships of soy proteins. In S Damodaran, A Paraf, eds, Food Proteins and Their Applications. Marcel Dekker, New York, pp 257–291
- Wu C-Y, Adachi T, Hatano T, Washida H, Suzuki A, Takaiwa F. Promoters of rice seed storage protein genes direct endosperm-specific gene expression in transgenic rice. Plant Cell Physiol. 1998a;39:885–889. [Google Scholar]
- Wu C-Y, Suzuki A, Washida H, Takaiwa F. The GCN4 motif in a rice glutelin gene is essential for endosperm-specific gene expression and is activated by Opaque-2 in transgenic rice plants. Plant J. 1998b;14:673–683. doi: 10.1046/j.1365-313x.1998.00167.x. [DOI] [PubMed] [Google Scholar]
- Yamagata H, Sugimoto T, Tanaka K, Kasai Z. Biosynthesis of storage proteins in developing rice seeds. Plant Physiol. 1982;70:1094–1100. doi: 10.1104/pp.70.4.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata H, Tanaka K. The site of synthesis and accumulation of rice storage proteins. Plant Cell Physiol. 1986;27:135–145. [Google Scholar]