Abstract
Selective inhibitors could help unveil the mechanisms by which inhibition of poly(ADP-ribose) polymerases (PARPs) elicits clinical benefits in cancer therapy. We profiled 10 clinical PARP inhibitors and commonly used research tools for their inhibition of multiple PARP enzymes. We also determined crystal structures of these compounds bound to PARP1 or PARP2. Veliparib and niraparib are selective inhibitors of PARP1 and PARP2; olaparib, rucaparib, and talazoparib are more potent inhibitors of PARP1 but are less selective. PJ34 and UPF1069 are broad PARP inhibitors; PJ34 inserts a flexible moiety into hydrophobic subpockets in various ADP-ribosyltransferases. XAV939 is a promiscuous tankyrase inhibitor and a potent inhibitor of PARP1 in vitro and in cells, whereas IWR1 and AZ-6102 are tankyrase selective. Our biochemical and structural analysis of PARP inhibitor potencies establishes a molecular basis for either selectivity or promiscuity and provides a benchmark for experimental design in assessment of PARP inhibitor effects.
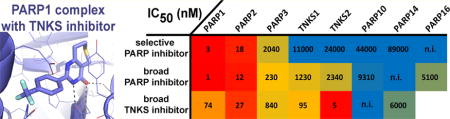
Graphical Abstract
INTRODUCTION
PARP inhibitors provide therapeutic opportunities in the treatment of various cancers as well as nononcologic conditions.1 Inhibition of poly(ADP-ribose) polymerase-1 and -2 (PARP1, PARP2) using olaparib is now clinical practice,2 and several other compounds are in late development.3,4 Several hallmarks of cancer are also affected by other PARP family members including PARP3,5,6 the tankyrases (TNKS1/PARP5a and TNKS2/PARP5b),7,8 PARP10,9 PARP13,10 and PARP14.11,12 Thus, there may be clinical application for inhibitors of PARP family members other than PARP1 and -2. Although it is clear that most cancer cells with defects in DNA damage response pathways are sensitive to PARP inhibitors, the mechanisms of cytotoxicity due to PARP inhibition are unknown.13,14 Thus, we do not understand whether, in therapeutic applications, broad inhibition of the PARP family is preferable over selective inhibition of single PARP enzymes. To be able to make full use of these chemical tools, we must gain a better understanding of PARP inhibitor effects, including off-target action.15
As a consequence of these considerations, development of compounds that inhibit PARP family members selectively is a priority. Good progress has been made with the tankyrases, and several cell-active, potent, and selective tankyrase inhibitors are now available.16–18 At the same time, attempts have been made to standardize in vitro assay technology and to characterize PARP inhibitors in terms of selectivity.19,20 However, most PARP inhibitors have yet been profiled only against a handful of family members, rarely including any representative of the mono-ADP-ribosyltransferase subfamily. Therefore, a more comprehensive, comparative analysis of widely used PARP and tankyrase inhibitors was needed to facilitate interpretation of experimental effects of these compounds including off-target effects within the PARP family.
Earlier, we have used Tm shift assays to investigate binding of 185 PARP inhibitors and chemically related compounds to 13 of the 17 human PARP family members.21 The resulting interaction map showed that most PARP inhibitors are rather unselective, in agreement with a recent study,19 but neither of these two investigations allowed precise potency ranking of compounds, and neither was based on enzymatic inhibition. To overcome these limitations, we characterized the enzymatic activities of those 11 PARP family members for which we detected ADP-ribosyltransferase activity and evaluated their inhibition by 10 important PARP and tankyrase inhibitors. Finally, using X-ray crystallography, we mapped the structural determinants for the potencies of these compounds.
RESULTS AND DISCUSSION
ADP-Ribosylation Assay
PARP inhibitor potencies have been estimated with a variety of methods including enzymatic inhibition, surface plasmon resonance, fluorescence polarization using custom-made probe compounds, and bead based assays. Our objective was to obtain a comparable measure of PARP inhibitor potencies for many PARP family members using an assay of enzymatic inhibition. First, we validated a previously published method22 to ensure unbiased addition of biotin-labeled NAD+ onto PARP substrate proteins and onto growing PAR polymers (Supporting Information, Figure S1). Next, we determined the KM values for NAD+ turnover by PARP1–4, the two tankyrases, and five members of the mono-ADP-ribosyltransferase subfamily (PARP6−16; Table 1, Supporting Information, Table S1 and Figure S2).
Table 1.
Kinetic Parameters of PARP Full Length Enzymes and Catalytic Domain Fragmentsa
| protein | construct | KMNAD+(μM) | KMNAD+ (literature) |
|---|---|---|---|
| PARP1 | FL | 0.78 ± 0.45 | 50 μM62 |
| PARP1 | ART | 94 ± 27 | 30−130 μM63–66 |
| PARP2 | FL | 1.9 ± 0.5 | 130 μM65 |
| PARP2 | ART | 159 ± 5 | |
| PARP3 | FL | 131 ± 57 | |
| PARP3 | ART | 2170 ± 645 | |
| PARP4 | ART | 92 ± 17 | |
| TNKS1 | ART | 31 ± 4 | |
| TNKS2 | ART | 251 ± 56 | |
| PARP10 | FL | 98 ± 11 | 50 μM67 |
| PARP10 | ART | 90 ± 27 | |
| PARP12 | ART | 299 ± 76 | |
| PARP14 | ART | 62 ± 7 | |
| PARP15 | ART | 11.0 ± 4.2 | |
| PARP16 | FL | 582 ± 196 | 280 μM68,69 |
Experimental data are shown in Supporting Information, Figure S2. FL, full length protein; ART, ADP-ribosyltransferase domain including the regulatory subdomain (in PARP1−4).
Catalytic Fragments of PARP1, -2, and -3 Have Lower Ligand Affinities than the Full Length Proteins
Many published inhibitor IC50 and Ki values have been obtained with PARP catalytic fragments rather than full length enzymes, and several commercial assay kits contain catalytic fragments. PARP1 crystal structures23,24 suggested that other domains might fold onto the catalytic domain, possibly stimulating enzymatic turnover by displacing the regulatory subdomain from its position blocking the NAD+-binding cleft, a mechanism that has recently been established.25 As most PARP family members are multidomain proteins, similar regulatory mechanisms likely exist, in particular for PARP2−4 with regulatory subdomains homologous to that of PARP1. Indeed, we find that the isolated catalytic domains of PARP1 and its nearest homologue, PARP2, have significantly lower enzymatic rates than the respective full length proteins (Figure 1A,B and Table 1). A similar result was obtained for PARP3, but not for PARP10, a mono-ADP-ribosyltransferase that lacks a regulatory subdomain (Figure 1B and Table 1).
Figure 1.

Enzymatic activities and inhibition of selected PARP enzymes and their isolated catalytic domains. (A) Nonlinear regression plots of the NAD+-dependent ADP-ribosylation rates of full length PARP1 and -2 and their catalytic domain fragments. (B) Domain arrangements of full length PARP1, -2, -3, and -10 proteins and the regions covered by the catalytic domain fragments. KM values (reported in detail in Table 1) are indicated by bars adjacent to each protein construct (black bars, full length proteins; gray bars, catalytic domain fragments). Domain designations: BRCT, BRCA1 carboxy terminal homology; PARP, ADP-ribosyltransferase domain; reg, regulatory subdomain; RRM, RNA recognition motif; SAM, sterile α motif domain; UIM, ubiquitin interacting motif; WGR, WGR-motif containing nucleic acid binding domain; ZnF, zinc finger domain. (C) Concentration response curves for in vitro inhibition of full length PARP1 and -2 and their catalytic fragments by olaparib. The IC50 value calculated from each data set is indicated. (D) Correlation of the IC50 values for olaparib, veliparib, rucaparib, and PJ34 determined using either full length enzymes or catalytic domain fragments of PARP1 (white), PARP2 (orange), and PARP10 (black). Full data are reported in Table 2 and Supporting Information, Figures S3−S8.
Having established that PARP1 and -2 catalytic domain fragments apparently have lower dinucleotide affinities than the full length enzymes, we asked whether their affinities for PARP inhibitors differ as well. Olaparib inhibited full length PARP1 with 10-fold higher potency than its catalytic fragment and full length PARP2 with 20-fold higher potency than its catalytic fragment (Figure 1C). Expanding this analysis on four different PARP inhibitors showed that this was generally true for PARP1 and -2 (Figure 1D). For PARP3, an inhibition analysis of the catalytic fragment was not meaningful owing to its very low activity. For PARP10, no correlation could be established, likely owing to low affinities of these PARP inhibitors for the mono-ADP-ribosyltransferase subfamily. We conclude that the use of PARP1, -2, and -3 catalytic fragments instead of full length enzymes may significantly underestimate PARP inhibitor potencies.
Veliparib and Niraparib are Selective Inhibitors of PARP1 and PARP2
We set up enzyme inhibition assays for each PARP enzyme in the presence of NAD+ at concentrations at or below their respective KM. Activity profiles were determined for the current clinical PARP inhibitors olaparib (AZD-2281, KU-0059436),26 veliparib (ABT-888),27 niraparib (MK-4827),28 rucaparib,29 talazoparib,30 the widely used research tools 1 (UPF1069)31 and PJ34,32 as well as the tankyrase inhibitors and effectors of Wnt signaling, XAV93933 and IWR-1.16 The results of this enzymatic profiling analysis are summarized in Figures 2, 3, and 4 and Supporting Information, and experimental data are reported in Supporting Information, Figures S3−S12. We went on to determine crystal structures of these inhibitors bound to either PARP1 or PARP2 (Figure 3 and 4 and Supporting Information, Table S3).
Figure 2.

In vitro potencies of PARP inhibitors. Schematic representation of PARP inhibitor potencies mapped on a phylogenetic tree of human PARP enzymes. Red sphere sizes are proportional, on a logarithmic scale, to IC50 values in the range 1 nM to 10 μM; red dots indicate IC50 values higher than 10 μM; black dots indicate no inhibition detected. Experimental data underlying these graphs are presented in Table 2 and Supporting Information, Figures S3−S9.
Figure 3.

Selective vs broad PARP inhibition and their structural basis. Concentration−response curves for PARP inhibitor dependent in vitro inhibition of full length PARP1,-2, -3, and-10 and the catalytic fragments of tankyrase-1 and -2. For clarity, plots are shown only for a selection of PARP family members; full details and IC50 values calculated from these data are reported in Table 2 and Supporting Information, Figures S3−S8. Symbol colors are explained in the legends and pertain to all panels. Homologous key side chains are shown in all structure panels to facilitate orientation. (A) Concentration−response curves for veliparib. (right panel) Crystal structures of veliparib bound to PARP1 (in pink; PDB 2RD6) and PARP2 (in green),37 with key side chain interactions indicated. A water mediated interaction with PARP2-E335 in the α-helical regulatory subdomain is conserved in PARP1 (involving D766) but not in any other PARP family member. A structure of PARP10 with veliparib shows the ligand bound in a similar orientation albeit without making interactions with the protein outside the nicotinamide pocket (Supporting Information, Figure S13). (B) Concentration−response curves for niraparib. (right panel) Crystal structure of niraparib bound to PARP1. Niraparib selectivity for PARP1 and -2 is rationalized by interactions with the regulatory subdomain via a hydrogen bond to D766 as for veliparib. (C) Concentration−response curves for olaparib. (right panel) Crystal structure of olaparib bound to PARP2. Olaparib forms several hydrogen bonds with backbone atoms in the catalytic domain including R444 and water mediated hydrogen bond with the D339 and van der Waals interactions with the aliphatic part of the E335 side chain in the regulatory domain. (D) Concentration−response curves for talazoparib. (right panel) Crystal structure of talazoparib bound to PARP1. Talazoparib efficiently fills the nicotinamide and N-ribose subpockets, forming a water mediated hydrogen bond with the catalytic residue E988 and van der Waals interactions on both sides of the cleft. (E) Concentration−response curves for rucaparib. (right panel) Crystal structure of rucaparib bound to PARP1 (pink) and TNKS2 (orange; PDB 4BJC). (F) Concentration−response curves for PJ34. (right panel) Crystal structure of PJ34 bound to PARP1 (pink) showing two conformations of the inhibitor’s dimethyl glycinamide moiety. PJ34 binding to PARP3 (cyan; PDB 3CE0), PARP15 (gray; PDB 3GEY), TNKS1 (yellow; PDB 3UH2), and TNKS2 (blue; PDB 4BJB) utilizes similar interactions in the nicotinamide subpocket but takes advantage of nonpolar features in the pocket by yet different conformations of the dimethyl glycinamide moiety.
Figure 4.

The potent tankyrase inhibitor XAV939 inhibits PARP1 with submicromolar potency in vitro and in cells. (A) Potencies of tankyrase inhibitors. Schematic representation of PARP inhibitor potencies mapped on a phylogenetic tree of human PARP enzymes. Data for IWR-1 inhibition of tankyrase-1 and all data for 3 were taken from previous publications.16,17 Experimental data underlying the remainder are presented in Table 3 and Supporting Information, Figures S10−S12. Red sphere sizes are proportional, on a logarithmic scale, to IC50 values in the range 1 nM to 10 μM; red dots indicate IC50 values higher than 10 μM; black dots indicate no inhibition detected. (B) Concentration−response curves for in vitro inhibition of selected PARP and tankyrase enzymes by XAV939 and IWR-1. (C) (left panel) Structural alignment of tankyrase-2 (gold; PDB 3KR8)47 and PARP1 (pink; reported here), both in complex with XAV939. Tankyrase-2 side chains that contribute to XAV939 interactions are shown. In the PARP1 α-helical regulatory domain (which is missing in the tankyrases), aliphatic side chains interacting with the trifluoromethyl group of XAV939 are shown. (right panel) Structural alignment of the tankyrase-2 IWR-1 complex70 (gold) with PARP1−XAV939 (pink). IWR-1 does not engage in interactions with the nicotinamide pocket but is bound in the adenine subsite. (D) Concentration−response fingerprints of PARP1 in HEK293 cells following preincubation with either XAV939 or olaparib. Both compounds engage PARP1 in cells. (E) Inhibition of PARP1 activity in H2O2-treated HEK293 cells preincubated with either XAV939 or PJ34.
One important finding of our inhibitor profiling analysis is the superior selectivity of veliparib (Figure 2 and Figure 3A) and niraparib (Figure 2 and Figure 3B) toward PARP1 and -2. With IC50 values >100-fold lower than those for other family members, veliparib is the only PARP1 and -2 inhibitor that meets chemical probe criteria.34,35 Because both veliparib and niraparib have been shown to be cell-active, this implies these compounds generate meaningful biological data pertaining to the functions of PARP1 and -2 activities. Both inhibitors make extensive interactions with the active sites of PARP1 and -2, including unique, both direct and water mediated hydrogen bonding, as well as the conserved interactions in the nicotinamide binding pocket shared with the vast majority of PARP inhibitors.36 Veliparib and niraparib form similar (water mediated respectively direct) interactions with the α-helical regulatory subdomains, involving either E763 or D766 of PARP1 and E335 of PARP2 (Figure 3A,B). These interactions form the basis for selectivity toward PARP1 and -2, as these carboxyl side chains are not conserved in any other PARP family member.37 We confirmed this by a crystal structure of the PARP10 complex with veliparib, in which the inhibitor is bound in the nicotinamide pocket without making further interactions with the protein (Supporting Information, Figure S13).
Olaparib and Talazoparib Are Potent but Unselective PARP1 Inhibitors
Consistent with previous studies,21,30,38,39 we find that olaparib and talazoparib are potent inhibitors of PARP1 (Figure 2 and Figure 3C,D). Olaparib and talazoparib are also somewhat selective toward PARP1 and -2, but only by a factor ~15−20 over other family members. Crystal structures explain the potencies of these compounds: Apart from conserved nicotinamide subpocket binding, olaparib forms several hydrogen bonds with backbone atoms in the catalytic domain and van der Waals interactions with the aliphatic part of the E335 side chain in the regulatory domain of PARP2 (Figure 3C). Talazoparib makes conserved interactions with the nicotinamide subpocket of PARP1 but also efficiently fills the rest of the dinucleotide pocket, forming a water mediated hydrogen bond with the catalytic residue E988 and van der Waals interactions on both sides of the cleft (Figure 3D). These results were corroborated by a very similar crystal structure.40
Rucaparib is the least selective clinical PARP1 inhibitor, and PJ34 is clearly the least selective PARP1 inhibitor overall in the panel (Figure 2 and Figure 3E,F). Comparison of our PARP1–rucaparib crystal structure with the previously published TNKS2 complex41 suggests flexibility in the terminal secondary amine tail that may facilitate different modes of interaction depending on the local environment (Figure 3E). A similar mechanism underlies the promiscuity of PJ34. In PARP1–PJ34 crystals, this inhibitor binds in slightly different orientations in the two molecules of the asymmetric unit, with the flexible dimethyl glycinamide moiety pointing into opposite directions (Figure 3F). This flexible moiety has been observed in yet a different conformation in PARP3,42 and binding of PJ34 to two distinct sites has been observed in tankyrase-2.43 We have solved a complex of PARP15−PJ34 earlier44 and a structure of Pseudomonas aeruginosa ExoA−PJ34 has also been published.45 Comparison of the crystal structure of the PARP1–PJ34 complex with those structures shows that the terminal dimethyl glycinamide moiety confers flexible van der Waals interaction propensity, enabling the compound to interact with nonpolar surfaces on either side of the NAD+ binding crevice. The mobility and physicochemical properties of the dimethyl glycinamide moiety enable PJ34 to interact with the local environments of various ADP-ribosyltransferases. Our results imply that PJ34 and rucaparib are particularly poor choices for probing the effects of selective inhibition of PARP1/2.
In agreement with previous analyses,19,46 we cannot confirm 1 as a selective inhibitor of PARP2 over PARP1 (Figure 2 and Supporting Information).
XAV939 Is Not a Specific Inhibitor of Tankyrases
The reportedly tankyrase selective compound XAV939 inhibited full length PARP1 and -2 in the midnanomolar concentration range (IC50 of 75 and 30 nM, respectively), which is very similar to its potency toward the tankyrases (IC50 of 95 and 5 nM, respectively; Figure 4 and Supporting Information). Comparison of the crystal structures of XAV939 in complex with PARP1 and tankyrase-2 reveals a similar anchoring to conserved glycines and orientation of the compound in the pockets (Figure 4C). The fluorinated aliphatic tail of XAV939 forms interactions with hydrophobic residues F1035, I1039, and Y1050 in tankyrase-2.47 Corresponding residues are missing in PARP1; instead, XAV939 interacts with aliphatic parts of E763 and D766 of the PARP1 regulatory subdomain.
These results underscore the importance of evaluating PARP family inhibitors using PARP1 full length protein: The discrepancy between the XAV939 potency determined here and that reported before is likely due to the fact that catalytic fragments were used for in vitro IC50 determinations.33 This interpretation is supported by recent determination of the affinities of XAV939 for PARP1 and -2 using a fluorescence displacement assay.19 In the original study, nanomolar inhibition of PARP2, the nearest homologue of PARP1, was reported.33 For cellular experiments, XAV939 was linked to a bulky dye,33 resulting in a derivative that might bind tankyrases but would most likely clash with the regulatory subdomain in PARP1 and its closest homologues.
The significance of PARP1 inhibition by XAV939 in cells was studied using a cellular thermal shift assay (CETSA) and an assay of inhibition of poly-ADP-ribosylation activity following DNA damage. A concentration−response fingerprint48 of XAV939 demonstrated target engagement for endogenous PARP1 in HEK293 cells (Figure 4D). Consistent with these CETSA results, we found that increasing concentrations of XAV939 inhibited poly-ADP-ribosylation induced by hydrogen peroxide, a potent inducer of DNA damage (Figure 4E). It is important to note that many studies use XAV939 at similar concentrations in cells to inhibit tankyrases. Therefore, we conclude that cellular effects of XAV939 cannot be interpreted in terms of tankyrase inhibition alone.
It is clear that selective inhibition of tankyrases can be achieved by leaving the nicotinamide pocket and expanding compounds into the adenosine subsite.41,49 Several compounds with these properties have been presented; for instance, IWR-1,16 2 (AZ-6102),18 and 3 (G007-LK; 4-[5-[(E)-2-[4-(2-chlorophenyl)-5-(5-methylsulfonylpyridin-2-yl)-1,2,4-triazol-3-yl]ethenyl]-1,3,4-oxadiazol-2-yl]benzonitrile)50 are highly selective over other PARP family members (Figure 4 and Supporting Information). Interestingly, these adenosine subsite binding ligands are unable to inhibit not only the regulatory subdomain containing PARP1−4 but also the mono-ADP-ribosyltransferases in the family. Targeting the adenine subsite with a different moiety, or targeting a different subsite altogether, may thus be a strategy toward selective mono-ADP-ribosyltransferase inhibitors.
CONCLUSIONS
Our results provide a benchmark for interpretation of the cellular effects of ten widely used PARP and tankyrase inhibitors. Previously, these effectors have regularly been used at micromolar concentrations, but effective cellular concentrations of PARP inhibitors are similar to our in vitro IC50 values.27,51–53 Thus, the results presented here provide relevant effector concentrations for experimental design to achieve either selective inhibition of PARP1 and -2 using veliparib, or broad inhibition of PARP enzymes using PJ34 or rucaparib. Moreover, they illustrate the importance of determining the relevant concentration regimes for PARP inhibitors used as chemical tools. Finally, previous findings based on the use of XAV939 in cells need to be re-evaluated in the light of our results revealing XAV939 as an inhibitor of PARP1 and -2.
EXPERIMENTAL SECTION
Recombinant Protein Production and Purification
Proteins were expressed in Escherichia coli strains BL21(DE3) pRARE or C41, as N- or C-terminal hexahistidine fusions. Full-length proteins for activity assays were purified by immobilized metal affinity chromatography (IMAC), and all other protein constructs were purified by IMAC followed by size exclusion chromatography as previously.21 Integrity of all protein batches was verified by LC-ESI-MS.
PARP Inhibitors
All compounds used were certified to be ≥95% pure based on NMR and HPLC analysis (see Supporting Information for details).
In Vitro ADP-Ribosylation Assay
PARP automodification or histone modification was measured essentially as described before,22 using a CLARIOstar (BMG Labtech) multiplate reader. For concentration–response experiments, NAD+ concentrations were adjusted to KM or lower for each enzyme construct. PARP inhibitors were added from stock solutions in DMSO followed by 15 min preincubation before addition of NAD+. The final concentration of DMSO was 1% in all reactions. DMSO solutions were stored at room temperature, and DMSO from the same batch was used in control reactions.
Cellular Thermal Shift Assay
CETSA was performed by Pelago Bioscience AB as described.54 In short, HEK293 cells were incubated with dilution series of compounds (or 1% DMSO as control). After 60 min incubation, cells were heated to 52 °C for 3 min and lysed, and lysates were cleared by centrifugation. Supernatants were analyzed by SDS-PAGE followed by Western Blotting using a mouse monoclonal HRP-conjugated anti-PARP1 antibody (Santa Cruz Biotechnology, sc-8007) for quantitation of endogenous full length PARP1 protein remaining in the soluble fraction.
Cellular PARP Activity Assay
HEK293 cells were treated with either DMSO vehicle or increasing doses of XAV939 or PJ34 for 30 min. Cells were then stimulated with H2O2 (500 μM, 15 min) to induce PARP1 activation. Western blots of cellular lysates were probed with either anti-PAR (10H) or antiactin antibody.
Data Analysis and Statistics
KM values were estimated using nonlinear regression analysis and curve fitting using the Michaelis–Menten function with no further constraints in Prism (GraphPad Software). We report means ± standard errors based on rate data measured with two or three technical replicates. IC50 values were estimated using three-parameter regression analysis and curve fitting with no further constraints using Prism. Experiments were carried out with two or three technical replicate series and repeated 1−5 times. We report best-fit values ± standard errors of representative experiments.
Protein Crystallization, Structure Determination, and Refinement
Crystallization of PARP catalytic domain fragments is described in Supporting Information materials and methods. Diffraction data (Supporting Information, Table S3) were indexed, integrated using XDS,55 scaled, and truncated using SCALA or XSCALE and the CCP4 suite of programs.56 BESSY synchrotron diffraction data were processed using XDSAPP.57 The structures were solved by molecular replacement with PHASER58 (Supporting Information, Table S3). All structures were refined using Refmac559 or Buster,60 and model building was done with Coot.61
Supplementary Material
Table 2.
In Vitro Potencies of PARP Inhibitorsa

|

|

|

|

|

|

|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Veliparib | Niraparib | Olaparib | Talazoparib | Rucaparib | PJ34 | 1 (UPF1069) | |||||||||
| Protein | Construct | pIC50 | IC50 | pIC50 | IC50 | pIC50 | IC50 | pIC50 | IC50 | pIC50 | IC50 | pIC50 | IC50 | pIC50 | IC50 |
| PARP1 | FL | 8.48±0.14 | 3.3 | 7.79±0.08 | 16.7 | 8.86±0.12 | 1.4 | 8.95±0.18 | 1.1 | 8.50±0.08 | 3.2 | 7.96±0.10 | 10.9 | 6.84±0.68 | 145 |
| PARP1 | Zn3-C | 6.45±0.07 | 354 | 7.16±0.10 | 18.9 | 6.52±0.08 | 301 | 6.32±0.07 | 481 | ||||||
| PARP1 | ART | 7.45±0.08 | 35.2 | 6.88±0.14 | 132 | 7.99±0.07 | 10 | 7.40±0.12 | 39.8 | 7.22±0.13 | 61 | ||||
| PARP2 | FL | 7.76±0.10 | 17.5 | 7.81±0.16 | 15.3 | 7.91±0.11 | 12.3 | 8.38±0.06 | 4.1 | 7.55±0.18 | 28.2 | 7.48±0.06 | 33.5 | 5.97±0.05 | 1000 |
| PARP2 | ART | 6.86±0.07 | 137 | 6.60±0.12 | 251 | 6.86±0.09 | 138 | 6.42±0.06 | 382 | ||||||
| PARP3 | FL | 5.67±0.11 | 2040 | 6.53±0.18 | 296 | 6.64±0.10 | 230 | 7.20±0.04 | 62.8 | 6.39±0.09 | 512 | 5.84±0.15 | 1430 | 5.89±0.21 | 1300 |
| PARP4 | ART | 5.46±0.09 | 3500 | 6.35±0.14 | 446 | 6.39±0.11 | 410 | 6.59±0.14 | 254 | 6.08±0.09 | 839 | 6.12±0.18 | |||
| TNKS1 | SAM-ART | 4.97±0.06 | 10740 | 6.49±0.13 | 322 | 6.84±0.05 | 144 | 5.51±0.12 | 3100 | ||||||
| TNKS1 | ART | 5.63±0.08 | 2355 | 5.91±0.06 | 1230 | 5.77±0.05 | 1690 | ||||||||
| TNKS2 | ART | 4.62±0.10 | 24000 | 5.29±0.96 | 5130 | 5.3±0.04 | 2340 | 6.97±0.07 | 108 | 6.05±0.05 | 890 | 5.53±0.10 | 2900 | ||
| PARP10 | FL | 5.72±0.15 | 1900 | 5.71±0.07 | 1900 | 5.03±0.09 | 9300 | 5.11±0.08 | 7800 | 6.25±0.11 | 570 | 5.23±0.10 | 5800 | 5.15±0.08 | 7100 |
| PARP10 | ART | 4.36±0.07 | 43800 | 5.48±0.04 | 3300 | 5.90±0.07 | 1250 | 5.64±0.05 | 2310 | 5.17±0.15 | 6800 | ||||
| PARP12 | ART | 4.23±0.27 | 59400 | 6.10±0.15 | 79 | 4.99±0.04 | 10050 | 5.18±0.15 | 6650 | 6.65±0.11 | 230 | ||||
| PARP14 | ART | 4.05±0.26 | 89000 | 5.76±0.08 | 17300 | n.i. | n.i. | 4.60±0.04 | 25500 | 5.05±0.05 | 8830 | 5.38±0.05 | 4200 | 5.24±0.09 | 5700 |
| PARP15 | ART | n.i. | n.i. | 4.54±0.16 | 29200 | 4.75±0.06 | 17600 | 4.32±0.16 | 47400 | 4.49±0.07 | 32600 | 5.05±0.15 | 9000 | ||
| PARP16 | FL | n.i. | n.i. | 5.29±0.17 | 5100 | 5.73±0.15 | 1900 | n.i. | n.i. | ||||||
IC50 values in nanomolar (nM)
Table 3.
In Vitro Potencies of Tankyrase Inhibitorsa

|

|

|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2 (AZ-6102) | IWR-1 | XAV939 | ||||||||
| Protein | Construct | pIC50 | IC50 | Lit.IC50 | pIC50 | IC50 | Lit.IC50 | pIC50 | IC50 | Lit.IC50 |
| PARP1 | FL | 5.79±0.23 | 1600 | 200018 | n.i. | n.i. | 7.13±0.05 | 74.1 | 12017 | |
| PARP1 | ART | 6.78±0.08 | 169 | 220033 | ||||||
| PARP2 | FL | 6.01±0.22 | 990 | 50018 | n.i. | n.i. | 7.57±0.10 | 26.9 | ||
| PARP2 | ART | 11433 | ||||||||
| PARP3 | FL | n.i. | n.i. | n.i. | n.i. | 6.08±0.11 | 839 | |||
| PARP4 | ART | n.i. | n.i. | 5.36±0.15 | 4390 | |||||
| TNKS1 | SAM-ART | 7.00±0.1 | 100 | 318 | 7.02±0.09 | 94.6 | 1117,33 | |||
| TNKS2 | ART | 9.32±0.09 | 0.6 | 118 | 6.87±0.08 | 135 | 18016 | 8.28±0.07 | 5.2 | 4–817,33 |
| PARP10 | FL | n.i. | n.i. | n.i. | n.i. | n.i. | n.i. | |||
| PARP14 | ART | 5.57±0.23 | 3400 | 5.23±0.12 | 5900 | |||||
| PARP15 | ART | 4.59±0.15 | 25600 | n.i. | n.i. | |||||
IC50 values in nanomolar (nM)
Acknowledgments
We thank the beamline staff at the Berliner Elektronenspeicher-ring-Gesellschaft für Synchrotronstrahlung (BESSY; Berlin, Germany) and the Diamond Light Source (Didcot, UK) for excellent support; Ezeogo Obaji and Lari Lehtiö for supplying the PARP2 cDNA; Lawrence Lum for supplying IWR1; the AstraZeneca Open Innovation Program for supplying 2; Emma Hansson for assistance with protein purification; Rozbeh Jafari and Daniel Martinez Molina for CETSA analysis; and the Protein Science Facility at Karolinska Institutet/SciLifeLab for molecular cloning. This work was financed by the IngaBritt och Arne Lundbergs Research Foundation (no. 403), the Swedish Cancer Society (2012/313; 2014/716), the Swedish Foundation for Strategic Research (RBc08-14), and the Swedish Research Council (621-2012-5247; 2015-04603) (to H.S.) and the NIH (1R01NS088629; to M.S.C.). Synchrotron data collection was supported by the European Community’s Seventh Framework Programme (FP7) under BioStruct-X (no. 283570). M.L. was a stipendiary of the Swedish Society for Medical Research.
Abbreviations
- ART
ADP-ribosyltransferase
- DMSO
dimethyl sulfoxide
- HRP
horseradish peroxidase
- IMAC
immobilized metal ion chromatography
- KM
Michaelis constant
- LC-ESI-MS
liquid chromatography−electrospray ionization mass spectrometry
- NAD+
nicotinamide adenine dinucleotide
- PARP
poly(ADP-ribose) polymerase
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- TNKS
tankyrase
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jmedchem.6b00990.
Figures detailing enzymatic characterization and enzyme inhibition analyses; crystallographic data collection and refinement statistics; extended materials and methods (PDF)
Molecular formula strings (CSV)
Accession Codes
Atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4R5W, 4R6E, 4RV6, 4TVJ, 4UND, 4UXB, and 5LX6). Authors will release the atomic coordinates and experimental data upon article publication.
ORCID
Herwig Schüler: 0000-0003-4059-3501
Author Contributions
A.-G.T., T.E., T.K., M.L., A.F.P, M.S.C, and H.S. designed and carried out research; T.K., L.T., M.M. and H.S. analyzed data; M.L. and H.S. wrote the manuscript. T.E., T.K., M.L., and A.F.P. contributed equally to this work
Notes
The authors declare no competing financial interest.
References
- 1.Curtin NJ, Szabo C. Therapeutic applications of PARP inhibitors: anticancer therapy and beyond. Mol Aspects Med. 2013;34(6):1217–1256. doi: 10.1016/j.mam.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim G, Ison G, McKee AE, Zhang H, Tang S, Gwise T, Sridhara R, Lee E, Tzou A, Philip R, Chiu HJ, Ricks TK, Palmby T, Russell AM, Ladouceur G, Pfuma E, Li H, Zhao L, Liu Q, Venugopal R, Ibrahim A, Pazdur R. FDA approval summary: Olaparib monotherapy in patients with deleterious germline BRCA-mutated advanced ovarian cancer treated with three or more lines of chemotherapy. Clin Cancer Res. 2015;21(19):4257–4261. doi: 10.1158/1078-0432.CCR-15-0887. [DOI] [PubMed] [Google Scholar]
- 3.Feng FY, de Bono JS, Rubin MA, Knudsen KE. Chromatin to clinic: the molecular rationale for PARP1 inhibitor function. Mol Cell. 2015;58(6):925–934. doi: 10.1016/j.molcel.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ledermann JA. PARP inhibitors in ovarian cancer. Ann Oncol. 2016;27(Suppl 1):i40–i44. doi: 10.1093/annonc/mdw094. [DOI] [PubMed] [Google Scholar]
- 5.Beck C, Boehler C, Guirouilh Barbat J, Bonnet ME, Illuzzi G, Ronde P, Gauthier LR, Magroun N, Rajendran A, Lopez BS, Scully R, Boussin FD, Schreiber V, Dantzer F. PARP3 affects the relative contribution of homologous recombination and non-homologous end-joining pathways. Nucleic Acids Res. 2014;42(9):5616–5632. doi: 10.1093/nar/gku174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fenton AL, Shirodkar P, Macrae CJ, Meng L, Koch CA. The PARP3- and ATM-dependent phosphorylation of APLF facilitates DNA double-strand break repair. Nucleic Acids Res. 2013;41(7):4080–4092. doi: 10.1093/nar/gkt134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lehtio L, Chi NW, Krauss S. Tankyrases as drug targets. FEBS J. 2013;280(15):3576–3593. doi: 10.1111/febs.12320. [DOI] [PubMed] [Google Scholar]
- 8.Riffell JL, Lord CJ, Ashworth A. Tankyrase-targeted therapeutics: expanding opportunities in the PARP family. Nat Rev Drug Discovery. 2012;11(12):923–936. doi: 10.1038/nrd3868. [DOI] [PubMed] [Google Scholar]
- 9.Kaufmann M, Feijs KL, Luscher B. Function and regulation of the mono-ADP-ribosyltransferase ARTD10. Curr Top Microbiol Immunol. 2014;384:167–188. doi: 10.1007/82_2014_379. [DOI] [PubMed] [Google Scholar]
- 10.Todorova T, Bock FJ, Chang P. Poly(ADP-ribose) polymerase-13 and RNA regulation in immunity and cancer. Trends Mol Med. 2015;21(6):373–384. doi: 10.1016/j.molmed.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bubici C, Papa S. JNK signalling in cancer: in need of new, smarter therapeutic targets. Br J Pharmacol. 2014;171(1):24–37. doi: 10.1111/bph.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camicia R, Winkler HC, Hassa PO. Novel drug targets for personalized precision medicine in relapsed/refractory diffuse large B-cell lymphoma: a comprehensive review. Mol Cancer. 2015;14:207. doi: 10.1186/s12943-015-0474-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Connor MJ. Targeting the DNA damage response in cancer. Mol Cell. 2015;60(4):547–560. doi: 10.1016/j.molcel.2015.10.040. [DOI] [PubMed] [Google Scholar]
- 14.Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer. 2016;16(2):110–120. doi: 10.1038/nrc.2015.21. [DOI] [PubMed] [Google Scholar]
- 15.Toniatti C, Jones P, Graham H, Pagliara B, Draetta G. Oncology drug discovery: planning a turnaround. Cancer Discovery. 2014;4(4):397–404. doi: 10.1158/2159-8290.CD-13-0452. [DOI] [PubMed] [Google Scholar]
- 16.Chen B, Dodge ME, Tang W, Lu J, Ma Z, Fan CW, Wei S, Hao W, Kilgore J, Williams NS, Roth MG, Amatruda JF, Chen C, Lum L. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol. 2009;5(2):100–107. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lau T, Chan E, Callow M, Waaler J, Boggs J, Blake RA, Magnuson S, Sambrone A, Schutten M, Firestein R, Machon O, Korinek V, Choo E, Diaz D, Merchant M, Polakis P, Holsworth DD, Krauss S, Costa M. A novel tankyrase small-molecule inhibitor suppresses APC mutation-driven colorectal tumor growth. Cancer Res. 2013;73(10):3132–3144. doi: 10.1158/0008-5472.CAN-12-4562. [DOI] [PubMed] [Google Scholar]
- 18.Johannes JW, Almeida L, Barlaam B, Boriack-Sjodin PA, Casella R, Croft RA, Dishington AP, Gingipalli L, Gu C, Hawkins JL, Holmes JL, Howard T, Huang J, Ioannidis S, Kazmirski S, Lamb ML, McGuire TM, Moore JE, Ogg D, Patel A, Pike KG, Pontz T, Robb GR, Su N, Wang H, Wu X, Zhang HJ, Zhang Y, Zheng X, Wang T. Pyrimidinone nicotinamide mimetics as selective tankyrase and wnt pathway inhibitors suitable for in vivo pharmacology. ACS Med Chem Lett. 2015;6(3):254–259. doi: 10.1021/ml5003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papeo G, Avanzi N, Bettoni S, Leone A, Paolucci M, Perego R, Quartieri F, Riccardi-Sirtori F, Thieffine S, Montagnoli A, Lupi R. Insights into PARP inhibitors’ selectivity using fluorescence polarization and surface plasmon resonance binding assays. J Biomol Screening. 2014;19(8):1212–1219. doi: 10.1177/1087057114538319. [DOI] [PubMed] [Google Scholar]
- 20.Papeo G, Posteri H, Borghi D, Busel AA, Caprera F, Casale E, Ciomei M, Cirla A, Corti E, D’Anello M, Fasolini M, Forte B, Galvani A, Isacchi A, Khvat A, Krasavin MY, Lupi R, Orsini P, Perego R, Pesenti E, Pezzetta D, Rainoldi S, Riccardi-Sirtori F, Scolaro A, Sola F, Zuccotto F, Felder ER, Donati D, Montagnoli A. Discovery of 2-[1-(4,4-Difluorocyclohexyl)piperidin-4-yl]-6-fluoro-3-oxo-2,3-dihydro-1H-isoind ole-4-carboxamide (NMS-P118): a potent, orally available, and highly selective PARP-1 inhibitor for cancer therapy. J Med Chem. 2015;58(17):6875–6898. doi: 10.1021/acs.jmedchem.5b00680. [DOI] [PubMed] [Google Scholar]
- 21.Wahlberg E, Karlberg T, Kouznetsova E, Markova N, Macchiarulo A, Thorsell AG, Pol E, Frostell A, Ekblad T, Oncu D, Kull B, Robertson GM, Pellicciari R, Schüler H, Weigelt J. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat Biotechnol. 2012;30(3):283–288. doi: 10.1038/nbt.2121. [DOI] [PubMed] [Google Scholar]
- 22.Langelier MF, Planck JL, Servent KM, Pascal JM. Purification of human PARP-1 and PARP-1 domains from Escherichia coli for structural and biochemical analysis. Methods Mol Biol. 2011;780:209–226. doi: 10.1007/978-1-61779-270-0_13. [DOI] [PubMed] [Google Scholar]
- 23.Ruf A, Mennissier de Murcia J, de Murcia G, Schulz GE. Structure of the catalytic fragment of poly(AD-ribose) polymerase from chicken. Proc Natl Acad Sci U S A. 1996;93(15):7481–7485. doi: 10.1073/pnas.93.15.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langelier MF, Planck JL, Roy S, Pascal JM. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science. 2012;336(6082):728–732. doi: 10.1126/science.1216338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eustermann S, Wu WF, Langelier MF, Yang JC, Easton LE, Riccio AA, Pascal JM, Neuhaus D. Structural basis of detection and signaling of DNA single-strand breaks by human PARP-1. Mol Cell. 2015;60(5):742–754. doi: 10.1016/j.molcel.2015.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O’Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JH, de Bono JS. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361(2):123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 27.Penning TD, Zhu GD, Gandhi VB, Gong J, Liu X, Shi Y, Klinghofer V, Johnson EF, Donawho CK, Frost DJ, Bontcheva-Diaz V, Bouska JJ, Osterling DJ, Olson AM, Marsh KC, Luo Y, Giranda VL. Discovery of the poly(ADP-ribose) polymerase (PARP) inhibitor 2-[(R)-2-methylpyrrolidin-2-yl]-1H-benzimidazole-4-carboxamide (ABT-888) for the treatment of cancer. J Med Chem. 2009;52(2):514–523. doi: 10.1021/jm801171j. [DOI] [PubMed] [Google Scholar]
- 28.Sandhu SK, Schelman WR, Wilding G, Moreno V, Baird RD, Miranda S, Hylands L, Riisnaes R, Forster M, Omlin A, Kreischer N, Thway K, Gevensleben H, Sun L, Loughney J, Chatterjee M, Toniatti C, Carpenter CL, Iannone R, Kaye SB, de Bono JS, Wenham RM. The poly(ADP-ribose) polymerase inhibitor niraparib (MK4827) in BRCA mutation carriers and patients with sporadic cancer: a phase 1 dose-escalation trial. Lancet Oncol. 2013;14(9):882–892. doi: 10.1016/S1470-2045(13)70240-7. [DOI] [PubMed] [Google Scholar]
- 29.Plummer R, Jones C, Middleton M, Wilson R, Evans J, Olsen A, Curtin N, Boddy A, McHugh P, Newell D, Harris A, Johnson P, Steinfeldt H, Dewji R, Wang D, Robson L, Calvert H. Phase I study of the poly(ADP-ribose) polymerase inhibitor, AG014699, in combination with Temozolomide in patients with advanced solid tumors. Clin Cancer Res. 2008;14(23):7917–7923. doi: 10.1158/1078-0432.CCR-08-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen Y, Rehman FL, Feng Y, Boshuizen J, Bajrami I, Elliott R, Wang B, Lord CJ, Post LE, Ashworth A. BMN 673, a novel and highly potent PARP1/2 inhibitor for the treatment of human cancers with DNA repair deficiency. Clin Cancer Res. 2013;19(18):5003–5015. doi: 10.1158/1078-0432.CCR-13-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moroni F, Formentini L, Gerace E, Camaioni E, Pellegrini-Giampietro DE, Chiarugi A, Pellicciari R. Selective PARP-2 inhibitors increase apoptosis in hippocampal slices but protect cortical cells in models of post-ischaemic brain damage. Br J Pharmacol. 2009;157(5):854–862. doi: 10.1111/j.1476-5381.2009.00232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abdelkarim GE, Gertz K, Harms C, Katchanov J, Dirnagl U, Szabo C, Endres M. Protective effects of PJ34, a novel, potent inhibitor of poly(ADP-ribose) polymerase (PARP) in in vitro and in vivo models of stroke. Int J Mol Med. 2001;7(3):255–260. [PubMed] [Google Scholar]
- 33.Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, Hild M, Shi X, Wilson CJ, Mickanin C, Myer V, Fazal A, Tomlinson R, Serluca F, Shao W, Cheng H, Shultz M, Rau C, Schirle M, Schlegl J, Ghidelli S, Fawell S, Lu C, Curtis D, Kirschner MW, Lengauer C, Finan PM, Tallarico JA, Bouwmeester T, Porter JA, Bauer A, Cong F. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461(7264):614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 34.Frye SV. The art of the chemical probe. Nat Chem Biol. 2010;6(3):159–161. doi: 10.1038/nchembio.296. [DOI] [PubMed] [Google Scholar]
- 35.Arrowsmith CH, Audia JE, Austin C, Baell J, Bennett J, Blagg J, Bountra C, Brennan PE, Brown PJ, Bunnage ME, Buser-Doepner C, Campbell RM, Carter AJ, Cohen P, Copeland RA, Cravatt B, Dahlin JL, Dhanak D, Edwards AM, Frederiksen M, Frye SV, Gray N, Grimshaw CE, Hepworth D, Howe T, Huber KV, Jin J, Knapp S, Kotz JD, Kruger RG, Lowe D, Mader MM, Marsden B, Mueller-Fahrnow A, Muller S, O’Hagan RC, Overington JP, Owen DR, Rosenberg SH, Roth B, Ross R, Schapira M, Schreiber SL, Shoichet B, Sundstrom M, Superti-Furga G, Taunton J, Toledo-Sherman L, Walpole C, Walters MA, Willson TM, Workman P, Young RN, Zuercher WJ. The promise and peril of chemical probes. Nat Chem Biol. 2015;11(8):536–541. doi: 10.1038/nchembio.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferraris DV. Evolution of poly(ADP-ribose) polymerase-1 (PARP-1) inhibitors. From concept to clinic. J Med Chem. 2010;53(12):4561–4584. doi: 10.1021/jm100012m. [DOI] [PubMed] [Google Scholar]
- 37.Karlberg T, Hammarstrom M, Schütz P, Svensson L, Schüler H. Crystal structure of the catalytic domain of human PARP2 in complex with PARP inhibitor ABT-888. Biochemistry. 2010;49(6):1056–1058. doi: 10.1021/bi902079y. [DOI] [PubMed] [Google Scholar]
- 38.Menear KA, Adcock C, Boulter R, Cockcroft XL, Copsey L, Cranston A, Dillon KJ, Drzewiecki J, Garman S, Gomez S, Javaid H, Kerrigan F, Knights C, Lau A, Loh VM, Jr, Matthews IT, Moore S, O’Connor MJ, Smith GC, Martin NM. 4-[3-(4-cyclopropanecarbonylpiperazine-1-carbonyl)-4-fluorobenzyl]-2H-phthalazin- 1-one: a novel bioavailable inhibitor of poly(ADP-ribose) polymerase-1. J Med Chem. 2008;51(20):6581–6591. doi: 10.1021/jm8001263. [DOI] [PubMed] [Google Scholar]
- 39.Yao H, Ji M, Zhu Z, Zhou J, Cao R, Chen X, Xu B. Discovery of 1-substituted benzyl-quinazoline-2,4(1H,3H)-dione derivatives as novel poly(ADP-ribose)polymerase-1 inhibitors. Bioorg Med Chem. 2015;23(4):681–693. doi: 10.1016/j.bmc.2014.12.071. [DOI] [PubMed] [Google Scholar]
- 40.Aoyagi-Scharber M, Gardberg AS, Yip BK, Wang B, Shen Y, Fitzpatrick PA. Structural basis for the inhibition of poly(ADP-ribose) polymerases 1 and 2 by BMN 673, a potent inhibitor derived from dihydropyridophthalazinone. Acta Crystallogr, Sect F: Struct Biol Commun. 2014;70(9):1143–1149. doi: 10.1107/S2053230X14015088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haikarainen T, Narwal M, Joensuu P, Lehtio L. Evaluation and structural basis for the inhibition of tankyrases by PARP Inhibitors. ACS Med Chem Lett. 2014;5(1):18–22. doi: 10.1021/ml400292s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lehtio L, Jemth AS, Collins R, Loseva O, Johansson A, Markova N, Hammarstrom M, Flores A, Holmberg-Schiavone L, Weigelt J, Helleday T, Schüler H, Karlberg T. Structural basis for inhibitor specificity in human poly(ADP-ribose) polymerase-3. J Med Chem. 2009;52(9):3108–3111. doi: 10.1021/jm900052j. [DOI] [PubMed] [Google Scholar]
- 43.Kirby CA, Cheung A, Fazal A, Shultz MD, Stams T. Structure of human tankyrase 1 in complex with small-molecule inhibitors PJ34 and XAV939. Acta Crystallogr, Sect F: Struct Biol Cryst Commun. 2012;68(2):115–118. doi: 10.1107/S1744309111051219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karlberg T, Klepsch M, Thorsell AG, Andersson CD, Linusson A, Schüler H. Structural basis for lack of ADP-ribosyltransferase activity in poly(ADP-ribose) polymerase-13/zinc finger antiviral protein. J Biol Chem. 2015;290(12):7336–7344. doi: 10.1074/jbc.M114.630160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yates SP, Taylor PL, Jorgensen R, Ferraris D, Zhang J, Andersen GR, Merrill AR. Structure-function analysis of water-soluble inhibitors of the catalytic domain of exotoxin A from Pseudomonas aeruginosa. Biochem J. 2005;385(3):667–675. doi: 10.1042/BJ20041480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sunderland PT, Woon EC, Dhami A, Bergin AB, Mahon MF, Wood PJ, Jones LA, Tully SR, Lloyd MD, Thompson AS, Javaid H, Martin NM, Threadgill MD. 5-Benzamidoisoquinolin-1-ones and 5-(omega-carboxyalkyl)isoquinolin-1-ones as isoform-selective inhibitors of poly(ADP-ribose) polymerase 2 (PARP-2) J Med Chem. 2011;54(7):2049–2059. doi: 10.1021/jm1010918. [DOI] [PubMed] [Google Scholar]
- 47.Karlberg T, Markova N, Johansson I, Hammarstrom M, Schütz P, Weigelt J, Schüler H. Structural basis for the interaction between tankyrase-2 and a potent Wnt-signaling inhibitor. J Med Chem. 2010;53(14):5352–5355. doi: 10.1021/jm100249w. [DOI] [PubMed] [Google Scholar]
- 48.Martinez Molina D, Jafari R, Ignatushchenko M, Seki T, Larsson EA, Dan C, Sreekumar L, Cao Y, Nordlund P. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341(6141):84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- 49.Ekblad T, Camaioni E, Schüler H, Macchiarulo A. PARP inhibitors: polypharmacology versus selective inhibition. FEBS J. 2013;280(15):3563–3575. doi: 10.1111/febs.12298. [DOI] [PubMed] [Google Scholar]
- 50.Voronkov A, Holsworth DD, Waaler J, Wilson SR, Ekblad B, Perdreau-Dahl H, Dinh H, Drewes G, Hopf C, Morth JP, Krauss S. Structural basis and SAR for G007-LK, a lead stage 1,2,4-triazole based specific tankyrase 1/2 inhibitor. J Med Chem. 2013;56(7):3012–3023. doi: 10.1021/jm4000566. [DOI] [PubMed] [Google Scholar]
- 51.Shultz MD, Kirby CA, Stams T, Chin DN, Blank J, Charlat O, Cheng H, Cheung A, Cong F, Feng Y, Fortin PD, Hood T, Tyagi V, Xu M, Zhang B, Shao W. [1,2,4]triazol-3-ylsulfanylmethyl)-3-phenyl-[1,2,4]oxadiazoles: antagonists of the Wnt pathway that inhibit tankyrases 1 and 2 via novel adenosine pocket binding. J Med Chem. 2012;55(3):1127–1136. doi: 10.1021/jm2011222. [DOI] [PubMed] [Google Scholar]
- 52.Li M, Yu X. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell. 2013;23(5):693–704. doi: 10.1016/j.ccr.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murai J, Huang SY, Renaud A, Zhang Y, Ji J, Takeda S, Morris J, Teicher B, Doroshow JH, Pommier Y. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol Cancer Ther. 2014;13(2):433–443. doi: 10.1158/1535-7163.MCT-13-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jafari R, Almqvist H, Axelsson H, Ignatushchenko M, Lundback T, Nordlund P, Martinez Molina D. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat Protoc. 2014;9(9):2100–2122. doi: 10.1038/nprot.2014.138. [DOI] [PubMed] [Google Scholar]
- 55.Kabsch W. XDS. Acta Crystallogr, Sect D: Biol Crystallogr. 2010;66(2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Evans P. Scaling and assessment of data quality. Acta Crystallogr, Sect D: Biol Crystallogr. 2006;62(1):72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 57.Krug M, Weiss MS, Heinemann U, Mueller U. XDSAPP: a graphical user interface for the convenient processing of diffraction data using XDS. J Appl Crystallogr. 2012;45:568–572. [Google Scholar]
- 58.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr, Sect D: Biol Crystallogr. 2005;61(4):458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 59.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr, Sect D: Biol Crystallogr. 2011;67(4):355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bricogne G, Blanc E, Brandl M, Flensburg C, Keller P, Paciorek W, Roversi P, Sharff A, Smart OS, Vonrhein C, Womack TO. BUSTER, version 2.10.2. Global Phasing Ltd.; Cambridge, United Kingdom: 2016. [Google Scholar]
- 61.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr, Sect D: Biol Crystallogr. 2010;66(4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Langelier MF, Servent KM, Rogers EE, Pascal JM. A third zinc-binding domain of human poly(ADP-ribose) polymerase-1 coordinates DNA-dependent enzyme activation. J Biol Chem. 2008;283(7):4105–4114. doi: 10.1074/jbc.M708558200. [DOI] [PubMed] [Google Scholar]
- 63.Kim H, Jacobson MK, Rolli V, Menissier-de Murcia J, Reinbolt J, Simonin F, Ruf A, Schulz G, de Murcia G. Photoaffinity labelling of human poly(ADP-ribose) polymerase catalytic domain. Biochem J. 1997;322(2):469–475. doi: 10.1042/bj3220469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simonin F, Poch O, Delarue M, de Murcia G. Identification of potential active-site residues in the human poly(ADP-ribose) polymerase. J Biol Chem. 1993;268(12):8529–8535. [PubMed] [Google Scholar]
- 65.Ame JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, Muller S, Hoger T, Menissier-de Murcia J, de Murcia G. PARP-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J Biol Chem. 1999;274(25):17860–17868. doi: 10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- 66.Ruf A, Rolli V, de Murcia G, Schulz GE. The mechanism of the elongation and branching reaction of poly(ADP-ribose) polymerase as derived from crystal structures and mutagenesis. J Mol Biol. 1998;278(1):57–65. doi: 10.1006/jmbi.1998.1673. [DOI] [PubMed] [Google Scholar]
- 67.Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW, Shilton BH, Luscher B. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell. 2008;32(1):57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 68.Di Paola S, Micaroni M, Di Tullio G, Buccione R, Di Girolamo M. PARP16/ARTD15 is a novel endoplasmic-reticulum-associated mono-ADP-ribosyltransferase that interacts with, and modifies karyopherin-ss1. PLoS One. 2012;7(6):e37352. doi: 10.1371/journal.pone.0037352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karlberg T, Thorsell AG, Kallas A, Schüler H. Crystal structure of human ADP-ribose transferase ARTD15/PARP16 reveals a novel putative regulatory domain. J Biol Chem. 2012;287(29):24077–24081. doi: 10.1074/jbc.M112.379289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Narwal M, Venkannagari H, Lehtio L. Structural basis of selective inhibition of human tankyrases. J Med Chem. 2012;55(3):1360–1367. doi: 10.1021/jm201510p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


