Abstract
Reverse transcription of the HIV-1 RNA genome into double-stranded DNA is a central step in infection1 and a common target of antiretrovirals2. The reaction is catalyzed by viral reverse transcriptase (RT)3,4 that is packaged in an infectious virion along with 2 copies of dimeric viral genomic RNA5 and host tRNALys3, which acts as a primer for initiation of reverse transcription6,7. Upon viral entry, initiation is slow and non-processive compared to elongation8,9. Despite extensive efforts, the structural basis for RT function during initiation has remained a mystery. Here we apply cryo-electron microscopy (cryo-EM) to determine a three-dimensional structure of the HIV-1 RT initiation complex. RT is in an inactive polymerase conformation with open fingers and thumb and with the nucleic acid primer-template complex shifted away from the active site. The primer binding site (PBS) helix formed between tRNALys3 and HIV-1 RNA lies in the cleft of RT and is extended by additional pairing interactions. The 5′ end of the tRNA refolds and stacks on the PBS to create a long helical structure, while the remaining viral RNA forms two helical stems positioned above the RT active site, with a linker that connects these helices to the RNase H region of the PBS. Our results illustrate how RNA structure in the initiation complex alters RT conformation to decrease activity, highlighting a potential target for drug action.
During the initiation phase of reverse transcription, RT must bind productively to the viral RNA-tRNALys3 complex and then navigate a highly-structured 5′ region of the HIV-1 genome10. Critical elements within the viral RNA and host tRNA that are necessary for efficient initiation have been identified9,11–18. RT pauses at discrete locations, is generally slowed during initiation compared to elongation8–10, and can bind the viral RNA-tRNALys3 primer site in different orientations19. A rich body of structural data on RT, a heterodimer of p51 and p66 subunits, have shown how its polymerase and RNase H domains interact with DNA/DNA and DNA/RNA duplexes in the absence and presence of antiviral drugs2. Lacking however, are structures that reflect initiation, showing how RT binds to a large bimolecular viral RNA-tRNALys3 complex.
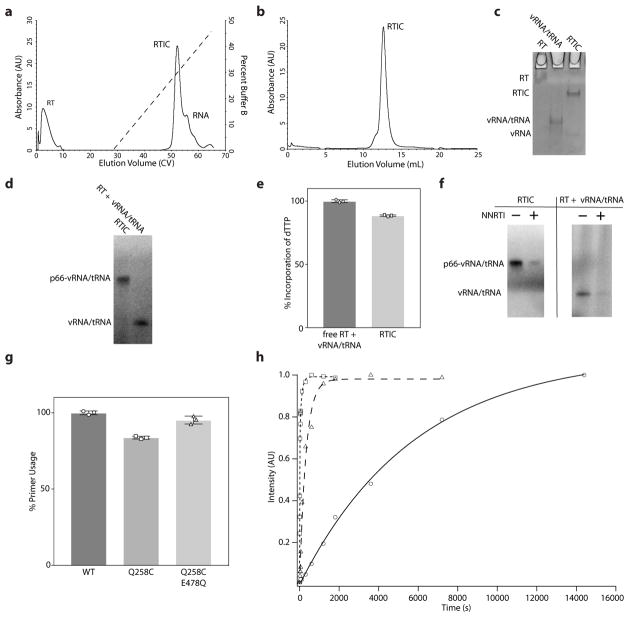
We applied cryo-EM complemented by biochemical and biophysical experiments to determine the molecular architecture of the HIV-1 reverse transcription initiation complex (RTIC). The RTIC was formed using a 101-nucleotide fragment of HIV-1 genomic RNA (vRNA) that encompasses the primer binding site and additional RNA elements required for efficient initation of reverse transcription (Figure 1a). A binary vRNA-tRNA complex was formed with human tRNALys3 that contained a specific cross-linkable nucleotide (“convertible G”) at position 71. The RTIC is kinetically labile undergoing rapid RT dissociation from the tRNA-vRNA complex8,9 with several distinct binding orientations19. To stabilize the RTIC for structural characterization, the vRNA-tRNA complex was specifically cross-linked to RT containing a Q258C mutation in the p66 subunit (Figure 1b), which interacts in the minor groove of RT-nucleic acid complexes2,20,21. After extending the tRNA primer one dideoxynucleotide for highest crosslinking efficiency, the cross-linked vRNA-tRNA-RT ternary complex was formed and purified from free RT and RNA (Figure 1c, Extended Data Figure 1a–c). Crosslinking did not affect the global activity of the RTIC. The final cross-linked HIV-1 RTIC has equivalent total activity in incorporating of the next dNTP as an un-crosslinked initiation complex with rates that are only 3-fold slower, and is strongly inhibited by nevirapine, a non-nucleoside RT inhibitor (NNRTI) that works through conformational modulation of RT22 (Extended Data Figure 1d–h). The RTIC studied structurally here thus represents an active functional state of reverse transcription initiation.
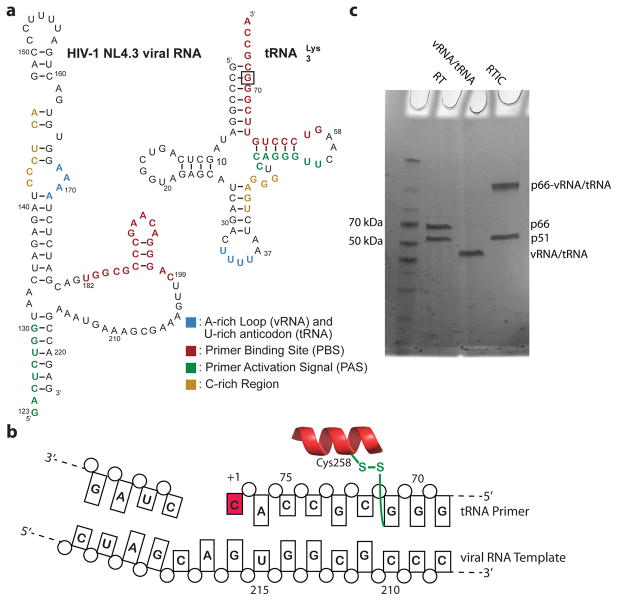
Figure 1. RTIC Constructs and Purification.
a, HIV-1 viral RNA (NL4.3) and tRNALys3 sequences used in RTIC formation. The viral RNA includes sequences complementary to the tRNA primer (colored). Interactions between the regions are reported to be involved in the regulation of initation. b, The crosslinking scheme used for purification of the RTIC. The N2-cystamine-dG was placed at position 71 of the tRNA primer. After extending the primer by one nucleotide, a ddCTP (red), a disulfide bond forms between G71 and mutated C258 on the RT p66 thumb subdomain. c, Non-reducing SDS-PAGE gel of the free vRNA/tRNA, RT, and crosslinked RTIC. RT runs as two bands corresponding to the two subunits. The annealed vRNA/tRNA complex runs as a single band on the gel. The purified RTIC runs as two bands corresponding to the p51 subunit and the crosslinked p66-vRNA/tRNA. Gel analysis was performed on all samples used in the manuscript (>10) and consistently exhibited similar results.
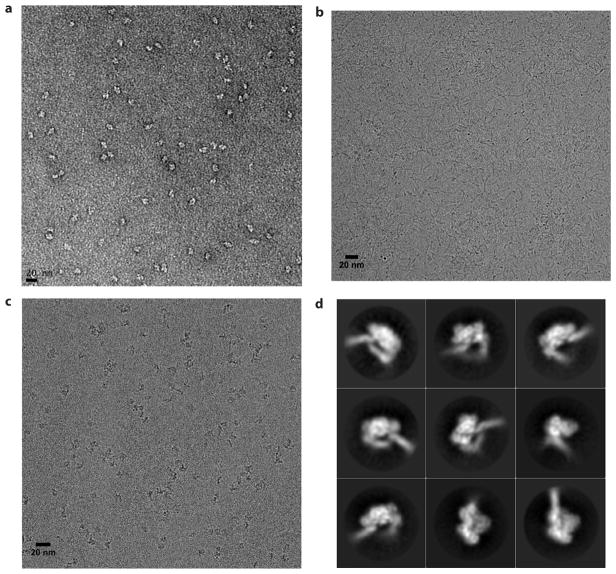
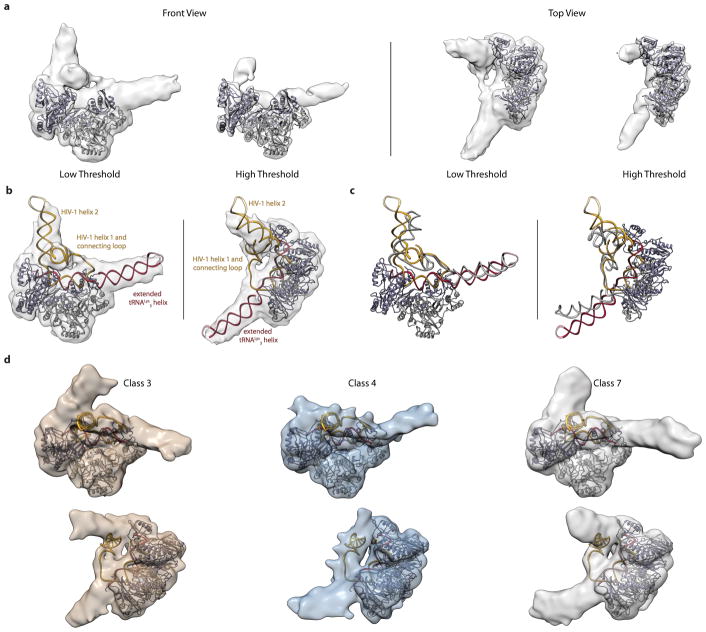
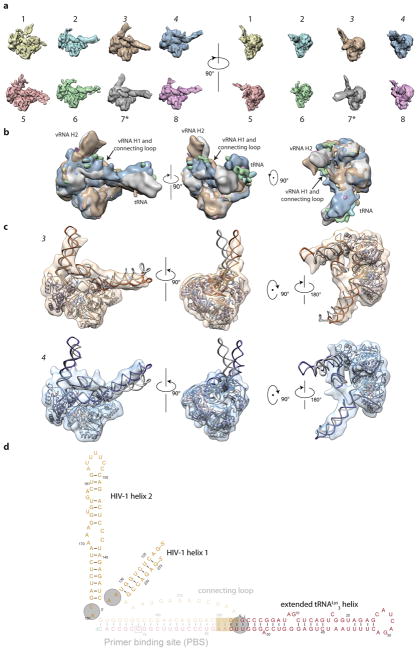
We first assessed the quality of the RTIC sample by negative stain EM23, which confirmed a homogenous RT-RNA complex (Extended Data Figure 2a). Upon cryo-EM preparation, however, the complex dissociated. This problem was alleviated by addition of beta-octyl glucoside (OG), which resulted in monodisperse single particles that we visualized by cryo-EM (Extended Data Figure 2b, c). Two-dimensional class averages of RTIC clearly showed the RT core as well as protruding RNA densities (Extended Data Figure 2d). Three-dimensional classification of particle projections revealed significant conformational variability in the apex of RNA densities (Extend Data Figures 3a). Due to this segmental flexibility we obtained a low-resolution reconstruction (8.0 Å) that best describes the global architecture of RTIC (Figure 2a, Extended Data Figure 3b). This EM-density map encompassing protein and all RNA regions was of sufficient quality to visualize the tRNA and vRNA, thereby enabling us to approximately position the RNA structures located outside of the RT binding cleft. In addition, we obtained a 4.5 Å map by masking out the dynamic peripheral RNA elements and focusing the particle classification and structure refinement on the RT, primer binding site (PBS) helix in the cleft, and additional helical tRNA density (Figure 3, Extended Data Figure 3). This higher resolution map allowed us to describe the conformation of RT and the RNA inside the binding cleft (Figure 3, Extended Data Figure 4). An independent 8.2 Å cryo-EM reconstruction of the RTIC was determined in low salt and Mg2+ and revealed a very similar global conformation for the complex (Extended Data Figure 5), suggesting that the RTIC architecture has limited salt dependence. Models were constructed using the 8.0 Å map to define the global RTIC architecture and the 4.5 Å map to define the structural features of the RTIC core and active site. While the 4.5 Å map provides sufficient resolution to orient the PBS helix of the RTIC, the orientation of the peripheral RNA helical elements of the vRNA and tRNA into the 8.0 Å map is more subjective and relied on iterative Rosetta24 modeling using an accepted secondary structure from past biochemical and biophysical data14,25 (for more details, see Methods).
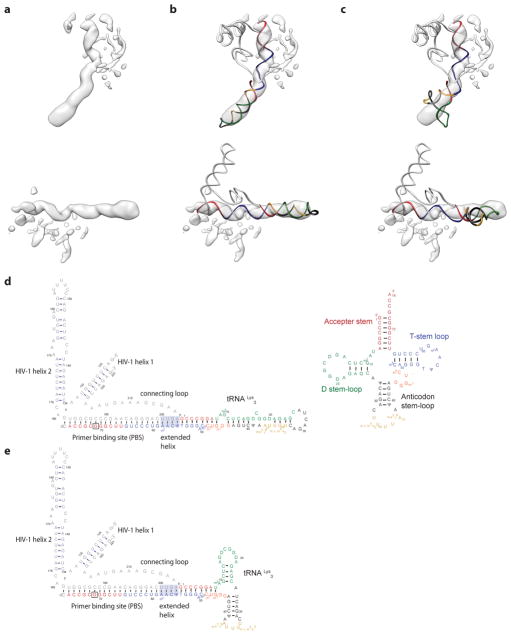
Figure 3. Structure of the RTIC core.
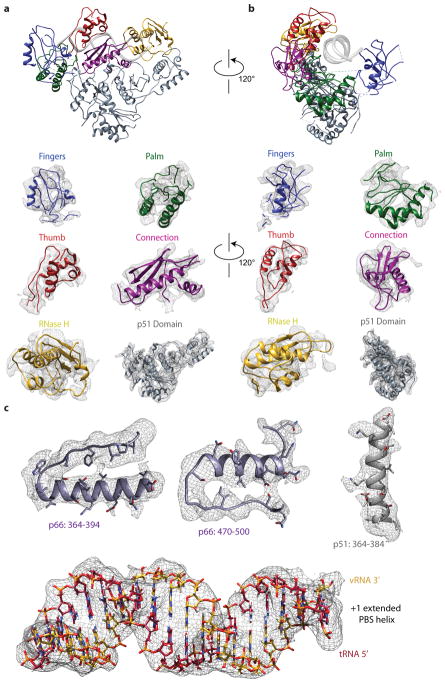
a, Cryo-EM density map of the RTIC core at 4.5 Å resolution. The helical PBS density is regular until it reaches the RNase H active site. Extra helical density for an additional 4 base pairs between the vRNA and tRNA is present. Density corresponding to the tRNA is located just outside the RNase H domain. The junction between the extended PBS and tRNA helix is distorted, possibly due to masking or flexibility in this region. This portion of the RTIC is helical in the global map (Figure 2). b, Representative model of the RTIC core that accounts for RT and the entire +1 extended PBS helix. While there is helical density that can accomodate an additional base-pairs between the vRNA and tRNA, we have not included it in this model. c, Fit of the polymerase active site region in the 4.5 Å map. The RNA and protein backbone are well modeled in this region of the map.
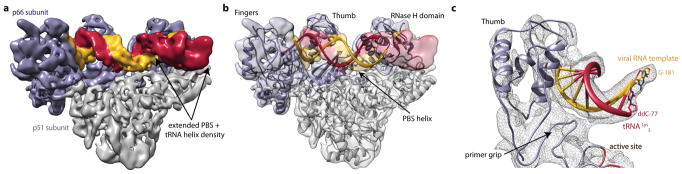
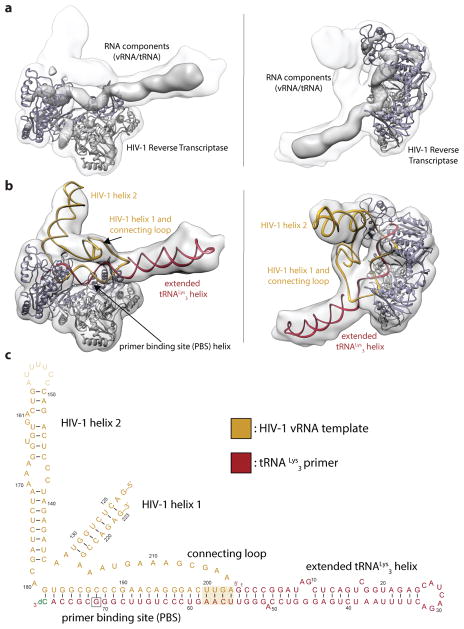
Figure 2. Global architecture of the RTIC.
a, Unmasked 3D reconstruction of the entire RTIC at a global resolution of 8.0 Å. A model of RT (p66: purple, p51: gray) is docked into the map. A low threshold map has been overlayed into the density to illustrate the helical nature of the bound RNA duplex. b, Global model of the RTIC with the vRNA-tRNA components. The EM density accounts for the majority of vRNA and tRNA structure. Density corresponding to the upper HIV helix 2 stem loop is missing, suggesting it is partially disordered. c, Proposed secondary structure of the vRNA-tRNA bound within the RTIC. The majority of vRNA helices are well accounted for in the density, with the exception of the apical portions of helix 2 (faded). Additional base-pairs (boxed) between the vRNA and the tRNA, which extend the PBS helix, are consistent with the continuous helix that spans the RT binding cleft. The tRNA has refolded and adopted an extended helical conformation.
The overall RTIC structure shows the RT core with RNA double-helical density within the binding cleft that spans from the active site to RNase H domain. The helical RNA in the cleft corresponds to the HIV-1 PBS helix formed between nucleotides (nts) 59–76 in the tRNA and nts 182–199 in the vRNA, with the addition of one ddCTP nucleotide needed to elongate the complex and allow efficient RT-RNA cross linking (Figure 2c). The helical density for this +1 extended PBS helix is further extended near the RNase H domain by formation of an additional 4 base pairs, likely between complementary tRNA nts 55–58 and viral RNA nts 200–203. The nucleotide identities of positions 201–203 are highly conserved among recorded HIV-1 sequences26, 70% for 201 and >96% for 202/203, suggesting that this is a common structural feature. In the 8.0 Å global map, a long continuous helical RNA density is observed to extend away from the RNase H domain (Figure 2b). Accordingly, we propose that the 5′ end of tRNA (nts 1–54) refolds to form a secondary structure with a contiguous helix (Figure 2c). Specifically, the D and anticodon stems from nucleotides 10–41 rearrange to form a continuous helical structure, which fits the observed density far better than the three-way junction27,28 observed in the free initiation complex (Extended Data Figure 6).
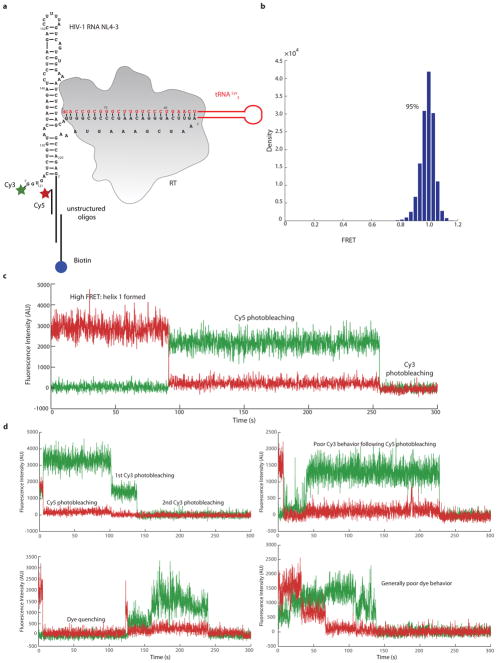
The helical refolded tRNA domain is connected by a single-stranded connection loop (CL) to a 7 base pair helix (H1) involving the 5′ (nts 125–131) and 3′ (nts 217–223) termini of the viral RNA construct (Figure 2b, c) and containing the conserved primer activation signal (PAS) sequence. H1 and CL form a bridge between the RNA located in the RNase H domain and that located near the active-site of RT. A three–way RNA junction is formed by the PBS, H1 and a second helical stem loop (H2) comprising nts 134–178 in HIV-1 viral RNA. Density consistent with single-stranded RNA connects H1 to the PBS in the active site. The relative strength, indicative of stability, of the EM density for H1, the CL, and the apical regions of H2 differs among several of our low-resolution classes, as does their orientations with respect to the base of H2 and the PBS (Extended Data Figure 7a, b). For classes that contain strong density of these RNA features, similar models fit these maps by treating the helical RNA elements as rigid units around flexible junction regions (Extended Data Figure 7c). The presence of helix H1 was confirmed by single-molecule FRET experiments, in which Cy3 dye was attached to the 5′ phosphate of the vRNA and a Cy5-labeled, biotinylated oligonucleotide was hybridized to an extension on the 3′end (Extended Data Figure 8a). In this experiment, observation of a high FRET state would indicate H1 formation. In the buffer conditions used for cryo-EM imaging, we find that >95% of RTIC molecules are in a stable high-FRET state, indicating H1 formation for a surface-immobilized RTIC at room temperature. (Extended Data Figure 8b, c).
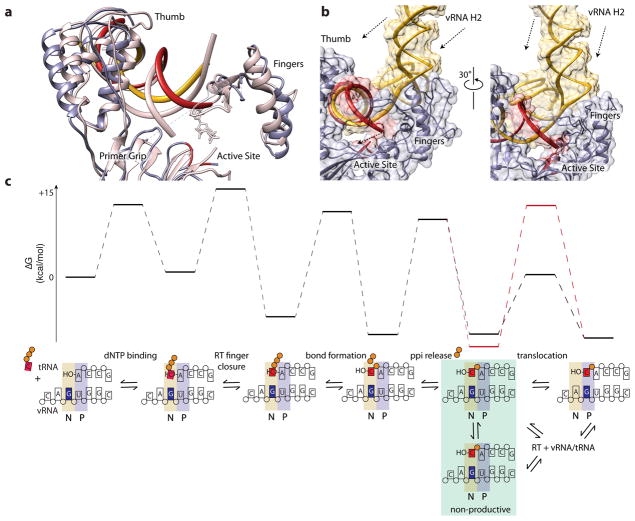
Although the RTIC is active in addition of the next dNTP (Extended Data Figure 1d, e, h), the complex adopts an inactive conformation in which the position of the tRNA primer terminus within the palm subdomain is shifted ~13 Å away from the active site of RT, reminiscent of nucleic acid-RT complexes bound with an NNRTI (Figure 4a, Extended Data Figure 9)22. As observed in RT structures with bound NNRTI, the primer grip (B12-B13-B14 sheet) is displaced towards the 3′ terminus of the primer strand22. The PBS helix is not translocated, with the dC77-G101 pair in the nucleotide acceptor site (N-site) (Figure 4). The PBS helix is also lifted ~6.0 Å away from the palm and connection domains. The path of the viral RNA template and the base of H2 come into close contact with the residues in the fingers domain. The fingers domain of RT adopts a semi-open conformation similar to that of RT structures bound to nucleic acids that lack an incoming nucleotide2,22 (Figure 4a, Extended Data Figure 9). Based on prior mechanistic studies of RT enzymology8,29, the RTIC here is blocked in a pre-translocation conformation for the primer-template complex (Figure 4c). Unlike NNRTI-bound complexes, the RTIC is functional and can incorporate the next dNTP, suggesting conformational plasticity within the RT active site. Although RT contacts the RNA substrate using similar domains as in previously determined RT-nucleic acid complexes2,20,22, the extent of these interactions appears different. The thumb and RNase H domains make the vast majority of observed RNA contact in the RTIC, with a substantial loss of potential interactions in the palm and connection subdomains, consistent with decreased RT-RNA affinity in the initiation complex. The loss of RT-RNA contacts in the palm subdomain20 arises from displacement of the tRNA primer terminus away from the active site (Figure 4a). Although the RTIC structure is not at sufficient resolution to identify specific protein-RNA contacts, there appear to be additional RT-RNA interactions involving the fingers domain from the vRNA template-strand and H2. The sterically bulky vRNA helices immediately adjacent to the fingers region form a wedge that hinders proper accommodation of the PBS into the cleft and leads to loss of RT-RNA contacts in the cleft and displacement of tRNA 3′ end (Figure 4b); this likely inhibits translocation of the PBS helix to allow efficient and rapid incorporation of the next dNTP.
Figure 4. The +1 RTIC adopts an inactive conformation.
a, Comparison of RTIC primer (red) and template (yellow) strands with RT-dsDNA complex (pink, 1RTD23) that has the 3′ primer terminus located in the P-site with N-site occupied by a nucleotide. The PBS helix of the RTIC must be translocated and shifted in order to reposition into a P-site conformation. The thumb is in an open conformation and the primer grip has shifted compared to the active structure. The fingers are in a semi-open conformation22. b, vRNA structure outside the RT active site may prevent proper translocation of RNA substrate during initiation. Two views of the RNA (vRNA: yellow, tRNA: red) near the active site of RT. The arrows indicate the direction the RNA must move in order for the PBS to reposition into the active site. The global structure here is represented in surface mode to highlight potential steric clashes. c, Reaction coordinate diagram and corresponding reaction scheme of HIV-1 reverse transcription on an RNA template using published values29. The +1 RTIC appears to be trapped in a pre-translocation state (green box). The increased energetic barrier for translocation (red) corresponds to the energetic requirement for melting base pairs in H2. We have included the possibility of a non-productive conformer at the pre-translocation step to acknowledge our inability to completely distinguish an on-pathway complex from one that is relaxed and off-pathway. There is also an alternative pathway in which the RTIC dissociates and reassociates to forgo translocation and reposition the primer terminus.
The architecture of the vRNA-tRNA complex in the initiation complex explains prior experimental results on the role of RNA in initiation8–10. The observed RNA conformation is consistent with chemical probing and enzymatic mapping on similar binary vRNA-tRNA and ternary complexes, which were previously interpreted in terms of tRNA-viral RNA pairings13,14,25. No additional interactions between the vRNA and tRNA occur beyond the extended PBS helix at the +1 stage of initiation (Figure 2b), consistent with biochemical results on similar HIV-1 subtype-B sequences13,14. Notably absent is any PAS/anti-PAS interaction between HIV-1 nts 123–131 and tRNA nts 48–55 (Figure 1a), which has been implicated in RT initiation and shown to form dynamically in the absence of RT11,12,27,30. The formation and positions of vRNA H1 and H2 are consistent with their proposed function as barriers during initiation13,14,19. The conserved CL, bridging RNA within the RNase H domain back to H1, may help position the vRNA helices in the proper orientation for RT binding to the tRNA 3′ terminus. The MAL isolate, commonly used in past initiation studies, maintains many of these sequence elements with an added 23nt insertion in the CL that may engage in additional interactions. Both H1 and H2 are required for efficient RT initiation, and their displacement and unfolding are required for RT to proceed9,11,15,17; melting of H2 during initiation occurs after addition of the 6th nucleotide19.
tRNALys3 in the RTIC forms an elongated helical structure compatible with an alternative predicted fold31 that involves an extended PBS structure stabilized by RT. This conformation is consistent with the presence of modified nucleotides in tRNALys3 (Extended Data Figure 6d), and is favored by the extended stacking and RT contacts around the RNase H domain. The RNA fold in the RTIC likely sequesters important sequences for vRNA-tRNA interactions, such as the PAS/anti-PAS and A-rich loop/anticodon sequence interactions9,11–18, which may subsequently form as RNAs rearrange in response to RT extension during initiation. The RNA tertiary conformation within the RTIC is clearly dynamic, as shown by prior single-molecule data19,27,30 and suggested by our cryo-EM data. We observe several conformations with variable orientations and density for the extended tRNA helix and three-way junction of H1, H2 and PBS (Extended Data Figure 7). Such plasticity is likely essential for the RTIC to proceed to elongation.
Our results suggest a model of RT initiation in which RNA structure regulates RT activity. tRNALys3 and vRNA form a dynamic RNA complex, in which the tRNA paired to HIV-1 RNA refolds to form a metastable conformation of its 5′ region. The ability to refold in this way could explain the use of tRNALys3 in HIV-1 initiation. Although RT contacts the PBS in the cleft, the disrupted palm subdomain contacts between the PBS and RT explain the poor affinity of RT for the vRNA/tRNA complex. Within the framework of the standard dNTP incorporation mechanism29, RT in this +1 initiation complex adopts a pre-translocation conformation with an open active site and improper positioning of nucleic acid for catalysis (Figure 4c). The vRNA helices, whose orientation hinders productive binding to RT, must be displaced and/or unfolded for the tRNA primer terminus to reposition within the active site such that the RT fingers can clamp down on an incoming nucleotide (Figure 4b). The dissociation of RT during initiation is rapid, and competes with forward polymerization reactions8,9. RT may dissociate and rebind to the vRNA/tRNA to reposition the primer terminus into the active site. In this pathway, RT rebinding could facilitate melting of downstream RNA structures that hinder translocation. The necessity for these rearrangements during early stages of initiation likely explains the low processivity of initiation and the observed pauses that control the start of HIV-1 replication8,9. The single-stranded, A-rich CL bridging the 3′ end of the vRNA PBS to H2 may serve to position the vRNA helices properly and allow conformational communication with the RT RNase H domain and refolded tRNA. As RT proceeds, structural rearrangements in vRNA and tRNA must occur to favor the transition to processive elongation. Thus, the initiation complex likely changes progressively as initiation proceeds, and may be specifically vulnerable to inhibition by drugs. Higher-resolution structural views of these different states, and dynamics to link them together will be needed to elucidate further the steps of initiation and underlying RNA conformations that regulate early steps in HIV-1 infection.
Online Methods
Sample Preparation
HIV-1 vRNA constructs were prepared by in vitro transcription with T7 RNA polymerase as previously described27,28. Transcripts were denatured in 8 M urea and purified on a sequencing PAGE gel. Gel extraction was performed using 0.3 M ammonium acetate. Following ethanol precipitation, the RNA was dissolved in 10 mM Bis-Tris propane, pH 7.0, 10 mM NaCl and stored at −20°C. The crosslinkable tRNALys3 construct was purchased from TriLink Biotechnologies. The crosslinkable RNA primer was chemically synthesized, PAGE purified, and analyzed by denaturing-PAGE and mass spectrometry. During synthesis, an N2-Cystamine-2′-deoxyguanosine was placed at the 71 position for crosslinking purposes.
vRNA-tRNA complexes were formed by mixing the vRNA and tRNA in a 1:1 molar ratio at 1 μM each in 10 mM Bis-Tris propane, pH 7.0, 10 mM NaCl. The mixture was heated to 90°C and slow cooled to room temperature. The vRNA-tRNA complex was purified away from higher order and unannealed monomer species using a Superdex 200 (26/60) gel filtration column with 10 mM Bis-Tris propane, pH 7.0, 100 mM NaCl. The presence of a single species was confirmed with native PAGE and samples were concentrated on a Vivaspin 20 10,000 MWCO concentrator. Samples were stored at −20°C and exhibited minimal aggregation over time.
HIV-RT was expressed in Escherichia coli strain BL21(DE3). Two expression vectors, one containing the p66 and ampicillin resistance and the other containing p51 and kanamycin resistance, were constructed. The COOH-terminus of p66 contains an unstructured linker and a six-histidine tag. A cysteine mutation for crosslinking was introduced into helix H of p66 (Q258C)20. The protein used in this study also had the C280S mutation, introduced in prior structural work and the E478Q mutation, introduced to eliminate RNase H activity as RT has been shown to cleave dsRNA when stalled for long periods of time20,32. Cell pellets were lysed through sonication and the enzyme was purified by gravity Ni-nitrilotriacetic acid affinity chromatography, followed by cation exchange chromatography using a Superdex 200 (26/600). The his-tag was cleaved by thrombin digestion overnight. The cleaved protein was re-applied to a Ni-NTA column to removed protein with uncleaved his-tag. This was followed by an additional final size-exclusion chromatography step. The protein was stored at 4°C in 300 mM NaCl, 50 mM Tris, pH 8.0, 5 mM β-met.
The RTIC was prepared by mixing RT and vRNA-tRNA complex at 2 and 1 μM, respectively, in a buffer containing 25 mM NaCl, 25 mM KCl, 5 mM MgCl2, 50 mM Tris, pH 7.5, 100 μM ddCTP (or dCTP if used for +2 incorporation assays). The mixture was allowed to crosslink overnight at room temperature. The complex was purified by anion-exchange chromatography with a linear gradient. This was followed by a size-exclusion chromatography step to remove any higher-molecular weight aggregates. The purity and homogeneity of the final complex was assessed by SDS-PAGE (under non-reducing conditions) and size-exclusion chromatography (Extended Data Figure 1).
Amino-GMP labeled viral RNA for single-molecule experimentation was transcribed as previously described but with nucleotide concentrations of 1 mM ATP, CTP, UTP and 0.5 mM GMP. The vRNA sequence is identical to that used in the cryo-EM experiments, but contains an additional unstructured sequence on the 3′ end for immobilization and oligonucleotide-hybridization purposes and an additional GU on the 5′ end for labeling purposes. 5′-Amino-G-Monophosphate, purchased from TriLink Biotechnologies, was added to the reaction at a final concentration of 1 mM. The reaction was incubated at 37°C for 4 hour. 5′-Amino-GMP labeled RNA was purified by phenol/chloroform extractions followed by a 10DG (Bio-Rad) desalting column in 10 mM Bis-Tris (ph 7.0), 75 mM NaCl. The RNA was then separated from template DNA free NTPs by size exclusion chromatography (ENRICH SEC 650 10x300) in 100 mM sodium phosphate buffer (pH 8.2), 75 mM NaCl. Purified amino-GMP labeled RNA was concentrated to 1 μM and labeled using NHS chemistry with 1000-fold excess cyanine dye (Lumiprobe). Excess dye was removed by passage over a 10DG desalting column follow by size exclusion chromatography (ENRICH SEC 650 10x300) purification to buffer exchange the labeled vRNA. Labeling efficiency was calculated by measuring the absorbance values of the labeled species at both 260 nm (RNA absorbance) and 550 nm (Cy3 absorbance). These absorbance values were used to calculate the concentrations of the RNA and the Cy3 dye. Using the ratio between these two values, we estimate that our 5′ labeling efficiency is approximately 70%.
Dye-labeled vRNA-tRNA complexes were heat-annealed and purified as previously described. The single-molecule RTIC complex was prepared as stated above, but with a several modifications. To simplify the purification, the his-tag was kept on the p66 subunit of RT. The RTIC was then applied to a Ni-NTA column and washed with 300 mM NaCl to remove the free vRNA-tRNA complexes. The RTIC was eluted from the column. Synthetic oligonucleotides with sequences 5′-GCGGGAGAUCAGGCAU(Am6)-Cyanine5-3′ and 5′-BiotinCUAUUCCCUAUCCdC-3′ (Trilink) were annealed to the complex at 37°C for 5 minutes in 10-fold molar excess. Excess oligonucleotides and free RT were rinsed away during TIRF slide preparation. The above protocol was also performed for a dye-labeled vRNA-tRNA only control, but skipped the RTIC complex formation, purification, and free RT rinse.
Single-Molecule FRET Experiments
Single-molecule FRET experiments were performed using a prism-based total internal reflection instrument with a diode-pumped solid-state 532 nm laser as previously described27,33–38. This includes the use of an oxygen scavenging system (protocatechuate 3,4-dioxygenase (PCD) and β-carboxy-cis,cis-muconic acid (PCA)) and a triplet state relaxer (6-Hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid (Trolox)) to reduce aberrant dye behaviors. The laser power measured 50 mW at the prism. The fluorescence signal was recorded with an exposure of 100 msec per frame for 5 minutes at room temperature. FRET traces were manually analyzed using home-written scripts in MATLAB (MathWorks)27. This analysis begins by employing a colocalization script to select only spots that exhibit both Cy3 donor and Cy5 acceptor fluorescence (under donor-only excitation conditions). Such colocalization allows us to eliminate partially labeled molecules. Next, FRET traces were manually inspected to eliminate cases with multiple single-dye photobleaching events (multiple molecules) or traces that exhibit poor dye photophysics (Extended Data Figure 8d). After this manual inspection, the final dataset used for analysis includes only traces where both dyes exhibit clear single photobleaching events to ensure reliable data (Extended Data Figure 8c). For our single-molecule experiment, 708 traces were selected through colocalization. After manual inspection and elimination of poor traces, 480 traces were used for the final analysis. In addition to the RTIC experiment, a control experiment using a dye-labeled vRNA-tRNA was performed to assess the FRET states without RT. We found that in the absence of cross-linked RT, a small population of low FRET state molecules with FRET efficiency 0.3 exists, but this state is not observed upon binding and cross-linking of RT (data not shown).
Negative Stain EM
3.5 μl of 0.1 μM RTIC sample was applied on glow discharged carbon-coated grids, blotted and stained with 1% uranyl formate according to standard protocols23. Negative stained grids were imaged on an FEI Morgagni at 100kV.
Cryo-EM Data Acquisition
RTIC complex in high monvalent salt buffer (300 mM NaCl, 10 mM Tris-HCl pH 8.0) containing 0.2–0.25% (w/v) OG was applied to glow discharged holey carbon grids (Quantifoil R2/2, 200 mesh) and subsequently vitrified using a FEI Vitrobot. Frozen hydrated samples were imaged on an FEI Titan Krios at 300 kV with a Gatan K2 Summit direct detection camera in counting mode with 200 ms exposure per frame. 40 frames per micrograph were collected at a magnification of 29,000, corresponding to 1 Å/pixel at the specimen level. In total, 4,209 micrographs were collected at defoci values ranging from -1.3 to −2.5 μm. The movie frames were motion-corrected and dose-weighted by MotionCor239 and CTF parameters were estimated by CTFFIND440.
RTIC complex in low monovalent salt buffer and Mg2+ (75 mM NaCl, 2 mM MgCl2, 10 mM Tris-HCl pH 8.0) containing 0.2% (w/v) OG was applied to glow discharged lacey carbon grids (EMS, 200 mesh, Copper) and subsequently vitrified using a Leica EM GP. Frozen hydrated samples were imaged on a Tecnai F20 at 200 kV with a Gatan K2 Summit direct detection camera in counting mode with 200 ms exposure per frame. 60 frames per micrograph were collected at a magnification of 29,000, which corresponds to 1.286 Å/pixel at the specimen plane. In total, 898 micrographs were collected at defoci values varying from −2.0 to −3.0 μm and dose rate 8.0 electrons/pixel/sec. The micrograph movies were motion-corrected, dose-weighted as above, and CTF parameters were estimated by GCTF41.
Cryo-EM Data Processing
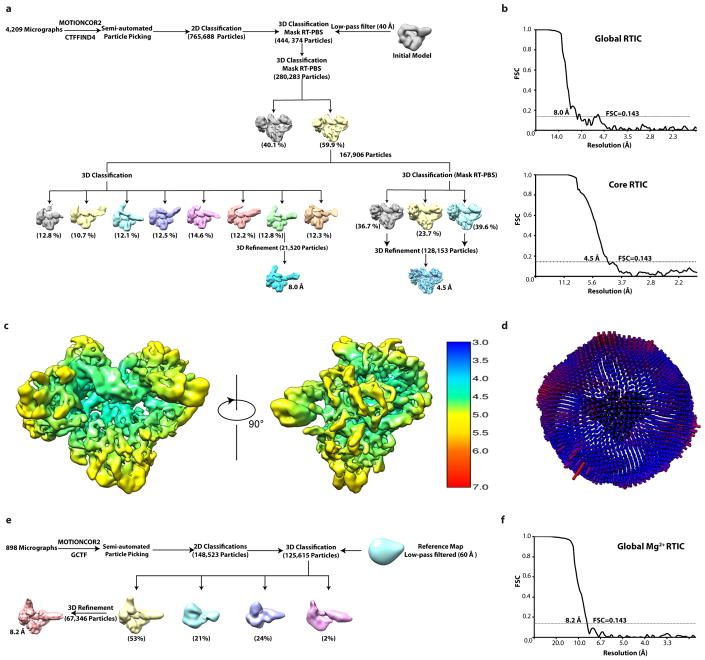
Cryo-EM data for the 8.0 and 4.5 Å maps were processed using Relion42–44. 765,688 particle projections were semi-automatically picked from the motion-corrected micrographs, and sorted through subsequent rounds of reference-free 2D classification. 444,374 particle projections belonging to classes with well-defined RT and RNA features were selected for further processing (Extended Data Figure 3a). An initial 3D model was obtained using VIPER45 based on the selected 2D classes, and used for 3D classification in Relion42,44(Extended Data Figure 3a). Because particle alignment was affected by the flexible protruding RNA, we employed a mask and focused the alignment on RT and PBS alone. 167,906 particle projections sorted to 3D classes displaying all features of RT and PBS were selected for subsequent 3D classifications. To further improve the quality of RT/PBS core, one more round of 3D classification with finer angular sampling was executed; particles from two classes with well-defined secondary structure densities were combined and the 3D structure was refined to a resolution of 4.5 Å. For global RTIC maps (including the flexible protruding RNA) a 3D classification without mask using the 167,906 particle projections subset was performed; eight classes obtained showed the tRNA and vRNA in various conformations. The class displaying most of the RNA protrusions was refined to a resolution of 8.0 Å. The resolution reported is according to 0.143 “gold standard” FSC criterion (Extended Data Figure 3b). The 4.5-Å and 8.0-Å maps were corrected for the modulation transfer function (MTF) of K2 direct detection camera at 300 kV and then sharpened using a B factor of −250 and −200 Å2, respectively, during the post-processing step (Extended Data Table 1). Local resolution was estimated using Relion (Extended Data Figure 3c).
Cryo-EM data for the 8.2 Å, Mg2+, map was processed using Relion. 148,523 particles projections were semi-automatically picked from motion-corrected micrographs and sorted through subsequent rounds of reference-free 2D classification. 125,615 particle projections belonging to classes with well-defined RT and RNA features were selected for further processing (Extended Data Figure 3e). An initial 3D model was obtained using EMAN2 based on selected 2D classes 46 and used for 3D classification. The resolution reported is according to the 0.143 “gold standard” FSC criterion (Extended Data Figure 3f). The maps were corrected for the modulation transfer function (MTF) of K2 direct detection camera at 200 kV and then sharpened using the B factor of −220 during the post-processing step (Extended Data Table 1).
Model Building and Refinement
The crystal structure of RT bound to a DNA/DNA duplex, with nucleic acid substrate removed, was used as a starting model for RT22. After manually fitting the main-chain backbone of RT into its distinct density, four regions of EM density corresponding to RNA are apparent. The most notable region of density is the well-formed RNA helix in the cleft of RT, which corresponds to the PBS helix of the vRNA/tRNA complex. The complete model of the vRNA/tRNA complex was built piecewise, and iteratively, using the Rosetta FARFAR method24. First, initial models of the first seven base pairs of the PBS helix (vRNA residues 181–186 and tRNA residues 71–76 with the extended dC, originally modeled as RNA for simplicity and later edited to a ddC in Coot47) were built with FARFAR, then clustered. Centers of the ten most populated clusters were fit into the density using the colores tool in the Situs package48. Resulting models were manually inspected and selected based on fit to the density and proximity of G71 to C258 on RT (base and residue involved in crosslinking). This helix was extended out to nineteen bases pairs (to include vRNA residues 187–199 and tRNA residues 59–70), models were again clustered, and the cluster centers were fit into the density. The model that best fit the 4.5 Å cryo-EM density was selected. This RT-PBS model, called the RTIC core, was then refined using Phenix real-space-refinement49 with secondary structure restraints in place for the RNA and protein. To further restrain the model during refinement, the N2-cystamine-deoxyguanosine was inserted into the tRNA sequence and a loose disulfide bond constraint with C258 was used during refinement (this was later reverted to a dG as there is no density for the linker atoms). The model was visually inspected and manually adjusted in Coot47. Protein residues lacking EM-density, the vast majority located in the fingers and palm subdomains, were removed after comparison to prior models of RT (Extended Data Figure 4). Due to insufficient resolution, large regions of RT did not exhibit reasonable density for sidechains. Therefore the RT model was truncated to a main-chain backbone before final inspection and submission to the PDB. The geometry of the final refined model was validated using Molprobity50. This refined RTIC core model (Figure 3) served as the anchor point for orienting the vRNA and tRNA portions of the global RTIC model.
Two additional regions of RNA density were located near the fingers subdomain of RT (Figure 2a). Since the crosslinking method used to form the complex harnesses RT polymerase activity, these regions of density correspond to the template vRNA helices. We traced the vRNA template strand out of the active site, allowing us to confidently orient the base of vRNA H2. Models of vRNA H2 (residues 134–178) were built with FARFAR24 based on the consensus secondary structure from past biochemical and biophysical data25,51. These models were clustered and fit into the density.
After positioning vRNA H2, only one region of RNA density near the fingers remained unaccounted. This density, which was continuous with H2 density, corresponds to vRNA H1 (residues 125–131 and 217–223). This density also connects to RNA located in the cleft of RT near the RNase H domain. This suggests that the connection loop (CL) may contact H1 and contribute to the density observed in this region. To confirm the presence of H1 in the RTIC, single-molecule experiments were performed in which a FRET pair was placed on the 5′ and 3′ ends of the helix. We find that in our imaging conditions, 85–95% of molecules exist in a stable high FRET, consistent with H1 formation. vRNA helix 1 was modeled as an ideal A-form helix, then fit into the density using UCSF Chimera52. This initial fit was later refined during the global model building (described below). The connecting loop was partially built in Coot47 starting from vRNA residues 216 and 204. After manual fitting of the first several bases into the density, the rest of connecting loop was built using Rosetta24 and minimized. A model with close fit to the density was chosen for later refinement.
Using the same approach as for the global orientation of the vRNA helices, we find that the fourth and final region of RNA density, located near the RNase H domain of RT, corresponds to remaining portion of tRNALys3. While both vRNA helices exhibited density consistent with past secondary structure models, the tRNA density appeared to differ. Instead of revealing expected density for the two independently folded anti-codon and D-stem loops of the tRNA, the RTIC global map showed density consistent with a continuous helix. Also notable was that this helix extends directly from the PBS helix. After re-examining the sequences of the vRNA and tRNALys3, we noted that it was possible for the vRNA and tRNALys3 to form four extra base pairs, which would extend the PBS from 18 to 22 bp. This extended PBS would be consistent with the continuous helical density in this region. Three out of the four pairs are conserved among subtype-B HIV-1 sequences, suggesting that this structural feature is common. The most variable position would pair with the m1A at position 50 of tRNALys3. Variability in this position is not unexpected, as a Watson-Crick pair would not be able to form. We generated a second, alternative fold for the remaining portions of the tRNALys3 using mFOLD. This secondary structure, differing from previously-observed free form secondary by a free energy of less than 1 kcal/mol, is consistent with a long helical structure and accounts for the density observed in the RTIC global map. In addition to being a good fit for the density, the bulges in this model are consistent with the locations of modified nucleotides that would exist in human tRNALys3. This model also sequesters the anticodon bases of the tRNA, in agreement with chemical mapping data that suggest that these bases are paired13–16,53,54. We note that the apical portion of the extended tRNA helix has very weak EM-density and is likely dynamic. This dynamic nature of the tRNA is illustrated by the wide variety of final conformations seen in the 3D classes of the RTIC. Models of the extended tRNA helix (residues 2–53) were built individually, clustered, and fit into the density.
The models of the nineteen base pair extended-PBS helix, vRNA helix 1, vRNA helix 2, and the extended tRNA helix were grafted together, with connecting regions built de novo with FARFAR24. Coordinate restraints were applied based on the initial fits to the density for each of these four regions. These penalties were applied for deviations in positions >10 Å. The best scoring models were fit into the density in Chimera and a single model was manually selected for further refinement. Regions with the worst agreement with the density, as observed by manual inspection, were subjected to further iterations of FARFAR24 rebuilding and density fitting. The final vRNA/tRNA model was merged with a poly-alanine backbone RT model and refined with one round of Phenix real-space-refinement49 using secondary structure restraints. Due to the inclusion of all vRNA and tRNA bases found in our RNA constructs into the model, the model building and refinement procedure may force potentially disordered regions to fit into the 8.0 Å cryo-EM density map. We stress that the global model presented in the manuscript is meant to aid in interpreting the orientation of the vRNA and tRNA helices with respect to RT and its active sites while showing that the density can encompass most RNA elements. The model should not be used to interpret individual base locations or conformations. For the creation of models for classes 3, 4, and the Mg2+; models of the vRNA H1, vRNA H2, and tRNA were taken from the global model described above. These helical regions were treated as rigid bodies and only the connecting hinge regions (Extended Data Figure 7c) were rebuilt using the protocol described above. All figures for the RTIC core and global models were prepared in Chimera52.
Activity Assays
For all activity assays, the RTIC, RT, and vRNA/tRNA were all purified as described above.
Timecourse Assay
RTIC (200 nM) was preincubated for 20 min at 37°C in 50 mM Tris-HCl, pH 8.0, 50 mM KCl, 2.5 mM MgCl2. Free vRNA/tRNA (200 nM) and RT (2 μM) were also preincubated for 20 min under the same conditions, but with dCTP in order to fully incorporate the first nucleotide prior to dTTP incorporation. Incorporation reactions were started by adding a mixture of α-32P-dTTP (50 nM), and dTTP (50 μM). Reactions were quenched at a range of times from 1 second to 4 hours with the addition of EDTA and SDS loading buffer. The reactions were run on an 4–20% SDS-PAGE gel, dried, and exposed 18 hours on a phosphoimager screen (Molecular Dynamics) and imaged with a Storm 860 (Molecular Dynamics). Bands were quantified using ImageQuant. Intensity was normalized to the highest band intensity for the individual time course assays after background subtraction (set to 1). All time course assays were reliably reproduced and the slow reactions required no special equipment8. Plotting and curve fitting was done using IgorPro. For NNRTI experiments, 1 μM nevirapine was added to the pre-reaction incubation mixture of the RTIC.
Relative Total Incorporation Assay
Reactions were performed as described above. RTIC reaction mixtures were quenched at 1 hour and the free RT + vRNA/tRNA were quenched at 30 min. Samples were quantified as described above. Incorporation was normalized to the average free RT + vRNA/tRNA band intensity (set to 100%). Relative total incorporation assays were done in triplicate.
Reverse Transcriptase Assay
vRNA/tRNA complexes were purified as described above using a tRNA that was labeled on the 5′ end with Cyanine3 dye. Reactions were pre-incubated at 37°C for 5 min in 50 mM Tris-HCl (pH 8.0), 50 mM KCl, 6 mM MgCl2, and 5 mM β-met at a vRNA/tRNA concentration of 200 nM and RT concentration of 3 μM. Reactions were initiated by the addition of a dNTP mixture that brought the final individual dNTP concentrations to 100 μM. Reactions were perfomed in triplicate and quenched at 30 min with EDTA (50 mM). Samples were denatured in a formamide loading buffer, heated for 5 min at 95°C, and loaded on an 8.5% polyacrylamide gel that was pre-run for 2 hours. Samples were run for 3 hours at 120W before imaging with a Typhoon Trio (Amersham Biosciences). Fully extended and unextended primer bands were quantified using ImageQuant. Percent primer extension was calculated and normalized to wild-type RT.
Data Availability
Cryo-EM maps of the global RTIC, the core of the RTIC, and the global RTIC w/MgCl2 have been deposited in the Electron Microscopy Data Bank under accession codes EMDB-7032, EMDB-7031, and EMDB-7540. The coordinates of the RTIC core model have been deposited in the Protein Data Bank under accession code 6B19. The global RTIC model is available as Supplementary Data. All other data are available from the corresponding author upon reasonable request.
Extended Data
Extended Data Figure 1. Purification and activity of RTIC.
a, Initial anion-exchange purification of the RTIC away from free RT and vRNA/tRNA. This purification was repeated for each sample (>10) used in the manuscript, with only slight variations in the chromatogram. b, Polishing step using size-exclusion chromatography purification of the RTIC after anion-exchange. This purification was repeated for each sample used in the manuscript (>10), with only slight variations in the chromatogram. c, A final 10% native TBE gel on the purified components. RT barely enters the gel for the running conditions. The RTIC runs as a single band, however trace amounts of free vRNA and/or vRNA/tRNA complex are sometimes present. This native gel is a representative result that was repeated independently for all purified RTIC samples used in the paper (>10). d, Autoradiograph image illustrating that the RTIC is capable of incorporating an incoming α-32P-dTTP nucleotide when extended and purified using dCTP instead of ddCTP. This gel is a representative result that was repeated independently for crosslinked and uncrosslinked samples (>6 independently prepared samples) used in dTTP incorporation assays. e, The RTIC incorporates α-32P-dTTP at roughly 89% efficiency compared to the free components after reaching a plateau. Values are mean ± s.d. (n=3 independent experiments) with normalization to total incorporation of free RT + vRNA/tRNA reactions. f, Autoradiograph image showing that the incorporation of dTTP is inhibited in the presence nevirapine (NNRTI). Images have been adjusted to allow for identification of the NNRTI-inhibited band. This gel is a representative result that was repeated independently for crosslinked and uncrosslinked samples (3 samples). g, Relative activities, judged by primer usage, of wild-type, Q258C, and Q258C/E478Q reverse transcriptase mutants used in this study. Values are mean ± s.d. (n=3 independent experiments) with normalization to the primer usage of wild-type RT. h, RTIC (triangles), RTIC w/NNRTI (circles) or vRNA/tRNA + excess RT (squares) reactions were initiated by addition of α-32P-dTTP and quenched at different time points. Data were fit using the relationship for the free vRNA/tRNA + RT reaction:
Data were fit using the relationship for the RTIC (with or without NNRTI) reaction:
where A and B represent the amplitude of the fast and slow processes respectively, kpol is the apparent extension rate constant, and kslow is the rate of the slow process. The second relationship was used for the RTIC data, as the slow process appears to dominate incorporation when the vRNA/tRNA substrate is crosslinked to RT. The best fits were obtained with: A = 0.7166 AU, kpol = 0.1078 s−1, B = 0.2754, kslow = 0.01002 s−1 for the vRNA/tRNA + excess RT; B = 0.9808, kslow = 0.003140 s−1 for the RTIC; and B = 1.095, kslow = 0.0001714 s−1 for the RTIC w/NNRTI. kslow is ~3.19 times slower for the crosslinked RTIC compared to the un-crosslinked components. Assays were independently repeated three times to ensure reproducibility.
Extended Data Figure 2. Representative negative stain EM images, cryo-EM images, and 2D averages of the RTIC.
a, Representative negative stain EM image of HIV reverse transcriptase initiation complex (RTIC) reveals a mono-disperse sample that is free of aggregates. Approximately a dozen were taken of each sample prior to cryo-EM grid preparation to ensure sample quality. b, Cryo-EM image of RTIC without beta-octyl glucoside (OG). The long chains correspond to RNA from the complex with very few particles resembling the protein. Results are reproducible in the absence of OG (>10 samples tested). c, Cryo-EM image of RTIC with OG. Single particles corresponding to the complex appear similar to the negative stain visualization. All 5,107 images used in both cryo-EM datasets have a similar appearance with slight differences in particle density. d, Representative 2D averages of RTIC complex from the cryo-EM data collected with OG. Both datasets exhibit very similar 2D classes.
Extended Data Figure 3. Data processing workflow for RTIC complex.
a, Data processing workflow for the 8.0 Å global and 4.5 Å core maps. b, Gold standard Fourier shell correlation (FSC) curve of RTIC core and global maps. c, The final 4.5 Å map is colored according to local resolution estimated by Relion. d, Angular distribution of particle projections. The length of each projection direction is proportional to the number of assigned particles. e, Data processing workflow for the 8.2 Å global Mg2+ map. f, Gold standard Fourier shell correlation (FSC) curve of RTIC Mg2+ global map.
Extended Data Fig 4. Quality of the cryo-EM density for the core RTIC map.
a, View of HIV-1 reverse transcriptase from the front. The subdomains of RT are colored. Underneath the main RTIC view, each subdomain of RT, plus the p51 subunit, is shown fit into the 4.5 Å map. b, View of HIV-1 reverse transcriptase from the polymerase active site side. The subdomains of RT are colored. Underneath the main RTIC view, each subdomain of RT, plus the p51 subunit, is shown fit into the 4.5 Å map. For a and b, Regions of protein, namely loops and linkers, which lacked sufficient density were removed after comparison with previously published structures of RT. These regions are indicated by dotted lines and are most commonly found in the finger and palm subdomains. c, Representative regions of 4.5 Å map fitted with protein secondary structure that display densities for side chains. A view of the PBS helix fit into the 4.5 Å map is also shown; phosphates of the RNA backbone are partially resolved. Regions are colored with respect to the main text models.
Extended Data Figure 5. Mg2+ global map views and structure comparison.
a, Side and top views of the 8.2 Å global map at different density thresholds. The orientation of the peripheral vRNA and tRNA elements is within the variability seen among the different RTIC conformers. b, A model of the RTIC built into the Mg2+ density using the main text global RTIC model. vRNA and tRNA helices were treated as rigid bodies derived from main text model (see Extended Data Figure 6 and Methods). c, Comparison of the global RTIC model RNA (gray) with the Mg2+ model RNA (colored). All three regions of RNA structure (H1, H2, and tRNA) differ in the Mg2+ model, but are adequately described by rigid body movements of the RNA helical elements taken from the global RTIC model. Both H1 and H2 represent a significant structural barrier to initiation. d, Partial accommodation of H1 into high monovalent salt classes 3, 4, and 7.
Extended Data Figure 6. Low resolution tRNA density and fold comparison.
a, Top and side views of the elongated helical tRNA density observed in the low resolution global map of the RTIC. b, Top and side views of the vRNA/tRNA model generated using the hypothesized elongated tRNA helical fold. The tRNA model fits the long helical density well. Corresponding secondary structure is in panel D. c, Top and side views of the vRNA/tRNA model generated using previously hypothesized tRNA secondary structures that have the anticodon and D-stem loops independently folded. Corresponding secondary structure is in panel E. d, Secondary structure depiction of the new vRNA/tRNA and canonical clover-leaf fold of the tRNA. The different domains are colored and correspond with the models in panels B–C. E, Secondary structure depiction of the old vRNA/tRNA fold with independent anticodon and D-stem loops. The domains are colored and correspond with the model in panel C and clover-leaf fold of the tRNA in panel D.
Extended Data Figure 7. Peripheral RNA heterogeneity of the RTIC conformers.
a, Tiled views of 8 conformations emerged from 3D classification of RTIC. Each class is numbered and class 7 was used for the global RTIC reconstruction. b, Superposition of the 8 classes from a. The main areas of RNA heterogeneity are focused on the orientations of vRNA H2, H1/CL, and the tRNA. With no stabilizing protein contacts, vRNA H2, H1, and the tRNA sample a wide range of conformations, limiting the resolution of the global map. c, Additional RTIC models built into classes 3 (tan) and 4 (blue). The models for the tRNA, vRNA H1, and vRNA H2 were all derived from the global RTIC model and treated as rigid bodies for model building. Connecting loop wasn’t built in these models as the density for this region wasn’t clear in these maps, though there is reasonable density to model a loop near H1. Junctions between the helices serve as hinges that allow for movement of the independent domains. The main text global RTIC model (gray) is included as a comparison. d, The vRNA and tRNA helices treated as rigid bodies for modeling are bolded. Hinge points for each helix are highlighted with gray circles and serve as points of flexibility for the RTIC.
Extended Data Figure 8. Single-molecule experimentation and analysis.
a, Secondary structure depiction of the vRNA/tRNA construct used for the single-molecule experimentation. The labeling scheme is shown, with the Cy3 dye located on the 5′ end of the vRNA helix 1 and Cy5 dye located on an oligonucleotide positioned near the 5′ end of helix 1. The vRNA/tRNA complex was crosslinked to RT for the experimentation. b, 95% of the RTIC complexes are in the high FRET, helix 1 formation, state (480 traces analyzed, see Methods). c, Example trace of the ones used for final FRET analysis. The high FRET state of the RTIC complex, which is attributed to helix 1 formation. Photobleaching events for both Cy5 and Cy3 are indicated. d, Examples of traces removed from final FRET analysis. Traces exhibit the presence of multiple molecules (multiple single-dye photobleaching events) or poor dye behavior (blinking and quenching).
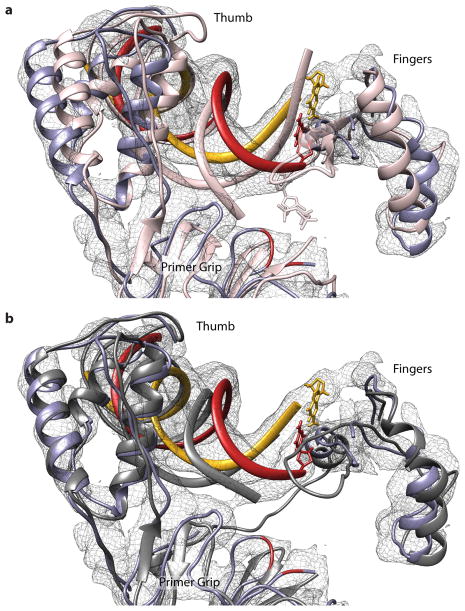
Extended Data Figure 9. Comparison with NNRTI bound and active RT/nucleic acid complexes in the cryo-EM map.
All alignments between structures and the RTIC were done using the p51 subunit. a, Comparison of an active conformation RT/nucleic acid structure (pink, 1RTD) with the RTIC core (RT:purple, tRNA primer:red, vRNA template:yellow). The EM map overlay shows the poor fit of the 1RTD model in the fingers, thumb, and primer grip of RT. Deviations of the nucleic acid primer and template of 1RTD away from the RTIC density is also apparent. b, Comparison of an NNRTI bound RT/nucleic acid structure (dark gray, 3V81) with the RTIC core. The EM map overlay shows the closer fit of the fingers and primer grip regions of RT in the 3V81 model. The thumb region also overlays well, but with slight deviations. Most noticeably, the nucleic acid primer/template in the 3V81 model deviates, although not as dramatically as in 1RTD, from the RTIC core EM density.
Extended Data Table 1.
Cryo-EM data collection, refinement, and validation statistics.
| RTIC Core (EMDB-7031) (PDB-6B19) | RTIC Global (EMDB-7032) (Supplementary Data file for model) | RTIC Global w/MgCl2 (EMDB-7540) | |
|---|---|---|---|
| Data collection and processing | |||
| Magnification (calibrated) | 50,000 | 50,000 | 38,880 |
| Voltage (kV) | 300 | 300 | 200 |
| Electron exposure (e–/Å2) | 60 and 85 | 60 and 85 | 75 |
| Defocus range (μm) | −1.3 to −2.5 | −1.3 to −2.5 | −2.0 to −3.0 |
| Pixel size (Å) | 1.0 | 1.0 | 1.286 |
| Symmetry imposed | C1 | C1 | C1 |
| Initial particle images (no.) | 765,688 | 765,688 | 148,523 |
| Final particle images (no.) | 128,153 | 21,520 | 67,346 |
| Map resolution (Å) | 4.5 | 8.0 | 8.2 |
| FSC threshold | 0.143 | 0.143 | 0.143 |
| Refinement | |||
| Initial model used (PDB code) | 3V81 | 3V81 | |
| Map sharpening B factor (Å2) | −250 | −200 | −200 |
| Model composition | |||
| Non-hydrogen atoms | 5,299 | 8,545 | |
| Protein residues | 909 | 962 | |
| RNA nucleotides | 38 | 178 | |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.023 | 0.003 | |
| Bond angles (°) | 1.603 | 0.83 | |
| Validation | |||
| MolProbity score | 2.41 | 1.92 | |
| Clashscore | 15.00 | 14.25 | |
| Poor rotamers (%) | N/A | N/A | |
| Ramachandran plot | |||
| Favored (%) | 80.46 | 96.15 | |
| Allowed (%) | 19.42 | 3.85 | |
| Disallowed (%) | 0.12 | 0.00 | |
Supplementary Material
Supplemental data list: 1. Global RTIC model
Acknowledgments
We thank A. Frost and L. Stryer for suggesting β-OG as an additive for cryo-EM, R. Kornberg, M. Levitt, P. Geiduschek and W. Sundquist for critically reading the manuscript, M. Levitt for discussion of alternative tRNA folds and general support, D. Herschlag for discussions, and N.R. Latorraca for discussions and assistance with the Sherlock cluster. Supported by NIH grant GM082545 to E.V.P., T32-GM008294 (Molecular Biophysics Training Program) to K.P.L., A.T.C, and K.K., NSF GRFP (DGE-114747) to A.T.C and K.K., and Gabilan Stanford Graduate Fellowship to K.K.. We would like to thank Stanford University and the Stanford Research Computing Center for providing the Sherlock cluster resources. Additional calculations were performed on the Stanford BioX3 cluster, supported by NIH Shared Instrumentation Grant 1S10RR02664701.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions K.P.L., Y.K.M., and D.H.C. acquired preliminary cryo-EM data and performed initial cryo-EM map calculations. Y.K.M. acquired cryo-EM data and obtained the 3D reconstructions shown in the main manuscript. K.P.L. acquired Mg2+ cryo-EM data and performed corresponding cryo-EM map calculations. A.C. purified the vRNA used for single-molecule experimentation and performed the single-molecule experiments. K.P.L., D.B., and L.M. performed all vRNA and RT sample preparations. K.P.L performed all α-32P-dTTP incorporation assays. D.B. performed the RT activity assays. K.P.L. designed the purification scheme and purified the RTIC used in all experimentation. K.K. performed the vRNA/tRNA model building with input from K.P.L. K.P.L. and Y.K.M. performed final RTIC model building and refinement. K.P.L., Y.K.M, G.S., J.D.P., and E.V.P. interpreted the data. K.P.L and E.V.P wrote the manuscript with input from J.D.P., Y.K.M. and G.S.
The authors declare no competing financial interests.
References
- 1.Gilboa E, Mitra SW, Goff S, Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979;18:93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- 2.Sarafianos SG, et al. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J Mol Biol. 2009;385:693–713. doi: 10.1016/j.jmb.2008.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970;226:1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- 4.Temin HM, Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970;226:1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- 5.Paillart JC, Shehu-Xhilaga M, Marquet R, Mak J. Dimerization of retroviral RNA genomes: an inseparable pair. Nat Rev Microbiol. 2004;2:461–472. doi: 10.1038/nrmicro903. [DOI] [PubMed] [Google Scholar]
- 6.Huang Y, et al. Incorporation of excess wild-type and mutant tRNA(3Lys) into human immunodeficiency virus type 1. J Virol. 1994;68:7676–7683. doi: 10.1128/jvi.68.12.7676-7683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ratner L, et al. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 8.Lanchy JM, Ehresmann C, Le Grice SF, Ehresmann B, Marquet R. Binding and kinetic properties of HIV-1 reverse transcriptase markedly differ during initiation and elongation of reverse transcription. EMBO J. 1996;15:7178–7187. [PMC free article] [PubMed] [Google Scholar]
- 9.Lanchy JM, et al. Contacts between reverse transcriptase and the primer strand govern the transition from initiation to elongation of HIV-1 reverse transcription. J Biol Chem. 1998;273:24425–24432. doi: 10.1074/jbc.273.38.24425. [DOI] [PubMed] [Google Scholar]
- 10.Isel C, Ehresmann C, Marquet R. Initiation of HIV Reverse Transcription. Viruses. 2010;2:213–243. doi: 10.3390/v2010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beerens N, Berkhout B. The tRNA primer activation signal in the human immunodeficiency virus type 1 genome is important for initiation and processive elongation of reverse transcription. J Virol. 2002;76:2329–2339. doi: 10.1128/jvi.76.5.2329-2339.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beerens N, Groot F, Berkhout B. Initiation of HIV-1 reverse transcription is regulated by a primer activation signal. J Biol Chem. 2001;276:31247–31256. doi: 10.1074/jbc.M102441200. [DOI] [PubMed] [Google Scholar]
- 13.Goldschmidt V, Ehresmann C, Ehresmann B, Marquet R. Does the HIV-1 primer activation signal interact with tRNA3(Lys) during the initiation of reverse transcription? Nucleic Acids Res. 2003;31:850–859. doi: 10.1093/nar/gkg187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldschmidt V, et al. Structural variability of the initiation complex of HIV-1 reverse transcription. J Biol Chem. 2004;279:35923–35931. doi: 10.1074/jbc.M404473200. [DOI] [PubMed] [Google Scholar]
- 15.Goldschmidt V, et al. Direct and indirect contributions of RNA secondary structure elements to the initiation of HIV-1 reverse transcription. J Biol Chem. 2002;277:43233–43242. doi: 10.1074/jbc.M205295200. [DOI] [PubMed] [Google Scholar]
- 16.Isel C, et al. Structural basis for the specificity of the initiation of HIV-1 reverse transcription. EMBO J. 1999;18:1038–1048. doi: 10.1093/emboj/18.4.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwatani Y, Rosen AE, Guo J, Musier-Forsyth K, Levin JG. Efficient initiation of HIV-1 reverse transcription in vitro. Requirement for RNA sequences downstream of the primer binding site abrogated by nucleocapsid protein-dependent primer-template interactions. J Biol Chem. 2003;278:14185–14195. doi: 10.1074/jbc.M211618200. [DOI] [PubMed] [Google Scholar]
- 18.Liang C, et al. The importance of the A-rich loop in human immunodeficiency virus type 1 reverse transcription and infectivity. J Virol. 1997;71:5750–5757. doi: 10.1128/jvi.71.8.5750-5757.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu S, Harada BT, Miller JT, Le Grice SF, Zhuang X. Initiation complex dynamics direct the transitions between distinct phases of early HIV reverse transcription. Nat Struct Mol Biol. 2010;17:1453–1460. doi: 10.1038/nsmb.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 21.Jacobo-Molina A, et al. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc Natl Acad Sci U S A. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das K, Martinez SE, Bauman JD, Arnold E. HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism. Nat Struct Mol Biol. 2012;19:253–259. doi: 10.1038/nsmb.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peisley A, Skiniotis G. 2D Projection Analysis of GPCR Complexes by Negative Stain Electron Microscopy. Methods Mol Biol. 2015;1335:29–38. doi: 10.1007/978-1-4939-2914-6_3. [DOI] [PubMed] [Google Scholar]
- 24.Das R, Karanicolas J, Baker D. Atomic accuracy in predicting and designing noncanonical RNA structure. Nat Methods. 2010;7:291–294. doi: 10.1038/nmeth.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watts JM, et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foley B, LT, Apetrei C, Hahn B, Mizarchi J, Rambaut A, Wolinsky S, Korber B. HIV Sequence Compendium 2013. Los Alamos National Laboratory; 2013. NM, 13-26007. [Google Scholar]
- 27.Coey A, Larsen K, Puglisi JD, Viani Puglisi E. Heterogeneous structures formed by conserved RNA sequences within the HIV reverse transcription initiation site. RNA. 2016;22:1689–1698. doi: 10.1261/rna.056804.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puglisi EV, Puglisi JD. Secondary structure of the HIV reverse transcription initiation complex by NMR. J Mol Biol. 2011;410:863–874. doi: 10.1016/j.jmb.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li A, Gong S, Johnson KA. Rate-limiting Pyrophosphate Release by HIV Reverse Transcriptase Improves Fidelity. J Biol Chem. 2016;291:26554–26565. doi: 10.1074/jbc.M116.753152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beerens N, et al. Role of the primer activation signal in tRNA annealing onto the HIV-1 genome studied by single-molecule FRET microscopy. RNA. 2013;19:517–526. doi: 10.1261/rna.035733.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gotte M, et al. HIV-1 reverse transcriptase-associated RNase H cleaves RNA/RNA in arrested complexes: implications for the mechanism by which RNase H discriminates between RNA/RNA and RNA/DNA. EMBO J. 1995;14:833–841. doi: 10.1002/j.1460-2075.1995.tb07061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marshall RA, Dorywalska M, Puglisi JD. Irreversible chemical steps control intersubunit dynamics during translation. Proc Natl Acad Sci U S A. 2008;105:15364–15369. doi: 10.1073/pnas.0805299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johansson M, Chen J, Tsai A, Kornberg G, Puglisi JD. Sequence-dependent elongation dynamics on macrolide-bound ribosomes. Cell Rep. 2014;7:1534–1546. doi: 10.1016/j.celrep.2014.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Leary SE, Petrov A, Chen J, Puglisi JD. Dynamic recognition of the mRNA cap by Saccharomyces cerevisiae eIF4E. Structure. 2013;21:2197–2207. doi: 10.1016/j.str.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aitken CE, Puglisi JD. Following the intersubunit conformation of the ribosome during translation in real time. Nat Struct Mol Biol. 2010;17:793–800. doi: 10.1038/nsmb.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J, Tsai A, Petrov A, Puglisi JD. Nonfluorescent quenchers to correlate single-molecule conformational and compositional dynamics. J Am Chem Soc. 2012;134:5734–5737. doi: 10.1021/ja2119964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheres SH. Semi-automated selection of cryo-EM particles in RELION-1.3. J Struct Biol. 2015;189:114–122. doi: 10.1016/j.jsb.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scheres SH. Processing of Structurally Heterogeneous Cryo-EM Data in RELION. Methods Enzymol. 2016;579:125–157. doi: 10.1016/bs.mie.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 45.Penczek PA, Grassucci RA, Frank J. The ribosome at improved resolution: new techniques for merging and orientation refinement in 3D cryo-electron microscopy of biological particles. Ultramicroscopy. 1994;53:251–270. doi: 10.1016/0304-3991(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 46.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 47.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 48.Wriggers W. Conventions and workflows for using Situs. Acta Crystallogr D Biol Crystallogr. 2012;68:344–351. doi: 10.1107/S0907444911049791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lavender CA, Gorelick RJ, Weeks KM. Structure-Based Alignment and Consensus Secondary Structures for Three HIV-Related RNA Genomes. PLoS Comput Biol. 2015;11:e1004230. doi: 10.1371/journal.pcbi.1004230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 53.Isel C, Ehresmann C, Keith G, Ehresmann B, Marquet R. Initiation of reverse transcription of HIV-1: secondary structure of the HIV-1 RNA/tRNA(3Lys) (template/primer) J Mol Biol. 1995;247:236–250. doi: 10.1006/jmbi.1994.0136. [DOI] [PubMed] [Google Scholar]
- 54.Isel C, et al. Specific initiation and switch to elongation of human immunodeficiency virus type 1 reverse transcription require the post-transcriptional modifications of primer tRNA3Lys. EMBO J. 1996;15:917–924. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental data list: 1. Global RTIC model
Data Availability Statement
Cryo-EM maps of the global RTIC, the core of the RTIC, and the global RTIC w/MgCl2 have been deposited in the Electron Microscopy Data Bank under accession codes EMDB-7032, EMDB-7031, and EMDB-7540. The coordinates of the RTIC core model have been deposited in the Protein Data Bank under accession code 6B19. The global RTIC model is available as Supplementary Data. All other data are available from the corresponding author upon reasonable request.