Summary
Binding of sweet, umami, and bitter tastants to G-protein coupled receptors (GPCR) in apical membranes of type II taste bud cells (TBCs) triggers action potentials that activate a voltage-gated nonselective ion channel to release ATP to gustatory nerves mediating taste perception. Although CALHM1 is necessary for ATP release, the molecular identification of the channel complex that provides the conductive ATP-release mechanism suitable for action potential-dependent neurotransmission remains to be determined. Here, we show that CALHM3 interacts with CALHM1 as a pore-forming subunit in a CALHM1/CALHM3 hexameric channel, endowing it with fast voltage-activated gating identical to that of the ATP-release channel in vivo. Calhm3 is co-expressed with Calhm1 exclusively in type II TBCs and its genetic deletion abolishes taste-evoked ATP release from taste buds and GPCR-mediated taste perception. Thus, CALHM3, together with CALHM1, is essential to form the fast voltage-gated ATP-release channel in type II TBCs required for GPCR-mediated tastes.
Graphical abstract
Ma et al. identify a CALHM1/CALHM3 hetero-hexameric ion channel as the mechanism by which type II taste bud cells release ATP as a neurotransmitter to gustatory neurons in response to GPCR-mediated tastes, including sweet, bitter and umami substances.
Introduction
Taste buds in the tongue and palate epithelium are the detectors of chemicals contained in foods and drinks, and transmit their taste information to the brain through afferent gustatory nerves. Most mammals, including human and mouse, detect sweetness, bitterness, saltiness, sourness and umami (“meaty” or “savory” taste of monosodium L-glutamate) as the five basic taste modalities, plus several less-well characterized tastes such as “fat”, “starch” and “calcium.” Taste perception mechanisms can be dichotomized into those involving ion channels and those involving G-protein coupled receptors (GPCRs) (Liman et al., 2014). The GPCRs are located in the apical membranes of type II taste bud cells (TBCs), where they detect sweet, umami, and bitter compounds (Kinnamon, 2011; Liman et al., 2014). GPCR activation triggers a signal transduction cascade involving activation of heterotrimeric G proteins and phospholipase C-β2 (PLCB2), production of InsP3, and InsP3-dependent Ca2+ release from the endoplasmic reticulum through InsP3 receptor type 3 (InsP3R3). The intracellular [Ca2+] rise activates monovalent cation-selective transient receptor potential M5 (TRPM5) channels in the basolateral plasma membrane, causing membrane depolarization that triggers Na+ action potential firing, and depolarization-induced release of ATP that in turn acts as the primary neurotransmitter to stimulate P2X receptors on afferent gustatory neurons (Finger et al., 2005; Kinnamon, 2013). Type II TBC neurotransmitter release is highly unusual in employing an ion-channel mechanism rather than classical vesicular exocytosis (Chaudhari, 2014; Kinnamon, 2011; Liman et al., 2014; Taruno et al., 2013). Type II cells lack classical synaptic structures, including synaptic vesicles and expression of genes involved in synaptic vesicle filling (Clapp et al., 2006; Clapp et al., 2004; DeFazio et al., 2006).
The bone fide channel complex of the ATP release channel remains unknown. Calcium homeostasis modulator 1 (CALHM1), a voltage-gated nonselective channel with a wide-pore diameter (Ma et al., 2012; Siebert et al., 2013), is an essential component of the channel mechanism that releases ATP in response to taste-evoked Na+ action potentials (Taruno et al., 2013). In its absence, taste compounds fail to stimulate ATP release, and mice lose perception of GPCR-mediated tastes despite intact type II cell signaling (Taruno et al., 2013; Tordoff et al., 2014). However, the voltage-dependent activation kinetics and pharmacological sensitivity of CALHM1 channels differ markedly from those of the neurotransmitter-release channels in situ (Chaudhari, 2014; Kinnamon, 2013; Ma et al., 2012). When expressed in Xenopus oocytes, CALHM1 channels are activated by membrane depolarization with kinetics (τ> 500 ms) (Ma et al., 2012) that are too slow to be activated by the Na+ action potentials of ∼3 ms half-width duration (Ma et al., 2017) that trigger ATP release (Murata et al., 2010; Taruno et al., 2013). Importantly, the activation kinetics of ATP-release channel currents in type II TBCs are considerably faster (τ∼10 ms (Ma et al., 2017; Romanov et al., 2008; Takeuchi et al., 2011) than those of heterologously-expressed CALHM1. Furthermore, ATP release by type II TBCs is inhibited by the nonspecific pannexin-1 and connexin hemichannel inhibitor carbenoxolone (CBX) (Dando and Roper, 2009; Huang et al., 2011; Huang et al., 2007; Murata et al., 2010), whereas CALHM1 currents in Xenopus oocytes are not (Ma et al., 2012). These results indicate that CALHM1 is a necessary component of the voltage-activated ATP-release channel in type II TBCs, but is itself insufficient to account for the properties of the endogenous channel in vivo (Chaudhari, 2014; Kinnamon, 2013). Although pannexins were suggested to play a role in ATP release, recent evidence indicates that they are not involved (Romanov et al., 2012; Tordoff et al., 2015; Vandenbeuch et al., 2015). Thus, the molecular identification of the ATP-release channel complex that provides the conductive ATP-release mechanism suitable for action potential-dependent neurotransmission in type II TBCs remains to be determined.
CALHM1 is gated by membrane voltage and by extracellular Ca2+ (Ma et al., 2012; Siebert et al., 2013). In heterologous expression systems, CALHM1 forms homo-hexameric channels with a wide pore diameter (∼14 Å), structural properties that it shares by convergent evolution with connexin and pannexin/innexin/LRRC8 channel families (Ma et al., 2012; Siebert et al., 2013). CALHM1 is a nonselective channel, with significant permeabilities to divalent cations as well as to anions, including ATP (Siebert et al., 2013). In vertebrates, five homologs of CALHM1 exist. CALHM genes, originally identified as the FAM26 gene family (Dreses-Werringloer et al., 2008), are present throughout vertebrates but they lack significant sequence homology to other known genes. Outside of vertebrates, CALHM1 homologs are absent in yeast and Drosophila, whereas Caenorhabditis elegans possesses a single homolog, clhm-1. Heterologous expression of CLHM-1 revealed it to be a functional ion channel with permeation properties and Ca2+o- and voltage-dependencies reminiscent of those of human and mouse CALHM1 (Tanis et al., 2017; Tanis et al., 2013). The functions of the other members of this gene family have not yet been identified.
Here we demonstrate that the Calhm1 homolog Calhm3 is enriched in type II TBCs and is a pore-forming subunit of a novel hexameric CALHM1/CALHM3 ion channel. Whereas CALHM3 expressed alone does not generate ionic currents, CALHM1/CALHM3 channels have fast voltage-dependent activation kinetics and pharmacological sensitivity identical to those of the ATP-release channel in type II TBCs. Genetic deletion of Calhm3 eliminates voltage-gated nonselective currents and taste-evoked ATP release in type II TBCs without affecting cell excitability or diminishing Calhm1 expression, and results in the loss of responses to sweet, umami and bitter tastes. Thus, CALHM3 is essential for action potential-dependent release of ATP from type II taste cells, and a voltage-gated CALHM1/CALHM3 channel is the ATP-release channel required for GPCR-mediated tastes.
Results
CALHM3 Accelerates Voltage-Activated Gating of CALHM1
ATP release by type II TBCs is triggered by action potentials whereas CALHM1 channels have activation kinetics that are too slow for them to be activated by action potentials. Accordingly, we reasoned that homo-hexameric CALHM1 channels cannot be the endogenous ATP-release channel and that additional components must be associated with the ATP-release channel complex that confer fast voltage activation. To identify candidate components, we first considered other homologs of CALHM1. Calhm2 and Calhm3 are also expressed in TBCs (Moyer et al., 2009), but only Calhm1 and Calhm3 are enriched there (Taruno et al., 2013) (Figure S1A). To determine whether these other CALHM1 homologs are also functional ion channels, we first used the Xenopus oocyte expression system. Expression of CALHM1 (Ma et al., 2012; Siebert et al., 2013) or the single C. elegans homolog (Tanis et al., 2013) in Xenopus oocytes generates voltage-gated currents that activate with slow kinetics. In contrast, expression of either CALHM2 or CALHM3, both of which localized to the oocyte plasma membrane (not shown), or both together failed to generate novel plasma membrane conductances (Figures 1A, S1B and S1C), suggesting that unlike CALHM1 or its C. elegans homolog, neither CALHM2 nor CALHM3 can form functional plasma membrane channels.
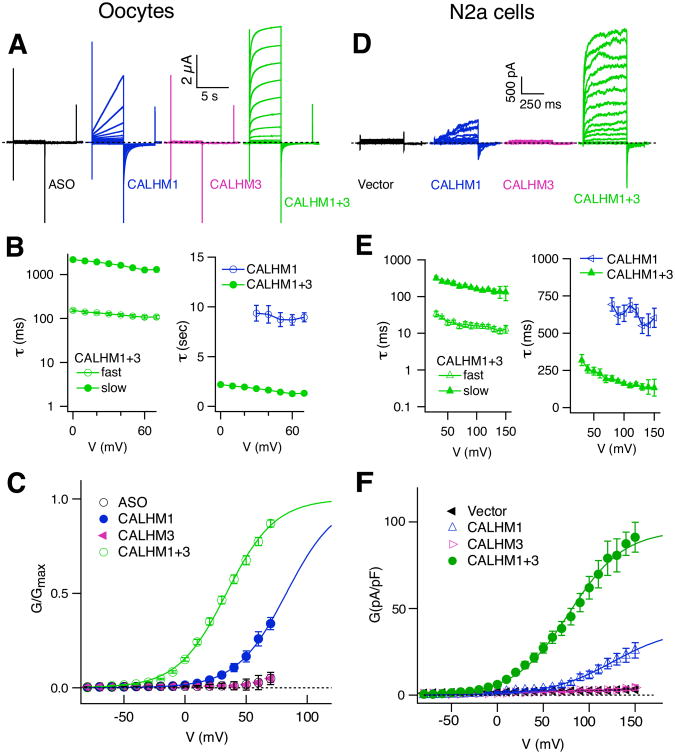
Figure 1. CALHM3 Enhances Voltage-dependent Gating of CALHM1 Channels.
(A-C) Biophysical features of CALHM currents in Xenopus oocytes. (A) Representative families of currents in control Xenopus oocytes (ASO) and oocytes expressing mouse CALHM1, CALHM3 or both (CALHM1+3), evoked by 5-s voltage pulses from -80 mV to +70 mV in 10-mV increments from holding potential of -40 mV every 30 s. Dashed line: zero-current level. (B) Activation time constants: left, fast (τfast) and slow (τslow) components of CALHM1+3 currents (n =19), obtained by fitting outward currents with a double exponential function; right, τ of CALHM1 currents (n = 13), obtained by fitting with a single exponential function, compared with τslow of CALHM1+3 currents. (C) Conductance-voltage (G-V) relations. See METHODS for normalization procedures. Solid lines: Boltzmann function fits with V0.5 and Z0: +77.6 ± 3.2 mV, 1.3 ± 0.1 e for CALHM1 (n = 13); +30.5 ± 1.9 mV, 1.4 ± 0.1 e for CALHM1+3 (n = 27). Mean ± s.e.m., two-tailed unpaired Student's t-test: P = 8.56E-16, t38 = 13.225 for V0.5; P = 0.154, t38 = 1.454 for Z0.
(D-F) Similar to (A-C), in N2a cells. (D) Representative families of currents in N2a cells expressing mouse CALHM1, CALHM3 or both or neither, evoked by 500-ms voltage pulses from -80 mV to +150 mV in 10-mV increments from holding potential of -40 mV every 10 s. (E) Activation time constants: left, τfast and τslow for currents in N2a cells co-expressing CALHM1+3 (n = 22) obtained by a double exponential fit; right, activation time constants for CALHM1 currents (n = 14) obtained by single exponential fitting, compared with τslow of CALHM1+3 currents. The activation time constants obtained from oocytes and N2a cells were different due to different duration of the depolarizing pulses and the slow voltage-clamp and larger capacitance of oocytes. (F) G-V relations obtained by measurement of inward currents at -80 mV evoked by 500-ms voltage pre-pulses, normalized to individual whole-cell capacitance. Whole-cell capacitances were not different (vector: 14.6 ± 1.2 pF, n = 14; CALHM1: 12.7 ± 0.7 pF, n =14; CALHM3: 14.3 ± 1.4 pF, n = 12; CALHM1+3: 13.7 ± 0.6 pF, n = 22 (Two-tailed unpaired Student's t-test: CALHM1, P = 0.163, t26 = 1.436; CALHM3: P = 0.872, t24 = 0.162; CALHM1+3, P = 0.469, t34 = 0.733). Solid lines: Boltzmann function fits with V0.5 and Z0: +148.8 ± 6.4 mV, 0.78 ± 0.06 e for CALHM1 (n=14); +79.5 ± 4.2 mV, 0.84 ± 0.03 e for CALHM1+3 (n = 22). (Two-tailed unpaired Student's t-test: P = 2.259E-11, t34 = 9.744 for V0.5; P = 0.365, t34 = 0.918 for Z0). The G-V relations of CALHM1+CALHM3 from Xenopus oocytes and N2a cells were different due to different depolarizing voltage-ranges, from -80 mV to +70 mV in oocytes and -80 mV to + 150 mV in N2a cells, respectively. See also Figures S1 and S2.
We next asked whether co-expressing either homolog with CALHM1 affected the CALHM1 channel currents. Co-expression of CALHM2 with CALHM1 resulted in currents with magnitudes and channel gating kinetics similar to those of CALHM1 alone (Figures S1B and S1C). In contrast, expressing CALHM3 with CALHM1 (CALHM1+CALHM3) generated large currents with 50-fold faster activation kinetics (τ ∼ 100 ms) than those of CALHM1 (τ > 5,000 ms) (Figures 1A and 1B), and shifted the voltage dependence of activation to more hyperpolarizing voltages (Figures 1C, S1C and S1D). Extracellular Ca2+ (Ca2+o) is an allosteric modulator of voltage-dependent gating of CALHM1 (Ma et al., 2012). Currents in CALHM1+CALHM3-expressing Xenopus oocytes retained sensitivity to Ca2+o (Figure S1D).
To determine the relevance of these findings for mammalian cells, we undertook similar studies by expressing CALHM isoforms in the N2a mouse neuroblastoma cell line. Unlike in Xenopus oocytes, expression of CALHM1 in N2a cells generated only small slowly-activating voltage-gated currents (Figures 1D and 1F). As in oocytes, expression of either CALHM2 or CALHM3 alone or together failed to generate novel plasma membrane conductances. Furthermore, co-expressing CALHM2 with CALHM1 resulted in currents similar to those of CALHM1 alone (not shown). However, expression of CALHM3 with CALHM1 resulted in profoundly altered currents. Voltage-activated current density was enhanced by ∼10-fold at +100 mV (Figures 1D and 1F), and the conductance-voltage (G-V) relationship was shifted to more hyperpolarizing voltages from an activation voltage of +70 mV to ∼0 mV (Figure 1F). Most notably, expression of CALHM3 with CALMH1 strongly accelerated the voltage-dependent activation kinetics by over an order of magnitude from τ > 500 ms to τ ∼10 ms (Figure 1E). Together, these results demonstrate that CALHM3 strongly affects voltage gating of CALHM1 channels.
To determine whether CALHM3 altered the permeability properties of CALHM1 channels, we measured reversal potentials in Xenopus oocytes expressing CALHM1 or CALHM1+CALHM3. CALHM3 had no significant effect on the relative permeabilities of Ca2+, Na+, K+ and Cl- (PCa:PNa:PK:PCl, 8.2:1:1.12:0.56 for CALHM1 and 8.1:1:1.08:0.54 for CALHM1+CALHM3, respectively; Figure S2). To address whether CALHM3 altered CALHM1 ATP permeability, ATP release from HeLa cells expressing CALHM3 and CALHM1 was measured by a luciferin-luciferase assay used previously to demonstrate the ability of CALHM1 to mediate ATP release (Taruno et al., 2013). CALHM3 alone failed to promote ATP release, consistent with its failure to generate an ion conductance, whereas it enhanced CALHM1-mediated ATP release (Figures 2 and S3).
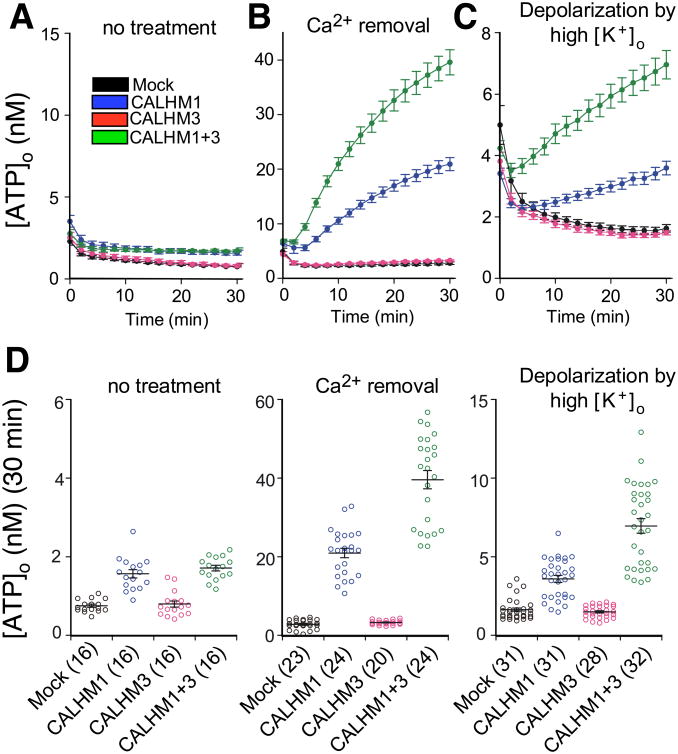
Figure 2. CALHM3 Accelerates CALHM1-mediated ATP Release.
(A-C) Time courses of extracellular-ATP levels due to release from mock-, CALHM1-, CALHM3- and CALHM1+3-transfected HeLa cells exposed to normal (1.9 mM; A) or essentially zero (17 nM; B) Ca2+o or to depolarization by exposure to high [K+]o (117.5 mM; C)
(D) Summary of total release over 30 min. Number of wells shown in parenthesis. Mean ± s.e.m. Statistical differences were calculated by one-way ANOVA (F(3,60)=41.74, p = 1.069E-14 for no treatment; F(3, 87) = 163.51, P = 1.207E-35 for Ca2+o removal; F(3,118)=81.37, p = 1.339E-28 for high [K+]o) with Bonferroni post-hoc test. *P < 0.0083; n.s., not significant. Exact P values of Mock vs CALHM1, Mock vs CALHM3, Mock vs CALHM1+3, and CALHM1 vs CALHM1+3 are respectively 5.4645E-10, 0.6969, 3.0590E-12, and 0.1912 for no treatment; 1.0156E-14, 0.8178, 3.5725E-33, and 2.7564E-16 for Ca2+o removal; and 2.0393E-6, 0.7282, 1.6021E-25, and 1.2008E-13 for high [K+]o. See also Figure S3.
These results suggest that CALHM3 strongly enhances voltage-dependent gating of CALHM1 without altering its permeability properties. Although CALHM proteins lack obvious voltage-sensing domains, the activation kinetics observed in N2a cells co-expressing CALHM1 and CALHM3 are similar to those of some voltage-gated K+ channels, suggesting that the CALHM1+CALHM3 currents could be activated by action potentials (Ma et al., 2017).
CALHM3 Interacts with CALHM1 As a Pore-Forming Subunit of a Novel Hexameric Ion Channel
To determine whether CALHM1 and CALHM3 interact in a single ion channel complex, we used heterologous expression because insufficient amounts of CALHM1 and CALHM3 proteins precluded biochemical analyses of CALHM proteins in taste bud tissues (see Methods). We first expressed epitope-tagged CALHM1 and CALHM3 alone or together in N2a cells to examine their biochemical interaction. When co-expressed, the two proteins co-localized (Figure 3A), and immunoprecipitation of one CALHM homolog co-precipitated the other (Figure 3B), indicating that they interact. In contrast, immunoprecipitation of CALHM1 failed to pull down Panx-1 or other membrane proteins (Figure S4), demonstrating the specificity of the interaction. Whereas CALHM1 was mainly localized in intracellular compartments when expressed alone (as in Dreses-Werringloer et al., 2008), co-expression of CALHM3 enhanced its plasma membrane (PM) localization (Figure 3C). To examine this more directly, we employed surface biotinylation of plasma membrane proteins. These studies suggested that co-expressed CALHM1 and CALHM3 reciprocally promoted their PM localization (Figures 3D-G). These results may account for the order-of-magnitude larger currents in N2a cells expressing both CALHM homologs compared with CALHM1 alone (Figures 1D and 1F).
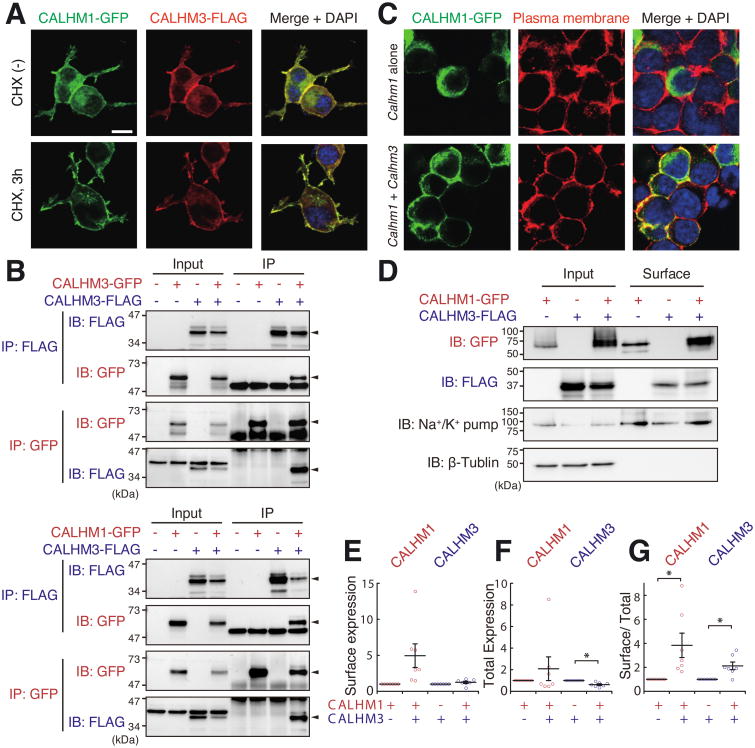
Figure 3. CALHM3 Co-localizes and Interacts with CALHM1.
(A) Epitope-tagged CALHM1 and CALHM3 co-localize in N2a cells transfected with CALHM1-GFP and CALHM3-FLAG (1:1 ratio), in absence and presence of cycloheximide (CHX; to minimize intracellular CALHM). DAPI was used as nuclear counterstain.
(B) CALHM3 co-immunoprecipitates with itself and with CALHM1. Left: CALHM3 physically interacts with CALHM3. N2a cells were transfected with CALHM3-GFP and/or CALHM3-FLAG. Right: CALHM3 physically interacts with CALHM1. N2a cells transfected with CALHM1-GFP and/or CALHM3-FLAG as indicated. Epitope-tagged CALHM proteins were immunoprecipitated 24 h after transfection and analyzed by western blotting. Input: whole-cell lysate; IP, immunoprecipitated sample.
(C) PM localization of CALHM1 is promoted by co-expression of CALHM3. Biotin immunoreactivity and DAPI were used as PM marker and nuclear counterstain, respectively.
(D) Cell surface proteins were biotinylated and pulled down with streptavidin beads. CALHM in whole-cell lysates (Input) and biotinylated-protein samples (Surface) detected by immunoblotting using anti-tag antibodies. Na+/K+-ATPase and β-tubulin were used as markers of PM and cytoplasm, respectively.
(E-G) Levels of CALHM proteins in PM (E), total cells (F), and the ratio of PM to total cells (G). CALHM proteins in PM fraction and whole cell lysates detected as in (D) were measured and normalized by the amounts of Na+/K+-ATPase (E, G) and β-tubulin (F, G). Data shown as fold-change caused by co-expression of other isoform. Surface/total expression ratios of CALHM1 and CALHM3 were both increased by co-expression of other isoform. Mean ± s.e.m.; *P < 0.05 (one sample t-test); n=7. P values for CALHM1 and CALHM3 are respectively 0.053 (t6 = 2.410) and 0.168 (t6 = 1.566) in (E); 0.359 (t6 = 0.994) and 0.002 (t6 = 5.099) in (F); 0.034 (t6 = 2.766) and 0.012, t6 = 3.547 (G).
See also Figure S4.
When expressed alone, CALHM1 forms a homo-hexameric channel complex as determined by native PAGE biochemistry and single molecule photobleaching (Siebert et al., 2013). CALHM3 could interact with CALHM1 in a channel complex as an accessory β-subunit or as a pore-forming subunit. To distinguish between these possibilities, we first performed Blue native (BN)-PAGE of N2a cell lysates from cells expressing tagged CALHM1 and CALHM3 proteins. BN-PAGE of N2a cell lysates revealed that CALHM1-FLAG and CALHM1-GFP exist in complexes of ∼630 (n=4) and ∼780 (n=3) kDa, respectively (Figure 4A). The greater than predicted molecular masses for hexameric CALHM1-FLAG and CALHM1-GFP (∼245 and ∼430 kDa, respectively) are likely due to anomalous slow migration of CALHM1 in BN-PAGE (Figure S5A). The molecular mass difference of ∼150 kDa between CALHM1-FLAG and CALHM1-GFP complexes is ∼6 times the molecular mass difference between GFP and FLAG, consistent with a homo-hexameric CALHM1 complex (see also Figure S5C). Importantly, co-expression of CALHM3-GFP increased the molecular mass of the CALHM1-associated complex by ∼110 kDa (n=3; Figures 4B and S5C), indicating that the two proteins interact in a single complex with each channel containing ≥1 CALHM3 subunit.
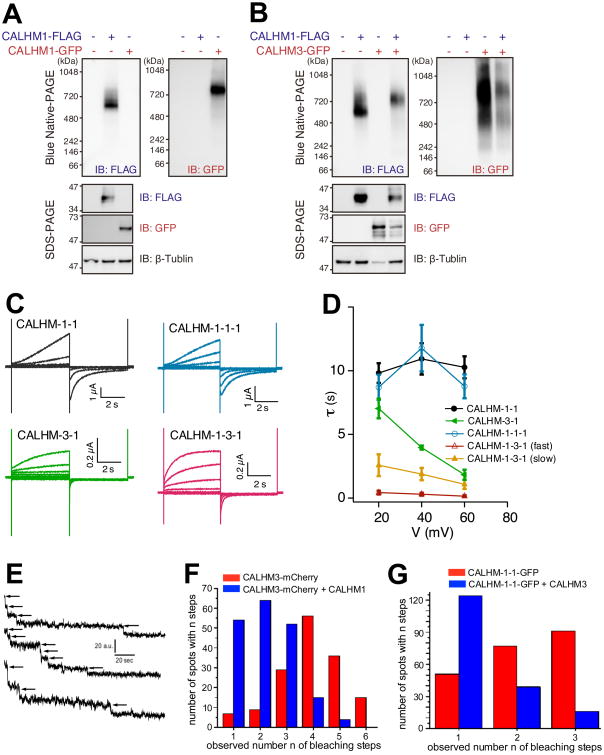
Figure 4. CALHM3 and CALHM1 Exist in a Single Hexameric Channel Complex.
(A) BN-PAGE analysis of CALHM1-FLAG and CALHM1-GFP in N2a cell lysates 24 h after transfection. Molecular-weight shift between CALHM1-FLAG and CALHM1-GFP complexes is in agreement with a CALHM1 homo-hexamer.
(B) CALHM3 is incorporated into the same protein complex with CALHM1 in lysates from N2a cells transfected 24 hrs earlier with CALHM1-FLAG and/or CALHM3-GFP. Whole-cell lysates analyzed by BN-PAGE (upper) and SDS-PAGE (lower). Co-expression of CALHM3-GFP slowed migration of the CALHM1-FLAG-associated complex (upper left panel).
(C) Representative families of whole-cell currents from Xenopus oocytes expressing CALHM-1-1, CALHM-3-1, CALHM-1-1-1 and CALHM-1-3-1 concatemers, evoked by 5-s voltage pulses every 30 s from -80 mV to +60 mV in 20-mV increments from a holding potential of -40 mV in bath containing 1.5 mM Ca2+ and 1 mM Mg2+. Dashed lines: zero-current level.
(D) Activation time constants obtained from a single-exponential function for CALHM-1-1 (n = 8), CALHM-3-1 (n = 5) and CALHM-1-1-1 (n = 10) currents, and from a double-exponential function for CALHM-1-3-1 currents (n = 8).
(E) Representative examples of single molecule bleaching records obtained from Xenopus oocytes expressing CALHM3-mCherry alone.
(F) Distribution of number of bleaching steps observed from CALHM3-mCherry expressing oocytes co-expressing (188 particles) or not (149 particles) untagged-CALHM1.
(G) Distribution of number of bleaching steps observed from CALHM1-CALHM1-GFP concatemers co-expressing (179 spots) or not (219 spots) untagged-CALHM3.
See also Figure S5.
A rigorous determination of whether CALHM3 interacts with the CALHM1-channel complex as a pore-forming subunit would ideally involve a demonstration that specific mutations in CALHM3 alter the relative ion selectivity of the channel. However, the channel formed by CALHM1+CALHM3 is non-selective, and furthermore neither the location of the ion-conduction pathway in CALHM1 nor its molecular determinants have been defined. Therefore, as an alternative approach we constructed a series of CALHM concatemers. Dimeric CALHM-1-1 and trimeric CALHM-1-1-1 concatemers were functional with currents having slow activation kinetics similar to those generated by expression of monomeric CALHM1 (Figures 4C and 4D), validating this approach. In contrast, dimeric CALHM-3-1 and trimeric CALHM-1-3-1 concatemers generated currents with faster activation kinetics resembling those generated by co-expression of CALHM1 and CALHM3 (Figures 4C and 4D). Furthermore, CALHM3 accelerated the activation kinetics of a CALHM-1-1 concatemer (Figure S5D). Importantly, CALHM1 and CALHM-1-1, CALHM-1-1-1 and CALHM-1-3-1 concatemers all ran at identical molecular weights in BN-PAGE (Figure S5B). Together, these data strongly suggest that CALHM3 associates with CALHM1 as a pore-forming subunit in a hexameric channel complex.
To obtain independent evidence of a heteroligomeric CALHM1/CALHM3 channel complex, we performed single-molecule photobleaching of plasma membrane-localized tagged-CALHM proteins expressed in Xenopus oocytes. Photobleaching of a functional homo-dimeric CALHM-1-1-GFP concatemer as well as CALHM3-mCherry indicated that each assembled into homo-hexamers (Figures 4E-G and Figures S5E and S5G). The number of bleaching steps of CALHM-1-1-GFP was reduced by co-expression of untagged CALHM3 (Figures 4G, S5G and S5H), and the number of photobleaching steps of CALHM3-mCherry was reduced by co-expression of untagged CALHM1 (Figures 4F, S5E and S5F), both supporting that conclusion that CALHM1 and CALHM3 assemble into a single hexameric channel complex.
Together, these results demonstrate that CALHM1 and CALHM3 interact as pore-forming subunits in a novel hetero-hexameric CALHM1/CALHM3 ion channel with voltage-dependent activation kinetics greatly accelerated compared with a homo-oligomeric CALHM1 channel. Because the activation kinetics of the hetero-oligomeric CALHM1/CALHM3 channel are fast enough to be activated by taste-evoked action potentials, these data suggested that a CALHM1/CALHM3 channel may be the endogenous voltage-gated ATP-release channel in type II TBCs.
CALHM3 is a Functional Component of the ATP-Release Channel in Type II TBCs
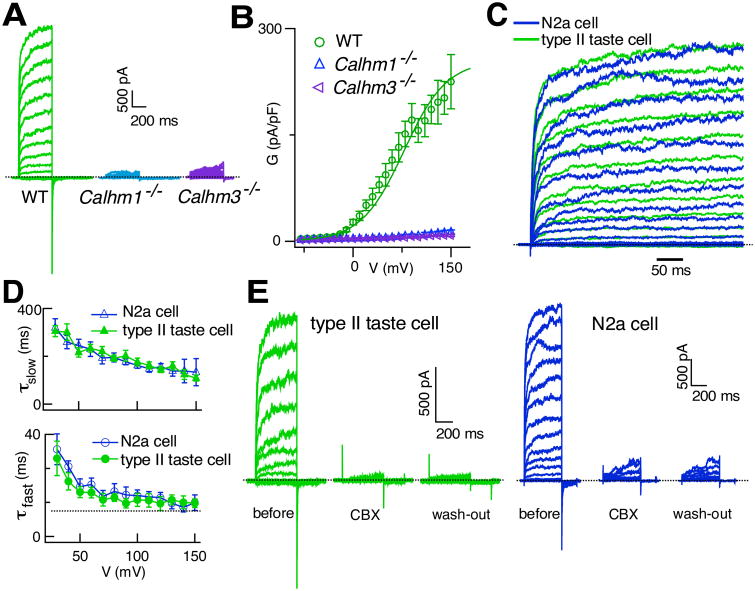
To test this hypothesis, we recorded whole-cell voltage-gated nonselective currents in TBCs isolated from mice with GFP expressed under the Trpm5 promoter specifically in type II TBCs (Clapp et al., 2006). To isolate nonselective voltage-gated currents from voltage-gated K+ and Na+ currents, 10 mM tetraethylammonium (TEA+) was included in the bath solution, and 2 mM TEA+ and 140 Cs+ were included in the pipette solution to block TEA+-sensitive K+ currents, and the whole-cell was clamped at a holding potential of -40 mV to inactivate the voltage-gated Na+ currents without adding TTX (Ma et al., 2012). The ATP-release channel currents are not activated or inactivated at this holding potential (Ma et al., 2017). Under these recording conditions, ATP release-channel currents can be recognized as large voltage-gated nonselective outward currents with inward tail currents at -80 mV (Ma et al., 2017) (Figures 5A and 5B). The voltage-dependent activation and deactivation kinetics and steady-state gating features of the voltage-activated nonselective currents from WT type II TBCs were remarkably similar to those of CALHM1/CALHM3 channels expressed in N2a cells (Figures 5C, 5D and S6). Furthermore, carbenoxolone (CBX, 10 μM), which was previously used to infer the involvement of pannexin-1 or connexin hemi-channels in peripheral taste perception, inhibited the CALHM1/CALHM3 channel currents in N2a cells as well as the voltage-gated nonselective currents in type II TBCs (Figures 5E and S7A-C). In contrast, CALHM1 currents in Xenopus oocytes (Ma et al., 2012) and the voltage-gated K+ and Na+ currents in type II TBCs (Figure S7D-G) were not inhibited by CBX. The similar gating and pharmacological properties of the currents in CALHM1+CALHM3-expressing N2a cells and the nonselective currents in type II cells strongly suggest that the ATP-release channel in type II TBCs is a CALHM1/CALHM3 heteroligomeric channel.
Figure 5. CALHM3 is an Essential Component of the Voltage-gated Non-selective ATP-release Channel in Type II TBCs.
(A) Representative families of whole-cell currents from WT, Calhm1–/– and Calhm3–/– type II TBCs, evoked by 500-ms voltage pulses from -80 mV to +150 mV in 10-mV increments from a holding potential of -40 mV. Dashed line: zero-current level. Non-selective voltage-gated currents are abolished in cells lacking either CALHM1 or CALHM3. Residual currents previously observed in Calhm1–/– type II TBC (Taruno et al., 2013) were non-specific leak currents likely caused by longer voltage pulses (1 s) used.
(B) G-V relations obtained by whole-cell capacitance-normalized currents at -80 mV evoked by 500-ms voltage pre-pulses. Solid line represents a Boltzmann function fit to WT data (V0.5 +78.0 ± 8.3 mV, Z0 0.88 ± 0.04 e, n = 21). Cell capacitance: 5.0 ± 0.4 pF (n = 21), 4.7 ± 0.7 pF (n = 10) and 4.9 ± 0.3 (n =20) for WT, Calhm1–/– and Calhm3–/– cells, respectively. Whole-cell capacitances are not different (two-tailed Student's unpaired t-test: Calhm1–/– versus WT, P = 0.633, t29 = 0.841; Calhm3–/– versus WT, P = 0.226, t39 = 0.226.
(C) Representative families of normalized outward currents from a WT type II TBC and a N2a cell co-expressing CALHM1+CALHM3, evoked by 500-ms voltage pulses from -80 mV to +120 mV in 10-mV increments from a holding potential of -40 mV. Dashed line: zero-current level.
(D) Activation-time constants τfast (lower panel) and τslow (upper panel) obtained from double-exponential fits to activation currents of WT type II TBC (n = 21) and N2a cells co-expressing CALHM1+CALHM3 (n = 22), respectively. Heterologous expression of CALHM1 with CALHM3 in N2a cells generated currents with voltage-dependent kinetic features identical to those of type II TBCs. Mean ± s.e.m.
(E) Carbenoxolone (CBX) inhibits non-selective voltage-gated currents in type II TBC (n = 9) (top) and CALHM1+3 currents in N2a cells (n = 14) (bottom). Representative families of whole-cell currents in a WT type II TBC before, after perfusion of 10 μM CBX in bath solution for ∼10 min, and after 5-min wash-out by normal bath solution, evoked by 500-ms voltage pulses from -80 mV to +100 mV in 10-mV increments from a holding potential of -40 mV. Dashed line: zero-current level. Representative families of whole-cell currents in a N2a cell co-expressing CALHM1+CALHM3 before, after perfusion with10 μM CBX for ∼10 min, and 5 min after wash-out by the normal bath solution, evoked by 500-ms voltage pulses from -80 mV to +130 mV in 10-mV increments from a holding potential of -40 mV. Dashed line: zero-current level.
See also Figures S6 and S7.
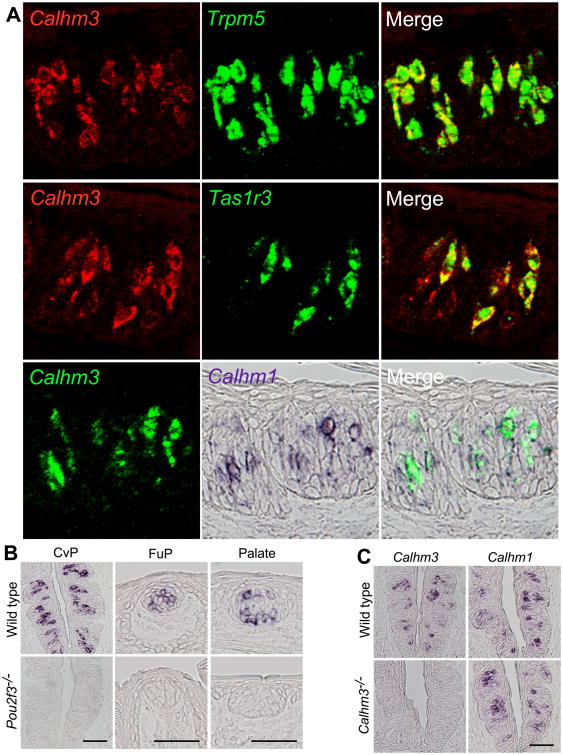
To test this hypothesis in vivo, we generated a constitutive Calhm3-/- mouse. We first confirmed the localization of Calhm3 expression in taste buds. In situ hybridization of taste epithelium revealed that Calhm3 was co-expressed with the type II TBC components Trpm5 and Tas1r3 (Figure 7A), and was absent in taste buds of Pou2f3–/– mice, which lack type II TBCs (Matsumoto et al., 2011) (Figure 7B). Double-label in situ hybridization revealed that Calhm1 and Calhm3 co-localized in the same cells (Figure 7A). Thus, Calhm3, like Calhm1, is expressed specifically in type II TBCs. Calhm3–/– mice lack Calhm3 expression without affecting the Calhm1 expression (Figure 7C), are viable and fertile, and had no overt taste-bud morphological abnormalities (Figures 7A and 7B). We recorded the voltage-activated nonselective currents in type II cells from mice with either CALHM1 or CALHM3 genetically deleted. The voltage-activated nonselective currents were completely absent in cells isolated from Calhm1-/- mice (Figures 5A and 5B). We previously observed residual nonselective voltage-gated residual currents in Calhm1-/- type II TBCs (Taruno et al., 2013), but here we established that those recordings suffered from a non-specific leak conductance. Thus, CALHM1 is essential for the voltage-activated nonselective currents in type II TBCs. Remarkably, the currents were also absent in cells from the Calhm3-/- mice (Figures 5A and 5B). In contrast, taste-evoked global [Ca2+ ]i signals were unaltered in isolated type II cells from Calhm3-/- mice (Figures 6A and 6B), indicating that loss of CALHM3 did not affect type II cell excitability. Nevertheless, taste-evoked ATP release from intact taste buds in circumvallate papillae in sheets of lingual epithelium was abolished in Calhm3–/– mice (Figure 6C). These results suggest that CALHM3 is essential for GPCR-mediated taste perception by regulating the voltage-gated ATP-release mechanism. Based on the effects of CALHM3 on CALHM1 channel gating, we hypothesized that CALHM3 is required for taste perception by contributing to an ion channel mechanism that enables action potentials to activate the ATP-release channel.
Figure 7. Calhm3 is Selectively Expressed in Type II TBCs.
(A-C) Double-label in situ hybridization of Calhm3 in gustatory tissues. Calhm3 mRNA is expressed in a subset of TBC of circumvallate (CvP) and fungiform (FuP) papillae and palate but is absent in taste buds of Pou2f3–/– mice lacking type II cells. (A) Fluorescence and immunohistochemical double-label in situ hybridization directly illustrates cellular co-expression of Calhm3 and Trpm5 in CvP taste buds. Calhm3 expression is absent from Trpm5-negative cells. Tas1r3 is expressed in a subset of Calhm3-expressing CvP TBCs. Calhm3 is expressed in the same cells that express Calhm1. (B) Calhm3 mRNA expressed in a subset of cells in taste buds of circumvallate (CvP) and fungiform (FuP) papillae and palate of WT mice is absent in taste buds of Pou2f3–/– mice lacking type II cells. (C) In CvP taste bud cells from Calhm3–/– mice, Calhm3 expression is absent, whereas Calhm1 expression is normal. Scale bar, 50 μm.
See also Figure S8.
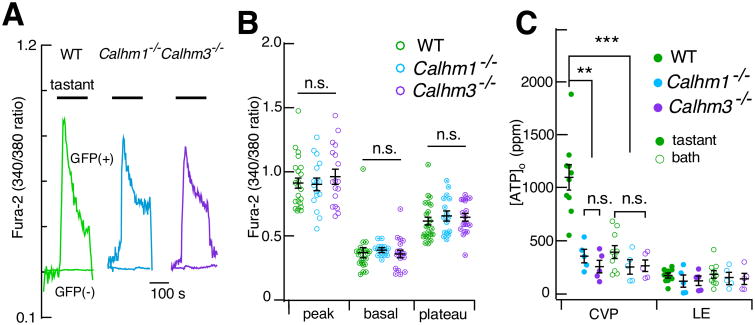
Figure 6. Type II TBC [Ca2+]i Signaling is Normal Whereas ATP Release is Abolished in Calhm1–/– and Calhm3–/– Mice.
(A,B) [Ca2+]i signaling is normal in type II TBC from Calhm1–/– and Calhm3–/– mice. (A) Representative fura-2 fluorescence ratios in single GFP-positive (type II) (upper traces) and GFP-negative (lower traces) cells in response to 3-min exposure to a cocktail of bitter and sweet compounds in WT, Calhm1–/– and Calhm3–/– mice. (B) Summary of fura-2 ratio responses for WT (n = 22, 4 experiments), Calhm1–/– (n = 16, 6 experiments) and Calhm3–/– (n = 18, 5 experiments) type II cells. No differences observed among genotypes (two-tailed Student's unpaired t-test, Calhm1–/– versus WT: P = 0.944, t36 = 0.071; P = 0.547, t36 = 0.607; P = 0.367, t36 = 0.915 and Calhm3–/– versus WT: P = 0.432, t38 = 0.794; P = 0.930, t38 = 0.088; P = 0.509, t38 = 0.667, for peak, basal and plateau Ca2+ signaling, respectively). The basal [Ca2+]i were calibrated as 97 ± 25 nM, 102 ± 7 nM and 87 ± 18 nM for WT, Calhm1–/– and Calhm3–/– mice, respectively. Data presented as mean ± s.e.m.; *P < 0.01, **P < 0.001, ***P < 0.0001.
(C) Tastant cocktail- and bath solution-evoked ATP release from gustatory CVP tissue and non-gustatory lingual epithelium (LE). Bitter/sweet taste mixture elicits marked ATP release from CVP versus LE in WT mice (n = 10; two-tailed Student's unpaired t-test, P = 0.0000003, t18 = 8.073). This is abolished in both Calhm3–/– (n = 5) and Calhm1–/– (n = 5) mice (two-tailed Student's unpaired t-test, Calhm3–/– versus WT: P = 0.00023, t13 = 5.023; Calhm1–/– versus WT, P = 0.00068, t13 = 4.433). ATP was significantly released from CVP tissue by bitter/sweet taste-mixture stimulation (n = 10, WT) compared with response to bath solution (n = 20, including Calhm1–/– and Calhm3–/– ; two-tailed Student's unpaired t-test, P = 0.000000001, t28 = 8.153, **P <0.01; ** P <0.001.
Genetic Deletion of Calhm3 Impairs Type II Cell GPCR-Mediated Taste Perception
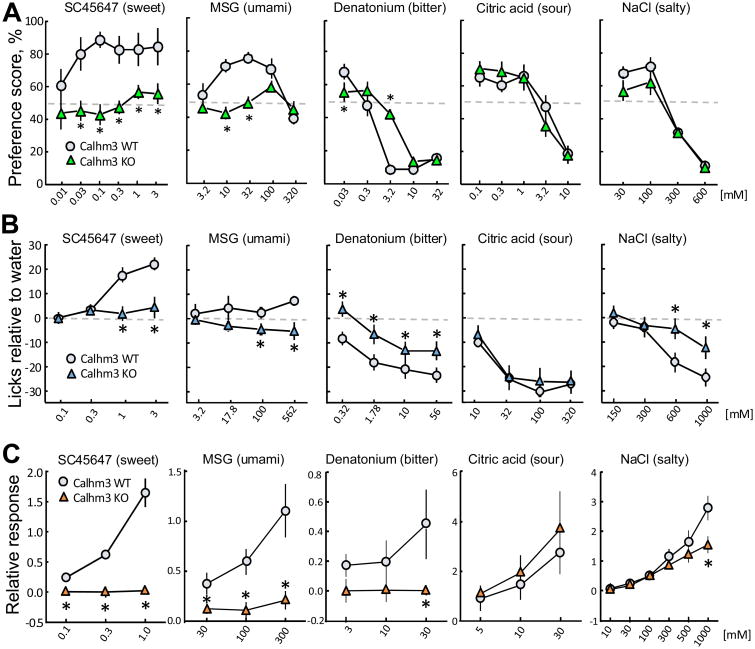
To examine whether CALHM3 is necessary for taste perception, we used two behavioral tests – 48-h two-bottle preferences and brief access lick rates (gustometry). Both methods revealed that Calhm3–/– mice had a marked reduction in avoidance of representative bitter compounds and a loss of avidity for representative sweet and umami compounds (Figures 8A, 8B, S8A and S8B and Table S1). Calhm3–/– mice also had reduced avoidance of CaCl2 and high concentrations of NaCl, which also involve GPCR-mediated taste transduction (Oka et al., 2013; Tordoff et al., 2014; Tordoff et al., 2008). In contrast, Calhm3–/– and wild-type (WT) mice responded similarly to low concentrations of NaCl and sour compounds, which are not detected by type II TBCs, and to capsaicin, an irritant mediated by somatosensory neurons (Figures 8A, 8B, S8A and S8B and Table S1). Thus, CALHM3 is required for GPCR-mediated taste perception. We then employed in vivo extracellular recordings of the whole chorda tympani nerve responses to various tastants. These results revealed strongly reduced responses to sweet, umami and bitter compounds as well as to high concentrations of NaCl in Calhm3–/– mice, whereas responses to citric acid and lower concentrations of NaCl were not different from WT controls (Figures 8C and S8C). Because Calhm3 is not expressed in the gustatory neurons in the cranial sensory ganglia (Figures S8D), the data indicate that the sensory defect in Calhm3–/– mice was due to loss of function in the peripheral taste system. Expression of Calhm1 (Figure 7C), Trpm5, Plcb2, Tas1r3 and Tas2r105+108 (Figure S8E) were qualitatively similar in taste buds from Calhm3–/– mice, indicating that the peripheral sensory defect was due specifically to loss of CALHM3. Thus, CALHM3 is required for GPCR-mediated taste perception in vivo. Together with our finding that the activation kinetics of the hetero-oligomeric CALHM1/CALHM3 channel are sufficiently fast to be activated by taste-evoked action potentials, these data suggest that a CALHM1/CALHM3 channel is the endogenous voltage-gated ATP-release channel in type II TBCs.
Figure 8. CALHM3 is essential for GPCR-mediated taste perception.
(A-C) Wild-type and Calhm3–/– mice preference scores during 48-h two-bottle choice tests (A) and lick rates during 5-sec brief-access tests relative to licking when water was presented (B), and chorda tympani nerve responses to sweet, umami, bitter, sour, and salty taste stimuli (C). Symbols depict means ± s.e.m. (n = 6 – 19 mice per group; see METHODS); *P < 0.05 (post hoc LSD or Tukey-Kramer tests). Chorda tympani nerve responses to NaCl were examined in the presence of 100 μM amiloride to expose activation of ENaC-insensitive salt transduction at high concentrations. See also Figure S8 and Table S1.
Discussion
Type II TBCs respond to sweet, bitter and umami substances by generating action potentials that activate a voltage-gated nonselective channel that releases ATP as the neurotransmitter to gustatory neurons (Chaudhari, 2014; Kinnamon, 2011; Liman et al., 2014). CALHM1 was previously shown to be a necessary component of the ATP-release channel in type II TBCs (Taruno et al., 2013). However, CALHM1 is a slow voltage-activated channel in heterologous expressing cells (Ma et al., 2012; Siebert et al., 2013), whereas endogenous voltage-gated nonselective currents and ATP release in vivo are activated with much faster kinetics in response to taste-evoked fast action potentials. Accordingly, it has been unclear how slow voltage-activated CALHM1 channels contribute to ATP release in vivo. Our study shows that the Calhm1 homolog Calhm3 is enriched in type II TBCs and interacts with CALHM1 by contributing as a pore-forming subunit in a novel CALHM1/CALHM3 ion channel that has rapid voltage-gated activation kinetics that are indistinguishable from the endogenous ATP-release channel in vivo. Furthermore, genetic deletion of CALHM3 abolishes ATP release and type II TBC-mediated taste perception. CALHM3 KO mice (tested here) and CALHM1 KO mice (Taruno et al., 2013) had similar behavioral taste phenotypes. We note that CALHM1 KO mice were indifferent to all concentrations of denatonium whereas CALHM3 mice avoided high concentrations of this bitter compound; this most likely was due to a “carry-over effect” artifact related to differences in the order the taste solutions were presented. Together, our results suggest that the voltage-gated ATP-release channel in type II TBCs is a novel hetero-hexameric ion channel composed of CALHM1 and CALHM3.
Our biochemical and single-molecule photobleaching studies of expressed monomeric and concatemeric CALHM proteins suggest that CALHM3 is integrated into a CALHM1 channel as a pore-forming subunit in a channel that remains a hexamer with permeability properties similar to those of a CALHM1 homo-hexameric channel. Despite the lack of a canonical voltage sensor in either CALHM1 or CALHM3, integration of CALHM3 into a CALHM1 channel confers to it voltage-dependent fast activation that is similar to that of traditional voltage-gated K+ channels (τ ∼10 ms) (Patel et al., 2004; Remillard and Leblanc, 1996). The molecular mechanisms by which CALHM3 switches slow gating of a CALHM1 channel to fast gating of the CALHM1/CALHM3 channel remain to be identified. Nevertheless, they play a critical role in enabling the release channel to respond to tastant-evoked action potentials by providing a channel gating mechanism to couple action potential firing to ATP release in type II TBCs.
An unexpected finding was that CALHM1 currents were completely absent in type II TBCs from Calhm3–/– mice. Qualitative mRNA determinations suggest that Calhm1 expression is not down-regulated in taste buds from Calhm3-/- mice. Preliminary immunohistochemistry experiments also suggest that CALHM1 protein levels are not reduced in TBCs lacking CALHM3 (not shown). It is notable that only small, slow voltage-activated currents could be elicited in CALHM1-expressing N2a cells, whereas much larger ones were generated in Xenopus oocytes. It is possible that CALHM3 is necessary for CALHM1 to function as an ion channel in mammalian cells, including type II TBC, possibly by maintaining CALHM1 protein levels in the plasma membrane, as suggested by our co-expression studies in heterologous cells. It is also possible that CALHM3 stabilizes functional CALHM1 in the plasma membrane of isolated cells, and that CALHM1 currents were absent in isolated Calhm3-/- type II TBCs because of CALHM1 channel rundown during single-cell preparation. These results suggest that additional features of the CALHM1/CALHM3 interaction in type II TBC remain to be elucidated. Nevertheless, the loss in Calhm3-/- mice of tastant-evoked ATP release from intact TBCs as well as perception of GPCR-mediated tastes suggests that even if CALHM1 is functional in type II cells in intact taste buds in vivo, it is insufficient to function as a voltage-gated ATP-release channel. It is interesting to note that whereas heterologously-expressed CALHM1/CALHM3 channels are Ca2+ permeable, global taste-evoked [Ca2+]i signals were not different between WT TBCs and those lacking CALHM1/CALHM3 channels, in agreement with our previous observations (Taruno et al., 2013). A CALHM1/CALHM3-mediated [Ca2+]i signal may have been undetected because it is highly spatially-localized or because the total CALHM1/CALHM3 Ca2+ permeability is small, possibly because CALHM1/CALHM3 is not expressed at high levels or that the channel is less Ca2+ permeable in vivo, possibly due to competition for permeation by ATP. In all cases, a CALHM1/CALHM3-mediated [Ca2+]i signal would be masked by the larger [Ca2+]i signal generated by Ca2+ release and entry though other pathways.
CALHM1 has been shown to influence cortical and hippocampal neuron functions (Cisneros-Mejorado et al., 2017; Ma et al., 2012; Vingtdeux et al., 2016), amyloid-beta metabolism (Dreses-Werringloer et al., 2008; Vingtdeux et al., 2015), airway ciliary beat frequency (Workman et al., 2017) and to be critical for taste perception (Hellekant et al., 2015; Taruno et al., 2013; Tordoff et al., 2014), whereas no physiological function has been known for CALHM3. Here we demonstrate that CALHM3 is an essential component of the voltage-activated ATP-release channel in type II TBCs, and that its expression is required for GPCR-mediated taste sensation. Whether CALHM3 is also involved in other functions attributed to CALHM1 remains to be determined. ATP is a signaling molecule regulating many physiological activities. Accordingly, CALHM1/CALHM3 channels may play a role in ATP release elsewhere in the body (Lazutkaite et al., 2017; Sana-Ur-Rehman et al., 2017; Workman et al., 2017). Previously, Ca2+-dependent exocytotic release of ATP was the only ATP-release pathway suitable for action potential-mediated rapid purinergic neurotransmission. However, with the discovery here of the rapidly-activating voltage-gated CALHM1/CALHM3 channel, we provide the first demonstration of an ion channel-mediated mechanism of fast purinergic neurotransmission. Whether this mechanism is unique to taste cells or has broader relevance remains to be determined.
Star Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, J. Kevin Foskett (foskett@pennmedicine.upenn.edu).
Experimental Model and Subject Details
Mouse lines and animal care
All animal experiments were conducted according to protocols approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania, the Monell Chemical Senses Center and Kyoto Prefectural University of Medicine.
Calhm3+/– mice were generated using CRISPR-Cas9 genome editing as described (Henao-Mejia et al., 2016). Briefly, 23 nucleotide guide sequences at the start of exon 1 (5′ ACGTGGCTACAGTCATGGATAGG 3′) and in the middle of exon 2 (5′ ATTTTGCTAATATGACCCCCAGG 3′) were selected and checked for potential off target effects using the CRISPR Design Tool(Ran et al., 2013). Two T7 promoter-guide RNA (gRNA) PCR products and a linearized T7 promoter-Cas9 plasmid were used for in vitro transcription. C57BL/6 zygotes were injected with the Cas9 RNA and gRNAs; resulting pups were genotyped with the primers 5′ GCCTAGTGATGCTTCGGTC 3′ (F), 5′ CAATGGGACTGGTGTGACC 3′ (R in deletion) and 5′ CATGGTGGCTCAAGACCATC 3′ (R) (WT, 865 bp; Calhm3 deletion, 632 bp). Calhm3+/– mice were crossed with C57BL/6 and a line that possessed germline transmission of a 2315-bp deletion in the Calhm3 locus was used for this study. All experiments were performed with wild-type and Calhm3–/– or Calhm1–/– (Taruno et al., 2013) littermates. Loss of Calhm3 expression was verified by in situ hybridization. TRPM5-GFP mice, a gift from Dr. R. Margolskee (Clapp et al., 2006), were crossed with Calhm1+/– or Calhm3+/– animals to mark type II TBCs with GFP for identification in whole-cell electrophysiology. Mice were housed in a pathogen-free, temperature- and humidity-controlled vivarium on a 12:12 h light/dark cycle. Diet consisted of standard laboratory chow and water. All experiments were performed with WT and Calhm3–/– or Calhm1–/– littermates of both sexes that were at least 52 days old. Mouse genotypes were determined by PCR (Transnetyx, Cordova, TN).
Cell culture
N2a (#CCL-131), HeLa (#CCL-2) and HEK293T (#CRL-3216) cells were purchased from American Type Culture Collection and were routinely checked for mycoplasma. HeLa cells were cultured in DMEM medium supplemented with 10% fetal bovine serum and ×1 Antibiotic-Antimycotic (Invitrogen) at 37°C in a humidified incubator with 5% CO2/air (Taruno et al., 2013). The N2a mouse neuroblastoma cell line was cultured in Eagle's minimum essential medium supplemented with 10% FBS and 0.5× penicillin/streptomycin (Invitrogen) at 37°C in a humidified incubator with 5% CO2 (Ma et al., 2012).
Method Details
In situ hybridization
Single- and double-label in situ hybridization procedures have been described previously (Ohmoto et al., 2008; Ohmoto et al., 2011; Taruno et al., 2013). In brief, oral epithelia containing taste buds and cranial sensory ganglia where cell bodies of gustatory neurons are located were dissected from adult mice deeply anesthetized with urethane, embedded in frozen O.C.T. Compound (Sakura Finetech USA, Torrance, CA) and sectioned to 8-μm thickness using a cryostat (CM1900, Leica Microsystems, Wetzlar, Germany). Fresh-frozen sections were fixed with 4% paraformaldehyde (PFA), treated with diethylpyrocarbonate, pre-hybridized with salmon sperm DNA for 2 h at 58°C, and hybridized with antisense riboprobe(s), except Calhm1 for 40 h at 58°C and unless stated otherwise. After hybridization, the sections were washed in 0.2 × SSC at 58°C and blocked with 0.5% blocking reagent (Roche Diagnostics, Basel, Switzerland) in Tris– buffered saline. Chromogenic signals were developed for one day using alkaline phosphatase–conjugated anti-digoxigenin antibody (1:500, 11093274910, Roche Diagnostics) and 4-nitro blue tetrazolium chloride and 5-bromo-4-chloro-3-indolyl-phosphate as chromogen substrate. To detect Calhm1 mRNA, hybridization and wash were done at 65°C and chromogenic signals were developed for two days. When Calhm3 riboprobe corresponding to the deleted sequence in Calhm3-targeted allele, which is about one-third as long as its full-length probe (see below), was used, chromogenic signals were developed for four days. Stained images were obtained with a Nikon eclipse 80i microscope (Nikon Instruments Inc., NY) equipped with a DXM1200C digital camera (Nikon). For double-label fluorescence in situ hybridization, fluorescent signals were developed using alkaline phosphatase–conjugated anti-digoxigenin antibody (1:500, Roche Diagnostics) followed by HNPP Fluorescent Detection Set (Roche Diagnostics) and biotin–conjugated anti-fluorescein antibody (1:500, BA-0601, Vector laboratories, Burlingame, CA) followed by avidin-biotin complex (Vector Laboratories), tyramide signal amplification biotin system (1:50, PerkinElmer, Waltham, MA), and Alexa fluor 488–conjugated streptavidin (4 μg/mL, Thermo Fisher Scientific, Waltham, MA). Fluorescent images were obtained using a Leica SP2 confocal scanning microscope with a pinhole size of 2.0 Airy Units. For double-labeling of Calhm1 and Calhm3, fluorescent signals of Calhm3 were first developed using biotin-conjugated anti-fluorescein antibody (Vector Laboratories) followed by avidin-biotin complex (Vector Laboratories), tyramide signal amplification biotin system (PerkinElmer), and Alexa fluor 488-conjugated streptavidin (Thermo Fisher Scientific). After capturing the fluorescent signals of Calhm3 with a Leica SP2 confocal microscope, Calhm1 signals were detected using alkaline phosphatase-conjugated anti-digoxigenin antibody and chromogen substrate as described previously (Taruno et al., 2013). After developing chromogenic signals for two days, stained images were obtained as described above. Fluorescent and stained images were processed with Photoshop (Adobe Systems) and analyzed on a computer screen. Digoxigenin- and/or fluorescein-labeled antisense RNAs were prepared using RNA labeling mix (Roche Diagnostics) and an RNA polymerase (Promega, Madison, WI) and fragmented under alkaline conditions to a length of about 150 bases before hybridization. RNA probes generated were to nucleotides 310-3491 of Trpm5 (accession no. AF228681), nucleotides 588-3123 of Plcb2 (accession no. BC145249), nucleotides 525-2725 of Tas1r3 (accession no. AF337039), nucleotides 1-903 of Tas2r105 (accession no. AF227147), nucleotides 1-894 of Tas2r108 (accession no. AF227148), nucleotides 1-1407 and 2148-2369 of Calhm1 cDNA fragment, which contains 1047 bases of the entire coding sequence, and 1322 bases of 3′-noncoding region, and nucleotides 1-528 (corresponding to the deleted sequence of Calhm3 targeted allele) or 1-1653 of Calhm3. Calhm3 cDNA was obtained from total RNA extracted from circumvallate papillae of C57BL/6J mice and used as a template for in vitro transcription of antisense Calhm3 riboprobes.
Quantitative real-time RT-PCR
Taste buds were isolated from 6-week-old C57BL/6 male mice as described below, morphologically selected, and collected into RNAlater (Qiagen). The isolated taste buds were then spun down at 3,600 rpm for 5 min before total RNA was extracted using RNeasy micro kits (Qiagen, Valencia, CA) and treated with RNase-free DNase I to avoid genomic DNA contamination according to the manufacturer's protocol. Similarly, RNA was extracted from lingual epithelial sheets devoid of taste buds that were cut out of the tongue epithelium obtained as described below and collected into RNAlater. First-strand cDNA was synthesized using Superscript III First-Strand Synthesis System for RT (Invitrogen). Taqman gene expression assay was performed with THUNDERBIRD Probe qPCR Mix (TOYOBO, Osaka, Japan) on a real-time PCR system (StepOnePlus, Applied Biosystems). Each sample was obtained from one mouse and each assay was run in triplicate. Expression levels were normalized to that of Calhm2 in taste buds after 2-ΔΔCt calculation, with β-actin as the endogenous control. The following Taqman assays were used: Calhm1 (Mm01207259_m1); Calhm2 (Mm00505271_m1); Actb (Mm01205647_g1); Calhm3 (5′ AGGCAGTGTCTCGGTACCT 3′ (F), 5′ CACCACTATCACCAGCAAGGTTAT 3′ (R) and 5′ CCAGCCGATGGCCTGT 3′ (reporter)). All assays were exon-exon boundary spanning.
Behavioral assays
Brief-access gustometer tests
Taste solution acceptance was assessed with brief-access tests using procedures similar to those reported earlier (Taruno et al., 2013). The tests were conducted with three cohorts of mice: Cohort 1 involved 14 Calhm3 WT mice (6 male, 8 female), and 11 Calhm3-/- (KO) littermates (4 male, 7 female); Cohort 2 involved 8 WT (5 male, 3 female) and 9 KO littermates (3 male, 6 female); Cohort 3 involved 8 WT (4 male, 4 female) and 7 KO littermates (3 male, 4 female). The mice were bred from heterozygous parents and weaned at 22 days, at which time tail tips were collected for genotyping. The mice were maintained under standard vivarium conditions [described online (Tordoff and Bachmanov, 2001)] with pelleted AIN-76A diet to eat and deionized water to drink. They were housed in same-sex groups until 7-10 days before testing began, when each mouse was housed alone in a plastic “tub” cage (26.5 cm × 17 cm × 12 cm) with a stainless-steel grid lid, and wood shavings scattered on the floor. At the beginning of testing, the mice ranged in age from 52 – 204 days and weight from 14.1 – 28.6 g. The knockout had no effect on body weight (mean ± s.e.m., g: WT males = 24.5 ± 1.1, WT females = 18.6 ± 1.3, KO males = 24.9 ± 1.7, KO females = 19.6 ± 1.5).
First, the mice were trained to lick taste solutions. To this end, they were water-deprived for 22.5 h and then placed into the chamber of a DiLog Instruments MS160 gustometer with its shutter open. The 14.5 × 30 × 15 cm test chamber of the gustometer provided access to the spout of a bottle of taste solution from behind a motorized shutter. The spout was part of a high-frequency alternating current contact circuit so that each lick the mouse made was detected and recorded. Each mouse had continuous access to water for 25 min from the time it first licked the drinking spout. It was then returned to its home cage and given water for 1 h. On the following two days, this procedure was repeated, except the shutter allowing access to water was closed 5 s after each time the mouse began to lick, and it was reopened after a 7.5-s interval. Once again, after 25 min, the mouse was returned to its home cage and given water for 1 h. During the 2nd test using these procedures, all mice had learned to obtain water during the 5-s access periods.
The first cohort of mice then received three series of tests with sucrose, quinine hydrochloride (QHCl), HCl and NaCl in the first cycle, saccharin, denatonium benzoate and calcium chloride in the second cycle, and Polycose [a soluble maltooligosaccharide (Sclafani, 2004)], citric acid and capsaicin in the third cycle. The second cohort of mice was tested with monosodium glutamate (MSG). The third cohort was tested with MSG, MSG mixed in 10 μM amiloride, inosine monophosphate (IMP), IMP mixed in 10 μM amiloride, and the artificial sweetener, SC45647.
To account for different proclivities to lick the different taste compounds, the deprivation regimen used to investigate the responses of mice in Cohort 1 to sucrose, saccharin and Polycose, and mice in Cohort 3 to the umami compounds and SC45647 differed from that used to investigate the response to the other taste compounds. Prior to a session with these “positively hedonic” taste compounds, each mouse received free access to food and water for 24 h. It then received 1 g of food and 2 mL of water, and the session began 24 h later. After these sessions, the mouse had a recovery day with free access to food and water for 24 h. Its water was then removed for 22.5 h to prepare it for the next session. Prior to sessions involving the other taste compounds, each mouse was water-deprived for 23 h. All sessions lasted 25 min.
During a test session, the mice could lick water or four concentrations of one taste compound. When positively hedonic taste compounds were being investigated, the session began with a single test of the highest concentration available in order to kindle the mouse's interest in the drinking spout. After this, repeated series of 5 concentrations (including water) were presented in a quasi-random order (a concentration could appear only once in a series of five tests). For each exposure, the shutter was open for 5 s during which licks of the drinking spout were counted. This was followed by 7.5 s with the shutter closed, during which a new taste solution was positioned ready for the next presentation. Tests involving the other taste compounds were similar except that 1-s “washout” trials with water were interposed between each test trial. Thus, a mouse received access to a taste solution for 5 s followed by 7.5 s with the shutter closed, then access to water for 1 s followed by 7.5 s with the shutter closed, followed by the next taste solution for 5 s, and so on. The brief washout trials with water were included in order to dissuade the mouse from quitting because it expects only bad-tasting solutions.
The mean number of licks in response to each taste solution concentration made by each mouse was obtained by averaging the results of identical exposures together. These values for individual mice were then used in mixed-design analyses of variance with factors of group (WT or KO) and concentration (Table S1). Mice that did not respond during presentation of a particular concentration of a taste compound were not included in statistical analyses of that compound. Post hoc Least Significant Difference (LSD) tests were used to assess differences between the groups in consumption of specific concentrations of taste solution and differences in response of each group to individual concentrations of each taste compound (Statistica 10, Stat Soft Inc, Tulsa, OK). All analyses were conducted using a criterion for significance of P < 0.05. No special provisions were made to blind the technician conducting the experiment; the risk of introducing an error due to misidentification of a mouse was greater than the potential risk of bias in this automated test.
Two-bottle choice tests
The methods for conducting two-bottle choice tests are described in detail on-line (Tordoff and Bachmanov, 2001). We investigated 11 taste solutions. This required five cohorts of Calhm3-/- mice and their WT littermate controls: Cohort 1, consisting of 15 WT and 10 Calhm3-/- mice, was tested with sucrose, QHCl, NaCl and HCl; Cohort 2, consisting of 10 WT and 10 Calhm3-/- mice, was tested with saccharin, denatonium benzoate; MSG, CaCl2, Polycose and capsaicin; Cohort 3, consisting of 11 WT and 13 Calhm3-/- mice, was tested with sucrose and citric acid; Cohort 4, consisting of 6 WT and 6 Calhm3-/- mice, was tested with citric acid and capsaicin (some taste compounds were tested twice). Preference tests for SC-45647 were conducted using the mice that had previously been used to assess brief-access lick rates to umami compounds (see above).
The mice received ascending concentrations of each taste solution, spanning the range from barely detectable to either strongly preferred or strongly avoided (by WT mice). For each 48-h test, each mouse was presented with two fluid-filled graduated drinking tubes, which allowed the contents to be measured to the nearest 0.1 mL. After 24 h, the positions of the drinking tubes were switched to control for any side preferences. After 48 h, intakes were recorded and the test of the next concentration of taste solution began. During the first test in a concentration-preference series, both drinking tubes contained deionized water. In subsequent tests, one drinking tube contained water and the other contained the taste solution. Mice received 2 – 4 days with only water to drink before starting a new concentration series.
The volumes of water and taste solutions consumed were recorded volumetrically to the nearest 0.1 mL. Solution preference scores were calculated based on the ratio of taste solution intake to total liquid intake, and expressed as a percentage. These values were analyzed by mixed-design analyses of variance with factors of group (WT or KO) and concentration (Table S1). Differences in responses of each group to individual concentrations of each taste compound were determined using post-hoc LSD tests. No special provisions were made to blind the technician conducting the experiment; the risk of introducing an error due to misidentification of a mouse was greater than the potential risk of bias.
Molecular biology
All Calhm cDNAs used in this study were from mouse, except in Figure S3 where human CALHMs were studied. For expression in Xenopus oocytes, Calhm1 was inserted between SalI and AgeI in pBF. For expression in N2a cells, Calhm1 was inserted between SalI and SacII in pIRES2.AcGFP1. Calhm3 in pBF and pIRES2.AcGFP1 were generated exactly as the Calhm1 constructs. Calhm3-mCherry in pBF was created by digestion of an existing plasmid that contained mCherry with SalI and XmaI and insertion of a Calhm3 PCR product; a short linker (PRARDP) separates Calhm3 from mCherry. To carboxyl-terminally fuse GFP or FLAG to Calhm1 and Calhm3 (Calhm1-GFP, Calhm1-FLAG, Calhm3-GFP and Calhm3-FLAG), Calhm sequences without the stop codon were amplified by PCR and cloned in frame into the mammalian expression vectors pEGFP-N1 or pFLAG. The XhoI and EcoRI sites were used for insertion of Calhm1; NheI and EcoRI were used for Calhm3.
Concatemeric CALHM constructs were generated for expression in N2a cells. A mouse CALHM1-CALHM1 concatemer tagged with 3×FLAG (CALHM-1-1-3×FLAG), in which the carboxyl-terminus of the first CALHM1 subunit was linked to the amino-terminus of the second CALHM1 subunit by a short flexible linker (PRGGSGGGSGTG) was created in pIRES2.AcGFP1 by triple ligation with a SalI site 5′ of the start codon, an AgeI site in the linker and a BamHI site 3′ of the stop codon. The mouse CALHM3-CALHM1 concatemer tagged with V5 (CALHM-3-1-V5) was generated exactly as the CALHM1-1-1-3× FLAG concatemer in pIRES2AcGFP1. To create the CALHM1-CALHM1-CALHM1 concatemer tagged with 3xFLAG, (CALHM-1-1-1-3×FLAG) in pIRES2.AcGFP1, the Calhm-1-1-3×FLAG concatemer was digested with SacII and AgeI in the linker and a third Calhm1 subunit was inserted. The linker between the first and second CALHM1 subunits (PRGGSGGGSGAL) contains unique sequence compared with the linker between the second and third CALHM1 subunits (ATGGSGGGSGTG) to enable sequencing. The CALHM1-CALHM3-CALHM1 construct tagged with 3×FLAG (CALHM-1-3-1-3×FLAG) was generated in pIRES2.AcGFP1 by digesting Calhm-1-1-3×FLAG with AgeI and inserting the Calhm3 subunit. The carboxyl-terminus of the first CALHM1 subunit was linked to the amino-terminus of CALHM3 by a flexible linker (PRGGSGGGSGTG) and the carboxyl-terminus of CALHM3 was linked to the amino-terminus of the other CALHM1 subunit by another flexible linker (TSGGSGGGSGTG). Correct directionality of Calhm3 was verified by restriction digest. To create the CALHM1-3×FLAG-sfGFP-sfGFP construct, codon optimized superfolder GFP (Addgene pET28a-sfGFP) was used. PCR was used to amplify sfGFP-sfGFP from an existing construct and the product was inserted between the PstI and BamHI sites in pIRES AcGFP1; a SmaI site was introduced between the PstI site and first sfGFP. Calhm-1-3×FLAG was PCR amplified and inserted between the EcoRI site (5′) and SmaI (3′) sites in the intermediate sfGFP-sfGFP construct. Calhm-1-3×FLAG was linked to the amino-terminus of the first sfGFP by the linker LRARGGSGENLYFQG and the linker LKPRARGGSGGGSGG separates the first and second sfGFP.
Concatemeric CALHM constructs were also generated in the Xenopus oocyte expression vector pBF. A mouse CALHM1- CALHM1 concatemer (CALHM-1-1), in which the carboxyl-terminus of the first CALHM1 subunit was linked to the amino-terminus of the second CALHM1 subunit by the linker PRARENLYFQG was created by overlap extension PCR, with a SalI site 5′ of the start codon, an XmaI site engineered into the linker and an AgeI site 3′ of the stop codon. A CALHM3-CALHM1 concatemer (CALHM-3-1) with a BamHI site 5′ of the Calhm3 start codon, an XmaI site engineered into the PRENLYFQGVD linker and an AgeI site 3′ of the Calhm1 stop codon was generated using the same method. The CALHM1-CALHM1 CALHM1 trimeric concatemer (CALHM-1-1-1) was created by inserting a Calhm1 subunit between the BamHI and SalI sites of the Calhm-1-1 concatemer; the linker between the first two subunits (PRENLYFQGVD) contained a unique sequence compared with the linker between the second and third subunits (PRARENLYFQG) to enable sequencing. The CALHM1-CALHM3-CALHM1 trimeric concatemer (CALHM-1-3-1) was generated by amplification of Calhm-1-3, then insertion of this PCR product between the BamHI and SalI sites in the Calhm1 in pBF construct. The linker following the first Calhm1 subunit (PRARENLYFQG) contains an XmaI site and the linker following the Calhm3 subunit contains a SalI site. To create the CALHM1-CALHM1 concatemer tagged with GFP (CALHM-1-1-GFP), overlap extension PCR was used to fuse Calhm1 and Calhm1-GFP, which were both amplified from existing constructs. The resulting PCR product, which has the linker PRAHQNLYFQG following the carboxyl-terminus of the first CALHM1 subunit was inserted between the SalI and BamHI sites in pBF. All constructs were confirmed by sequencing.
Co-immunoprecipitation
Insufficient amounts of CALHM1 and 3 proteins in taste bud tissues precluded biochemical analyses of CALHM proteins in taste cells. We attempted to generate CALHM antibodies multiple times with unsatisfactory results, most likely reflecting low-expression level of CALHM1 proteins, as suggested by qRT-PCR (large Ct values compared to that of Actb, data not shown). To detect CALHM1 proteins for biochemical analyses, we also generated a knock-in mouse in which endogenous CALHM1 proteins are C-terminally tagged with a V5-epitope tag (reported in Taruno et al., 2017). Although CALHM1-V5 was detected with a commercially available highly-sensitive anti-V5 antibody, it required taste-bud tissues collected from at least 4 mice and a harsh lysis condition with a high concentration of SDS (4%) which is not compatible for co-immunoprecipitation.
Accordingly, we explored the biochemical interaction of CALHM1 and CALHM3 in a heterologous expression system. N2a cells (2.2 × 106 cells) were plated on 6-cm dishes (Corning) and transfected on the following day with empty vectors, 5 μg mouse CALHM1-GFP, 5 μg CALHM3-FLAG, or 5 μg CALHM1-GFP + 5 μg CALHM3-FLAG. The total amount of DNA (10 μg) was adjusted with appropriate control vectors (pEGFP-N1 and pFLAG). In the experiments shown in Figure S4, cells were transfected with CALHM1-GFP + CALHM3-FLAG, Panx1-FLAG, CD4ΔC-FLAG, or CD74ΔN-FLAG. cDNAs of Panx1, carboxyl-terminus truncated CD4 (CD4ΔC), and amino-terminus truncated CD74 (CD74ΔN) were kindly provided by Dr. Val Shestopalov (University of Miami), Dr. Gerald W Zamponi (University of Calgary) and Dr. Gergely Lukacs (McGill University). 24 h later, cells were washed twice with ice-cold PBS and harvested in 1 mL lysis buffer (PBS containing 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, and 1× protease inhibitor cocktail (P8340, Sigma, St. Louis, MO, USA)). After centrifugation (20,000 × g for 10 min at 4°C), supernatants were collected as whole-cell lysates. Whole-cell lysates (900 μg) were incubated with 40 μL of Protein G agarose resin (Thermo Fisher Scientific, Waltham, MA, USA) coupled to mouse anti-FLAG M2 antibody (#200472, Stratagene, San Diego, CA, USA) or Protein A agarose resin (Thermo Fisher Scientific) coupled to rabbit anti-GFP antibody (#SP3005P, Novus Biologicals, Littleton, CO, USA) in 1 mL of the IP buffer (PBS containing 1 % Triton X-100 and 1 mM phenylmethylsulfonyl fluoride) with head-over-tail rotation at 4°C overnight. The next day, the resins were washed 3× with the ice-cold IP buffer and incubated in 30 μL of 1× Laemmli sample buffer at 95°C for 5 min to elute proteins from the resins. The whole cell lysates (30 μg) (Input) and 20 % of the eluate samples (IP) were subjected to SDS-PAGE/Western blotting analysis.
Surface biotinylation
N2a cells were seeded onto T-75 flasks (Corning) at 7.5 × 106 cells/well the day before transfection. Cells were transfected with 7.5 μg mouse CALHM1-GFP cDNA and/or 37.5 μg CALHM3-FLAG. The total amount of DNA (45 μg) was adjusted with appropriate control vectors (pEGFP-N1 and pFLAG). 24 h later, cells were washed twice with ice-cold PBS and incubated with 0.25 mg/mL EZ-link Sulfo-NHS-SS-Biotin (Thermo Fisher Scientific) in 12 mL of PBS containing 2 mM CaCl2 for 30 min at 4°C. The biotinylation reaction was stopped by addition of 600 μL of 2 M glycine in PBS, followed by sequential washes with PBS containing 100 mM glycine (twice) and PBS (twice). After the final wash, cells were collected, lysed in 1 mL of lysis buffer (PBS containing 1 % Triton X-100, 1 mM phenylmethylsulfonyl fluoride, and 1× protease inhibitor cocktail), and centrifuged at 20,000 × g at 4°C for 10 min. The supernatants were collected as the whole-cell lysates. Biotinylated proteins in 1.5 mg of the whole-cell lysates were pulled down by 200 μL of Pierce NeutrAvidin agarose resin (Thermo Fisher Scientific) and eluted by incubating the resin in 150 μL of 1× Laemmli sample buffer at 37°C for 1 h. To enable quantitative analysis, the amount of NeutrAvidin beads was determined so that no biotinylated proteins were detected in the flow-through samples. The whole-cell lysates (25 μg) denatured in 1× Laemmli sample buffer (Total) and 50 μL of the eluate samples (Surface) were subjected to SDS-PAGE/Western blotting analysis.
SDS-PAGE/western blotting
Proteins were separated in 10 % polyacrylamide gels by SDS-PAGE and transferred to nitrocellulose membranes using standard protocols. The membranes were blocked with 5 % (w/v) non-fat dry milk in Tris-buffered saline (50 mM Tris/HCl, 150 mM NaCl; pH 7.6) containing 0.1 % Tween 20 (5 % milk-TBST) at room temperature for 1 h and immunoblotted with mouse anti-GFP (#632381, Clontech, Mountain View, CA, USA), rabbit anti-FLAG (#2044, Cell Signaling Technology, Danvers, MA, USA), mouse anti-FLAG (#F3165, Sigma), mouse anti-Na+/K+-ATPase (#ab7671, Abcam, Cambridge, UK) or anti-β-tuublin antibody (#32-2600,Thermo Fisher Scientific) in 5 % milk-TBST at room temperature for 1 h or at 4°C overnight. The membranes were then washed with TBST for 3 × 5 min and incubated with horseradish peroxidase-conjugated goat anti-rabbit or mouse IgG antibody in 5 % milk-TBST at room temperature for 1 h. After 3 × 5 min washes with TBST, immunoreactive bands were detected using Amersham ECL chemiluminescent reagent (GE Healthcare Bio-Sciences, Piscataway, NJ, USA) and ChemiDoc XRS+ system with Image Lab software (Bio-Rad, Hercules, CA, USA). Band intensities were quantified using Image J software (https://imagej.nih.gov/ij/) after background correction. To investigate more than one protein on the same blot, membranes were stripped in stripping buffer (Restore Western blot stripping buffer, Thermo Fisher Scientific) at 42°C for 60 min with gentle agitation, and then re-probed with a different antibody.
Blue native PAGE
Blue Native PAGE (BN-PAGE) western blotting was performed according to Invitrogen's recommended protocols using their NativePAGE™ reagents. For experiments shown in Figure 4B, N2a cells (8 × 105 cells) were plated on 6-well plates (Corning) and, on the following day, transfected with empty vectors, 1 μg CALHM1-FLAG and/or 2.7 μg CALHM3-GFP. The total amount of DNA (3.7 μg) was adjusted with appropriate control vectors (pEGFP-N1 and pFLAG). For the experiments shown in Figure S5A, HEK293 cells (8 × 105 cells) plated on 6-well plates were transfected with 2 μg of CALHM1-GFP or human Pannexin 1-GFP. For experiments shown in Figures 4B and 4C, 10 μg of cDNAs for CALHM1-FLAG, CALHM1-FLAG-2×xGFP, CALHM-1-1-FLAG, CALHM-1-1-1-FLAG, CALHM-1-3-1-FLAG, and 50 μg cDNA for CALHM3-V5, were transiently transfected into N2a cells (3 × 107 cells in 100 mm plate). Transiently-transfected N2a or HEK293 cells were lysed in NativePAGE™ Sample Buffer containing 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, and 1× protease inhibitor cocktail. Appropriate volume of NativePAGE™ G-250 Sample Additive was introduced immediately before loading on a NativePAGE™ Novex® 4-16% Bis-Tris Gel (20 μg/well). NativeMark™ Unstained Protein Standard was added to the gel to estimate molecular mass. The gel was run at 150 V for the first 30 min in the Dark Blue Cathode Buffer, and then at 150V for the remainder of the time (120 min) in the Light Blue Cathode Buffer. Following electrophoresis, proteins were transferred to PVDF membranes using NuPAGE® Transfer Buffer at 100 V for 53 min. The proteins were fixed on the membrane by shaking the membrane in 8% acetic acid for 15 min. Acetic acid was removed by rinsing the membrane in deionized water. Background dye was removed by shaking in methanol followed by washing in water. The molecular mass markers were visualized with ponceau S staining and imaged using a ChemiDoc XRS+ system with Image Lab software (Bio-Rad). Standard western blotting techniques, described above, were used to detect CALHM proteins. The same lysates (25 μg) were denatured in 1× Laemmli sample buffer at 95°C for 5 min and analysed by SDS-PAGE/western blotting as described above.
Single-molecule photobleaching
Total internal reflection microscopy (TIRFM) of single CALHM3-mCherry and concatemeric CALHM-1-1-GFP molecules expressed in Xenopus oocytes was accomplished using a custom-built system (Demuro et al., 2011) based around an Olympus IX71 microscope equipped with a ×60, NA 1.45 TIRF objective. Defolliculated stage VI oocytes were injected with 10 nL mixtures of cRNAs at final concentrations ranging from 0.001 to 0.1 μg/μL and incubated for 14-44 h before imaging experiments. Devitellinated oocytes were allowed to settle on a cover glass forming the base of the recording chamber and were bathed in Ca2+-free Ringer's solution (in mM: 120 NaCl, 2 KCl, 5 MgCl2, 1 EGTA, 5 HEPES, pH 7.4). Tagged molecules were excited by total internal reflection of a laser beam (532 nm for mCherry; 488 nm for GFP) incident through the microscope objective. Images (128 × 128 pixel; 1 pixel = 0.33 μm) were acquired at 10 frames s-1 by a Cascade 128+ electron multiplying CCD camera (Roper Scientific). Resulting image stacks (∼2,000 frames; 100 s) were processed in MetaMorph (Molecular Devices) by averaging every two consecutive frames, followed by subtraction of a heavily smoothed (7×7 pixel) copy of each frame to correct for bleaching of autofluorescence and other background signals. Fluorescent spots corresponding to individual tagged CALHM channels were identified by visual inspection, and fluorescence traces were obtained from 3 × 3 pixel regions of interest centered on these spots. The number of bleaching steps in each trace was determined by visual inspection, with measurements restricted to those spots that showed complete bleaching and where fluorescence steps could be clearly resolved. Data were plotted and fitted by binominal distributions using Origin 9.0 (www.originlab.com).
Immunohistochemistry
For the experiments shown in Figure 3A, N2a cells on poly-L-lysine-coated glass coverslips placed in 24-well plates were transfected with 0.5 μg mouse CALHM1-GFP cDNA and 0.5 μg CALHM3-FLAG cDNA and treated with 100 μg/mL cycloheximide or vehicle (0.1 % water) 3 h before fixation at 24 h post-transfection. For the experiments shown in Figure 3C, N2a cells were transfected with 0.5 μg CALHM1-GFP cDNA with or without 2.5 μg CALHM3-FLAG cDNA. Twenty-four h after transfection, cells were washed twice with ice-cold PBS and incubated in 1 mL of PBS containing 2 mM CaCl2 and 0.25 mg/mL EZ-link Sulfo-NHS-SS-Biotin (Thermo Fisher Scientific) at 4°C for 30 min. To quench the excess biotin reagent, 50 μL of PBS containing 2 M glycine was added to the cells. After washing twice with ice-cold PBS, cells were fixed in PBS containing 4 % paraformaldehyde at room temperature for 20 min, washed in PBS for 3 × 3 min, blocked for 1 h at room temperature with the blocking buffer (PBS containing 3 % normal goat serum and 0.1 % Triton X-100), and incubated at 4°C overnight with primary antibodies diluted in the blocking buffer. The next day, cells were washed with PBS at room temperature for 3 × 10 min, incubated in the dark at room temperature for 1 h with Alexa Fluor-conjugated secondary antibodies (Thermo Fisher Scientific) diluted in the blocking buffer, and washed with PBS in the dark at room temperature for 3 × 10 min before coverslips were mounted on slides in VectaShield with 4′,6-diamidino-2-phenylindole (DAPI) (H-1500, Vector Laboratories, Burlingame, CA, USA). Primary and secondary antibodies used in this study were: mouse anti-GFP (Clontech), rabbit anti-FLAG (Cell Signaling Technology), rabbit anti-biotin (#ab53494, Abcam), Alexa fluor 488-conjugated goat anti-mouse IgG, and Alexa fluor 546-conjugated goat anti-rabbit IgG. Images of stained cells were captured with a LSM510 confocal scanning microscope (Carl Zeiss, Oberkochen, Germany) using an EC Plan-Neofluar 40×/1.30 NA Oil objective. Images show single optical sections, collected with the pinhole set to 1 Airy Unit for the red channel and adjusted to give the same optical slice thickness in the green channel. DAPI (blue channel) was excited with two photons using a MaiTai titanium-sapphire laser tuned to 780 nm (Spectra-Physics, Santa Clara, CA, USA).
ATP release
ATP release from cultured cells
Extracellular ATP released from HeLa cells was measured by the luciferin–luciferase reaction as we previously described (Taruno et al., 2013). HeLa cells were seeded at a density of 20,000 cells per well in 96-well microplates (Corning Costar) a day before transfection with 0.1 μg mouse or human CALHM1 cDNA and/or 0.1 μg CALHM3 cDNA, where total amount of DNA (0.2 μg) was adjusted with empty vector (pIRES2.AcGFP1). Media was replaced with the normal bath solution after 20 - 30 h, and cells were incubated for 30 min before 75 μL of the 100-μL bath solution containing was replaced with an equal volume of the Ca2+-free or high-K+ solutions to remove free Ca2+ or increase [K+]o, respectively. The plate was placed in a microplate luminometer (Centro LB960, Berthold Technologies, Germany) immediately after 10 μL of ATP assay solution (FLAA-1KT, Sigma-Aldrich) was dispensed into each well. Luminescence was measured every 2 min. ATP concentration was calculated from a standard curve in each plate. The normal bath contained (in mM) 150 NaCl, 5 KCl, 2 CaCl2, 1 MgCl2, 10 HEPES and 10 glucose, pH 7.4, adjusted with NaOH. The Ca2+-free solution contained (in mM) 150 NaCl, 5 KCl, 5 EGTA, 1 MgCl2, 10 HEPES and 10 glucose, pH 7.4, adjusted with NaOH. The high-K+ depolarizing solution contained (in mM) 155 KCl, 2 CaCl2, 1 MgCl2, 10 HEPES and 10 glucose, pH 7.4, adjusted with NaOH. The final [Ca2+]o and [K+]o were respectively ∼17 nM and 117.5 mM.
ATP release from intact taste buds
Taste-evoked ATP release from intact taste buds with sheets of lingual epithelium was measured as we described previously (Taruno et al., 2013). Tongue epithelium was cut into pieces containing the CvP or lingual epithelium devoid of taste buds, and mounted in an Ussing-type chamber with the serosal side exposed to 130 μL Tyrode's solution. The apical surface was exposed to a mixture of tastants (1 mM denatonium benzoate, 10 μM cycloheximide and 2 mM saccharin sodium) in the Tyrode's solution for 3 min. Chamber solution (100 μL) was collected and added to an equal volume of ATP assay solution FLAA-1KT, Sigma-Aldrich) to determine ATP concentration by the luciferin-luciferase assay with a microplate luminometer (Synergy 2, BioTek). The Tyrode's solution contained (in mM) 140 NaCl, 5 KCl, 2 CaCl2, 1 MgCl2, 10 glucose, 5 Na-pyruvate and 10 HEPES, pH 7.4 with NaOH.
Taste bud cell (TBC) isolation
Animals were sacrificed by CO2 inhalation and cervical dislocation. The circumvallate taste epithelium was enzymatically delaminated, taste buds were collected from peeled epithelium, and dissociated single TBCs were collected as detailed previously (Ma et al., 2017; Taruno et al., 2013). Briefly, 0.5 mL of a mixture of enzymes containing Dispase II (2 mg/mL; Roche), collagenase A (1 mg/mL; Roche) and trypsin inhibitor (1 mg/mL; Sigma) diluted in a Ca2+-Tyrode's solution (in mM: 140 NaCl, 5 KCl, 2 CaCl2, 1 MgCl2, 10 glucose, 5 Na-pyruvate, and 10 Hepes, pH adjusted to 7.4 with NaOH.) was injected under the lingual epithelium. After 30 min in Ca2+-Tyrode's solution at room temperature, the epithelium was peeled off and incubated in Ca2+-free Tyrode's solution (in mM: 140 NaCl, 5 KCl, 5 EGTA, 10 glucose, 5 Na-pyruvate, and 10 Hepes, pH adjusted to 7.4 with NaOH) for 15 min. For isolation of single TBCs, 0.2 mg/mL elastase (Sigma) and 10 μg/mL DNase I (Roche) were added to the above enzyme mixture. Gentle suction with a glass capillary pipette removed cells from the circumvallate taste buds. Isolated cells were placed on poly-L-lysine-coated coverslips and allowed to settle for ∼60 min before electrophysiological recordings.
Single-cell calcium imaging
Isolated TBCs on coverslips were loaded with 2.5 μM fura-2AM (Thermo/Molecular Probes) for 45 min at room temperature in a bath containing (in mM) 140 NaCl, 5 KCl, 2 CaCl2, 1 MgCl2, 10 glucose, 5 Na-pyruvate, 10 HEPES, pH 7.4 with NaOH. Cells were imaged on the stage of an inverted microscope (IX-83, Olympus, Tokyo Japan; 20× 0.8 NA Plan Apo objective) with continuous perfusion (Warner Instruments RC-26) and stimulated for 2-4 min by perfusion of a taste mix (2 mM saccharin, 1 mM denatonium benzoate, 10 mM cycloheximide, pH 7.4). Fura-2 was alternately illuminated at 340 and 380 nm (Lambda LS, Sutter Instruments, Novato CA) and emitted fluorescence (510 nm; filter set 79002, Chroma Technologies, Rockingham, VT) was collected with a sCMOS camera (Hamamatsu, Tokyo Japan) under control of MetaFluor software (Molecular Devices, Sunnyvale CA). Images of GFP fluorescence (470/40 ex, 525/40 em, Chroma FITC filter set 49002) were acquired every 10 frames to identify GFP-positive (+) and GFP-negative (-) cells. Background fluorescence was subtracted during analysis. Changes in [Ca2+]i are presented as changes in fluorescence excitation ratio (340/380).
Electrophysiology
Whole-chorda tympani nerve recordings
The mice (6 mice of each genotype) were anesthetized with an intraperitoneal injection of a mixture of 4.28 mg/mL ketamine, 0.86 mg/mL xylazine, and 0.14 mg/mL acepromazine in saline (5 μL/g body weight). Anesthesia was maintained with additional injections of this chemical mixture. The mouse was fixed with a head holder after its trachea was cannulated, and its chorda tympani nerve (CTn) was dissected free from its junction with the lingual nerve to the tympanic bulla, where the central part of the CTn was cut and placed on a platinum wire recording electrode. An indifferent electrode touched the walls of the wound. The taste stimuli were delivered to the tongue with an open flow system controlled by a computer under conditions of constant flow and temperature (25°C). Each stimulation lasted for 30 s with a 60-s rinse time between stimulations. Care was exercised to ensure that the flow was directed over the fungiform papillae. The nerve impulses were fed into a custom-made amplifier, monitored over a loudspeaker and an oscilloscope, and recorded (PowerLab/sp4; AD Instruments, Colorado Springs, CO). The integrated response during stimulation was calculated by subtracting the area of nerve activity preceding stimulation from that during stimulation. Thus, the data reflect the level of activity during taste stimulation. The responses to all compounds were expressed relative to the responses to 0.1 M NH4Cl in each mouse. The averages in each animal and group were calculated, and two-way repeated measure ANOVAs were used to determine the effects of genotype using Prism 6 software (GraphPad Software, Inc., La Jolla, CA). When a significant interaction was detected between genotype and taste solution concentration, Tukey-Kramer multiple comparison tests were conducted to determine which pairs of means differed significantly.
N2a and Type II TBC electrophysiology
All recordings were performed at room temperature (20 ∼ 22°C). Data were acquired with an Axopatch 200B amplifier at 5 kHz. Currents were filtered by an eight-pole Bessel filter at 1 kHz and sampled at 5 kHz with an 18-bit A/D converter. Electrode capacitance was compensated electronically, and 60% of series resistance was compensated with a lag of 10 μs. Electrodes were made from thick-walled PG10150-4 glass (World Precision Instruments). HEKA Pulse software (HEKA Eletronik, Germany) was used for data acquisition and stimulation protocols. Igor Pro was used for graphing and data analysis (WaveMetrics, Inc.). Leak subtractions were not applied in the current study. Whole-cell currents were recorded in N2a cells 16-24 h after transfection as we previously described (Ma et al., 2012). The pipette solution contained (in mM) 140 Cs+ 2 TEA+, 1 Ca2+, 11 EGTA, 30 CI-, and 10 Hepes, pH 7.3 adjusted by methanesulfonate, ∼ 310 mOsm. Bath solution contained (in mM) 140 Na+, 5.4 K+, 10 TEA+, 1.5 Ca2+, 1 Mg2+, 150 Cl-, 20 Sucrose, and 10 Hepes, pH 7.4 adjusted by methanesulfonate, ∼330 mOsm. The electrode resistance was 1.5-2 MΩ in these recording conditions.
Whole-cell currents were recorded in single isolated GFP-expressing type II TBCs dissociated from circumvallate papillae from Trpm5-GFP; Calhm1–/–, Trpm5-GFP; Calhm3–;–, Trpm5-GFP and control littermates (Trpm5-GFP; Calhm1+/+; Calhm3+/+) as previously described (Ma et al., 2012). Electrodes had a resistance of 4 - 7 MΩ, with solutions used the same as those used in N2a cells. The cells were voltage-clamped at a holding potential of -40 mV to inactivate Na+ channels without adding tetrodotoxin. For recording Na+ and K+ currents, the pipette solution contained (in mM) 140 K+, 6 Na+, 1 Mg2+, 1 Ca2+, 30 Cl−, 11 EGTA, 3 ATP3-, 0.3 Tris1GTP, and 10 Hepes, pH 7.3 adjusted by methanesulfonate, -290 mOsm. The bath solution contained (in mM) 150 Na+, 5.4 K+, 1.5 Ca2+, 1 Mg2+, 150 Cl−, 20 glucose, and 10 Hepes, pH 7.4 adjusted by methanesulfonate, ∼330 mOsm. For experiments with carbenoxolone (CBX), fresh aliquots of 10 μl of 10 mM CBX (Sigma, in distilled H2O and stored at -20°C) were added to 10 ml of bath solution in each experiment.
Xenopus oocyte electrophysiology
Two-electrode voltage clamp recordings were performed as previously described (Ma et al., 2012). cRNAs for mouse CALHM1, CALHM2, and CALHM3 were in vitro transcribed from linearized plasmids with the mMessage mMachine kit (Ambion). Oocytes were defolliculated with collagenase (Worthington Biochemical Corp.). At least 2 h after collagenase treatment, 1 ng (CALHM1) or 5 ng (CALHM2, CALHM3) cRNA was injected into oocytes with 10 ng of Xenopus connexin-38 antisense oligonucleotide to inhibit endogenous Cx38 currents (Ma et al., 2012). When co-expressing homologs, the same amounts of cRNAs were used. Oocytes were kept at 16°C in ND96 solution (in mM): 96 NaCl, 2 KCl, 1.8 CaCl2, 1 MgCl2, 2.5 Na-pyruvate, 1× penicillin-streptomycin, and pH 7.6 adjusted by NaOH. Recordings were performed at room temperature (20-22°C) 2-5 days after injection. Oocytes were injected with a 50 nL mixture of 20 mM BAPTA and 10 mM Ca2+ at least 30 min prior to recording to clamp [Ca2+]i to ∼100 nM and minimize activation of endogenous Ca2+-activated Cl- currents (Ma et al., 2012). Currents were recorded in a bath containing (in mM) 100 Na+, 100 Cl-, 2 K+, 10 HEPES, pH 7.2 adjusted with methanesulfonate, with either 1.5 mM Ca2+ and 1 mM Mg2+ or 0 Ca2+ (0.5 EGTA and 0.5 EDTA). Data were acquired with an OC-725C amplifier (Warner Instrument Corp.) at 1 kHz with 16-bit A/D converter (Instrutech ITC-16). Electrodes were filled with 3 M KCl, connected by 3 M KCl agar bridges to bath solution.
Relative permeabilities of Ca2+, Na+, K+ and Cl- (PCa:PNa:PK:PCl) were estimated from reversal potentials (Erev) using the Goldman-Hodgkin-Katz (GHK) constant field equation as we previously described (Ma et al., 2012; Siebert et al., 2013; Tanis et al., 2013).
Conductance-voltage (G-V) relations of CALHM1 were normalized to conductance evoked by a pre-pulse to +40 mV in 0-Ca2+o to define Gmax for an individual oocyte. G-V relations of control and CALHM3 (in Figure 1C) and CALHM2, CALHM2+3, CALHM1+2 (in Figure S1C), were normalized to average Gmax from CALHM1-expressing oocytes in 0-Ca2+o from the same batch of oocytes. G-V relations of CALHM1+3 (in Figure 1D) and CALHM1+2+3 (in Figure S1C) were normalized to individual oocyte Gmax, obtained from a Boltzmann function fitting to individual oocytes. Currents recorded at -80 mV in 1 mM Gd3+ were used for leak subtraction.
Current activation time constants were best fitted with a double exponential function (τfast and τslow) for CALHM1+3 (in Figures 1B and 1E), and concatemer CALHM-1-3-1, whereas activation currents of CALHM1 (in Figures 1B and 1E) and concatemers CALHM-1-1, CALHM-3-1 and CALHM-1-1-1 (in Figure 4D) could be fitted with only a single exponential. CALHM1 activation currents failed to reach a plateau during the long depolarization pulses (5 s for oocytes and 500 ms for N2a cells). However, longer depolarization pulses were not possible because of destabilization of whole-cell patches in N2a cells and appearance of large endogenous currents in oocytes. Thus, our study here has focused on the ability of CALHM3 to switch this apparent slow single activation kinetics of CALHM1 currents to double exponential kinetics of CALHM1+CALHM3 currents.
Quantification and Statistical Analysis
All of the statistical details of experiments, including definitions of significance, the statistical methods used and methods used to determine whether data met assumptions of the statistical approach can be found in the figure legends, figures, Results, and METHOD DETAILS. Most statistical analyses employed Student t- tests, as stated in Figure legends. More complicated statistical tests were required for the behavioral assay data, described in detail in METHOD DETAILS.
Supplementary Material
Highlights.
Type II taste bud cells release ATP as a neurotransmitter by an ion-channel mechanism
CALHM1 is a component but cannot account for properties of the native release channel
A hexameric CALHM1/CALHM3 channel has properties identical to the native channel
Genetic deletion of CALHM3 abolishes sweet, bitter and umami taste perception
Acknowledgments
We thank Tiffany Weiss and Hillary Ellis for their adept technical assistance with behavioral tests, and Drs. G. Lukacs, V. Shestopalov and G. W. Zamponi for cDNAs. This work was supported by NIH R01DC012538 (Z.M. and J.K.F.), R37GM048071 (I.P.), RO1AG053988 (A.D.), R01DC015491 (I.M.), R01EY015537 (C.H.M.) and R03DC014328 (J.E.T.), JSPS KAKENHI 26713008 and 16K15181 (A.T.), JSPS KAKENHI 25670111 and 15K15034 (Y.M.), Salt Science 1542 (A.T.), Society for Research on Umami Taste (A.T.).
Footnotes
Author Contributions: Z.M., A.T., M.O., M.J., J.C.L., H.M., N.N., R.J.L., R.P., H.H., J.E.T., I.M., and M.G.T designed and performed experiments. J.H.-M. and J.E.T. designed and generated expression constructs and the Calhm3–/– mice. I.P. and A.D. performed and analyzed single-molecule bleaching experiments. Y.M., C.H.M. and J.K.F. designed experiments and helped with data interpretation. Z.M., A.T., M.G.T., I.M. and J.K.F. wrote the manuscript.
Competing Financial Interests: The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Chaudhari N. Synaptic communication and signal processing among sensory cells in taste buds. J Physiol. 2014;592:3387–3392. doi: 10.1113/jphysiol.2013.269837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisneros-Mejorado A, Gottlieb M, Ruiz A, Chara JC, Perez-Samartin A, Marambaud P, Matute C. Blockade and knock-out of CALHM1 channels attenuate ischemic brain damage. J Cereb Blood Flow Metab. 2017 doi: 10.1177/0271678X17713587. 271678X17713587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapp TR, Medler KF, Damak S, Margolskee RF, Kinnamon SC. Mouse taste cells with G protein-coupled taste receptors lack voltage-gated calcium channels and SNAP-25. BMC Biol. 2006;4:7. doi: 10.1186/1741-7007-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapp TR, Yang R, Stoick CL, Kinnamon SC, Kinnamon JC. Morphologic characterization of rat taste receptor cells that express components of the phospholipase C signaling pathway. J Comp Neurol. 2004;468:311–321. doi: 10.1002/cne.10963. [DOI] [PubMed] [Google Scholar]
- Dando R, Roper SD. Cell-to-cell communication in intact taste buds through ATP signalling from pannexin 1 gap junction hemichannels. J Physiol. 2009;587:5899–5906. doi: 10.1113/jphysiol.2009.180083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFazio RA, Dvoryanchikov G, Maruyama Y, Kim JW, Pereira E, Roper SD, Chaudhari N. Separate populations of receptor cells and presynaptic cells in mouse taste buds. J Neurosci. 2006;26:3971–3980. doi: 10.1523/JNEUROSCI.0515-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuro A, Penna A, Safrina O, Yeromin AV, Amcheslavsky A, Cahalan MD, Parker I. Subunit stoichiometry of human Orai1 and Orai3 channels in closed and open states. Proc Natl Acad Sci U S A. 2011;108:17832–17837. doi: 10.1073/pnas.1114814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreses-Werringloer U, Lambert JC, Vingtdeux V, Zhao H, Vais H, Siebert A, Jain A, Koppel J, Rovelet-Lecrux A, Hannequin D, et al. A polymorphism in CALHM1 influences Ca2+ homeostasis, Abeta levels, and Alzheimer's disease risk. Cell. 2008;133:1149–1161. doi: 10.1016/j.cell.2008.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger TE, Danilova V, Barrows J, Bartel DL, Vigers AJ, Stone L, Hellekant G, Kinnamon SC. ATP signaling is crucial for communication from taste buds to gustatory nerves. Science. 2005;310:1495–1499. doi: 10.1126/science.1118435. [DOI] [PubMed] [Google Scholar]
- Hellekant G, Schmolling J, Marambaud P, Rose-Hellekant TA. CALHM1 deletion in mice affects glossopharyngeal taste responses, food intake, body weight, and life span. Chemical senses. 2015;40:373–379. doi: 10.1093/chemse/bjv009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henao-Mejia J, Williams A, Rongvaux A, Stein J, Hughes C, Flavell RA. Generation of genetically modified mice using the CRISPR-Cas9 genome-editing sSystem. Cold Spring Harbor protocols. 2016;2016 doi: 10.1101/pdb.prot090704. pdb prot090704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YA, Stone LM, Pereira E, Yang R, Kinnamon JC, Dvoryanchikov G, Chaudhari N, Finger TE, Kinnamon SC, Roper SD. Knocking out P2X receptors Reduces transmitter secretion in taste bBuds. J Neurosci. 2011;31:13654–13661. doi: 10.1523/JNEUROSCI.3356-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YJ, Maruyama Y, Dvoryanchikov G, Pereira E, Chaudhari N, Roper SD. The role of pannexin 1 hemichannels in ATP release and cell-cell communication in mouse taste buds. Proc Natl Acad Sci U S A. 2007;104:6436–6441. doi: 10.1073/pnas.0611280104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnamon SC. Taste receptor signalling - from tongues to lungs. Acta Physiol (Oxf) 2011;204:158–168. doi: 10.1111/j.1748-1716.2011.02308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnamon SC. Neurosensory transmission without a synapse: new perspectives on taste signaling. BMC Biol. 2013;11:42. doi: 10.1186/1741-7007-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazutkaite G, Solda A, Lossow K, Meyerhof W, Dale N. Amino acid sensing in hypothalamic tanycytes via umami taste receptors. Mol Metab. 2017;6:1480–1492. doi: 10.1016/j.molmet.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liman ER, Zhang YV, Montell C. Peripheral coding of taste. Neuron. 2014;81:984–1000. doi: 10.1016/j.neuron.2014.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Saung WT, Foskett JK. Action potentials and ion conductances in wild-type and CALHM1-knockout type II taste cells. J Neurophysiol. 2017;117:1865–1876. doi: 10.1152/jn.00835.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Siebert AP, Cheung KH, Lee RJ, Johnson B, Cohen AS, Vingtdeux V, Marambaud P, Foskett JK. Calcium homeostasis modulator 1 (CALHM1) is the pore-forming subunit of an ion channel that mediates extracellular Ca2+ regulation of neuronal excitability. Proc Natl Acad Sci U S A. 2012;109:E1963–1971. doi: 10.1073/pnas.1204023109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto I, Ohmoto M, Narukawa M, Yoshihara Y, Abe K. Skn-1a (Pou2f3) specifies taste receptor cell lineage. Nat Neurosci. 2011;14:685–687. doi: 10.1038/nn.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer BD, Hevezi P, Gao N, Lu M, Kalabat D, Soto H, Echeverri F, Laita B, Yeh SA, Zoller M, Zlotnik A. Expression of genes encoding multi-transmembrane proteins in specific primate taste cell populations. PLoS One. 2009;4:e7682. doi: 10.1371/journal.pone.0007682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y, Yasuo T, Yoshida R, Obata K, Yanagawa Y, Margolskee RF, Ninomiya Y. Action potential-enhanced ATP release from taste cells through hemichannels. J Neurophysiol. 2010;104:896–901. doi: 10.1152/jn.00414.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmoto M, Matsumoto I, Yasuoka A, Yoshihara Y, Abe K. Genetic tracing of the gustatory and trigeminal neural pathways originating from T1R3-expressing taste receptor cells and solitary chemoreceptor cells. Mol Cell Neurosci. 2008;38:505–517. doi: 10.1016/j.mcn.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Ohmoto M, Okada S, Nakamura S, Abe K, Matsumoto I. Mutually exclusive expression of Gαia and Gα14 reveals diversification of taste receptor cells in zebrafish. J Comp Neurol. 2011;519:1616–1629. doi: 10.1002/cne.22589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka Y, Butnaru M, von Buchholtz L, Ryba NJ, Zuker CS. High salt recruits aversive taste pathways. Nature. 2013;494:472–475. doi: 10.1038/nature11905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SP, Parai R, Parai R, Campbell DL. Regulation of Kv4.3 voltage-dependent gating kinetics by KChIP2 isoforms. J Physiol. 2004;557:19–41. doi: 10.1113/jphysiol.2003.058172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remillard CV, Leblanc N. Mechanism of inhibition of delayed rectifier K+ current by 4-aminopyridine in rabbit coronary myocytes. J Physiol. 1996;491(Pt 2):383–400. doi: 10.1113/jphysiol.1996.sp021223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov RA, Bystrova MF, Rogachevskaya OA, Sadovnikov VB, Shestopalov VI, Kolesnikov SS. The ATP permeability of pannexin 1 channels in a heterologous system and in mammalian taste cells is dispensable. J Cell Sci. 2012;125:5514–5523. doi: 10.1242/jcs.111062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov RA, Rogachevskaja OA, Khokhlov AA, Kolesnikov SS. Voltage dependence of ATP secretion in mammalian taste cells. J Gen Physiol. 2008;132:731–744. doi: 10.1085/jgp.200810108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sana-Ur-Rehman H, Markus I, Moore KH, Mansfield KJ, Liu L. Expression and localization of pannexin-1 and CALHM1 in porcine bladder and their involvement in modulating ATP release. Am J Physiol Regul Integr Comp Physiol. 2017;312:R763–R772. doi: 10.1152/ajpregu.00039.2016. [DOI] [PubMed] [Google Scholar]
- Sclafani A. The sixth taste? Appetite. 2004;43:1–3. doi: 10.1016/j.appet.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Siebert AP, Ma Z, Grevet JD, Demuro A, Parker I, Foskett JK. Structural and functional similarities of Calcium Homeostasis Modulator 1 (CALHM1) ion channel with connexins, pannexins, and innexins. J Biol Chem. 2013;288:6140–6153. doi: 10.1074/jbc.M112.409789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K, Seto Y, Ohtubo Y, Yoshii K. Dye-permeable, voltage-gated channel on mouse fungiform taste bud cells. Brain Res. 2011;1373:17–24. doi: 10.1016/j.brainres.2010.12.019. [DOI] [PubMed] [Google Scholar]
- Tanis JE, Ma Z, Foskett JK. NH2 terminus regulates voltage-dependent gating of CALHM ion channels. American Journal of Physiology. 2017 doi: 10.1152/ajpcell.00318.2016. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanis JE, Ma Z, Krajacic P, He L, Foskett JK, Lamitina T. CLHM-1 is a functionally conserved and conditionally toxic Ca2+-permeable ion channel in Caenorhabditis elegans. J Neurosci. 2013;33:12275–12286. doi: 10.1523/JNEUROSCI.5919-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taruno A, Sun H, Nakajo K, Murakami T, Ohsaki Y, Kido MA, Ono F, Marunaka Y. Post-translational palmitoylation controls the voltage gating and lipid raft association of the CALHM1 channel. J Physiol. 2017;595:6121–6145. doi: 10.1113/JP274164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taruno A, Vingtdeux V, Ohmoto M, Ma Z, Dvoryanchikov G, Li A, Adrien L, Zhao H, Leung S, Abernethy M, et al. CALHM1 ion channel mediates purinergic neurotransmission of sweet, bitter and umami tastes. Nature. 2013;495:223–226. doi: 10.1038/nature11906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tordoff MG, Aleman TR, Ellis HT, Ohmoto M, Matsumoto I, Shestopalov VI, Mitchell CH, Foskett JK, Poole RL. Normal Taste Acceptance and Preference of PANX1 Knockout Mice. Chemical senses. 2015;40:453–459. doi: 10.1093/chemse/bjv025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tordoff MG, Bachmanov AA. Monell Chemical Senses Center; 2001. Monel mouse taste phenotyping project. www.monell.org/MMTPP. [Google Scholar]
- Tordoff MG, Ellis HT, Aleman TR, Downing A, Marambaud P, Foskett JK, Dana RM, McCaughey SA. Salty taste deficits in CALHM1 knockout mice. Chemical senses. 2014;39:515–528. doi: 10.1093/chemse/bju020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tordoff MG, Shao H, Alarcon LK, Margolskee RF, Mosinger B, Bachmanov AA, Reed DR, McCaughey S. Involvement of T1R3 in calcium-magnesium taste. Physiological genomics. 2008;34:338–348. doi: 10.1152/physiolgenomics.90200.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbeuch A, Anderson CB, Kinnamon SC. Mice Lacking Pannexin 1 Release ATP and Respond Normally to All Taste Qualities. Chemical senses. 2015;40:461–467. doi: 10.1093/chemse/bjv034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vingtdeux V, Chandakkar P, Zhao H, Blanc L, Ruiz S, Marambaud P. CALHM1 ion channel elicits amyloid-beta clearance by insulin-degrading enzyme in cell lines and in vivo in the mouse brain. J Cell Sci. 2015;128:2330–2338. doi: 10.1242/jcs.167270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vingtdeux V, Chang EH, Frattini SA, Zhao H, Chandakkar P, Adrien L, Strohl JJ, Gibson EL, Ohmoto M, Matsumoto I, et al. CALHM1 deficiency impairs cerebral neuron activity and memory flexibility in mice. Scientific reports. 2016;6:24250. doi: 10.1038/srep24250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman AD, Carey RM, Chen B, Saunders CJ, Marambaud P, Mitchell CH, Tordoff MG, Lee RJ, Cohen NA. CALHM1-Mediated ATP Release and Ciliary Beat Frequency Modulation in Nasal Epithelial Cells. Scientific reports. 2017;7:6687. doi: 10.1038/s41598-017-07221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.