Abstract
The regulatory sequences of many genes encoding seed storage proteins have been used to drive seed-specific expression of a variety of proteins in transgenic plants. Because the levels at which these transgene-derived proteins accumulate are generally quite low, we investigated the utility of the arcelin-5 regulatory sequences in obtaining high seed-specific expression in transgenic plants. Arcelin-5 is an abundant seed protein found in some wild common bean (Phaseolus vulgaris L.) genotypes. Seeds of Arabidopsis and Tepary bean (Phaseolus acutifolius A. Gray) plants transformed with arcelin-5 gene constructs synthesized arcelin-5 to levels of 15% and 25% of the total protein content, respectively. To our knowledge, such high expression levels directed by a transgene have not been reported before. The transgenic plants also showed low plant-to-plant variation in arcelin expression. Complex transgene integration patterns, which often result in gene silencing effects, were not associated with reduced arcelin-5 expression. High transgene expression was the result of high mRNA steady-state levels and was restricted to seeds. This indicates that all requirements for high seed-specific expression are cis elements present in the cloned genomic arcelin-5 sequence and trans-acting factors that are available in Arabidopsis and Phaseolus spp., and thus probably in most dicotyledonous plants.
Seeds, especially those of legumes and cereals, contain large quantities of protein and are a major source of plant dietary protein, consumed by humans and livestock. Most seeds, however, have nutritional shortcomings, such as a deficiency in one or more essential amino acids and the presence of antinutritional factors. Gene transfer techniques can be used to alter the amino acid composition of seed proteins and to improve the nutritional quality of seeds (Tabe and Higgins, 1998). Aside from being a source of dietary protein, seeds can also be used as “bioreactors” for the production of pharmaceutically or industrially important products (Goddijn and Pen, 1995). For all of these purposes, seed-specific expression of transgenes at sufficiently high levels is required.
The high levels at which many seed storage proteins accumulate make their regulatory sequences excellent tools with which to achieve this goal. As illustrated in Table I, many seed storage proteins and their expression signals have been studied in transgenic plants. In general, transcription and intron splicing occur correctly in heterologous plants, and the introduced genes are spatially and temporally expressed in a way similar to that in the plant species from which the regulatory sequences were originally derived. In most cases, the protein products show normal processing and intracellular transport in the developing seeds, indicating that different plant species have similar mechanisms of gene regulation and protein processing (Sun and Larkins, 1993; Habben and Larkins, 1995). Therefore, the flanking regulatory regions of genes encoding seed storage proteins could be used in chimeric gene constructs to ensure the effective organ-specific synthesis of novel or heterologous proteins in transgenic plants. However, the level at which the introduced proteins accumulate is generally low, usually not more than a few percent (Table I). This could be due to many factors, such as degradation of the foreign protein and promoters that function less efficiently in heterologous seeds.
Table I.
Examples of heterologous expression of genes encoding seed storage proteins
| Protein/Gene | Source Plant | Host Plant | Promoter | Levela | Reference |
|---|---|---|---|---|---|
| 2S1 Albumin | Arabidopsis | Nicotiana tabacum | Own | 0.1 | De Clercq et al. (1990) |
| 2S Albumin | Bertholletia excelsa | Arabidopsis | at2S1 | 1.0–2.0 | De Clercq et al. (1990) |
| B. napus | at2S1 | 1.0–2.0 | De Clercq et al. (1990) | ||
| Soybean lectin | 0.02–0.06 | Guerche et al. (1990a) | |||
| β-Phaseolin | 1.7–4.0 | Altenbach et al. (1992) | |||
| N. tabacum | β-Phaseolin | 3.0–8.0 | Altenbach et al. (1989) | ||
| at2S1 | 1.0–2.0 | De Clercq et al. (1990) | |||
| Vicia narbonensis | CaMV 35S | 0.0–0.01 | Saalbach et al. (1994) | ||
| Legumin B4 | 1.0–4.8 | Pickardt et al. (1995) | |||
| 2S Albumin | Helianthus annuus | Lupinus angustifolius | Pea vicilin | 5.0 | Molvig et al. (1997) |
| β-Conglycinin | Glycine max | Petunia hybrida | Own | 0.1–1.0 | Beachy et al. (1985) |
| Soybean lectin | N. tabacum | Own | 0.2 | Okamuro et al. (1986) | |
| β-Phaseolin | P. vulgaris | N. tabacum | Own | 1.0 | Sengupta-Gopalan et al. (1985) |
| Oryza sativa | Glutelin Gt1 | 0.0–4.0 | Zheng et al. (1995) | ||
| Phytohemagglutinin-L | P. vulgaris | N. tabacum | Own | 0.02–0.05 | Sturm et al. (1988); Voelker et al. (1989) |
| α-Amylase inhibitor | P. vulgaris | Pisum sativum | PHA-L | 1.0–3.0 | Schroeder et al. (1995) |
| Psl lectin | P. sativum | N. tabacum | Own | 0.2–0.9 | de Pater et al. (1996) |
| Legumin A | P. sativum | Nicotiana plumbaginifolia | Own | 0.2 | Ellis et al. (1988) |
| O. sativa | Glutelin Gt1 | 0.0–4.2 | Sindhu et al. (1997) | ||
| Vicilin | P. sativum | N. tabacum | Own | 0.5 | Higgins et al. (1988) |
| 19-kD Zein | Zea mays | P. hybrida | Own | 0.0 | Ueng et al. (1988) |
| 15-kD Zein | Z. mays | N. tabacum | β-Phaseolin | 0.02–1.6 | Hoffman et al. (1987) |
| CaMV 35S | 1.0–2.0 | Bagga et al. (1995) |
Protein product, as a percentage of total extractable protein in transgenic seeds.
In this respect, arcelin genes could represent an interesting alternative. Arcelins are seed proteins found in some genotypes of wild common bean (Phaseolus vulgaris L.) and are thought to be involved in the high resistance levels of these genotypes to the bruchid pest Zabrotes subfasciatus (Osborn et al., 1988). Arcelin genes are genetically closely linked with and related to the phytohemagglutinin and α-amylase inhibitor genes (Chrispeels and Raikhel, 1991). Seven arcelin variants have been identified, of which we have characterized in detail the arcelin-5 variant present in the wild P. vulgaris genotype G02771 (Goossens et al., 1994). This genotype contains two arcelin-5 genes: the arc5-I gene that encodes the Arc5a protein and the arc5-II gene that encodes Arc5b and a minor nonglycosylated isoform, Arc5c. The sequence similarity between the arc5-I and arc5-II transcribed regions is more than 98%.
Arcelin 5 is a very abundant protein (30%–40% of the total seed protein content), yet it is encoded by only two genes per haploid genome (Goossens et al., 1994). In contrast, phaseolin, which is the common seed storage protein present in all P. vulgaris genotypes, is encoded by a multigene family with seven to nine genes per haploid genome (Slightom et al., 1985). Phaseolin normally accounts for up to 60% of the total protein of P. vulgaris seeds. It is not known whether all copies contribute equally to the observed expression levels, but this suggests that the amount of arcelin per gene copy is much higher than that of phaseolin.
Recently, we isolated an arcelin 5-I genomic clone (Goossens et al., 1995) that contains an actively expressed arcelin-5 gene (Dillen et al., 1997). To investigate the potential of the arcelin-5 expression signals for gene engineering, we have introduced various fragments of the arc5-I genomic clone into Tepary bean (Phaseolus acutifolius A. Gray) and Arabidopsis (L.) Heynh plants. Expression analysis showed that in both species arcelin-5 proteins accumulate to high levels, suggesting that the arcelin-5 regulatory regions could be used in chimeric gene constructs to ensure high accumulation of heterologous proteins in seeds of transgenic plants.
MATERIALS AND METHODS
Transformation of Phaseolus acutifolius with arcelin-5 Genes
Tepary bean (Phaseolus acutifolius A. Gray, genotype NI576) was transformed as described by Dillen et al. (1997). Gene transfer was achieved with the Agrobacterium tumefaciens strain C58C1RifR containing the helper plasmid pMP90 (Koncz and Schell, 1986) and harboring the binary vector pATARC3-B1b or pATARC3-B52b. These vectors are derived from the binary vector pATAG3 (Fig. 1) that contains between the T-DNA borders the nptII (neomycin phosphotransferase II) and the uidA (GUS) genes. To construct pATARC3-B1b, a BamHI-fragment of the arc5-I gene (Fig. 2) was inserted into the unique BamHI site of pATAG3. pATARC3-B52b is identical to pATARC3-B1b except that the arc5-I-coding region is replaced by the arc5-II-coding region. This chimeric gene thus comprises the arc5-II coding region between the arc5-I regulatory sequences and was constructed because a genomic arc5-II clone was not available.
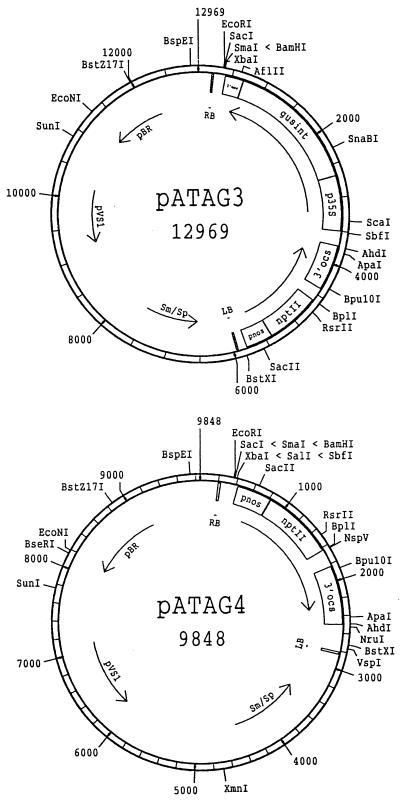
Figure 1.
Plasmid maps of pATAG3 and pATAG4. These binary plasmids contain between the T-DNA borders the nptII gene under control of the nopaline synthase (pnos) promoter and the octopine synthase 3′ termination and polyadenylation signals (3′ocs) (pATAG3 and pATAG4) and the Escherichia coli gus gene (Jefferson, 1987) with the potato st-l1 intron (gusint; Vancanneyt et al., 1990) under the control of the CaMV 35S promoter (p35S) and the nopaline synthase 3′ processing and polyadenylation signals (3′nos) (pATAG3 only). pBR, Origin of replication; pVS1, stability and replication functions of the Pseudomonas aeruginosa pVS1 plasmid (Deblaere et al., 1987); Sm/Sp, spectinomycin and streptomycin resistance locus; RB and LB, right and left border repeat of the T-DNA. Single-cutting restriction enzymes are shown outside of the plasmids.
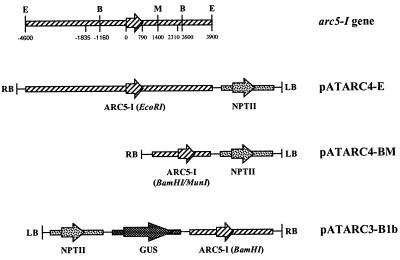
Figure 2.
Schematic representation of the arc5-I genomic clone and the T-DNA of the binary vectors pATARC4-E, pATARC4-BM, and pATARC3-B1b. Coding regions are indicated by arrows and 5′ and 3′ flanking regulatory sequences as blocks. Numbers correspond with positions in the arc5-I gene relative to the translation start site. Regions upstream of position −1,835 and downstream of position +2,310 were not sequenced. E, EcoRI site; B, BamHI site; M, MunI-site; RB and LB, right and left border repeat of the T-DNA; NPTII, pnos-nptII-3′ocs chimeric gene; GUS, the p35S-gusint-3′nos chimeric gene. pATARC3-B1b contains the BamHI fragment of the arc5-I genomic clone, whereas pATARC4-E and pATARC4-BM contain the complete genomic fragment and the BamHI/MunI fragment, respectively.
Primary transformants were assessed for the number of transgene loci by segregation analysis of their progeny. To this end, GUS assays were performed on small pieces of dry seed cotyledon tissue. Tissue was incubated in staining buffer (100 mm NaPO4, 2 mm 5-bromo-4-chloro-3-indolyl-β-d-GlcUA cyclohexaylammonium salt, 0.1% (v/v) β-mercaptoethanol, and 0.1% (v/v) Triton X-100, pH 7.2) for 2 h at 37°C. The number of integrated T-DNAs was determined by Southern blot analysis of primary transformants using the Gene Images kit (Amersham). Total leaf DNA used in Southern blot analysis was prepared as described by Goossens et al. (1994).
Detection and Quantification of the Arcelin-5 Protein in Transgenic P. acutifolius Seeds
Crude seed protein samples were obtained by two successive extractions of ground cotyledon tissue in 10 mm NaCl and 50 mm Gly, pH 2.4 for 30 min at room temperature under continuous shaking. After centrifugation for 10 min at 20,000g, the pellet was removed and supernatants were pooled. Protein concentrations of the crude extracts were determined by measuring the UV-A280. Proteins were separated by SDS-PAGE (Hames and Rickwood, 1990) and visualized by Coomassie Blue staining. Expression levels of arcelin-5 proteins in transgenic P. acutifolius seeds were estimated (as a percentage of total extractable seed protein) by indirect ELISA (as described by Harlow and Lane, 1988) using a rabbit polyclonal anti-arcelin-5 antiserum. Arcelin-5 proteins purified from the wild Phaseolus vulgaris L. genotype G02771 mixed with total seed proteins of the nontransgenic P. acutifolius genotype NI576 were used to construct a standard curve. Each transgenic line was examined at least three times for arcelin-5 expression levels (one–two seeds per assay).
Transformation of Arabidopsis with arcelin-5 Genes
Arabidopsis (L.) Heynh genotype Columbia-0 was transformed according to the protocol of Bechtold et al. (1993). Vacuum infiltration was carried out with the Agrobacterium tumefaciens strain C58C1RifR containing the helper plasmid pMP90 (Koncz and Schell, 1986) and harboring the binary vector pATARC4-BM or pATARC4-E. These vectors are derived from the binary vector pATAG4 (Fig. 1) into which a BamHI/MunI or an EcoRI fragment of the arc5-I gene (Fig. 2) were inserted, respectively.
Transgenic seedlings (T1 generation) were selected on growth medium (Valvekens et al., 1988) containing 50 μg mL−1 kanamycin (Sigma) and 200 μg mL−1 cefotaxime (Claforan, Hoechst, Frankfurt). The T2 segregation was analyzed under the same conditions. The number of integrated T-DNAs was determined by Southern blot analysis of T2 seedlings with the Gene Images kit (Amersham). Total seedling DNA was prepared as described by Barthels et al. (1997).
Detection and Quantification of the Arcelin-5a Protein in Transgenic Arabidopsis Seeds
Crude seed protein extracts were obtained according to a modified extraction protocol of van der Klei et al. (1993). Ground seeds were extracted twice with hexane to remove lipids. The residue was lyophilized and subsequently extracted twice with 50 mm NaCl and 50 mm Gly, pH 2.4, for 15 min at room temperature under continuous shaking. To prevent protein degradation, a protease inhibitor mix (2× CØmplete, Roche Diagnostics, Brussels) was added to the extraction buffer. The pellet was removed by centrifugation at 20,000g and supernatants were pooled. Total protein quantity in the crude extracts was determined by the Lowry method using the DC protein assay (Bio-Rad) with BSA as a standard. Proteins were separated on SDS-PAGE and visualized by Coomassie Blue staining. Expression levels of arcelin-5 proteins in transgenic Arabidopsis seeds were estimated (as a percentage of total extractable seed protein) by western blot analysis (as described by Harlow and Lane, 1988) using a rabbit polyclonal anti-arcelin-5 antiserum. Arcelin-5 proteins purified from the P. vulgaris genotype G02771 were used to construct a standard curve. Estimations of Arc5a expression levels were conducted at least two times for each transgenic line (approximately 500 seeds per assay).
Detection and Quantification of arc5-I and at2S mRNA in Transgenic Arabidopsis Siliques
Siliques at stages D, E, and DS—stages at which the highest mRNA steady-state levels of seed protein genes are observed (Guerche et al., 1990b)—were harvested and pooled. Total RNA was prepared following the method described by Shirzadegan et al. (1991). The presence and size of arc5-I transcripts in different plant organs was verified by northern blot analysis (Sambrook et al., 1989). mRNA steady-state levels were determined by slot blot analysis as described by Guerche et al. (1990b) except that a nonradioactive detection method was used (Gene Images, Amersham). Levels of both arc5-I and the endogenous 2S albumin transcripts were estimated using an arcelin-5 DNA probe (covering the complete coding sequence of the arc5-I gene; Goossens et al., 1995) or a 2S2 DNA probe (covering the complete coding sequence of the Arabidopsis at2S2 gene; Krebbers et al., 1988), respectively.
The at2S2 probe used is probably not specific to the at2S2 transcripts alone, but might also hybridize with transcripts from the other endogenous 2S albumin genes (Guerche et al., 1990b). Therefore, the term “at2S transcripts” will be used instead of “at2S2 transcripts.” Unlabeled sense RNA (arc5-I or at2S2) synthesized with an in vitro transcription system (Riboprobe combination system SP6/T7, Promega), was used to generate the standard curve. After hybridization and detection, each signal on the film was quantified by densitometry scanning using imaging software (Imagemaster VDS, Pharmacia). Each RNA preparation was examined at least three times for arc5-I or at2S2 steady-state levels.
RESULTS
Sequence Analysis of the arc5-I Gene and Design of arcelin-5 Constructs
Analysis of the sequenced fragment of the arc5-I genomic clone (Fig. 2; Goossens et al., 1995) revealed the presence of a large number of putative regulatory elements in the 5′ and 3′ flanking sequences of the arc5-I gene (data not shown). Among these are cis-regulatory elements thought to be involved in (quantitative) seed-specific expression (see Thomas, 1993, and refs. therein). The analysis showed that most of the seed-specific motifs found in sequences of diverse legume globulins were also encountered in the 5′ flanking sequence of the arc5-I gene. The majority of these elements was clustered between positions −500 to −50 upstream from the translation start site. In contrast, elements specific for monocotyledonous seed storage proteins could not be detected in the arc5-I sequence.
Aside from these putative seed-specific regulatory elements, computer analysis also showed the presence of multiple potential MARs (for a review, see Breyne et al., 1994; Holmes-Davis and Comai, 1998) in the arc5-I gene. Three clusters of potential MAR sequences were detected: two 5′ MARs (located at positions −1,800 to −1,500 and −1,000 to −500 upstream from the translation start site) and one 3′ MAR (located at position 1,150–1,600 downstream from the translation start site). Analogous situations were found in the regulatory sequences of other abundantly expressed seed storage proteins of leguminous species, such as the P. vulgaris phaseolin (Slightom et al., 1983) and the broad bean legumin B4 (Bäumlein et al., 1986). In arc5-I, motifs were detected only on the basis of sequence similarity; no experiments were conducted to prove their functionality.
In P. acutifolius transformation experiments, T-DNA constructs were used that contained the BamHI fragment of the arc5-I gene (introduced into pATARC3-B1b; Fig. 2) or a chimeric gene containing the arc5-II coding region between the arc5-I 5′ and 3′ regulatory sequences (introduced into pATARC3-B52b). For Arabidopsis transformation experiments, two constructs were designed with arc5-I gene fragments of different sizes (Fig. 2) to assess which part of the genomic clone suffices to obtain high seed-specific expression and/or low plant-to-plant variation in transgenic plants. The BamHI/MunI fragment was introduced into the first construct (pATARC4-BM) and contained potential TATA and CCAT boxes, the majority of the potential cis-regulatory elements for seed-specific expression, and also one cluster of 5′ MAR sequences. The second construct (pATARC4-E) contained the EcoRI fragment, which represented the largest arc5-I fragment available from the genomic clone. This fragment harbored the other potential MARs (see above) and possibly additional regulatory elements in the nonsequenced part of the EcoRI fragment (Fig. 2).
Detection of Arc5 Proteins in Transgenic Plants
The Arc5 protein was detected both by Coomassie Blue staining and western blotting (Figs. 3 and 4) in transgenic seeds. Apart from the additional band representing Arc5a, Arc5b, or Arc5c, no major alterations were obvious in the total protein profile of transgenic P. acutifolius (Fig. 3A) or Arabidopsis (Fig. 3B) seeds. The arc5-I and arc5-II genes both encode a precursor protein of 261 amino acids with a signal peptide of 21 amino acids. In P. vulgaris, this signal peptide is removed from the precursor, to which no (Arc5c), one (Arc5b), or two (Arc5a) glycan chains of the complex fucosylated type are subsequently attached (Goossens et al., 1994). SDS-PAGE and western blot analysis showed that the Arc5 proteins from transgenic P. acutifolius and Arabidopsis seeds co-migrated with the Arc5 proteins that were purified from P. vulgaris seeds (Figs. 3 and 4), indicating that the protein was processed correctly. In arc5-II-expressing P. acutifolius lines, the Arc5c protein could not be distinguished on SDS-PAGE because of its low abundance and because of the presence of a background of P. acutifolius proteins with a similar electrophoretic mobility. The expected minor levels of Arc5c could, however, clearly be observed by western blot analysis (data not shown). No Arc5 degradation products were detected in transgenic seeds of either species.
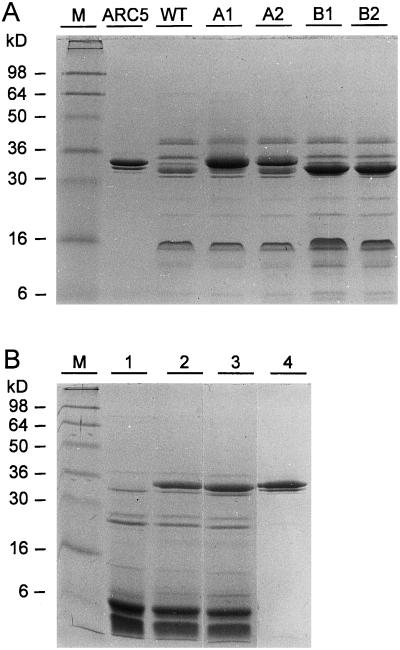
Figure 3.
SDS-PAGE on crude protein extracts of transgenic P. acutifolius and Arabidopsis seeds. Proteins were visualized with Coomassie Blue staining. A, P. acutifolius seed proteins extracted from nontransgenic (wild-type) NI576 (lane WT) plants, transgenic plants containing arc5-I (lanes A1 and A2), and transgenic plants containing arc5-II (lanes B1 and B2). Lane ARC5 contains arcelin-5 proteins purified from the P. vulgaris genotype G02771 (from top to bottom, Arc5a, Arc5b, and Arc5c, respectively), and lane M contains the marker proteins. The molecular masses are indicated on the left in kD. B, Crude protein extracts of Arabidopsis seeds of the nontransformed Columbia-0 genotype (lane 1) and of transgenic lines BM-410 (lane 2) or E-103 (lane 3). Lane 4 contains arcelin-5 proteins purified from the P. vulgaris genotype G02771, and lane M contains the marker proteins. The molecular masses are indicated on the left in kD.
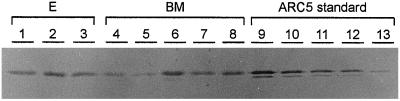
Figure 4.
Quantification of Arc5a by western blot analysis in transgenic Arabidopsis seeds. Lanes contain 1 μg of protein of crude extracts of seeds of transgenic lines harboring the T-DNA of pATARC4-E (E; lanes 1–3) or the T-DNA of pATARC4-BM (BM; lanes 4–8). Lanes 9, 10, 11, 12, and 13 contain a dilution series of purified arcelin-5 proteins: 150, 100, 75, 50, and 25 ng were loaded, respectively.
Quantification of Arc5 Accumulation Levels in Transgenic Seeds
Arc5 accumulation levels were determined as the percentage of total extractable seed protein. The three most used methods to measure total protein concentrations (i.e. UV-A280, the Bradford method, and the Lowry method) gave substantially different values for the same protein extract of seeds of either P. acutifolius or Arabidopsis. Measured values differed up to 7-fold (for P. acutifolius seed extracts) or even 20-fold (for Arabidopsis seed extracts) depending on the method used. This discrepancy could be explained by the fact that these quantification methods rely on the recognition of only a few amino acids and that the bulk of the seed protein pool is made up of a small number of different proteins in many plant species.
Therefore, for seeds of each species a large-scale protein extraction was performed. Salts (from the extraction buffer) and low-Mr seed compounds were removed by gel filtration (NAP-10, Pharmacia) and extracts were subsequently lyophilized. Total protein levels were then determined by weighing. In this way, it was possible to verify that reliable results were obtained with UV-A280 for P. acutifolius and with the Lowry method for Arabidopsis. Additionally, Arc5 accumulation levels were assayed after extraction with different buffer systems and at different pHs and were independent of the buffer system or pH used to generate the extract (data not shown).
Transgenic P. acutifolius and Arabidopsis plants were selected for the presence of one transgenic locus with intact T-DNA inserts. Arc5 protein was quantified in seeds of hemizygous and homozygous progeny of the selected lines (Table II). We will primarily discuss the situation in homozygous plants, so expression levels mentioned in the text refer to levels in seeds of homozygous progeny.
Table II.
Characterization of transgenic plants harboring one transgenic locus
| Line | Copies | Arc5 Protein
Level
|
mRNA Ratio arc5-I/at2S | |
|---|---|---|---|---|
| Hemizygous | Homozygous | |||
| n | ||||
| P. acutifolius | ||||
| B1b-27a | 2 (IRR) | 5.8 ± 1.5 | 16.2 ± 0.6 | |
| B1b-8a | 2 (IRR) | 5.6 ± 0.6 | 23.8 ± 2.6 | |
| B52b-44.4 | 1 | 6.3 ± 0.6 | 15.7 ± 1.4 | |
| B52b-44.X | 1 | 6.7b | 25.4 ± 7.0 | |
| Arabidopsis | ||||
| BM-406 | 1 | ND | 5.3 ± 1.0 | 2.3 |
| BM-301 | 1 | 6.8 ± 0.8 | 5.4 ± 1.6 | ND |
| BM-409 | 1 | 5.2 ± 0.2 | 6.2 ± 1.0 | ND |
| BM-410 | 1 | 6.5 ± 1.2 | 7.7 ± 1.0 | 10.5 |
| BM-306 | 1 | 6.0 ± 0.1 | 8.1 ± 2.2 | ND |
| BM-304 | 2 (IRR) | 1.3 ± 0.3 | 1.0 ± 0.1 | ND |
| BM-414 | 2 (IRR) | 2.2 ± 0.1 | 1.2 ± 0.6 | 0.02 |
| BM-402 | 2 (IRR) | 1.2 ± 0.1 | 0.8 ± 0.6 | ND |
| BM-416 | ≥3 | 3.5 ± 0.7 | 1.3 ± 0.5 | ND |
| BM-401 | ≥3 | 2.8 ± 0.0 | 2.5 ± 2.1 | ND |
| BM-303 | ≥3 | 8.8 ± 1.5 | 5.0 ± 0.5 | ND |
| BM-412 | ≥3 | 11.7 ± 0.0 | 14.3 ± 0.9 | 6.6 |
| E-307 | 1 | 7.9 ± 0.8 | 8.6 ± 2.0 | ND |
| E-402 | 1 | ND | 9.7 ± 0.2 | ND |
| E-313 | 1 | 7.3 ± 0.6 | 9.8 ± 2.3 | 2.7 |
| E-105 | 1 | 9.5 ± 0.9 | 11.9 ± 0.1 | ND |
| E-103 | 1 | 8.1 ± 0.3 | 14.7 ± 0.1 | 2.0 |
| E-305 | 2 (IRR) | 9.6 ± 0.4 | 14.1 ± 2.7 | 2.3 |
| E-411 | ≥3 | 1.6 ± 1.0 | 3.2b | ND |
| E-102 | ≥3 | 4.9 ± 0.3 | 10.8 ± 4.2 | 2.0 |
| E-306 | ≥3 | 7.3 ± 2.4 | 13.0 ± 1.2 | ND |
Transgenic lines marked with B1b, B52b, BM, and E contain T-DNA inserts from pATARC3-B1b, pATARC3-B52b, pATARC4-BM, and pATARC4-E, respectively. In case of multiple copies, the organization (when known) is indicated between brackets. IRR, Inverted repeat over the right T-DNA border. The Arc5 protein level is indicated as percentage of total extractable protein of seeds of hemizygous or homozygous transgenic plants. Values are followed by the sd. ND, Not determined due to limited amounts of seeds available. In case of P. acutifolius, measurements were performed on aliquots of one hemizygous seed and thus really reflect the Arc5 level in a hemizygous seed. In contrast, in Arabidopsis, measurements were performed on an aliquot of approximately 500 seeds harvested from a hemizygous plant, thus representing the level in a mixture of homozygous transformed, homozygous nontransformed and hemizygous seeds.
These lines showed small deletions at the left and right border of the T-DNA inserts.
Due to the limited number of seeds available quantification was only performed once and thus no sd could be calculated.
The highest arc5 expression levels were found in the P. acutifolius lines, in which Arc5 accumulation levels ranged from 15% to 25% of total seed protein. In line B1b-8 the high expression level in seeds of homozygous transgenic plants was inherited through three successive generations (for other lines only one homozygous transgenic generation was available). A clear gene dosage effect could be observed when hemizygous and homozygous plants were compared: transgene copy doubling resulted in higher transgene expression levels.
Arabidopsis lines also exhibited high Arc5 protein accumulation in the seeds, with levels ranging from 1% to 15% of total seed protein. Among Arabidopsis plants transformed with the same T-DNA construct, relatively low plant-to-plant variation (less than 15-fold) was observed. Moreover, when only transgenic lines containing one T-DNA copy were taken into account, variation was less than 2-fold. This low variation was obtained for both the pATARC4-BM and the pATARC4-E constructs. The range of Arc5 accumulation levels was also similar for the two constructs. However, lines containing the largest arc5-I genomic fragment (EcoRI lines in Table II) generally showed the highest expression levels. This was most obvious when only lines harboring one T-DNA copy were considered: single-copy BM lines showed expression levels from 5.3% to 8.1%, whereas single-copy E lines had expression levels ranging from 8.6% to 14.7%. No gene dosage effect was consistently observed in BM or E lines, as a consequence of multiple T-DNA integration, nor after the transition from hemi- to homozygous plants.
Detection and Quantification of arc5-I Transcripts in Transgenic Arabidopsis
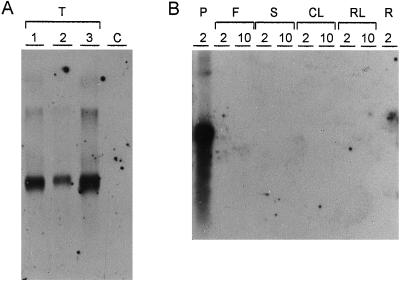
Northern blot analysis of total RNA from siliques of transgenic Arabidopsis lines confirmed the presence of arc5-I transcripts with a size of approximately 1,100 nu-cleotides (Fig. 5), corresponding to the size expected from the cDNA sequence (Goossens et al., 1994). No arc5-I transcripts were detected in total RNA preparations from flowers, cauline or rosette leaves, stems, or roots from transgenic Arabidopsis plants (Fig. 5), indicating that expression of the arc5-I gene was restricted to seeds in Arabidopsis.
Figure 5.
Northern blot analysis of different plant organs of transgenic Arabidopsis. A, Total RNA extracted from siliques of transgenic plants (lanes T1–T3) and of a nontransformed Columbia-0 plant (lane C) hybridized with an arc5-I probe. B, Total RNA extracted from different organs of a transgenic plant harboring the pATARC4-E T-DNA hybridized to an arc5-I probe. Total RNA (2 and 10 μg) of siliques (P), flowers (F), stems (S), cauline leaves (CL), rosette leaves (RL), and roots (R) were loaded.
Steady-state levels of arc5-I mRNA were quantified in developing siliques of some homozygous transgenic plants with one transgenic locus. In parallel, mRNA levels of the most abundant Arabidopsis seed proteins, the 2S albumins, were determined. The data are presented as the molar ratio of arc5-I mRNA to at2S mRNA (Table II). This analysis indicated that lines that produced high amounts of Arc5a polypeptides (>5% of total protein) generally showed high steady-state levels of arc5-I mRNA. Moreover, in these lines the transgene mRNA steady-state level was significantly higher than that of endogenous 2S albumin transcripts.
DISCUSSION
Seed storage proteins generally accumulate to very high quantities in developing seeds. So far, efforts to transfer the high expression levels directed by these seed storage protein genes to a heterologous system have had limited success, although various protein-coding regions and heterologous host plants have been used. This can be explained in part by the fact that seed storage proteins are encoded by multigene families, and an individual gene therefore only contributes to a fraction of the total seed storage protein. However, even when this is taken into account, expression levels are often lower than expected (see Table I). The regulatory sequences used in these studies to drive seed-specific expression may therefore lack essential cis elements. Alternatively (or additionally), trans-acting factors may not be present in the appropriate amounts or at the appropriate time in the heterologous plant. The highest expression levels (up to 8%) directed by a transgene were obtained in chimeric gene constructs with the promoter of the P. vulgaris seed storage protein β-phaseolin (Altenbach et al., 1989, 1992). Phaseolin, which is encoded by a multigene family with seven to nine genes per haploid genome (Slightom et al., 1985), accounts for up to 60% of the total protein in P. vulgaris seeds. Romero Andreas et al. (1986) described a novel seed storage protein in some wild genotypes of P. vulgaris called arcelin. Of the seven arcelin variants identified so far, we have previously characterized the arcelin-5 variant (Goossens et al., 1994). This protein is encoded by two functional gene copies per haploid genome and accumulates to levels of 30% to 40% of the total protein content in the wild P. vulgaris genotype G02771. In the present study we demonstrate that, in contrast to the results obtained for many other genes encoding seed storage proteins, high arcelin-5 accumulation levels can also be obtained in a heterologous system.
The arcelin-5 gene was introduced into the legume species P. acutifolius and into the crucifer Arabidopsis. In seeds from transgenic Arabidopsis, high accumulation levels were found, i.e. up to 15% of the total seed protein content. In P. acutifolius, a species more closely related to P. vulgaris from which the arc5-I gene was isolated, expression was even higher and ranged from 15% to 25% of the total seed protein content. This is similar to the levels obtained by introgression of the arcelin-5 locus by backcross breeding into P. vulgaris cultivars (A. Goossens and C. Cardona, unpublished results), although still somewhat lower than the levels found in the wild P. vulgaris genotype, in which arcelin-5 was originally identified. The genetic background may thus be important in modulating arcelin-5 expression.
Northern analysis indicated that the high arcelin protein levels are the result of high mRNA steady-state levels. In transgenic Arabidopsis plants that produce high amounts of arcelin protein, the transgene mRNA expression level is higher than the level of endogenous 2S albumin transcripts. Whether the high arcelin mRNA steady-state levels are primarily due to a high transcription rate or a high stability of the arcelin-5 transcript has not yet been established. To answer this question, chimeric genes that contain heterologous coding regions driven by the arc5-I regulatory sequences are now being constructed. Notwithstanding the high arc5-I/at2S mRNA ratio, the major storage proteins in seeds of these transgenic lines are the 2S albumins (>50% of total protein based on densitometry scanning of Coomassie Blue-stained SDS-PAGE; data not shown). This phenomenon could be caused by a difference in protein stability, by a difference in translational efficiency for the different transcripts, and/or by a suboptimal timing of arcelin expression. Northern analysis also showed that arcelin-5 expression in transgenic plants is restricted to seeds. This and the high accumulation levels observed indicate that all of the elements necessary for high seed-specific expression are cis elements present in the genomic arc5-I clone and trans-acting factors that may be generally present in dicotyledonous plants.
Another remarkable characteristic of plants transformed with the arc5 gene is the low plant-to-plant variation in transgene expression. This could be a major advantage of the use of the arcelin-5 expression signals in chimeric gene constructs, as it would limit the number of transformants that need to be generated and analyzed. When only transgenic lines with one T-DNA copy are considered, variation is less than 2-fold. Moreover, even in plants with a more complex T-DNA integration pattern (inverted repeats and three or more linked copies), arcelin expression remains high, especially in the P. acutifolius lines and the Arabidopsis E lines. This is surprising, as complex integration patterns, and particularly inverted repeats, are very often hallmarks of gene silencing (Hobbs et al., 1993; Depicker and Van Montagu, 1997; Stam et al., 1997). On the other hand, most plants transformed with a construct containing the arc5-I gene and a 2S albumin antisense gene under control of the arc5-I 5′ and 3′ regulatory sequences in a tandem array accumulate very low levels of arcelin-5, suggesting a form of silencing in this particular gene configuration (A. Goossens and G. Angenon, unpublished results).
It could be speculated that certain genes have evolved a mechanism to avoid silencing effects. Such a mechanism could be particularly useful for seed protein genes, which are often organized as clustered, highly expressed, and highly homologous members of a multigene family. The exact genomic configuration of the arcelin genes is not known; however, they are probably in close physical proximity to each other and to the phytohemagglutinin genes, as is the case with two phytohemagglutinin genes of P. vulgaris cv Tendergreen (Hoffman and Donaldson, 1985). Whether features protecting against silencing are present in the arc5-I gene and whether these have been disturbed in some arc5 gene constructs remains to be studied.
To dissect the signals in the arc5-I gene necessary for the high seed-specific expression, we introduced two different fragments of the arc5-I genomic clone into Arabidopsis plants. The first construct (pATARC4-BM) contains 1,160 bp of 5′ and 610 bp of 3′ flanking sequences of the arc5-I gene and harbors potential TATA and CCAT boxes, most of the potential cis-regulatory elements for high seed-specific expression, and one region of 5′ MAR sequences. A similar construct (but with 1,800 bp of 3′ flanking sequences) was also introduced into P. acutifolius. The second construct (pATARC4-E) contains the arc5-I EcoRI fragment, the largest fragment available from the genomic clone that possesses additional potential regulatory elements.
Although the maximum accumulation level obtained with the pATARC4-BM and pATARC4-E constructs was similar, the E lines showed on average a higher expression than the BM lines. It remains to be determined whether this was caused by specific transcription factor-binding elements, a stabilizing effect of the increasing length of the arc5-I-flanking sequences, additional MAR sequences, or other, unknown factors. MARs have attracted attention because of their perceived capacity to increase levels of transgene expression, reduce transformant-to-transformant variation of transgene expression, and confer copy number dependence to transgene expression. These properties are a consequence of a possible role for MARs as boundary elements or chromatin-regulatory elements (Holmes-Davis and Comai, 1998). MAR sequences are also present in the arc5-I gene and may thus be important elements in the low variation and the high expression levels observed for all arc5-I constructs and the overall higher expression levels in the E lines.
The work presented here indicates that the 5′ and 3′ flanking sequences (≥1.1 and ≥0.6 kb, respectively) of the arc5-I seed storage protein gene contain most, if not all, of the essential information for correct developmental and spatial regulation and exceptionally high accumulation of the arcelin-5 protein in transgenic plants. Moreover, these expression signals appear to function efficiently in two different plant species that are taxonomically not closely related. Therefore, arcelin-5 expression signals could provide a powerful tool for engineering seed characteristics in various plant species.
ACKNOWLEDGMENTS
The authors wish to thank Hilde Dhuyvetter for excellent technical assistance, Ann Depicker and Geert De Jaeger for helpful comments, and Martine De Cock, Karel Spruyt, Rebecca Verbanck, and Christiane Germonprez for help with the preparation of the manuscript and the figures.
Abbreviation:
- MAR
matrix attachment region
Footnotes
This work was supported in part by a grant from the Algemeen Bestuur voor Ontwikkelingssamenwerking. A.G. and J.D.C. both received fellowships from the Vlaams Instituut voor de bevordering van het Wetenschappelijk-Technologisch Onderzoek in de Industrie.
LITERATURE CITED
- Altenbach SB, Kuo C-C, Staraci LC, Pearson KW, Wainwright C, Georgescu A, Townsend J. Accumulation of a Brazil nut albumin in seeds of transgenic canola results in enhanced levels of seed protein methionine. Plant Mol Biol. 1992;18:235–245. doi: 10.1007/BF00034952. [DOI] [PubMed] [Google Scholar]
- Altenbach SB, Pearson KW, Meeker G, Staraci LC, Sun SSM. Enhancement of the methionine content of seed proteins by the expression of a chimeric gene encoding a methionine-rich protein in transgenic plants. Plant Mol Biol. 1989;13:513–522. doi: 10.1007/BF00027311. [DOI] [PubMed] [Google Scholar]
- Bagga S, Adams H, Kemp JD, Sengupta-Gopalan C. Accumulation of 15-kilodalton zein in novel protein bodies in transgenic tobacco. Plant Physiol. 1995;107:13–23. doi: 10.1104/pp.107.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthels N, van der Lee FM, Klap J, Goddijn OJM, Karimi M, Puzio P, Grundler FMW, Ohl SA, Lindsey K, Robertson L. Regulatory sequences of Arabidopsis drive reporter gene expression in nematode feeding structures. Plant Cell. 1997;9:2119–2134. doi: 10.1105/tpc.9.12.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäumlein H, Wobus U, Pustell J, Kafatos FC. The legumin gene family: structure of a B type gene of Vicia fabaand a possible legumin gene specific regulatory element. Nucleic Acids Res. 1986;14:2707–2720. doi: 10.1093/nar/14.6.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beachy RN, Chen Z-L, Horsch RB, Rogers SG, Hoffmann NJ, Fraley RT. Accumulation and assembly of soybean β-conglycinin in seeds of transformed petunia plants. EMBO J. 1985;4:3047–3053. doi: 10.1002/j.1460-2075.1985.tb04044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thalianaplants. CR Acad Sci Ser III Life Sci. 1993;316:1194–1199. [Google Scholar]
- Breyne P, Van Montagu M, Gheysen G. The role of scaffold attachment regions in the structural and functional organization of plant chromatin. Transgenic Res. 1994;3:195–202. doi: 10.1007/BF01973987. [DOI] [PubMed] [Google Scholar]
- Chrispeels MJ, Raikhel NV. Lectins, lectin genes, and their role in plant defense. Plant Cell. 1991;3:1–9. doi: 10.1105/tpc.3.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deblaere R, Reynaerts A, Höfte H, Hernalsteens J-P, Leemans J, Van Montagu M (1987) Vectors for cloning in plant cells. In R Wu, L Grossman, eds, Recombinant DNA: Part D. Methods in Enzymology, Vol 153. Academic Press, New York, pp 277–292
- De Clercq A, Vandewiele M, Van Damme J, Guerche P, Van Montagu M, Vandekerckhove J, Krebbers E. Stable accumulation of modified 2S albumin seed storage proteins with higher methionine contents in transgenic plants. Plant Physiol. 1990;94:970–979. doi: 10.1104/pp.94.3.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pater S, Pham K, Klitsie I, Kijne J. The 22 bp W1 element in the pea lectin promoter is necessary and, as a multimer, sufficient for high gene expression in tobacco seeds. Plant Mol Biol. 1996;32:515–523. doi: 10.1007/BF00019103. [DOI] [PubMed] [Google Scholar]
- Depicker A, Van Montagu M. Post-transcriptional gene silencing in plants. Curr Opin Cell Biol. 1997;9:373–382. doi: 10.1016/s0955-0674(97)80010-5. [DOI] [PubMed] [Google Scholar]
- Dillen W, De Clercq J, Goossens A, Van Montagu M, Angenon G. Agrobacterium-mediated transformation of Phaseolus acutifoliusA. Gray. Theor Appl Genet. 1997;94:151–158. [Google Scholar]
- Ellis JR, Shirsat AH, Hepher A, Yarwood JN, Gatehouse JA, Croy RRD, Boulter D. Tissue-specific expression of a pea legumin gene in seeds of Nicotiana plumbaginifolia. Plant Mol Biol. 1988;10:203–214. doi: 10.1007/BF00027397. [DOI] [PubMed] [Google Scholar]
- Goddijn OJM, Pen J. Plants as bioreactors. Trends Biotechnol. 1995;13:379–387. [Google Scholar]
- Goossens A, Ardiles Diaz W, De Keyser A, Van Montagu M, Angenon G. Nucleotide sequence of an arcelin 5-I genomic clone from wild Phaseolus vulgaris (accession no. Z50202) (PGR 95-075) Plant Physiol. 1995;109:722. [Google Scholar]
- Goossens A, Geremia R, Bauw G, Van Montagu M, Angenon G. Isolation and characterisation of arcelin-5 proteins and cDNAs. Eur J Biochem. 1994;225:787–795. doi: 10.1111/j.1432-1033.1994.0787b.x. [DOI] [PubMed] [Google Scholar]
- Guerche P, De Almeida ERP, Schwarztein MA, Gander E, Krebbers E, Pelletier G. Expression of the 2S albumin from Bertholletia excelsa in Brassica napus. Mol Gen Genet. 1990a;221:306–314. doi: 10.1007/BF00259393. [DOI] [PubMed] [Google Scholar]
- Guerche P, Tiré C, Grossi de Sa F, De Clercq A, Van Montagu M, Krebbers E. Differential expression of the Arabidopsis2S albumin genes and the effect of increasing gene family size. Plant Cell. 1990b;2:469–478. doi: 10.1105/tpc.2.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habben JE, Larkins BA (1995) Improving protein quality in seeds. In J Kigel, G Galili, eds, Seed Development and Germination. Marcel Dekker, New York, pp 791–810
- Hames BD, Rickwood D. Gel Electrophoresis of Proteins: A Practical Approach, Ed 2. Oxford: Oxford University Press; 1990. [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Higgins TJV, Newbigin EJ, Spencer D, Llewellyn DJ, Craig S. The sequence of a pea vicilin gene and its expression in transgenic tobacco plants. Plant Mol Biol. 1988;11:683–695. doi: 10.1007/BF00017468. [DOI] [PubMed] [Google Scholar]
- Hobbs SLA, Warkentin TD, DeLong CMO. Transgene copy number can be positively or negatively associated with transgene expression. Plant Mol Biol. 1993;21:17–26. doi: 10.1007/BF00039614. [DOI] [PubMed] [Google Scholar]
- Hoffman LM, Donaldson DD. Characterization of two Phaseolus vulgarisphytohemagglutinin genes closely linked on the chromosome. EMBO J. 1985;4:883–889. doi: 10.1002/j.1460-2075.1985.tb03714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman LM, Donaldson DD, Bookland R, Rashka K, Herman EM. Synthesis and protein body deposition of maize 15-kd zein in transgenic tobacco seeds. EMBO J. 1987;6:3213–3221. doi: 10.1002/j.1460-2075.1987.tb02638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes-Davis R, Comai L. Nuclear matrix attachment regions and plant gene expression. Trends Plant Sci. 1998;3:91–97. [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- Koncz C, Schell J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacteriumbinary vector. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- Krebbers E, Herdies L, De Clercq A, Seurinck J, Leemans J, Van Damme J, Segura M, Gheysen G, Van Montagu M, Vandekerckhove J. Determination of the processing sites of an Arabidopsis2S albumin and characterization of the complete gene family. Plant Physiol. 1988;87:859–866. doi: 10.1104/pp.87.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molvig L, Tabe LM, Eggum BO, Moore AE, Craig S, Spencer D, Higgins TJV. Enhanced methionine levels and increased nutritive value of seeds of transgenic lupins (Lupinus angustifoliusL.) expressing a sunflower seed albumin gene. Proc Natl Acad Sci USA. 1997;94:8393–8398. doi: 10.1073/pnas.94.16.8393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro JK, Jofuku KD, Goldberg RB. Soybean seed lectin gene and flanking nonseed protein genes are developmentally regulated in transformed tobacco plants. Proc Natl Acad Sci USA. 1986;83:8240–8244. doi: 10.1073/pnas.83.21.8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn TC, Alexander DC, Sun SSM, Cardona C, Bliss FA. Insecticidal activity and lectin homology of arcelin seed protein. Science. 1988;240:207–210. doi: 10.1126/science.240.4849.207. [DOI] [PubMed] [Google Scholar]
- Pickardt T, Saalbach I, Waddell D, Meixner M, Müntz K, Schieder O. Seed specific expression of the 2S albumin gene from Brazil nut (Bertholletia excelsa) in transgenic Vicia narbonensis. Mol Breed. 1995;1:295–301. [Google Scholar]
- Romero Andreas J, Yandell BS, Bliss FA. Bean arcelin. 1. Inheritance of a novel seed protein of Phaseolus vulgarisL. and its effect on seed composition. Theor Appl Genet. 1986;72:123–128. doi: 10.1007/BF00261467. [DOI] [PubMed] [Google Scholar]
- Saalbach I, Pickardt T, Machemehl F, Saalbach G, Schieder O, Müntz K. A chimeric gene encoding the methionine-rich 2S albumin of the Brazil nut (Bertholletia excelsaH.B.K.) is stably expressed and inherited in transgenic grain legumes. Mol Gen Genet. 1994;242:226–236. doi: 10.1007/BF00391017. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed. 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schroeder HE, Gollasch S, Moore A, Tabe LM, Craig S, Hardie DC, Chrispeels MJ, Spencer D, Higgins TJV. Bean α-amylase inhibitor confers resistance to the pea weevil (Bruchus pisorum) in transgenic peas (Pisum sativumL.) Plant Physiol. 1995;107:1233–1239. doi: 10.1104/pp.107.4.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta-Gopalan C, Reichert NA, Barker RF, Hall TC, Kemp JD. Developmentally regulated expression of the bean β-phaseolin gene in tobacco seed. Proc Natl Acad Sci USA. 1985;82:3320–3324. doi: 10.1073/pnas.82.10.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirzadegan M, Christie P, Seemann JR. An efficient method for isolation of RNA from tissue cultured plant cells. Nucleic Acids Res. 1991;19:6055. doi: 10.1093/nar/19.21.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindhu AS, Zheng Z, Murai N. The pea seed storage protein legumin was synthesized, processed, and accumulated stably in transgenic rice endosperm. Plant Sci. 1997;130:189–196. [Google Scholar]
- Slightom JL, Drong RF, Klassy RC, Hoffman LM. Nucleotide sequences from phaseolin cDNA clones: the major storage proteins from Phaseolus vulgarisare encoded by two unique gene families. Nucleic Acids Res. 1985;13:6483–6498. doi: 10.1093/nar/13.18.6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slightom JL, Sun SM, Hall TC. Complete nucleotide sequence of a French bean storage protein gene: phaseolin. Proc Natl Acad Sci USA. 1983;80:1897–1901. doi: 10.1073/pnas.80.7.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam M, de Bruin R, Kenter S, van der Hoorn RAL, van Blokland R, Mol JNM, Kooter JM. Post-transcriptional silencing of chalcone synthase in Petuniaby inverted transgene repeats. Plant J. 1997;12:63–82. [Google Scholar]
- Sturm A, Voelker TA, Herman EM, Chrispeels MJ. Correct glycosylation, Golgi-processing, and targeting to protein bodies of the vacuolar protein phytohemagglutinin in transgenic tobacco. Planta. 1988;175:170–183. doi: 10.1007/BF00392425. [DOI] [PubMed] [Google Scholar]
- Sun SSM, Larkins BA. Transgenic plants for improving seed storage proteins. In: Kung S-d, Wu R., editors. Transgenic Plants, Vol 1: Engineering and Utilization. San Diego: Academic Press; 1993. pp. 339–372. [Google Scholar]
- Tabe L, Higgins TJV. Engineering plant protein composition for improved nutrition. Trends Plant Sci. 1998;3:282–286. [Google Scholar]
- Thomas TL. Gene expression during plant embryogenesis and germination: an overview. Plant Cell. 1993;5:1401–1410. doi: 10.1105/tpc.5.10.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueng P, Galili G, Sapanara V, Goldsbrough PB, Dube P, Beachy RN, Larkins BA. Expression of a maize storage protein gene in petunia plants is not restricted to seeds. Plant Physiol. 1988;86:1281–1285. doi: 10.1104/pp.86.4.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens D, Van Montagu M, Van Lijsebettens M. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thalianaroot explants by using kanamycin selection. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vancanneyt G, Schmidt R, O'Connor-Sanchez A, Willmitzer L, Rocha-Sosa M. Construction of an intron-containing marker gene: splicing of the intron in transgenic plants and its use in monitoring early events in Agrobacterium-mediated plant transformation. Mol Gen Genet. 1990;220:245–250. doi: 10.1007/BF00260489. [DOI] [PubMed] [Google Scholar]
- van der Klei H, Van Damme J, Casteels P, Krebbers E. A fifth 2S albumin isoform is present in Arabidopsis thaliana. Plant Physiol. 1993;101:1415–1416. doi: 10.1104/pp.101.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker TA, Herman EM, Chrispeels MJ. In vitro mutated phytohemagglutinin genes expressed in tobacco seeds: role of glycans in protein targeting and stability. Plant Cell. 1989;1:95–104. doi: 10.1105/tpc.1.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Sumi K, Tanaka K, Murai N. The bean seed storage protein β-phaseolin is synthesized, processed, and accumulated in the vacuolar type-II protein bodies of transgenic rice endosperm. Plant Physiol. 1995;109:777–786. doi: 10.1104/pp.109.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]