SUMMARY
Objective
Juvenile Myoclonic Epilepsy (JME) is a common adolescent-onset genetic generalized epilepsy (GGE) syndrome. Multiple linkage and association studies have found that BRD2 influences the expression of JME. The BRD2-JME connection is further corroborated by our murine model: Brd2 haplo-insufficiency produces characteristics that typify the clinical hallmarks of JME. Neither we, nor several large-scale studies of JME, found JME-related BRD2 coding mutations. Therefore, we investigated non-coding BRD2 regions, seeking the origin of BRD2’s JME influence. BRD2’s promoter harbors a JME-associated SNP (rs3918149) and a CpG (C-phosphate-G dinucleotides) island (CpG76), making it a potential “hotspot” for JME-associated epigenetic variants. Methylating promoter CpG sites causes gene silencing, often resulting in reduced gene expression. We tested for differences in DNA methylation at CpG76 in three different subgroups: 1) JME patients vs. their unaffected family members, 2) JME vs. patients with other forms of GGE, and 3) Caucasian vs. non-Caucasian JME patients.
Methods
We used DNA pyrosequencing to analyze the methylation status of 10 BRD2 promoter CpG sites in lymphoblastoid cells from JME patients of Caucasian and non-Caucasian origin, unaffected family members, and also non-JME GGE patients. We also measured global methylation levels and DNA methyl transferase 1 (DNMT1) transcript expression in JME families by standard methods.
Results
CpG76 is highly methylated in JME patients compared to unaffected family members. In families with non-JME GGE, we found no relationship between promoter methylation and epilepsy. In non-Caucasian JME families, promoter methylation was mostly not associated with epilepsy. This makes the BRD2 promoter a JME-specific, ethnicity-specific, differentially methylated region. Global methylation was constant across groups.
Significance
BRD2 promoter methylation in JME, and the lack of methylation in unaffected relatives, in non-JME GGE patients, and in non-Caucasian JME, demonstrates that methylation specificity is a possible seizure susceptibility motif in JME risk and suggests JME therapeutics targeting BRD2.
Keywords: Juvenile Myoclonic Epilepsy, Genetic Generalized Epilepsy, BRD2, DNA methylation, linkage analysis, association analysis, gene expression, ethnicity
INTRODUCTION
Juvenile Myoclonic Epilepsy (OMIM 608816), the most common of the adolescent onset generalized epilepsy syndromes, has a prevalence of ~1 in 1000 people worldwide (National Institutes of Health)1. JME is characterized clinically by involuntary myoclonic jerks (mostly on awakening). The interictal electroencephalogram (EEG) in untreated patients shows bursts of generalized spikes-and waves, often at a frequency of 4–6 Hz 2; 3. Although most patients respond to pharmacological treatment, the frequency of symptom recurrence is high when antiepileptic medication is discontinued 4.
JME is highly familial. Our group originally reported strong linkage of a JME-related gene locus at 6p21, the region containing BRD2 5. Association analysis pointed to the BRD2 gene as the best-supported candidate 6–8. The JME-6p21 linkage was confirmed multiple times 8–10. Other studies showed the region (and later the BRD2 gene) to be related to the expression of abnormal EEG patterns 11. 6p21 shows linkage to the photoparaxysmal response 12 and follow-up studies showed association of SNPs in BRD2 with photosensitivity 13. It is important to note that the BRD2-JME linkage appeared to be ethnicity dependent 8; 10 and specific to Caucasians. Recently, Santos et al., (2017), in a systematic review on genetic associations in JME using Prisma guidelines, recognized BRD2 as a significant JME causative/susceptibility gene 14.
The JME-BRD2 link in humans is strongly corroborated by studies of the JME mouse model. This model, in which one Brd2 allele is non-functional (Brd2+/−) recapitulates the JME phenotype with remarkable fidelity. The mouse data show that Brd2 haplo-insufficiency causes increased seizure susceptibility, spontaneous seizures, abnormal interictal EEGs, and other JME hallmarks 15. The proximate neuro-anatomical cause appears to be related to a deficit of specific GABAergic neurons in certain seizure-related brain regions in the basal ganglia pathway i.e. neocortex and striatum 15. That an insufficiency of Brd2 in mice causes a JME-like phenotype implies that, in humans, it is a deficit of BRD2 that contributes to JME susceptibility. However, we have not found disease-related exonic mutations in human BRD2 that are associated with JME or even that affect BRD2 gene expression, nor have any been reported [Genetic Testing Registry, accessed on January 5, 2018].
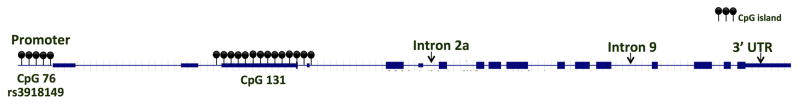
Since no JME-related exonic mutations have been reported, and JME is a common epileptic syndrome, it is likely that sequence variants leading to seizure susceptibility are in the non-coding, control regions of BRD2. We have identified one such non-coding region within the BRD2 promoter, one that harbors a JME-associated SNP (rs3918149) and a CpG (C-phosphate-G dinucleotides) island (Figure 1). CpG islands are clusters of CpG dinucleotides; they are usually non-methylated. But methylation of CpG islands in the promoter of a gene modulates the gene’s expression. DNA methylation is known to play a critical role in brain function and behavior, and dysregulated methylation is a cause of disease 16. Since DNA methylation leads to altered gene expression, it may underlie changes in BRD2 expression that lead to epileptogenesis in JME via changes in neuronal excitability or changes in neuronal populations (as suggested by our mouse model studies). A growing number of human and murine studies have shown the importance of DNA methylation in epilepsy susceptibility (reviewed in 17). These epigenetic DNA methylation alterations could contribute to JME susceptibility and development/maintenance of JME.
Figure 1.
Schematic of BRD2 locus showing positions of CpG islands, SNP and other landmark features
Experiments have shown that methylation of the promoter CpG sites in BRD2 can lead to gene silencing. During adipocyte differentiation, DNA methylation status of the CpG island in BRD2’s promoter changes BRD2 expression 18. Since the CpG island in the BRD2 promoter also harbors a JME-associated promoter SNP (rs3918149) 6, the BRD2 promoter is a “hotspot” for possible JME-associated epigenetic variants.
In the present study, we tested whether DNA methylation of the BRD2 promoter (and therefore a possible deficit of BRD2 transcription) is related to the JME phenotype. We asked the following questions: 1) Do JME patients show altered BRD2 promoter methylation compared to family members without JME? 2) Do BRD2 promoter methylation changes within families co-segregate with JME? 3) Do patients diagnosed with GGEs that are not JME have altered BRD2 promoter methylation patterns? 4) Do non-Caucasian JME patients show the same BRD2 promoter methylation pattern as Caucasian JME patients?
METHODS
Participants
Our study included the GGE cohort, including JME and non-JME GGE patients, together with their participating family members 8. These subjects were ascertained in North America, mostly from the East Coast cities of the USA, and collected between 1992 and 2005. The JME probands were diagnosed in accordance with international classification guidelines (Commission on Classification and Terminology of the International League Against Epilepsy 1989) as described by Greenberg et al., 2000 8. A cohort of non-JME GGEs was collected at the same time from the same sources, as described elsewhere 8.
DNA preparation
The blood samples were collected from the patients and family members. Buffy coats were transformed to establish B-cell lymphoblastoid cell lines and kept frozen in liquid nitrogen. DNA was extracted from established cultures of lymphoblastoid cell lines using Wizard Genomic DNA purification kit (Promega).
DNA Pyrosequencing
1 μg DNA was subjected to bisulfite treatment using EZ DNA methylation-Gold kit (Zymo Research) following the manufacturer’s protocol. For pyrosequencing-based methylation analysis of CpG76, the bisulfite modified DNA was PCR amplified using the biotinylated primers (Qiagen) shown in table 1. The biotinylated PCR products were purified using the sepharose beads and denatured using 0.2 M NaOH solution using the pyrosequencing vacuum prep tool. Subsequently, 0.3 μM pyrosequencing primer (Table 1) was annealed to the purified single stranded PCR product and sequencing was carried out using MD96 pyrosequencing System (Qiagen). Quantitation of cytosine methylation was carried out using the PyroMark96 software. The methylated and unmethylated human control DNA (Qiagen) were used to verify bisulfite conversion.
Table 1.
Pyrosequencing primers
| Assay | Forward | Reverse | Sequencing | Number of CpG |
|---|---|---|---|---|
| 1 | GAAGTTGGGGATATGGATAAGT | TTAACCCCCAACCCCTCC | AGTTATAGAGATATGGGTT | 4 |
| 2 | GTTTGAGGTAGTTATGTTGAAT | ACCACTAAAACCAACTTTCC | TATGGAAAAGAAGAAGAGTTATA | 3 |
| 3 | AAGTTAGTAGTATTTAGATTGGTTGAT | TACTCCTACTCTATATATATTCCTA | TGTTTAGAAATTTAGA | 3 |
Global methylation
Global methylation levels of lymphoblastoid cell DNA was measured using the Methylflash Global DNA Methylation ELISA Easy kit (Epigentek) following the manufacture’s recommendations. Briefly, 100 ng of genomic DNA was used for 5-methyl-cytosine (5-Mc) quantitation. The input DNA was washed and incubated with a capture antibody. The wells were then washed and detection antibody was applied. Use of enhancer solution and development solution created a color change proportional to the quantity of 5-mC, and the samples were read colorimetrically on an automated plate reader at 450-nmabsorbance. The use of a standard curve enabled the quantification of 5-mC based on absorbance measurements.
Quantitative RT-PCR
Total RNA was extracted from patient-derived lymphoblastoid cells using RNeasy mini kit and converted into cDNA by iScript cDNA synthesis kit (BioRad) according to manufacturer’s recommendations. qRT-PCR analysis was performed using Taqman probes for DNMT1-Hs00154749 and GAPDH-Hs02758991_g1 (Applied Biosystems) on Realplex Mastercycler (Eppendorf). Expression relative to GAPDH was calculated using 2ΔΔCt
Statistical Methods
The data analyses were performed using Microsoft Excel 2011, Prism 7 (version 7.0a), and R software. Pyrosequencing data in triplicates are expressed in percent methylation and differences in percent methylation were tested for statistical significance at the 1% level based on a Student’s t-test /one-way Analysis of Variance (ANOVA), and a non-parametric bootstrap procedure 19. Because biologically related individuals are not independent, standard statistical procedures that falsely assume independence can have inflated type 1 error. To circumvent this potential problem, we bootstrapped over the independent families to obtain p-values for statistically significant results. Real time PCR data in triplicates are expressed as means and standard errors.
RESULTS
CpG76 is methylated in the DNA from lymphoblasts of Caucasian JME patients
Using pyrosequencing, we tested the hypothesis that the JME phenotype is related to DNA methylation of the BRD2 promoter by measuring the level of CpG76 methylation in the lymphoblastoid cells from Caucasian JME patients and from unaffected family members. We initially tested specifically Caucasian JME families for BRD2 promoter methylation based on the evidence that the BRD2-JME connection exists (primarily) in families of European origin 8. We designed three pyrosequencing assays covering ten CpG sites at CpG76 within the BRD2 promoter (Figure 1 and Table 1). We pyrosequenced 23 JME patients, some of whom were related, and 23 of the unaffected family members from those families.
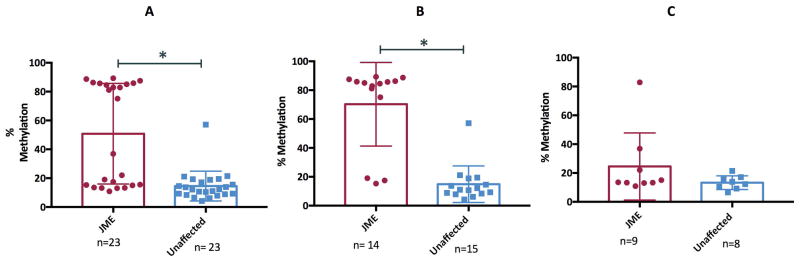
The pyrosequencing analysis revealed a markedly bimodal distribution for percent methylation in JME patients (n=23) [Figure 2A(red dots)], and an apparently unimodal distribution for their unaffected family members (n=23)[Figure 2A(blue dots)]. This suggests that BRD2 is influencing JME in some (but not all) of our JME patients, which is consistent with our previous findings 20 of heterogeneity in JME. To identify those families where BRD2 is likely involved, we stratified our affected families into two groups based on whether there was increase sharing of genetic material (i.e., positive linkage evidence) among affected individuals. In the linked families, we found that the percent difference in methylation between JME patients and their unaffected family members was statistically significant (Figure 2B; p<0.0001). Conversely, in the unlinked families, the difference in percent methylation was not statistically significant (Figure 2C). These data indicate that JME segregates together with DNA methylation status in the BRD2 promoter region, and that JME is a heterogeneous disease.
Figure 2. Status of DNA methylation of the BRD2 promoter in human families identified through a JME patient.
2A–C: A box plot showing CpG 76 methylation levels in Caucasian JME patients and unaffected family members (A), Caucasian JME patients with positive evidence of linkage to BRD2 and unaffected family members (B) and Caucasian JME patients without positive linkage evidence to BRD2 and unaffected family members (C). Data represented as means ± SEM. * indicates significant comparison.
CpG76 is unmethylated in non-JME GGE patients
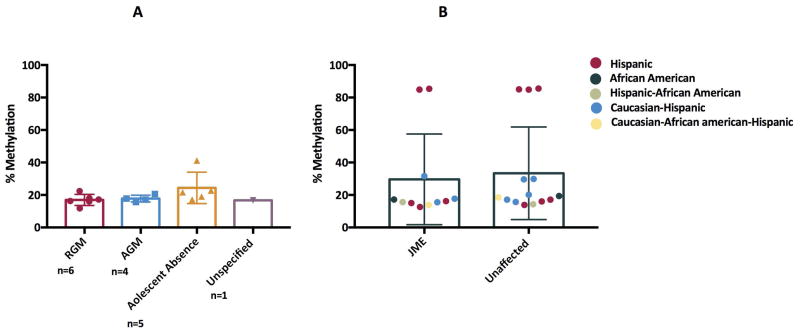
Our previous data showed that non-JME forms of GGE are not linked or associated with the BRD2 locus 21. We analyzed methylation of CpG76 in the BRD2 promoter in patients with Epilepsy with Tonic-Clonic Seizures (or Random Grand Mal (RGM)), Awakening Grand Mal (AGM) and Juvenile Absence Epilepsy (JAE) patients. These patients did not have myoclonic jerks as part of their clinical presentation, according to their medical records, their own report, and those of their family members. Pyrosequencing analysis of 7 RGM, 4 AGM and 5 JAE patients revealed methylation levels similar to those of unaffected JME family members, i.e., there was no evidence of JME-level methylation at the BRD2 promoter (Figure 3A). Non-JME GGE families also showed little evidence of segregation of genetic markers with affectedness 20. These latest data corroborate earlier findings that BRD2 is not linked to non-JME forms of GGE and supports the finding that seizure-related changes in the BRD2 promoter are JME-specific.
Figure 3A–B.
DNA methylation analysis of CpG76 by pyrosequencing on DNA from lymphoblastoid cells in Caucasian non-JME Genetic Generalized Epilepsy patients (A) and non-Caucasian JME families (B)
CpG76 is unmethylated in non-Caucasian JME patients
Our earlier studies revealed that a SNP (rs3918149) located in CpG76 showed a significant association with JME in Caucasian Europeans 6, 7 but no association in Tunisian 22 or Southern Indian populations. We therefore asked whether the JME-specific BRD2 promoter methylation we observed is associated with ethnicity. CpG76 methylation was examined in Hispanic, African-American (AA) and mixed-ethnicity JME families (such as Hispanic-AA, Caucasian-Hispanic and Caucasian-Hispanic-AA) all diagnosed according to the same clinical criteria as the Caucasian JME patients. There was no difference in methylation patterns comparing non-Caucasian JME patients and their unaffected family members (Figure 3B). This finding supports our past work showing that BRD2 is linked to JME only in Caucasians 8. That five of the 25 non-Caucasian JME patients, all Hispanic, showed the high levels of methylation similar to Caucasian JME patients further underlines the importance of genetic heterogeneity within GGE 23.
Global methylation levels are similar in Caucasian JME patients and family members
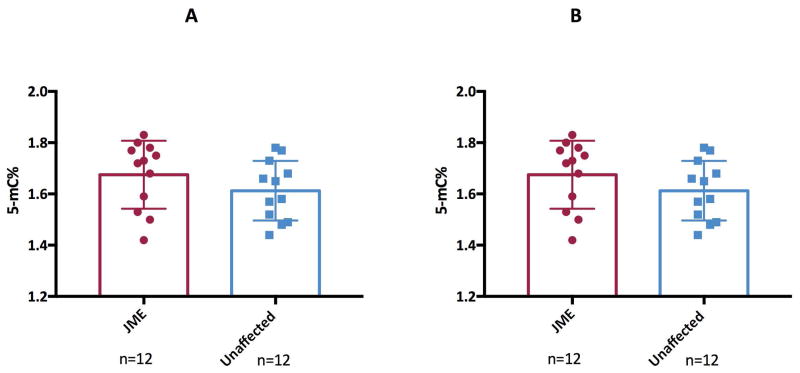
Altered methylation levels of the BRD2 promoter in Caucasian JME patients could be due to alterations in global methylation of B-lymphoblastoid cells. Global methylation patterns are maintained by an enzyme DNA methyl transferase 1 (DNMT1). Hence, we measured global methylation levels using a 5-mC based immunoassay platform and DNMT1 transcripts by qPCR. No significant difference in genome wide 5-methyl cytosine levels (Figure 4A) and DNMT1 transcripts (Figure 4B) was observed between JME patients and unaffected family members.
Figure 4. Global DNA methylation in JME patients and unaffected family members.
4a: A box plot showing global DNA methylation levels (represented by 5-mC%) by ELISA on DNA from Caucasian JME patients and unaffected family members.
4b: Normalized DNMT1 gene expression was measured by qRT-PCR in lymphoblasts from JME patients and unaffected family members. Data were normalized to GAPDH.
DISCUSSION
JME was the first form of epilepsy for which a genetic locus was confirmed, demonstrating JME’s genetic origin 24 and linkage and association analyses from different research groups over the past three decades have established BRD2 as a major genetic component for JME. (See 14 for a review.).
In this section, we expand on the following:
What id BRD2
BRD2 Methylation and Suppression of Protein Expression
TheEffect of Ethnicity, Type of GGE, and Co-segregation of JME and BRD2
Valproic Acid Affects Methylation
The JME-Methylation Finding is Not because of Changes in Global Methylation Levels
Conclusion
1. What is BRD2?
BRD2, a bromodomain containing transcriptional activator, is involved in cell cycle, growth, proliferation, and apoptosis. It is also an essential developmental gene; Brd2’s absence in mice (Brd2−/ −) leads to profound neurodevelopmental failure and early in-utero demise 25; 26. Heterozygous mice (BRD2+/−) are viable and display a startlingly JME-like phenotype, including spontaneous generalized seizures, increased seizure susceptibility to fluorothyl after puberty, female preponderance, and JME-associated behavioral traits, as well as brain-region and sex-specific GABAergic system anomalies 15. The compelling mouse-human phenotype similarities also suggest that JME is, at its core, a neurodevelopmental disorder.
2. BRD2 Methylation and Suppression of Protein Expression
Our current study shows that JME segregates together with a BRD2 methylation variant in the promoter region, a variant that probably leads to reduced BRD2 production. DNA methylation of gene promoters is tightly correlated with transcription suppression, yet the mechanism for such suppression is still unclear 27; 28. Multiple studies have shown a negative association between the level of DNA methylation in promoters and the expression of downstream genes18; 29. Aberrant promoter methylation at the BRD2 locus would most likely result in a BRD2 deficit. We hypothesize that the BRD2 deficit leads to seizure susceptibility due to a BRD2-deficit-caused decrease in specific GABAergic neuronal populations in the brain, as suggested by our mouse model. Also, the linkage evidence 5; 8–10 shows that, at the BRD2 locus, JME is dominantly inherited. This supports the idea that one of the inherited alleles has reduced production of the BRD2 protein.
Growing evidence from the human and murine studies has highlighted the role of DNA methylation in epilepsy susceptibility (reviewed in 17). For instance, Kobow et al. 30 2009 found that increased levels of DNA methylation in the promoter region of Reelin are associated with the pathogenesis of temporal lobe epilepsy patients. Increased methylation at the CPA6 promoter has also been found in patients with focal epilepsy and in febrile seizures31. Furthermore, the neuron restrictive silencing factor, which regulates neuronal gene expression and recruits methylating enzymes, is involved in seizure development and progression32. Recently, genome-wide as well as gene-specific alterations in DNA methylation have been reported in epilepsy pathogenesis (reviewed in 17). To our knowledge, this is the first study to report that aberrant DNA methylation is associated with a GGE phenotype. The dynamics of the DNA methylation mechanism may provide an explanation for some features of JME including adolescent onset timing and possible maternal inheritance 8; 30.
As we noted, the JME-associated SNP rs3918149 is located in the CpG76 island within the BRD2 promoter region. SNPs associated with promoter CpG islands show correlation with gene expression 34. Hence, it is possible that methylation of rs3918149 itself in CpG76 has a role regulating BRD2 transcript abundance in brain.
3. The Effect of Ethnicity, Type of GGE, and Co-Segregation of JME and BRD2
We observed aberrant methylation of the BRD2 promoter only in Caucasian JME patients and then only in families that show positive evidence of linkage to BRD2, that is, co-segregation of JME with genetic markers in the region. We did not see methylation in patients from families that showed evidence against linkage to the region, nor in patients from JME families who were not of Caucasian background. It is well documented that DNA methylation profiles differ in different populations 32. Such differences may contribute to ethnically-based disease susceptibilities differences 35. The underlying reason for racial differences in DNA methylation, at BRD2 or, indeed, anywhere in the genome, is not known. The ethnicity-based methylation differences we observed may reflect variation in brain environment, brain metabolism or genomic signals for methylation. Irrespective of mechanism, we hypothesize that methylated BRD2 in Caucasian JME patients may indicate an ethnicity-specific etiologic pathway for development of epilepsy.
The diagnosis-dependent differences we observed also emphasize the importance of carefully differentiating different forms of disease in order to identify the specific genes involved in disease etiology. The recent trend in studying the genetic origins of many common diseases is to classify all forms of disease as though they had the same genetic basis. This classification approach has led to false starts and wasted resources 20; 23. Our work on methylation also shows that the non-coding regions of the genome can also contribute to common epilepsy susceptibility. While approaches to understanding how the non-coding regions can affect disease expression are only beginning to be developed, our work shows that gaining such understanding is essential for understanding disease etiology.
4. Valproic Acid Affects Methylation
In a majority patients with JME, seizures are well controlled with valproic acid (VPA) with a response rate of up to 80% 36 (although, due to the side-effects associated with VPA use, the popularity of the drug has decreased over time as alternatives have become available). It may not be a coincidence that VPA is a histone deacetylase inhibitor and regulates DNA methylation 37. While VPA has been shown to cause DNA demethylation 35 (thus hypothetically counteracting the highly methylated BRD2 promoter in JME patient), the Caucasian JME patients in our cohort who received VPA treatment still showed high methylation levels of the promoter. Thus, it is unlikely that medication status affected our results.
5. The JME-Methylation Finding is Not because of Changes in Global Methylation Levels
A previous study also suggested a link between BRD2 and DNA methylation. Depletion of BRD2 in HeLa cells by small interfering RNA resulted in ~2.5-fold increased expression of DNA methyl transferase 1 (DNMT1) 38. DNMT1 has an important role in the maintenance of the tissue-specific DNA methylation patterns 39. The aberrant methylation signature in some JME patients of Caucasian origin prompted us to analyze DNMT1 expression and global methylation. We did not observe any change in global methylation level and DNMT1 expression in the lymphoblasts between JME patients and unaffected family members. Thus, the loss of methylation at the BRD2 promoter in the lymphoblasts of JME patients is locus-specific and associated with JME expression.
The brain tissue from well-diagnosed JME patients is unavailable so we cannot determine if human JME patients have a brain-localized BRD2 deficit. Therefore, we have used patient-derived lymphoblastoid lines for DNA methylation evaluation of the BRD2 promoter. Several studies have reported that disease-associated DNA methylation abnormalities can be detected in different tissues 40. One question is: How much do methylation patterns change in different tissues? There are tissue-specific variations in DNA methylation patterns 33; 41, so we sought to answer the question as to whether changes in lymphocyte methylation patterns mirror changes in the brain. We correlated promoter CpG sites in the lymphocytes with those in brain in-silico, using a web-based BECon tool 42. We found that 6 out of 20 promoter CpG sites were correlated in the two tissues (correlation >0.3) in at least one of the three examined brain regions (Table 2), indicating conservation of epigenetic function. These results suggest there is a proportion of CpGs for which methylation estimations in blood are representative of methylation in brain and BRD2 promoter methylation in the blood could be used as a surrogate for brain tissue.
Table 2.
Blood-brain correlations of the promoter CpG sites
| Probe ID | Correlation coefficient of DNA methylation between blood and brain regions | Associated brain regions |
|---|---|---|
| cg08491668 | 0.3 | BA7 |
| cg01641778 | 0.41; 0.58 | BA10; BA7 |
| cg13224077 | 0.52 | BA10 |
| cg20471890 | 0.39 | BA7 |
| cg01393792 | 0.64 | BA7 |
| cg18610053 | 0.4 | BA10 |
Abbreviations: BA7, Brodmann Area 7 (Parietal cortex)
BA10, Brodmann Area 10(Anterior prefrontal cortex)
6. Conclusions
The results of the present study show evidence that BRD2 promoter methylation variation is a cause of JME in Caucasians, and perhaps in other ethnicities. Identification of the variants that affect BRD2 production might be used to predict JME susceptibility, as well as improving the ability to diagnose JME.
KEY POINT BOX.
DNA methylation status at the BRD2 promoter, as well as global methylation, was analyzed in lymphoblastoid cells from JME patients, and from unaffected family members, of JME families of both Caucasian and non-Caucasian origin, and also from non-JME GGE patients.
JME segregates together with BRD2 promoter methylation in Caucasians.
Non-JME GGE and non-Caucasian JME families do not show such segregation.
Global methylation levels do not differ in JME patients compared to unaffected family members, showing that BRD2 promoter methylation is specifically related to JME expression.
Acknowledgments
This work is supported by Nationwide Children’s Hospital, Columbus, OH.
Footnotes
DISCLOSURE OF CONFLICTS OF INTERESTS
The authors declare no competing interests. We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
BIBLIOGRAPHY
- 1.Camfield CS, Striano P, Camfield PR. Epidemiology of juvenile myoclonic epilepsy. Epilepsy Behav. 2013;28(Suppl 1):S15–17. doi: 10.1016/j.yebeh.2012.06.024. [DOI] [PubMed] [Google Scholar]
- 2.Janz D. The idiopathic generalized epilepsies of adolescence with childhood and juvenile age of onset. Epilepsia. 1997;38:4–11. doi: 10.1111/j.1528-1157.1997.tb01073.x. [DOI] [PubMed] [Google Scholar]
- 3.Kasteleijn-Nolst Trenite DG, Schmitz B, Janz D, et al. Consensus on diagnosis and management of JME: From founder’s observations to current trends. Epilepsy Behav. 2013;28(Suppl 1):S87–90. doi: 10.1016/j.yebeh.2012.11.051. [DOI] [PubMed] [Google Scholar]
- 4.Pavlovic M, Jovic N, Pekmezovic T. Antiepileptic drugs withdrawal in patients with idiopathic generalized epilepsy. Seizure. 2011;20:520–525. doi: 10.1016/j.seizure.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Greenberg DA, Delgado-Escueta AV, Maldonado HM, et al. Segregation analysis of juvenile myoclonic epilepsy. Genet Epidemiol. 1988;5:81–94. doi: 10.1002/gepi.1370050204. [DOI] [PubMed] [Google Scholar]
- 6.Pal DK, Evgrafov OV, Tabares P, et al. BRD2 (RING3) is a probable major susceptibility gene for common juvenile myoclonic epilepsy. Am J Hum Genet. 2003;73:261–270. doi: 10.1086/377006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavalleri GL, Walley NM, Soranzo N, et al. A multicenter study of BRD2 as a risk factor for juvenile myoclonic epilepsy. Epilepsia. 2007;48:706–712. doi: 10.1111/j.1528-1167.2007.00977.x. [DOI] [PubMed] [Google Scholar]
- 8.Greenberg DA, Durner M, Keddache M, et al. Reproducibility and complications in gene searches: linkage on chromosome 6, heterogeneity, association, and maternal inheritance in juvenile myoclonic epilepsy. Am J Hum Genet. 2000;66:508–516. doi: 10.1086/302763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weissbecker KA, Durner M, Janz D, et al. Confirmation of linkage between juvenile myoclonic epilepsy locus and the HLA region of chromosome 6. Am J Med Genet. 1991;38:32–36. doi: 10.1002/ajmg.1320380109. [DOI] [PubMed] [Google Scholar]
- 10.Sander T, Bockenkamp B, Hildmann T, et al. Refined mapping of the epilepsy susceptibility locus EJM1 on chromosome 6. Neurology. 1997;49:842–847. doi: 10.1212/wnl.49.3.842. [DOI] [PubMed] [Google Scholar]
- 11.Durner M, Sander T, Greenberg DA, et al. Localization of idiopathic generalized epilepsy on chromosome 6p in families of juvenile myoclonic epilepsy patients. Neurology. 1991;41:1651–1655. doi: 10.1212/wnl.41.10.1651. [DOI] [PubMed] [Google Scholar]
- 12.Tauer U, Lorenz S, Lenzen KP, et al. Genetic dissection of photosensitivity and its relation to idiopathic generalized epilepsy. Ann Neurol. 2005;57:866–873. doi: 10.1002/ana.20500. [DOI] [PubMed] [Google Scholar]
- 13.Lorenz S, Taylor KP, Gehrmann A, et al. Association of BRD2 polymorphisms with photoparoxysmal response. Neurosci Lett. 2006;400:135–139. doi: 10.1016/j.neulet.2006.02.026. [DOI] [PubMed] [Google Scholar]
- 14.Santos BPD, Marinho CRM, Marques T, et al. Genetic susceptibility in Juvenile Myoclonic Epilepsy: Systematic review of genetic association studies. PLoS One. 2017;12:e0179629. doi: 10.1371/journal.pone.0179629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Velisek L, Shang E, Veliskova J, et al. GABAergic neuron deficit as an idiopathic generalized epilepsy mechanism: the role of BRD2 haploinsufficiency in juvenile myoclonic epilepsy. PLoS One. 2011;6:e23656. doi: 10.1371/journal.pone.0023656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cholewa-Waclaw J, Bird A, von Schimmelmann M, et al. The Role of Epigenetic Mechanisms in the Regulation of Gene Expression in the Nervous System. J Neurosci. 2016;36:11427–11434. doi: 10.1523/JNEUROSCI.2492-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobow K, Blumcke I. The emerging role of DNA methylation in epileptogenesis. Epilepsia. 2012;53(Suppl 9):11–20. doi: 10.1111/epi.12031. [DOI] [PubMed] [Google Scholar]
- 18.Sun R, Wu Y, Wang Y, et al. DNA methylation regulates bromodomain-containing protein 2 expression during adipocyte differentiation. Mol Cell Biochem. 2015;402:23–31. doi: 10.1007/s11010-014-2310-1. [DOI] [PubMed] [Google Scholar]
- 19.Hastie T, Friedman JH, Tibshirani R. The elements of statistical learning : data mining, inference, and prediction. 2nd ed. New York: Springer-Verlag; 2009. [Google Scholar]
- 20.Greenberg DA, Stewart WC. How should we be searching for genes for common epilepsy? A critique and a prescription. Epilepsia. 2012;53(Suppl 4):72–80. doi: 10.1111/j.1528-1167.2012.03616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greenberg DA, Delgado-Escueta AV. The chromosome 6p epilepsy locus: exploring mode of inheritance and heterogeneity through linkage analysis. Epilepsia. 1993;34(Suppl 3):S12–18. doi: 10.1111/j.1528-1167.1993.tb06255.x. [DOI] [PubMed] [Google Scholar]
- 22.Layouni S, Buresi C, Thomas P, et al. BRD2 and TAP-1 genes and juvenile myoclonic epilepsy. Neurol Sci. 2010;31:53–56. doi: 10.1007/s10072-009-0190-z. [DOI] [PubMed] [Google Scholar]
- 23.Greenberg DA, Stewart WL. Remind me again what disease we are studying? A population genetics, genetic analysis, and real data perspective on why progress on identifying genetic influences on common epilepsies has been so slow. Prog Brain Res. 2014;213:199–221. doi: 10.1016/B978-0-444-63326-2.00011-9. [DOI] [PubMed] [Google Scholar]
- 24.Greenberg DA, Delgado-Escueta AV, Widelitz H, et al. Juvenile myoclonic epilepsy (JME) may be linked to the BF and HLA loci on human chromosome 6. Am J Med Genet. 1988;31:185–192. doi: 10.1002/ajmg.1320310125. [DOI] [PubMed] [Google Scholar]
- 25.Shang E, Wang X, Wen D, et al. Double bromodomain-containing gene Brd2 is essential for embryonic development in mouse. Dev Dyn. 2009;238:908–917. doi: 10.1002/dvdy.21911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gyuris A, Donovan DJ, Seymour KA, et al. The chromatin-targeting protein Brd2 is required for neural tube closure and embryogenesis. Biochim Biophys Acta. 2009;1789:413–421. doi: 10.1016/j.bbagrm.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walsh CP, Bestor TH. Cytosine methylation and mammalian development. Genes Dev. 1999;13:26–34. doi: 10.1101/gad.13.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gutierrez-Arcelus M, Lappalainen T, Montgomery SB, et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. Elife. 2013;2:e00523. doi: 10.7554/eLife.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weber M, Hellmann I, Stadler MB, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 30.Kobow K, Jeske I, Hildebrandt M, et al. Increased reelin promoter methylation is associated with granule cell dispersion in human temporal lobe epilepsy. J Neuropathol Exp Neurol. 2009;68:356–64. doi: 10.1097/NEN.0b013e31819ba737. [DOI] [PubMed] [Google Scholar]
- 31.Belhedi N, Perroud N, Karege F, et al. Increased CPA6 promoter methylation in focal epilepsy and in febrile seizures. Epilepsy Res. 2014;108:144–8. doi: 10.1016/j.eplepsyres.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 32.Garriga-Canut M, Schoenike B, Qazi R, et al. 2-Deoxy-D-glucose reduces epilepsy progression by NRSF-CtBP-dependent metabolic regulation of chromatin structure. Nat Neurosci. 2006;9:1382–7. doi: 10.1038/nn1791. [DOI] [PubMed] [Google Scholar]
- 33.Pal DK, Durner M, Klotz I, et al. Complex inheritance and parent-of-origin effect in juvenile myoclonic epilepsy. Brain Dev. 2006;28:92–98. doi: 10.1016/j.braindev.2005.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hannon E, Spiers H, Viana J, et al. Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat Neurosci. 2016;19:48–54. doi: 10.1038/nn.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fraser HB, Lam LL, Neumann SM, et al. Population-specificity of human DNA methylation. Genome Biol. 2012;13:R8. doi: 10.1186/gb-2012-13-2-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olden K, Lin YS, Gruber D, et al. Epigenome: biosensor of cumulative exposure to chemical and nonchemical stressors related to environmental justice. Am J Public Health. 2014;104:1816–1821. doi: 10.2105/AJPH.2014.302130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mantoan L, Walker M. Treatment options in juvenile myoclonic epilepsy. Curr Treat Options Neurol. 2011;13:355–370. doi: 10.1007/s11940-011-0131-z. [DOI] [PubMed] [Google Scholar]
- 38.Milutinovic S, D’Alessio AC, Detich N, et al. Valproate induces widespread epigenetic reprogramming which involves demethylation of specific genes. Carcinogenesis. 2007;28:560–571. doi: 10.1093/carcin/bgl167. [DOI] [PubMed] [Google Scholar]
- 39.Hnilicova J, Hozeifi S, Stejskalova E, et al. The C-terminal domain of Brd2 is important for chromatin interaction and regulation of transcription and alternative splicing. Mol Biol Cell. 2013;24:3557–3568. doi: 10.1091/mbc.E13-06-0303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 41.Dempster EL, Pidsley R, Schalkwyk LC, et al. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet. 2011;20:4786–4796. doi: 10.1093/hmg/ddr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davies MN, Volta M, Pidsley R, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13:R43. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edgar RD, Jones MJ, Meaney MJ, et al. BECon: a tool for interpreting DNA methylation findings from blood in the context of brain. Transl Psychiatry. 2017;7:e1187. doi: 10.1038/tp.2017.171. [DOI] [PMC free article] [PubMed] [Google Scholar]