Abstract
To understand how genomic heterogeneity of glioblastoma contributes to the poor response to therapy characteristic of this disease, we performed DNA and RNA sequencing on GBM tumor samples and the neurospheres and orthotopic xenograft models derived from them. We used the resulting data set to show that somatic driver alterations including single nucleotide variants, focal DNA alterations, and oncogene amplification on extrachromosomal DNA (ecDNA) elements were in majority propagated from tumor to model systems. In several instances, ecDNAs and chromosomal alterations demonstrated divergent inheritance patterns and clonal selection dynamics during cell culture and xenografting. We infer that ecDNA inherited unevenly between offspring cells, a characteristic that affects the oncogenic potential of cells with more or fewer ecDNAs. Longitudinal patient tumor profiling found that oncogenic ecDNAs are frequently retained throughout the course of disease. Our analysis shows that extrachromosomal elements allow rapid increase of genomic heterogeneity during glioblastoma evolution, independent of chromosomal DNA alterations.
Keywords: glioblastoma, double minute, extrachromosomal DNA, tumor evolution
INTRODUCTION
Cancer genomes are subject to continuous mutagenic processes in combination with an insufficient DNA damage repair 1. Somatic genomic variants that are acquired prior to and throughout tumorigenesis may provide cancer cells with a competitive advantage over their neighboring cells in the context of a nutrition- and oxygen-poor microenvironment, resulting in increased survival and/or proliferation rates 2. The Darwinian evolutionary process results in intratumoral heterogeneity in which single cancer-cell-derived tumor subclones are characterized by unique somatic alterations 3. Chemotherapy and ionizing radiation may enhance intratumoral evolution by eliminating cells lacking the ability to deal with increased levels of genotoxic stress, while targeted therapy may favor subclones in which the targeted vulnerability is absent 4,5. Increased clonal heterogeneity has been associated with tumor progression and mortality 6. Computational methods that analyze the allelic fraction of somatic variants identified from high throughput sequencing data sets are able to infer clonal population structures and provide insights into the level of intratumoral clonal variance 7.
Glioblastoma (GBM), a WHO grade IV astrocytoma, is the most prevalent and aggressive primary central nervous system tumor. GBM is characterized by poor response to standard post-resection radiation and cytotoxic therapy, resulting in dismal prognosis with a 2 year survival rate around 15% 8. The genomic and transcriptomic landscape of GBM has been extensively described 9–11. Intratumoral heterogeneity in GBM has been well characterized, in particular with respect to somatic alterations affecting receptor tyrosine kinases 12–14. To evaluate how genomically heterogeneous tumor cell populations are affected by selective pressures arising from the transitions from tumor to culture to xenograft, we performed a comprehensive genomic and transcriptomic analysis of thirteen GBMs, the glioma-neurosphere forming cultures (GSC) derived from them, and orthotopic xenograft models (PDX) established from early passage neurospheres. Our results highlight the evolutionary process of GBM cells, placing emphasis on the diverging dynamics of chromosomal DNA alterations and extrachromosomally amplified DNA elements in tumor evolution.
RESULTS
Genomic profiling of glioblastoma, derived neurosphere and PDX samples
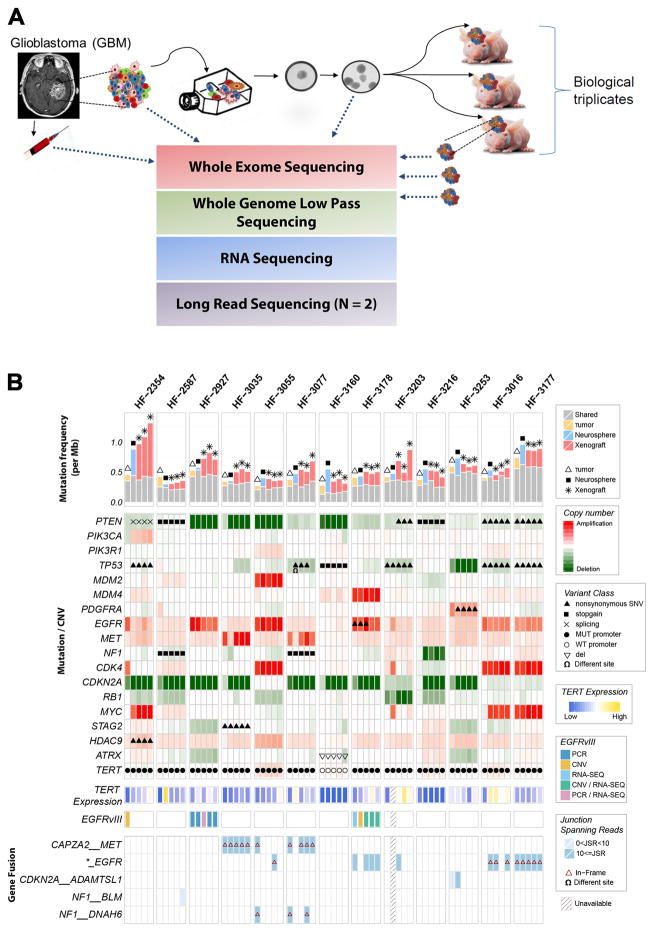
We established neurosphere cultures from 12 newly diagnosed and one matched recurrent GBM (Supplementary Table 1). Neurosphere cultures between 7 and 18 passages were used for molecular profiling and engrafting orthotopically into nude mice. The sample cohort included one pair of primary (HF3016) and matching recurrent (HF3177) GBM. A schematic overview of our study design is presented in Fig. 1a. To determine whether model systems capture the somatic alterations that are thought to drive gliomagenesis, and whether there is selection for specific driver genes, we performed whole genome sequencing at a median depth of 6.5× to determine genome wide DNA copy number as well as exome sequencing on all samples. DNA copy number was generally highly preserved between tumor and derived model systems (Supplementary Fig. 1). Whole chromosome 7 gain and chromosome 10 loss were retained in model systems when detected in the tumor, consistent with their proposed role as canonical GBM lesions that occur amongst the earliest events in gliomagenesis 15. The global DNA copy number resemblance between xenografts and the GBM from which they were derived confirms that PDXs recapitulate the majority of molecular properties found in the original tumor.
Figure 1. Comprehensive comparison of GBM, derived neurospheres and PDX models.
Genomic and transcriptomic characterization were performed on 13 patient tumors, their derivative neurosperes and xenograft models. Long read PacBio sequencing was performed on two xenograft tumors. A. Schematic study overview. B. Somatic driver alterations compared between GBM tumors and derivative model systems.
We compared mutation and DNA copy number status of genes previously found to be significantly mutated, gained, or lost in GBM 9,11. We found that 100% of homozygous deletions and somatic single nucleotide variants (sSNVs) affecting GBM driver genes in tumor samples were propagated to the neurospheres and xenografts, including non-coding variants in the TERT promoter (Fig. 1b). Genomic amplifications showed greater heterogeneity. In two cases, MYC amplification was not detected in the parental tumor, but presented in the derivative neurospheres and maintained in xenografts, consistent with its role in glioma stem cell maintenance 16,17. Other genes showing variable representation across tumor and model systems included MET in HF3035 and HF3077, and EGFR and PIK3CA in HF2354. The HF2354 derived model systems were considerably less similar compared to the primary tumor than other cases which coincided with HF2354 being the only case subjected to neoadjuvant carmustine treatment. Whole chromosome gains of chromosome 1, 14 and 21, and one copy loss of chromosome 3, 8, 13, 15 and 18 were acquired in the neurosphere culture and propagated to the xenograft models (Supplementary Fig. 1). At the gene level, this resulted in newly detected mutations in PTEN and TP53, focal amplification of MYC (also in HF3016), and absence of CDK4 and EGFR amplification in the neurosphere and xenografts relative to the tumor sample (Fig. 1b).
Extrachromosomal elements are frequently found in glioblastoma
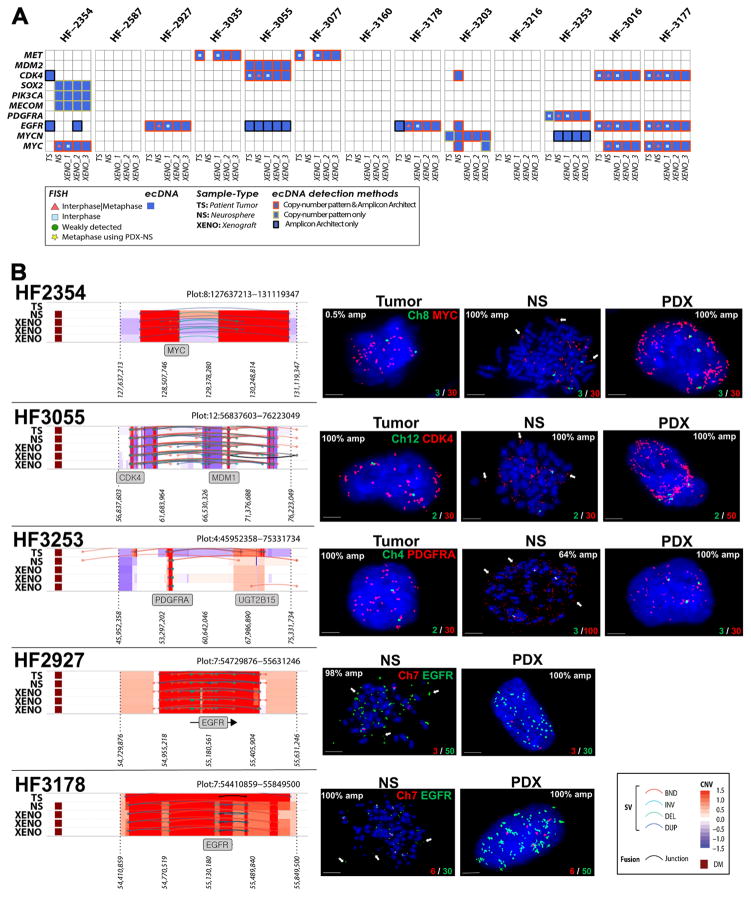
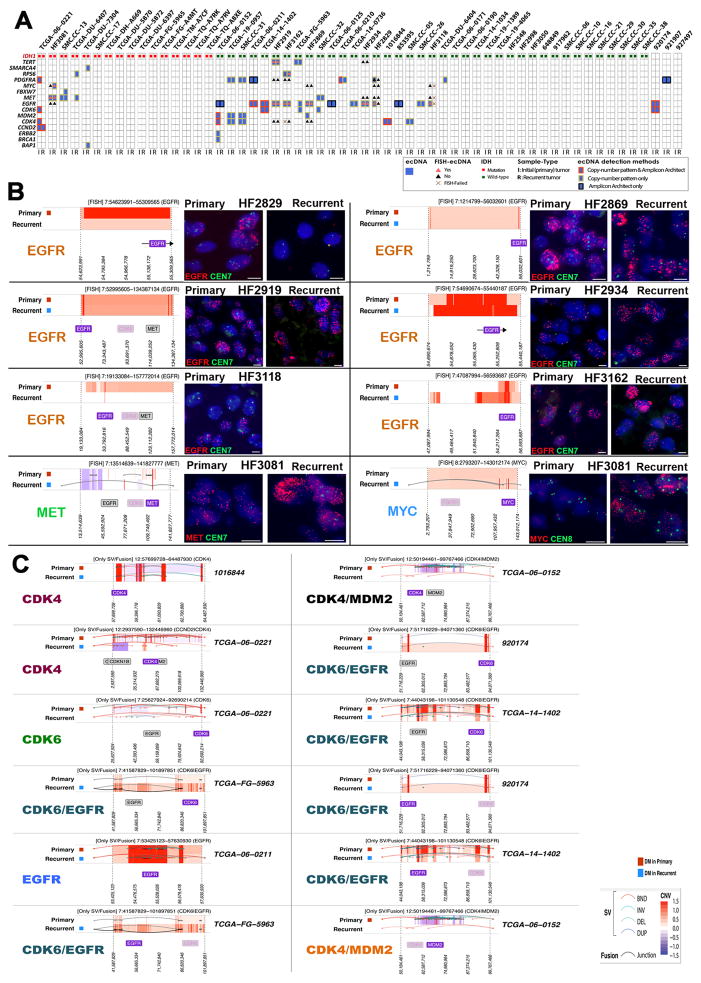
Cytogeneticists have since long recognized that DNA in cancer can be amplified as part of chromosomal homogenously staining regions (HSR) and as extrachromomal minute bodies 18. An early example of the importance of extrachromosomal DNA elements (ecDNA) in cancer was the discovery of double minutes carrying the oncogene N-MYC in neuroblastoma 19. A recent survey of a compendium of cancer cells and cell lines highlighted the frequent presence of ecDNA in glioblastoma, among other cancer types, 20, confirming previous studies 21–23. We searched our data set for complex patterns of DNA copy number amplification and rearrangement that are suggestive of ecDNA elements (Supplementary Fig. 2). In addition, we ran the AmpliconArchitect algorithm which detects ecDNAs in an unsupervised manner on the basis of sequencing reads connecting amplified DNA segments 20. On the basis of the union of AmpliconArchitect predictions and DNA copy number patterns we predicted 93 ecDNAs originating from 79 unique genomic loci which were distributed over 49 of the thirteen patient tumors and their derived model systems (Supplementary Table 2). The predicted ecDNA elements contained oncogenes including MYC, MYCN, EGFR, PDGFRA, MET, the MECOM/PIK3CA/SOX2 gene cluster and the CDK4/MDM2 gene cluster. In total, 22 of the 25 unique oncogene carrying ecDNAs were detected in more than one sample, i.e. in neurospheres and matching PDX or in tumor sample and matching neurosphere or PDX (Fig. 2a). We performed interphase FISH on tumor samples and PDX, and metaphase FISH on neurospheres to validate 34 predicted ecDNA amplifications, including of EGFR (HF2927, HF3178, HF3016 and HF3177), MYC (HF2354, HF3016 and HF3177), CDK4 (HF3055, HF3016 and HF3177), MET (HF3035 and HF3077), MDM2 (HF3055) and PDGFRA (HF3253). In all interphase FISH experiments we observed a highly variable number of fluorescent signals per nucleus, ranging from two to 100 (Fig. 2b, Supplementary Table 3). This heterogeneity was strongly suggestive of differences in the number DNA copies of the targeted gene per cell and thereby of an extrachromosomal DNA amplification. Metaphase FISH on neurosphere cells validated the extrachromosomal status in all cases (Fig. 2b). Our analysis showed that oncogene amplification frequently resided on extrachromosomal DNA elements.
Figure 2. ecDNA in hGBM samples and FISH validation.
A. Heatmap of samples versus driver genes predicted to reside on extrachromosomal DNA elements that were detected with either copy number based or Amplicon Architect methods. Only ecDNAs with markers were validated using FISH. B. Left panel: DNA copy number and genomic rearrangements at ecDNA loci that were predicted with the copy number based approach. Right panel: Representative FISH images showing amplification of MYC, CDK4, PDGFRA in tumor (T), neurospheres (NS, metaphase spread) and PDXs (red) and control chromosomal probes (green). EGFR amplification in neurospheres and PDX (green) and Chr7 control are shown. Arrows in metaphase FISH images mark extrachromosomally DNA elements. A minimum of 100 nuclei for each tumor, NS and two PDX biological replicates were counted. The percentage of nuclei presenting each oncogene amplification is shown. Scale bars, 3 μm.
Extrachromosomal MET DNA elements mark a distinct tumor subclone
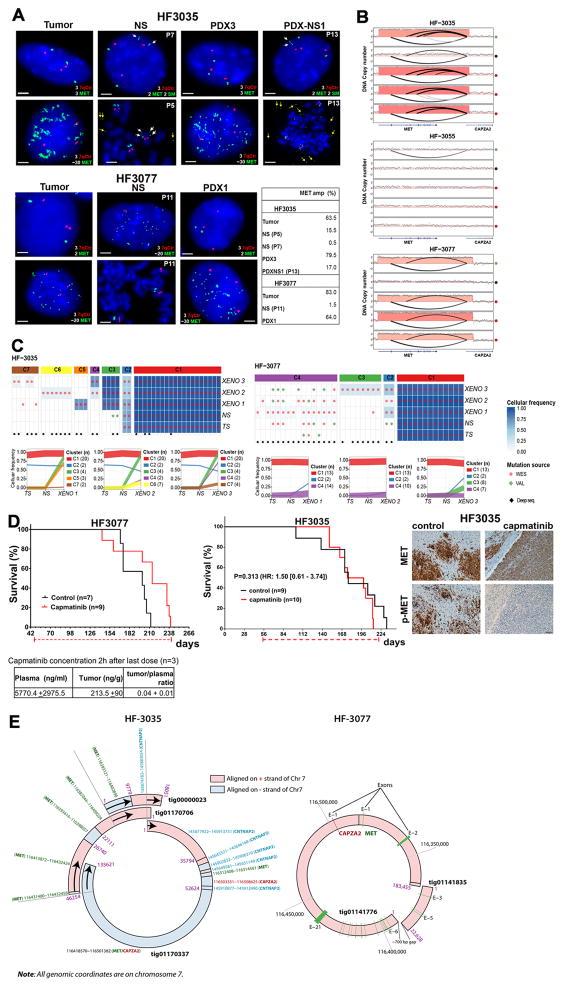
Among the identified oncogene carrying ecDNA elements, two cases of extrachromosomal MET amplification stood out due to their variable presence across the parental tumor (high frequency), neurosphere (low frequency) and xenograft triplicates (high frequency) (Fig. 3a). In both cases, the MET amplification associated with a transcript fusion with neighboring gene CAPZA2 (Fig. 3b, Supplementary Fig. 3a). Additional details on the CAPZA2-MET fusions are provided in the Supplementary Note. The pattern of undetectable and re-appearing MET rearrangements may result from clonal selection of glioblastoma cells with a competitive advantage for proliferation in vivo. This hypothesis is strengthened by the observation that the breakpoints of the lesions were identical across samples from the same parental origin (Supplementary Fig. 3b). MET is a growth factor responsive cell surface receptor tyrosine kinase and may provide context dependent proliferative signals 24. We reasoned that evolutionary patterns resulting in such dominant clonal selection would likely be replicated by sSNVs tracing the cells carrying the MET amplicon. To evaluate clonal selection patterns, we determined variant allele fractions of all sSNVs identified across HF3035 and HF3077 samples. To increase our sensitivity to detect mutations present in small numbers of cells, we corroborated the exome sequencing data using high coverage (>1,400x) targeted sequencing. All mutations detected in the HF3035 GBM were recovered in the neurosphere and xenografts. The mutational profile of HF3035 suggested that a subclone developed in the xenografts that was not present in parental GBM and neurosphere and revealed a subclone that was present at similar frequencies in all samples (Fig. 3c). Only a single and very low frequency LAMB1 mutation (variant allele fraction in tumor = 0.003) present in the HF3077 primary tumor, but not detected in its derived neurosphere, resurfaced in one of three xenografts with a 0.04 variant allele fraction. A low frequency subclone (C2) developed in the neurosphere which was transmitted to xenografts (Fig. 3c). Subclonal heterogeneity as recovered by the mutation profiles thus suggested a very different clonal selection trend compared to to the disappearing and resurfacing MET amplifications and associated transcript fusions. EcDNAs are thought to inherit through random distribution over the two daughter cells25, possibly through a binomial model26, but much is unknown with respect to the propagation of ecDNA through cancer cell populations. The disjointed propagation of chromosomal SNVs and extrachromosomal MET ecDNAs indicate that they are marking different tumor subclones and suggest alternative modes of tumor evolution. While sSNVs are copied to daughter cells during mitosis such that both cells inherit the full spectrum of chromosomal alterations present in the parental cell, ecDNA elements likely randomly segregate and end up in the daughter cells in uneven numbers.
Figure 3. Extrachromosomal MET DNA.
A. Representative FISH images for MET (green) and chromosome 7 control probes (7qCtr, red) labeling of HF3035 and HF3077 tumor, neurosphere (NS), xenografts (PDX), and NS established from HF3035 xenograft tumors (PDX-NS1). Passage numbers are indicated for neurosphere cultures. White arrows point to 2 fragmented MET signals in one chromosome in HF3035 samples (2SM). Yellow arrows point to extrachromosomal MET in metaphase nuclei of HF3035 neurospheres. The percentage of nuclei presenting MET amplification for each sample is shown. Scale bars, 2 μm. B. DNA copy number and chromosomal rearrangement of the 7q31 locus in three sets of GBM tumors and derivate models. C. Top panel: Coverage-controlled sSNVs detected using exome and deep validation sequencing Color reflects cellular frequency estimates. Bottom panel: Clonal tracing from HF3035 and HF3077 parent tumor to neurospheres and xenografts. Each line represents a group of mutations computationally inferred to reflect a subclone. D. Top panel: Treatment with single agent capmatinib (30 mg/kg, daily oral doses) increases survival of HF3077 PDX, but not of HF3035. Kaplan-Meier survival curves were compared by log-rank (Mantel-Cox) test, significance set at P<0.05 (*), HR [95% CI], treatment schedule (doted red line) and number of mice in each arm (n) are shown. Representative images for MET and p-MET immunohistochemistry of 5 control and 5 capmatinib-treated HF3035 PDXs show complete inhibition of p-MET and unaltered MET expression in treated tumors. Scale, 40 μm. Bottom panel: Capmatinib concentration in the plasma and tumor tissue collected 2h after the last dose was determined by LC-MS/MS for HF3077 PDX, results are expressed as mean ± SE for 3 samples E. Double minute structures containing the chromosome 7q31 locus including the MET and CAPZA2 genes in HF3035 and HF3077 xenografts, predicted from long read sequencing.
MET expressing cells exhibited MET activation and were selected early during tumor formation in the orthotopic xenografts (Supplementary Fig. 3c), suggesting that MET activity was driving selection for MET amplified cells in vivo. Treatment of HF3077 PDX with ATP-competitive MET inhibitor capmatinib (INCB28060) 27 at a daily oral dose of 30 mg/kg showed a significant survival benefit, despite the relatively low concentration of drug in the brain tumor as assessed by LC-MS/MS (Fig. 3d). In contrast, capmatinib treatment of HF3035 PDX did not increase survival nor decrease MET expression but resulted in decrease of phospho-MET in treated tumors. This may reflect MET functions that are independent of the kinase activity in these tumors, as previously proposed 28,29. These results demonstrate that targeting MET in GBM harboring MET ecDNA amplification has therapeutic potential, but further work is needed to establish the factors that determine sensitivity of MET-amplified tumors to single agent ATP-competitive inhibitor treatment.. Comparable to the orthotopic xenografts, subcutaneous PDX tumors formed from implant of HF3035 neurosphere cells were dominated by MET-amplified cells accompanied by robust MET expression (Supplementary Fig. 3c). The increase in the frequency of MET-amplification in HF3035 cells in vivo are therefore not dependent on factors uniquely present in the brain microenvironment.
Different genetic origins for ecDNA have been postulated, with evidence for post-replicative excision of chromosomal fragments and non-homologous end joining 30. Interphase FISH analysis in the parental HF3077 tumor identified a small percentage of nuclei with 3 copies of chromosome 7 but only 2 copies of MET. The frequency of cells with one deleted copy of MET in Ch 7 increased significantly in HF3077 neurospheres and decreased in the xenografts (Supplementary Table 3). The observed gene deletion in one copy of chromosome 7 is suggestive of the post-replication segregation-based model of double minute formation30. To precisely define the genomic contents and structure of the predicted ecDNAs, we generated long read (Pacific Biosciences) DNA sequencing from a single xenograft of each HF3035 and HF3077, and performed de novo assembly. In HF3035, seven assembled contigs (range: 6,466 ~ 135,621 bp) were identified to have sequence fragments (at least 1,000 bp long) aligned on the MET-CAPZA2 region of hg19 chromosome 7. Interestingly, analysis of the aligned sequence fragments from the seven contigs revealed a more complex structural rearrangement than expected from the analysis of short read sequencing data. For example, the 135kb tig01170337 contig consisted of 8 sequence framents that were nonlinearly aligned on alternating strands of the MET-CAPZA2 and CNTNAP2 regions. Other contigs such as tig01170699, tig01170325, and tig00000023 also showed nonlinear alignment, suggesting that these contigs resulted from chromosomal structural variations. We performed pairwise sequence comparison of the contigs to search for sequence fragments (at least 5,000 bp long) shared among them, and we found four contigs each of which shared sequence fragments with one of the contigs. Interestingly, three of them could be connected in a circular form using the shared sequence fragments (Fig. 3e; Supplementary Fig. 4a), revealing a circular structure that may represent the full ecDNA. In HF3077, only two contigs were detected to be aligned on the MET-CAPZA2 region of hg19 chromosome 7 (Fig. 3e; Supplementary Fig. 4a). Presence of only two aligned contigs in HF3077 might be related to the lower sequence coverage of the ecDNA structure, compared to HF3035 (34× vs 405×, respectively) (Supplementary Fig. 4b). The longest contig, tig01141776 (183,455 bp long), consisted of two segment framents that were nonlinearly aligned over exon 1 of CAPZA2 and all except exons 3–5 of MET, suggesting that it resulted from structural variations. The second short contig, tig01141835 (22,628 bp long), was aligned as a whole over exon 3–5 of MET. Interestingly, connecting the two contigs created a circular DNA segment. Through analysis of PacBio sequencing, we were able to detect and reconstruct the predicted extrachromosomal element structures.
Multiple ecDNA elements are longitudinally preserved in a patient GBM and its derivative model systems
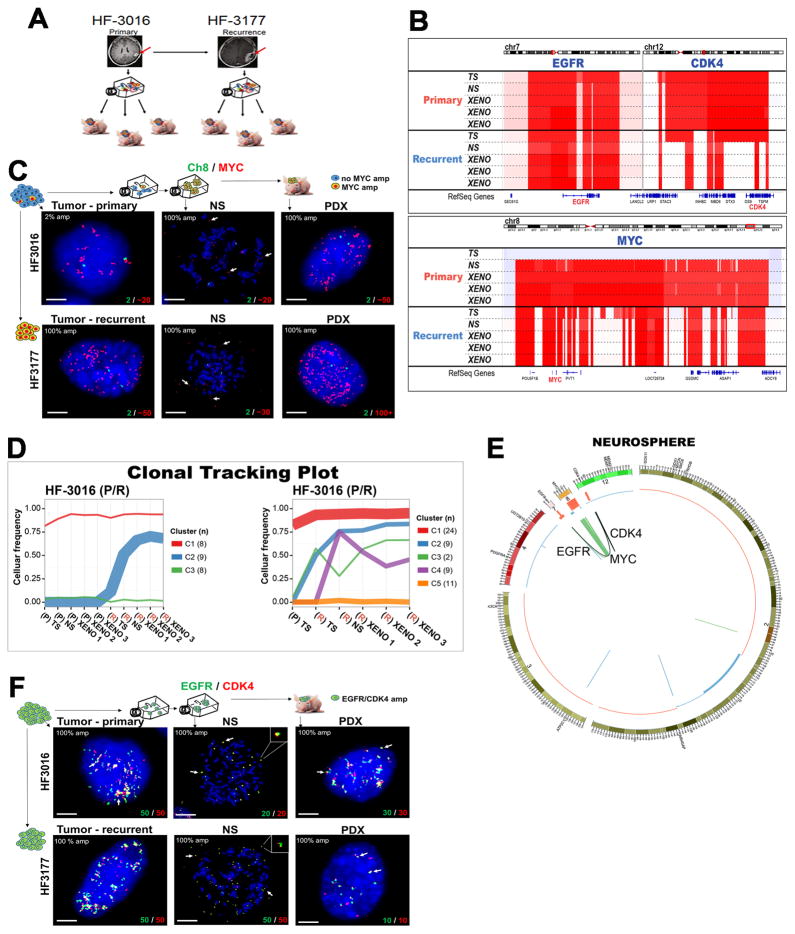
Analysis of a pair of primary and recurrent GBM included in our cohort, respectively HF3016 and HF3177, showed that chromosomal and extrachromosomal elements jointly orchestrated complex evolutionary dynamics (Fig. 4a). Primary and recurrent tumor were globally very similar (Fig. 1b, Supplementary Fig. 1). While the HF3016 primary tumor showed diploid MYC DNA copy numbers, a focal MYC amplification was detected in the neurosphere and PDXs derived from this tumor, and the same MYC amplification was identified in all samples from the recurrent tumor (Fig. 4b). Interestingly, FISH analysis showed that MYC amplification was present in low frequency (2%) in the initial HF3016 tumor, and was enriched to 100% of nuclei in the neurospheres and in the recurrent tumor (Fig. 4c, Supplementary Table 3). Metaphase FISH analysis confirmed extrachromosomal MYC amplification in both HF3016 and HF3177 neurospheres (Fig. 4c). The sSNV based clonal tracking plots for the paired patient samples identified two subclones in the HF3177 recurrence (Fig. 4d) that were not detected in the HF3016 neurosphere/PDX models, suggesting that these were independent of the MYC ecDNA element. Of note, a 0.5% cell frequency amplification was also detected in the parental tumor sample of HF2354, which increased to high levels in the derived neurosphere. DNA copy number analysis detected parallel EGFR and CDK4 amplifications in the HF3016 primary GBM that were retained in HF3177 GBM recurrence as well as all model systems. Sequencing reads connecting the two amplifications and suggesting a complex structural variant were detected in the HF3016 neurosphere, the HF3016 PDXs, all HF3177 samples, but not the HF3016 primary GBM (Fig. 4e). Metaphase FISH on HF3016 neurosphere and HF3177 neurosphere confirmed that the CDK4 and EGFR amplifications were part of the same ecDNA element (Fig. 4f). The genomic and extrachromosomal characteristics of these two tumor samples, their derived neurosphere cultures and xenografts provide an example of how multiple ecDNA elements are able to be preserved during tumor progression while in parallel acquiring new tumor subclones marked by sets of chromosomal sSNVs.
Figure 4. Extrachromosomal DNA marks subclones driving tumor progression in patient tumors and derived model systems.
A. Establishing neurosphere cultures and PDX models from a paired primary/recurrent GBM. B. DNA copy number analysis shows co-amplification of EGFR (chr7)/CDK4 (chr 12) is detected in primary GBM HF3016 which is sustained in both neurosphere and xenografts derived from this primary tumor, as well as the recurrent GBM HF3177, and the neurosphere/xenografts thereof. The HF3016 primary tumor is not MYC amplified. The HF3016 neurosphere, as well as all HF3177 samples, show focal MYC amplification. C. Representative FISH images from 50 metaphase and 100 interphase nuclei for MYC (red) and Ch8 marker (green) show that a small fraction (2%) of the cells in HF3016 tumor presents MYC amplification, while 100% of nuclei in the remaining samples present MYC amplification, which is clearly extrachromosomal (white arrows) in the metaphase spreads (NS). D. Clonal tracing of a pair of primary-recurrent GBM, their matching neurospheres, and xenografts. Each line represents a group of mutations computationally inferred to reflect a subclone. E. Starting in the neurosphere of the primary tumor, a complex structural variant is identified that connects the CDK4 locus to the EGFR locus. The MYC locus is not part of this variant. The EGFR/CDK4 variant is detected in HF3016 PDXs as well as all HF3177 samples. F. EGFR (green) and CDK4 (red), detected by FISH, are amplified in 100% of nuclei for every sample from this patient, with identical copy numbers in each nucleus (bottom of the panels). Overlapping dots show that EGFR/CDK4 co-localize (white arrows) and metaphase FISH (NS) shows extrachromosomal co-amplification in the same double minute (inserts). Images are representative of 50 metaphase and 100 interphase nuclei per group. Scale bars, 3 μm.
Longitudinal maintenance of extrachromosomal DNA in patient tumors
Large, megabase sized extrachromosomal elements, typically described as double minutes, are frequently found in glioblastoma and can be identified using whole genome sequencing and DNA copy number data 21–23. To determine whether extrachromosomal DNA can survive therapeutical barriers, we evaluated the DNA copy number profiles of 58 matching pairs of primary and recurrent glioma for the presence of ecDNAs. Evidence supporting the presence of ecDNA was found in 33 primary and 33 recurrent tumors spanning 38 patients and of these, ecDNA elements targeting cancer driver genes 31 were predicted in 25 primary tumors (Fig. 5a; Supplementary Table 2). The most frequently targeted gene was EGFR which was identified in 14 primary tumors, in agreement with previous reports22. CDK4, PDGFRA were detected in six and seven primary tumors, respectively. Crossreferencing the list of ecDNA predictions with a manually curated list of oncogenes suggested that 60% of ecDNAs target an oncogene. Consistent with oncogene amplification frequencies, IDH wild type tumor contained relatively more ecDNAs than IDH mutants (median 2 respectively 1), but the fraction of samples with at least a single ecDNA was comparable (0.5 vs 0.6, IDH wild type and IDH mutant, respectively). We corroborated our computational predictions through interphase FISH analyses of 18 predicted ecDNAs and 30 non-altered loci across 7 primary/recurrent tumor pairs. Sixteen out of 17 genomic amplifications showed the highly variable number of DNA signals that is strongly suggestive of the extrachromosomal nature of the DNA locus (Fig. 5b, Supplementary Fig. 5a) whereas the 26 control DNA regions predicted to be non-amplified were confirmed as such (Supplementary Table 4). EGFR harboring ecDNA was preserved in the recurrent tumor in 11 out 13 pairs, half of which carried an EGFRvIII mutation, including the HF2934 recurrent tumor analyzed after treatment with EGFR inhibitor dacomitinib (Fig. 5b, Supplementary Table 4). One tumor lost EGFR ecDNA and vIII mutation upon recurrence (HF2829), after treatment with the standard of care (radiation and temozolomide). In one case MET ecDNA was present in the primary tumor and maintained in the recurrence, while MYC ecDNA emerged upon recurrence, similar to what we reported above for the HF3016/HF3177 pair. To corroborate 55 DNA copy number predicted ecDNAs, we analysed whole genome and RNA sequencing data, which identified sequencing reads connecting adjacent focally amplified DNA segments (Fig. 5c and Supplementary Fig. 5b) supporting the predictions. After disease recurrence, 23 of 25 tumors preserved at least one cancer driver ecDNA, supporting the notion that ecDNA can prevail following the selective pressure imposed by anti-cancer therapy. We did not detect any significant correlations between somatic mutations and the presence of ecDNA. We observed a significantly shorter time to second surgery (log rank test, p = 0.018) for patients whose primary tumor sample was predicted to carry at least one ecDNA, relative to patients with a primary tumor that contained no predicted ecDNAs (Supplementary Figure 5d). The presence of an IDH mutation in glioma associates with relatively favorable prognosis. The number of IDH mutant cases was evenly balanced between the ecDNA+ (19 of 62) and ecDNA- (13 of 35) groups, suggesting that the significant outcome difference is independent of ecDNA status. There was no significant difference in time to progression when performing the analysis for the IDH mutant (log rank test p-value 0.14) and IDH wild type (log rank test p-value 0.12) separately. This analysis was limited by the cohort size and the low sensitivity of the current technology in detecting ecDNA.
Figure 5. Copy number variant driver genes located on the potential double minute (DM) regions.
A. 66 tumors (33 P, 33 R) from 38 patients were predicted to contain at least one ecDNA that was detected with either copy number based or Amplicon Architect methods. Amongst these, 44 driver gene harboring ecDNAs were predicted in 25 primary tumors, of which 32 were also detected in the matching recurrent tumors. B. Left panel: DNA copy number and genomic rearrangements at predicted ecDNA loci that were predicted with the copy number based approach. Right panel: Representative FISH images in FFPE tissue sections showing amplification of EGFR, MET and MYC in (red) and control chromosomal probes (green). Fifty nuclei were examined per sample. Scale bars, 5 μM. C. DNA copy number based predictions of extrachromosomal DNA segments validated using whole genome or RNA sequencing.
DISCUSSION
Glioblastoma is a heterogeneous disease that is highly resistant to chemo- and radiotherapy. New modalities for treatment are urgently needed. Modeling of tumors through cell culture and orthotopic xenotransplantation are essential approaches for preclinical therapeutic target screening and validation, but in GBM have yet to result in novel treatments. To what extent these models truthfully recapitulate the parental tumor is a topic of active discussion. Here, we showed that neurosphere and orthotopic xenograft tumor models are genomically similar, capturing over 80% of all genomic alterations detected in the parental tumors.
EcDNA were discovered decades ago and have been incidently found to play important roles in tumorigenesis and gliomagenesis in particular 18,19,21–23,30. Only recently have we started to understand the magnitude and frequency of these somatic alterations, and their impact on tumor evolution20. Our results provide direct evidence that extrachromosomal amplification of oncogenic elements enhances genomic diversity during tumor evolution. We showed how ecDNA elements can mark major clonal expansions in otherwise stable genomic backgrounds and related ecDNA presence to tumor progression. Jointly, these findings change our views on the evolution of glioma, with potentially translation to other ecDNA carrying cancer types20. Little is known about the mechanism through which these elements arise and how they become fixed across a cancer cell population. Our analysis provides a comprehensive study of the fate of chromosomal SNVs and ecDNA oncogene amplifications in GBM in a panel of tumors and derivative models. We further demonstrated the widespread presence of ecDNA driven oncogene amplification through extensive FISH analysis on sets of paired primary and recurrent tumor samples. Focal gene amplifications have traditionally been recognized as homogeneously staining regions (HSR) and these may originate from chromosomal insertions of ecDNA 25. We did not observe HSR-like staining patterns for the amplified genes in the metaphase FISH images in this study. HSRs have been observed in glioblastomas 20 and are thought to be less frequent than ecDNA 32. We captured the early stages of MYC ecDNA expansion in the HF3016 and HF2354 tumors with 0.5–2% of cells presenting amplification (<30 copies/nucleus), with no evidence of chromosomal based gene amplification, while in all derived models, as well as the HF3016 recurrence (HF3177), the frequency of MYC amplification increased to 100% of cells with up to 100 copies/nucleus. These results are consistent with an origin through excision of a MYC containing chromosomal DNA segment and end-joining into a circular ecDNA, with subsequent amplification of the ecDNA30, followed by selection of MYC-amplified cells in vitro and in the recurrent tumor. Spindle assembly and chromosome segregation during mitosis lead to genetically identical daughter cells, containing similar sets of chromosomal sSNVs and DNA copy number alterations. Double minutes/ecDNAs are replicated during S-phase, but lack the centromeres that dictate the organization of the mitotic spindle, and as a result are randomly distributed across the daughter cells during mitosis. EcDNA elements are thus inherited in a radically different fashion than chromosomes. This divergence in inheritance mechanism may explain for example why the evolution of the MET event was not similarly captured by sSNVs (Fig. 6), and shows that extrachromosomal elements play a key role in increasing genomic diversity during tumor evolution. Previous studies have found that extrachromosomal bodies can provide a reservoir for therapeutically targetable genomic alterations 33. Targeted MET inhibition of MET amplified GBMs has shown clinical promise 34, although the variable responses to MET inhibition recorded in our data suggest that single MET inhibiting agent efficacy is influenced by other factors. Our observations extend recent findings that ecDNA are frequently detected in cancer 20,22 and demonstrate that detection of point mutations alone is insufficient to accurately delineate tumor evolutionary process. In analogy to the loss of ecDNA carrying MET following neurospheroid culturing, loss of EGFR amplification in traditional serum containing glioblastoma cultures has been observed 35,36. The loss of ecDNA EGFR and PDGFRA amplification have also been reported for neurosphere cultures grown in the presence of EGF and bFGF 23,37, but not consistently 14 suggesting that these oncogenes are sometimes but not always indispensable. Withdrawing EGF from the media was shown to promote maintenance of EGFR amplification 37. Preservation of EGFR and PDGFRA amplifications in glioblastoma tumors propagated in mouse subcutaneous xenograft tumors has been previously reported36,38. Our results show that ecDNA carrying amplification of MYC, CDK4, EGFR, and PDGFRA were maintained in neurosphere cultures supplemented with EGF and bFGF, at least up to passage 18. The ecDNA amplifications were subsequently maintained in the intracranial xenograft tumors originated by the neurosphere implants. Unlike the other ecDNA-amplified oncogenes, we observed that MET ecDNA amplification dramatically decrease in frequency in the neurosphere cultures and surprisingly re-emerged in high frequency after intracranial implants.
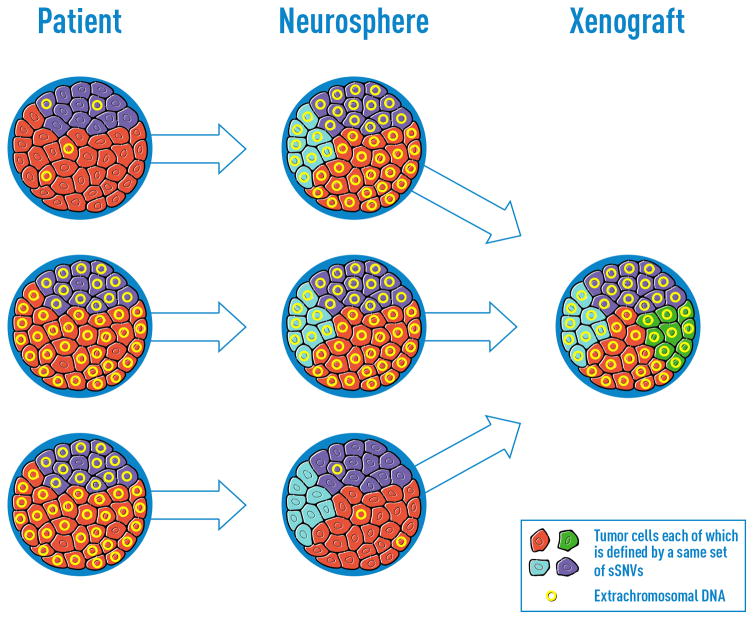
Figure 6. Schematic illustration of extrachromosomal DNA element contribution to clonal evolution in GBM patient derived models.
The proliferation patterns in GBM tumors and models in which ecDNAs provide a dominant evolutionary force.
Double minutes and other ecDNAs have been reported in 10–40% of GBM 21–23. These lesions involved frequently amplified oncogenes such as MYC, EGFR, PDGFRA and a region on chromosome 12p that includes CDK4 and MDM2. EcDNAs reported to date span up to several megabases in size, and some but not all ecDNAs can be recognized by an intermittent amplification-deletion DNA copy number pattern21,22. DNA copy number and short read sequencing technology are limited in their ability to sensitively and specifically detect ecDNAs in particular with respect to samples that additionally harhor overlapping chromosomal alterations and we therefore only have incomplete knowledge on the frequence and structure of extrachromosomal DNA elements 14,20–23. This is reflected by the incomplete overlap in ecDNA predictions from supervised review and the unsupervised AmpliconArchitect method. Long read technology such as used in the single molecule zero-mode waveguides approach from Pacific Biosciences or nanopore sequencing from Oxford Nanopore Technologies may offer advantages for ecDNA detection. Whether ecDNA size and structure affects the mechanism of tumorigenesis is unclear and is another reflection of our lack of knowledge of extrachromosomal DNA, in particular as an understudied domain in cancer. Our analysis emphasizes the importance of this genomic alteration mechanism for gliomagenesis. Future studies that specifically target the formation of episomal events may lead to therapies to prevent this process from happening. The models we described here may play a pivotal role in evaluating the potential of such approaches.
ONLINE METHODS
Tumor sample collection and cell culture
Resected brain tumor specimens were collected at Henry Ford Hospital (Detroit, MI) with written informed consent from patients, under a protocol approved by the Henry Ford Hospital Institutional Review Board, and graded pathologically according to the WHO criteria. This work was performed in compliance with all relevant ethical regulations for research using human specimens.
A portion of each tumor specimen was snap frozen and stored in liquid nitrogen. An adjacent portion was used for cell culture. Tumors are dissociated enzymatically and neurospheres enriched in cancer stem-like cells (CSC) were cultured, as described in detail 39,40. Neurosphere cultures were serially passaged in vitro. No mycoplasma contamination was identified in the subset of samples tested. Cells with passages between 7 and 18 were used for mouse implants and molecular analysis, except for those designed “high passage”, where passage 40 was used.
Patient derived xenografts (PDX)
Orthotopic xenografts
In compliance with all relevant ethical regulations for animal research under a protocol approved by the Henry Ford Hospital Institutional Animal Care and Use Committee (IACUC)GBM neurosphere cell suspensions were implanted into 8-week old female nude mice (NCRNU, Taconic Farms) as described41. A minimum of 8 mice were implanted with each neurosphere line. Animals were anesthetized with a mixture of ketamine and xylazine. Dissociated neurosphere cells (3×105) were injected using a Hamilton syringe at a defined intracranial location: AP+1.0, ML+2.5, DV-3.0. Animals were monitored daily by an observer blinded to the group allocation and sacrificed upon first signs of neurological deficit or weight loss greater than 20%. Brains were harvested, placed in a coronal matrix for 2 mm sections, with the first cut across the implant site. Brain sections were alternately frozen in dry ice and embedded in OCT for storage at −80°C, or formalin fixed and paraffin embedded (FFPE).
Subcutaneous xenografts
Dissociated neurosphere cells (1×106) were injected in the flank of nude mice. Animals were sacrificed and tumors excised when diameter reached 10 mm.
Drug Treatment
HF3077 PDXs were randomized to control and treatment groups. Mice were treated with capmatinib (purchased from Matrix Scientific Products (Columbia, SC)) suspensions in 0.5% methylcellulose/0.1% Tween 80 prepared every week and administered by oral gavage using a 20g ×1.5″ gavage needle (Cadence) at a dose of 30 mg/kg once a day (5 days/week) until the end of the study. Control animals received vehicle only mock gavage. Forty-five days after implant, animals were randomized to control or treatment groups. Each mouse was followed until death with no censoring and mean survival differences were estimated using a t-distribution to estimate 95% confidence intervals. With a sample size of 9 mice per group, a two-sided 95% confidence interval for the difference in mean survival would extend 0.92SD from the observed difference in mean survival, assuming the CI is based on large sample z statistic. Equivalently we expected 80% power to detect a difference in mean survival of 1.4SD, for the common standard deviation, when n=9 animals per group and alpha=0.05. Animals were monitored daily and sacrificed upon first signs of neurological deficit or weight loss greater than 20%. Control animals were administered vehicle. Kaplan-Meier Survival curves were compared by log-rank test.
To evaluate brain penetrance of capmatinib
2h after administration of the last capmatinib 30 mg/kg dose, blood samples were drawn, animals were sacrificed and brains were harvested and 2mm coronal sections were frozen in OCT. Tumor tissue was dissected from the frozen blocks. Capmatinib concentration in homogenized tumor tissue and plasma was determined for 3 treated animals and one control was quantified by LC-MS/MS.
Xenograft tumor macrodissection of frozen tissue
Brain samples of 3 randomly selected animals per xenograft line were used. Frozen 2mm coronal sections were transferred to a cryostat (Cryotome E, ThermoElectronCorporation) set to −16°C. Six μm sections were cut and stained with hematoxylin, to locate the tumor. Tumor tissue was excised from the frozen block with a scalpel into a pre-chilled microtube and stored at −80°C.
Nucleic Acids isolation
Genomic DNA was isolated from frozen tumor samples, macrodissected xenograft tumor (3 biological replicates), and neurosphere cultures using QIAamp DNA mini Kit (Qiagen #51304), with on column RNase A digestion, following manufacturer instructions. DNA was isolated from blood using DNA QIAamp Blood kit (Qiagen).
Total RNA was extracted from frozen tumor samples, macrodissected xenograft tumor (3 biological replicates), and neurosphere culture using MirVana (Ambion # AM1560), followed by DNAse treatment using DNA-free (Ambion AM 1906).
Fluorescence in situ hybridization (FISH)
FISH on matching tumor samples/neurospheres/PDX
FISH probes were prepared from purified BAC clones (BACPAC Resource Center; Supplementary Table 5). Probes were labeled with Orange-dUTP or with Green-dUTP (Abbott Molecular Inc., Abbott Park, IL), by nick translation.
Metaphase slides were prepared from neurosphere cell cultures that were harvested and fixed in methanol:acetic acid (3:1), according to standard cytogenetic procedures. Tumor touch preparations were prepared by imprinting thawed tumor tissue onto positively-charged glass slides and fixing them in methanol:acetic acid (3:1) for 30 min then air-dried. Frozen tumor and macrodissected xenograft tumor samples were prepared as described 42. The FISH probes were denatured at 75 °C for 5 min and held at 37 °C for 10–30 min until 10 ul of probe was applied to each sample slide. Slides were coverslipped and hybridized overnight at 37 °C in the ThermoBrite hybridization system (Abbott Molecular Inc.). The posthybridization wash was with 2X SSC/0.2% TWEEN 20 at 73 °C for 3 min followed by a brief water rinse. Slides were air-dried and then counterstained with VECTASHIELD mounting medium with 4′-6-diamidino-2-phenylindole (DAPI) (Vector Laboratories Inc., Burlingame, CA).
Image acquisition was performed at 1000× system magnification with a COOL-1300 SpectraCube camera (Applied Spectral Imaging-ASI, Vista, CA) mounted on an Olympus BX43 microscope. Images were analyzed using FISHView v7 software (ASI) and 100 – 200 interphase nuclei were scored for each sample in addition to analysis of 50 – 100 metaphase spreads for each cell line.
FISH on paired primary/recurrent FFPE gliomas
Fluorescence in situ assay was performed using RPS6/Con 9, CDK4/Con 12, EGFR/con 7, MYC/con 8, PDGFRA/con 4, C-Met/con 7, TERT/Con 5 FISH probes from Empire Genomics (Buffalo, N.Y.). The slides were hybridized with the FISH probes according to the manufacturer’s instructions with slight modifications. The slides were then examined under fluorescence microscope (Nikon 80i) equipped with multiple filters and signals were manually counted in 50 cells for each slide.
Immunohistochemistry
Sections of formalin fixed, paraffin embedded human glioma surgical samples, tumor xenografts, or multicellular spheroids were deparaffinized with xylene and rehydrated through graded alcohol into in phosphate buffered saline. Antigens were unmasked by 10 min incubation in boiling in citrate buffer and sections stained with anti-Met rabbit monoclonal antibody (D1C2) (Cell signaling #8198) or anti-phospho-Met (Tyr1234/1235) rabbit monoclonal antibody (D26) (Cell signaling #3077) and visualized with Betazoid DAB (Biocare BDB 2004) and counterstained with Envision Flex Hematoxylin (Dako K8008). Images were captured using a Eclipse E800M microscope equipped with a Nikon DS-Fi2 color digital camera (Nikon).
Reverse Transcription and PCR
cDNA was prepared from 1 μg DNAseI-treated total RNA isolated from tumor, neurosphere and xenografts using Superscript III Reverse Transcriptase and oligo dT (Thermo Fisher Scientific). cDNA was used as a template for PCR reaction in a iCycler instrument (BioRad), using Platinum Taq DNA Polymerase (Thermo Fisher Scientific) and the oligos described on Supplementary Table 6.
LC-MS/MS Quantitation of Capmatinib and Crizotinib in Mouse Plasma and Tumor
For mouse plasma sample analysis, 25 μL of each sample was precipitated with 200 μL of acetonitrile. This suspension was vortexed for 30 min and centrifuged at 4k rpm for 15 min, after which 100 μL of the extract was aliquoted and mixed with 200 μL of acetonitrile/water (1/2, v/v) prior to LC-MS/MS analysis. The extracted plasma samples were analyzed on a Waters Acquity UPLC system coupled with a Waters Xevo TQ-S triple quadrupole mass spectrometer operated at positive mode. The capillary voltage was set to 0.5 kv and collision energy to 32 ev. Capmatinib (purchased from Matrix Scientific Products (Columbia, SC)) and crizotinib (purchased from LC Laboratories (Woburn, MA)) were separated using a Waters Acquity UPLC BEH C18 column (1.7 μm, 2.1 × 30 mm) and detected by a multiple reaction monitoring transition, m/z 413.04>354.07 for capmatinib and m/z 450.04>260.18 for crizotinib, respectively. The mobile phase A was 0.1% acetic acid/water and B was 0.1% acetic acid/acetonitrile. The LC gradient was 10% B (0–0.3 min), 10–95% B (0.3–1.3 min), 95% B (1.3–1.7 min), 10% B (1.7–2.0 min) and the flow rate was 0.5 mL/min. The column temperature was 40 °C. The injection volume was 2 μL. Under these conditions, the retention time was 0.85 min for capmatinib and 0.74 min for crizotinib. The method was validated with an analytical range of 1 – 1000 ng of capmatinib and crizotinib in untreated CD-1 mouse plasma, respectively.
Mouse tumor tissue samples were homogenized in methanol:water (80:20, v/v) to a concentration of 100 mg (tissue)/mL. The homogenates were vortexed for 10 min and centrifuged at 15k rpm for 5 min, then 100 μL of the supernatant was transferred into an HPLC vial for LC-MS/MS analysis. The tissue homogenates were analyzed by using the same method as described above. The method was validated with an analytical range of 1 – 1000 ng/mL of capmatinib and crizotinib in untreated mouse tumor tissue homogenates, respectively.
Whole Exome Sequencing
Library Construction and Sequencing
The sequencing libraries were prepared using the KAPA library prep protocol (catalog number KK8234, KAPA Biosystems, Wilmington, MA). The exomes were captured using the SureSelect XT Human All Exon V5 kit (Agilent Technologies, Santa Clara, CA). Samples were then sequenced 2×100 bp to about 340× depth on the Illumina HiSeq 2000.
BAM File Generation
The raw output (BCL) files of an Illumina sequencer were converted to FASTQ files using Illumina’s offline basecalling software CASAVA version 1.8.2. The FASTQ files were then aligned to the reference genome (hg19 for human) using BWA version 0.7.0 43 for DNA samples with parameters suitable for a given aligner. The aligned BAM files are subjected to mark duplication, re-alignment, and re-caliberation using Picard version 1.112 and GATK version 1.5 44 when applicable before any downstream analysis are conducted.
Whole Genome Low Pass Sequencing
Library Construction and Sequencing
The Illumina compatible libraries were prepared using KAPA DNA Library preparation kit (Catalog No. KK8232) as per the manufacturer’s protocol. In brief, DNA was fragmented to a median size of 200bp by sonication. Fragmented DNA ends were polished and 5′-phosphorylated. After addition of 3′-A to the ends, indexed Y-adapters were ligated and the samples were PCR amplified. The resulting DNA libraries were quantified and validated by qPCR, and sequenced on Illumina’s HiSeq 2000 in a paired-end read format for 76 cycles. The resulting BCL files containing the sequence data were converted into “.fastq.gz” files and individual sample libraries were demultiplexed using CASAVA version_1.8.2 with no mismatches.
RNA Sequencing
Library Construction and Sequencing
The Illumina compatible libraries were prepared using Illumina’s TruSeq RNA Sample Prep kit v2, as per the manufacturer’s protocol. In brief, Poly-A RNA was enriched using Oligo-dT beads. Enriched Poly-A RNA was fragmented to a median size of 150bp using chemical fragmentation and converted into double stranded cDNA. Ends of the double stranded cDNA were polished, 5′-phosphorylated, and 3′-A tailed for ligation of the Y-shaped indexed adapters. Adapter ligated DNA fragments were PCR amplified, quantified and validated by qPCR, and sequenced on Illumina’s HiSeq 2000 in a paired-end read format for 76 cycles. The resulting BCL files containing the sequence data were converted into “.fastq.gz” files & individual sample libraries were demultiplexed using CASAVA version_1.8.2 with no mismatches.
BAM File Generation
RNA sequencing BAM files were generated and analyzed using the Pipeline for RNAseq Data Analysis (PRADA ver 1.1) 45. In brief, PRADA uses Burroughs-Wheeler alignment, Samtools, and Genome Analysis Toolkit to align RNAseq reads to a reference database composed of whole genome sequences (hg19) and transcriptome sequences (Ensembl64). Details of the PRADA pipeline are described in its manuscript.
Targeted Resequencing
Library Construction and Sequencing
The Illumina compatible libraries were prepared using KAPA DNA Library preparation kit (Catalog No. KK8232) as per the manufacturer’s protocol. In brief, DNA was fragmented to a median size of 200bp by sonication. Fragmented DNA ends were polished and 5′-phosphorylated. After addition of 3′-A to the ends, indexed Y-adapters were ligated and the samples were PCR amplified. The resulting DNA libraries were enriched for targeted regions using NimbleGen SeqCap EZ Choice Library 4 RXN (Catalog No. 06740251001) and NimbleGen SeqCap EZ Reagent Kit Plus v2 (Catalog No. 06953247001) as per the manufacturer’s protocol. The enriched libraries were quantified and validated by qPCR, and sequenced on Illumina’s HiSeq 2000 in a paired-end read format for 76 cycles. The resulting BCL files containing the sequence data were converted into “.fastq.gz” files and individual sample libraries were demultiplexed using CASAVA version_1.8.2 with no mismatches.
BAM File Generation
Sequencing FASTQ files were aligned to the reference genome (hg19 for human) and processed to BAM files by the same pipeline as in whole exome sequencing.
Pacific Biosciences (PacBio) Long Read Sequencing
Library Construction and Sequencing
The DNA libraries were prepared following the Pacific Biosciences 20 kb Template Preparation Using BluePippin Size-Selection System protocol. No DNA shearing was performed since the samples were already fragmented. The sheared DNA was size selected on a BluePippin system (Sage Science Inc., Beverly, MA, USA) using a cutoff range of 7 kb to 50 kb. The DNA Damage repair, End repair and SMRT bell ligation steps were performed as described in the template preparation protocol with the SMRTbell Template Prep Kit 1.0 reagents (Pacific Biosciences, Menlo Park, CA, USA). The sequencing primer annealing and the P6 polymerase binding reactions were prepared according to the BindingCalculator (Pacific Biosciences BindingCalculator-master_v2.3.1.1). The libraries were sequenced on a PacBio RSII instrument at a loading concentration (on-plate) of 80pM, 90pM and 100pM using the MagBead OneCellPerWell v1 collection protocol, DNA sequencing kit 4.0, SMRT cells v3 and 4 hours movies.
Filtering the sequencing reads
Reads and subreads were filtered based on their length and quality values, using smrtpipe.py from the SMRT-Analysis package.
Structural Variation Analysis
Canu (version 1.2) 46 was used for assembling the filtered PacBio sequence subreads with the parameters suggested for low coverage data. The assembled contigs were aligned by nucmer (version 3.23) 47 to the human genome reference (hg19) and the contigs having sequence fragments aligned to the MET-CAPZA2 region of chromosome 7 were selected for structural variation analysis. For the selected contigs, we performed a blastn search 48 against mouse genome using the sequence fragments aligned to the MET-CAPZA2 region of hg19 in order to make sure that they originated from human (Supplementary Table 7). Sequence framents shared by two contigs were identified with pairwise alignment of the contigs using the nucmer. Two contigs were considered to be connected only if they shared a sequence fragment which was at least 5,000 bp long with the minimum 99% identity. The high confident shared sequence fragments were used for connecting the contigs into a circular form in the HF3035. In HF3077, only two contigs (tig01141776 and tig01141835) were aligned to the MET-CAPZA2 region of chromosome 7, and the two contigs shared 621 bp long sequence with 95.6% identity between the 3′ end of tig01141776 and the 5′ end of tig01141835.
Gene Fusion and Gene Expression Analysis
To detect transcript fusions, PRADA aligned RNAseq reads to a reference database composed of whole genome sequences (hg19) and transcriptome sequences (Ensembl64). Two lines of evidence were required for identification of a gene fusion: 1) a minimum of two discordant read pairs mapping to a candidate gene pair; 2) a minimum of one junction spanning read mapping to a junction that connected exons between the candidate gene pair, with its pair mate mapping to the either of the two genes. Several filters were applied to remove false positives and artifacts, of which the most prominent is based on significant sequence similarity between the two fusion genes (using BLASTN, Expect value = 0.01). Gene expression was measured as ‘reads per kilobase per million’ (RPKM) to normalize for gene length and library size. Specific details of the PRADA pipeline are described elsewhere 45.
Structural Variant Detection
To detect structural variants, we applied SpeedSeq 49 (with default parameters) to whole genome sequencing from both tumor and matching normal samples. We filtered somatic variants by requiring at least 4 reads supporting evidence in a tumor and no reads in its matching normal.
EGFR Intragenic Rearrangement
General User dEfined Supervised Search for intragenic fusion (GUESS-if), a module of PRADA, was also used to search for EGFR intragenic rearrangements, as previously described 11. In brief, using the same rationale as in PRADA gene fusion identification, GUESS-if looked for spanning reads for abnormal junctions that were not present in known transcripts. To assure a high accuracy, we required at least 10 reads spanning exon 1–8 of EGFR.
Validation of Somatic Single Nucleotide Variants
To validate our somatic single nucleotide mutation calls, we performed targeted resequencing at high coverage (>1,400x). We selected 792 unique bases, which had been found to be mutated in tumor, neurosphere, or xenografts but not in all of them. These sites corresponded to 1368sSNVs. In total, 1287of 1368mutations called from the exome sequencing data were detected in the high coverage data, resulting in a true positive validation rate of 94%. Evidence for recovered somatic mutation was observed in 1001 of 2646 wild type nucleotides. The variant allelic fractions (VAFs), i.e. the number of reads harboring the variant allele divided by all reads covering to that base, of exome and validation sequencing were highly correlated (Pearson correlation = 0.92).
Somatic Single Nucleotide Variant Calling
Somatic single nucleotide variants (sSNVs) from tumor and patient-matched normal samples were detected by using MuTect algorithm (version 1.14) with default parameters 50. The search for somatic small insertion/deletions (Indels) was performed by using Pindel 51 with tumor and patient-matched normal samples. All sSNVs and small indels were annotated by ANNOVAR (version 2012-10-23) 52. Only exonic or splicing sSNVs were selected for analysis. Mutation counts for individual samples are available in Supplementary Table 8.
Inference of Cellular Frequency and Mutational Clusters
We defined cellular frequency of a mutation as the fraction of cells harboring the mutation. Estimation of cellular frequency was performed using PyClone version 0.12.7 7. For each set of patient, neurosphere, and xenograft samples, PyClone was run on the somatic mutations whose sites were covered over all the samples using multi-sample joint analysis mode with PyClone beta binomial density and parental copy number priors. Allelic copy numbers were estimated by applying Sequenza 53 to exome sequencing data. Default options for PyClone were used. To avoid potential artifacts from sequencing coverage, we limited the analysis to the mutations at the sites covered with at least 50× over all samples from a same patient. PyClone inferred clusters of mutations whose cellular frequencies co-vary over samples. Our analysis was limited only to mutation clusters with at least two mutations.
Removing Putative Mouse Reads in Short Read Sequencing Data
Sequencing reads derived from xenograft samples are a mixture of reads from human and mouse. We utilized Xenome 54 to select sequencing reads arising from human. Then, the selected human reads selected were aligned to the human genome using the same pipeline as in patient and neurosphere samples.
Identification of Copy Numbers from Low Pass Sequence Data
We used NBICSeq version 0.5.2 55 with bin size 1000 bps and BIC penalty 3 to estimate somatic copy number alterations in low pass sequencing data from tumor and patient-matched normal samples.
Detecting TERT Promoter Mutations
We evaluated whole genome low pass sequencing and whole exome sequencing for the presence of TERT mutations in a supervised way using GATK pileup. We required minimum 2 variant alleles (combined from WGS and WES) for detection of TERT promoter mutations. C228T mutation on 5:1295228-1295228 was detected in 7 patients, and C250T mutation on 5:1295250-1295250 was detected in 5 patients.
Detecting ATRX Indels
Indels were called using Pindel (Version 0.2.4t) with the default parameters except maximum allowed mismatch rate being 0.1 51. Somatic indels were further filtered to require a minimum 5 supporting tumor reads.
Analysis of B-allele-frequency segments
B-allele-frequency segments were inferred by applying Sequenza (Version 2.1.1) 53 to whole exome sequencing data with the default parameters. Analysis of B-allele fractions using whole genome sequencing in our sample cohort revealed loss of heterozygosity (LOH) of chromosome 10 in two cases with diploid chromosome 10, suggesting these cases had first lost a single copy of the chromosome which was subsequently duplicated (Supplementary Fig. 1). We evaluated chromosome 10 LOH using Affymetrix SNP6 profiles from 320 IDH-wildtype TCGA glioblastoma 11, and found that 27 of 52 tumors with diploid chromosome 10 similarly showed LOH, underscoring the importance of aberrations in chromosome 10 in gliomagenesis and evolution (Supplementary Fig. 6).
Data used for longitudinal analysis in glioma patient tumors
Segmented copy number profiles for thirteen TCGA GBM patients and fourteen TCGA LGG patients were were obtained from the TCGA portal. Copy number profiles for ten patients from MD Anderson Cancer Center (MDACC) and fourteen patients from either Samsung Medical Center (SMC) or Seoul National University Hospital (SNUH) were previously processed 4,56. Additional copy number data for seven patients from MD Anderson were generated by applying NBICseq version 0.5.255 to low pass whole genome sequencing (WGS). For fusion detection and structural variant calling, the same pipelines as described in the corresponding method subsections were applied for unaligned RNA sequencing files and whole genome sequencing BAM files from TCGA GBM (n=6 for RNAseq; n=10 for WGS), TCGA LGG (n=14 for RNAseq; n=13 for WGS), and MD Anderson patients (n=9 for RNAseq; n=7 for WGS). Sequencing data for the TCGA cohort were downloaded from CGHub. Fusion calls for Samsung Medical Center cohort patients were previously processed 56.
Predicting extrachromosomal DNA (ecDNA) candidates
After visualizing segmented copy numbers in the Integrative Genomics Viewer (IGV) 57, we manually scrutinized potential extrachromosomal DNA candidate regions by searching for intermittent patterns of DNA copy number amplification. In cases where structural variations and gene fusions were available, we projected those variation breakpoints onto the copy number IGV view plots to corroborate our DNA copy number based predictions to get additional evidence on presence of our predicted ecDNAs.
In order to avoid biases of our method such as the presence of multiple adjacent amplifications in oncogene regions, we additionally applied the AmpliconArchitect method58 to 125 samples with available whole genome sequencing data (65 samples from our hGBM cohort and 60 longitudinal glioma samples) in order to identify ecDNAs in an unsupervised manner. We processed 46 TCGA glioma samples through the Institute for Systems Biology Cancer Genomics Cloud that provides a cloud-based platform for TCGA data analysis. We used the processed segmented copy number profiles (described in the previous section) to identify interval(s) of interest that are required for the input to AmpliconArchitect. Default parameters and reference files were used for all other settings. The ecDNAs predicted by AmpliconArchitect were filtered by only selecting amplicons with at least 6 amplified amplicon copy count that resulted in relatively balanced numbers of ecDNAs between low pass sequencing cases (a median depth of 6.5X) and TCGA whole genome cases. The AmpliconArchitect-predicting ecDNAs further merged with those predicted by our method in cases where those ecDNAs overlap each other.
To identify tumor driver genes carried by our predicted ecDNAs, we used a list of copy number driver genes (CNA_drivers_per_tumor_type.tsv file) downloaded from Integrative OncoGenomics31 and glioblastoma frequently-altered genes from the TCGA study 11, then intersecting those gene regions with the predicted ecDNA regions. AmpliconArchitect also had an internal function on identifying oncogenes (from 522 oncogenes from the COSMIC database (Aug 2014) 59) covered by the predicted ecDNA, and we included those oncogenes into a list of ecDNA carrying driver genes. Details on how to run AmpliconArchitect have been described in the corresponding manuscript 58 and its source code depository.
Statistical Analysis
Survival curves were estimated with the Kaplan-Meier method, and comparison of survival curves between groups was performed with the log-rank test in either GraphPad Prism 7 or R survival package.
All other statistical computations were performed with R (The R Project for Statistical Computing)
DATA AVAILABILITY
The datasets in form of BAM files from exome sequencing, low pass whole genome sequencing and RNA sequencing generated during the current study are available in the the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under accession number EGAS00001001878.
URLs
ISB-CGC: https://isb-cgc.appspot.com/; TCGA Public Data Access: https://portal.gdc.cancer.gov/; PRADA: http://sourceforge.net/projects/prada/; European Genome-phenome Archive; http://www.ebi.ac.uk/ega/; BACPAC Resource Center: https://bacpacresources.org; Picard: http://picard.sourceforge.net; AmpliconArchitect: https://github.com/virajbdeshpande/AmpliconArchitect; Integrative OncoGenomics: http://www.intogen.org
Supplementary Material
Acknowledgments
The authors would like to thank our colleagues at Henry Ford Hospital: Dr. N. Lehman and Dr. C. Hao for contributions to pathology reviews; L. Scarpace for clinical information; S. Irtenkauf, L. Hasselbach, K. Nelson, K. Bergman, and S. Sobiechowski for cell culture and animal work; A. Transou, Y. Meng, and E. Carlton for histology. We are indebted to Matt Wimsatt (JAX) for the creative design in figure 6. We thank G. Geneau, S. Roland, and Pac Bio platform personnel of the Génome Québec/Genome Canada-funded Innovation Centre for providing Pacific Biosciences sequencing. AmpliconArchitect analysis of TCGA was made possible through the Cancer Genomics Cloud of the Institute for Systems Biology (ISB-CGC). This work was supported by the LIGHT Research Program at the Hermelin Brain Tumor Center (ACD, TM); grants from the National Institutes of Health R01 CA190121 (RGWV); Cancer Center Support Grant P30CA034196; the Cancer Prevention & Research Institute of Texas (CPRIT) R140606 (RGWV), and The National Brain Tumor Society (RGWV). This work was also supported by a grant of the Korea Health Technology R&D project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI14C3418). We are hugely indebted to the patients who provided biospecimens for the purpose of this study.
Footnotes
AUTHOR CONTRIBUTIONS
A.C.D., H.K., and R.G.W.V. led the study and wrote the manuscript. T.M. obtained the patient samples that made the study possible. A.C.D. supervised the establishment of primary cultures and xenografts, prepared samples for molecular profiling, designed all in vitro and in vivo experiments and performed data analysis. H.K. designed, supervised, and performed all bioinformatic analyses. L.M.P., S.Z., S.S., and J.Z. performed data pre-processing and data analysis. T.M. and L.M.P. collected clinical data. J.K. and A.M. performed FISH experiments. Y.J. performed liquid chromatography–mass spectrometry. A.P. supervised and performed all Illumina sequencing studies, including whole-genome, exome and RNA-seq library preparation and sequencing experiments. M.F. provided clinical and pathology reviews. M.E.W., C.M., D.C., E.F.P., and L.C. provided valuable input regarding study design, data analysis, and interpretation of results. D.H.N., T.M. and R.G.W.V. provided validation datasets. T.M., L.C. and R.G.W.V. provided financial and technical infrastructure and oversaw the project.
COMPETING INTERESTS
The authors declare no competing financial interests.
Supplementary Note, Figures and Tables are available as supplementary data.
References
- 1.Roos WP, Thomas AD, Kaina B. DNA damage and the balance between survival and death in cancer biology. Nat Rev Cancer. 2016;16:20–33. doi: 10.1038/nrc.2015.2. [DOI] [PubMed] [Google Scholar]
- 2.Yap TA, Gerlinger M, Futreal PA, Pusztai L, Swanton C. Intratumor heterogeneity: seeing the wood for the trees. Sci Transl Med. 2012;4:127ps10. doi: 10.1126/scitranslmed.3003854. [DOI] [PubMed] [Google Scholar]
- 3.Aparicio S, Caldas C. The implications of clonal genome evolution for cancer medicine. N Engl J Med. 2013;368:842–51. doi: 10.1056/NEJMra1204892. [DOI] [PubMed] [Google Scholar]
- 4.Kim H, et al. Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome Res. 2015;25:316–27. doi: 10.1101/gr.180612.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sequist LV, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andor N, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med. 2016;22:105–13. doi: 10.1038/nm.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roth A, et al. PyClone: statistical inference of clonal population structure in cancer. Nat Methods. 2014;11:396–8. doi: 10.1038/nmeth.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolecek TA, Propp JM, Stroup NE, Kruchko C. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro Oncol. 2012;14(Suppl 5):v1–49. doi: 10.1093/neuonc/nos218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ceccarelli M, et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell. 2016;164:550–63. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–77. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snuderl M, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20:810–7. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 13.Sottoriva A, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110:4009–14. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szerlip NJ, et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci U S A. 2012;109:3041–6. doi: 10.1073/pnas.1114033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ozawa T, et al. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell. 2014;26:288–300. doi: 10.1016/j.ccr.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, et al. c-Myc is required for maintenance of glioma cancer stem cells. PLoS One. 2008;3:e3769. doi: 10.1371/journal.pone.0003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Annibali D, et al. Myc inhibition is effective against glioma and reveals a role for Myc in proficient mitosis. Nat Commun. 2014;5:4632. doi: 10.1038/ncomms5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cox D, Yuncken C, Spriggs AI. Minute Chromatin Bodies in Malignant Tumours of Childhood. Lancet. 1965;1:55–8. doi: 10.1016/s0140-6736(65)90131-5. [DOI] [PubMed] [Google Scholar]
- 19.Kohl NE, et al. Transposition and amplification of oncogene-related sequences in human neuroblastomas. Cell. 1983;35:359–67. doi: 10.1016/0092-8674(83)90169-1. [DOI] [PubMed] [Google Scholar]
- 20.Turner KM, et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature. 2017;543:122–125. doi: 10.1038/nature21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanborn JZ, et al. Double minute chromosomes in glioblastoma multiforme are revealed by precise reconstruction of oncogenic amplicons. Cancer Res. 2013;73:6036–45. doi: 10.1158/0008-5472.CAN-13-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng S, et al. A survey of intragenic breakpoints in glioblastoma identifies a distinct subset associated with poor survival. Genes Dev. 2013;27:1462–72. doi: 10.1101/gad.213686.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikolaev S, et al. Extrachromosomal driver mutations in glioblastoma and low-grade glioma. Nat Commun. 2014;5:5690. doi: 10.1038/ncomms6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Organ SL, Tsao MS. An overview of the c-MET signaling pathway. Ther Adv Med Oncol. 2011;3:S7–S19. doi: 10.1177/1758834011422556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Storlazzi CT, et al. Gene amplification as double minutes or homogeneously staining regions in solid tumors: origin and structure. Genome Res. 2010;20:1198–206. doi: 10.1101/gr.106252.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lundberg G, et al. Binomial mitotic segregation of MYCN-carrying double minutes in neuroblastoma illustrates the role of randomness in oncogene amplification. PLoS One. 2008;3:e3099. doi: 10.1371/journal.pone.0003099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X, et al. A novel kinase inhibitor, INCB28060, blocks c-MET-dependent signaling, neoplastic activities, and cross-talk with EGFR and HER-3. Clin Cancer Res. 2011;17:7127–38. doi: 10.1158/1078-0432.CCR-11-1157. [DOI] [PubMed] [Google Scholar]
- 28.Tesfay L, Schulz VV, Frank SB, Lamb LE, Miranti CK. Receptor tyrosine kinase Met promotes cell survival via kinase-independent maintenance of integrin alpha3beta1. Mol Biol Cell. 2016;27:2493–504. doi: 10.1091/mbc.E15-09-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arena S, Pisacane A, Mazzone M, Comoglio PM, Bardelli A. Genetic targeting of the kinase activity of the Met receptor in cancer cells. Proc Natl Acad Sci U S A. 2007;104:11412–7. doi: 10.1073/pnas.0703205104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogt N, et al. Molecular structure of double-minute chromosomes bearing amplified copies of the epidermal growth factor receptor gene in gliomas. Proc Natl Acad Sci U S A. 2004;101:11368–73. doi: 10.1073/pnas.0402979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rubio-Perez C, et al. In silico prescription of anticancer drugs to cohorts of 28 tumor types reveals targeting opportunities. Cancer Cell. 2015;27:382–96. doi: 10.1016/j.ccell.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 32.Bigner SH, Mark J, Bigner DD. Cytogenetics of human brain tumors. Cancer Genet Cytogenet. 1990;47:141–54. doi: 10.1016/0165-4608(90)90024-5. [DOI] [PubMed] [Google Scholar]
- 33.Nathanson DA, et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science. 2014;343:72–6. doi: 10.1126/science.1241328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chi AS, et al. Rapid radiographic and clinical improvement after treatment of a MET-amplified recurrent glioblastoma with a mesenchymal-epithelial transition inhibitor. J Clin Oncol. 2012;30:e30–3. doi: 10.1200/JCO.2011.38.4586. [DOI] [PubMed] [Google Scholar]
- 35.Humphrey PA, et al. Amplification and expression of the epidermal growth factor receptor gene in human glioma xenografts. Cancer Res. 1988;48:2231–8. [PubMed] [Google Scholar]
- 36.Pandita A, Aldape KD, Zadeh G, Guha A, James CD. Contrasting in vivo and in vitro fates of glioblastoma cell subpopulations with amplified EGFR. Genes Chromosomes Cancer. 2004;39:29–36. doi: 10.1002/gcc.10300. [DOI] [PubMed] [Google Scholar]
- 37.Schulte A, et al. Glioblastoma stem-like cell lines with either maintenance or loss of high-level EGFR amplification, generated via modulation of ligand concentration. Clin Cancer Res. 2012;18:1901–13. doi: 10.1158/1078-0432.CCR-11-3084. [DOI] [PubMed] [Google Scholar]
- 38.Giannini C, et al. Patient tumor EGFR and PDGFRA gene amplifications retained in an invasive intracranial xenograft model of glioblastoma multiforme. Neuro Oncol. 2005;7:164–76. doi: 10.1215/S1152851704000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hasselbach LA, et al. Optimization of High Grade Glioma Cell Culture from Surgical Specimens for Use in Clinically Relevant Animal Models and 3D Immunochemistry. J Vis Exp. 2014;83:e51088. doi: 10.3791/51088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.deCarvalho AC, et al. Gliosarcoma stem cells undergo glial and mesenchymal differentiation in vivo. Stem Cells. 2010;28:181–90. doi: 10.1002/stem.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Irtenkauf SM, et al. Optimization of Glioblastoma Mouse Orthotopic Xenograft Models for Translational Research. Comp Med. 2017;67:300–314. [PMC free article] [PubMed] [Google Scholar]
- 42.Graveel C, et al. Activating Met mutations produce unique tumor profiles in mice with selective duplication of the mutant allele. Proc Natl Acad Sci U S A. 2004;101:17198–203. doi: 10.1073/pnas.0407651101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Torres-Garcia W, et al. PRADA: pipeline for RNA sequencing data analysis. Bioinformatics. 2014;30:2224–6. doi: 10.1093/bioinformatics/btu169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berlin K, et al. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat Biotechnol. 2015;33:623–30. doi: 10.1038/nbt.3238. [DOI] [PubMed] [Google Scholar]
- 47.Delcher AL, et al. Alignment of whole genomes. Nucleic Acids Res. 1999;27:2369–76. doi: 10.1093/nar/27.11.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 49.Chiang C, et al. SpeedSeq: ultra-fast personal genome analysis and interpretation. Nat Methods. 2015;12:966–8. doi: 10.1038/nmeth.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–71. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Favero F, et al. Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann Oncol. 2015;26:64–70. doi: 10.1093/annonc/mdu479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Conway T, et al. Xenome--a tool for classifying reads from xenograft samples. Bioinformatics. 2012;28:i172–8. doi: 10.1093/bioinformatics/bts236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xi R, et al. Copy number variation detection in whole-genome sequencing data using the Bayesian information criterion. Proc Natl Acad Sci U S A. 2011;108:E1128–36. doi: 10.1073/pnas.1110574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim J, et al. Spatiotemporal Evolution of the Primary Glioblastoma Genome. Cancer Cell. 2015;28:318–28. doi: 10.1016/j.ccell.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 57.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turner KM, et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature. 2017 doi: 10.1038/nature21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Forbes SA, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–11. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets in form of BAM files from exome sequencing, low pass whole genome sequencing and RNA sequencing generated during the current study are available in the the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under accession number EGAS00001001878.