Summary
OBJECTIVE
Focal cortical dysplasia (FCD) is a major pathology in patients undergoing surgical resection to treat pharmacoresistant epilepsy. MRI post-processing methods may provide essential help for detection of FCD. In this study, we utilized surface-based MRI morphometry and machine learning for automated lesion detection in a mixed cohort of patients with FCD type II from three different epilepsy centers.
METHODS
Sixty-one patients with pharmacoresistant epilepsy and histologically proven FCD type II were included in the study. The patients had been evaluated at three different epilepsy centers using 3 different MRI scanners. T1-volumetric sequence was used for post-processing. A normal database was constructed with 120 healthy controls. We also included 35 healthy test controls and 15 disease test controls with histologically confirmed hippocampal sclerosis to assess specificity. Features were calculated and incorporated into a nonlinear neural network classifier which was trained to identify lesional cluster. We optimized the threshold of the output probability map from the classifier by performing ROC analyses. Success of detection was defined by overlap between the final cluster and the manual labeling. Performance was evaluated using k-fold cross-validation.
RESULTS
The threshold of 0.9 showed optimal sensitivity of 73.7% and specificity of 90.0%. The area under the curve for the ROC analysis was 0.75 which suggests a discriminative classifier. Sensitivity and specificity were not significantly different for patients from different centers, suggesting robustness of performance. Correct detection rate was significantly lower in patients with initially normal MRI than patients with unequivocally positive MRI. Subgroup analysis showed the size of training group and normal control database impacted classifier performance.
SIGNIFICANCE
Automated surface-based MRI morphometry equipped with machine learning showed robust performance across cohorts from different centers and scanners. The proposed method may be a valuable tool to improve FCD detection in presurgical evaluation for patients with pharmacoresistant epilepsy.
Keywords: epilepsy, surgery, FCD, MRI post-processing
Introduction
Focal cortical dysplasia (FCD) type II is a major cause of pharmacoresistant epilepsy in patients undergoing surgical resection.1 Typical MRI features of FCD type II include cortical thickening, blurring of gray-white matter junction, hyperintense signal on T2 or FLAIR sequences and the “transmantle sign”.2,3 Despite improvements in MRI resolution, some FCD type II lesions are too subtle to be detected by conventional visual analysis of MRI scans,4 especially when noninvasive data do not point to a specific brain region. Discovering a previously missed lesion can have practical clinical impact by refocusing the surgical hypothesis, and lead to improved postoperative seizure outcomes. Post-operative seizure outcomes of patients with positive MRI are significantly better than those with negative MRI.1
MRI post-processing methods have been used to improve detection of FCD lesions.5–8 Our previous studies used morphometric analysis program (MAP), a voxel-based morphometry post-processing method, which showed favorable results in detecting FCD.9 Although very practical and robust to implement, VBM methods have some inherent limitations. They do not contain spatial relationships across the cortical surface, and any errors in registration can result in missing subtle lesions.10 Additionally, reaffirmation by an experienced MAP reader or a neuroradiologist is still necessary to conclude the MAP results as per the previous studies;8,9,11 therefore, the yield and diagnostic confidence can depend on the reader’s experience. To overcome these inherent weaknesses and further increase yield, a multivariate surface-based morphometry (SBM) approach may be beneficial. Recently, a few studies have reported multivariate SBM approaches combined with machine learning, with high accuracy for automated FCD detection in adult and pediatric patients (some had negative MRI by visual analyses)12–14. However, the previous studies were all based on single centers, using a small cohort of patients with histologically confirmed or radiologically defined FCD,12–14 and only one study assessed the false positive rate in healthy controls.12
To test the potential clinical value of any MRI post-processing method, it is necessary to evaluate its robustness in a large data set from different epilepsy centers using different MRI scanners. Herein, to test the diagnostic value of SBM and machine learning in patients with histologically proven FCD type II, we chose a mixed cohort from different epilepsy centers using different 3T MRI scanners. Additionally, receiver operator curve (ROC) analysis was performed to obtain the optimal threshold for the classifier output probability maps for automated lesion detection.
Materials and methods
Patient and Normal Control selection
Patients with pharmacoresistant epilepsy and histologically proven FCD type II from three different epilepsy centers were included in the study [Second Affiliated Hospital of Zhejiang University (SAHZU), China; Beijing Tiantan Hospital of Capital Medical University (BTH), China; Cleveland Clinic Foundation (CCF), USA]. Exclusion criteria were patients under five years old and low image quality due to motion, noise, or other image artifacts. These patients were evaluated using three different 3T MRI scanners (GE discovery MR750, Siemens Verio, Siemens Trio, respectively). The normal database used for inter-subject normalization was constructed using scans from 120 normal controls obtained from four different scanners. To assess specificity, we additionally included two test groups: (1) healthy test group: 35 healthy subjects free of neurological disease; (2) disease test group: 15 patients with histopathologically confirmed hippocampal sclerosis, no temporal lobe FCD, and who became seizure-free at one year following temporal lobectomy. For subgroup analysis, patient’s MRI scans were classified as “MRI negative” (unremarkable abnormalities) or “MRI positive” by official radiology report. This study was approved by the institutional review board ethical guidelines of three hospitals (SAHZU, BTH and CCF).
MRI acquisition
MRI scans from SAHZU were performed on a 3.0-T scanner (MR750, GE Healthcare, USA) including 3D T1 sagittal brain volume imaging (BRAVO) sequence (TR/TE= 8.2/3.2 ms, TI= 450 ms, flip angle=12 degrees, slice thickness= 1 mm, no gap, matrix=256 × 256, voxel size= 0.47×0.47×1mm3). Patients from BTH were scanned on a 3.0T Siemens Verio scanner (Siemens Medical system, South Iselin, NJ) including 3D T1 sagittal Magnetization Prepared Rapid Gradient Echo sequence (MPRAGE) (TR/TE=1900ms/2.53ms, TI=900 ms, flip angle=12 degrees, slice thickness = 1 mm, no gap, matrix=256×256, voxel size= 0.98×0.98×1mm3). MRI scans from CCF were performed on a 3.0-T Siemens Trio scanner (Siemens Medical system, Erlangen, Germany) including 3D T1 coronal MPRAGE (TR/TE= 1,860ms/3.4ms, TI=1,100 ms, flip angle= 10 degrees, slice thickness= 0.94 mm, no gap, matrix=256×256, isotropic voxels=0.94 mm). Normal controls and healthy test group were included from SAHZU, CCF and the Pediatric Imaging, Neurocognition and Genetic Study (PING) in which MRI was performed on nine 3T scanners from three manufacturers (Siemens, GE, Philips Medical, Andover, MA, USA), including the ones matching the patient scanners above. Detailed parameters available on the PING website (http://ping.chd.ucsd.edu/).
Cortical reconstruction
We used standard processes in FreeSurfer software v5.3 (http://surfer.nmr.mgh.harvard.edu/) for cortical reconstruction.15–17 In brief, the processing involves (1) segmentation of white matter, (2) tessellation of the gray/white matter boundary, (3) inflation of the folded surface tessellation, and (4) automatic correction of topological defects. These steps have been described in detail elsewhere.14 Reconstruction results of each subject were inspected visually and any inaccuracies due to imaging artifacts were manually corrected.
Lesion labels
The lesion masks were created manually in Freesurfer, on the T1-weighted volumetric sequence, informed by post-operative MRI, FLAIR and T2-weighted images (so that it is possible to create lesion masks for the cases initially thought to be MRI-negative by report). The lesion masks were then registered onto the cortical surface reconstructions. Each vertex in the training dataset was given one of two response values: lesional (one) if within the lesion mask, or normal (zero) if outside of the lesion mask.
Measures of morphological and intensity features
Six cortical features were acquired at each vertex of the 3D cortical reconstruction: cortical thickness, gray-white matter intensity contrast, curvature, sulcal depth, “doughnut” maps and local cortical deformation (LCD).14 Cortical thickness was calculated as follows. First, for each point on the white matter surface, the shortest distance to the pial surface was measured. Second, the shortest distance from each point on the pial surface to the white matter surface was computed. Cortical thickness at each vertex was computed as the average of the two values.15 Gray-white matter intensity contrast was estimated by calculating the ratio of the gray matter signal intensity to the white matter signal intensity.18 The gray and white matter signal intensities were sampled at a distance of 30% of the cortical thickness above the gray-white boundary and 1 mm below the gray-white boundary, respectively. Compared to healthy cortex, lesions with blurring of the grey-white matter boundary were expected to have low gray-white matter intensity contrast values. Mean curvature was measured as 1/r, where r is the radius of an inscribed circle and is equal to the average of the principal curvatures k1 and k2.19 Sulcal depth was calculated by the dot product of the movement vector of the cortical surface during inflation. Shallow gyral areas of the brain move inwards during inflation and have a negative value, whereas deep sulcal areas move outwards and have a positive value.14 “Doughnut” maps were assessed by measuring cortical thickness and gray-white matter intensity within a 6 mm radius circle and within the doughnut, where the circle was centered on a vertex on the inflated surface and the doughnut was a surrounding region around the circle.14 LCD was computed by the sum of intrinsic curvature within a 25 mm radius ring (gray circle).14 For every individual, cortical thickness, gray-white matter intensity contrast and “doughnut” maps were smoothed using a 10 mm FWHM Gaussian kernel and then these features underwent two normalization procedures: 1) within-subject z-scoring, 2) between-subject z-scoring by the population of 120 controls. All feature maps were registered to an average space (fsaverage_sym) that had an identical number of vertices for each hemisphere. For cortical thickness, gray-white matter intensity contrast and LCD, interhemispheric asymmetry was calculated. The right hemisphere vertex values for each feature were subtracted from the left hemisphere values to create a left hemisphere asymmetry map and visa versa for the right hemisphere. For those asymmetry maps of each hemisphere, positive values indicated greater ipsilateral feature values while negative indicated greater contralateral feature values. More details can be found elsewhere14 and all code is freely available at https://github.com/kwagstyl/FCDdetection.
Evaluation of effectiveness of individual morphological feature
The effectiveness of all the features was evaluated individually for each patient, and then for the entire cohort, by comparing kernel density plots of feature values within the lesion mask and the contralateral, homotopic healthy cortex.
Machine learning classification and validation
Automated lesion detection was performed using an artificial neural network classifier implemented in MATLAB R2015b (The MathWorks, Natick, MA, U.S.A.). The classifier was trained using all the aforementioned morphological and intensity features, as well as their corresponding interhemispheric asymmetry. Separate neural networks were also trained using individual features to evaluate the discriminatory value of each feature.
A k-fold cross-validation strategy (k=5) was used to validate the performance of the classifier. The top five percent vertices were identified and grouped into neighbor-connected clusters. The final cluster is considered as the highest mean probability value. In addition, a threshold was set (and tested in the next section with ROC analysis) so that the vertices with values above threshold were identified as lesional, and vertices with values below threshold were considered as normal. Successful detection was defined by overlap between the final cluster (classifier output) and the manual label.
ROC analysis
We varied the threshold values of the classifier output probability map to evaluate the sensitivity and specificity of the classifier. Any degree of overlap between the final cluster and manual lesion label was defined as correctly detected. The percentage of overlap is calculated by (number of overlapping vertices between the final cluster and the manual lesion label/total number of vertices in the final cluster) × 100%. Sensitivity was calculated as the proportion of patients in whom the final cluster overlapped with the manual lesion label. Specificity was defined as the proportion of the subjects/patients in the healthy/disease test group who had no supra-threshold clusters. Youden index was calculated to get the optimal threshold (Youden index= sensitivity + specificity − 1). The area under the ROC was calculated to further quantify the performance of the classifier.
Factors impacting classifier performance
Three factors were additionally tested to evaluate their effects on classifier performance: number of training cases, size of normal control database and scanner type.
Firstly, to assess the effect of the number of training cases, patients form SAHZU, BTH and CCF were defined as separate training groups (11 patients, 16 patients and 34 patients respectively). Keeping all the other factors the same, the classifier was trained on the 3 patient groups separately, and then performance was evaluated.
Secondly, to evaluate the influence of the size of normal database used for inter-subject normalization of features, 120 normal controls were divided into three groups (SAHZU, 22 controls; CCF 24 controls; and PING 74 controls). Keeping all the other factors the same, the classifier was normalized by the 3 controls groups separately, and then performance was evaluated.
Thirdly, to test the role of scanner type, patients form SAHZU and CCF were normalized by SAHZU and CCF normal controls, respectively. Keeping all the other factors the same, performance was evaluated. We could not include patients from BTH in this analysis, because no normal scans were acquired from the scanner used in BTH.
Statistical analysis
Descriptive statistics were used for each variable. If continuous variables (age, age at seizure onset, disease duration, sensitivity, specificity) were normally distributed, 2-sample t tests or one-way analysis of variance (ANOVA) was used. If not, Mann-Whitney U-test was used. Fisher’s exact test was used for categorical variables (sex, children or adults). Statistical significance was set at the 5% level.
Results
Patient Demographics and clinical information
A total of 61 pharmacoresistant epilepsy patients with histologically proven FCD type II from three epilepsy centers (11 patients from SAHZU; 16 patients from BTH; 34 patients from CCF) were included (32 males, 33 children, mean age ± SD = 20.43 ± 13.36). A total of 17 patients (27.9%) were MRI-negative by initial radiology report; in all of the 17 patients, subtle FCD lesions were identified at the patient management conference, which was aided by multimodal localization from semiology, EEG, PET, SPECT and magnetic source imaging. Seizure-freedom was achieved in 72.1% one year after surgery. Detailed profile of patients, control subjects, and test subjects can be found in Table 1.
Table 1.
Patients and controls demographics
| Patients (Total=61) |
Normal database (Total=120) |
Healthy test group (Total=35) |
Disease test group (Total=16) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SAHZU Total=11 |
BTH Total=16 |
CCF Total=34 |
SAHZU Total=22 |
CCF Total=24 |
PING study Total=74 |
CCF Total=13 |
PING Total=22 |
SAHZU Total=4 |
CCF Total=11 |
|
| Male, N(%) | 3(27.3%) | 10 (62.5%) | 19(55.9%) | 11(50%) | 11(45.8%) | 32(43.2%) | 7 (53.8%) | 10(45.5%) | 2 (50.0%) | 5 (45.4%) |
| Age at MRI scan, year | 20.3 (5–23) | 20.5 (5–36) | 20.7 (5–58) | 34.5 (23–49) | 27.3 (23–40) | 13.8 (5–21) | 36.5 (25–42) | 13.8 (5–21) | 38.0 (25–51) | 36.1 (15–55) |
| Children (age ≤18 years) | 7(63.6%) | 9(56.3%) | 17(50%) | 0 | 0 | 51 | 0 | 14 | ||
| Age at onset, year | 6.8 (0.17–19) | 6.90 (0.42–12) | 6.45 (0.5–35) | – | – | – | – | – | 17 (13–19) | 13.5 (1–48) |
| Epilepsy duration, month | 163.9 (12–204) | 167.4 (12–324) | 165.8 (24–540) | – | – | – | – | – | 252 (144–384) | 272.7 (84–516) |
| MRI scanners (all 3T) | GE discovery MR750 | Siemens Verio | Siemens Trio | GE discovery MR750 | Siemens Trio | GE discovery MR750(12); GE Signa HDX(15); Siemens Trio(39); Phillips Achieva(8) | Siemens Trio | GE discovery MR750 (1); GE Signa HDX(4); Siemens Trio (12); Phillips Achieva(5) | GE discovery MR750 | Siemens Trio |
PING: Pediatric Imaging, Neurocognition, and Genetics.
ROC analyses
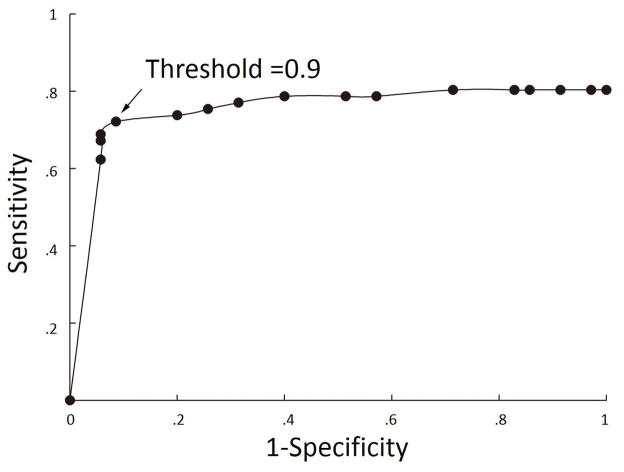
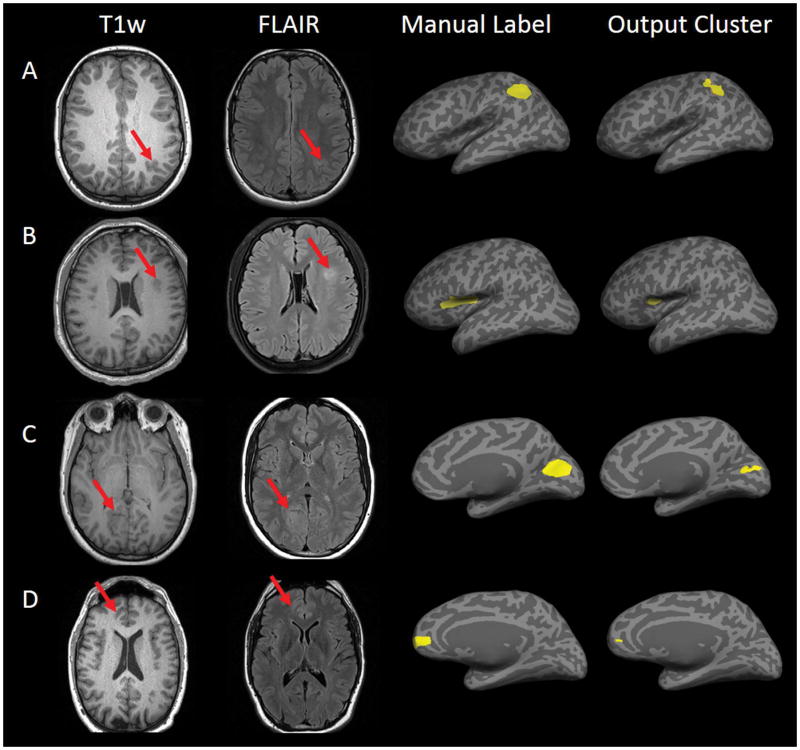
The value with which the output probability map from the neural network was thresholded had a marked effect on classifier sensitivity and specificity (Table 2). ROC analysis showed optimal overall performance at threshold of 0.9, where sensitivity of the whole group was 73.7% and specificity was 90.0% (91.4% specificity in healthy test group and 86.7% specificity in disease test group). No statistically significance was seen in the specificity of the healthy test group and disease test group across the analysis of factors impacting classifier performance. Hence, we combined the healthy test group and disease test group into one group in the following analysis. The area under the curve for the ROC analysis was 0.75, which suggested a good discriminative classifier (Figure 1). Figure 2 shows examples of successful detection. For the 45 patients with correctly identified lesions at the optimal threshold of 0.9, the mean percentage of overlap was 88.8% (SD=22.9%, range=1.6%–100%).
Table 2.
Sensitivity and specificity at different thresholds in controls (normal test controls and disease test controls) and patients showing the optimal threshold at 0.9
| Classifier threshold | Two test groups | FCD detected (Total=61) | Sensitivity | Specificity | Youden index | |
|---|---|---|---|---|---|---|
|
| ||||||
| Healthy test group detected (Total=35) | Disease test group detected(Total=15) | |||||
| 0.1 | 35 | 15 | 49 | 80.3% | 0 | −19.7% |
| 0.2 | 35 | 15 | 49 | 80.3% | 0 | −19.7% |
| 0.3 | 34 | 14 | 49 | 80.3% | 4.0% | −15.7% |
| 0.4 | 32 | 144 | 49 | 80.3% | 8.0% | −11.7% |
| 0.5 | 29 | 12 | 49 | 80.3% | 18.0% | −1.7% |
| 0.6 | 20 | 11 | 48 | 78.7% | 38.0% | 16.7% |
| 0.7 | 14 | 9 | 48 | 78.7% | 54.0% | 32.7% |
| 0.8 | 9 | 5 | 46 | 75.4% | 72.0% | 47.4% |
| 0.9 | 3 | 2 | 45 | 73.7% | 90.0% | 63.7% |
| 0.95 | 2 | 2 | 42 | 68.9% | 92.0% | 60.8% |
| 0.96 | 2 | 2 | 42 | 68.9% | 92.0% | 60.8% |
| 0.97 | 2 | 1 | 41 | 67.2% | 94.0% | 61.2% |
| 0.98 | 2 | 1 | 38 | 62.3% | 94.0% | 56.3% |
Figure 1.
Receiver Operator Characteristics (ROC) curve showing effects of classifier output threshold on sensitivity and specificity.
Figure 2.
Examples of 4 patients with a correctly detected lesion. First column: presurgical 3D T1-weighted images which were used as input to the processing. Second column: T2-weighted fluid-attenuated inversion recovery images on the same or closest slice. Third column: manual labels shown on inflated cortical surface. Fourth column: classifier cluster output shown on inflated cortical surface.
We further analyzed the 16 patients in whom the classifier did not correctly identify the lesions. In 8 patients the classifier did not output a probability map at the threshold of 0.9, i.e., the results were negative. For the other 8 patients, the lesion in one patient was detected as the 5th cluster, and in seven patients their lesions were not detected in any of the top 5 clusters.
Subgroup analyses
Sensitivity at the optimal threshold (0.9) showed similar results among three different centers (P=0.990), at 72.7% for SAHZU, 75% for BTH, 73.5% for CCF, respectively. In the pediatric group (<=18 years old), the sensitivity was 69.7% (23 of 33), which was lower than the sensitivity of the adult group (78.5%, 22/28), but did not reach statistical significance (p=0.562). Additionally, for the 44 MRI-positive patients, the detected clusters co-localized with the manual lesion in 36, yielding a sensitivity of 81.8%; for the 17 MRI-negative patients, a significantly lower proportion of the lesions were correctly detected (9/17, 52.9%, P=0.048).
Effectiveness of morphological features
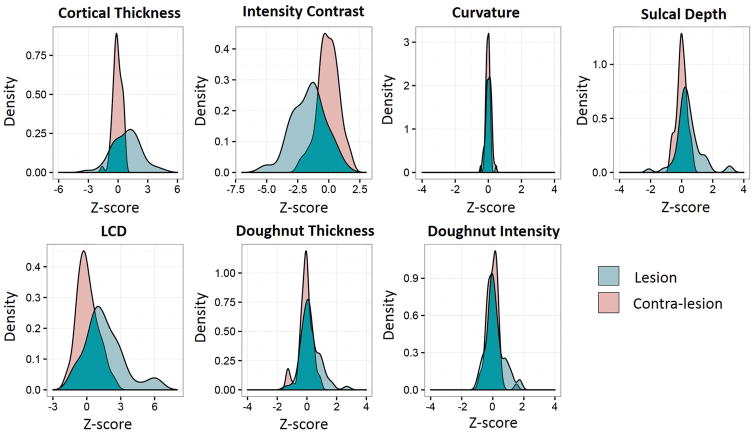
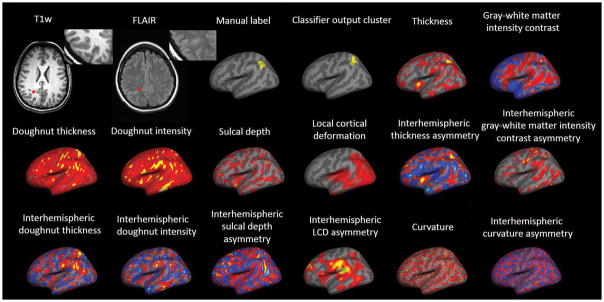
As shown in Figure 3, correct classification was largely based on gray-white matter intensity contrast, LCD and cortical thickness, which showed the most group-level difference as compared to the normal control group. Figure 4 shows an example of successful detection with illustration of all the features used for classification by the neural network classifier. In this particular case, thickness and gray-white matter intensity contrast features were the most helpful for the lesion detection.
Figure 3.
Quantitative evaluation of features in lesions as compared to healthy, homotopic cortex group. Kernel density plots showing the distribution of mean feature values in lesional and homotopic cortices across the cohort of FCD patients. Green = lesion profile. Pink = Contra-lesion profile (homotopic cortex). Homotopic cortex profile was calculated from 120 normal controls.
Figure 4.
Example patient showing measures generated by the surface-based morphometry approach in the parietal lobe. Note the multifocal appearance of each individual feature map. Information from each individual feature map was integrated by machine learning to generate a final cluster showing excellent concordance with expert labeling. Gray-white matter intensity measures blurring at the gray-white boundary; local cortical deformation (LCD) measures folding complexity; doughnut thickness measures local thickness variability; doughnut intensity measures local intensity variability at the gray-white boundary; interhemispheric gray-white matter intensity asymmetry measures the difference of bihemispheric blurring at the gray-white boundary; interhemispheric LCD asymmetry measures the difference of bihemispheric folding complexity; interhemispheric doughnut thickness measures the difference of bihemispheric local thickness variability; interhemispheric doughnut intensity measures the difference of bihemispheric local intensity variability at the gray-white boundary.
Impact of the number of training cases
When patients from SAHZU, BTH and CCF were used separately as training group, sensitivity was 54.5%, 75.0% and 61.7%, respectively; specificity was 76.0%, 60.0%, 86.0%, respectively. Optimal thresholds were all at 0.98 for all three groups.
Impact of the size of normal databases
When patient feature maps were normalized by smaller number of controls, i.e., SAHZU, CCF and PING separately, sensitivity was 50.8%, 50.8% and 68.9%, respectively; specificity was 94.0%, 94.0%, 92.0%, respectively. Optimal thresholds were 0.97, 0.97 and 0.90 for the three groups.
Evaluation of the role of scanner-specific normal database
When the training group (patients from SAHZU and CCF) was normalized by its corresponding SAHZU and CCF normal controls, sensitivity was 57.8% and specificity was 78.0% based on the best threshold (0.9).
Discussion
Detection of FCD lesions is crucial for epilepsy presurgical evaluation, as postoperative seizure outcomes in patients with a visible MRI lesion concordant with the clinical semiology and scalp EEG findings can be much improved compared to those without visible lesions.1 However, in clinical practice, one may miss some FCD type II lesions when localizing clinical semiology and scalp EEG findings are lacking. Automated MRI post-processing techniques to identify putative lesion locations can be essential in these situations. Our retrospective study reveals the usefulness of a surface based post-processing and machine learning method in automatically detecting FCD type II lesions (73.7% sensitivity), while maintaining a low probability of false positives (90.0% specificity). The robustness of this methodology was seen by similar performances based on MRI data sets from three different centers using three different 3T MRI scanners.
Contribution to literature
Recently, Hong et al. used a surface-based method coupled with multivariate approach which showed a high sensitivity (14 of 19, 74%) in automatic classifying FCD type II lesions in patients with MRI-negative epilepsy.12 No lesional vertices were identified in healthy and disease controls (patients with temporal lobe epilepsy), demonstrating excellent specificity.12 In the study by Ahmed et al. who reported another surface-based method,13 14 out of 24 MRI-negative patients with histologically proven FCD were correctly and automatically identified. In both studies, only a small group of adult patients were studied and pediatric patients were not included. Moreover, no differentiation between the FCD subtypes was provided in the latter study. Adler et al. delineated the SBM and neural network methods used in this current study14, and reported successful detection rate of 73% in 22 pediatric patients with radiological diagnosis of FCD; however, specificity could not be tested. In the present study, we evaluated the diagnostic value of an adapted version of the methods by Adler et al.14 in a large group of patients with histologically proven FCD type II including both children and adults. By systematically investigating the effects of classifier thresholds on sensitivity and specificity, ROC analysis was performed to identify an optimal classifier threshold (0.9). Based on this threshold, our methods showed high sensitivity of 73.7% in patients with FCD type II, while maintaining a high specificity (90.0%); both results were similar to previous studies.12–14 Moreover, our study tested the robustness of the methods on scans from different epilepsy centers and different MRI scanners. Subgroup analysis showed similar high sensitivity among three different epilepsy centers, and there was no statistically significant difference between children and adults. Overall, our findings provide evidence that the fully automated SBM and machine learning approach could offer a substantial gain of FCD detection in the presurgical evaluation for pharmacoresistant patients. The demonstrated robustness is key to the clinical application of our methods.
Size of the Training Group and Normal Control Database
Our results showed the sensitivity and specificity was largely influenced by the size of patients as training group and normal control database. The lower number of patients included in the training group, the lower sensitivity and specificity tended to be. This is intuitive because the classifier performance should improve as it learns idiosyncratic features in each training case, especially when the lesions are located in different brain lobes. Similarly, the lower number of normal controls included, the lower sensitivity and specificity tended to be. A larger normal control database would bring more normally distributed vertex feature values with lower standard deviations, which would likely help improve the accuracy of identifying abnormal vertices. This data highlights the importance of setting up a reasonably sized normal database and training database before clinical application of the methods.
Role of scanner-specific normal database
When we used scanner-specific normal database for the patients, the sensitivity was lower than using the average normal database. When using scanner specific normal database, one would typically expect increased sensitivity and specificity. We speculate that our findings are due to the reduced number of training cases and reduced size of normal database when the scanners needed to be matched, which masked the benefits brought by having the same scanner. This finding suggests that one should not be discouraged by not having a scanner-specific normal database before starting to use the proposed methods. A large combined training group and normal database, made publicly available, may be used instead.11
Effectiveness of Feature maps
In line with the previous study by Adler et al.,14 gray-white matter intensity contrast, LCD and cortical thickness were more sensitive to detect FCD than other features. These results can be explained by the fact that blurring of the gray-white matter junction, gyration shape and cortical thickness are typical MRI characteristic of FCD type II.2,3 Notably, multifocal appearance of individual features does not preclude the machine learning algorithm to successfully detect the lesion, as exemplified in Figure 4.
“MRI-negative” Cases
Additionally, about 30% of patients in our study were MRI-negative by initial visual inspection, but the lesions were picked up at the multimodal patient management conference by experienced team of experts; this team setup is not always available elsewhere. Our current data showed good sensitivity (52.9%) in this group of patients where the MRI was negative by initial visual inspection, suggesting that the use of type II lesions that are MRI-positive (obvious lesions) to train the classifier did benefit detection of the lesions that are initially not visually detected (subtle lesions), although with a lower sensitivity. It remains to be tested whether the more challenging type I lesions can be effectively detected with the current methods and training set. Given that the type I and type II FCD share some common radiological characteristics but not all30, significant method development is likely needed. This is further complicated by the fact that lesion labels, used as the “gold standard” for lesional vertices, are difficult to create with type I FCD. Even on pathological examination, previous studies showed low inter-rater agreement on the existence and subtype classification of type I FCD29.
False Positive Findings
In the present study, one challenge was the presence of positive clusters in normal test controls and extralesional clusters in patients (false positives). For the three healthy test controls in whom abnormal clusters were detected, two were located in the same region (right inferior temporal), the other was located in left mesial frontal lobe. For the two disease test controls who had false positive clusters, both clusters were located in the temporal lobe ipsilateral to the hippocampal sclerosis. In the 8 patients with FCD type II where clusters outside of the manual lesion masks were detected, five were located in the contralateral hemisphere, 3 were detected in the ipsilateral hemisphere but distant from the known lesions. The following factors could cause the existence of false positives: (1) errors could be made due to registration inaccuracy, motion artifact or bias field artifact; (2) frequent seizures could result in subtle abnormalities (e.g., atrophy) that may be difficult to distinguish from developmental aberrations;20 (3) structurally abnormal but dormant lesions were not uncommonly seen in epileptic brains.21–25 Thus, the findings of the post-processing methods should always be interpreted in conjunction with electroclinical characteristics. Future studies incorporating intracranial EEG could be used to determine whether there are abnormal electrophysiological characteristics associated with these “false-positive” regions.
Limitations and Future Directions
This study demonstrates the ability of automated tools to aid in the detection of focal cortical dysplasias. One limitation with the current study is that it is not ideally suited to assess the extent to which the lesions are correctly delineated. Accurate delineation of lesions would be invaluable to presurgical planning. In some patients, the histopathological changes may extend beyond visible MRI changes used to outline the lesions, in others the surgical resection which guided manual lesion delineation might exceed the lesion extent. Thus, we acknowledge that there is subjectivity and the potential for error in the manual lesion delineations. Due to the absence of a ground truth for each lesion label, it is impossible to assess the extent to which discrepancies between manual and automated segmentations are due to errors in the former or the latter. Careful validation with a cohort where comprehensive post-surgical histopathological analysis and coregistration with pre-operative MRI would be required to assess the extent to which the automated method is correctly identifying the lesion borders.
We did not use FLAIR data as a multivariate input, because of the unavailability of FLAIR data in our control subjects. Patients whose lesions only exhibit subtle signal change on FLAIR images may therefore have false negative results. Further sensitivity and specificity can be achieved by incorporating 3D FLAIR scans or normalized 2D FLAIR scans, as FLAIR intensity was reported to be the most discriminatory feature for detecting lesional vertices.14,26
It would be important for future studies to compare the effectiveness of various postprocessing methods reported in the literature through a multi-center data-sharing platform where FCD cases and control cases can be shared and tested. This will allow comparison of yields, sensitivity and specificity of the various postprocessing methods, as well as lesion characteristics. To this end, quantitative MRI maps that are more specific to tissue microstructure and can provide neuroimaging markers of tissue properties such as myelin, water and iron content would be useful.27,28 Future work incorporating features from quantitative MRI maps under the framework of machine learning is likely to improve automated lesion detection particularly in the case of subtle, MRI negative lesions.30
Conclusion
We demonstrated the usefulness of a surface-based MRI morphometry with machine learning using the largest-to-data cohort of pharmacoresistant patients with FCD type II, which show robust performance across cohorts from different centers and scanners. This freely available method can be a valuable tool to improve noninvasive presurgical evaluation for patients with pharmacoresistant epilepsy.
Acknowledgments
JB and SW were supported by the National Natural Science Foundation of China (81671282; 81671283; 91332202). SA received funding from the Rosetrees Trust. KW received funding from Neuroscience in Psychiatry Network (Wellcome Trust 095844/Z/11/Z).
Data collection and sharing for this project was partly funded by the Pediatric Imaging, Neurocognition and Genetics Study (PING) (National Institutes of Health Grant RC2DA029475). PING is funded by the National Institute on Drug Abuse and the Eunice Kennedy Shriver National Institute of Child Health & Human Development. PING data are disseminated by the PING Coordinating Center at the Center for Human Development, University of California, San Diego.
Footnotes
Conflicts of interest
None of the authors has any conflict of interest to disclose.
Ethical Publication Statement
We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
References
- 1.Lerner JT, Salamon N, Hauptman JS, et al. Assessment and surgical outcomes for mild type I and severe type II cortical dysplasia: A critical review and the UCLA experience. Epilepsia. 2009;50:1310–1335. doi: 10.1111/j.1528-1167.2008.01998.x. [DOI] [PubMed] [Google Scholar]
- 2.Krsek P, Maton B, Korman B, et al. Different features of histopathological subtypes of pediatric focal cortical dysplasia. Ann Neurol. 2008;63:758–769. doi: 10.1002/ana.21398. [DOI] [PubMed] [Google Scholar]
- 3.Sisodiya SM, Fauser S, Cross JH, et al. Focal cortical dysplasia type II: biological feature and clinical perspectives. Lancet Neurol. 2009;8:830–843. doi: 10.1016/S1474-4422(09)70201-7. [DOI] [PubMed] [Google Scholar]
- 4.Tassi L, Garbelli R, Colombo N, et al. Electroclinical, MRI and surgical outcomes in 100 epileptic patients with FCD type II. Epileptic Disord. 2012;14:257–266. doi: 10.1684/epd.2012.0525. [DOI] [PubMed] [Google Scholar]
- 5.Bernasconi A, Antel SB, Collins DL, et al. Texture analysis and morphological processing of magnetic resonance imaging assist detection of focal cortical dysplasia in extra-temporal partial epilepsy. Ann Neurol. 2001;49:770–775. [PubMed] [Google Scholar]
- 6.Antel SB, Bernasconi A, Bernasconi N, et al. Computational models of MRI characteristics of focal cortical dysplasia improve lesion detection. Neuroimage. 2002;17:1755–1760. doi: 10.1006/nimg.2002.1312. [DOI] [PubMed] [Google Scholar]
- 7.Colliot O, Bernasconi N, Khalili N, et al. Individual voxel-based analysis of gray matter in focal cortical dysplasia. Neuroimage. 2006;29:162–171. doi: 10.1016/j.neuroimage.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 8.Wagner J, Weber B, Urbach H, et al. Morphometric MRI analysis improves detection of focal cortical dysplasia type II. Brain. 2011;134:2844–2854. doi: 10.1093/brain/awr204. [DOI] [PubMed] [Google Scholar]
- 9.Wang ZI, Jones SE, Jaisani Z, et al. Voxel-based morphometric magnetic resonance imaging (MRI) postprocessing in MRI-negative epilepsies. Ann Neurol. 2015;77:1060–1075. doi: 10.1002/ana.24407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thesen T, Quinn BT, Carlson C, et al. Detection of epileptogenic cortical malformations with surface-based MRI morphometry. Plos One. 2011;6:e16430. doi: 10.1371/journal.pone.0016430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huppertz HJ, Wellmer J, Staack AM, et al. Voxel-based 3D MRI analysis helps to detect subtle forms of subcortical band hetertopia. Epilepsia. 2008;49:772–785. doi: 10.1111/j.1528-1167.2007.01436.x. [DOI] [PubMed] [Google Scholar]
- 12.Hong SJ, Kim H, Schrader D, et al. Automated detection of cortical dysplasia type II in MRI-negative epilepsy. Neurology. 2014;83:48–55. doi: 10.1212/WNL.0000000000000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmed B, Brodley CE, Blackmon KE, et al. Cortical feature analysis and machine learning improves detection of “MRI-negative” focal cortical dysplasia. Epilepsy Behav. 2015;48:21–28. doi: 10.1016/j.yebeh.2015.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adler S, Wagstyl K, Gunny R, et al. Novel surface features for automated detection of focal cortical dysplasias in paediatric epilepsy. Neuroimage Clin. 2016;14:18–27. doi: 10.1016/j.nicl.2016.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A. 2000;97:11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischl B, van der Kouwe A, Destrieux C, et al. Automatically parcellating the human cerebral cortex. Cereb Cortex. 2004;14:11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- 17.Fischl B, Salat DH, Busa E, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 18.Salat DH, Lee SY, van der Kouwe AJ, et al. Age-associated alterations in cortical gray and white matter signal intensity and gray to white matter contrast. Neuroimage. 2009;48:21–28. doi: 10.1016/j.neuroimage.2009.06.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pienaar R, Fischl B, Caviness V, et al. A methodology for analyzing curvature in the developing brain from preterm to adult. Int J imaging Syst Technol. 2008;18:42–68. doi: 10.1002/ima.v18:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fauser S, Sisodiya SM, Martinian L, et al. Multi-focal occurrence of cortical dysplasia in epilepsy patients. Brian. 2009;132:2079–2090. doi: 10.1093/brain/awp145. [DOI] [PubMed] [Google Scholar]
- 21.Tellez-Zenteno JF, Hernandez Ronquillo L, Moien-Afshari F, et al. Surgical outcomes in lesional and non-lesional epilepsy: a systematic review and meta-analysis. Epilepsy Res. 2010;89:310–318. doi: 10.1016/j.eplepsyres.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 22.McDonald CR, Hagler DJ, Ahmadi ME, et al. Regional neocortical thinning in mesial temporal lobe epilepsy. Epilepsia. 2008;49:794–803. doi: 10.1111/j.1528-1167.2008.01539.x. [DOI] [PubMed] [Google Scholar]
- 23.Mueller SG, Laxer KD, Barakos J, et al. Widespread neocortical abnormalities in temporal lobe epilepsy with and without mesial sclerosis. Neuroimage. 2009;46:353–359. doi: 10.1016/j.neuroimage.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Najm I, Jehi L, Palmini A, et al. Temporal patterns and mechanisms of epilepsy surgery failure. Epilepsia. 2013;54:772–782. doi: 10.1111/epi.12152. [DOI] [PubMed] [Google Scholar]
- 25.Labate A, Cerasa A, Aguglia U, et al. Neocortical thinning in “benign” mesial temporal lobe epilepsy. Epilepsia. 2011;52:712–717. doi: 10.1111/j.1528-1167.2011.03038.x. [DOI] [PubMed] [Google Scholar]
- 26.Cardinale F, Francione S, Gennari L, et al. Surface-projected fluid-attenuation-inversion-recovery analysis: a novel tool for advanced imaging of epilepsy. World Neurosurg. 2017;98:715–726. doi: 10.1016/j.wneu.2016.11.100. [DOI] [PubMed] [Google Scholar]
- 27.Weiskopf N, Suckling J, Williams G, et al. Quantitative multi_parameter mapping of R1, PD(*), MT, and R2(*) at 3T: a multi-center validation. Front Neurosci. 2013;7:95. doi: 10.3389/fnins.2013.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deoni SC, Dean DC, 3rd, Remer J, et al. Cortical maturation and myelination in healthy toddlers and young children. Neuroimage. 2015;115:147–161. doi: 10.1016/j.neuroimage.2015.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chamberlain WA, Cohen ML, Gyure KA, et al. Interobserver and intraobserver reproducibility in focal cortical dysplasia (malformations of cortical development) Epilepsia. 2009;50(12):2593–2598. doi: 10.1111/j.1528-1167.2009.02344.x. [DOI] [PubMed] [Google Scholar]
- 30.Adler S, Lorio S, Jacques TS, et al. Towards in vivo focal cortical dysplasia phenotyping using quantitative MRI. Neuroimage Clin. 2017;15:95–105. doi: 10.1016/j.nicl.2017.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]