Abstract
Like Arabidopsis thaliana, the flowering of the legume Medicago truncatula is promoted by long day (LD) photoperiod and vernalization. However, there are differences in the molecular mechanisms involved, with orthologs of two key Arabidopsis thaliana regulators, FLOWERING LOCUS C (FLC) and CONSTANS (CO), being absent or not having a role in flowering time function in Medicago. In Arabidopsis, the MADS-box transcription factor gene, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (AtSOC1), plays a key role in integrating the photoperiodic and vernalization pathways. In this study, we set out to investigate whether the Medicago SOC1 genes play a role in regulating flowering time. Three Medicago SOC1 genes were identified and characterized (MtSOC1a–MtSOC1c). All three MtSOC1 genes, when heterologously expressed, were able to promote earlier flowering of the late-flowering Arabidopsis soc1-2 mutant. The three MtSOC1 genes have different patterns of expression. However, consistent with a potential role in flowering time regulation, all three MtSOC1 genes are expressed in the shoot apex and are up-regulated in the shoot apex of plants in response to LD photoperiods and vernalization. The up-regulation of MtSOC1 genes was reduced in Medicago fta1-1 mutants, indicating that they are downstream of MtFTa1. Insertion mutant alleles of Medicago soc1b do not flower late, suggestive of functional redundancy among Medicago SOC1 genes in promoting flowering.
Keywords: flowering time, Medicago, photoperiod, vernalization, legume, genome evolution
Introduction
In annual plants, the transition from vegetative growth to flowering, termed floral induction, is regulated by environmental and endogenous cues to promote flowering in spring time (Srikanth and Schmid, 2011; Letswaart et al., 2012; Romera-Branchat et al., 2014). In the Brassica Arabidopsis thaliana, the key environmental cues which promote flowering are exposure to a prolonged period of cold (vernalization), followed by long day (LD) photoperiods. Endogenous signals such as carbohydrate status, gibberellin metabolism, developmental stage, and the autonomous floral promotion pathway also interact to promote flowering (Andres and Coupland, 2012; McClung et al., 2016; Cheng et al., 2017). In Arabidopsis, floral induction is repressed in non-inductive conditions by the MADS-box transcription factors FLOWERING LOCUS C (FLC) and SHORT VEGETATIVE PHASE (SVP) (Andres and Coupland, 2012). These floral repressors bind to regulatory elements of FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) and other genes, such as those involved in gibberellin metabolism (Hepworth et al., 2002; Searle et al., 2006; Tao et al., 2012; Mateos et al., 2015). Vernalization downregulates FLC expression by epigenetic and non-coding RNA-based mechanisms (Whittaker and Dean, 2017). SVP represses flowering under short day (SD) photoperiods but has reduced expression under LDs in the inflorescence meristem and under high ambient temperatures (Hartmann et al., 2000; Lee et al., 2007). Flowering is then promoted in LDs by the stabilization of the zinc finger transcription factor CONSTANS (CO) protein. CO is expressed in leaf phloem companion cells and activates the expression of the FT gene, which encodes a mobile florigen signal protein (Kardailsky et al., 1999; Wigge et al., 2005; Corbesier et al., 2007; Notaguchi et al., 2008). Studies in rice have shown that FT protein moves to the shoot apical meristem (SAM), whereupon it interacts with bZIP transcription factor FLOWERING D (FD) via a 14-3-3 protein-mediated complex (Abe et al., 2005; Taoka et al., 2011) to activate expression of the MADS box genes SOC1 and floral meristem identity (FMI) gene APETALA 1 (AP1) (Abe et al., 2005; Wigge et al., 2005; Yoo et al., 2005), thereby inducing the floral transition.
In Arabidopsis, SOC1 is a ‘floral integrator’ gene that perceives inputs from the vernalization, LD photoperiodic, and gibberellin pathways to promote flowering (Lee et al., 2000; Onouchi et al., 2000; Samach et al., 2000; Hepworth et al., 2002; Moon et al., 2003; Yoo et al., 2005; Helliwell et al., 2006; Searle et al., 2006; Sheldon et al., 2006; Jung J.H. et al., 2012; Hou et al., 2014). SOC1 is also regulated post-transcriptionally (Song et al., 2009; Kim et al., 2013). Upon activation at the shoot apex, the SOC1 protein binds its own regulatory sequences (Immink et al., 2012; Tao et al., 2012) and interacts with AGAMOUS-LIKE 24 (AGL24), for translocation to the nucleus, thereby providing positive feedback from inductive floral cues (Lee et al., 2008; Liu et al., 2008; Torti and Fornara, 2012). Proper integration of inductive floral cues by SOC1 is, therefore, an important step in regulating floral induction in Arabidopsis.
Despite strong conservation of flowering time genes between Arabidopsis and legume species, differences in the regulation and function of genes controlling flowering in legumes are becoming progressively more apparent (Weller and Ortega, 2015). Medicago truncatula (Medicago, Mt) and the garden pea (Pisum sativum, Ps), are vernalization-responsive, long-day annual legume species which are emerging models for flowering time studies in the agronomically important Fabaceae family (Clarkson and Russell, 1975; Laurie et al., 2011; Putterill et al., 2013; Weller and Ortega, 2015). The most striking difference is the absence of a clear FLC ortholog in legume species (Hecht et al., 2005). Expansions of flowering time genes are typical, such that SVP, CO-LIKE (COL), FT, SOC1, and FRUITFUL (FUL) orthologs typically occur in multi-gene families in Medicago, garden pea, and soybean (Glycine max, Gm) (Hecht et al., 2005; Kim et al., 2012; Jung C.H. et al., 2012; Putterill et al., 2013). Yet, differences in gene function seem to have arisen – MtSVP genes do not delay flowering when over-expressed in transgenic Medicago (Jaudal et al., 2014), and MtCOL genes do not appear to regulate photoperiodic flowering (Wong et al., 2014). Insights have been gained into the function and regulation of garden pea and Medicago FT genes by inductive seasonal cues. PsFTa1/GIGAS is a LD photoperiod inducible and graft-transmissible mobile signal which promotes flowering in garden pea (Hecht et al., 2011). In Medicago, MtFTa1 is the sole target of the vernalization pathway and is rapidly up-regulated in leaves in response to LD photoperiod (Laurie et al., 2011). Transgenic Medicago over-expressing MtFTa1 flowers very early, while loss-of-function fta1-1 mutants flower late and no longer respond to vernalization (Laurie et al., 2011). Medicago spring mutants flower early without vernalization and have elevated levels of MtFTa1 when grown in LD photoperiods (Jaudal et al., 2013; Yeoh et al., 2013). However, it is unclear how vernalization or LD activates Medicago FT expression and how FT triggers flowering.
Here, we examined the role of three Medicago SOC1 homologs (MtSOC1a, MtSOC1b, and MtSOC1c). Our study indicates that all three MtSOC1s are up-regulated by favorable seasonal cues in the shoot apex, via both MtFTa1-dependent and MtFTa1-independent pathways, and likely play a role in the regulation of Medicago flowering.
Materials and Methods
Sequence and Phylogenetic Analyses
MtSOC1 cDNA sequences were obtained as described previously (Hecht et al., 2005). AtSOC1 was used as a query for a BLASTP search of GenBank. Predicted SOC1 amino acid sequences belonging to Rosid species were aligned using the MUSCLE algorithm (Edgar, 2004) in Geneious software version 10.0.2 (Biomatters). A neighbor-joining phylogenetic tree was constructed using aligned amino acid residues 61–170, which excluded the variable C-terminal domain and the conserved MADS DNA-binding domain.
Plant Materials and Growth Conditions
Medicago truncatula wild-type accessions Jester and R108 were grown as previously described (Laurie et al., 2011). fta1-1 (R108 background) was described by Laurie et al. (2011). Tnt1 retroelement insertions in the R108 background at the MtSOC1b locus (Medtr8g033250) were sourced from the Noble Foundation collection (Tadege et al., 2008). Primers for genotyping and quantitative real-time RT-PCR (qRT-PCR) are listed in Supplementary Table 1. Flowering time was scored by counting the number of nodes to the first flower on the main stem. Arabidopsis thaliana soc1-2 (Columbia) mutants were described in Lee et al. (2000). Arabidopsis plants were grown and flowering time scored by counting the total number of rosette leaves at flowering (Laurie et al., 2011).
Generation of Binary Constructs and Arabidopsis Transformation
The DNA was amplified by PCR using Platinum High Fidelity Taq DNA polymerase (Invitrogen) and cloned into binary expression constructs by Gateway® LR recombination technology. Construct integrity was verified by Sanger sequencing and restriction digest. Arabidopsis soc1-2 (Col) (Lee et al., 2000) was transformed in accordance with Narusaka et al. (2010) with Agrobacterium tumefaciens GV3101 harboring the constitutive expression (CaMV 35S) construct pB2GW7 (Karimi et al., 2002), incorporating the coding sequences of AtSOC1, MtSOC1a, MtSOC1b, and MtSOC1c. Transgenic plants were selected on soil by spraying with 0.02% BASTA (glufosinate).
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was isolated from Medicago and Arabidopsis and 1 μg used for cDNA synthesis, as described previously (Laurie et al., 2011). qRT-PCR was performed in 10 μL reactions using 3 μL of cDNA diluted 1/30, with Roche SYBR Green I master mix, in the Roche LightCycler 480 instrument. Multi-well plates were loaded using a CAS1200 PCR robot. Calculation of the relative gene expression levels was based on the ΔΔCt method (Pfaffl, 2001). In Medicago, the data were normalized to the reference gene, PROTODERMAL FACTOR 2 (PDF2) (Kakar et al., 2008). In Arabidopsis, the data were normalized to ACTIN2 (Lee et al., 2013). See Supplementary Table 1 for qRT-PCR primer sequences.
Statistical Analyses
Differences between means for treatments in gene expression data, and for flowering times of transgenic lines compared to the Arabidopsis soc1-2 mutant, were assessed by one-way ANOVA at the 0.05 significance level, with corrections for multiple comparisons as stated, using GraphPad Prism version 7.0.
Results
An Ancient Genome Duplication Has Resulted in Two Distinct Classes of SOC1 Genes in Legumes
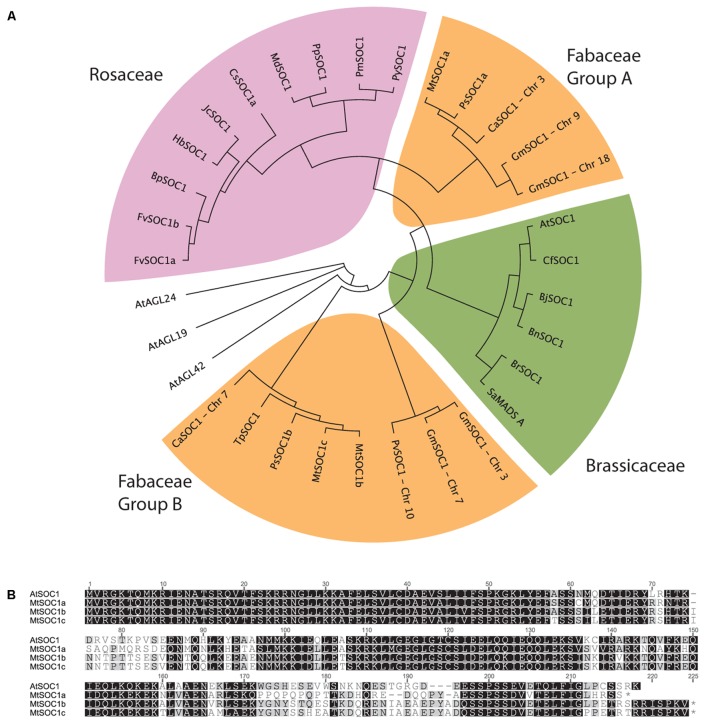
The full-length AtSOC1 amino acid sequence was used as a BLASTP query to search for predicted SOC1 orthologs in GenBank. Sequences from 21 Rosid species were retrieved and aligned using the MUSCLE algorithm. A region 110 amino acids long corresponding to positions 61–170 (spanning the Intervening and Keratin domains, excluding the highly conserved MADS DNA-binding domain and highly variable C-terminal domain) was used to generate a neighbor-joining phylogenetic tree (Figure 1A). As a control, related AGL sequences (AGL19, AGL24, and AGL42) from Arabidopsis were included and did not cluster with any of the predicted SOC1 orthologs. Two distinct classes of SOC1 proteins were observed for Fabaceae members. We have named them Fabaceae Group A and B SOC1s (Figure 1A). Medicago has three SOC1 genes, MtSOC1a (Medtr07g075870), MtSOC1b (Medtr08g033250), and MtSOC1c (Medtr08g033220) (Hecht et al., 2005). All three MtSOC1 (c.v. R108) genes encode proteins sharing 65–67% amino acid identity with AtSOC1 and were identical over the N-terminal MADS domain that is important for DNA-binding activity (Figure 1B). MtSOC1b and MtSOC1c proteins are highly similar to one another, sharing 93% identity (Figure 1B).
FIGURE 1.
Phylogenetic relationships among SOC1 proteins. (A) Phylogeny of Rosid SOC1 proteins showing two distinct clades of Fabaceae SOC1, based upon a neighbor-joining tree from a MUSCLE alignment of residues 61–170, comprising a region spanning the Intervening and Keratin domains. Related A. thaliana AGAMOUS-LIKE sequences were used as outgroups. Species abbreviations: At: Arabidopsis thaliana, Bj: Brassica juncea, Bn: Brassica napus, Bp: Betula platyphylla, Br: Brassica rapa, Ca: Cicer arietinum, Cf: Cardamine flexuosa, Cs: Citrus sinensis, Fv: Fragaria vesca, Glyma: Glycine max, Hb: Hevea brasiliensis, Jc: Jatropha curcas, Md: Malus x domestica, Mt: Medicago truncatula, Phvul: Phaseolus vulgaris, Pm: Prunus mume, Pp: Pyrus pyrifolia, Py: Prunus x yedoensis, Ps: Pisum sativum, Sa: Sinapis alba, Tp: Trifolium pratense. (B) Alignment of full-length Arabidopsis thaliana (Col-0) and Medicago (R108 accession) SOC1.
To investigate the evolution of the two groups of legume SOC1 genes, we examined their genomic regions in diverse legume species. This revealed that a genomic region containing approximately 20 genes had been duplicated (Supplementary Figure 1). The synteny of this duplicated region has remained largely conserved. However, there are differences which can be used to determine which region likely contains the Group A or B SOC1 genes. For example, the region containing MtSOC1b also includes a predicted deacetylase gene (Medtr08g033340) that is not present in the region with the MtSOC1a locus (a Group A SOC1). All the legume species we examined possessed this duplication, with the duplication existing in diverse taxonomic clades of papilionoid legumes; the phaseoloid or warm season legumes (such as soybean, common bean, cowpea, and pigeon pea), the galegoid or temperate legumes (Medicago, garden pea, and chickpea), and in lupin and peanut species, which belong to more distantly related clades (Genistoids and Dalbergioids, respectively). This indicates that the duplication likely occurred during the early evolution of the legume family, likely as a result of a whole genome duplication (WGD) event that occurred about 58 million years ago during the early evolution of the papilionoid legumes (Bertioli et al., 2009). Also in M. truncatula, there is a duplication of the Group B SOC1 genes, resulting in MtSOC1b and MtSOC1c genes (Figure 1A, Supplementary Figure 1). This duplication is also present in M. sativa (Ms, alfalfa) (Supplementary Figure 2), but is absent in the other legumes, indicating that it occurred more recently.
The MtSOC1 Genes Can Partially Complement an Arabidopsis soc1-2 Mutant
To investigate the function of the three MtSOC1 genes, the ability of each gene to complement the Arabidopsis late-flowering soc1-2 (Col) mutant (Lee et al., 2000) was examined. A series of transgenic plant lines expressing the coding sequence of each MtSOC1 gene under the control of the constitutive CaMV 35S promoter were generated, in the soc1-2 (Col) mutant background. For each MtSOC1 gene, multiple transgenic lines were identified that promoted the flowering of soc1-2 mutant to varying extents, as inferred by flowering times statistically significantly different to soc1-2 (Figures 2A,B). As a control, the Arabidopsis SOC1 gene was also re-introduced into the soc1-2 mutant. Only one of seven lines (line 1) expressing AtSOC1 completely rescued the soc1-2 late-flowering mutant phenotype, meanwhile three lines partially rescued (lines 4, 6, and 7 flowered statistically significantly earlier than soc1-2) (Figure 2C). To investigate the possibility that transgene expression level contributed to the variation in flowering time observed, we isolated RNA from seedling tissues of additional transgenic lines containing 35S:MtSOC1 cassettes in the soc1-2 background and examined transgene expression levels by qRT-PCR. Generally, the earlier flowering lines had higher levels of SOC1 expression (Supplementary Figure 3). Overall, these results indicate that all three MtSOC1 genes can promote flowering and are functionally equivalent to the Arabidopsis SOC1 gene.
FIGURE 2.
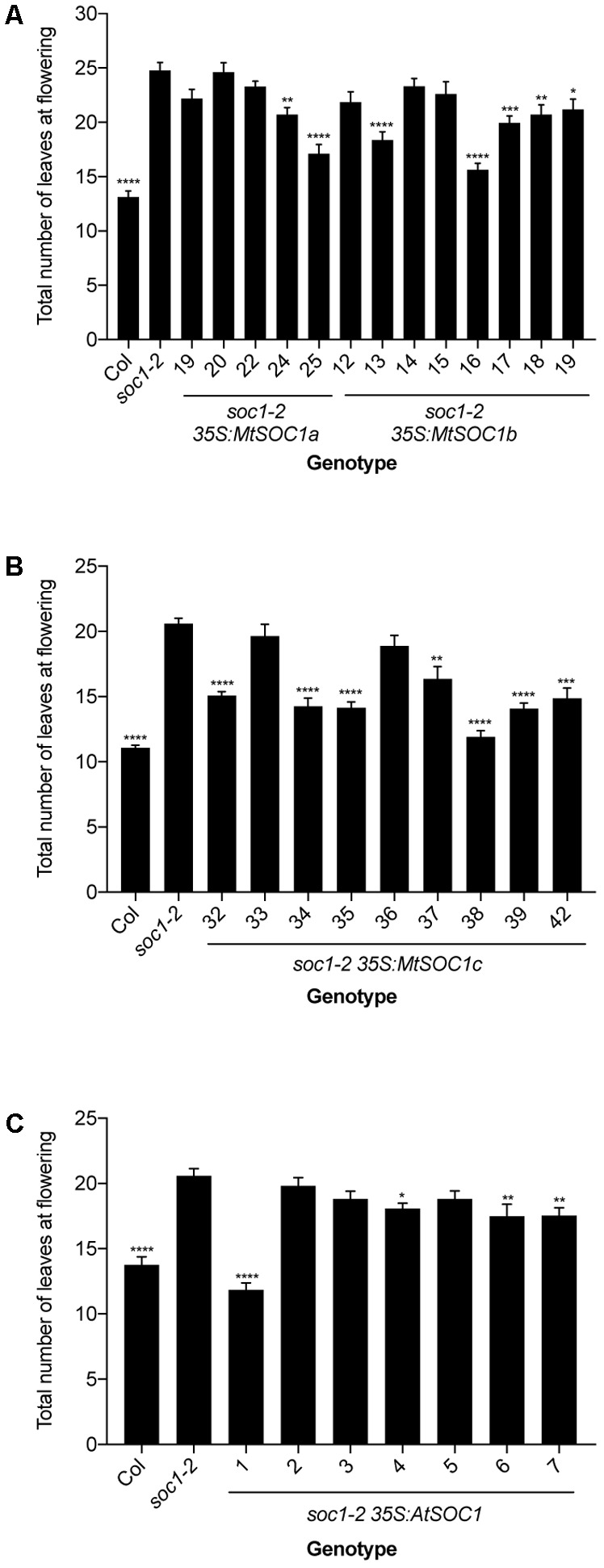
Complementation of late-flowering Arabidopsis soc1-2 (Col) mutants by Medicago SOC1 genes. Flowering time phenotypes of genetically independent transgenic plant lines ectopically expressing SOC1 transgenes in the soc1-2 mutant background. (A) Flowering time of homozygous T3 transgenic lines harboring 35S:MtSOC1a or 35S:MtSOC1b cassettes. (B) Flowering time of homozygous T3 transgenic lines harboring a 35S:MtSOC1c cassette. (C) Flowering time of T2 transgenic lines harboring a 35S:AtSOC1 cassette. Data are mean ± SD of at least 12 plants grown under 16 h illumination at ∼120 μE.m2.s-1 light intensity at 22°C day time/18°C night time. Statistically significant differences between means of soc1-2 versus each other genotype were determined by one-way ANOVA. Asterisks, where annotated, denote P-values. ∗P ≤ 0.05, ∗∗P ≤ 0.01, ∗∗∗P ≤ 0.001, ∗∗∗∗P ≤ 0.0001.
The MtSOC1 Genes Have Distinct Expression Profiles
Next, the expression pattern of the three MtSOC1 genes was analyzed by qRT-PCR using primers specific for each MtSOC1 gene (Supplementary Figure 4). All three MtSOC1 genes are expressed (Figure 3). MtSOC1a is expressed in vegetative tissues (leaves, stem, apical nodes), but is almost undetectable in reproductive tissues (floral buds, flowers, and seed pods) (Figure 3A). In contrast, MtSOC1b and MtSOC1c have a similar but not identical pattern of expression, and both are detected in vegetative tissues, in floral buds, and in flowers (Figures 3B,C). Consistent with a potential role in floral induction, all three MtSOC1 genes are expressed under inductive environmental conditions in apical node tissue, containing the shoot apical meristem (SAM) (Figure 3). All three MtSOC1 genes are also expressed in stem internode tissue, with MtSOC1b and MtSOC1c being expressed at the highest level in this tissue.
FIGURE 3.
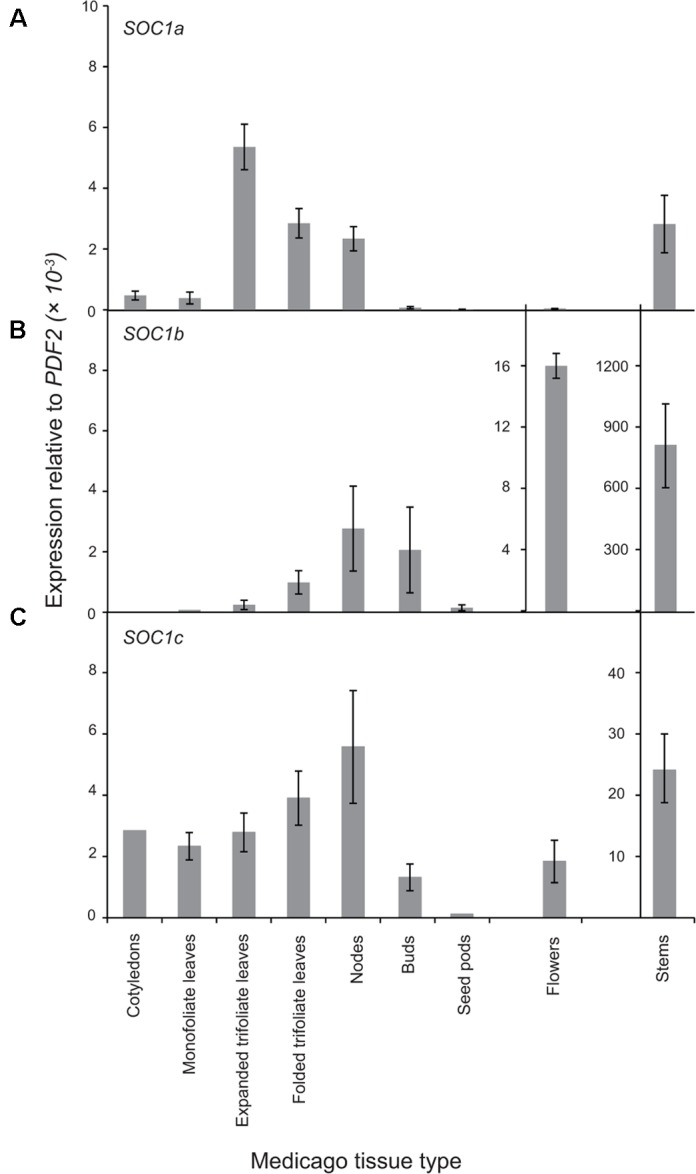
Expression pattern of MtSOC1 genes. (A) MtSOC1a, (B) MtSOC1b, and (C) MtSOC1c. RNA was isolated from various tissues of WT c.v. Jester plants grown under inductive conditions. Vegetative tissues were harvested 15 days after sowing and reproductive tissues 35 days after sowing in soil. Data are the mean ± SE of three biological replicates and transcripts were normalized to PDF2. All tissues were harvested at ZT 2.
To determine whether the Group A and B SOC1 genes also have distinct patterns of expression in other plants, we first examined available RNAseq data from Medicago sativa (The Alfalfa Gene Index and Expression Atlas Database1). This data also revealed that the three alfalfa SOC1 genes are expressed at their highest levels in stem internodes. MsSOC1a was expressed at relatively low levels in leaves compared with the MsSOC1b and MsSOC1c genes. For all three MsSOC1 genes, low or no expression was detected in developing flowers, nor root nodules, while modest expression was found in roots (Supplementary Figure 5A). Next, we examined the expression of the soybean SOC1 genes in the RNA Seq Atlas database available at https://soybase.org/ (Severin et al., 2010). This revealed that GmSOC1b from chromosome 3 is only expressed at low levels in the tissues examined (young leaves, flowers, developing seeds, roots, and nodules), whereas the other GmSOC1b (from chromosome 7) and the Group A GmSOC1s from chromosomes 9 and 18 had somewhat similar patterns of expression, with low levels of expression in developing seeds, and modest expression in the other tissues (Supplementary Figure 5B).
Given that the legume SOC1 genes have similar patterns of expression, we examined their promoters with the aim of identifying conserved regulatory elements. Short regions of high conservation were identified within the various legume promoters (Supplementary Figure 6A). Examination of these conserved regions revealed potential transcription factor-binding sites. Notably, there were multiple MADS-box transcription factor-binding sites (Supplementary Figure 6B). In Arabidopsis, the SOC1 promoter is bound by the MADS-box proteins, FLC, SVP, and SOC1 itself (Searle et al., 2006; Immink et al., 2012; Tao et al., 2012; Mateos et al., 2015). Potential SPL, WRKY, and bHLH transcription factor-binding sites were also found in the legume SOC1 promoter regions (Supplementary Figure 6B).
MtSOC1b and MtSOC1c Up-regulation in Response to Inductive Conditions Requires MtFTa1
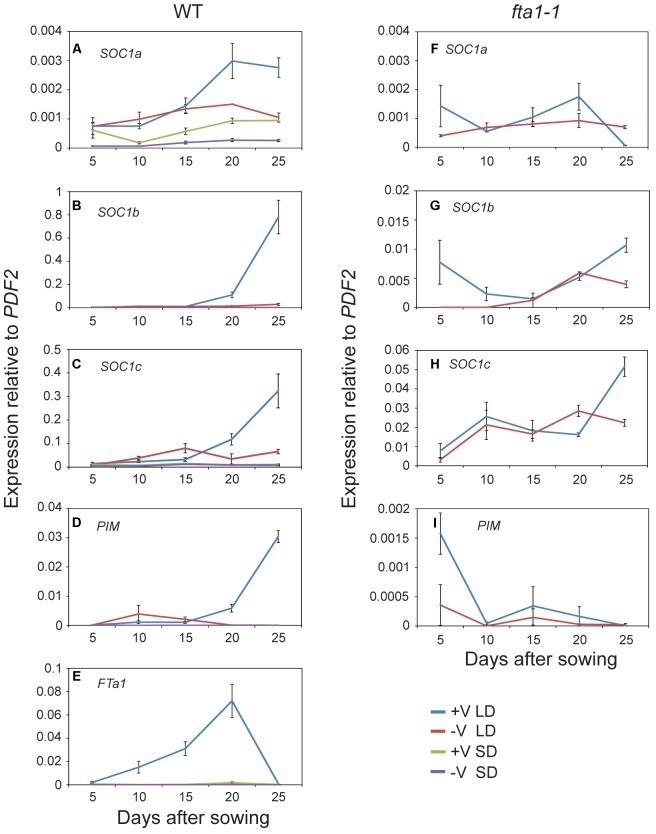
Given the potential role of the MtSOC1 genes in flowering time, we examined their expression during development under different environmental conditions. As Medicago flowering is promoted by both vernalization and LD photoperiods, wild-type plants (accession R108) were grown with and without vernalization (± V) under LD (16 h light:8 h dark) and SD (8 h light:16 dark) photoperiods. Germinated seeds of all treatments were sown into soil at the same time. Gene expression was examined in aerial tissues (excluding cotyledons) for samples collected at days 5–15 after sowing, and in apical nodes at days 20 and 25 (Figure 4 and statistical analysis of this data is shown in Supplementary Figure 7). MtSOC1a was the lowest expressed MtSOC1 gene throughout the experiment, but was expressed at the highest levels in the vernalized LD samples at days 20 and 25 (Figure 4A). MtSOC1b was the most strongly expressed gene under inductive conditions in WT and was induced by day 20 (Figure 4B), with high expression by day 25. In contrast, MtSOC1b was relatively weakly expressed under SD (Figure 4B). However, it did show higher expression under LD photoperiod even without vernalization, compared to vernalized and non-vernalized SD treatments (Supplementary Figure 8). MtSOC1c behaved in a similar fashion to MtSOC1b, being strongly up-regulated under inductive conditions in WT, although to a lesser extent (Figure 4C).
FIGURE 4.
Time course of MtSOC1 expression during development. (A–E) Expression of indicated genes in wild-type R108 and (F–I) in fta1-1 (R108 background). Wild-type plants were grown under LD and SD photoperiod, with and without vernalization. fta1-1 plants were grown under LD alone, with and without vernalization. All treatments were sown into soil at the same time. Aerial tissues of plants excluding cotyledons were harvested for days 5–15 and apical nodes only for days 20 and 25. Data are the mean ± SE of 2–4 biological replicates and transcripts were normalized to PDF2. All samples were harvested at ZT 2.
The role of MtFTa1 in the induction of the MtSOC1 genes was also addressed in this experiment, by examining the expression of MtSOC1 in the late-flowering and vernalization insensitive fta1-1 (R108) mutant background (Laurie et al., 2011). In fta1-1 mutants grown under LD, both MtSOC1b and MtSOC1c were expressed at similarly low levels irrespective of vernalization, and expression was dramatically attenuated compared to vernalized wild-type plants (for MtSOC1b and MtSOC1c approximately 100-fold, Figures 4G,H), suggesting both these genes are common targets of the vernalization pathway and are downstream of MtFTa1.
Expression of the FMI gene, MtPIM (Benlloch et al., 2006), was examined in all conditions tested (Figures 4D,I). Similar to MtSOC1 genes in WT plants grown under inductive conditions, MtPIM was up-regulated in apical nodes by day 20, thereby heralding the commitment to flowering, and confirming the timing of MtSOC1 up-regulation is consistent with these genes having a role in the floral transition at the apex. Furthermore, only vernalized LD conditions resulted in a gradual up-regulation of MtFTa1 in WT plants (Figure 4E), which is necessary to rapidly promote flowering under favorable environmental conditions (Laurie et al., 2011; Jaudal et al., 2013). In wild-type, MtFTa1 induction is concomitant with MtSOC1b and MtSOC1c activation in apical nodes, in both vernalization and LD photoperiod conditions, over the duration of this experiment.
Regulation of MtSOC1 by Photoperiod Shift
As the MtSOC1 genes are up-regulated under inductive LD photoperiods (Figure 4, Supplementary Figure 7), we sought to examine in greater detail the effect of photoperiod on MtSOC1 expression in apical nodes. Since exposure of SD-raised vernalized WT (accession R108) plants to just 3 LD is sufficient to commit Medicago to flowering (Laurie et al., 2011), we investigated whether MtSOC1 is induced in the SAM after this period of exposure to LD photoperiod, and the role of MtFTa1 in this activation (Figure 5 and statistical analysis of this data is shown in Supplementary Figure 9). In plants permanently grown under LD, which were harvested at the conclusion of the experiment (the equivalent of 3 SD time point), MtSOC1a and MtSOC1b were expressed at higher levels in WT than fta1-1 (Figures 5A,B). All three MtSOC1 genes were relatively weakly expressed in SD before shift (SD BS) conditions (Figures 5A–C). Upon transfer of WT plants into LD, MtSOC1a and MtSOC1b were up-regulated and remained high when transferred back to SDs (Figures 5A,B). An increase in MtSOC1a and MtSOC1b expression was also evident when fta1-1 mutant plants were shifted into LDs, and generally this increase was smaller than for WT plants (although none of the changes in expression were statistically significant; Supplementary Figure 9). Although MtSOC1c is expressed at higher levels in LD grown plants, compared with SD grown plants (Figure 4C), no statistically significant up-regulation in MtSOC1c expression was observed over the six days of this experiment (Figure 5C). To check the LD exposure was sufficient to induce markers of the floral transition, we examined the expression of the FMI genes, MtPIM and MtFULc (Benlloch et al., 2006; Jaudal et al., 2015), in WT of pre-shift SD samples, and in samples harvested at the conclusion of the experiment, which had seen both 3 LD and subsequently 3 SD (Figure 5D). MtPIM expression remained unchanged between these time points, while MtFULc was activated following return to SD, suggesting that MtFULc is likely activated before MtPIM.
FIGURE 5.
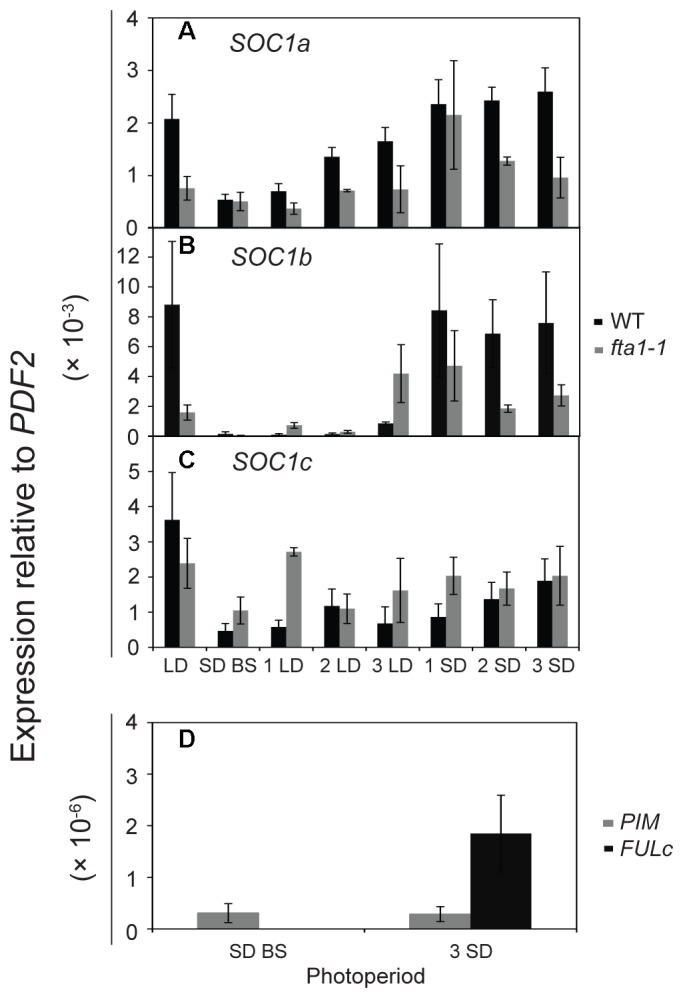
Regulation of MtSOC1 in apical nodes by changes in photoperiod. Expression of (A) MtSOC1a, (B) MtSOC1b, (C) MtSOC1c, and (D) MtPIM and MtFULc. Vernalized WT (c.v. R108, black) and fta1-1 (R108, gray) plants were grown under SD photoperiod until the monofoliate leaf had appeared (∼6 days). Plants were sampled [SD-BS refers to SD sample collected before shifting (BS) into LDs] and then transferred to LD photoperiod for sampling for 3 days (1 LD, 2 LD, and 3 LD). The remaining plants were then returned to SD for 3 days and sampled each day (1 SD, 2 SD, and 3 SD). LD control plants were grown permanently under LD and harvested at the conclusion of the experiment. Samples are the mean ± SE of 3 biological replicates and transcripts were normalized to PDF2. All tissues were harvested at ZT 4 in both light regimes.
Overall, our results indicated that LDs promote the expression of the three MtSOC1 genes (Figures 4, 5). However, the dynamics of this LD response varied between the MtSOC1 genes, with MtSOC1b showing the largest increase in expression after exposure to three LDs, MtSOC1a showing a moderate increase and MtSOC1c showing little response (Figure 5). This rapid induction of MtSOC1a and MtSOC1b is not eliminated in the fta1-1 mutant (Figure 5), likely due to the involvement of other LD-inducible MtFT genes, such as MtFTb1(Laurie et al., 2011). In contrast, the large increase in MtSOC1a and MtSOC1b expression is probably associated with floral induction (as indicated by the up-regulation of the FMI gene, MtPIM) and is not seen in the fta1-1 mutants which flower late and are yet to express MtPIM (Figure 4). Consistent with this idea, MtSOC1b expression begins to increase at later in development in the fta1-1 mutant (Supplementary Figure 8).
Medicago soc1b Mutants Do Not Flower Late
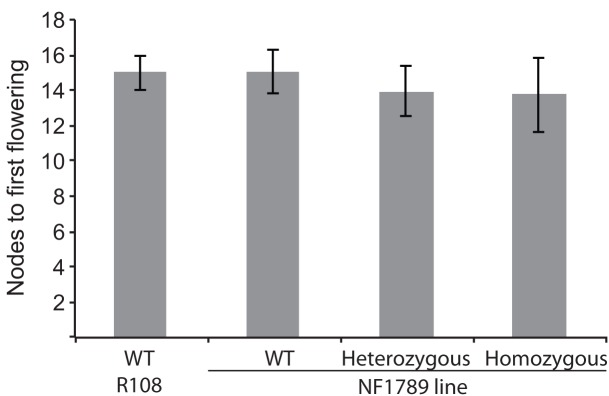
To identify mutations within the MtSOC1 genes, a reverse genetics approach was employed to screen for Tnt1 retrotransposon-tagged insertion mutants, sourced from the Noble Foundation collection (Tadege et al., 2008) by PCR. Two independent lines in the c.v. R108 background were found to have Tnt1 insertions within exon 7 of MtSOC1b (Supplementary Figure 10A). Homozygous plants for an insertion annotated as NF1789 were identified using PCR primers binding within the MtSOC1b gene and the Tnt1 insertion. No qRT-PCR products were detected using primers across the insertion site, indicating that the Tnt1 sequences prevent a correctly spliced mRNA from being produced (Supplementary Figure 10B). Homozygous NF1789 plants flowered at a similar time to wild-type (R108), when grown under inductive conditions (Figure 6). Moreover, a second independent insertion mutant, line NF9471 (also with a Tnt1 insertion within exon 7), also flowered at a similar time to wild-type (Supplementary Figure 10C).
FIGURE 6.
The soc1b mutant does not flower late. Flowering time of vernalized wild-type R108 and insertion mutant line NF1789 grown under LD photoperiod. Individual plants of a segregating NF1789 line were genotyped to determine if they were wild-type, heterozygous or homozygous mutants. Data are mean ± SE of 8 or more plants. There is no statistically significant difference between the flowering times of the lines.
Discussion
While detailed knowledge of how flowering time is regulated exists for Arabidopsis, a member of the Brassica family, less is known about how flowering is regulated in other plant families. There are several differences between Arabidopsis and legumes relating to flowering time control, with evolutionary and genetic data indicating the likely involvement of legume-specific genes and mechanisms. For example, legumes lack a clear ortholog of the key Arabidopsis vernalization-responsive gene, FLC (Hecht et al., 2005) and the ability to respond to vernalization has likely evolved independently in legumes and other plant families (Bouché et al., 2017). Orthologs of the key gene in the Arabidopsis photoperiodic flowering pathway, CONSTANS (CO) do not function to control flowering of Medicago (Wong et al., 2014). Here, we set out to investigate the SOC1 genes of Medicago truncatula and their role in the control of flowering time.
The SOC1 gene plays an important role in Arabidopsis flowering by integrating multiple floral cues within the apex to promote flowering at the appropriate time. We have found that legumes contain two classes of SOC1 genes, which we refer to as Group A and Group B SOC1s, as a result of a duplication of a region containing ∼20 genes. This duplication is present in diverse legumes and likely a result of the WGD that occurred during the early evolution of the papilionoid legumes about 58 million years ago (Bertioli et al., 2009). This WGD has been important in the evolution of various legume-specific traits allowing the duplicated genes to acquire new functions, while the other gene copy maintains its original function (Young and Bharti, 2012). In the case of the two groups of SOC1 genes, they differ in their amino acid sequences and patterns of expression, indicating that they might have evolved distinct functions.
Despite the duplication that resulted in two groups of legume SOC1s and the more recent Medicago-specific tandem duplication, all three MtSOC1 genes are able to promote the flowering of the Arabidopsis soc1-2 mutant. Similar phenotypes have also been reported for the two Group A GmSOC1 genes, with GmGAL1/GmSOC1 (Glyma18g224500) being able to partially complement the late-flowering soc1-1 (Ler) mutant, and causing early flowering when constitutively expressed in a wild-type Arabidopsis (Col) background (Zhong et al., 2012). GmSOC1-like, the other Group A member (Glyma09g266200), also resulted in early-flowering when heterologously expressed (Na et al., 2013). Similar to Arabidopsis and GmSOC1 genes (Lee et al., 2000; Samach et al., 2000; Zhong et al., 2012; Na et al., 2013), Medicago SOC1 genes are expressed in a range of tissues, though at differing levels. All three MtSOC1 genes are expressed in apical node tissue, consistent with a potential role in control of flowering time, as reported for GmSOC1 genes under inductive SD conditions (Na et al., 2013). MtSOC1b and MtSOC1c are also expressed at relatively high levels in stem tissue, indicating that they might also play a role in other aspects of Medicago development, such as internode elongation.
All three MtSOC1 genes were up-regulated by inductive conditions of LD and vernalization, in a spatio-temporal manner concomitant with MtFTa1 and MtPIM induction in leaves and shoot apices, respectively. Medicago fta1-1 mutants do not respond to vernalization (Laurie et al., 2011), indicating that MtFTa1 is an important and perhaps the sole direct target of the vernalization process. Consistent with this hypothesis, there are no clear differences between the expressions of the different MtSOC1 genes in an fta1-1 mutant compared to wild-type grown without vernalization. This suggests MtSOC1 genes can be placed genetically downstream of MtFTa1 in the vernalization pathway. Supporting this conclusion, studies on early-flowering Medicago spring mutants, which show LD-conditional ectopic MtFTa1 expression, have higher levels of MtSOC1a expression in both leaves and apices, compared to wild-type (Jaudal et al., 2013). The requirement for MtFTa1 for vernalization, as well as MtSOC1 induction, has been further highlighted in Medicago vrn2 mutants that flower early under LD photoperiod without vernalization(Jaudal et al., 2016). vrn2-1 mutants show elevated levels of MtFTa1 under LD photoperiod, accompanied by increased levels of all three MtSOC1 genes in leaf tissue. vrn2-1 fta1-1 double mutants flowered late like fta1-1 single mutants, which was accompanied by a suppression of MtSOC1 expression in the double mutant back to levels approaching wild-type (Jaudal et al., 2016). Taken together, these data from us and others clearly demonstrate that both MtFTa1 and vernalization are essential for maximal MtSOC1 induction, in response to favorable seasonal cues.
MtSOC1 expression is also up-regulated by LD photoperiod in the absence of vernalization (therefore with low MtFTa1 expression) (Figure 4 and Supplementary Figure 7). We have previously shown that shifting Medicago plants from non-inductive SDs into inductive LDs for just 3 days, is sufficient to both up-regulate two functional MtFT genes, MtFTa1 and MtFTb1, and to commit to flowering in plants that are subsequently returned back SD photoperiod (Laurie et al., 2011). MtFTa1 and MtFTb1 are both rapidly up-regulated in the leaves, while MtFTa1 is also up-regulated in the shoot apex. The expression of MtFTa1 and MtFTb1 rapidly declines upon returning plants to SD photoperiod (Laurie et al., 2011). Here, we show that 3 days of MtFTa1 expression is sufficient to induce the expression of MtSOC1a and MtSOC1b genes (Figures 5A,B). MtSOC1a and MtSOC1b expression also remains at higher levels when the plants are shifted back into SDs. Consistent with both MtFTa and MtFTb genes having a role in LD induction of flowering in legumes (Hecht et al., 2011; Laurie et al., 2011), this induction was reduced, but not eliminated, in the fta1-1 mutant background. The rapid induction of MtSOC1a and MtSOC1b suggests that they might be early targets of MtFT genes. In Arabidopsis, SOC1 is also rapidly up-regulated during the floral transition (Samach et al., 2000). The FMI gene, MtFULc (Jaudal et al., 2015), was also induced six days after plants were first transferred into LDs, however, the Medicago ortholog of AP1, MtPIM, remained unchanged. MtSOC1c also did not show strong up-regulation upon exposure to LD in the photoperiod shift experiment, whereas in the time course experiment both MtSOC1c and MtPIM were up-regulated (Figure 4). This raises the possibility that these two genes could be more important slightly later during the Medicago floral transition. Following flowering, the expression of both MtSOC1b and MtSOC1c remained at high levels in floral nodes, indicating a potential role in floral development.
Overall, the results of this study indicate that Medicago SOC1 genes act downstream of the FT genes and likely act redundantly to promote flowering. Obtaining double and triple mutants’ combinations for the three SOC1 genes will be needed to determine their importance in flowering and potentially other aspects of Medicago development.
Author Contributions
JF, REL, and RM designed the experiments. JF, RHL, REL, and JLW performed the experiments. KM and JW identified the Medicago soc1b mutants. JF and RM wrote and edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by the New Zealand Marsden Fund (RM) and the Australian Research Council (JLW). The Medicago truncatula plants utilized in this research project, which are jointly owned by the Centre National de la Recherche Scientifique, were obtained from Noble Research Institute, LLC, United States, and were created through research funded, in part, by a grant from the National Science Foundation, NSF-0703285.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00496/full#supplementary-material
References
- Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., et al. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309 1052–1056. 10.1126/science.1115983 [DOI] [PubMed] [Google Scholar]
- Andres F., Coupland G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13 627–639. 10.1038/nrg3291 [DOI] [PubMed] [Google Scholar]
- Benlloch R., d’Erfurth I., Ferrandiz C., Cosson V., Beltran J. P., Canas L. A., et al. (2006). Isolation of MtPIM proves Tnt1 a useful reverse genetics tool in Medicago truncatula and uncovers new aspects of AP1-like functions in legumes. Plant Physiol. 142 972–983. 10.1104/pp.106.083543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertioli D. J., Moretzsohn M. C., Madsen L. H., Sandal N., Leal-Bertioli S. C., Guimaraes P. M., et al. (2009). An analysis of synteny of Arachis with Lotus and Medicago sheds new light on the structure, genomes stability and evolution of legume. BMC Genomics 10:45. 10.1186/1471-2164-10-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché F., Woods D. P., Amasino R. M. (2017). Winter memory throughout the plant kingdom: different paths to flowering. Plant Physiol. 173 27–35. 10.1104/pp.16.01322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J. Z., Zhou Y. P., Lv T. X., Xie C. P., Tian C. E. (2017). Research progress on the autonomous flowering time pathway in Arabidopsis. Physiol. Mol. Biol. Plants 23 477–485. 10.1007/s12298-017-0458-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson N., Russell J. (1975). Flowering responses to vernalization and photoperiod in annual medics (Medicago spp.). Austr. J. Agric. Res. 26 831–838. 10.1071/AR9750831 [DOI] [Google Scholar]
- Corbesier L., Vincent C., Jang S. H., Fornara F., Fan Q. Z., Searle I., et al. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316 1030–1033. 10.1126/science.1141752 [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann U., Hohmann S., Nettesheim K., Wisman E., Saedler H., Huijser P. (2000). Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J. 21 351–360. 10.1046/j.1365-313x.2000.00682.x [DOI] [PubMed] [Google Scholar]
- Hecht V., Foucher F., Ferrandiz C., Macknight R., Navarro C., Morin J., et al. (2005). Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 137 1420–1434. 10.1104/pp.104.057018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V., Laurie R. E., Vander Schoor J. K., Ridge S., Knowles C. L., Liew L. C., et al. (2011). The pea GIGAS gene is a FLOWERING LOCUS T homolog necessary for graft-transmissible specification of flowering but not for responsiveness to photoperiod. Plant Cell 23 147–161. 10.1105/tpc.110.081042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell C. A., Wood C. C., Robertson M., Peacock W. J., Dennis E. S. (2006). The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 46 183–192. 10.1111/j.1365-313X.2006.02686.x [DOI] [PubMed] [Google Scholar]
- Hepworth S. R., Valverde F., Ravenscroft D., Mouradov A., Coupland G. (2002). Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J. 21 4327–4337. 10.1093/emboj/cdf432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Zhou J., Liu C., Liu L., Shen L., Yu H. (2014). Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat. Commun. 5:4601. 10.1038/ncomms5601 [DOI] [PubMed] [Google Scholar]
- Immink R. G. H., Pose D., Ferrario S., Ott F., Kaufmann K., Valentim F. L., et al. (2012). Characterization of SOC1’s central role in flowering by the identification of its upstream and downstream regulators. Plant Physiol. 160 433–449. 10.1104/pp.112.202614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaudal M., Monash J., Zhang L., Wen J., Mysore K. S., Macknight R., et al. (2014). Overexpression of Medicago SVP genes causes floral defects and delayed flowering in Arabidopsis but only affects floral development in Medicago. J. Exp. Bot. 65 429–442. 10.1093/jxb/ert384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaudal M., Yeoh C. C., Zhang L., Stockum C., Mysore K. S., Ratet P., et al. (2013). Retroelement insertions at the Medicago FTa1 locus in spring mutants eliminate vernalisation but not long day requirements for early flowering. The Plant Journal 76 580–591. 10.1111/tpj.12315 [DOI] [PubMed] [Google Scholar]
- Jaudal M., Zhang L., Che C., Hurley D. G., Thomson G., Wen J., et al. (2016). MtVRN2 is a Polycomb VRN2-like gene which represses the transition to flowering in the model legume Medicago truncatula. Plant J. 86 145–160. 10.1111/tpj.13156 [DOI] [PubMed] [Google Scholar]
- Jaudal M., Zhang L., Che C., Putterill J. (2015). Three Medicago MtFUL genes have distinct and overlapping expression patterns during vegetative and reproductive development and 35S:MtFULb accelerates flowering and causes a terminal flower phenotype in Arabidopsis. Frontiers in Genetics 6:50. 10.3389/fgene.2015.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C. H., Wong C. E., Singh M. B., Bhalla P. L. (2012). Comparative genomic analysis of soybean flowering genes. PLoS One 7:e38250. 10.1371/journal.pone.0038250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J. H., Ju Y., Seo P. J., Lee J. H., Park C. M. (2012). The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J. 69 577–588. 10.1111/j.1365-313X.2011.04813.x [DOI] [PubMed] [Google Scholar]
- Kakar K., Wandrey M., Czechowski T., Gaertner T., Scheible W.-R., Stitt M., et al. (2008). A community resource for high-throughput quantitative RT-PCR analysis of transcription factor gene expression in Medicago truncatula. Plant Methods 4:18. 10.1186/1746-4811-4-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I., Shukla V. K., Ahn J. H., Dagenais N., Christensen S. K., Nguyen J. T., et al. (1999). Activation tagging of the floral inducer FT. Science 286 1962–1965. 10.1126/science.286.5446.1962 [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. 10.1016/s1360-1385(02)02251-3 [DOI] [PubMed] [Google Scholar]
- Kim H.-S., Abbasi N., Choi S.-B. (2013). Bruno-like proteins modulate flowering time via 3′ UTR-dependent decay of SOC1 mRNA. New Phytol. 198 747–756. 10.1111/nph.12181 [DOI] [PubMed] [Google Scholar]
- Kim M. Y., Shin J. H., Kang Y. J., Shim S. R., Lee S. H. (2012). Divergence of flowering genes in soybean. J. Biosci. 37 857–870. 10.1007/s12038-012-9252-0 [DOI] [PubMed] [Google Scholar]
- Laurie R., Diwadkar P., Jaudal M., Zhang L., Hecht V., Wen J., et al. (2011). The Medicago FLOWERING LOCUS T homolog, MtFTa1, is a key regulator of flowering time. Plant Physiol. 156 2207–2224. 10.1104/pp.111.180182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Suh S. S., Park E., Cho E., Ahn J. H., Kim S. G., et al. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14 2366–2376. 10.1101/gad.813600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Oh M., Park H., Lee I. (2008). SOC1 translocated to the nucleus by interaction with AGL24 directly regulates LEAFY. Plant J. 55 832–843. 10.1111/j.1365-313X.2008.03552.x [DOI] [PubMed] [Google Scholar]
- Lee J. H., Yoo S. J., Park S. H., Hwang I., Lee J. S., Ahn J. H. (2007). Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 21 397–402. 10.1101/gad.1518407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R., Baldwin S., Kenel F., McCallum J., Macknight R. (2013). FLOWERING LOCUS T genes control onion bulb formation and flowering. Nat. Commun. 4:2884. 10.1038/ncomms3884 [DOI] [PubMed] [Google Scholar]
- Letswaart R., Wu Z., Dean C. (2012). Flowering time control: another window to the connection between antisense RNA and chromatin. Trends Genet. 28 445–453. 10.1016/j.tig.2012.06.002 [DOI] [PubMed] [Google Scholar]
- Liu C., Chen H., Er H. L., Soo H. M., Kumar P. P., Han J. H., et al. (2008). Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135 1481–1491. 10.1242/dev.020255 [DOI] [PubMed] [Google Scholar]
- Mateos J. L., Madrigal P., Tsuda K., Rawat V., Richter R., Romera-Branchat M., et al. (2015). Combinatorial activities of SHORT VEGETATIVE PHASE and FLOWERING LOCUS C define distinct modes of flowering regulation in Arabidopsis. Genome Biol. 16:31. 10.1186/s13059-015-0597-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung C. R., Lou P., Hermand V., Kim J. A. (2016). The importance of ambient temperature to growth and the induction of flowering. Front. Plant Sci. 7:1266. 10.3389/fpls.2016.01266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J., Suh S. S., Lee H., Choi K. R., Hong C. B., Paek N. C., et al. (2003). The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 35 613–623. 10.1046/j.1365-313X.2003.01833.x [DOI] [PubMed] [Google Scholar]
- Na X., Jian B., Yao W., Wu C., Hou W., Jiang B., et al. (2013). Cloning and functional analysis of the flowering gene GmSOC1-like, a putative SUPPRESSOR OF OVEREXPRESSION CO1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in soybean. Plant Cell Rep. 32 1219–1229. 10.1007/s00299-013-1419-0 [DOI] [PubMed] [Google Scholar]
- Narusaka M., Shiraishi T., Iwabuchi M., Narusaka Y. (2010). The floral inoculating protocol: a simplified Arabidopsis thaliana transformation method modified from floral dipping. Plant Biotechnol. 27 349–351. 10.5511/plantbiotechnology.27.349 [DOI] [Google Scholar]
- Notaguchi M., Abe M., Kimura T., Daimon Y., Kobayashi T., Yamaguchi A., et al. (2008). Long-distance, graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant Cell Physiol. 49 1645–1658. 10.1093/pcp/pcn154 [DOI] [PubMed] [Google Scholar]
- Onouchi H., Igeno M. I., Perilleux C., Graves K., Coupland G. (2000). Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12 885–900. 10.1105/tpc.12.6.885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J., Zhang L., Yeoh C. C., Balcerowicz M., Jaudal M., Gasic E. V. (2013). FT genes and regulation of flowering in the legume Medicago truncatula. Funct. Plant Biol. 40 1199–1207. 10.1071/FP13087 [DOI] [PubMed] [Google Scholar]
- Romera-Branchat M., Andres F., Coupland G. (2014). Flowering responses to seasonal cues: what’s new? Curr. Opin. Plant Biol. 21C, 120–127. 10.1016/j.pbi.2014.07.006 [DOI] [PubMed] [Google Scholar]
- Samach A., Onouchi H., Gold S. E., Ditta G. S., Schwarz-Sommer Z., Yanofsky M. F., et al. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288 1613–1616. 10.1126/science.288.5471.1613 [DOI] [PubMed] [Google Scholar]
- Searle I., He Y. H., Turck F., Vincent C., Fornara F., Krober S., et al. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20 898–912. 10.1101/gad.373506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin A. J., Woody J. L., Bolon Y.-T., Joseph B., Diers B. W., Farmer A. D., et al. (2010). RNA-Seq Atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol. 10:160. 10.1186/1471-2229-10-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon C. C., Finnegan E. J., Dennis E. S., Peacock W. J. (2006). Quantitative effects of vernalization on FLC and SOC1 expression. Plant J. 45 871–883. 10.1111/j.1365-313X.2006.02652.x [DOI] [PubMed] [Google Scholar]
- Song H. R., Song J. D., Cho J. N., Amasino R. M., Noh B., Noh Y. S. (2009). The RNA binding protein ELF9 directly reduces SUPPRESSOR OF OVEREXPRESSION OF CO1 transcript levels in Arabidopsis, possibly via nonsense-nediated mRNA decay. Plant Cell 21 1195–1211. 10.1105/tpc.108.064774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth A., Schmid M. (2011). Regulation of flowering time: all roads lead to Rome. Cell. Mol. Life Sci. 68 2013–2037. 10.1007/s00018-011-0673-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M., Wen J. Q., He J., Tu H. D., Kwak Y., Eschstruth A., et al. (2008). Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 54 335–347. 10.1111/j.1365-313X.2008.03418.x [DOI] [PubMed] [Google Scholar]
- Tao Z., Shen L., Liu C., Liu L., Yan Y., Yu H. (2012). Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. Plant J. 70 549–561. 10.1111/j.1365-313X.2012.04919.x [DOI] [PubMed] [Google Scholar]
- Taoka K., Ohki I., Tsuji H., Furuita K., Hayashi K., Yanase T., et al. (2011). 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476 332–335. 10.1038/nature10272 [DOI] [PubMed] [Google Scholar]
- Torti S., Fornara F. (2012). AGL24 acts in concert with SOC1 and FUL during Arabidopsis floral transition. Plant Signal. Behav. 7 1251–1254. 10.4161/psb.21552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller J. L., Ortega R. (2015). Genetic control of flowering time in legumes. Front. Plant Sci 6:207. 10.3389/fpls.2015.00207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker C., Dean C. (2017). The FLC locus: a platform for discoveries in epigenetics and adaptation. Annu. Rev. Cell Dev. Biol. 33 555–575. 10.1146/annurev-cellbio-100616-060546 [DOI] [PubMed] [Google Scholar]
- Wigge P. A., Kim M. C., Jaeger K. E., Busch W., Schmid M., Lohmann J. U., et al. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309 1056–1059. 10.1126/science.1114358 [DOI] [PubMed] [Google Scholar]
- Wong A. C., Hecht V. F., Picard K., Diwadkar P., Laurie R. E., Wen J., et al. (2014). Isolation and functional analysis of CONSTANS-LIKE genes suggests that a central role for CONSTANS in flowering time control is not evolutionarily conserved in Medicago truncatula. Front. Plant Sci. 5:486. 10.3389/fpls.2014.00486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoh C. C., Balcerowicz M., Zhang L., Jaudal M., Brocard L., Ratet P., et al. (2013). Fine mapping links the FTa1 flowering time regulator to the dominant spring1 locus in Medicago. PLoS One 8:e53467. 10.1371/journal.pone.0053467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S. K., Chung K. S., Kim J., Lee J. H., Hong S. M., Yoo S. J., et al. (2005). CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 139 770–778. 10.1104/pp.105.066928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young N. D., Bharti A. K. (2012). Genome-enabled insights into legume biology. Annu. Rev. Plant Biol. 63 283–305. 10.1146/annurev-arplant-042110-103754 [DOI] [PubMed] [Google Scholar]
- Zhong X., Dai X., Xv J., Wu H., Liu B., Li H. (2012). Cloning and expression analysis of GmGAL1, SOC1 homolog gene in soybean. Mol. Biol. Rep. 39 6967–6974. 10.1007/s11033-012-1524-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.