Abstract
Accumulating evidence has indicated that microRNA (miRNA) dysregulation contributes to hepatocellular carcinoma (HCC) progression. miR-337-3p is downregulated in gastric cancer and neuroblastoma; however, its biological function and underlying mechanism in HCC remain unclear. In this study, we showed that the expression level of miR-337-3p was significantly decreased in HCC, and was associated with several clinicopathological characteristics, including tumor multiplicity, histological differentiation, and Barcelona Clinic Liver Cancer stage. Low expression level of miR-337-3p was associated with poor survival outcomes in HCC patients. Upregulation of miR-337-3p suppressed cell proliferation, migration, and invasion in HCC. Dual luciferase assay demonstrated that JAK2 was a direct downstream target of miR-337-3p. JAK2 reintroduction restored the inhibited proliferation, migration, and invasion of miR-337-3p overexpressed HCC cells. miR-337-3p functioned as a tumor suppressor to modulate the JAK2/STAT3 signaling pathway. The present findings indicate that miR-337-3p could be used as a prognostic predictor and therapeutic candidate for HCC.
Keywords: Hepatocellular carcinoma, miR-337-3p, JAK2, proliferation, invasion
Introduction
Hepatocellular carcinoma (HCC) ranked sixth for cancer incidence and fourth for cancer deaths in 2015, with 854,000 incident cases and 810,000 deaths globally [1]. Although substantial achievements have been made in HCC treatment, including molecule-targeted drugs and liver transplantation, the long-term outcome of HCC patients remains unfavorable [2,3]. Previous studies have reported that inactivation of tumor suppressors and abnormal regulation of signaling pathways were closely related to the pathogenesis of HCC; however, the underlying mechanism remains to be clarified.
MicroRNAs (miRNAs) are a class of highly conserved small RNAs, exerting crucial effects on post-transcriptional regulation of gene expression. Aberrant patterns of miRNA expression are associated with multiple developmental and pathological processes. miRNAs regulate many cellular activities through binding 3’-untranslated regions (UTRs) of their target mRNA sequences to inhibit their translation [4]. Recent studies have shown that a variety of miRNAs are involved in carcinogenesis, and function as oncogenes or tumor suppressors in HCC [5]. For instance, miR-233 plays a critical role in inhibiting the tumorigenesis and promoting the apoptosis of HCC through mTOR signaling pathway [6]. miR-487a promotes proliferation and metastasis of HCC by PIK3R1 and SPRED2 binding [7]. Therefore, it is important to investigate the role of miRNAs in HCC progression, providing a theoretical basis for diagnosis and treatment.
miR-337-3p has been indicated to be an important regulator in liver development [8] and chondrocyte growth [9]. Altered miR-337-3p expression plays a key role in tumor proliferation and metastasis. In gastric cancer, the level of miR-337-3p is downregulated in metastatic gastric cancer tissues, and ectopic miR-337-3p expression suppressed gastric cancer cell invasion [10]. In addition, miR-337-3p sensitizes lung cancer cells to paclitaxel treatment [11]. Although emerging evidence has indicated the involvement of miR-337-3p in several kinds of human cancers, its biological role and molecular mechanism in HCC are poorly understood.
In the present study, we found that miR-337-3p dysregulation was significantly correlated with clinicopathological features and survival outcomes of HCC patients. miR-337-3p regulated HCC cell proliferation, migration, and invasion through directly binding Janus kinase 2 (JAK2). Our results may provide a novel diagnostic and therapeutic candidate for HCC.
Materials and methods
Cell lines and cell culture
Human HCC cell lines (Focus, HepG2, Hep3B, MHCC-LM3, and SMMC7721) and human normal liver cell line QSG7701 were purchased from the Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). All cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM; Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) at 37°C in a humidified incubator containing 5% CO2.
Patients and tissue samples
A total of 50 matched fresh HCC specimens and adjacent normal tissues were obtained from HCC patients who underwent hepatic resection in the First Affiliated Hospital of Wannan Medical College. None of the patients had received radiotherapy or chemotherapy before surgery. All patients were consented, and the present study was approved by the Ethics Committee of the First Affiliated Hospital of Wannan Medical College.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from cell lines and tumor specimens using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. cDNA synthesis was performed using the PrimeScript RT Reagent Kit (TaKaRa, Dalian, China). Stem-loop specific primers for miR-337-3p and U6 were synthesized by RiboBio (Guangzhou, China). qRT-PCR was performed with the SYBR Premix Ex Taq II (TaKaRa) in an ABI 7900HT system (Applied Biosystems, Carlsbad, CA, USA). GAPDH and U6 were used as endogenous controls for mRNA and miRNA, respectively. The following primers were used for qRT-PCR detection: JAK2: 5’-GGGAGGTGGTCGCTGTAAAA-3’ (forward), 5’-ACCAGCACTGTAGCACACTC-3’ (reverse); GAPDH: 5’-TGTGGGCATCAATGGATTTGG-3’ (forward); 5’-ACACCATGTATTCCGGGTCAAT-3’ (reverse). Relative expression levels of target gene were calculated according to the 2-ΔΔCt method.
Lentivirus and cell transfection
Lentiviruses overexpressing miR-337-3p and the corresponding control were purchased from GenePharma (Shanghai, China). For rescue assays, the lentiviruses overexpressing JAK2 and the corresponding control were purchased from GenePharma. MHCC-LM3 and SMMC7721 cells were infected with lentiviruses plus 5 mg/ml of polybrene (GenePharma).
Cell counting kit-8 (CCK-8) assay
Cell viability was assessed by CCK-8 assay. Cells were seeded into 96-well plates at a density of 103 cells per well, and cultured for 1, 2, 3, 4, and 5 days. At the indicated time points, 10 μl of CCK-8 reagent (Dojindo Laboratories, Kumamoto, Japan) was added to each well. The absorbance at 450 nm was measured after incubation for 2 h at 37°C. The experiments were performed in triplicate.
Colony formation assay
All cells were seeded in a six-well plate at a density of 500 cells per well. After culturing for two weeks, the colonies on the plates were fixed with absolute methanol for 5 min and stained with 0.1% crystal violet for 30 min. The experiments were conducted in triplicate.
Cell migration and invasion assays
Cell migration and invasion were assessed using the transwell chamber assay (Corning, New York, NY, USA). For cell migration, 2 × 104 cells were seeded into the upper chamber. For invasion assay, a total of 2 × 104 cells suspended with serum-free medium were plated in the upper chamber coated with Matrigel (BD Biosciences, San Jose, CA, USA), and the lower chamber was maintained with DMEM containing 10% FBS. After incubation for 24 h, the inserts were washed with phosphate-buffered saline, fixed with methanol, and stained with crystal violet. The cells in the upper chamber were carefully removed using a cotton tip, and the migrated or invaded cells were photographed using a bright field microscope. Five random fields of view were analyzed for each chamber.
Western blotting
Total protein was separated using sodium dodecyl sulfate polyacrylamide gel electrophoresis, transferred to polyvinylidene fluoride membrane, and blocked with 5% skim milk. Membranes were probed with GAPDH, JAK2, signal transducer and activator of transcription 3 (STAT3) or phospho-STAT3 primary antibodies (Cell Signaling Technology, Danvers, MA, USA) at 4°C overnight, followed by incubation with horseradish peroxidase-conjugated secondary antibody, using GAPDH as the internal control. Proteins were visualized with enhanced chemiluminescence reagents (EMD Millipore, Billarica, MA, USA).
Luciferase reporter assay
For the luciferase assay, cells were seeded in 96-well plates 24 h before transfection and co-transfected with the JAK2 wild-type (WT) or mutant (MUT) 3’-UTR reporter vector using Lipofectamine 2000 (Invitrogen). Luciferase activities were determined with the Dual-Luciferase Reporter System (Promega, Madison, WI, USA) according to the manufacturer’s instructions.
Xenograft tumor model
SMMC7721 overexpressing miR-337-3p or the control cells (2 × 106) were injected subcutaneously into the flakes of nude mice (four weeks of age, n = 3 per group). The tumor volume was calculated every five days, using the following equation: volume = (width2 × length)/2. Mice were sacrificed one month after injection. All animal experiments were performed with the approval of the First Affiliated Hospital of Wannan Medical College. Tissue samples were used for immunohistochemistry.
Statistical analysis
All data are presented as mean ± standard deviation. A two-tailed Student’s t test was used to assess the differences between different groups. The Spearman correlation test was used to examine the correlation between miR-337-3p and JAK2 expression. All statistical analyses were performed using SPSS 23.0 (IBM SPSS software, NY, USA) and Prism 7.00 (GraphPad Software, La Jolla, CA, USA). Data were considered statistically significant when P < 0.05.
Results
miR-337-3p is downregulated in HCC tissues and cell lines
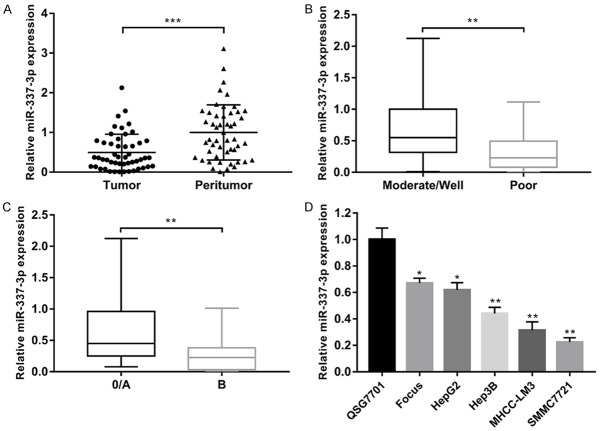
The expression level of miR-337-3p was examined in 50 pairs of HCC samples and matched peritumor tissues using qRT-PCR. We found that miR-337-3p expression was significantly decreased in HCC samples compared to that in the non-tumor counterparts (Figure 1A). Correlation between the clinicopathologic features of 50 HCC patients and the expression of miR-337-3p is shown in Table 1. The expression level of miR-337-3p was significantly associated with tumor multiplicity (P = 0.010), histological differentiation (P = 0.024), and Barcelona Clinic Liver Cancer (BCLC) stage (P = 0.011). After dividing patients into two groups based on histological differentiation, a lower expression level of miR-337-3p was observed in the poorly differentiated group than in the moderate/well differentiation group (Figure 1B). Compared to the BCLC 0/A stage group, patients with BCLC B stage had a lower miR-337-3p expression level (Figure 1C). Next, the expression level of miR-337-3p was determined in five human HCC cell lines (Focus, HepG2, Hep3B, MHCC-LM3, and SMMC7721) and one human hepatic cell line QSG7701. Consistent with the expression in tissue samples, miR-337-3p was downregulated in all five HCC cell lines (Figure 1D).
Figure 1.
The expression of miR-337-3p was significantly decreased in hepatocellular carcinoma (HCC) tissues and HCC cell lines. A. The expression of miR-337-3p in 50 pairs of HCC and peritumor tissues was examined by qRT-PCR. miR-337-3p expression was significantly decreased in HCC tissues compared with the peritumor tissues (P < 0.001). B. A lower expression level of miR-337-3p was observed in patients with poor histological differentiation (P < 0.01). C. The level of miR-337-3p was lower in the BCLC B stage group than in the BCLC 0/A stage group (P < 0.01). D. The level of miR-337-3p in HCC cell lines (Focus, HepG2, Hep3B, MHCC-LM3, and SMMC7721) and normal liver cell line QSG7701 was investigated. miR-337-3p levels were markedly downregulated in HCC cell lines compared with QSG7701 (P < 0.05). Data are shown as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
Table 1.
Correlation between miR-337-3p expression and clinicopathological features of HCC (n = 50)
| Clinicopathological features | High miR-337-3p expression (n = 25) | Low miR-337-3p expression (n = 25) | P value* |
|---|---|---|---|
| Age | 0.239 | ||
| < 60 years | 14 | 18 | |
| ≥ 60 years | 11 | 7 | |
| Gender | 0.725 | ||
| Male | 21 | 19 | |
| Female | 4 | 6 | |
| Hepatitis B | 0.667 | ||
| Negative | 2 | 4 | |
| Positive | 23 | 21 | |
| Tumor size | 0.556 | ||
| < 5 cm | 15 | 17 | |
| ≥ 5 cm | 10 | 8 | |
| Tumor multiplicity | 0.010 | ||
| Single | 15 | 6 | |
| Multiple | 10 | 19 | |
| Histological differentiation | 0.024 | ||
| Moderate/Well | 16 | 8 | |
| Poor | 9 | 17 | |
| α-fetoprotein level | 0.508 | ||
| < 20 ng/ml | 7 | 5 | |
| ≥ 20 ng/ml | 18 | 20 | |
| BCLC stage | 0.011 | ||
| 0/A | 18 | 9 | |
| B | 7 | 16 |
P < 0.05 was considered to be statistically significance.
Decreased miR-337-3p expression is correlated with poor survival outcomes
We further evaluated the impact of miR-337-3p on the survival outcomes of HCC patients. The overall survival (OS) of patients with low miR-337-3p expression was poorer than that of patients with high miR-337-3p expression (Figure 2A). In addition, decreased miR-337-3p expression was correlated with higher recurrence probability (Figure 2B). As shown in Table 2, univariate analysis identified 4 prognostic factors for OS: miR-337-3p expression (P = 0.028), tumor multiplicity (P = 0.036), histological differentiation (P = 0.041), and BCLC stage (P < 0.001). To further determine the independent predictive factors for OS, we performed the multivariate Cox regression analysis, including only variables which were significant in the univariate analysis. miR-337-3p expression (P = 0.004), histological differentiation (P = 0.004), and BCLC stage (P < 0.001) were found to be independent prognostic factors for OS of HCC patients. These findings showed that decreased miR-337-3p may play a role in the pathogenesis and development of HCC.
Figure 2.
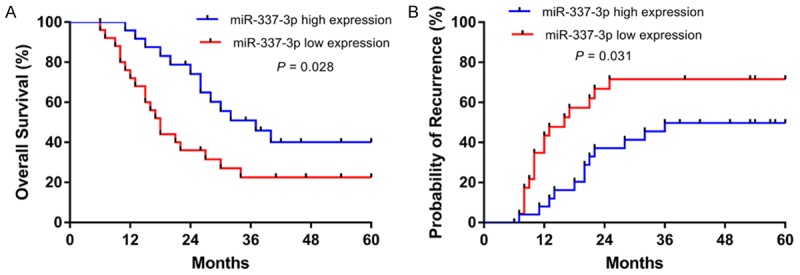
Kaplan-Meier analysis was performed to examine the overall survival and recurrence of HCC patients with different miR-337-3p expression levels. A. Low miR-337-3p expression predicted unfavorable overall survival (P = 0.028). B. The probability of recurrence was higher in patients with low miR-337-3p expression (P = 0.031).
Table 2.
Prognostic factors for overall survival in hepatocellular carcinoma patients
| Clinicopathological features | Overall Survival | ||||
|---|---|---|---|---|---|
|
| |||||
| Univariate | Multivariate | ||||
|
| |||||
| Log-rank | P | HR | 95% CI | P value* | |
| Age | 0.013 | 0.909 | |||
| < 60 years | |||||
| ≥ 60 years | |||||
| Gender | 0.003 | 0.953 | |||
| Male | |||||
| Female | |||||
| Hepatitis B | 1.140 | 0.286 | |||
| Negative | |||||
| Positive | |||||
| Tumor size | 2.267 | 0.132 | |||
| < 5 cm | |||||
| ≥ 5 cm | |||||
| Tumor multiplicity | 4.383 | 0.036 | Not included | ||
| Single | |||||
| Multiple | |||||
| Histological differentiation | 4.183 | 0.041 | |||
| Moderate/Well | Reference | ||||
| Poor | 3.336 | 1.468-7.583 | 0.004 | ||
| α-fetoprotein level | 1.851 | 0.174 | |||
| < 20 ng/ml | |||||
| ≥ 20 ng/ml | |||||
| BCLC stage | 21.333 | < 0.001 | |||
| 0/A | Reference | ||||
| B | 33.333 | 7.463-142.857 | < 0.001 | ||
| miR-337-3p expression level | 4.844 | 0.028 | |||
| High | Reference | ||||
| Low | 6.926 | 1.883-25.473 | 0.004 | ||
P < 0.05 was considered to be statistically significance.
Overexpressed miR-337-3p inhibits the proliferation, migration, and invasion of HCC cells
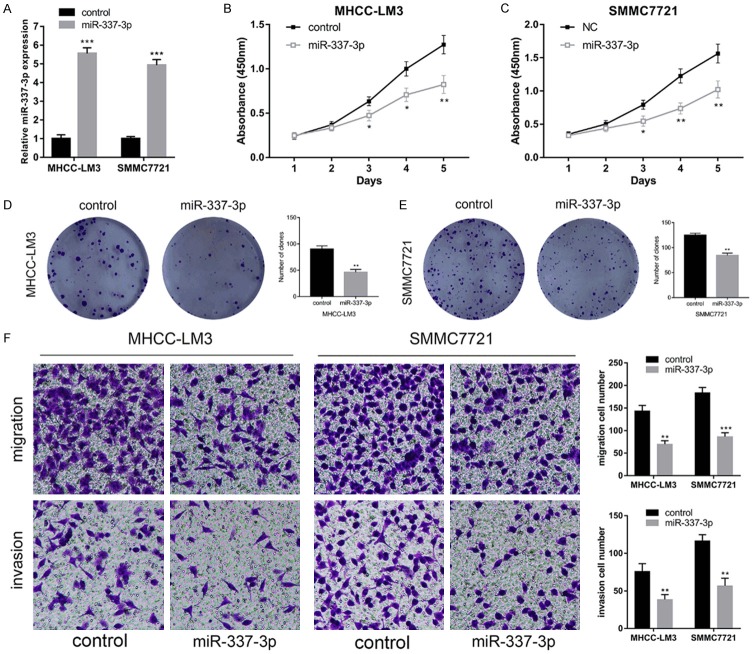
To assess the biological function of miR-337-3p in HCC, we transfected lentiviruses overexpressing miR-337-3p or control into MHCC-LM3 and SMMC7721 cells. Figure 3A shows approximate 5.5-fold and 5.0-fold increase in MHCC-LM3 cells and SMMC7721 cells, respectively. Results of CCK-8 assays indicated that miR-337-3p overexpression significantly suppressed the proliferation of HCC cells (Figure 3B and 3C). Moreover, colony formation assays confirmed the inhibitory effect of miR-337-3p on MHCC-LM3 and SMMC7721 cells (Figure 3D and 3E). We next explored the effects of miR-337-3p on the migration and invasion of HCC cells using transwell assays. It was demonstrated that the number of migration and invasive clones was significantly lower in the miR-337-3p overexpression group than in the control group (Figure 3F). These results indicated that miR-337-3p could significantly inhibit the proliferation, migration, and invasion of HCC cells.
Figure 3.
Overexpression of miR-337-3p inhibited MHCC-LM3 and SMMC7721 proliferation, migration, and invasion. A. The transfection of lentivirus- overexpressing miR-337-3p increased the expression of miR-337-3p in MHCC-LM3 and SMMC7721 cells. B and C. CCK-8 assays were performed to measure the cell viability in miR-337-3p overexpressed MHCC-LM3 and SMMC7721 cells. miR-337-3p overexpression inhibited HCC cell proliferation (P < 0.05). D and E. Colony formation assays indicated that upregulated miR-337-3p suppressed HCC cell proliferation (P < 0.01). F. The transfection of miR-337-3p lentivirus suppressed MHCC-LM3 and SMMC7721 cell migration and invasion, as detected by transwell assays (P < 0.01). Data are shown as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
JAK2 is a direct target gene of miR-337-3p
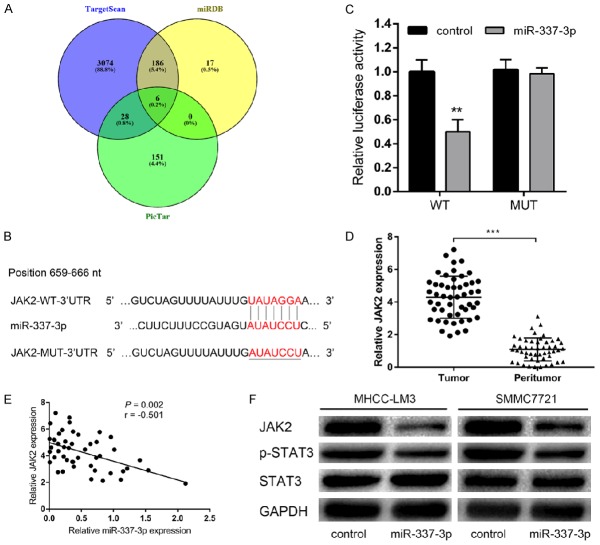
We used multiple databases (TargetScan, miRDB, and PicTar) to predict the target gene of miR-337-3p, and JAK2 was predicted by all three databases (Figure 4A). Complementary sequences can be observed between miR-337-3p and JAK2 3’-UTR (Figure 4B). JAK2 was involved in cancer progression [12], and was previously reported to be the target of various miRNAs [13-15]. To investigate whether JAK2 was a direct target of miR-337-3p, we performed a luciferase reporter assay. When co-transfected with JAK2 3’-UTR luciferase reporter plasmid, miR-337-3p led to a significant decrease in the luciferase activity of JAK2, while the luciferase activity was unaffected by miR-337-3p-MUT (Figure 4C). qRT-PCR results further verified the upregulation of JAK2 expression in 50 pairs of HCC tissues compared with their matched peritumor tissues (Figure 4D). Further analysis revealed that the level of miR-337-3p was inversely associated with JAK2 mRNA levels (Figure 4E). Consistently, we observed that JAK2 protein levels in the miR-337-3p overexpressing cells were lower than those in controls (Figure 4F). Thus, these results indicated that JAK2 was a novel direct target of miR-337-3p in HCC.
Figure 4.
JAK2 was a direct target gene of miR-337-3p. A. TargetScan, miRDB, and PicTar databases were used to predict the target genes of miR-337-3p, and JAK2 was predicted by all three databases. B. The putative binding sequence of miR-337-3p in the 3’-UTR of JAK2. C. The dual luciferase reporter assay showed that miR-337-3p bound to the 3’-UTR of JAK2 (P < 0.01). D. qRT-PCR analysis of JAK2 mRNA expression in 50 pairs of HCC and peritumor tissues. The expression of JAK2 was obviously increased in HCC tissues compared with the peritumor tissues (P < 0.001). E. A significant inverse correlation between miR-337-3p and JAK2 mRNA was observed in HCC tissues (P = 0.002). F. Overexpression of miR-337-3p reduced the expression of JAK2 and p-STAT3 in MHCC-LM3 and SMMC7721 cells. Data are shown as mean ± SD. **P < 0.01, ***P < 0.001.
Given that JAK2 is an important regulator in the JAK2/STAT3 signaling pathway, we next examined whether this signaling pathway was involved in miR-337-3p-associated HCC progression. Western blotting results showed that the levels of JAK2 and p-STAT3 were decreased in SMMC7721 cells overexpressing miR-337-3p (Figure 4F). The data showed that the JAK2/STAT3 signaling pathway was the downstream of miR-337-3p in HCC.
Alteration of JAK2 expression influences the effects of miR-337-3p on HCC cells
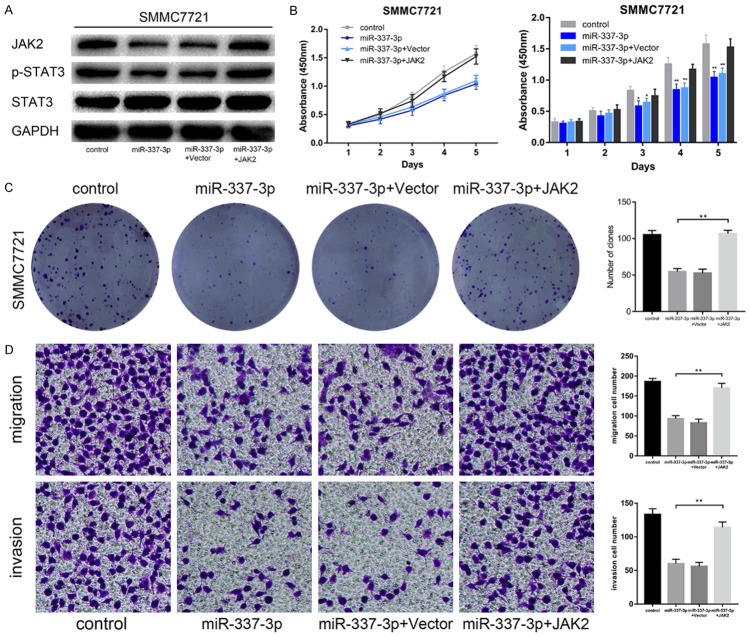
Next, we investigated whether JAK2 mediated the effects of miR-337-3p on HCC progression. SMMC7721 cells overexpressing miR-337-3p were transfected with JAK2 lentivirus. The overexpression of JAK2 was confirmed by western blotting. JAK2 upregulation led to an increased p-STAT3 level in SMMC7721 cells overexpressing miR-337-3p (Figure 5A). The results of CCK-8, colony formation assays, and transwell assays showed that elevated JAK2 expression restored the suppressed cell proliferation, migration, and invasion (Figure 5B-D). These results supported that JAK2 is a downstream functional effector of miR-337-3p.
Figure 5.
Alteration of JAK2 expression affected the influence of miR-337-3p on HCC cells. A. Western blotting assays were performed to detect JAK2 and p-STAT3 expression levels in miR-337-3p-overexpressing SMMC7721 cells with or without JAK2 upregulation. B. CCK-8 assays were used to evaluate the effect of JAK2 upregulation on cell proliferation in miR-337-3p overexpressed SMMC7721 cells. Upregulation of JAK2 restored the suppressed growth in miR-337-3p overexpressed SMMC7721 cells (P < 0.01). C. Colony formation assays showed that JAK2 upregulation reversed the inhibitory effect of miR-337-3p overexpression in SMMC7721 cells (P < 0.01). D. Transwell assays were performed to examine the effect of JAK2 upregulation on cell migration and invasion in miR-337-3p-overexpressed SMMC7721 cells (P < 0.01). Data are shown as mean ± SD. *P < 0.05, **P < 0.01.
miR-337-3p overexpression inhibits HCC proliferation in vivo
To validate the in vitro data that overexpression of miR-337-3p suppresses HCC proliferation, an in vivo xenograft tumor model was constructed. SMMC7721 cells stably overexpressing miR-337-3p and control cells were inoculated into the nude mice (Figure 6A). Consistent with the in vitro results, miR-337-3p overexpression resulted in decreased tumor size and weight (Figure 6B and 6C). Additionally, immunohistochemistry showed that JAK2 and Ki-67 expression levels were reduced in the miR-337-3p-overexpressed group (Figure 6D). These results further verified the suppressive effect of miR-337-3p on HCC proliferation.
Figure 6.
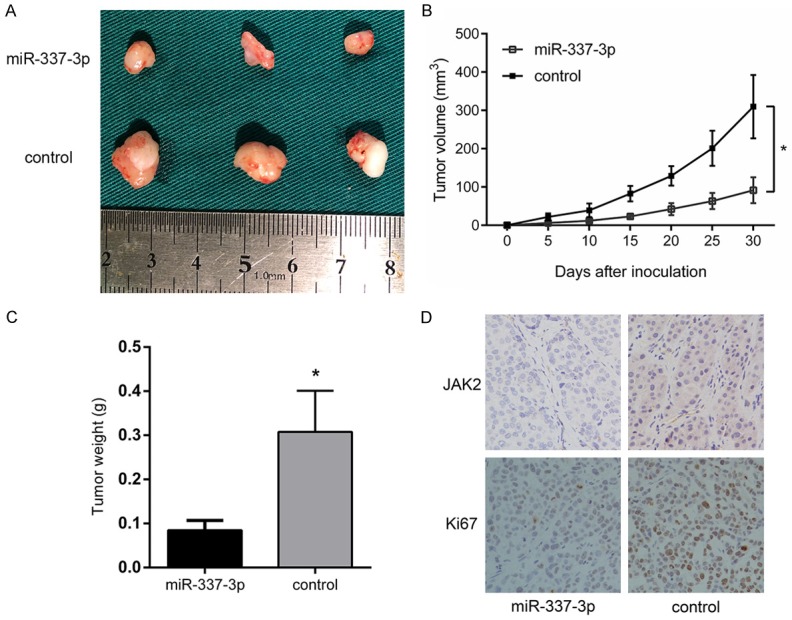
Overexpression of miR-337-3p inhibited tumorigenicity. A. Representative tumor tissues extracted from mice inoculated with SMMC7721 cells overexpressing miR-337-3p or control. B. Measurement of tumor volumes at the indicated time points (P < 0.05). C. Tumor weights were calculated (P < 0.05). D. Representative JAK2 and Ki67 immunostaining of xenograft tumors. Data are shown as mean ± SD. *P < 0.05.
Discussion
Emerging evidence has shown that various miRNAs are involved in the pathological process of HCC [16,17]. In the current study, we attempted to elucidate the relationship between miR-337-3p dysregulation and HCC. We demonstrated the decreased expression of miR-337-3p in HCC tissues compared with peritumor tissues. The expression levels of miR-337-3p were downregulated in five HCC cell lines. Moreover, our results showed that the expression level of miR-337-3p was significantly correlated with tumor multiplicity, histological differentiation, and BCLC stage. Decreased miR-337-3p expression predicted unfavorable OS of HCC patients. Further studies confirmed that the overexpression of miR-337-3p evidently inhibited the proliferation, migration, and invasion of MHCC-LM3 and SMMC7721 cell lines; this indicated that miR-337-3p served as a tumor suppressor in HCC.
The hsa-miR-337 gene is localized at chromosome 14q32.2. Previous studies have reported that miR-337-3p is differentially expressed and presented as a tumor suppressor in various human cancers through binding downstream targets. miR-337-3p directly binds the matrix metallopeptidase 14 (MMP-14) 3’-UTR to repress myeloid zinc finger 1 (MZF1)-facilitated MMP-14 expression, thus inhibiting the progression of gastric cancer [18]. In colorectal cancer, miR-337-3p promotes the senescence of HCT116 cells by suppressing the expression of casein kinase II [19]. In order to clarify the molecular mechanism of miR-337-3p in HCC progression, we performed bioinformatics analysis, and found that miR-337-3p could target the 3’UTR of JAK2. qRT-PCR analysis revealed that the expression of JAK2 was negatively correlated with the level of miR-337-3p in tumor samples. Dual luciferase assay confirmed that JAK2 was a direct downstream target of miR-337-3p. In addition, inhibited cell proliferation and invasion could be rescued by overexpressing JAK2, suggesting that miR-337-3p dampened HCC progression via JAK2.
JAK2 is a member of the Janus family of non-receptor protein tyrosine kinases, and regulates several cellular processes by inducing cytoplasmic signaling pathways [20]. JAK2 acts as an oncogene and is involved in the proliferation, invasion, and drug resistance of tumor cells. In myeloproliferative leukemia, JAK2 regulates megakaryocytic proliferation and differentiation in both normal and pathological conditions [21]. STAT3 phosphorylation is correlated with p53 mutation and patient survival. Loss of p53 function activates JAK2 signaling, which promotes tumor growth and induces resistance to gemcitabine in pancreatic cancer [22]. Reportedly, JAK2 serves as a direct downstream target of multiple miRNAs in human cancers. Ding et al. revealed that miR-375 functions as a tumor suppressor to inhibit gastric cancer proliferation by targeting JAK2 [13]. As suggested by Wang et al., miR-101 could suppress cell proliferation and induce breast cancer cell apoptosis through binding JAK2 [15]. In the present study, JAK2 was demonstrated to be a direct target of miR-337-3p in HCC.
In addition, we found that miR-337-3p is involved in the dysregulated JAK2/STAT3 signaling pathway in HCC. The JAK2/STAT3 signaling pathway is identified to play an important role in cancer progression, metastasis, and angiogenesis. JAK2/STAT3 transmits signals from cell membrane to nucleus in response to extracellular growth factors and cytokines. Wang et al. showed that lnc-BM and JAK2 promote breast cancer brain metastases by mediating communication between breast cancer cells and the brain microenvironment and activating the lnc-BM/JAK2/STAT3 pathway [23]. In colorectal cancer, human colorectal cancer-derived mesenchymal stem cells increase the migration and invasion of cancer cells through IL-6/JAK2/STAT3 signaling [24]. Stimulation of the B-cell receptor activates the JAK2/STAT3 pathway in chronic lymphocytic leukemia [25]. Our results revealed that JAK2/STAT3 signaling was regulated by miR-337-3p in HCC progression.
In conclusion, our study presented an association of miR-337-3p with JAK2 in HCC. Downregulation of miR-337-3p and overexpression of JAK2 were observed in HCC. miR-337-3p significantly inhibited HCC cell proliferation, migration, and invasion through the JAK2/STAT3 signaling pathway. Low miR-337 expression level predicted poor OS and increased recurrence probability. The miR-337-3p/JAK2/STAT3 axis provides a novel insight into the molecular mechanisms underlying HCC progression, and may be a promising target for HCC therapeutic strategies.
Disclosure of conflict of interest
None.
References
- 1.Global Burden of Disease Cancer Collaboration. Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O, Dandona R, Dandona L, Fleming T, Forouzanfar MH, Hancock J, Hay RJ, Hunter-Merrill R, Huynh C, Hosgood HD, Johnson CO, Jonas JB, Khubchandani J, Kumar GA, Kutz M, Lan Q, Larson HJ, Liang X, Lim SS, Lopez AD, MacIntyre MF, Marczak L, Marquez N, Mokdad AH, Pinho C, Pourmalek F, Salomon JA, Sanabria JR, Sandar L, Sartorius B, Schwartz SM, Shackelford KA, Shibuya K, Stanaway J, Steiner C, Sun J, Takahashi K, Vollset SE, Vos T, Wagner JA, Wang H, Westerman R, Zeeb H, Zoeckler L, Abd-Allah F, Ahmed MB, Alabed S, Alam NK, Aldhahri SF, Alem G, Alemayohu MA, Ali R, Al-Raddadi R, Amare A, Amoako Y, Artaman A, Asayesh H, Atnafu N, Awasthi A, Saleem HB, Barac A, Bedi N, Bensenor I, Berhane A, Bernabe E, Betsu B, Binagwaho A, Boneya D, Campos-Nonato I, Castaneda-Orjuela C, Catala-Lopez F, Chiang P, Chibueze C, Chitheer A, Choi JY, Cowie B, Damtew S, das Neves J, Dey S, Dharmaratne S, Dhillon P, Ding E, Driscoll T, Ekwueme D, Endries AY, Farvid M, Farzadfar F, Fernandes J, Fischer F, G/Hiwot TT, Gebru A, Gopalani S, Hailu A, Horino M, Horita N, Husseini A, Huybrechts I, Inoue M, Islami F, Jakovljevic M, James S, Javanbakht M, Jee SH, Kasaeian A, Kedir MS, Khader YS, Khang YH, Kim D, Leigh J, Linn S, Lunevicius R, El Razek HMA, Malekzadeh R, Malta DC, Marcenes W, Markos D, Melaku YA, Meles KG, Mendoza W, Mengiste DT, Meretoja TJ, Miller TR, Mohammad KA, Mohammadi A, Mohammed S, Moradi-Lakeh M, Nagel G, Nand D, Le Nguyen Q, Nolte S, Ogbo FA, Oladimeji KE, Oren E, Pa M, Park EK, Pereira DM, Plass D, Qorbani M, Radfar A, Rafay A, Rahman M, Rana SM, Soreide K, Satpathy M, Sawhney M, Sepanlou SG, Shaikh MA, She J, Shiue I, Shore HR, Shrime MG, So S, Soneji S, Stathopoulou V, Stroumpoulis K, Sufiyan MB, Sykes BL, Tabares-Seisdedos R, Tadese F, Tedla BA, Tessema GA, Thakur JS, Tran BX, Ukwaja KN, Uzochukwu BS, Vlassov VV, Weiderpass E, Wubshet Terefe M, Yebyo HG, Yimam HH, Yonemoto N, Younis MZ, Yu C, Zaidi Z, Zaki ME, Zenebe ZM, Murray CJ, Naghavi M. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3:524–548. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao L, Wang X, Tang Y, Huang S, Hu CA, Teng Y. FGF19/FGFR4 signaling contributes to the resistance of hepatocellular carcinoma to sorafenib. J Exp Clin Cancer Res. 2017;36:8. doi: 10.1186/s13046-016-0478-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muaddi H, Al-Adra DP, Beecroft R, Ghanekar A, Moulton CA, Doyle A, Selzner M, Wei A, McGilvray ID, Gallinger S, Grant DR, Cattral MS, Greig PD, Kachura J, Cleary SP, Sapisochin G. Liver transplantation is equally effective as a salvage therapy for patients with hepatocellular carcinoma recurrence following radiofrequency ablation or liver resection with curative intent. Ann Surg Oncol. 2018;25:991–999. doi: 10.1245/s10434-017-6329-x. [DOI] [PubMed] [Google Scholar]
- 4.Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell. 2013;153:516–519. doi: 10.1016/j.cell.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Wong CM, Tsang FH, Ng IO. Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat Rev Gastroenterol Hepatol. 2018;15:137–151. doi: 10.1038/nrgastro.2017.169. [DOI] [PubMed] [Google Scholar]
- 6.Dong Z, Qi R, Guo X, Zhao X, Li Y, Zeng Z, Bai W, Chang X, Hao L, Chen Y, Lou M, Li Z, Lu Y. MiR-223 modulates hepatocellular carcinoma cell proliferation through promoting apoptosis via the Rab1-mediated mTOR activation. Biochem Biophys Res Commun. 2017;483:630–637. doi: 10.1016/j.bbrc.2016.12.091. [DOI] [PubMed] [Google Scholar]
- 7.Chang RM, Xiao S, Lei X, Yang H, Fang F, Yang LY. miRNA-487a promotes proliferation and metastasis in hepatocellular carcinoma. Clin Cancer Res. 2017;23:2593–2604. doi: 10.1158/1078-0432.CCR-16-0851. [DOI] [PubMed] [Google Scholar]
- 8.Demarez C, Gerard C, Cordi S, Poncy A, Achouri Y, Dauguet N, Rosa DA, Gunning PT, Manfroid I, Lemaigre FP. MicroRNA-337-3p controls hepatobiliary gene expression and transcriptional dynamics during hepatic cell differentiation. Hepatology. 2018;67:313–327. doi: 10.1002/hep.29475. [DOI] [PubMed] [Google Scholar]
- 9.Huang Z, Zhang N, Ma W, Dai X, Liu J. MiR-337-3p promotes chondrocytes proliferation and inhibits apoptosis by regulating PTEN/AKT axis in osteoarthritis. Biomed Pharmacother. 2017;95:1194–1200. doi: 10.1016/j.biopha.2017.09.016. [DOI] [PubMed] [Google Scholar]
- 10.Wang Z, Wang J, Yang Y, Hao B, Wang R, Li Y, Wu Q. Loss of has-miR-337-3p expression is associated with lymph node metastasis of human gastric cancer. J Exp Clin Cancer Res. 2013;32:76. doi: 10.1186/1756-9966-32-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du L, Subauste MC, DeSevo C, Zhao Z, Baker M, Borkowski R, Schageman JJ, Greer R, Yang CR, Suraokar M, Wistuba II, Gazdar AF, Minna JD, Pertsemlidis A. miR-337-3p and its targets STAT3 and RAP1A modulate taxane sensitivity in non-small cell lung cancers. PLoS One. 2012;7:e39167. doi: 10.1371/journal.pone.0039167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pasquier F, Cabagnols X, Secardin L, Plo I, Vainchenker W. Myeloproliferative neoplasms: JAK2 signaling pathway as a central target for therapy. Clin Lymphoma Myeloma Leuk. 2014;14(Suppl):S23–35. doi: 10.1016/j.clml.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 13.Ding L, Xu Y, Zhang W, Deng Y, Si M, Du Y, Yao H, Liu X, Ke Y, Si J, Zhou T. MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res. 2010;20:784–793. doi: 10.1038/cr.2010.79. [DOI] [PubMed] [Google Scholar]
- 14.Hou BH, Jian ZX, Cui P, Li SJ, Tian RQ, Ou JR. miR-216a may inhibit pancreatic tumor growth by targeting JAK2. FEBS Lett. 2015;589:2224–2232. doi: 10.1016/j.febslet.2015.06.036. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Li L, Guo R, Li X, Lu Y, Guan X, Gitau SC, Wang L, Xu C, Yang B, Shan H. miR-101 promotes breast cancer cell apoptosis by targeting Janus kinase 2. Cell Physiol Biochem. 2014;34:413–422. doi: 10.1159/000363010. [DOI] [PubMed] [Google Scholar]
- 16.Zhao G, Han C, Zhang Z, Wang L, Xu J. Increased expression of microRNA-31-5p inhibits cell proliferation, migration, and invasion via regulating Sp1 transcription factor in HepG2 hepatocellular carcinoma cell line. Biochem Biophys Res Commun. 2017;490:371–377. doi: 10.1016/j.bbrc.2017.06.050. [DOI] [PubMed] [Google Scholar]
- 17.Kabir TD, Ganda C, Brown RM, Beveridge DJ, Richardson KL, Chaturvedi V, Candy P, Epis M, Wintle L, Kalinowski F, Kopp C, Stuart LM, Yeoh GC, George J, Leedman PJ. A microRNA-7/growth arrest specific 6/TYRO3 axis regulates the growth and invasiveness of sorafenib-resistant cells in human hepatocellular carcinoma. Hepatology. 2018;67:216–231. doi: 10.1002/hep.29478. [DOI] [PubMed] [Google Scholar]
- 18.Zheng L, Jiao W, Mei H, Song H, Li D, Xiang X, Chen Y, Yang F, Li H, Huang K, Tong Q. miRNA-337-3p inhibits gastric cancer progression through repressing myeloid zinc finger 1-facilitated expression of matrix metalloproteinase 14. Oncotarget. 2016;7:40314–40328. doi: 10.18632/oncotarget.9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim SY, Lee YH, Bae YS. MiR-186, miR-216b, miR-337-3p, and miR-760 cooperatively induce cellular senescence by targeting alpha subunit of protein kinase CKII in human colorectal cancer cells. Biochem Biophys Res Commun. 2012;429:173–179. doi: 10.1016/j.bbrc.2012.10.117. [DOI] [PubMed] [Google Scholar]
- 20.Neradugomma NK, Subramaniam D, Tawfik OW, Goffin V, Kumar TR, Jensen RA, Anant S. Prolactin signaling enhances colon cancer stemness by modulating Notch signaling in a Jak2-STAT3/ERK manner. Carcinogenesis. 2014;35:795–806. doi: 10.1093/carcin/bgt379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Besancenot R, Roos-Weil D, Tonetti C, Abdelouahab H, Lacout C, Pasquier F, Willekens C, Rameau P, Lecluse Y, Micol JB, Constantinescu SN, Vainchenker W, Solary E, Giraudier S. JAK2 and MPL protein levels determine TPO-induced megakaryocyte proliferation vs differentiation. Blood. 2014;124:2104–2115. doi: 10.1182/blood-2014-03-559815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wormann SM, Song L, Ai J, Diakopoulos KN, Kurkowski MU, Gorgulu K, Ruess D, Campbell A, Doglioni C, Jodrell D, Neesse A, Demir IE, Karpathaki AP, Barenboim M, Hagemann T, Rose-John S, Sansom O, Schmid RM, Protti MP, Lesina M, Algul H. Loss of P53 function activates JAK2-STAT3 signaling to promote pancreatic tumor growth, stroma modification, and gemcitabine resistance in mice and is associated with patient survival. Gastroenterology. 2016;151:180–193. e112. doi: 10.1053/j.gastro.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 23.Wang S, Liang K, Hu Q, Li P, Song J, Yang Y, Yao J, Mangala LS, Li C, Yang W, Park PK, Hawke DH, Zhou J, Zhou Y, Xia W, Hung MC, Marks JR, Gallick GE, Lopez-Berestein G, Flores ER, Sood AK, Huang S, Yu D, Yang L, Lin C. JAK2-binding long noncoding RNA promotes breast cancer brain metastasis. J Clin Invest. 2017;127:4498–4515. doi: 10.1172/JCI91553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Hu F, Li G, Li G, Yang X, Liu L, Zhang R, Zhang B, Feng Y. Human colorectal cancer-derived mesenchymal stem cells promote colorectal cancer progression through IL-6/JAK2/STAT3 signaling. Cell Death Dis. 2018;9:25. doi: 10.1038/s41419-017-0176-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rozovski U, Wu JY, Harris DM, Liu Z, Li P, Hazan-Halevy I, Ferrajoli A, Burger JA, O’Brien S, Jain N, Verstovsek S, Wierda WG, Keating MJ, Estrov Z. Stimulation of the B-cell receptor activates the JAK2/STAT3 signaling pathway in chronic lymphocytic leukemia cells. Blood. 2014;123:3797–3802. doi: 10.1182/blood-2013-10-534073. [DOI] [PMC free article] [PubMed] [Google Scholar]